Abstract
The FOXP2 gene, located on human 7q31 (at the SPCH1 locus), encodes a transcription factor containing a polyglutamine tract and a forkhead domain. FOXP2 is mutated in a severe monogenic form of speech and language impairment, segregating within a single large pedigree, and is also disrupted by a translocation in an isolated case. Several studies of autistic disorder have demonstrated linkage to a similar region of 7q (the AUTS1 locus), leading to the proposal that a single genetic factor on 7q31 contributes to both autism and language disorders. In the present study, we directly evaluate the impact of the FOXP2 gene with regard to both complex language impairments and autism, through use of association and mutation screening analyses. We conclude that coding-region variants in FOXP2 do not underlie the AUTS1 linkage and that the gene is unlikely to play a role in autism or more common forms of language impairment.
Autism is a neurodevelopmental disorder characterized by deficits in reciprocal social interaction and communication, accompanied by repetitive and stereotyped behaviors and interests (World Health Organization 1993; American Psychiatric Association 1994).
Specific language impairment (SLI) is defined as a significant deficit in language development that exists despite adequate educational opportunity and normal nonverbal intelligence. A diagnosis of SLI is made after ruling out the presence of other conditions, such as autism (Tomblin et al. 1996).
Although autism and SLI are generally accepted to be clinically distinct, the boundaries between the two conditions are not always clear, and there remains a group of children who show social and/or language difficulties yet fail to meet strict diagnostic criteria for either disorder. Some have argued for the formation of a “semantic-pragmatic” classification for these “borderline autistic/language impaired” individuals (Rapin and Allen 1983; Bishop and Rosenbloom 1987).
Language deficits form a major component of the autism diagnostic criteria, and, in general, autistic individuals tend to experience more-severe linguistic impairments than are associated with SLI alone (Lord et al. 1994). Autistic children typically make few spontaneous remarks, produce stereotyped utterances, and make only minimal use of gesture (Tager-Flusberg et al. 2001). Although some autistic children may develop acceptable skills in terms of vocabulary, grammar, and phonology, they invariably retain fundamental difficulties with the use of language in a social context (i.e., pragmatics) (Mawhood et al. 2000). A substantial proportion of autistic children completely fail to develop language at all (Rapin 1997; Tager-Flusberg et al. 2001). In contrast, the types of language problems seen in SLI tend to be more heterogeneous. Children affected by SLI show a wide range and severity of deficits with respect to the articulation of speech sounds, verbal expression, and comprehension of speech (Bishop 1994; Conti-Ramsden et al. 1997). Pragmatic impairments are usually absent or mild.
There is now a large amount of evidence, from family and twin studies, indicating a strong role for genetic factors in both autism (Folstein and Rutter 1977; Steffenburg et al. 1989; Bolton et al. 1994; Bailey et al. 1995) and SLI (Lewis and Thompson 1992; Bishop et al. 1995; Tomblin and Buckwalter 1998). However, it is accepted that each of these conditions is complex in nature, with several loci interacting to produce a genetic liability to disease onset (Pickles et al. 1995).
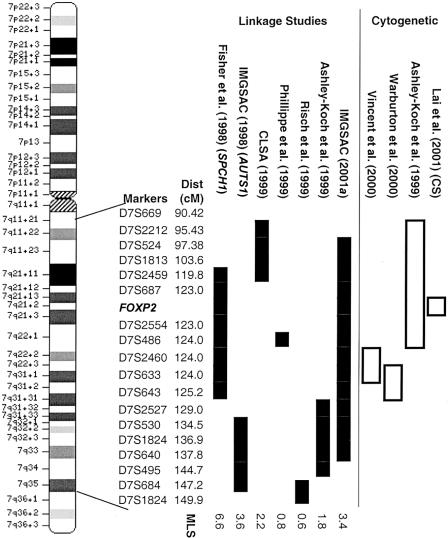
Recent advances in technology and statistical genetics have allowed the completion of several genomewide scans using sibling pairs affected by autism. The first of these studies yielded a maximum LOD score (MLS) of 3.55 in a 40-cM region on the long arm of human chromosome 7, between markers D7S530 and D7S684, in a subset of families from the United Kingdom (the AUTS1 locus [MIM 209850]; fig. 1) (International Molecular Genetic Study of Autism Consortium [IMGSAC] 1998). The involvement of this AUTS1 locus in autism is further supported by several independent linkage investigations with differing degrees of significance and varying chromosomal locations. (fig. 1) (Ashley-Koch et al. 1999; Collaborative Linkage Study of Autism 1999; Phillippe et al. 1999; Risch et al. 1999; IMGSAC 2001a).
Figure 1.
Autism and language studies of chromosome 7q. The chromosome 7 ideogram shows the order and map distance of markers used in various studies. Blackened boxes show approximate positions of regions highlighted by linkage studies. Unblackened boxes represent approximate positions of breakpoints in cytogenetic studies. Each MLS shown was obtained in the region highlighted for the appropriate linkage study. The methods of LOD estimation varied between studies. Note that the SPCH1 linkage has been directly attributed to the FOXP2 gene.
