Abstract
A major challenge of modern Biology is elucidating the functional consequences of natural mutations. Although we have a good understanding of the effects of laboratory-induced mutations on the molecular- and organismal-level phenotypes, the study of natural mutations has lagged behind. In this work, we explore the phenotypic space and the evolutionary history of a previously identified adaptive transposable element insertion. We first combined several tests that capture different signatures of selection to show that there is evidence of positive selection in the regions flanking FBti0019386 insertion. We then explored several phenotypes related to known phenotypic effects of nearby genes, and having plausible connections to fitness variation in nature. We found that flies with FBti0019386 insertion had a shorter developmental time and were more sensitive to stress, which are likely to be the adaptive effect and the cost of selection of this mutation, respectively. Interestingly, these phenotypic effects are not consistent with a role of FBti0019386 in temperate adaptation as has been previously suggested. Indeed, a global analysis of the population frequency of FBti0019386 showed that climatic variables explain well the FBti0019386 frequency patterns only in Australia. Finally, although FBti0019386 insertion could be inducing the formation of heterochromatin by recruiting HP1a (Heterochromatin Protein 1a) protein, the insertion is associated with upregulation of sra in adult females. Overall, our integrative approach allowed us to shed light on the evolutionary history, the relevant fitness effects, and the likely molecular mechanisms of an adaptive mutation and highlights the complexity of natural genetic variants.
Keywords: transposable elements, selective sweep, gene regulation, fitness, adaptation
Introduction
Understanding the functional consequences of naturally occurring mutations remains a largely open question in Biology. Most of our knowledge on the effect of mutations comes from the analyses of laboratory-induced mutations. However, it is not clear whether laboratory mutations are representative of mutations that arise and persist in natural populations (Kolaczkowski et al. 2011; Rose et al. 2011). First, most laboratory mutations studied are loss-of-function mutations that are most likely rare in natural populations and/or their effects are masked by the presence of buffering mechanisms (Landry and Rifkin 2012). Additionally, laboratory-induced mutations tend to be highly pleiotropic and it is difficult to infer which of the phenotypes might be targets of selection in nature (Kolaczkowski et al. 2011).
The recent explosion in the number of studies aimed at identifying natural adaptive mutations in several organisms allows us to study the effect of natural genetic variants at an unprecedented scale (González et al. 2008; Turner et al. 2010; Jones et al. 2012; Huang et al. 2014; Tobler et al. 2014). These studies are revealing that mapping genotype to phenotype is even more complex than previously thought due to the prevalence of gene-by-environment interactions, gene-by-gene interactions, and pleiotropy (Rockman 2012; Lehner 2013; Mackay 2014). First, being able to map a putatively adaptive mutation to its relevant phenotypic effect depends partly on finding the particular environmental conditions in which the mutation is adaptive (Paaby and Schmidt 2008; Storz and Wheat 2010). Thus, taking into account environmental information of the populations where putative adaptive mutations are identified should help mapping the mutation to its relevant phenotype. Second, epistatic interactions also affect the phenotypic outcome of mutations. The phenotypic effect of mutations could be enhanced or suppressed depending on the background being analyzed (Huang et al. 2012). Additionally, several backgrounds should be analyzed to discard the effect of other mutations and reliably attribute the identified phenotypic effect to the candidate mutation (Burnett et al. 2011). Third, many genes are linked to several traits (Paaby and Rockman 2013). In some cases, mutations can have antagonistic effects, that is, beneficial effects in a trait/environment and deleterious effects on a different trait/environment. Pleiotropic mutations can also have beneficial effects on two different traits (McGee et al. 2014). Tradeoffs are prevalent when selection acts on a single trait, whereas payoffs arise when multiple traits are selected for simultaneously (McGee et al. 2014). Thus, if we want to fully characterize the effects of a given natural mutation, several phenotypes need to be studied (Mackay 2010; Guio et al. 2014).
Finally, a comprehensive understanding of adaptation goes beyond identifying fitness consequences of adaptive mutations. Pinpointing the molecular mechanisms underlying adaptation is needed to provide conclusive support for the adaptive role of the mutation (Storz and Wheat 2010). Additionally, elucidating the evolutionary history of adaptive variation for fitness traits allows to start answering long-standing questions on the genetic basis of adaptation (Orr 2005).
In this work, we focused on characterizing the functional effects, the molecular mechanism, and the evolutionary history of a natural transposable element (TE)-induced mutation in Drosophila melanogaster: FBti0019386 belonging to the invader4 retrotransposon family (González et al. 2008, 2010; St Pierre et al. 2014). FBti0019386 has been identified as a candidate adaptive TE insertion based on its population dynamics (González et al. 2008). González et al. (2010) further reported that FBti0019386 shows parallel clinal frequency patterns in North America and Australia suggesting that it is involved in adaptation to temperate environments. FBti0019386 is inserted in the 5′-untranslated region (UTR) intron of sarah (sra) and 2.5 kb upstream of Bicoid-interacting protein 1 (Bin1) in the 3 R chromosomal arm (St Pierre et al. 2014). sra laboratory mutants affect several biological processes, such as egg activation, female meiosis, and long-term memory among others (Ejima et al. 2001, 2004; Chang et al. 2003; Horner et al. 2006; Takeo et al. 2006; Sakai and Aigaki 2010; Nakai et al. 2011). In most cases, these phenotypes are the result of the deregulation of calcineurin, which is inhibited by sra (Takeo et al. 2006; Sakai and Aigaki 2010; Nakai et al. 2011). Laboratory-induced mutations affecting Bin1, a highly conserved transcriptional corepressor, play a role during environmental stress response in Arabidopsis (Song and Galbraith 2006) and in Drosophila (Costa et al. 2011). Thus, to identify the phenotypic consequences of FBti0019386 mutation, we explored several candidate phenotypes previously associated with sra and Bin1 mutants in different developmental stages, in different environmental conditions, and in flies with different genetic backgrounds.
Our results showed that FBti0019386 increased in frequency in out-of-Africa populations due to positive selection and is associated with shorter developmental time (DT) and increased sensitivity to cold-stress. These two phenotypic effects together with the lack of correlation between FBti0019386 frequency and latitude in European populations raised doubts about the role of FBti0019386 in temperate adaptation. Finally, although FBti0019386 insertion could be inducing pi-RNA mediated heterochromatin assembly, the insertion is associated with upregulation of sra in adult females.
Results
FBti0019386 Flanking Regions Show Signatures of Positive Selection
We tested whether the region flanking FBti0019386 showed signatures of positive selection (see Materials and Methods for a description of the different tests used). We found an extreme decrease of nucleotide diversity (π) in strains with FBti0019386 insertion compared with strains without the insertion, which was accompanied by a decrease in Tajima’s D statistic (table 1, supplementary fig. S1A and B and table S1, Supplementary Material online) (Hudson et al. 1992; Tajima 1989). The Composite Likelihood (CL) test, specifically designed to detect selective sweeps (Nielsen et al. 2005), was higher in flies with FBti0019386 insertion compared with flies without the insertion, as expected if flies with the insertion show signatures of a selective sweep in the analyzed region (table 1). We confirmed that values of π, Tajima’s D, and CL were statistically different from neutral simulated scenarios in flies with FBti0019386 insertion but not in flies without the insertion (table 1 and supplementary table S2, Supplementary Material online).
Table 1.
Summary of the Analyses Showing Evidence of Positive Selection in the 1-kb Region around FBti0019386 Insertion.
| Observed |
Neutral Simulations |
Resampling of Strains |
||||||
|---|---|---|---|---|---|---|---|---|
| Mean (CI 95%) |
P value |
Mean (CI 95%) | P Value | |||||
| P | A | P | A | P | A | |||
| π | 0.43 | 4.51 | 3.92 (1.32, 7.81) | 4.20 (1.33, 8.04) | 0.001 | >0.05 | 3.35 (2.78, 3.87) | <0.001 |
| Tajima’s D | −1.77 | 0.68 | −0.11 (−1.46, 1.62) | −0.04 (−1.41, 1.64) | 0.007 | >0.05 | 0.4 (−0.19, 1.02) | <0.001 |
| CL (log) | −5.95 | −18.15 | −18.69 (−29.67, −8.80) | −15.20 (−25.89, −6.82) | 0.006 | >0.05 | −12.18 (−15.23, −8.81) | <0.001 |
Note.—Neutral simulations were performed with MS program using the parameter theta = 4. For simulations with theta = 5, please see supplementary table S2, Supplementary Material online. P, data set of strains with FBti0019386 insertion; A, data set of strains without FBti0019386 insertion.
To test whether the observed differences were due to the FBti0019386 insertion, we estimated the three statistics in random samples of the strains (see Materials and Methods). None of the randomized data sets had lower π, lower Tajima’s D, or higher CL value compared with the data set of strains with FBti0019386 insertion (table 1 and supplementary table S3, Supplementary Material online). Finally, we performed the Composite Likelihood Ratio (CLR; Nielsen et al. 2005) test comparing strains with and without the FBti0019386 insertion, and we found that it was significant: CLR = 24.40 P value = 7.82 × 10−7. Moreover, this CLR value is three times bigger than any of the CLR values calculated in a random sample of 1,000 1-kb-long regions from 3 R chromosome, where FBti0019386 is located (supplementary table S4, Supplementary Material online). Note that estimates of π and Tajima’s D in these 1,000 regions also showed that these two statistics did not significantly differ between strains with and without FBti0019386 insertion (supplementary fig. S1C and D, Supplementary Material online).
Note that we checked whether polymorphisms other than TE were present in the flanking regions analyzed. No other polymorphisms were found that could potentially confound the results of our tests of selection suggesting that the TE is the causative mutation.
Overall, we found evidence of positive selection in the region flanking FBti0019386 insertion suggesting that FBti0019386 is an adaptive insertion.
Exploring the Fitness Space of FBti0019386
To explore the phenotypic space of FBti0019386 insertion, we investigated several traits related to the phenotypic effects of nearby genes: Fecundity and egg hatchability associated with sra mutant alleles. Related to egg hatchability, we also investigated egg hatching time, egg-to-adult viability, and DT. Additionally, we investigated cold stress, osmotic stress, and starvation stress as Bin1 mutants have been shown to play a role in stress resistance.
