Abstract
It has been proposed that positive selection may be associated with protein functional change. For example, human and macaque have different outcomes to HIV infection and it has been shown that residues under positive selection in the macaque TRIM5α receptor locate to the region known to influence species-specific response to HIV. In general, however, the relationship between sequence and function has proven difficult to fully elucidate, and it is the role of large-scale studies to help bridge this gap in our understanding by revealing major patterns in the data that correlate genotype with function or phenotype. In this study, we investigate the level of species-specific positive selection in innate immune genes from human and mouse. In total, we analyzed 456 innate immune genes using codon-based models of evolution, comparing human, mouse, and 19 other vertebrate species to identify putative species-specific positive selection. Then we used population genomic data from the recently completed Neanderthal genome project, the 1000 human genomes project, and the 17 laboratory mouse genomes project to determine whether the residues that were putatively positively selected are fixed or variable in these populations. We find evidence of species-specific positive selection on both the human and the mouse branches and we show that the classes of genes under positive selection cluster by function and by interaction. Data from this study provide us with targets to test the relationship between positive selection and protein function and ultimately to test the relationship between positive selection and discordant phenotypes.
Keywords: innate immune evolution, species-specific responses, adaptive evolution, protein functional shift, predicting phenotypic response
Introduction
Immunology relies predominately on the mouse as a model organism. However, there are inconsistencies between humans and model organisms in terms of immune response (Mestas and Hughes 2004). For example, protein-coding sequence differences in Toll-like receptor 4 (TLR4) have been shown to produce nickel sensitivity in human but not mouse (Schmidt et al. 2010). We define these divergent phenotypic responses as discordant phenotypes. This discordance in immune response between humans and model organisms is particularly problematic in clinical trials; for example, the anti-inflammatory drug TGN1412 reached clinical trial in 2006 following promising results from testing in crab-eating macaques, but in humans the drug elicited a cytokine storm and multiple organ failure (Stebbings et al. 2007). Therefore discordant phenotypic responses to immune system challenge are evident even between closely related species.
Indeed discordant immune responses between human and model organisms may not be all that surprising considering the divergence times of human and mouse for example and indeed their diverse environments (Mestas and Hughes 2004). Also, we know that the protein-coding regions of immune system genes are among the most rapidly evolving of all genes in the vertebrate genome, with specific innate immune-related genes such as those involved in complement-mediated immunity being among the most rapidly evolving (Kosiol et al. 2008). The “Red Queen hypothesis” describes the evolutionary arms race between host and pathogen (Van Valen 1973), a dynamic that results in fixation of selectively advantageous amino acid substitutions (i.e., positive selection) in immune system genes (Sawyer et al. 2005). Understanding the relationship between genotype and discordant phenotypes remains a major goal of modern evolutionary and biomedical research (Kumar et al. 2011).
There are a number of individual cases where positive selection has been associated with functional shift, for example the SPRY domain of TRIM5α functions as a HIV-1 restriction factor in rhesus macaque but not in human (Stremlau et al. 2005) and signatures of positive selection in the rhesus macaque TRIM5α gene in an 11- to 13-amino acid segment of the SPRY domain have been functionally characterized as providing this HIV-1 restriction capability (Sawyer et al. 2005). Previously we have shown that using rational mutagenesis of positively selected residues in the myeloperoxidase (MPO) protein—an innate immune enzyme produced by neutrophils of mammals revealed that these residues governed the unique chlorination function of the MPO enzyme (Loughran et al. 2012). Although these individual gene-based studies provide support for a correlation between predicted positive selection at the sequence level and protein functional shift, it is clear that further large-scale studies and complementary in vitro forward mutation of ancestral proteins are needed to advance our understanding of the molecular nature of adaptation (Yokoyama et al. 2008; Yokoyama 2013).
We currently have a large number of phylogenetically diverse and high-quality vertebrate genomes available (Flicek et al. 2014). Although species level studies have been conducted on various vertebrate genomes to identify evidence of positive selection, concerns have been raised that they may contain false positives due to recombination (Anisimova et al. 2003), alignment error (Fletcher and Yang 2010), and possible sequence and annotation error (Schneider et al. 2009). Accounting for such potential sources of error in selective pressure analyses is essential for results of biological relevance. We now have population genomic data for human from the 1000 human genomes project, and for mouse from the 17 mouse genomes project (Keane et al. 2011; Abecasis et al. 2012). Population level data has been successfully applied to identifying proteins associated with adaptation to high-altitudes in Tibetans (Simonson et al. 2010). Recently, we have also seen a marked increase in the quality and number of Neanderthal genomes available (Green et al. 2010). The combination of population level data, ancestral genome data, and species level comparisons for selective pressure variation enables us to assess whether putatively positively selected residues are fixed or variable within their respective populations and potentially allows us to determine if and when a selective sweep may have happened (Sabeti et al. 2006).
Combined, these data provide us with a unique opportunity to compare the evolution of the innate immune system between human and mouse from two complementary perspectives: first, from the species comparison level and then from the population level comparison. At both levels, we are using estimates of positive selection as a proxy for potential changes in protein function for further study. For the species level comparison, we have applied lineage-specific codon-based models of evolution to estimate the heterogeneity in selective pressures across species and across sites in a protein (Yang 2007). Essentially this approach employs the ratio of replacement substitutions per replacement site (Dn) over silent substitutions per silent site (Ds) to estimate selective pressures in protein-coding sequences (Yang 2007). The lineage-specific codon models employed have been shown to have low levels of false positive detection using experimental conditions we have adhered to in our study (Yang and dos Reis 2011). In addition, we have incorporated filters for recombination and alignment error to further reduce false positive discovery rates (Anisimova et al. 2003; Fletcher and Yang 2010). The population level approach determined if the positively selected residues of the putative positively selected genes were fixed or variable in the corresponding population, specifically we tested the hypothesis that the putative positive selection event occurred recently by assessing if the substitution processes within the population were neutral or nonneutral (Nei and Li 1979; Tajima 1989).
Teasing apart the relationship between sequence and function is essential for us to better model human disease and drugs. Our findings show that there is evidence for species-specific positive selection in the innate immune system, and that it is more frequent in mouse than human. We find that a number of the putative positively selected genes are clustered into pathways and are associated with known functional discordance between human and mouse providing us with essential targets for the exploration of the relationship between genotype, phenotype, and functional discordance. Positive selection may also result in advantageous alleles becoming fixed within a population (Haldane 1927). Indeed, we observe the positively selected residues to be fixed (for the most part) in their respective modern populations, lending further support to the hypothesis that they may be under positive selection. The antagonistic coevolutionary relationship between hosts and pathogens is reported to result in a heightened rate of molecular evolution and greater genetic divergence of interacting pairs of proteins (Paterson et al. 2010). In our analysis, we find that the genes under positive selection are clustered into functional pathways and are part of protein–protein interactions, indicating that potential coevolutionary relationships may in fact be a major driving force among innate immune pathway genes. Our analysis does not include regulatory related differences and neutral/nearly neutral processes that most likely play a role in phenotypic discordance (McLean et al. 2011). Our analysis is conservative and only considers proteins with strong signatures for selective pressure, therefore selective forces acting at the pathway level only will not be detected (Daub et al. 2013). Nonetheless, this large-scale study of the evolution of innate immune genes provides the community with an overview of the pathways, proteins, and specific residues that we predict have undergone different selective pressures in the independent evolution of human and mouse lineages. We present key molecular targets of genes associated with known species-specific discordance for further study.