Concurrent studies of the KE family, a unique three-generation pedigree with a severe monogenic speech and language disorder, independently yielded strong evidence for linkage to a similar region of 7q31, between markers D7S2459 and D7S643 (the SPCH1 locus [MIM 602081]; fig. 1) (Fisher et al. 1998; Lai et al. 2000). The KE phenotype is characterized by severe orofacial dyspraxia, which impedes complex articulatory movement, accompanied by extreme impairments in both expressive and receptive language skills. There is also evidence of nonverbal deficits in some individuals (Vargha-Khadem et al. 1995). Although the affected members of the KE family show no autistic features and do not meet strict diagnostic criteria for SLI, the overlap between the SPCH1 and AUTS1 loci raised the question of whether a single gene on 7q might be involved in both autism and SLI (Folstein and Mankoski 2000). Such a hypothesis is strengthened by cytogenetic studies of individuals with chromosome 7 abnormalities (fig. 1). Ashley-Koch et al. (1999) described a family with a pericentric inversion of the long arm of chromosome 7 (inv[7][q22-q31.2]), transmitted from an unaffected mother to all three of her children. Interestingly, two of the three siblings in this family were affected by autism, and the third presented with a severe expressive-language disorder. Vincent et al. (2000) characterized a translocation transmitted from an unaffected mother to an autistic child (t[7;13][q31.2;q21]) and mapped the breakpoint within a highly conserved, brain-expressed gene of unknown function (RAY1) between markers D7S2460 and D7S633. Warburton et al. (2000) described two unrelated individuals, one with autism and a second with a severe expressive-language impairment, both of whom showed de novo abnormalities involving breakpoints on chromosome 7q31 (inv[7][p12.2;q31.3] and t[2;7][p23;q31.3], respectively). Finally, Lai et al. (2001) described a child (referred to as “CS”) affected by a severe orofacial dyspraxia and language deficits similar to those seen in the KE family, with a de novo translocation (t[5;7][q22;q31.2]) mapping to the SPCH1 region.
Recently, the gene mutated in the KE family was identified as FOXP2 (MIM 605317) (Lai et al. 2001). The FOX genes encode a large family of transcription factors, all of which possess a winged-helix—or forkhead box (“fox”)—DNA-binding domain. The known sequence of FOXP2, as reported by Lai et al. (2001), is organized into 19 exons (2 of which are alternatively spliced), and the major splice form encodes a 715-residue protein containing a characteristic fox domain (exons 12–14) and a 40-residue polyglutamine tract (exons 5 and 6). The polyglutamine repeat is encoded by a mixture of CAG and CAA codons and has been demonstrated to be stable in normal individuals (Lai et al. 2001). The mutation identified in the KE family is a G→A transition in exon 14 that cosegregates with the speech and language disorder in the KE pedigree. This nonsynonymous change results in an arginine-to-histidine substitution at a highly conserved residue within the fox domain (Lai et al. 2001). Furthermore, the FOXP2 gene was directly disrupted by the chromosomal breakpoint of the unrelated translocation patient, CS. Lai et al. (2001) suggested that the KE and CS phenotypes may be caused by haploinsufficiency of FOXP2 at a key stage of embryogenesis, which results in the abnormal development of neural structures important for speech and language.
Clearly, there is strong support for the role of chromosome 7q31 in the etiology of both autism and language disorders. However, questions remain with regard to the relevance of FOXP2 within more common and genetically complex forms of language impairment, and it is still a matter of debate as to whether the phenotypic and genetic overlaps between autism and SLI are caused by the same or by different loci. The present study therefore presents the characterization of FOXP2 within samples of patients with SLI and autism, with two aims. The first is to assess the relevance of the FOXP2 gene within forms of language impairment more common than those found in the KE family and in the translocation patient CS, and the second is to directly evaluate the hypothesis that the overlap in SPCH1 and AUTS1 mapping data reflects the involvement of a single gene—that is, FOXP2.
For the present study, 169 multiplex families with autism (857 individuals) were selected. These individuals represent the complete IMGSAC cohort (IMGSAC 2001b) and thus include those used in the original identification of AUTS1 (IMGSAC 1998). Families were collected over four successive stages, and affection status was assessed using a variety of standardized tests. In brief, parents from families identified through an initial screen were administered the Autism Diagnostic Interview–Revised (ADI-R [Lord et al. 1994]) and the Vineland Adaptive Behaviour Scales (Sparrow et al. 1984). Potential cases were assessed using the Autism Diagnostic Observation Schedule (ADOS [Lord et al. 1994]) or the ADOS-Generic (ADOS-G [Lord et al. 2000]). Where possible, psychometric evaluation was conducted using Raven’s progressive matrices (Raven 1989) or the Mullen Scales of Early Learning (Mullen 1995), as well as the British Peabody Picture Vocabulary Test III (Dunn and Dunn 1997) or an appropriate translation. A physical examination was undertaken and allowed the exclusion of children with signs of tuberous sclerosis. Where possible, affected individuals were karyotyped, and those found to have any chromosome abnormalities, including fragile X, were excluded.
In addition, we selected 43 families with language impairment (210 individuals) who form a subset of the SLI Consortium (SLIC) genome-screen sample (SLIC 2002). Probands were recruited by the Newcomen Centre at Guy’s Hospital, London, through three special schools for language disorders and through Afasic, a support organization for people with developmental and language impairments. All probands, either currently or in the past, had language skills >1.5 SD below the normative mean for their chronological age, on the receptive and/or expressive scales of the Clinical Evaluation of Language Fundamentals (CELF-R) battery (Semel et al. 1992). Any proband or sibling found to have a nonverbal IQ of <80 was excluded from the sample. Additional exclusion criteria included an International Classification of Diseases–10th Revision/Diagnostic and Statistical Manual of Mental Disorders–4th Edition diagnosis of childhood autism.