Because FBti0019386 is located 242.4 kb away from the distal breakpoint of In(3 R)Payne inversion and inversions are known to be under selection, we checked whether this inversion was present in any of the six strains used to perform the different phenotypic analyses (see Materials and Methods). We found that none of the strains used in our analyses carries In(3 R)Payne inversion.
We also checked whether polymorphisms other than the FBti0019386 insertion were present in the genomic region including sra and Bin1 genes. We did not find any polymorphism linked to the FBti0019386 that could potentially confound the results of the phenotypic assays performed.
FBti0019386 Insertion Does Not Affect Fecundity or Egg Hatching
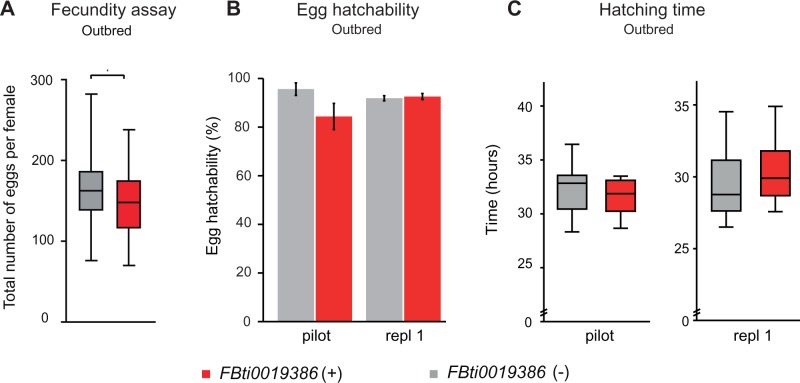
Laboratory mutant flies in which sra is underexpressed lay less eggs than wild-type flies and most of the eggs do not hatch (Horner et al. 2006). To check whether FBti0019386 insertion has an effect on fecundity, we compared the number of eggs laid per female in outbred populations with and without the insertion (see Materials and Methods). Our results showed that, on average, flies without the insertion laid slightly more eggs than flies with the insertion (t-test, P = 0.047) (fig. 1A). However, the size effect of the mutation was not significant (table 2). We also tested whether differences in fecundity were present early in life, as has been reported by Paaby et al. (2014). Although the mean number of eggs laid by flies with the insertion in the first 48 h of egg laying was bigger than the number laid by flies without the insertion (3.95 vs. 2.33 eggs), these differences were not statistically significant (t-test, P = 0.06).
Fig. 1.
FBti0019386 does not affect fecundity (A), egg hatchability (B), or hatching time (C) in outbred populations. (A) Average number of eggs laid by outbred females without FBti0019386 insertion (FBti0019386 (−)) and with FBti0019386 insertion (FBti0019386 (+)). (B) Percentage of hatched embryos. (C) Average hatching time. In all cases, error bars represent standard error of the mean (SEM).
Table 2.
Odds Ratios (OR) and Confidence Intervals (CI) for Phenotypic Experiments Performed with Embryos with and without FBti0019386.
| Experiment | Strain | OR (CI) |
|---|---|---|
| Fecundity | Outbred | 1.05 (0.67–1.64) |
| Hatching time in cold | Outbred pilot | 7.07 (3.37–14.83) |
| Outbred replica 1 | 2.21 (1.49–3.26) | |
| DT | Outbred pilot | 5.69 (2.72–11.94) |
| Outbred replica 1 | 2.62 (1.88–3.66) | |
| Outbred replica 2 | 2.60 (1.94–5.88) | |
| Individual DGRP | 1.95 (1.30–2.92) |
We then checked whether outbred flies with and without FBti0019386 differed in egg hatchability and/or hatching time. We first performed a pilot experiment using 150 embryos per strain and we found that flies with the insertion did not show significant differences compared with flies without the insertion in egg hatchability (t-test, P > 0.05) (fig. 1B) or hatching time (t-test, P > 0.05) (fig. 1C). Although differences were not significant, flies with the insertion showed a lower number of hatched eggs (fig. 1B) and a shorter hatching time (fig. 1C). We thus repeated the experiments using 500 embryos per strain and we found that flies with and without FBti0019386 did not differ in egg hatchability (t-test, P > 0.05) (fig. 1B) or hatching time (t-test, P > 0.05) (fig. 1C).
Overall, we did not find significant differences in fecundity, egg hatchability, or egg hatching time in flies with and without FBti0019386 insertion. These results suggest that FBti0019386 does not have a significant effect on these phenotypes.
FBti0019386 Insertion Does Not Affect Egg Hatching or Egg-To-Adult Viability under Cold Stress Conditions
As mentioned above, Bin1 plays a role in general environmental stress response in Drosophila (Costa et al. 2011). We thus screened several phenotypes in embryos under cold stress conditions: Egg hatching, egg hatching time, and egg-to-adult viability.
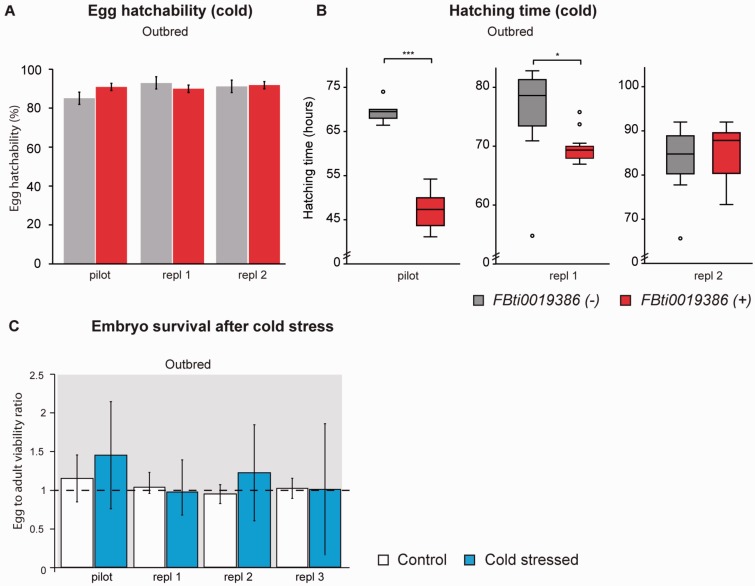
We performed egg hatchability and egg-hatching time assays in outbred populations under repeated cold stress exposure (see Materials and Methods). We did not detect differences in egg hatchability between flies with and without the insertion in any of the three replicas performed (t-test, P > 0.05) (fig. 2A). However, flies with FBti0019386 insertion from the pilot experiment and the first replica hatched significantly before flies without the element (t-test, P << 0.001 and P = 0.011, respectively) (table 2) whereas no differences were observed in the second replica (t-test, P > 0.05) (fig. 2B).
Fig. 2.
FBti0019386 does not affect embryo hatching or survival in cold stress conditions in outbred populations. (A) Percentage of embryos that hatched during cold-stress periods (see Materials and Methods). (B) Average egg hatching time. (C) Egg-to-adult survival after a single cold stress period during embryonic stage (cold stressed) and under control conditions (control). Bars represent the survival ratio between flies with FBti0019386 and flies without FBti0019386 and error bars represent SEM.
We further tested whether flies with and without FBti0019386 differed in the egg-to-adult viability after exposing outbred flies to a single cold-stress period during early embryo stages. Our results showed that there are no differences in survival between flies with and without the insertion in control conditions or under cold-stress (two-way ANOVA [analysis of variance], P > 0.05, fig. 2C).
Overall, and although variability in hatching time was observed in some of the experiments performed, our results suggest that FBti0019386 insertion does not affect cold-tolerance during the embryo stage.
FBti0019386 Is Associated with Increased Sensitivity to Cold Stress in Adults
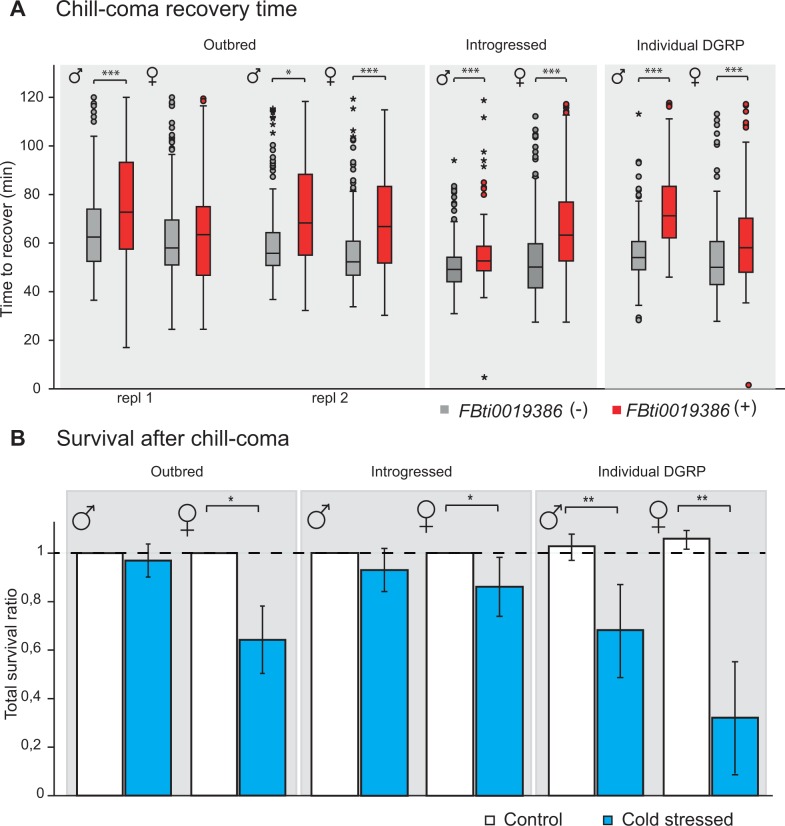
Because we could not find any significant difference between strains with and without FBti0019386 in embryonic stage, we decided to test whether differences between the two strains were present in adult flies. We first tested whether adult flies with and without FBti0019386 insertion differed in chill-coma recovery time (CCRT) and survival after cold stress. CCRT is used as a reliable measure of cold tolerance in Drosophila (Gibert et al. 2001; Macdonald et al. 2004). We observed that flies with the insertion showed significantly longer recovery time compared with flies without the insertion suggesting that they were more sensitive to cold stress (Mann–Whitney test, P << 0.001) (fig. 3A and table 3). We replicated this result in flies with the same genetic background (Mann–Whitney test, P < 0.05) and in flies with two other genetic backgrounds: The introgressed strains generated in our laboratory (Mann–Whitney test, P << 0.001) and a couple of inbred strains from the DGRP (Drosophila Genetic Reference Panel) project (Mann–Whitney test, P << 0.001) (fig. 3A and table 3) (see Materials and Methods).