Results
Evidence for Species-Specific Adaptation in Mouse and Human Innate Immune Genes
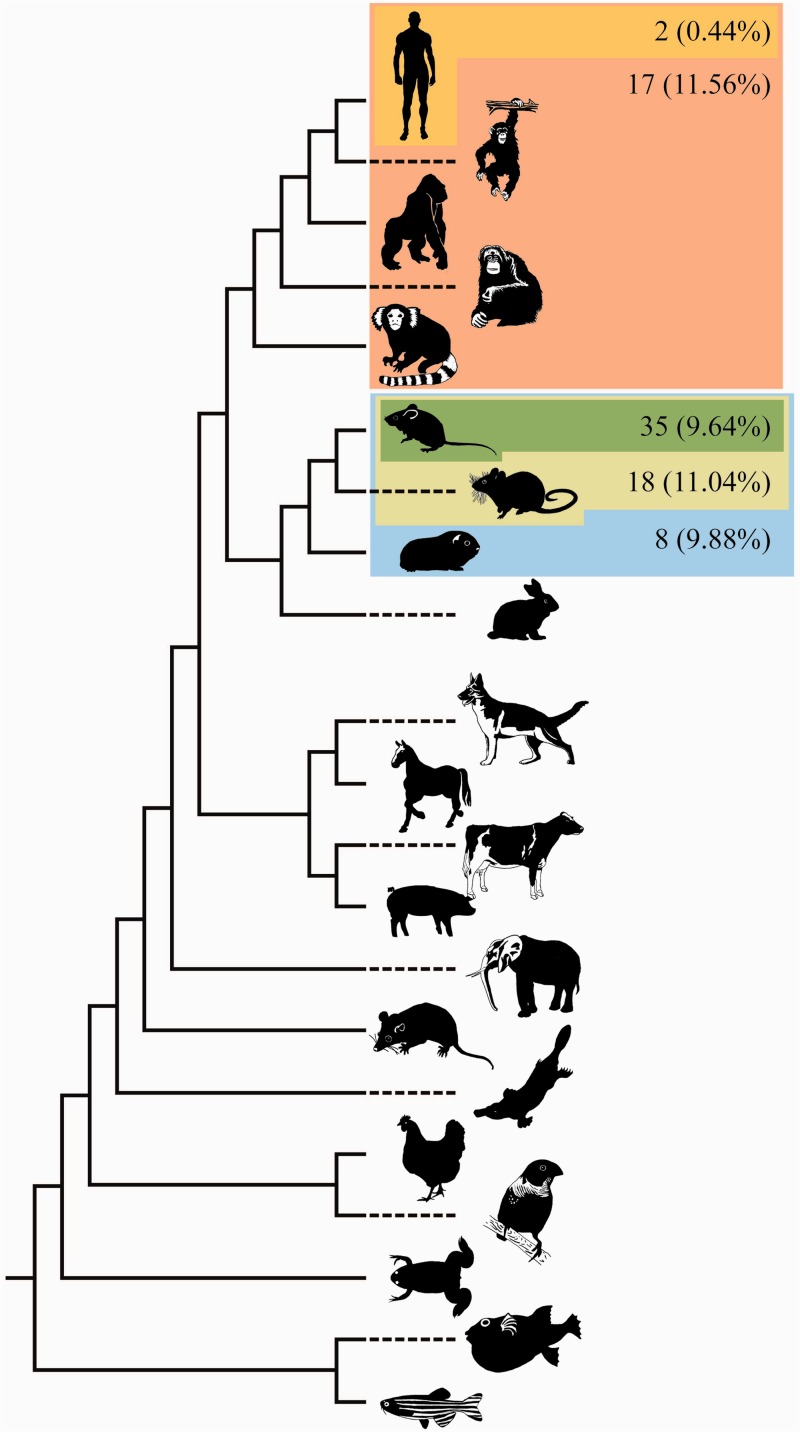
The 456 protein-coding gene families were identified using a data set of human genes reported to be involved in the innate immune response (Lynn et al. 2008) as query against a data set of 21 vertebrate genomes (see Materials and Methods). Species-specific paralogs were permitted within these gene families, and a gene family had to contain a minimum of seven sequences to be included in the selective pressure analyses (Anisimova et al. 2003). Following likelihood ratio tests (LRTs) of nested codon models (Yang 2007), we identified a number of candidate genes exhibiting signatures of positive selection particular to given lineages (supplementary table S1, Supplementary Material online). False positive predictions of positive selection have been shown to be associated with recombination (Anisimova et al. 2003). To ameliorate the frequency of false positives any candidate gene that had evidence of recombination proximal to a positively selected site was removed from our list of positively selected genes. Following the filter for recombination, our predictions were as follows: two candidate genes exhibiting lineage-specific positive selection (CARD6 and IRF9) in the human lineages (these two candidate genes were from the total of 456 gene families that contained a human ortholog), and 35 in the mouse lineage (these 35 genes were from the total of 363 gene families that contained a mouse ortholog; table 1 and fig. 1).
Table 1.
Sample of the Genes under Positive Selection in Human and Mouse and Their Parameter Estimates.
| Gene Name | Positively Selected Sites | P-Value of Positively Selected Sites |
|---|---|---|
| Genes under positive selection in human | ||
| CARD6 | 264, 346, 382, 750, 767, 805, 818, 903, 916, 937, 998, 1010, and 1031 | 13 > 0.50, 0 > 0.95, 0 > 0.99 |
| IRF3 | 119, 129, and 333 | 3 > 0.50, 0 > 0.95, 0 > 0.99 |
| Genes under positive selection in mouse | ||
| Adipoq | 25, 27, 29, and 82 | 4 > 0.50, 0 > 0.95, 0 > 0.99 |
| Atg9a | 634 and 662* | 2 > 0.50, 1 > 0.95, 0 > 0.99 |
| C1inh | 332*, 365, 468, 479 | 4 > 0.50, 1 > 0.95, 0 > 0.99 |
| C1ra | 468, 520, 574, 631, 633, and 634* | 6 > 0.50, 1 > 0.95, 0 > 0.99 |
| C6 | 220, 233, 319, 353, 378, 408, 419, 430, 554, 655, 681, 703, 792, and 930 | 14 > 0.50, 0 > 0.95, 0 > 0.99 |
| C8b | 242*, 263, 278, 383*, and 488 | 5 > 0.50, 2 > 0.95, 0 > 0.99 |
| Card6 | 394, 501, and 702 | 3 > 0.50, 0 > 0.95, 0 > 0.99 |
| Cd200 | 129 and 177 | 2 > 0.50, 0 > 0.95, 0 > 0.99 |
| Cd63 | 31, 118, 143, 184*, 194, and 203 | 6 > 0.50, 1 > 0.95, 0 > 0.99 |
| Cfh | 209, 243, 474, 767, 1005, 1068, 1074, 1104, 1181, and 1227 | 10 > 0.50, 0 > 0.95, 0 > 0.99 |
| Ecsit | 10, 12, 75, 82, 176, 325, 330**, 348, and 371 | 9 > 0.50, 1 > 0.95, 1 > 0.99 |
| Eif2ak2 | 136, 155, 181, 182*, 344, and 345 | 6 > 0.50, 1 > 0.95, 0 > 0.99 |
| F12 | 45, 65, 166, 243**, and 454 | 5 > 0.50, 1 > 0.95, 1 > 0.99 |
| Grn | 18, 101, 198, 303, 375*, 382, 411, 549, and 597 | 9 > 0.50, 1 > 0.95, 0 > 0.99 |
| Ifit2 | 191, 402, and 420 | 3 > 0.50, 0 > 0.95, 0 > 0.99 |
| Il1rapl2 | 566, 628, and 666* | 3 > 0.50, 1 > 0.95, 0 > 0.99 |
| Il2rb | 4, 13, 31, 55, 174, 202, 347*, 402, 418, 491, 496, and 516 | 12 > 0.50, 1 > 0.95, 0 > 0.99 |
| Il4ra | 47, 67, 308, 330, and 626 | 5 > 0.50, 0 > 0.95, 0 > 0.99 |
| Irf5 | 232, 259, and 262 | 3 > 0.50, 0 > 0.95, 0 > 0.99 |
| Lbp | 24, 40 and 329 | 3 > 0.50, 0 > 0.95, 0 > 0.99 |
| Lgals3 | 22, 92, 94, and 260 | 4 > 0.50, 0 > 0.95, 0 > 0.99 |
| Lrrfip1 | 328, 449, 468, 480, and 571 | 5 > 0.50, 0 > 0.95, 0 > 0.99 |
| Ltb4r1 | 53, 101, and 175 | 3 > 0.50, 0 > 0.95, 0 > 0.99 |
| Nlrp14 | 77, 79*, 186, 212, 219, 254, 257, 263, 272, 281, 284, 291, 294, 315, 319, 333, 358, 393, 415, 424, 453, 465, 530, 549, 552, 553, 584, 613, 657, 679, 684, 685, 687, 696, 782, 810, 814, 829, 846, 848, 902, 908, 912, 931, 953, 956, 958, 978, 982, 984, and 986 | 51 > 0.50, 0 > 0.95, 0 > 0.99 |