The SLIC study used this sample in an investigation of three quantitative measures of language abilities, none of which showed linkage to chromosome 7q (SLIC 2002). A comprehensive description of the relevant cohorts can be found in reports by IMGSAC (2001b) and SLIC (2002).
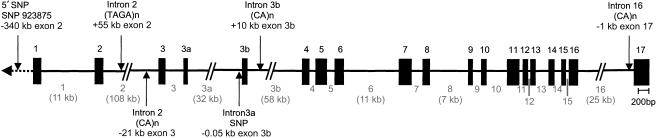
To begin, we used RepeatMasker software (BCM Search Launcher Web site) to identify four novel intronic polymorphic microsatellites (fig. 2) that lie within BACs (NH0095P09, RG250D13, and NH563O05) covering the FOXP2 sequence. Fluorescently labeled primers were designed (sequences are available from the authors on request) and were used to amplify all four microsatellites in available members of the families described above. The products were genotyped on ABI377 sequencing machines (PE Applied Biosystems), as described elsewhere (SLIC 2002), and were tested for association, as follows.
Figure 2.
Schematic of FOXP2 (adapted with permission from Lai et al. [2001]). Numbers in black indicate exon numbers. Numbers in grey indicate intron numbers as used in table 3. All exons are shown to scale. Introns are shown to scale with each other, and the sizes of all introns >5 kb are given in brackets (in kb). Positions of all microsatellites and SNPs used for association analysis are indicated by arrows, and distances (in kb) are given from the nearest coding exon. Exons 5 and 6 contain a polyglutamine encoding tract; exons 12–14 contain the forkhead (fox) domain; exons 3a and 3b are alternatively spliced; the KE mutation is found in exon 14; the CS translocation breakpoint is between exon 3b and exon 4.
Children from the autistic families were classified as “affected” or “unknown” and were tested for association through use of a transmission/disequilibrium test (TDT) within the sib_tdt program from the ASPEX package version 2.2 (The ASPEX Linkage Analysis Package ftp site). This program calculates probabilities for χ2 statistics by permuting parental alleles while fixing the identity-by-descent status of siblings within a family, thereby allowing the use of multiple siblings within a nuclear family.
We found no evidence for association within the autism group at the marker level (P>.05; table 1). Although allele-specific TDT indicated a weak association (P=.01) between the 237-bp allele of the intron 16 microsatellite and autistic disorder, the excess of nontransmitted alleles in this case (22 transmitted and 49 nontransmitted; see table 1) corresponds to a protective effect. Furthermore, after appropriate correction for multiple testing, this association was rendered nonsignificant.
Table 1.
Association of FOXP2 with Intronic Microsatellites: TDT within Autistic Families[Note]
|
No. of Alleles |
||||||||||
| Paternal |
Maternal |
Combined |
||||||||
| Micosatelliteaand Allele(Allele Sizein bp) | Frequencyb(%) | Transmitted | NotTransmitted | χ2 | Transmitted | NotTransmitted | χ2 | Transmitted | NotTransmitted | χ2 |
| Intron 2: (TAGA)n: | ||||||||||
| 3 (458) | 50.7 | 41 | 52 | 1.30 | 56 | 47 | .79 | 113 | 115 | .02 |
| 2 (454) | 25.9 | 48 | 36 | 1.71 | 38 | 42 | .20 | 101 | 93 | .33 |
| 4 (462) | 16.5 | 30 | 25 | .45 | 37 | 38 | .01 | 70 | 66 | .12 |
| Other | 6.8 | 14 | 20 | 10 | 14 | 24 | 34 | |||
| Intron 2: (CA)n: | ||||||||||
| 7 (195) | 39.3 | 48 | 60 | 1.33 | 56 | 75 | 2.76 | 109 | 140 | 3.86 |
| 2 (185) | 23.7 | 44 | 30 | 2.65 | 50 | 54 | .15 | 96 | 86 | .55 |
| 3 (187) | 22.8 | 35 | 41 | .47 | 48 | 37 | 1.42 | 86 | 81 | .15 |
| Other | 14.2 | 37 | 33 | 41 | 29 | 78 | 62 | |||
| Intron 3b: (CA)n: | ||||||||||
| 7 (435) | 30.2 | 46 | 44 | .04 | 42 | 48 | .4 | 93 | 97 | .08 |
| 8 (437) | 21.1 | 31 | 28 | .15 | 36 | 36 | .00 | 70 | 67 | .07 |
| 9 (439) | 14.9 | 20 | 34 | 3.63 | 26 | 21 | .53 | 49 | 58 | .76 |
| 6 (433) | 13.4 | 28 | 19 | 1.72 | 23 | 19 | .38 | 51 | 38 | 1.90 |
| Other | 20.7 | 37 | 37 | 36 | 39 | 74 | 77 | |||
| Intron 16: (CA)n: | ||||||||||
| 2 (225) | 45.5 | 71 | 51 | 3.28 | 42 | 54 | 1.50 | 116 | 108 | .29 |
| 8 (239) | 27.2 | 38 | 37 | .01 | 34 | 24 | 1.72 | 75 | 64 | .87 |
| 7 (237)c | 10.9 | 10 | 31 | 10.76 | 12 | 18 | 1.20 | 22 |
49 |
10.27 |
| Other | 16.2 | 36 | 36 | 35 | 27 | 73 | 65 | |||
Note.— Empirical P values for transmission of all alleles was >.1 for alleles of all microsatellites, with the exception of intron 3b ([CA]n), for which P=.08.