Fig. 3.
Flies with FBti0019386 insertion are more sensitive to cold stress. (A) Average time to recover after chill coma in adult flies from outbred populations, introgressed strains, and inbred DGRP strains (RAL-857 and RAL-802). (B) Survival ratio between flies with FBti0019386 insertion and flies without the insertion after chill coma exposure (cold stress) and in control conditions (control) in the three genetic backgrounds. Error bars represent SEM.
Table 3.
Odds Ratios (OR) and Confidence Intervals (CI) for Phenotypic Experiments Performed with Male and Female Adult Flies with and without FBti0019386.
| Experiment | Strain | Males OR (CI) | Females OR (CI) |
|---|---|---|---|
| CCRT | Outbred replica 1 | 3.44 (2.31–5.18) | N/Aa |
| Outbred replica 2 | 3.79 (2.54–5.67) | 5.18 (3.43–7.82) | |
| Introgressed | 2.44 (1.64–3.62) | 4.16 (2.69–6.41) | |
| Individual DGRP | 11.63 (6.79–19.93) | 2.26 (1.54–3.33) | |
| Survival after chill-coma | Outbred | N/A | 7.80 (3.27–18.60) |
| Introgressed | N/A | 1.89 (0.99–3.62) | |
| Individual DGRP | 9.94 (5.49–18) | 6.88 (3.43–13.82) | |
| Osmotic stress | Outbred | N/A | 1.61 (1.21–2.13) |
| Starvation stress | Outbred | 1.52 (1.15–2.01) | N/A |
aN/A (OR was estimated when differences between flies with and without FBti0019386 were statistically significant).
In accordance with this increased cold sensitivity, flies with the insertion also showed an increased mortality following chill-coma exposure, although these differences were not always significant (fig. 3B and table 3).
Finally, we also tested whether flies with FBti0019386 insertion were more sensitive to osmotic stress and starvation stress. We found that outbred females with the insertion were more sensitive to high salt concentrations (Kaplan–Meyer, log rank P < 0.001) (supplementary fig. S2A, Supplementary Material online, and table 3), and outbred males with the insertion were more sensitive to starvation stress (Kaplan–Meyer, log rank P < 0.001) (supplementary fig. S2B, Supplementary Material online, and table 3).
Overall, longer CCRT and lower cold-stress survival in flies with FBti0019386 insertion across backgrounds suggested that this mutation is negatively affecting adult cold-stress response. This high sensitivity to cold stress likely represents the cost of selection of this TE mutation. Furthermore, preliminary results are suggestive but not conclusive of a negative role of FBti0019386 in general response to stress.
FBti0019386 Insertion Is Associated with Shorter DT
During the course of the experiments, we noticed that flies with FBti0019386 showed a shorter DT than flies without the insertion. Because DT is relevant to fitness in all organisms, and especially for those such as D. melanogaster that occupy ephemeral habitats (Chippindale et al. 1997), we tested this observation. We found that outbred flies (Mann–Whitney test, pilot experiment P = 0.006 and replica 1 and 2 P < 0.001) and inbred DGRP flies (t-test, P = 0.02) with the insertion developed faster compared with flies without the TE insertion (fig. 4 and table 2). On average, flies with FBti0019386 insertion developed 9.4–17.9 h before compared with flies without the insertion. However, we could not detect significant DT differences in the introgressed strains differing by the presence/absence of FBti0019386 (t-test, P > 0.05) (fig. 4), suggesting that polymorphisms other than the TE influence DT in this background. Note that the effect size of the mutation on the other phenotypes studied also varies depending on the background being analyzed (tables 2 and 3). This suggests that polymorphisms other than FBti0019386 play a role not only in DT but also in other phenotypes.
Fig. 4.
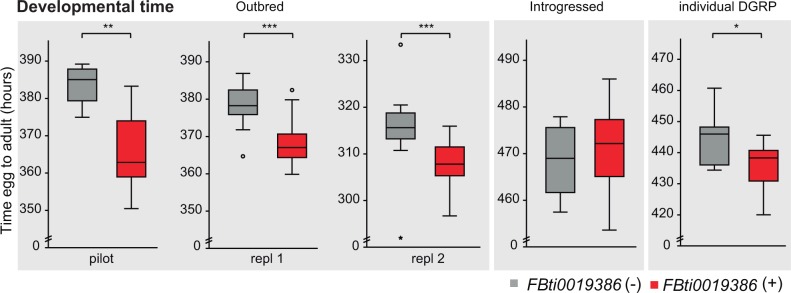
FBti0019386 is associated with shorter DT. Average egg-to-adult DT in populations without FBti0019386 insertion and with the insertion. Error bars represent SEM.
FBti0019386 Frequency Showed Clinal Patterns in North America and Australia but No Correlation between Frequency and Latitude Is Found in Europe
Shorter DT and increased sensitivity to cold stress are not consistent with a role of FBti0019386 in temperate adaptation (González et al. 2010). However, previous evidence for a role in temperate adaptation was based on the analysis of only two North American and five Australian populations (González et al. 2010). To further test these results, we estimated FBti0019386 frequencies in additional populations from North America, Australia, Europe, and Africa (supplementary table S5, Supplementary Material online) using T-lex2 pipeline (Fiston-Lavier et al. 2014). We found that FBti0019386 insertion is present at 10% frequency in a Rwanda population confirming its low frequency in Africa (supplementary table S5, Supplementary Material online). We confirmed that the TE is present at intermediate to high frequencies in 15 additional out-of-Africa populations (fig. 5 and supplementary table S5, Supplementary Material online). We also confirmed that the TE frequency varies clinally with latitude in North America and Australia (Pearson correlation P = 0.011 and P = 0.002, respectively; supplementary table S6, Supplementary Material online). However, when we analyzed the FBti0019386 frequency in six European populations we did not find any significant correlation between frequency and latitude (Pearson correlation P = 0.313; supplementary table S6, Supplementary Material online).
Fig. 5.
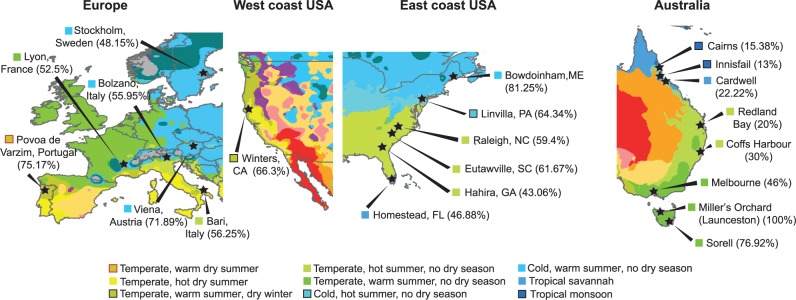
Climate map with Drosophila melanogaster population samples analyzed with T-lex2. The frequency of FBti0019386 in each population is shown in brackets. Climate maps are modified from Peel et al. (2007).
Besides latitude, we also tested whether other geographical and climatic variables showed significant correlations with FBti0019386 frequency. We found significant correlations between frequency and temperature-related variables in North America and between frequency and both temperature-related and precipitation-related variables in Australia (supplementary table S6, Supplementary Material online). No significant correlation was found in Europe (supplementary table S6, Supplementary Material online). Because most of the climatic variables are significantly correlated among them and with latitude (supplementary table S7, Supplementary Material online), we performed a Principal Component Analysis (PCA) to disentangle the relationships between the variables. In North America, climate variables were grouped in two components, in Australia in three and in Europe in two (supplementary table S8, Supplementary Material online). As expected based on the correlation analyses, only in North America and in Australia, some of the climatic variables grouped with latitude and frequency (supplementary fig. S3A, Supplementary Material online). In North America, the first component accounted for 46% of climatic variation (supplementary table S9, Supplementary Material online) and explained 54% of the variation in FBti0019386 frequency (supplementary fig. S3B, Supplementary Material online). In Australia, the first component accounted for 68% of climatic variation (supplementary table S9, Supplementary Material online) and explained 86% of the frequency variation (supplementary fig. S3B, Supplementary Material online). Finally in Europe, the first principal component explained 54% of the climatic variation (supplementary table S9, Supplementary Material online) but was not significantly correlated with FBti0019386 frequency (supplementary fig. S3B, Supplementary Material online).
Overall, although we were able to confirm the clinal pattern of FBti0019386 in North America and Australia, our results did not provide evidence for the presence of a clinal pattern in Europe. In Australia, the clinal pattern is well explained by the observed climatic variation, whereas in North America climatic variation did not fully explain the observed correlation between FBti0019386 frequency and latitude, suggesting that other factors might be involved in the observed clinal pattern. As expected, none of the climatic variables significantly correlated with TE frequency in Europe.
FBti0019386 Is Associated with Upregulation of sra in Female Flies
To shed light on the molecular mechanism of FBti0019386 insertion, we measured the expression of sra and Bin1 in nonstress conditions in embryos and in nonstress and cold-stress conditions in female flies with and without FBti0019386 insertion.