| Nlrp6 | 22, 25, 72, 77, 80, 81, 85, 96, 101, 113, 114, 190, 192, 251, 260**, 329, 344, 479, 488, 515, 553, 571, 628, 657, 727, 737, 739, 744, 771, 775*, 776, 793, 807, 865, 877, and 880 | 36 > 0.50, 2 > 0.95, 1 > 0.99 |
| Oas2 | 55, 56, 139, 171, 199, 211, 221, 298, 481, 549, and 711 | 11 > 0.50, 0 > 0.95, 0 > 0.99 |
| Plcg2 | 461 and 594* | 2 > 0.50, 1 > 0.95, 0 > 0.99 |
| Ptpn2 | 166, 206, 319, 321, and 329 | 5 > 0.50, 0 > 0.95, 0 > 0.99 |
| Rnf31 | 203, 431, and 1025 | 3 > 0.50, 0 > 0.95, 0 > 0.99 |
| Sirt1 | 107, 537, 698, and 701 | 4 > 0.50, 0 > 0.95, 0 > 0.99 |
| Snap23 | 109, 133, and 197 | 3 > 0.50, 0 > 0.95, 0 > 0.99 |
| Stat2 | 21, 130, 149, 157, 195, 205, 218, 354, 623*, 869, 871, 874, 876, and 877 | 14 > 0.50, 1 > 0.95, 0 > 0.99 |
| Tcf4 | 139 | 1 > 0.50, 0 > 0.95, 0 > 0.99 |
| Tlr3 | 266, 297, and 603 | 3 > 0.50, 0 > 0.95, 0 > 0.99 |
| Trif | 18, 327, 338, 388, 482, 556, and 711 | 8 > 0.50, 0 > 0.95, 0 > 0.99 |
Note.—For each gene the positively selected sites are displayed and the associated posterior probability is indicated as follows: * >0.95, ** >0.99 and no asterisk implies 0.50 < PP < 0.95.
Fig. 1.
Phylogeny of species included in this study and summary of lineage-specific positive selection results. The lineages that were tested for species-specific selective pressure variation are highlighted. The number and percentage of genes displaying evidence of species-specific positive selection are also shown.
A Subset of Mouse Innate Immune Pathways Are Enriched for Adaptive Evolution
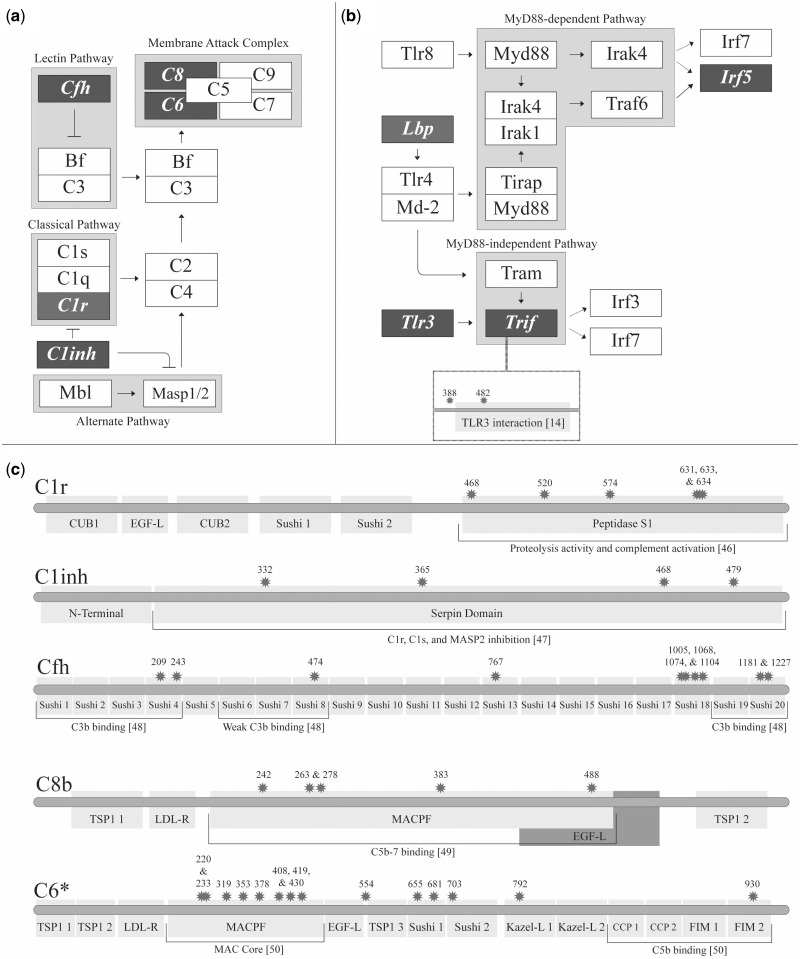
The 35 genes under positive selection in mouse included a notable number of proteins involved in direct interactions with one another (fig. 2) and in the same pathways: 5 genes in the complement cascade (C1ra, C1inh, C6, C8b, and Cfh), 4 genes in the TLR signaling pathway (Irf5, Lbp, Tlr3, and Trif), 3 genes in the Janus kinase-signal transducer an activator of transcription (JAK-STAT) pathway (Stat2, Il2rb, and Il4ra), and a 1 gene in the mitogen activated protein kinase (MAPK) pathway signaling pathway (Ecsit). Of particular interest was the positive selection predicted to occur on the Trif protein at the interaction interface with Tlr3 (fig. 2; Oshiumi et al. 2003).
Fig. 2.
Innate immune pathways incorporating multiple positively selected genes. The positively selected genes in (a) the complement system and (b) the TLR signaling pathway are illustrated as darkened rectangles. Signaling cascades are depicted as arrows and inhibitors are depicted as blunt-ended lines. Defined pathways and complexes are highlighted in gray boxes with the given name. (c) Positively selected sites of the complement system alongside information on domain structure. Information on the function of these domains is also given.
Ancestral Nodes Have Unique Subsets of Genes under Positive Selection
As positive selection could have arisen within the ancestral lineages separating modern human and mouse, we tested all ancestral lineages across the Euarchontoglires clade (fig. 1), that is, the ancestral primate, the most recent common ancestor (MRCA) of rat and mouse (murinae), and the MRCA of all rodents in the data set.
We found 18 genes (11.56% of testable genes) with evidence of species-specific positive selection in the ancestral primate lineage. These genes clustered into three categories relative to their function: Nod-like receptors (NLRP1, NLRP5, and NLRP8), TRIM receptors (TRIM5 and TRIM25), and interferon gamma receptors (IFNGR1 and IFNGR2; fig. 1 and supplementary table S1, Supplementary Material online). Within this set of genes there is one known protein–protein interaction between IFNGR1 and IFNGR2 associated with the JAK-STAT pathway (Kanehisa and Goto 2000). The JAK-STAT pathway mediates cellular responses including proliferation, differentiation, migration, apoptosis, and cell survival and is the target for development of therapeutics for a variety of immune disorders.