For positions of microsatellites, see figure 2.
Alleles with a frequency <10% are grouped as “other.”
Allele-specific TDT for allele 7 combined (underlined): P=.01 (uncorrected).
The heterogeneity of the SLI phenotype rendered the derivation of a consistent affection status impractical. We therefore employed three quantitative measurements of language abilities to examine FOXP2 association within the SLI cohort. The CELF-R was used to derive scores of expressive and receptive language abilities (ELS and RLS, respectively) (Semel et al. 1992). Each score is calculated from performance on three subtests designated to be primarily receptive or expressive in nature. The exact combination of individual tests used is dependent upon subject age. Additive raw scores from each segment are transformed to derive standardized scores with a mean of 100 and an SD of 15 in the general population calibration sample (Semel et al. 1992).
In addition, a test of nonword repetition (NWR) was used to assess phonological short-term memory (Gathercole et al. 1994). In this test, subjects are required to repeat tape-recorded nonsensical words of increasing length and complexity (e.g., “brufid” and “contramponist”). Studies show that individuals with current language impairments, as well as those who had language difficulties in early childhood which later resolved, perform poorly on this test (Gathercole et al. 1994; Bishop et al. 1999). In addition, it has been suggested that performance on the nonword repetition task is the best index of disorder in the KE pedigree (Vargha-Khadem et al. 1998).
Association was evaluated by the QTDT (quantitative transmission/disequilibrium test) program (Abecasis et al. 2000; QTDT Home Page), which employs a variance-components model that partitions association into between- and within-family components. The QTDT program includes a permutation framework, which allows the derivation of empirical P values for the sample being evaluated. This corrects for small sample sizes or deviations of quantitative traits from multivariate normality. We performed 1,000 simulations for each of the above traits and again found no evidence for association (P>.05) (table 2).
Table 2.
Association of FOXP2 with Intronic Microsatellites: QTDT within Language-Impaired Families[Note]
|
χ2 |
|||
| Microsatellite | ELS | RLS | NWR |
| Intron 2: (TAGA)n | 2.38 | .95 | 7.88 |
| Intron 2: (CA)n | 7.14 | 9.60 | 8.89 |
| Intron 3b: (CA)n | 8.23 | 10.18 | 7.69 |
| Intron 16: (CA)n | 2.58 | 1.30 | 3.11 |
Note.— All P values were >.1.
Despite the lack of positive findings from the association analyses, it remained possible that some individuals may harbor FOXP2 mutations that were indiscernible at the whole-sample level. We therefore initiated a mutation screen of the FOXP2 coding sequence by means of denaturing high performance liquid chromatography (DHPLC) (Kuklin et al. 1997).
DHPLC analysis was performed for all 43 probands of the families with SLI described above. For the autism cohort, a subset of 48 affected individuals were selected who contributed to the linkage peak on 7q. This enriched for individuals who are likely to carry etiological variants at the AUTS1 locus.
Primers, taken from the Lai et al. (2001) study and available on request, were used to amplify all 19 FOXP2 exons and surrounding intron-exon boundaries, by means of a touchdown PCR protocol (PCR Protocol for WAVE Machine Web site). DHPLC analysis was performed using the WAVE DNA Fragment Analysis System (Transgenomic), and fragments that showed a variant elution pattern were directly sequenced.
We identified a total of 11 sequence variants, all of which were single-base substitutions within intronic regions (table 3). No changes were seen within exons 12–14, which contain the fox domain.
Table 3.
Polymorphisms Detected in FOXP2
|
Frequency inIndividuals withc |
|||
| Intron/Exonaand Positionb | Change | Autism(n=48) | SLI(n=43) |
| Intron 3: | |||
| −102 bp from exon 3a | T/A | 9 | 11 |
| Intron 3a: | |||
| −32 bp from exon 3b | A/G | 0 | 1 |
| −48 bp exon3bd | T/C | 15 | 19 |
| −68 bp from exon3b | G/A | 1 | 0 |
| Intron 5: | |||
| +17 bp from exon5 | T/G | 8 | 10 |
| Intron 5/Exon 6: | |||
| … | Ins CAGCAG | 0 | 1 |
| Intron 11: | |||
| +9 bp from exon11 | T/C | 1 | 1 |
| −80 bp from exon12 | A/G | 0 | 1 |
| Intron 13: | |||
| +30 bp from exon13 | C/G | 1 | 2 |
| Intron 14: | |||
| +24 bp from exon14 | T/C | 0 | 1 |
| −44 bp from exon15 | T/G | 2 | 0 |
| −58 bp from exon15 | T/C | 3 | 0 |
For intron numbers, see figure 2.
Position is given in relation to nearest exon; “−” denotes that the SNP is found 5′ to the exonic sequence, and “+” denotes that the SNP is found 3′ to the exonic sequence.
Frequency of heterozygotes within the sample tested.
Polymorphism used to type individuals for association analysis.