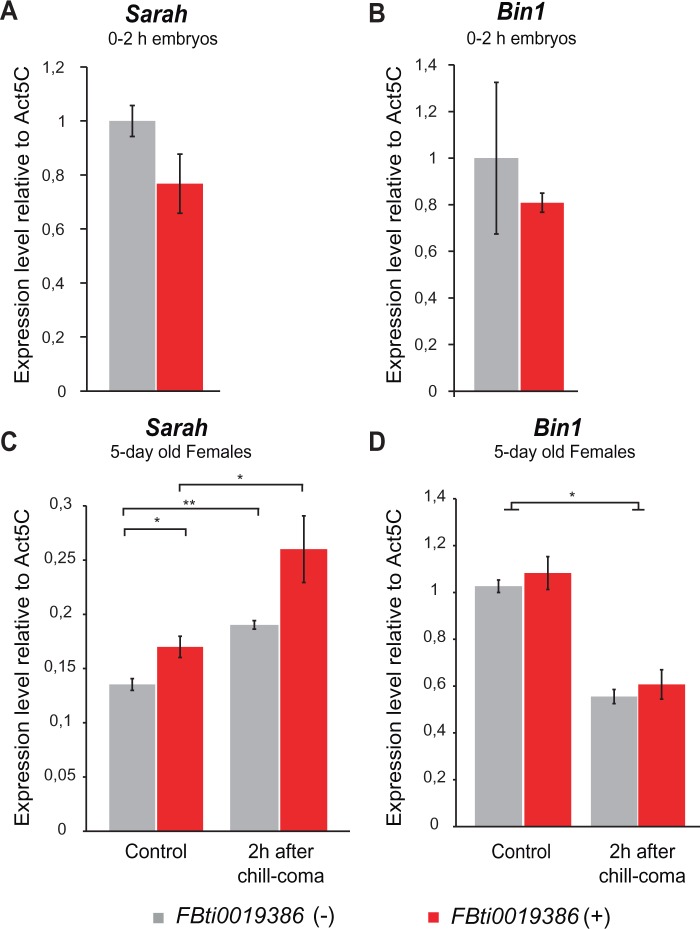
We did not observe significant differences in sra or Bin1 expression in embryos differing by the presence/absence of FBti0019386 insertion (t-test, P > 0.05) (fig. 6A and B). However, we observed that adult female flies with FBti0019386 insertion showed an increase of sra expression compared with flies without the insertion both in control conditions and after cold-stress conditions, although results were only significant under control conditions (t-test, P = 0.03) (fig. 6C). On the other hand, no significant differences in expression level between flies with and without FBti0019386 were observed for Bin1 (t-test, P > 0.05) (fig. 6D).
Fig. 6.
Flies with FBit0019386 insertion showed sra upregulation. Real-time polymerase chain reaction quantification of sra and Bin1 transcript levels in outbred flies without FBti0019386 insertion and with FBti0019836 insertion. We represented the average expression level of sra (A and C) and Bin1 (B and D) relative to Act5C with SEM error bars for three biological replicates in 0–2 h embryos and in 5-day-old females. Normalized expression measured 2 h after chill-coma for sra and Bin1 is depicted in (C) and (D), respectively.
Interestingly, we observed a change in sra and Bin1 expression after cold stress in flies with and without FBti0019386 insertion: sra is upregulated in cold stress conditions (t-test, P < 0.05 in both cases) (fig. 6C) whereas Bin1 is downregulated (t-test, P < 0.05 in both cases) (fig. 6D).
Overall, we did not observe any change in expression of sra and Bin1 in embryos, in agreement with the lack of phenotypic consequences of FBti0019386 in this developmental stage. However, we observed a upregulation of sra in flies with FBti0019386 insertion that was significant under nonstress conditions. Moreover, we showed that both sra and Bin1 changed their expression in response to cold stress.
FBti0019386 Could Be Affecting gene Expression by Ectopically Assembling Heterochromatin
TEs from the invader4 family contain sites with homology to PIWI interacting RNAs (piRNAs) that act as cis-acting targets for heterochromatin assembly by recruiting Heterochromatin Protein 1 a (HP1a) (Sentmanat and Elgin 2012). Specifically, these piRNA binding sites are located in the long terminal repeat (LTR) sequences. Because FBti0019386 is a 347-bp solo-LTR, we hypothesized that it could be inducing the ectopic assembly of heterochromatin. We analyzed the 14.6-kb region containing Bin1, sra, and FBti0019386 and found that both sense and antisense piRNAs bind specifically to FBti0019386 (fig. 7A) (see Materials and Methods). Second, we tested whether there is evidence for the presence of HP1a binding to FBti0019386 sequence. We found that HP1a specifically binds to FBti0019386 sequence (fig. 7B) (see Materials and Methods). Thus, these results suggest that FBti0019386 could be affecting gene expression by inducing the ectopic assembly of heterochromatin.
Fig. 7.
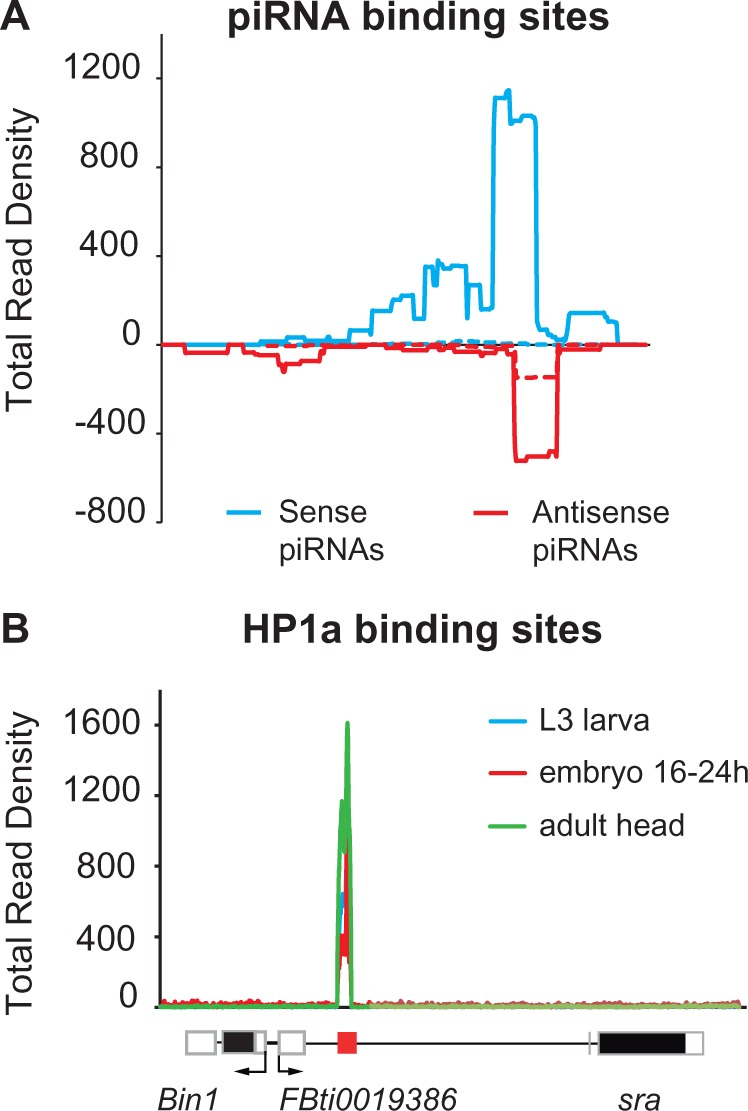
FBti0019386 could bind piRNA and HP1a protein. (A) Mapping of piRNA sense and antisense RNA-seq reads against FBti0019386 sequence. Data from Li et al. (2009) are depicted in dashed lines and data from Satyaki et al. (2014) are represented in continuous lines. (B) Mapping of reads coming from HP1a ChIP-Seq experimental data against the genome region containing Bin1, FBti0019386, and sra. Experimental data from L3 larva, 16–24 h embryo, and adult heads are given.
Discussion
In this work, we explored the plausible phenotypic space of the putatively adaptive FBti0019386 insertion in different developmental stages, embryo and adult, and in different environmental conditions, nonstress conditions and cold, osmotic, and starvation stress conditions. Overall, we found that FBti0019386 mediates sensitivity to cold stress conditions (fig. 3) and is associated with faster DT (fig. 4). These two phenotypic effects have plausible fitness consequences in nature that could explain why the mutation increased in frequency in natural populations but has not reached fixation. Increased sensitivity to cold stress conditions is likely to reduce fitness of the flies that carry FBti0019386 insertion, and may represent the cost of selection of this mutation. On the other hand, faster DT is likely to increase the fitness of flies with FBti0019386 insertion. In nature, quick development favors D. melanogaster individuals for several reasons. First, larvae feed on rotting fruits that are ephemeral. Thus, quick development allows larvae to pupate before the food source is exhausted. Second, competition increases as more and more eggs are laid on a piece of fruit, also favoring individuals with faster DT (Nunney 1990). Third, breeding sites in nature can be destroyed by physical factors and predation, individuals that develop faster are thus more likely to escape microhabitat destruction. And fourth, faster DT accelerates the age of first breeding, which is relevant for the organism if most reproduction happens in expanding populations. This is the case for D. melanogaster populations that expand their population size every spring. Thus, it is plausible that FBti0019386 increased in frequency in natural populations because of its positive effect on DT whereas it did not reach fixation because of its negative effect on cold-stress resistance. Our results emphasize the importance of exploring different phenotypes to fully characterize the effects of natural mutations, as have been suggested before (Mackay 2010; Guio et al. 2014). Although our results provide a plausible explanation for the effect of FBti0019386 insertion in natural populations, experiments under natural conditions are needed to unequivocally identify the effect of this insertion in nature.