Analyses of the ancestral murinae and rodent branches identified 18 (11.04%) and 8 (9.88%) genes, respectively, as under species-specific positive selection (fig. 1 and supplementary table S1, Supplementary Material online). There was limited functional and pathway information on these genes and there were no reports of direct protein–protein interactions in either ancestral lineage. The C1inh and C9 genes from the complement cascade featured in the murinae lineage with sites under selection unique to that ancestral lineage. There was also evidence for positive selection unique to the murinae lineage in the TRAF6 protein, a gene known to interact with the TLR signaling pathway (Kanehisa and Goto 2000).
Our results emphasize the heterogeneity in selective pressures in the innate immune systems of different lineages and species across the Euarchontoglires clade. These putative positively selected genes are prime targets for in vitro assessment of functional consequences of mutating these residues.
Positively Selected Residues Map to Essential Functional Domains and Are Predicted to Affect Structural Stability
Assessment of the 35 genes under positive selection uniquely in the mouse lineage identified clusters of positive selection in the complement and the TLR signaling cascades (fig. 2a and b; supplementary table S1, Supplementary Material online). To determine the potential impact of positively selected residues on immune function we employed data from SwissProt (UniProt Consortium 2014). The residues under positive selection in these proteins were concentrated in regions central to function and in regions known to interact with one another (fig. 2).
In the absence of in vitro functional assays for each of these positively selected residues and genes, we performed predictions on overall protein structure and stability based on in silico structural modeling and rational mutagenesis to further investigate these putative positively selected residues and to predict their potential impact. The dynamic flexibility index (dfi) measures the contribution of each residue to the overall structural dynamics and stability of the protein (Gerek et al. 2013). The dfi approach uses perturbation response scanning on a 3D elastic network model (Atilgan and Atilgan 2009; Atilgan et al. 2010) to measure the spatial fluctuation of each residue in response to perturbing residues along the peptide. dfi values indicate the resilience of each residue to perturbations, a low dfi indicates a residue essential for dynamic stability as they absorb the transfer of perturbation (i.e., these residues are structurally inflexible) whereas high dfi implies the residue is prone to perturbation (i.e., these residues are structurally flexible; Gerek et al. 2013). dfi values are reported to significantly correlate with known neutral variants displaying high dfi values and residues strongly linked with genetic disease having low dfi values (Gerek et al. 2013). In comparison to the solvent accessible surface area metric used to assess functional significance, the correlation of dfi to biological function has been reported to have a greater ability to discern functionally critical from noncritical sites (Gerek et al. 2013). In addition, in an analysis of 100 genes across 46 eukaryote genomes, dfi values were found to positively correlate with the rate of evolution observed over approximately 1 billion years of evolution showing that this approach is robust to large evolutionary distances (Gerek et al. 2013).
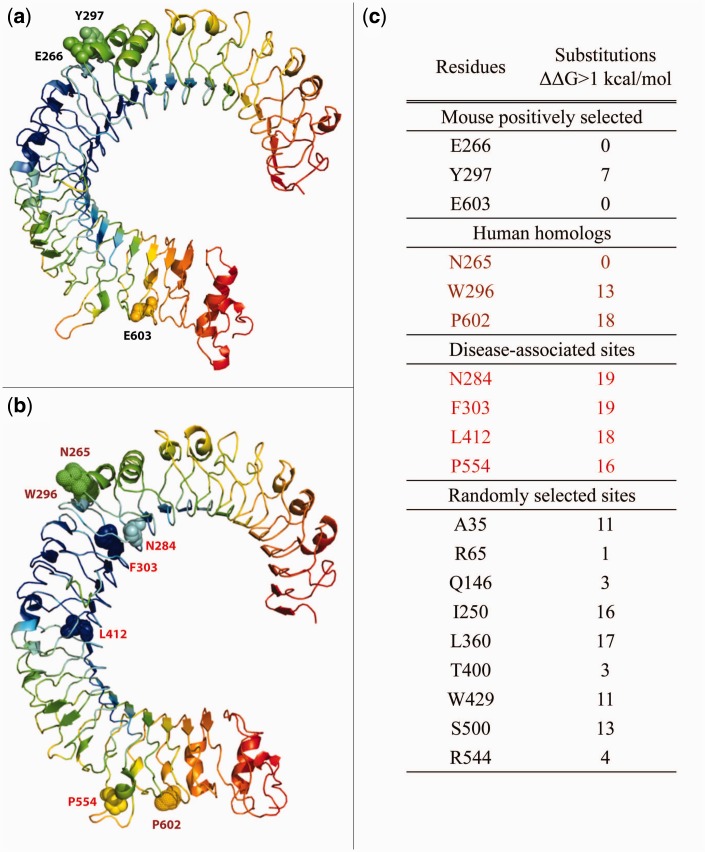
TLR3 was chosen for dfi analysis because it has a signature of positive selection in mouse but not human, and also because TLR3 is well characterized and there are crystal structures for it’s ligand-binding ectodomain available for in silico mutation (PDB identifier for Human TLR3 is 2A0Z, and for Mouse is 3CIG). First, structural flexibility was assessed. We assessed the dfi values for four well known human TLR3 disease-associated residues: N284 and P554 that alter the activity of TLR3 (Ranjith-Kumar et al. 2007), F303 which results in TLR3 deficiency (Zhang et al. 2007), and L412 involved in encephalopathy (Hidaka et al. 2006). All of these disease associated sites were found to have low dfi values (Stenson et al. 2003; fig. 3). Conversely, the positively selected residues from mouse TLR3 (i.e., E266, Y297, and E604) and their counterparts in human (i.e., N265, W296, and P602) had moderate to high dfi values, indicating structural flexibility, perhaps this flexibility is related to their role in ligand binding of the TLR3 ectodomain (fig. 3). Our results areconsistent with disease-associated sites being structurally inflexible in comparison to other sites (Hurst andSmith 1999; Liu and Bahar 2012; Gerek et al. 2013).
Fig. 3.
dfi of human TLR3 ectodomain. Ribbon diagrams of the crystal structure of the TLR3 ectodomain of (a) human (PDB id: 2A0Z) and (b) mouse (PDB id: 3CIG) colored with a spectrum of red–yellow–green–cyan–blue with respect to dfi. Red indicates the highest dfi values whereas blue indicates the lowest values. (c) The stability change for all possible substitutions was computed for: the positively selected sites in mouse (E266, Y297, and E603), their human homologs (N265, W296 and P602), known human disease-associated sites (N284, F303, L412, and P554; Stenson et al. 2003, and randomly selected sites. Except for the randomly selected sites, sites have been colored and indicated on the respective ribbon diagrams.
Next, we assessed the potential impact of the positively selected residues on protein stability by calculating the change in protein folding free energy (ΔΔG) in the crystal structures of the human and mouse TLR3 ectodomains (fig. 3). ΔΔG calculations are typically used to determine if mutations are potentially deleterious due to protein stability perturbations, with harmful mutations being over a threshold (usually set to >1 kcal/mol; Yue et al. 2005). To determine the tolerance of each putative positively selected position to mutation, we calculated ΔΔG for all possible amino acid substitutions at each of these specific positions. We did not predict a destabilizing effect for the three positively selected residues in the ectodomain of mouse TLR3 (E266, Y297, and E603; fig. 3), indicating that these positions in mouse are tolerant to mutation and are structurally flexible directly supporting the results from the dfi analysis. When we mutated the homologous residues in human (N265, W296, and P602), we found two sites (W296 and P602) exhibiting an effect on folding stability. More specifically, of the 19 possible mutations for the two positions W296 and P602, we observe destabilizing effects in 13 and 18 substitutions, respectively (fig. 3). Substitutions of W296 and P602 exhibit predicted changes in protein stability comparable to the human disease-associated sites (i.e., N284, P554, F303, and L412). Mutating randomly chosen residues in TLR3 predicted no such impact on protein stability (fig. 3). Taken together, these data suggest that two of the positively selected residues in mouse TLR3 and their homologous positions in human TLR3 potentially contribute differently to overall protein stability in these two species.