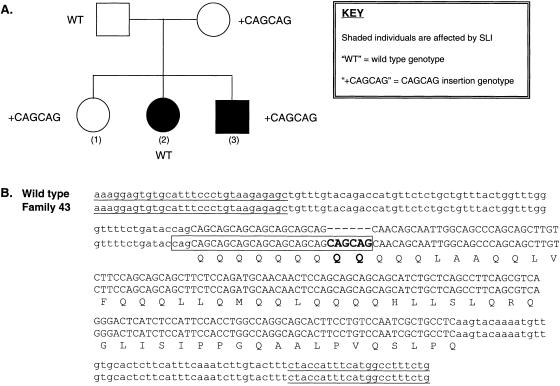
In one family with SLI (family 43), we identified a CAGCAG insertion within a polyglutamine stretch at the intron/exon border of exon 6 (fig. 3). This region represents the longest stretch of pure CAG repeats within the polyglutamine region and thus is the most likely place for an expansion to occur. We genotyped all available members of this family, but the expansion did not cosegregate with language impairment (fig. 3). In addition, the insertion may actually fall within intronic sequence—the exact position of the insertion remains unclear, because of the repetitive nature of this stretch of DNA—and it does not alter the reading frame of the sequence. It therefore appears likely that this expansion simply represents a rare polymorphism that is not relevant to the SLI phenotype.
Figure 3.
FOXP2 exon 6. A, Family 43 pedigree. Both the mother and father have no reported history of language problems. Child 1 is clinically normal, child 2 is clinically affected, and child 3 has a reported language delay but is too young to test formally. B, The CAGCAG insertion in FOXP2 exon 6. The boxed area represents the possible site for CAGCAG insertion. Underlined bases represent primers for exon 5. Exonic sequence is represented by capital letters; intronic sequence is represented by lowercase letters.
The DHPLC analysis identified a common single-nucleotide polymorphism (SNP) within intron 3a of the FOXP2 gene (fig. 2). This intron 3a polymorphism involves a T→C transition that destroys an AflII site within the sequence and therefore could be directly typed via a restriction enzyme assay. One hundred sixty-nine families with autism were again tested through use of the sib_tdt program from the ASPEX package (v2.2), and 43 families with SLI were tested within the QTDT program (Abecasis et al. 2000), as described above. We found no evidence for association with this SNP, within either the autism or the SLI groups (tables 4 and 5).
Table 4.
SNP Association Analyses of FOXP2: TDT within Autistic Families[Note]
|
No. of Alleles |
||||||||||
| Paternal |
Maternal |
Combined |
||||||||
| SNPa | Frequency(%) | Transmitted | NotTransmitted | χ2 | Transmitted | NotTransmitted | χ2 | Transmitted | NotTransmitted | χ2 |
| Intron 3a | 55.6 | 54 | 45 | .82 | 58 | 57 | .01 | 137 | 127 | .38 |
| 5′ (923875) | 48.5 | 31 | 39 | .91 | 35 | 23 | 2.48 | 91 | 87 | .09 |
Note.— All P values were >.1
For SNP positions, see figure 2.
Table 5.
SNP Association Analyses of FOXP2: QTDT within Language-Impaired Families[Note]
|
χ2 |
||||
| SNPa | Frequency(%) | ELS | RLS | NWR |
| Intron 3a | 51.0 | .07 | .81 | .65 |
| 5′ (923875) | 40.6 | .28 | 2.03 | .03 |
Note.— All P values were >.1
For SNP positions, see figure 2.
Although the Lai et al. (2001) article presented the entire coding region of the FOXP2 gene, the transcription start site could not be defined, and northern analyses indicated the existence of additional 5′ and/or 3′ untranslated exons that remain uncharacterized. Given the wide expression pattern of FOXP2 (Lai et al. 2001), we postulated that it was likely to represent a housekeeping gene and therefore initiated a search for CpG islands in the genomic sequence immediately upstream of the FOXP2 gene (CpG Islands Web site). Complementary bioinformatic analyses (e.g., Nix [MRC Human Genome Mapping Project Resource Center Web site] and Promoter Inspector [Genomatix Web site], and Ensembl [Ensembl Genome Server]) indicated that the closest CpG island lay 340 kb upstream of the present FOXP2 coding sequence.
Similarity searches demonstrated that this sequence was highly homologous (83% identity) to a CpG-rich region upstream of the mouse Foxp2 gene. Furthermore, this murine sequence was directly linked to the Foxp2 coding region in three independent ESTs (AW490098, BB660527, and BB656124), indicating that it is transcribed as part of the mouse Foxp2 mRNA.
As a final verification step, we therefore typed an SNP adjacent to this CpG island (SNP 923875) (dbSNP Home Page), through use of a restriction enzyme assay. We found no evidence for association at this 5′ SNP, in either the autism or the SLI groups (tables 4 and 5).
In the absence of any mutation or association evidence to suggest otherwise, we must therefore conclude that FOXP2 is unlikely to play a major role in the onset of autism or SLI. As a corollary, since the autism cases studied here included those originally used in the detection of the AUTS1 locus and the sample was enriched for individuals who showed linkage to 7q31, we can conclude that the SPCH1 and AUTS1 loci are attributable to different genes that, coincidentally, lie in similar positions on chromosome 7q.
Finally, it would appear that the role of FOXP2 in speech and language disorders does not generalize to more common and genetically complex forms of language impairment within our SLIC cohort. However, it is worth noting that although the probands with SLIC were chosen to represent a diverse range of impairments spread over many linguistic domains, it remains possible that FOXP2 variations may be involved in specific and distinct forms of speech and language impairments not represented within our sample.