By combining several tests that capture different signatures of selection at the DNA level, we demonstrate that FBti0019386 shows signatures of positive selection suggesting that it is an adaptive mutation (table 1). However, our results are not entirely consistent with a role of FBti0019386 in temperate adaptation as has been previously proposed (González et al. 2010). First, adaptation to temperate climates has been associated with increased stress resistance, increased DT, and decreased fecundity (Stanley and Parsons 1981; Hoffmann et al. 2003; Schmidt et al. 2005; Folguera et al. 2008; Schmidt and Paaby 2008; but see also James and Partridge 1995; James et al. 1997; Trotta et al. 2006). However, we found that FBti0019386 is associated with increased sensitivity to cold stress (fig. 3), with shorter DT (fig. 4) and does not significantly affect fecundity (fig. 1). Thus, the phenotypic effects of FBti0019386 are not consistent with a role of this insertion in temperate adaptation. Second, our global analyses of FBti0019386 population frequency showed that FBti0019386 frequency correlates with latitude and with climatic variables in North America and in Australia but not in Europe (fig. 5 and supplementary table S6, Supplementary Material online). We suggest that the clinal frequency patterns in North America and in Australia could be due to the dual colonization of these two continents by European and African populations rather than to the operation of spatially varying selection (Caracristi and Schlotterer 2003; Rouault et al. 2004; Duchen et al. 2013; Bergland et al. 2014). The lack of clinal frequency patterns in Europe would support this conclusion. However, it is also possible that phenotypic effects of FBti0019386 not yet characterized could be consistent with a role of this natural mutation in temperate adaptation. Additionally, although there is evidence for the presence of clinal variation in European populations (David et al. 1985, 1986, 1989; Costa et al. 1992), other works have shown that clines are weaker in Europe compared with other continents (Oakeshott, Chambers, et al. 1983; Oakeshott, Gibson, et al. 1983). This could be partly due to differences in the latitudinal ranges spanned by populations analyzed in the different continents. In this work, the latitudinal range spanned by North American (25.82°–45.06°) and Australian (−16.88° to −42.83°) populations is larger than the range spanned by European populations (41.13°–59.33°). In any case, genome-wide scan studies that identify loci that are differentiated between populations should be taken as a first step toward the identification of loci that are subject to spatially varying selection (González et al. 2010; Kolaczkowski et al. 2011; Fabian et al. 2012; Reinhardt et al. 2014). Further functional validation should be gathered before concluding that the candidate loci are under spatially varying selection (Bergland et al. 2014).
Our results also shed light on the molecular processes that lead from genotype to phenotype. We found that FBti0019386 is associated with upregulation of sra (fig. 6C). As previously described for other elements from the invader4 family, we showed that FBti0019386 has piRNA binding sites (fig. 7A) (Sentmanat and Elgin 2012). We also showed that HP1a binds specifically to the FBti0019386 sequence, further suggesting that FBti0019386 could be inducing the ectopic assembly of heterochromatin (fig. 7B). These results highlight the potential role of TE remnants as silencing signals to be used by piRNAs to direct heterochromatin formation (Sentmanat et al. 2013). Although we observed an upregulation of sra in adult females, we can not discard that heterochromatin assembly induced by FBti0019386 could be affecting gene expression in other developmental stages and/or specific tissues.
A recent update of FlyBase, the database of Drosophila genes and genomes, annotated two new Bin1 transcripts that have their transcription start site inside FBti0019386 (St Pierre et al. 2014). As a consequence, these two new transcripts would only be produced in strains with the insertion, and could contribute to differences in the level of Bin1 expression in flies with and without the insertion. Although we did not find differences in Bin1 expression, we cannot discard that differences in the level of expression of Bin1 are present in developmental stages, tissues, or environmental conditions that we have not investigated.
Although sra and Bin1 have not been associated with DT, both genes play important roles during development and have been associated with a wide range of processes (Chang et al. 2003; Ejima et al. 2004; Horner et al. 2006; Takeo et al. 2006, 2010; Chang and Min 2009; Matyash et al. 2009; Costa et al. 2011; Nakai et al. 2011). A genome-wide screening looking for genes influencing DT in D. melanogaster has shown that the many candidate genes were involved in a wide range of biological processes such as cellular metabolic processes, organismal development, and response to stress (Mensch et al. 2008). More recently, developmental timing in insects has been associated with hormonal and circadian control (Di Cara and King-Jones 2013; Yadav et al. 2014). Interestingly, sra is regulated by Shaggy/GSK-3β (sgg), a Ser–Thr kinase involved in the regulation of circadian rhythmicity (Martinek et al. 2001). On the other hand, both Bin1 and sra are stress-response genes: Bin1 is upregulated in response to stress and sra is downregulated (fig. 6). Bin1 is a known key player in transcriptional response to environmental stress (Costa et al. 2011). Although there was no previous evidence for a direct role of sra in response to stress, sra could be affecting stress response through its role in the calcium pathway (Takeuchi et al. 2009; Teets et al. 2013; Davies et al. 2014). sra inhibits calcineurin, a highly conserved protein in eukaryotes that has the ability to sense calcium (Hogan et al. 2003). Although it is not deeply understood, calcium pathways play a role during general cell-stress response including cold stress response (Takeuchi et al. 2009; Teets et al. 2013; Davies et al. 2014). Note that many genes that affect complex traits in Drosophila had well-characterized roles in early development and were not previously annotated to affect adult quantitative traits (Mackay 2010).
FBti0019386 adds to the growing list of TE-induced adaptive mutations that have been linked to their fitness effects and their underlying molecular mechanisms (Schmidt et al. 2010; Magwire et al. 2011; Guio et al. 2014; Mateo et al. 2014; Sun et al. 2014). Overall, these examples highlight the variety of mechanisms underlying adaptive mutations and point toward a significant role of TEs in response to stress (Casacuberta and González 2013). However, the number of characterized mutations is still too small to obtain an overall picture of adaptation. In depth, characterization of a representative set of adaptive mutations in natural populations will allow us to start answering long-standing questions in the field such as which traits are more relevant for adaptation? What is the effect-size distribution of adaptive mutations? and What evolutionary processes underlie adaptive evolution?
Materials and Methods
Sequence Analysis of the FBti0019386 Flanking Regions
Single nucleotide polymorphism (SNP) data were downloaded from the DGRP2 webpage (https://www.hgsc.bcm.edu/arthropods/drosophila-genetic-reference-panel) in vcf format. Strains with (N = 65) and without (N = 38) FBti0019386 insertion were filtered using vcftools v_0.1.10 (http://vcftools.sourceforge.net/).
We used three different statistics to detect positive selection: Nucleotide diversity (π), Tajima’s D, and the CL of SNPs. Positive selection results in the elimination of standing genetic variation that is linked to the adaptive mutation. Thus, if FBti0019386 has increased in frequency due to positive selection, we expect a decrease in π in flies with the insertion compared with flies without the insertion. π is calculated as the mean number of pairwise differences between two given sequences (Hudson et al. 1992). Tajima’s D statistic is calculated as the ratio between the mean number of pairwise differences and the number of segregating sites (Tajima 1989). This ratio is expected to be 0 in a neutrally evolving population whereas negative values of Tajima’s D can be taken as evidence of positive selection (Tajima 1989). Finally, CL test is calculated by multiplying the marginal likelihoods for each site along the studied sequences (Nielsen et al. 2005).
π, Tajima’s D, and CL were calculated for the two sets of sequences, with and without the insertion, using the PopGenome package in R (Pfeifer et al. 2014). Sliding windows analyses were performed for 200-bp-size windows spanning 1 and 2-kb regions flanking the insertion. Differences between strains with and without the insertion were more drastic for the 1-kb region flanking the insertion; therefore, we focused our analysis in this region.
Simulations were performed using the MS program (Hudson 2002). Theta values were estimated using the 205 DGRP2 strains for the 2-kb region around FBti0019386 (theta = 4.77/kb) and for the 3 R chromosomal arm (theta = 4.5/kb). Thus, simulations were performed for theta values of 4/kb and 5/kb, which are frequently used as neutral values in D. melanogaster.
Ad hoc perl scripts were used for the resampling analyses. In total, 1,000 random samples of the 103 DGRP strains analyzed were obtained keeping the same proportion as in the original present and absent data sets (60%/40%, respectively) and a sample size of nearly 50% of the total data set.
We also computed CLR as 2*(log CL (present) − log CL (absent)), for a 1-kb region around the TE insertion. Because demography could produce similar patterns as positive selection, we performed a random sampling of 1,000 1-kb-long regions from the 3 R chromosome for the absent and present data sets and calculated π, Tajima’s D, CL, and CLR tests in each one of them.
Fly Strains
Outbred Strains
We selected six inbred strains from the Drosophila Genetic Reference Panel (Mackay et al. 2012; Huang et al. 2014) homozygous for the presence of FBti0019386 insertion (RAL-21, RAL-40, RAL-177, RAL-402, RAL-405, and RAL-857). We placed ten virgin females and ten males of each strain in a fly chamber to create an outbred population sharing the TE insertion. We also selected six inbred strains without the insertion (RAL-75, RAL-138, RAL-383, RAL-461, RAL-822, and RAL-908) and created an outbred strain following the same procedure explained above. Each outbred population was maintained by random mating (N ≈ 800 flies per generation) for at least ten generations before starting the experiments.
Introgressed Strains
We selected two DGRP strains: One homozygous for the presence of FBti0019386 insertion (RAL-177) and one homozygous for the absence (RAL-802). We crossed RAL-177 virgin females with RAL-802 males and backcrossed the virgin females that carry FBti0019386 insertion from the following generations with RAL-802 males for 12 generations. After that, we did brother–sister crosses until we obtained homozygous strains for the absence and homozygous strains for the presence of FBti0019386.
Individual DGRP Strains
We used a couple of individual DGRP strains differing by the presence/absence of FBti0019386 insertion to perform our phenotypic assays. We used RAL-857 (homozygous for the presence of FBti0019386 insertion) and RAL-802 (homozygous for the absence).
Presence/Absence of In(3 R)Payne in the Analyzed Strains
To discard the effect of In(3 R)Payne inversion on FBti0019386 phenotypic effects, we genotyped the strains analyzed to detect the presence/absence of this inversion: The two outbred, the two introgressed, and the two individual DGRP strains. We used the primer sequences described in Matzkin et al. (2005). As a positive control, we used a strain that was previously genotyped in our laboratory and that carries the In(3 R)Payne inversion.
Phenotypic Assays
All experiments were performed using outbred populations. Additionally, we used introgressed and individual DGRP strains to perform CCRT assay, survival after chill-coma, and DT assays.
Fecundity
In total, 40 virgin females from each strain were placed individually in vials with one male from the same strain. During 17 days flies were moved to new vials every 2 days and the number of eggs laid per female during that period was counted. Total fecundity, that is, average of the total number of eggs laid per female during the 17 days, and early fecundity, that is, average of the total number of eggs laid per female during the first 48 h of egg laying, was compared between flies with and without FBti0019386.
Egg Hatchability and Hatching Time
In total, 800 4 - to 8-day-old flies were allowed to lay eggs for 3 h on apple juice-agar medium with fresh yeast. Embryos were separated in groups of 20 or 50 and placed into food vials. Vials were kept at room temperature (19–22°C) and checked during the following hours for hatched eggs (2–5 times per day). We analyzed the average time over the midpoint of each successive interval in order to estimate the hatching time. Two experiments were performed following this protocol: A first pilot experiment with 150 embryos per strain, and one replica with 500 embryos per strain.