The Majority of Positively Selected Residues Is Fixed within Human and Mouse Species
Positive selection can rapidly drive advantageous alleles to fixation within a population (Sabeti et al. 2006). Depending on factors such as on the age of the event, effective population size (Ne) of the species, and the strength of selection, positively selected sites may be fully fixed within a population or may have some degree of variability (Sabeti et al. 2006). We sought to determine if the positively selected residues identified in human and mouse are fixed or variable in their respective populations (table 2). We gathered all available single nucleotide polymorphism (SNP) data for all positively selected genes, that is, two human candidate genes and 27 mouse candidates. The majority (95–97%) of positively selected sites in both human and mouse was entirely fixed in their lineage. The exceptions were the CARD6 gene in human and six mouse genes (C6, C8b, Ecsit, Il4ra, Nlrp14, and Stat2), in total there were eight variable sites across these seven genes (table 2). Of these eight variable positions, four sites had substitutions to the ancestral allele present at the homologous position in other species, they were as follows: human CARD6 (G264E) and mouse Ecsit (S75L); Nlrp6 (R744K); and Stat2 (L874M). In addition, there were two substitutions at positively selected residues in mouse genes that resulted in amino acids with similar physicochemical properties as the homologous position in another species: C6 (R554L) and Il4ra (D626G; table 2).
Table 2.
Population Level Analysis of Positively Selected Sites in Human and Mouse Genes.
| Genes Tested | Number of Positively Selected Sites | Number of coding SNPs tested | Unfixed Sites |
|---|---|---|---|
| Genes under positive selection specifically in the human lineage | |||
| CARD6 | 13 | 38 | G264E |
| IRF9 | 3 | 9 | None |
| Genes under positive selection specifically in the mouse lineage | |||
| Adipoq | 4 | 9 | None |
| Atg9a | 1 | 3 | None |
| C1ra | 6 | 16 | None |
| C6 | 14 | 53 | R554L |
| C8b | 5 | 36 | M263I |
| Card6 | 3 | 4 | None |
| Cd63 | 6 | 6 | None |
| Cfh | 10 | 8 | None |
| Ecsit | 9 | 1 | S75L |
| Grn | 9 | 16 | None |
| Ifit2 | 3 | 14 | None |
| Il1rapl2 | 3 | 11 | None |
| Il4ra | 5 | 51 | F47S & D626G |
| Irf5 | 3 | 14 | None |
| Lbp | 2 | 33 | None |
| Lgals3 | 4 | 15 | None |
| Lrrfip1 | 5 | 49 | None |
| Ltb4r1 | 3 | 16 | None |
| Nlrp14 | 51 | 16 | A613S |
| Nlrp6 | 36 | 53 | None |
| Plcg2 | 1 | 51 | None |
| Rnf31 | 3 | 24 | None |
| Snap23 | 3 | 2 | None |
| Stat2 | 14 | 58 | L874M |
| Tcf4 | 1 | 20 | None |
| Tlr3 | 3 | 4 | None |
| Trif | 7 | 3 | None |
note.—Assessment of the fixation of positively selected sites was only possible on the genes shown above. For each gene, the number of positively selected sites and the number of coding SNPs tested are given. If a coding SNP within a positively selected codon resulted in a nonsynonymous substitution, the resulting unfixed variant is shown above as a mutation at their respective site (e.g., G264E).
The positively selected residues in human CARD6 and IRF9 genes were compared with the homologous positions in the recently released Neanderthal genomes (Green et al. 2010), and the same positively selected residues are fixed in Neanderthal as in modern human.
Population Data Show No Ongoing Selective Sweep in Modern Humans
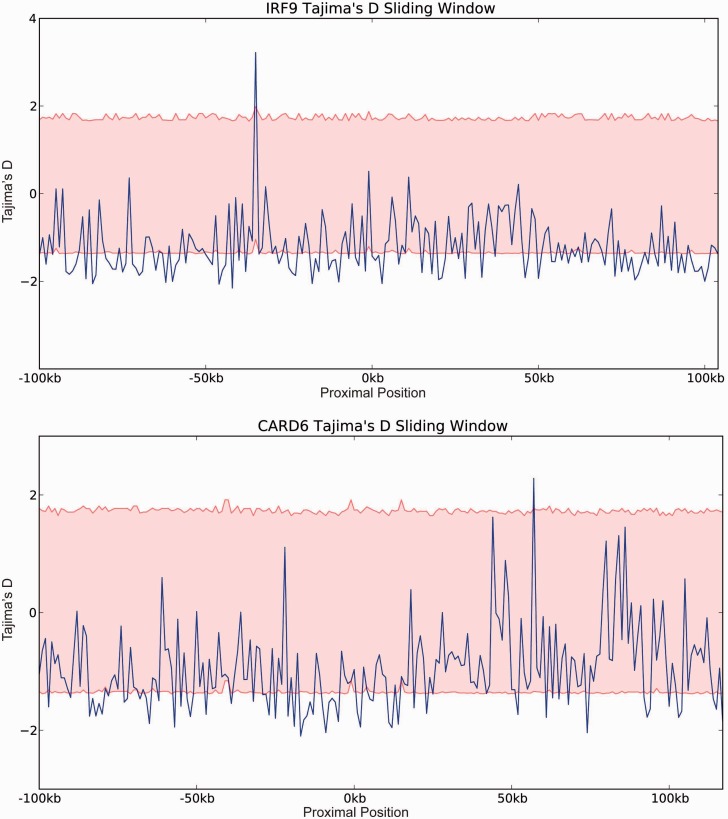
To determine if our two positively selected genes from human were under ongoing selective pressure, we tested for evidence of reduced variability in 1 kb sliding windows of allele frequency encompassing 100 kb up- and downstream for each of the two genes. Variability and departure from neutrality were measured in each window (Tajima 1989; fig. 4). The analysis of CARD6 and IRF9 identified comparable levels of variation for all windows assessed. The results of the CARD6 and IRF9 analyses are consistent with a sweep to fixation of the positively selected residues prior to the divergence of human and Neanderthal, and a relaxation of selective pressure in the current human population.
Fig. 4.
Neutrality tests for positively selected genes in the human lineage. Sliding window analysis of Tajima’s D of the positively selected genes identified in human. The analysis was conducted using a window size of 1 kb within 100 kb upstream and downstream of each gene. The 95% confidence interval is shown as red highlighted region.
Discussion
We have shown that the innate immune systems of human and mouse have experienced distinct species-specific selective forces. Signatures of positive selection have been correlated with alteration to protein function (Sawyer et al. 2005; Loughran et al. 2012) and the identification of such distinct patterns alongside known discordant immune responses provides the community with a set of potential molecular targets for further testing.
The positively selected genes in mouse are components of well-known innate immune pathways, and are involved upstream and downstream in these pathways. In pathways enriched for positively selected members (e.g., TLR signaling pathways and complement system), more than half the components are involved directly or indirectly with initiation (Cfh, C1r, C1inh, Tlr3, and Lbp), indicating that upstream genes may be more prone to positive selective pressure. Recent studies have reported a relaxation in selective constraint as you progress downstream through a pathway (Ramsay et al. 2009), whereas other reports detail patterns similar to those presented here (Alvarez-Ponce et al. 2009). The enrichment for positive selection that we observe at the start of the pathways is most likely because many of these genes directly interact with pathogens (C6, C8, and Lbp), bind to pathogens for immunological defense (Cfh), or are pathogen recognition receptors (Tlr3) and are most likely under selective pressure from the pathogen (Van Valen 1973). The absence of human-specific positively selected genes within the TLR signaling pathways and complement system may be due to the episodic pattern of primate TLR evolution (Wlasiuk and Nachman 2010).