Acknowledgments
Consortium members are as follows: United Kingdom—Centre for Social, Genetic and Developmental Psychiatry and Department of Child and Adolescent Psychiatry, Institute of Psychiatry, London: Sarah Palferman, Nicola Matthews, Martha Turner, Janette Moore, Amaia Hervas, Anne Aubin, Simon Wallace, Janine Michelotti, Catherine Wainhouse, Alina Paul, Elaine Thompson, Ramyani Gupta, Claire Garner, Marianne Murin, Christine Freitag, Nicola Ryder, Emily Cottington, Jeremy Parr, Greg Pasco, Andrew Pickles, Michael Rutter, and Anthony Bailey; Wellcome Trust Centre for Human Genetics, University of Oxford: Janine A. Lamb, Gabrielle Barnby, Pat Scudder, Elena Bonora, Angela Marlow, and Anthony P. Monaco; Newcomen Centre, Guys Hospital, London: Gillian Baird and Anthony Cox; Regional Genetics Centre, Division of Medical and Molecular Genetics, Guys Hospital, London: Zoe Docherty, Pamela Warburton, Elizabeth P. Green, and Stephen J. Abbs; Flemming Nuffield Unit, Newcastle: Ann Le Couteur, Helen R. McConachie, and Tom Berney; Neuropsychology Department, Newcastle General Hospital: Thomas P. Kelly; Developmental Psychiatry Section, University of Cambridge Clinical School: Petrus J. De Vries, Emma Gaitonde, and Patrick F. Bolton; Booth Hall Childrens Hospital, Manchester: Jonathan Green, Anne Gilchrist, and Jane Whittacker; and European Centre for Collection of Animal Cell Cultures, Salisbury: Bryan Bolton and Ros Packer. Italy—Dipartimento di Biologia, Universita di Bologna: Elena Maestrini, Francesca Blasi, and Elena Bacchelli. The Netherlands—AZU, Department of Child and Adolescent Psychiatry, Utrecht: Herman Van Engeland, Maretha V. De Jonge, Chantal Kemner, and Judith Timp. Germany—Deutsches Krebsforschungszentrum, Department of Molecular Genome Analysis: Sabine M. Klauck, Kim S. Beyer, Sabine Epp, and Annemarie Poustka; Deutsches Krebsforschungszentrum, Department of Biostatistics: Axel Benner; and J.W. Goethe–Universität, Department of Child and Adolescent Psychiatry, Frankfurt: Fritz Poustka, Dorothea Rühl, Gabriele Schmötzer, Sven Bölte, and Sabine Feineis-Matthews. France—Unité de Diagnostic et Evaluation de l'Autisme, Hôpital la Grave, Toulouse: Eric Fombonne, Bernadette Rogé, Jeanne Fremolle-Kruck, Catherine Pienkowski, and Marie-Thérèse Tauber. Denmark—Videnscenter Og Centre For Autisme, Virum: Lennart Pedersen, Torben Isager, Gunna Eriksen, and Demetrious Haracopos; John F. Kennedy Instituttet, Glostrup: Karen Brondum-Nielsen; Biophysics Group, Danish Technical University: Rodney M. J. Cotterill. Greece—Department of Psychological Paediatrics, Agia Sophia Childrens Hospital, Athens: John Tsiantis and Katerina Papanikolaou. United States—Department of Psychiatry, University of Chicago, Chicago: Catherine Lord, Christina Corsello, Stephen Guter, Bennett Leventhal, and Edwin Cook; UCLA Center for Neurobehavioural Genetics, Los Angeles: Susan L. Smalley, Stanley F. Nelson, Amy Liu, Janet Miller, Martha Dedricks, Lisa Chrzanowski, Julia Bailey, James McGough, and Jennifer Levitt; Child Study Center, Sterling Hall of Medicine, Yale University, New Haven: David Pauls, Fred Volkmar, Joel Bregman, Ami Klin, and John Alsobrook; and Department of Human Genetics, University of Pittsburgh, Pittsburgh: Daniel E. Weeks.