Egg hatchability and egg hatching time were also analyzed under cold stress conditions. Embryos were placed at 1°C overnight for 14 h and at 18°C during the day, and this cycle was maintained until all the eggs had hatched. We performed a pilot experiment with 100 embryos per strain and additional experiments with 240 and 160 embryos per strain, respectively.
Cold Stress in Embryos
In total, 800 7 - to 10-day-old flies were allowed to lay eggs for 3 h on apple juice-agar medium with fresh yeast. Embryos were collected following the methodology described in Schou (2013), and placed into food vials in groups of 50. When embryos were 3–6 h old, vials were placed at 1°C for 14 h, and maintained at 18°C until adult emergence. Simultaneously, control vials were always maintained at 18°C and not cold-exposed to control for other variables affecting egg to adult survival. We performed a first pilot experiment using 280 embryos per strain and three biological replicas using 350 embryos per strain (replica 1) and 750 embryos per strain (replica 2 and replica 3, respectively). In all cases, we analyzed egg to adult survival after all the adults had emerged.
Chill-Coma Recovery Time
In total, 500 3 - to 5-day-old flies were separated by sex and by strain and placed into five empty vials in groups of 50. We allowed flies to recover from CO2 anesthesia for 1 h and then vials were put in ice and kept in a 4°C chamber for 16 h as described in David et al. (1998). After the cold shock, adults were transferred to Petri dishes at room temperature (22–24°C), and recovery time was monitored for successive intervals of 30 s during 2 h. We considered as recovered flies those that were able to stand on their legs. As a control, we monitored survival of flies that were kept at room temperature: Three vials of 20 flies each, by sex and strain.
Survival after Chill-Coma
In total, 400 5 - to 8-day-old flies were separated by sex and strain and placed into six food vials in groups of 20. We allowed flies to recover from CO2 anesthesia for at least 2 days. After that, flies were changed to empty food vials and were put in ice, and kept in a 4°C chamber for 16 h. When adults were recovered from chill-coma, we transferred them to food vials and we monitored mortality during the next 5 days. As a control, we monitored survival of flies that were kept at room temperature: Three vials of 20 flies each, by sex and strain.
Osmotic Stress
In total, 2,000 4 - to 7-day-old flies were separated by sex and strain and placed in groups of 20 into 20 food vials containing 3% of NaCl, and into five vials with normal food as a control. Flies were maintained at room temperature (22–24°C) and dead flies were counted every 12–24 h until all the treated flies were dead.
Starvation Stress
In total, 2,000 3 - to 4-day-old flies per strain were separated by sex and strain and placed in groups of 20 into 20 food vials containing only 1.5% agar, and into five vials with normal food as a control. Flies were maintained at room temperature (22–24°C) and dead flies were counted three times a day until all the treated flies were dead.
Developmental Time
In total, 800 7 - to 10-day-old flies were allowed to lay eggs for 3 h. A total of 500 embryos per strain were collected and distributed in groups of 50 per food vial and were maintained at 18°C. Vials were checked every 6–8 h for emerging adults until all flies had emerged. We estimated the average DT over the midpoint of each successive interval.
Statistical Analyses of the Phenotypic Assays
Analyses were performed with SPSS v21. We first tested whether data followed a normal distribution by performing Kolmogorov–Smirnov test. t-Test was performed for normal data and Mann–Whitney test for nonnormal data. Survival curves were compared with log-rank test. When the statistical test was significant, we estimated the size effect of the mutation by calculating the odds-ratio and its confidence interval.
FBti0019386 Frequency Estimation for Natural Populations
To obtain FBti0019386 frequency, we run T-lex2 (Fiston-Lavier et al. 2014) using Drosophila whole-genome sequences available from a total of 23 populations from North America, Australia, Europe, and Africa (supplementary table S5, Supplementary Material online).
The accuracy of TE frequency estimates using T-lex2 is affected by coverage. However, coverage for all samples was higher than 20× except for Lyon (France) and California (USA), which had 8× and 4.7× coverage respectively, suggesting that overall frequency estimates are accurate.
Correlation Analysis of FBti0019386 Frequency with Geographic and Climate Variables
We analyzed whether the frequency of FBti0019386 insertion correlated with different geographical and climate variables in North America, Australia, and Europe using Pearson product–moment correlations. We also performed a PCA to disentangle the relationships between the climatic variables using Statistica (v8.0, StatSoft, Inc. 2007). Climatic data were obtained from the weather stations adjacent to collection sites of each population, available in Peel et al. (2007). When necessary, data were transformed as described in Sokal and Rohlf (2012) (see pages 411–422).
mRNA Transcript Levels Analysis (quantitative reverse transcription polymerase chain reaction)
Total RNA was extracted from three biological samples of 40 adult females (5-day old) from outbred populations differing by the presence/absence of FBti0019386 insertion using Trizol reagent and PureLink RNA Mini kit (Ambion). RNA was treated on-column with DNase I (Trizol) and after RNA purification. Reverse transcription was carried out using 1 µg of total RNA, Anchored-oligo(dT) primer, and Transcription First Strand cDNA Synthesis Kit (Roche). The resulting cDNA was used for quantitative reverse transcription polymerase chain reaction with SYBR Green (BioRad) on an iQ5 Thermal cycler. sra total expression was measured using a pair of primers specific to a 124-bp cDNA amplicon spanning the 5′-UTR/exon junction of the gene (5′-ACAACAACGGTGGAGAAGAGCCGT-3′ and 5′-GGTGCATCGGCGGACGCATTG-3′). For Bin1, we measured the 66-bp cDNA amplicon spanning the 5′-UTR/exon junction using specific primers (5′-TGTCGTCCCGTAGAGCAGAA-3′ and 5′-CAAGCAGATTGACCGCGAGA-3′). In both cases, we normalized the expression with Act5C (5′-GCGCCCTTACTCTTTCACCA-3′ and 5′-ATGTCACGGACGATTTCACG-3′). Expression was measured in nonstress conditions and in cold-stress conditions: 16 h at 4°C and 2 h at room temperature to allow flies to recover.
We also analyzed the expression of both genes in 0–2 h embryos using the same procedure. We collected the embryos from population cages containing approximately 800 flies from outbred populations differing by the presence/absence of FBti0019386 insertion. Briefly, 4 - to 8-day-old flies were allowed to lay eggs for 2 h on apple juice-agar medium with fresh yeast. Then, embryos were collected using a small brush and cleaned with water. Embryos were dechorionized by submerging them for 5 min in 50% bleach. After that, embryos were placed in a microcentrifuge tube, the excess of water was eliminated, and the samples were froze at −80°C until RNA extraction.
Detection of piRNA Reads Binding to FBti0019386 Sequence
We used small RNA sequencing data to check whether piRNAs reads mapped to FBti0019386 sequence, following a methodology similar to that described in Sentmanat and Elgin (2012). Briefly, we obtained the small RNA reads from Oregon R ovaries (accession number SRP000458) (Li et al. 2009), and from wild type ovaries (accession number: SRX470700) (Satyaki et al. 2014). We aligned the reads by using BWA-MEM package version 0.7.5 a-r405 (Li 2013) to the 14.6-kb sequence obtained from Drosophila reference genome, containing Bin1 and sra genes, and FBti0019386 (release five chromosomal coordinates 3 R: 12,010,721–12,025,306). Then, we used samtools and bamtools (Barnett et al. 2011) to index and filter by sense/antisense reads. Finally, we obtained the total read density using R (Rstudio v0.98.507).
Detection of HP1a Protein Binding in FBti0019386 Sequence
We downloaded all available raw data from modEncode HP1a protein ChIP-Seq experiments: Embryos (ID 3391 and 3392), third instar larvae (ID 4936), and adult heads (ID 5592) (http://data.modencode.org). Then, we mapped the reads against the 14.6-kb region described above. We performed the alignments following the same methodology as for the piRNA reads analysis.
Supplementary Material
Supplementary tables S1–S9 and figures S1–S3 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank Maite G. Barrón, Lain Guio, and Miriam Merenciano for comments on the manuscript. A.U. is an FPI fellow (BES-2012-052999) and J.G. is a Ramón y Cajal fellow (RYC-2010-07306). This work was supported by grants from the European Comission (Marie Curie CIG PCIG-2011-293860) and from the Spanish Government (Fundamental Research Projects Grant BFU-2011-24397) to J.G.