TLR3 is responsible for detecting double stranded RNA in viral infections. The species-specific positive selection we detected in TLR3 together with in silico predicted disparity in stability between human and mouse residues suggests that there may be potential protein structural differences between the human and mouse orthologs of TLR3. The positively selected residues predicted for TLR3 are of particular interest given the restricted antiviral role of human TLR3 in comparison to mouse Tlr3, and LPS upregulation of Tlr3 in mouse but not human macrophages (Ariffin and Sweet 2013). Further investigation is warranted to determine if these sites in TLR3 have functional impact and if that is related to the observed phenotypic discordance between human and mouse. The use of in silico modeling approaches such as the dfi method applied here can help to prioritize targets from a selective pressure analysis for in vitro testing.
The complement cascade is reported to neutralize herpes simplex virus in rat, mouse, and human. However, complement activation proceeds uniquely for each species: via the lectin pathway in mouse, via the alternative and lectin pathways in rat, and, in human via the classical pathway (Wakimoto et al. 2002). The exclusive use of the lectin pathway in mouse may in part be due to the species-specific functional shift we have identified in the alternative and classical pathways. In response to Acanthamoeba infection, both human and mouse are reported to initiate the complement cascade by the alternative pathway leading to Acanthamoeba binding with C9 of the membrane attack complex (MAC; Pumidonming et al. 2011). In contrast, mouse MAC is unable to lyse Acanthamoeba (Pumidonming et al. 2011). Here, we show that the C6 and C8b proteins of the complement cascade are under positive selection in mouse but not in human. These selected residues provide us with potential targets for the discordant response between human and mouse MAC. More generally, knowledge of the full plethora of genes under positive selection in human, mouse, or their ancestral lineages provide us with molecular markers that may contribute to our understanding of the potential difficulties in modeling human innate immunity (Mestas and Hughes 2004; Seok et al. 2013).
Although signatures of positive selection were identified in CARD6 and IRF9 in the lineage leading to modern human and indeed Neanderthal, there is no evidence of an ongoing selective sweep in modern human in these genes (Tajima 1989), it is important to note that this approach is sensitive to the age of the adaptive event with the outer limit for human of approximately 250,000 years ago (Sabeti et al. 2006). The conservation of these positively selected residues between modern human and Neanderthal may be due to shared ancestry dating these adaptive events to between approximately 400,000 and 600,000 years before present (ybp) (Scally and Durbin 2012), alternatively an introgressive event (such as that reported for the HLa locus (Abi-Rached et al. 2011)) would date the event between 40,000 and 400,000 ybp (Pinhasi et al. 2011).
The majority (96%) of positively selected sites was fixed in the modern human or mouse populations. However, it is important to note that the mouse population data are based on 21 artificially selected laboratory mouse strains, and therefore does not represent true population structure (Keane et al. 2011). The single unfixed positively selected site in human was position G264E of CARD6. This replacement SNP (rs61757657) is documented in only 3% of Africans and 1% of Americans (2% of Puerto Ricans; Kinsella et al. 2011; Abecasis et al. 2012). Therefore, our predictions of positive selection in human and mouse are for residues that we determined to be at high frequency in these species.
Our novel candidates for positive selection have direct utility for the study of human and mouse innate immunology. Such applications of evolutionary theory to immunology have the potential to contribute to more precise modeling of the human immune response in the future.
Materials and Methods
Generating the Data Set for Analysis
The innate immune data set consisted of 725 human innate immune response genes downloaded from InnateDB (Lynn et al. 2008). Ortholog identification was achieved using standalone BLASTp (v2.2.23+; Altschul et al. 1990) on the canonical transcripts of 21 high coverage (>6×) genomes: chicken, chimpanzee, cow, dog, elephant, fugu, gorilla, guinea pig, horse, human, marmoset, mouse, opossum, orangutan, pig, platypus, rabbit, rat, xenopus, zebra finch, and zebrafish. Genomes were downloaded from Ensembl Gene 60 on Ensembl BioMart (Kinsella et al. 2011). Orthologous groups were identified utilizing a threshold of less than e-150 and above 85% sequence alignment length to ensure high-quality alignments. Families were only analyzed if they contained six or more members.
Sequence Alignment and Selection
Amino acid multiple sequence alignment (MSA) was carried out using MUSCLE (v3.8.31; Edgar 2004) and PRANK (v100802; Loytynoja and Goldman 2005). The alignments from MUSCLE and PRANK were compared using MetAl (v1.1.0; Blackburne and Whelan 2012) and NoRMD (v1.2; Thompson et al. 2001), and a single alignment was selected for each family.
Protein Model Selection
Substitution models were selected for the MSA based on the results of ProtTest 3.0 (Darriba et al. 2011). Matrices were also assayed in the presence of invariable sites (+I), variable rate categories (+G), or a combination of these two factors (+I+G). The model of best fit for each MSA was chosen using the resulting Bayesian information criterion scores ([Darriba et al. 2011] and references therein).
Phylogenetic Reconstruction
Phylogenetic reconstruction was carried out using MrBayes (Ronquist and Huelsenbeck 2003). MSA input was provided by the results of either MetAl or NoRMD using the substitution model provided by ProtTest (Thompson et al. 2001; Darriba et al. 2011; Blackburne and Whelan 2012). For each MSA, four Markov chain Monte Carlo chains were run for a minimum of 106 generations or until convergence was reached. Chains were sampled every 200 generations with a burn-in fraction of 0.25. Under these settings a minimum of 3,750 samples were assayed for a posterior probability distribution for each gene alignment.
Selective Pressure Analysis
Selective pressure analyses were performed utilizing codeML from the PAML software package (v4.4e; Yang 2007). CodeML examines nested codon-based models of evolution in a maximum likelihood framework to determine the ratio of nonsynonymous substitutions per nonsynonymous site (Dn) to synonymous substitutions per synonymous site (Ds), or ω (Yang 2007). We employed branch-site specific models because we wished to scan for positive selection that is unique to a specific foreground lineage/clade and that is not present elsewhere on the tree (Yang and dos Reis 2011). Our analysis uses in-house software for implementing LRTs (available from authors on request). The LRT test statistic approximates the chi-squared (χ2) distribution critical value with degrees of freedom equal to the number of additional free parameters in the alternative model.
If models passed LRT and positive selection is inferred, the posterior probability of the positively selected site is estimated using two calculations: Naïve Empirical Bayes (NEB) or Bayes Empirical Bayes (BEB; Yang 2007). If both BEB and NEB are predicted, we utilized the BEB results as they have been reported to be more robust ([Yang 2007] and references therein).
Detection of Recombination
We tested if recombination had occurred in the genes of the single gene orthologous data set using RDP3 (Martin et al. 2010). This includes the following methods for detecting recombination: BOOTSCAN, GENECONV, MAXCHI, CHIMAERA, SISCAN, and 3SEQ (see [Martin et al. 2010] and references therein). Evidence of recombination was reported if an event was significant for a phylogenetic-based method (BOOTSCAN or SISCAN) as well as a substitution-based method (CHIMAERA, MAXCHI, GENECONV, or 3SEQ) (see [Martin et al. 2010] and references therein).
Structural Analysis of TLR3
We modeled 3D structures of mouse and human TLR3 ectodomain. We obtained 100% sequence identity between the target sequences and mouse (PDB id: 3CIG) and human (PDB id: 2A0Z) template structures. After having modeled structures, we first computed the dfi (Gerek et al. 2013) that quantifies the dynamic properties of individual residues in any protein structure and then the stability change caused by mutating each positively selected site in the structure (Gerek et al. 2013). Overall, due to 3D structure and the residue interaction network within a protein, some positions are more susceptible to these perturbations, showing high fluctuation responses and high dfi values, whereas other positions with low dfi values are stable and do not deviate significantly from their original position upon perturbation. Therefore, higher dfi means greater chance that this is a nonfunction-altering mutation.