We would like to thank all the families who have participated in the study, as well as the professionals who continue to make this study possible. Thanks to all at the Newcomen Centre, for their assistance with data collection and management, and to all members of the Monaco group, for their support and advice. This work has been funded by The Wellcome Trust, the U.K. Medical Research Council, BIOMED 2 grant CT-97-2759, European Commission Fifth Framework Grant QLG2-CT-1999-0094, Telethon-Italy grant E.1007, the Janus Korczak Foundation, Deutsche Forschungsgemeinschaft, Foundation France Télécom, Conseil Régional Midi-Pyrénées, Danish Medical Research Council, Sofiefonden, the Beatrice Surovell Haskells Fond for Child Mental Health Research of Copenhagen, Danish Natural Science Research Council grant 9802210, National Institute of Child Health and Development grant 5-P01-HD-35482, and National Institutes of Health grants MO1 RR06022 GCRC NIH, NIH K05 MH01196, and K02 MH01389. D.F.N. is funded by a Medical Research Council Studentship, E.B. is funded by a University of Oxford Graduate Prize Studentship, and A.P.M. is a Wellcome Trust Principal Research Fellow.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- ASPEX Linkage Analysis Package, The, ftp://lahmed.stanford.edu/pub/aspex/index.html
- BCM Search Launcher, http://searchlauncher.bcm.tmc.edu:9331/seq-util/seq-util.html (for RepeatMasker)
- CpG Islands, http://www.ebi.ac.uk/emboss/cpgplot/
- dbSNP Home Page, http://www.ncbi.nlm.nih.gov/SNP/
- Ensembl Genome Server, http://www.ensembl.org/Homo_sapiens/
- Genomatix: PromoterInspector, http://anthea.gsf.de/cgi-bin/promoterinspector/promoterinspector.pl
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for AUTS1 [MIM 209850], SPCH1 [MIM602081], and FOXP2 [MIM605317])
- PCR Protocol for WAVE Machine, http://www.well.ox.ac.uk/genomics/wave.html (for DHPLC PCR protocol)
- QTDT Home Page, http://www.well.ox.ac.uk/asthma/QTDT/index.html
- U.K. MRC Human Genome Mapping Project Resource Center, http://www.hgmp.mrc.ac.uk/ (for Nix)
References
- Abecasis GR, Cardon LR, Cookson WOC (2000) A general test of association for quantitative traits in nuclear families. Am J Hum Genet 66:279–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Psychiatric Association (1994) Diagnostic and statistical manual of mental disorders (DSM-IV). American Psychiatric Association, Washington, DC [Google Scholar]
- Ashley-Koch A, Wolpert CM, Menold MM, Zaeem L, Basu S, Donnelly SL, Ravan SA, Powell CM, Qumsiyeh MB, Aylsworth AS, Vance JM, Gilbert JR, Wright HH, Abramson RK, DeLong GR, Cuccaro ML, Pericak-Vance MA (1999) Genetic studies of autistic disorder and chromosome 7. Genomics 61:227–236 [DOI] [PubMed] [Google Scholar]
- Bailey A, Le Couteur A, Gottesman I, Bolton P, Simonoff E, Yuzda E, Rutter M (1995) Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med 25:63–77 [DOI] [PubMed] [Google Scholar]
- Bishop DV (1994) Is specific language impairment a valid diagnostic criteria? Genetic and psycholinguistic evidence. Phil Trans R Soc Lond B Biol Sci 346:105–111 [DOI] [PubMed] [Google Scholar]
- Bishop DVM, Bishop SJ, Bright P, James C, Delaney T, Tallal P (1999) Different origin of auditory and phonological processing problems in children with language impairment: evidence from a twin study. J Speech Lang Hear Res 42:155–168 [DOI] [PubMed] [Google Scholar]
- Bishop DVM, North T, Donlan C (1995) Genetic basis of specific language impairment: evidence from a twin study. Dev Med Child Neurol 37:56–71 [DOI] [PubMed] [Google Scholar]
- Bishop DVM, Rosenbloom L (1987) Classification of childhood language disorders. In: Yule W, Rutter M (eds) Language development and disorders. MacKeith Press, London, pp 16–41 [Google Scholar]
- Bolton P, Macdonald H, Pickles A, Rios P, Goode S, Crowson M, Bailey A, Rutter M (1994) A case-control family history study of autism. J Child Psychol Psychiat 35:877–900 [DOI] [PubMed] [Google Scholar]
- Collaborative Linkage Study of Autism (1999) An autosomal genome screen for autism. Am J Med Genet 88:609–615 [DOI] [PubMed] [Google Scholar]
- Conti-Ramsden G, Crutchley A, Botting N (1997) The extent to which psychometric tests differentiate subgroups of children with SLI. J Speech Lang Hear Res 40:765–777 [DOI] [PubMed] [Google Scholar]
- Dunn LM, Dunn LM (1997) Peabody picture vocabulary test. American Guidance Service, Circle Pines, MN [Google Scholar]
- Fisher SE, Vargha-Khadem, Watkins KE, Monaco AP, Pembrey ME (1998) Localisation of a gene implicated in a severe speech and language disorder. Nat Genet 18:168–170 [DOI] [PubMed] [Google Scholar]
- Folstein S, Rutter M (1977) Infantile autism: a genetic study of 21 twin pairs. J Child Psychol Psychiatry 18:297–321 [DOI] [PubMed] [Google Scholar]
- Folstein SE, Mankoski RE (2000) Chromosome 7q: where autism meets language disorder? Am J Hum Genet 67:278–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gathercole SE, Willis C, Baddeley AD, Emslie H (1994) The children’s test of nonword repetition: a test of phonological working memory. Memory 2:103–127 [DOI] [PubMed] [Google Scholar]
- International Molecular Genetic Study of Autism Consortium (1998) A full genome screen for autism with evidence for linkage to a region on chromosome 7q. Hum Mol Genet 7:571–578 [DOI] [PubMed] [Google Scholar]
- ——— (2001a) Further characterisation of the autism susceptibility locus AUTS1 on chromosome 7q. Hum Mol Genet 10:973–982 [DOI] [PubMed] [Google Scholar]