References
- Barnett DW, Garrison EK, Quinlan AR, Stromberg MP, Marth GT. BamTools: a C++ API and toolkit for analyzing and managing BAM files. Bioinformatics. 2011;27:1691–1692. doi: 10.1093/bioinformatics/btr174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergland AO, Tobler R, González J, Schmidt PS, Petrov DA. Secondary contact and local adaptation contribute to genome-wide patterns of clinal variation in Drosophila melanogaster. bioRxiv. 2014 doi: 10.1111/mec.13455. doi: 10.1101/009084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett C, Valentini S, Cabreiro F, Goss M, Somogyvari M, Piper MD, Hoddinott M, Sutphin GL, Leko V, McElwee JJ, et al. Absence of effects of Sir2 overexpression on lifespan in C. elegans and Drosophila. Nature. 2011;477:482–485. doi: 10.1038/nature10296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caracristi G, Schlotterer C. Genetic differentiation between American and European Drosophila melanogaster populations could be attributed to admixture of African alleles. Mol Biol Evol. 2003;20:792–799. doi: 10.1093/molbev/msg091. [DOI] [PubMed] [Google Scholar]
- Casacuberta E, González J. The impact of transposable elements in environmental adaptation. Mol Ecol. 2013;22:1503–1517. doi: 10.1111/mec.12170. [DOI] [PubMed] [Google Scholar]
- Chang KT, Min KT. Upregulation of three Drosophila homologs of human chromosome 21 genes alters synaptic function: implications for Down syndrome. Proc Natl Acad Sci U S A. 2009;106:17117–17122. doi: 10.1073/pnas.0904397106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang KT, Shi YJ, Min KT. The Drosophila homolog of Down's syndrome critical region 1 gene regulates learning: implications for mental retardation. Proc Natl Acad Sci U S A. 2003;100:15794–15799. doi: 10.1073/pnas.2536696100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chippindale AK, Alipaz JA, Chen HW, Rose RM. Experimental evolution of accelerated development in Drosophila. 1. Developmental speed and larval survival. Evolution. 1997;51:1536–1551. doi: 10.1111/j.1558-5646.1997.tb01477.x. [DOI] [PubMed] [Google Scholar]
- Costa E, Beltran S, Espinas ML. Drosophila melanogaster SAP18 protein is required for environmental stress responses. FEBS Lett. 2011;585:275–280. doi: 10.1016/j.febslet.2010.11.058. [DOI] [PubMed] [Google Scholar]
- Costa R, Peixoto AA, Barbujani G, Kyriacou CP. A latitudinal cline in a Drosophila clock gene. Proc Biol Sci. 1992;250:43–49. doi: 10.1098/rspb.1992.0128. [DOI] [PubMed] [Google Scholar]
- David J, Capy P, Payant V, Tsakas S. Thoracic trident pigmentation in Drosophila melanogaster: differentiation of geographical populations. Genet Sel Evol. 1985;17:211–224. doi: 10.1186/1297-9686-17-2-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David J, Mercot H, Capy P, McEvey S, Van Herrewege J. Alcohol tolerance and Adh gene frequencies in European and African populations of Drosophila melanogaster. Genet Sel Evol. 1986;18:405–416. doi: 10.1186/1297-9686-18-4-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David JR, Alonso-Moraga A, Borai F, Capy P, Mercot H, McEvey SF, Munoz-Serrano A, Tsakas S. Latitudinal variation of Adh gene frequencies in Drosophila melanogaster: a Mediterranean instability. Heredity (Edinb) 1989;62(Pt 1):11–16. doi: 10.1038/hdy.1989.2. [DOI] [PubMed] [Google Scholar]
- David RJ, Gibert P, Pla E, Petavy G, Karan D, Moreteau B. Cold stress tolerance in Drosophila: analysis of chill coma recovery in D. melanogaster. J Therm Biol. 1998;23(5):291–299. [Google Scholar]
- Davies SA, Cabrero P, Overend G, Aitchison L, Sebastian S, Terhzaz S, Dow JA. Cell signalling mechanisms for insect stress tolerance. J Exp Biol. 2014;217:119–128. doi: 10.1242/jeb.090571. [DOI] [PubMed] [Google Scholar]
- Di Cara F, King-Jones K. How clocks and hormones act in concert to control the timing of insect development. Curr Top Dev Biol. 2013;105:1–36. doi: 10.1016/B978-0-12-396968-2.00001-4. [DOI] [PubMed] [Google Scholar]
- Duchen P, Zivkovic D, Hutter S, Stephan W, Laurent S. Demographic inference reveals African and European admixture in the North American Drosophila melanogaster population. Genetics. 2013;193:291–301. doi: 10.1534/genetics.112.145912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejima A, Nakayama S, Aigaki T. Phenotypic association of spontaneous ovulation and sexual receptivity in virgin females of Drosophila melanogaster mutants. Behav Genet. 2001;31:437–444. doi: 10.1023/a:1012794421980. [DOI] [PubMed] [Google Scholar]
- Ejima A, Tsuda M, Takeo S, Ishii K, Matsuo T, Aigaki T. Expression level of sarah, a homolog of DSCR1, is critical for ovulation and female courtship behavior in Drosophila melanogaster. Genetics. 2004;168:2077–2087. doi: 10.1534/genetics.104.029934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian DK, Kapun M, Nolte V, Kofler R, Schmidt PS, Schlotterer C, Flatt T. Genome-wide patterns of latitudinal differentiation among populations of Drosophila melanogaster from North America. Mol Ecol. 2012;21:4748–4769. doi: 10.1111/j.1365-294X.2012.05731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiston-Lavier AS, Barrón M, Petrov DA, González J. T-lex2: genotyping, frequency estimation and re-annotation of transposable elements using single or pooled next-generation sequencing data. Nucleic Acids Res. 2014;43(4):e22. doi: 10.1093/nar/gku1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folguera G, Ceballos S, Spezzi L, Fanara JJ, Hasson E. Clinal variation in developmental time and viability, and the response to thermal treatments in two species of Drosophila. Biol J Linn Soc Lond. 2008;95:233–245. [Google Scholar]
- Gibert P, Moreteau B, Petavy G, Karan D, David JR. Chill-coma tolerance, a major climatic adaptation among Drosophila species. Evolution. 2001;55:1063–1068. doi: 10.1554/0014-3820(2001)055[1063:cctamc]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- González J, Karasov TL, Messer PW, Petrov DA. Genome-wide patterns of adaptation to temperate environments associated with transposable elements in Drosophila. PLoS Genet. 2010;6:e1000905. doi: 10.1371/journal.pgen.1000905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González J, Lenkov K, Lipatov M, Macpherson JM, Petrov DA. High rate of recent transposable element-induced adaptation in Drosophila melanogaster. PLoS Biol. 2008;6:e251. doi: 10.1371/journal.pbio.0060251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guio L, Barrón MG, González J. The transposable element Bari-Jheh mediates oxidative stress response in Drosophila. Mol Ecol. 2014;23:2020–2030. doi: 10.1111/mec.12711. [DOI] [PubMed] [Google Scholar]
- Hill T, Lewicki P. Tulsa (OK): StatSoft; 2007. STATISTICS: Methods and Applications. [Google Scholar]
- Hoffmann AA, Sorensen JG, Loeschcke V. Adaptation of Drosophila to temperature extremes: bringing together quantitative and molecular approaches. J Therm Biol. 2003;28:175–216. [Google Scholar]
- Hogan PG, Chen L, Nardone J, Rao A. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 2003;17:2205–2232. doi: 10.1101/gad.1102703. [DOI] [PubMed] [Google Scholar]
- Horner VL, Czank A, Jang JK, Singh N, Williams BC, et al. The Drosophila calcipressin sarah is required for several aspects of egg activation. Curr Biol. 2006;16:1441–1446. doi: 10.1016/j.cub.2006.06.024. [DOI] [PubMed] [Google Scholar]
- Huang W, Massouras A, Inoue Y, Peiffer J, Ramia M, et al. Natural variation in genome architecture among 205 Drosophila melanogaster Genetic Reference Panel lines. Genome Res. 2014;24:1193–1208. doi: 10.1101/gr.171546.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Richards S, Carbone MA, Zhu D, Anholt RR, et al. Epistasis dominates the genetic architecture of Drosophila quantitative traits. Proc Natl Acad Sci U S A. 2012;109:15553–15559. doi: 10.1073/pnas.1213423109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics. 2002;18:337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- Hudson RR, Slatkin M, Maddison WP. Estimation of levels of gene flow from DNA sequence data. Genetics. 1992;132:583–589. doi: 10.1093/genetics/132.2.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James AC, Azevedo RB, Partridge L. Genetic and environmental responses to temperature of Drosophila melanogaster from a latitudinal cline. Genetics. 1997;146:881–890. doi: 10.1093/genetics/146.3.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James AC, Partridge L. Thermal evolution of rate of larval development in Drosophila melanogaster in laboratory and field populations. J Evol Biol. 1995;8:315–330. [Google Scholar]
- Jones FC, Grabherr MG, Chan YF, Russell P, Mauceli E, et al. The genomic basis of adaptive evolution in threespine sticklebacks. Nature. 2012;484:55–61. doi: 10.1038/nature10944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolaczkowski B, Kern AD, Holloway AK, Begun DJ. Genomic differentiation between temperate and tropical Australian populations of Drosophila melanogaster. Genetics. 2011;187:245–260. doi: 10.1534/genetics.110.123059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry CR, Rifkin SA. The genotype-phenotype maps of systems biology and quantitative genetics: distinct and complementary. Adv Exp Med Biol. 2012;751:371–398. doi: 10.1007/978-1-4614-3567-9_17. [DOI] [PubMed] [Google Scholar]
- Lehner B. Genotype to phenotype: lessons from model organisms for human genetics. Nat Rev Genet. 2013;14:168–178. doi: 10.1038/nrg3404. [DOI] [PubMed] [Google Scholar]
- Li C, Vagin VV, Lee S, Xu J, Ma S, et al. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell. 2009;137:509–521. doi: 10.1016/j.cell.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. bioRxiv doi: 1303.3997v2.