The stability change is measured by calculating the protein folding free energy (ΔΔG). ΔΔG value for each amino acid substitution (mutant) is calculated from ΔGmutant − ΔGwt where ΔGwt is the wild type free energy of unfolding. To compute the ΔΔG values for each amino acid substitution, we applied the FoldX method (Guerois et al. 2002; Schymkowitz et al. 2005) that utilizes empirical potential combining both physical force fields and free parameter fitted with known experimental data. If the value of ΔΔG is greater than 0, the mutation has a destabilizing effect on the protein structure, while ΔΔG < 0, the mutation is stabilizing. In our analysis, we used 1 kcal/mol as a threshold. First, we computed the stability change of positively selected residues in the mouse structure and then we selected positively selected sites in the human sequence from the alignment information. Finally, we estimated ΔΔG for all possible amino acid substitutions in the human structure. In addition, we computed stability changes corresponding to sites that cause disease in humans. These disease-associated sites were obtained from the Human Gene Mutation Database (Stenson et al. 2003). To determine if there are patterns of “protein stability” are unique to positively selected residues compared with all other residues in the protein including disease sites, we estimated the ΔΔG for all possible amino acid substitutions of randomly selected sites as well as all positively selected and known disease-associated sites.
Fixation of Positively Selected Sites in Population SNP Data
To determine the frequency at which positively selected sites were fixed within the human population, we downloaded validated SNPs for protein-coding regions from Ensembl Biomart (Ensembl Variation 72; Kinsella et al. 2011). Using in-house software we mapped a total of 559 SNPs onto the protein-coding regions of 26 positively selected innate immune genes (3 human and 23 mouse). We also mapped our putative positively selected sites to this alignment.
Assessing Positively Selected Genes for Evidence of Selection within Human Population Data
To determine if population data corroborated the species-level findings for positive selection in human lineage, we downloaded variation data from the 1000 Genomes Project website, with each individual consisting of two chromosomal samples (Abecasis et al. 2012). To establish evidence of a selective sweep, we created population alignments for our human positively selected genes, including 100 kb upstream and downstream DNA (where possible). Using DnaSP (Librado and Rozas 2009) we calculated Tajima’s D (Tajima 1989) and nucleotide diversity (Nei and Li 1979) for each gene in 1 kb windows. To determine the significance of Tajima’s D, 10,000 coalescence simulations were conducted for each gene (Hudson 2002).
Supplementary Material
Supplementary table S1 is available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
This work was funded by Science Foundation Ireland Research Frontiers Programme grant (EOB2673) awarded to M.J.O'C. M.J.O'C. would also like to acknowledge the Fulbright commission for support. The authors wish to acknowledge the DJEI/DES/SFI/HEA Irish Centre for High-End Computing (ICHEC) for the provision of computational facilities and support.
References
- Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abi-Rached L, Jobin MJ, Kulkarni S, McWhinnie A, Dalva K, Gragert L, Babrzadeh F, Gharizadeh B, Luo M, Plummer FA, et al. The shaping of modern human immune systems by multiregional admixture with archaic humans. Science. 2011;334:89–94. doi: 10.1126/science.1209202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Alvarez-Ponce D, Aguade M, Rozas J. Network-level molecular evolutionary analysis of the insulin/TOR signal transduction pathway across 12 Drosophila genomes. Genome Res. 2009;19:234–242. doi: 10.1101/gr.084038.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimova M, Nielsen R, Yang Z. Effect of recombination on the accuracy of the likelihood method for detecting positive selection at amino acid sites. Genetics. 2003;164:1229–1236. doi: 10.1093/genetics/164.3.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariffin JK, Sweet MJ. Differences in the repertoire, regulation and function of Toll-like Receptors and inflammasome-forming Nod-like Receptors between human and mouse. Curr Opin Microbiol. 2013;16:303–310. doi: 10.1016/j.mib.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Atilgan C, Atilgan AR. Perturbation-response scanning reveals ligand entry-exit mechanisms of ferric binding protein. PLoS Comput Biol. 2009;5:e1000544. doi: 10.1371/journal.pcbi.1000544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atilgan C, Gerek ZN, Ozkan SB, Atilgan AR. Manipulation of conformational change in proteins by single-residue perturbations. Biophys J. 2010;99:933–943. doi: 10.1016/j.bpj.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburne BP, Whelan S. Measuring the distance between multiple sequence alignments. Bioinformatics. 2012;28:495–502. doi: 10.1093/bioinformatics/btr701. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daub JT, Hofer T, Cutivet E, Dupanloup I, Quintana-Murci L, Robinson-Rechavi M, Excoffier L. Evidence for polygenic adaptation to pathogens in the human genome. Mol Biol Evol. 2013;30:1544–1558. doi: 10.1093/molbev/mst080. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher W, Yang Z. The effect of insertions, deletions, and alignment errors on the branch-site test of positive selection. Mol Biol Evol. 2010;27:2257–2267. doi: 10.1093/molbev/msq115. [DOI] [PubMed] [Google Scholar]
- Flicek P, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, et al. Ensembl 2014. Nucleic Acids Res. 2014;42:D749–D755. doi: 10.1093/nar/gkt1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerek Z, Kumar S, Banu Ozkan S. Structural dynamics flexibility informs function and evolution at a proteome scale. Evol Appl. 2013;6:423–433. doi: 10.1111/eva.12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MH, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerois R, Nielsen JE, Serrano L. Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J Mol Biol. 2002;320:369–387. doi: 10.1016/S0022-2836(02)00442-4. [DOI] [PubMed] [Google Scholar]
- Haldane JB. A mathematical theory of natural and artificial selection. V: selection and mutation. Mathematical Proceedings of the Cambridge Philosophical Society. 1927;Vol. 23:838–844. [Google Scholar]
- Hidaka F, Matsuo S, Muta T, Takeshige K, Mizukami T, Nunoi H. A missense mutation of the Toll-like receptor 3 gene in a patient with influenza-associated encephalopathy. Clin Immunol. 2006;119:188–194. doi: 10.1016/j.clim.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Hudson RR. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics. 2002;18:337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- Hurst LD, Smith NG. Do essential genes evolve slowly? Curr Biol. 9. 1999:747–750. doi: 10.1016/s0960-9822(99)80334-0. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane TM, Goodstadt L, Danecek P, White MA, Wong K, Yalcin B, Heger A, Agam A, Slater G, Goodson M, et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–294. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsella RJ, Kahari A, Haider S, Zamora J, Proctor G, Spudich G, Almeida-King J, Staines D, Derwent P, Kerhornou A, et al. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database. 2011;2011:bar030. doi: 10.1093/database/bar030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosiol C, Vinar T, da Fonseca RR, Hubisz MJ, Bustamante CD, Nielsen R, Siepel A. Patterns of positive selection in six Mammalian genomes. PLoS Genet. 2008;4:e1000144. doi: 10.1371/journal.pgen.1000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Dudley JT, Filipski A, Liu L. Phylomedicine: an evolutionary telescope to explore and diagnose the universe of disease mutations. Trends Genet. 2011;27:377–386. doi: 10.1016/j.tig.