- ——— (2001b) A genomewide screen for autism: strong evidence for linkage to chromosomes 2q, 7q, and 16p. Am J Hum Genet 69:570–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuklin A, Munson K, Gjerde D, Haefele R, Taylor P (1997) Detection of single-nucleotide polymorphisms with the WAVE DNA fragment analysis system. Genet Test 1:201–206 [DOI] [PubMed] [Google Scholar]
- Lai CSL, Fisher SE, Hurst JA, Levy ER, Hodgson S, Fox M, Jeremiah S, Povey S, Curtis Jamison D, Green ED, Vargha-Khadem F , Monaco AP (2000) The SPCH1 region on human 7q31: genomic characterization of the critical interval and localization of translocations associated with speech and language disorder. Am J Hum Genet 67:357–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CSL, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP (2001) A novel forkhead-domain gene is mutated in a severe speech and language disorder. Nature 413:519–523 [DOI] [PubMed] [Google Scholar]
- Lewis BA, Thompson LA (1992) A study of developmental speech and language disorders in twins. J Speech Hear Res 35:1086–1094 [DOI] [PubMed] [Google Scholar]
- Lord C, Risi S, Lambrecht L, Cook EH Jr, Leventhal BL, DiLavore PC, Pickles A, Rutter M (2000) The autism diagnostic observation schedule-generic: a standard measure of social and communication deficits associated with the spectrum of autism. J Autism Dev Disord 30:205–223 [PubMed] [Google Scholar]
- Lord C, Rutter M, Le Couteur A (1994) Autism Diagnostic Interview–Revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J Aut Dev Disord 24:659–685 [DOI] [PubMed] [Google Scholar]
- Mawhood L, Howlin P, Rutter M (2000) Autism and developmental receptive language disorder—a comparative follow-up in early adult life I: cognitive and language outcomes. J Child Psychol Psychiat 41:547–559 [DOI] [PubMed] [Google Scholar]
- Mullen EM (1995) Mullens scales of early learning. American Guidance Service, Circle Pines, MN [Google Scholar]
- Philippe A, Martinez M, Guilloud-Bataille M, Gillberg C, Rastam M, Sponheim E, Coleman M, Zappella M, Aschauer H, Van Maldergem L, Penet C, Feingold J, Brice A, Leboyer M, and the Paris Autism Research International Sibpair Study (1999) Genome-wide scan for autism susceptibility genes. Hum Mol Genet 8:805–812 [DOI] [PubMed] [Google Scholar]
- Pickles A, Bolton P, Macdonald H, Bailey A, Le Couteur A, Sim CH, Rutter M (1995) Latent-class analysis of recurrence risks for complex phenotypes with selection and measurement error: a twin and family history study of autism. Am J Hum Genet 57:717–726 [PMC free article] [PubMed] [Google Scholar]
- Rapin I (1997) Current concepts: autism. N Engl J Med 337:97–104 [DOI] [PubMed] [Google Scholar]
- Rapin I, Allen D (1983) Developmental language disorders: nosologic considerations. In: Kirk U (ed) Neuropsychology of language, reading and spelling. Academic Press, New York, pp 155–184 [Google Scholar]
- Raven J (1989) Standard progressive matrices. Australian Council for Educational Research, Victoria [Google Scholar]
- Risch N, Spiker D, Lotspeich L, Nouri N, Hinds D, Hallmayer J, Kalaydjieva L, et al (1999) A genomic screen of autism: evidence for a multilocus etiology. Am J Hum Genet 65:493–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semel EM, Wiig EH, Secord W (1992) Clinical evaluation of language fundamentals–revised. Psychological Corporation, San Antonio [Google Scholar]
- The SLI Consortium (2002) A genomewide scan identifies two novel loci involved in specific language impairment. Am J Hum Genet 70:384–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparrow S, Balla D, Cicchetti D (1984) Vineland adaptive behaviour scales. American Guidance Service, Circle Pines, MN [Google Scholar]
- Steffenburg S, Gillberg C, Hellgren L, Andersson L, Gillberg IC, Jakobsson G, Bohman M (1989) A twin study of autism in Denmark, Finland, Iceland, Norway and Sweden. J Child Psychol Psychiat 30:405–416 [DOI] [PubMed] [Google Scholar]
- Tager-Flusberg H, Joseph R, Folstein S (2001) Current directions in research on autism. Ment Retard Dev Disabil Res Rev 7:21-29 [DOI] [PubMed] [Google Scholar]
- Tomblin JB, Buckwalter PR (1998) Heritability of poor language achievement among twins. J Speech Lang Hear Res 41:188–199 [DOI] [PubMed] [Google Scholar]
- Tomblin JB, Records NL, Zhang X (1996) A system for the diagnosis of specific language impairment in kindergarten children. J Speech Hear Res 39:1284–1294 [DOI] [PubMed] [Google Scholar]
- Vargha-Khadem F, Watkins K, Alcock KJ, Fletcher P, Passingham RE (1995) Praxic and nonverbal cognitive deficits in a large family with a genetically transmitted speech and language disorder. Proc Natl Acad Sci USA 92:930–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargha-Khadem F, Watkins K, Price CJ, Ashburner J, Alcock KJ, Connelly A, Frackowiak RSJ, Friston KJ, Pembrey ME, Mishkin M, Gadian DG, Passingham RE (1998) Neural basis of an inherited speech and language disorder. Proc Natl Acad Sci USA 95:12695–12700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent JB, Herbrick J-A, Gurling HMD, Bolton PF, Roberts W, Scherer SW (2000) Identification of a novel gene on chromosome 7q31 that is interrupted by a translocation breakpoint in an autistic individual. Am J Hum Genet 67:510–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton P, Baird G, Chen W, Morris K, Jacobs BW, Hodgson S, Docherty Z (2000) Support for linkage of autism and specific language impairment to 7q3 from two chromosome rearrangements involving band 7q31. Am J Med Genet 96:228–234 [DOI] [PubMed] [Google Scholar]
- World Health Organization (1993) The ICD-10 classification for mental and behavioural disorders: diagnostic criteria for research. World Health Organization, Geneva [Google Scholar]