- Macdonald SS, Rako L, Batterham P, Hoffmann AA. Dissecting chill coma recovery as a measure of cold resistance: evidence for a biphasic response in Drosophila melanogaster. J Insect Physiol. 2004;50:695–700. doi: 10.1016/j.jinsphys.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Mackay TF. Mutations and quantitative genetic variation: lessons from Drosophila. Philos Trans R Soc Lond B Biol Sci. 2010;365:1229–1239. doi: 10.1098/rstb.2009.0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TF. Epistasis and quantitative traits: using model organisms to study gene-gene interactions. Nat Rev Genet. 2014;15:22–33. doi: 10.1038/nrg3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TF, Richards S, Stone EA, Barbadilla A, Ayroles JF, et al. The Drosophila melanogaster Genetic Reference Panel. Nature. 2012;482:173–178. doi: 10.1038/nature10811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magwire MM, Bayer F, Webster CL, Cao C, Jiggins FM. Successive increases in the resistance of Drosophila to viral infection through a transposon insertion followed by a Duplication. PLoS Genet. 2011;7:e1002337. doi: 10.1371/journal.pgen.1002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinek S, Inonog S, Manoukian AS, Young MW. A role for the segment polarity gene shaggy/GSK-3 in the Drosophila circadian clock. Cell. 2001;105:769–779. doi: 10.1016/s0092-8674(01)00383-x. [DOI] [PubMed] [Google Scholar]
- Mateo L, Ullastres A, González J. A transposable element insertion confers xenobiotic resistance in Drosophila. PLoS Genet. 2014;10:e1004560. doi: 10.1371/journal.pgen.1004560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matyash A, Singh N, Hanes SD, Urlaub H, Jackle H. SAP18 promotes Kruppel-dependent transcriptional repression by enhancer-specific histone deacetylation. J Biol Chem. 2009;284:3012–3020. doi: 10.1074/jbc.M806163200. [DOI] [PubMed] [Google Scholar]
- Matzkin LM, Merritt TJ, Zhu CT, Eanes WF. The structure and population genetics of the breakpoints associated with the cosmopolitan chromosomal inversion In(3R)Payne in Drosophila melanogaster. Genetics. 2005;170:1143–1152. doi: 10.1534/genetics.104.038810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee LW, Aitchison EW, Caudle SB, Morrison AJ, Zheng L, Yang W, Rokyta DR. Payoffs, not tradeoffs, in the adaptation of a virus to ostensibly conflicting selective pressures. PLoS Genet. 2014;10:e1004611. doi: 10.1371/journal.pgen.1004611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mensch J, Lavagnino N, Carreira VP, Massaldi A, Hasson E, Fanara JJ. Identifying candidate genes affecting developmental time in Drosophila melanogaster: pervasive pleiotropy and gene-by-environment interaction. BMC Dev Biol. 2008;8:78. doi: 10.1186/1471-213X-8-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai Y, Horiuchi J, Tsuda M, Takeo S, Akahori S, Matsuo T, Kume K, Aigaki T. Calcineurin and its regulator sra/DSCR1 are essential for sleep in Drosophila. J Neurosci. 2011;31:12759–12766. doi: 10.1523/JNEUROSCI.1337-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Williamson S, Kim Y, Hubisz MJ, Clark AG, Bustamante C. Genomic scans for selective sweeps using SNP data. Genome Res. 2005;15:1566–1575. doi: 10.1101/gr.4252305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunney L. Drosophila on oranges: colonization, competition and coexistance. Ecology. 1990;71:1904–1915. [Google Scholar]
- Oakeshott JG, Chambers GK, Gibson JB, Eanes WF, Willcocks DA. Geographic variation in G6pd and Pgd allele frequencies in Drosophila melanogaster. Heredity (Edinb) 1983;50(Pt 1):67–72. doi: 10.1038/hdy.1983.7. [DOI] [PubMed] [Google Scholar]
- Oakeshott JG, Gibson JB, Willcocks DA, Chambers GK. Latitudinal variation in octanol dehydrogenase and acid phosphatase allele frequencies in Drosophila melanogaster. Theor Appl Genet. 1983;65:191–196. doi: 10.1007/BF00308064. [DOI] [PubMed] [Google Scholar]
- Orr HA. The genetic theory of adaptation: a brief history. Nat Rev Genet. 2005;6:119–127. doi: 10.1038/nrg1523. [DOI] [PubMed] [Google Scholar]
- Paaby AB, Bergland AO, Behrman EL, Schmidt PS. A highly pleiotropic amino acid polymorphism in the Drosophila insulin receptor contributes to life-history adaptation. Evolution. 2014;68:3395–3409. doi: 10.1111/evo.12546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paaby AB, Rockman MV. The many faces of pleiotropy. Trends Genet. 2013;29:66–73. doi: 10.1016/j.tig.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paaby AB, Schmidt PS. Functional significance of allelic variation at methuselah, an aging gene in Drosophila. PLoS One. 2008;3:e1987. doi: 10.1371/journal.pone.0001987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peel MC, Finlayson BL, McMahon TA. Updated world map of the Köppen-Geiger climate classification. Hydrol Earth Syst Sci. 2007;11:1633–1644. [Google Scholar]
- Pfeifer B, Wittelsburger U, Ramos-Onsins SE, Lercher MJ. PopGenome: an efficient Swiss army knife for population genomic analyses in R. Mol Biol Evol. 2014;31:1929–1936. doi: 10.1093/molbev/msu136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt JA, Kolaczkowski B, Jones CD, Begun DJ, Kern AD. Parallel geographic variation in Drosophila melanogaster. Genetics. 2014;197:361–373. doi: 10.1534/genetics.114.161463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman MV. The QTN program and the alleles that matter for evolution: all that's gold does not glitter. Evolution. 2012;66:1–17. doi: 10.1111/j.1558-5646.2011.01486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MR, Mueller LD, Burke MK. New experiments for an undivided genetics. Genetics. 2011;188:1–10. doi: 10.1534/genetics.111.128900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault JD, Marican C, Wicker-Thomas C, Jallon JM. Relations between cuticular hydrocarbon (HC) polymorphism, resistance against desiccation and breeding temperature; a model for HC evolution in D. melanogaster and D. simulans. Genetica. 2004;120:195–212. doi: 10.1023/b:gene.0000017641.75820.49. [DOI] [PubMed] [Google Scholar]
- Sakai T, Aigaki T. The Drosophila calcineurin regulator, Sarah, is involved in male courtship. Neuroreport. 2010;21:985–988. doi: 10.1097/WNR.0b013e32833eaade. [DOI] [PubMed] [Google Scholar]
- Satyaki PR, Cuykendall TN, Wei KH, Brideau NJ, Kwak H, Aruna S, Ferree PM, Ji S, Barbash DA. The Hmr and Lhr hybrid incompatibility genes suppress a broad range of heterochromatic repeats. PLoS Genet. 2014;10:e1004240. doi: 10.1371/journal.pgen.1004240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt JM, Good RT, Appleton B, Sherrard J, Raymant GC, et al. Copy number variation and transposable elements feature in recent, ongoing adaptation at the Cyp6g1 locus. PLoS Genet. 2010;6:e1000998. doi: 10.1371/journal.pgen.1000998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt PS, Paaby AB. Reproductive diapause and life-history clines in North American populations of Drosophila melanogaster. Evolution. 2008;62:1204–1215. doi: 10.1111/j.1558-5646.2008.00351.x. [DOI] [PubMed] [Google Scholar]
- Schmidt PS, Paaby AB, Heschel MS. Genetic variance for diapause expression and associated life histories in Drosophila melanogaster. Evolution. 2005;59:2616–2625. [PubMed] [Google Scholar]
- Schou MF. Fast egg collection method greatly improves randomness of egg sampling in Drosophila melanogaster. Fly (Austin) 2013;7:44–46. doi: 10.4161/fly.22758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sentmanat M, Wang SH, Elgin SC. Targeting heterochromatin formation to transposable elements in Drosophila: potential roles of the piRNA system. Biochemistry (Mosc) 2013;78:562–571. doi: 10.1134/S0006297913060023. [DOI] [PubMed] [Google Scholar]
- Sentmanat MF, Elgin SC. Ectopic assembly of heterochromatin in Drosophila melanogaster triggered by transposable elements. Proc Natl Acad Sci U S A. 2012;109:14104–14109. doi: 10.1073/pnas.1207036109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal RR, Rohlf FJ. Biometry: the principles and practice of statistics in biological research. 4th ed. New York: W. H. Freeman and Co; 2012. [Google Scholar]
- Song CP, Galbraith DW. AtSAP18, an orthologue of human SAP18, is involved in the regulation of salt stress and mediates transcriptional repression in Arabidopsis. Plant Mol Biol. 2006;60:241–257. doi: 10.1007/s11103-005-3880-9. [DOI] [PubMed] [Google Scholar]
- St Pierre SE, Ponting L, Stefancsik R, McQuilton P, FlyBase C. FlyBase 102—advanced approaches to interrogating FlyBase. Nucleic Acids Res. 2014;42:D780–D788. doi: 10.1093/nar/gkt1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley SM, Parsons PA. The response of the cosmopolitan species Drosophila melanogaster to ecological gradients. Proc Ecol Soc Aust. 1981;11:121–130. [Google Scholar]
- Storz JF, Wheat CW. Integrating evolutionary and functional approaches to infer adaptation at specific loci. Evolution. 2010;64:2489–2509. doi: 10.1111/j.1558-5646.2010.01044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Shen YH, Han MJ, Cao YF, Zhang Z. An adaptive transposable element insertion in the regulatory region of the EO gene in the domesticated silkworm, Bombyx mori. Mol Biol Evol. 2014;31(12):3302–3313. doi: 10.1093/molbev/msu261. [DOI] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeo S, Hawley RS, Aigaki T. Calcineurin and its regulation by Sra/RCAN is required for completion of meiosis in Drosophila. Dev Biol. 2010;344:957–967. doi: 10.1016/j.ydbio.2010.06.011. [DOI] [PubMed] [Google Scholar]
- Takeo S, Tsuda M, Akahori S, Matsuo T, Aigaki T. The calcineurin regulator sra plays an essential role in female meiosis in Drosophila. Curr Biol. 2006;16:1435–1440. doi: 10.1016/j.cub.2006.05.058. [DOI] [PubMed] [Google Scholar]
- Takeuchi K, Nakano Y, Kato U, Kaneda M, Aizu M, et al. Changes in temperature preferences and energy homeostasis in dystroglycan mutants. Science. 2009;323:1740–1743. doi: 10.1126/science.1165712. [DOI] [PubMed] [Google Scholar]
- Teets NM, Yi SX, Lee RE, Jr, Denlinger DL. Calcium signaling mediates cold sensing in insect tissues. Proc Natl Acad Sci U S A. 2013;110:9154–9159. doi: 10.1073/pnas.1306705110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobler R, Franssen SU, Kofler R, Orozco-Terwengel P, Nolte V, Hermisson J, Schlotterer C. Massive habitat-specific genomic response in D. melanogaster populations during experimental evolution in hot and cold environments. Mol Biol Evol. 2014;31:364–375. doi: 10.1093/molbev/mst205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotta V, Calboli FC, Ziosi M, Guerra D, Pezzoli MC, David JR, Cavicchi S. Thermal plasticity in Drosophila melanogaster: a comparison of geographic populations. BMC Evol Biol. 2006;6:67. doi: 10.1186/1471-2148-6-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner TL, Bourne EC, Von Wettberg EJ, Hu TT, Nuzhdin SV. Population resequencing reveals local adaptation of Arabidopsis lyrata to serpentine soils. Nat Genet. 2010;42:260–263. doi: 10.1038/ng.515. [DOI] [PubMed] [Google Scholar]
- Yadav P, Thandapani M, Sharma VK. Interaction of light regimes and circadian clocks modulate timing of pre-adult developmental events in Drosophila. BMC Dev Biol. 2014;14:19. doi: 10.1186/1471-213X-14-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.