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Liu Y, Bahar I. Sequence evolution correlates with structural dynamics. Mol Biol Evol. 2012;29:2253–2263. doi: 10.1093/molbev/mss097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughran NB, Hinde S, McCormick-Hill S, Leidal KG, Bloomberg S, Loughran ST, O'Connor B, O'Fágáin C, Nauseef WM, O'Connell MJ. Functional consequence of positive selection revealed through rational mutagenesis of human myeloperoxidase. Mol Biol Evol. 2012;29:2039–2046. doi: 10.1093/molbev/mss073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loytynoja A, Goldman N. An algorithm for progressive multiple alignment of sequences with insertions. Proc Natl Acad Sci U S A. 2005;102:10557–10562. doi: 10.1073/pnas.0409137102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn DJ, Winsor GL, Chan C, Richard N, Laird MR, Barsky A, Gardy JL, Roche FM, Chan TH, Shah N, et al. InnateDB: facilitating systems-level analyses of the mammalian innate immune response. Mol Syst Biol. 2008;4:218. doi: 10.1038/msb.2008.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DP, Lemey P, Lott M, Moulton V, Posada D, Lefeuvre P. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics. 2010;26:2462–2463. doi: 10.1093/bioinformatics/btq467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Reno PL, Pollen AA, Bassan AI, Capellini TD, Guenther C, Indjeian VB, Lim X, Menke DB, Schaar BT, et al. Human-specific loss of regulatory DNA and the evolution of human-specific traits. Nature. 2011;471:216–219. doi: 10.1038/nature09774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestas J, Hughes CC. Of mice and not men: differences between mouse and human immunology. J Immunol. 2004;172:2731–2738. doi: 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- Nei M, Li WH. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci U S A. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshiumi H, Matsumoto M, Funami K, Akazawa T, Seya T. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-beta induction. Nat Immunol. 2003;4:161–167. doi: 10.1038/ni886. [DOI] [PubMed] [Google Scholar]
- Paterson S, Vogwill T, Buckling A, Benmayor R, Spiers AJ, Thomson NR, Quail M, Smith F, Walker D, Libberton B, et al. Antagonistic coevolution accelerates molecular evolution. Nature. 2010;464:275–278. doi: 10.1038/nature08798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinhasi R, Higham TF, Golovanova LV, Doronichev VB. Revised age of late Neanderthal occupation and the end of the Middle Paleolithic in the northern Caucasus. Proc Natl Acad Sci U S A. 2011;108:8611–8616. doi: 10.1073/pnas.1018938108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pumidonming W, Walochnik J, Dauber E, Petry F. Binding to complement factors and activation of the alternative pathway by Acanthamoeba. Immunobiology. 2011;216:225–233. doi: 10.1016/j.imbio.2010.05.002. [DOI] [PubMed] [Google Scholar]
- Ramsay H, Rieseberg LH, Ritland K. The correlation of evolutionary rate with pathway position in plant terpenoid biosynthesis. Mol Biol Evol. 2009;26:1045–1053. doi: 10.1093/molbev/msp021. [DOI] [PubMed] [Google Scholar]
- Ranjith-Kumar CT, Miller W, Sun J, Xiong J, Santos J, Yarbrough I, Lamb RJ, Mills J, Duffy KE, Hoose S, et al. Effects of single nucleotide polymorphisms on Toll-like receptor 3 activity and expression in cultured cells. J Biol Chem. 2007;282:17696–17705. doi: 10.1074/jbc.M700209200. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Sabeti PC, Schaffner SF, Fry B, Lohmueller J, Varilly P, Shamovsky O, Palma A, Mikkelsen TS, Altshuler D, Lander ES, et al. Positive natural selection in the human lineage. Science. 2006;312:1614–1620. doi: 10.1126/science.1124309. [DOI] [PubMed] [Google Scholar]
- Sawyer SL, Wu LI, Emerman M, Malik HS. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc Natl Acad Sci U S A. 2005;102:2832–2837. doi: 10.1073/pnas.0409853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scally A, Durbin R. Revising the human mutation rate: implications for understanding human evolution. Nat Rev Genet. 2012;13:745–753. doi: 10.1038/nrg3295. [DOI] [PubMed] [Google Scholar]
- Schmidt M, Raghavan B, Muller V, Vogl T, Fejer G, Tchaptchet S, Keck S, Kalis C, Nielsen PJ, Galanos C, et al. Crucial role for human Toll-like receptor 4 in the development of contact allergy to nickel. Nat Immunol. 2010;11:814–819. doi: 10.1038/ni.1919. [DOI] [PubMed] [Google Scholar]
- Schneider A, Souvorov A, Sabath N, Landan G, Gonnet GH, Graur D. Estimates of positive Darwinian selection are inflated by errors in sequencing, annotation, and alignment. Genome Biol Evol. 2009;1:114–118. doi: 10.1093/gbe/evp012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schymkowitz JW, Rousseau F, Martins IC, Ferkinghoff-Borg J, Stricher F, Serrano L. Prediction of water and metal binding sites and their affinities by using the Fold-X force field. Proc Natl Acad Sci U S A. 2005;102:10147–10152. doi: 10.1073/pnas.0501980102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, Richards DR, McDonald-Smith GP, Gao H, Hennessy L, et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A. 2013;110(9):3507–3512. doi: 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonson TS, Yang Y, Huff CD, Yun H, Qin G, Witherspoon DJ, Bai Z, Lorenzo FR, Xing J, Jorde LB, et al. Genetic evidence for high-altitude adaptation in Tibet. Science. 2010;329:72–75. doi: 10.1126/science.1189406. [DOI] [PubMed] [Google Scholar]
- Stebbings R, Findlay L, Edwards C, Eastwood D, Bird C, North D, Mistry Y, Dilger P, Liefooghe E, Cludts I, et al. "Cytokine storm" in the phase I trial of monoclonal antibody TGN1412: better understanding the causes to improve preclinical testing of immunotherapeutics. J Immunol. 2007;179:3325–3331. doi: 10.4049/jimmunol.179.5.3325. [DOI] [PubMed] [Google Scholar]
- Stenson PD, Ball EV, Mort M, Phillips AD, Shiel JA, Thomas NS, Abeysinghe S, Krawczak M, Cooper DN. Human Gene Mutation Database (HGMD): 2003 update. Hum Mutat. 2003;21:577–581. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- Stremlau M, Perron M, Welikala S, Sodroski J. Species-specific variation in the B30.2(SPRY) domain of TRIM5alpha determines the potency of human immunodeficiency virus restriction. J Virol. 2005;79:3139–3145. doi: 10.1128/JVI.79.5.3139-3145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Plewniak F, Ripp R, Thierry JC, Poch O. Towards a reliable objective function for multiple sequence alignments. J Mol Biol. 2001;314:937–951. doi: 10.1006/jmbi.2001.5187. [DOI] [PubMed] [Google Scholar]
- UniProt Consortium. Activities at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2014;42:D191–D198. doi: 10.1093/nar/gkt1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Valen L. A new evolutionary law. Evol Theory. 1973;1:1–30. [Google Scholar]
- Wakimoto H, Ikeda K, Abe T, Ichikawa T, Hochberg FH, Ezekowitz RA, Pasternack MS, Chiocca EA. The complement response against an oncolytic virus is species-specific in its activation pathways. Mol Ther. 2002;5:275–282. doi: 10.1006/mthe.2002.0547. [DOI] [PubMed] [Google Scholar]
- Wlasiuk G, Nachman MW. Adaptation and constraint at Toll-like receptors in primates. Mol Biol Evol. 2010;27:2172–2186. doi: 10.1093/molbev/msq104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang Z, dos Reis M. Statistical properties of the branch-site test of positive selection. Mol Biol Evol. 2011;28:1217–1228. doi: 10.1093/molbev/msq303. [DOI] [PubMed] [Google Scholar]
- Yokoyama S. Synthetic biology of phenotypic adaptation in vertebrates: the next frontier. Mol Biol Evol. 2013;30:1495–1499. doi: 10.1093/molbev/mst075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S, Tada T, Zhang H, Britt L. Elucidation of phenotypic adaptations: molecular analyses of dim-light vision proteins in vertebrates. Proc Natl Acad Sci U S A. 2008;105:13480–13485. doi: 10.1073/pnas.0802426105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue P, Li Z, Moult J. Loss of protein structure stability as a major causative factor in monogenic disease. J Mol Biol. 2005;353:459–473. doi: 10.1016/j.jmb.2005.08.020. [DOI] [PubMed] [Google Scholar]
- Zhang SY, Jouanguy E, Ugolini S, Smahi A, Elain G, Romero P, Segal D, Sancho-Shimizu V, Lorenzo L, Puel A, et al. TLR3 deficiency in patients with herpes simplex encephalitis. Science. 2007;317:1522–1527. doi: 10.1126/science.1139522. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.