Abstract
In most eukaryotes, centromere is determined by the presence of the centromere-specific histone variant CenH3. Two types of chromosome morphology are generally recognized with respect to centromere organization. Monocentric chromosomes possess a single CenH3-containing domain in primary constriction, whereas holocentric chromosomes lack the primary constriction and display dispersed distribution of CenH3. Recently, metapolycentric chromosomes have been reported in Pisum sativum, representing an intermediate type of centromere organization characterized by multiple CenH3-containing domains distributed across large parts of chromosomes that still form a single constriction. In this work, we show that this type of centromere is also found in other Pisum and closely related Lathyrus species, whereas Vicia and Lens genera, which belong to the same legume tribe Fabeae, possess only monocentric chromosomes. We observed extensive variability in the size of primary constriction and the arrangement of CenH3 domains both between and within individual Pisum and Lathyrus species, with no obvious correlation to genome or chromosome size. Search for CenH3 gene sequences revealed two paralogous variants, CenH3-1 and CenH3-2, which originated from a duplication event in the common ancestor of Fabeae species. The CenH3-1 gene was subsequently lost or silenced in the lineage leading to Vicia and Lens, whereas both genes are retained in Pisum and Lathyrus. Both of these genes appear to have evolved under purifying selection and produce functional CenH3 proteins which are fully colocalized. The findings described here provide the first evidence for a highly dynamic centromere structure within a group of closely related species, challenging previous concepts of centromere evolution.
Keywords: centromere, CenH3, chromosome, centromere drive, adaptive evolution, gene duplication
Introduction
The centromere is a functional chromosomal domain that is essential for faithful chromosome segregation during cell division. It can be reliably identified by the presence of kinetochore proteins, one of which is the centromere-specific variant of histone H3 known as CenH3 (or CENP-A in mammals; Black and Bassett 2008). CenH3 replaces canonical histone H3 in centromeric nucleosomes, representing one of a few kinetochore proteins that are directly associated with DNA. This protein is encoded by a single-copy gene in most diploid genomes, including those that have undergone whole-genome duplication, suggesting that one copy of the duplicated gene is generally lost (Hirsch et al. 2009; Malik 2009). Contrary to histone H3, which is nearly invariant across eukaryotes, CenH3 shows a high degree of sequence variability (Malik 2009; Torras-Llort et al. 2009; Tek et al. 2011). The most diverse portion of the protein is the N-terminal tail, which can differ considerably in both length and sequence, even among closely related species, and lacks similarity between distantly related species. The remaining C-terminal portion consists of a histone fold domain (HFD), which is similar in all eukaryotes and shares significant similarity also with H3. This domain is crucial for nucleosome assembly and targeting of CenH3 to centromeres (Black et al. 2004).
Attempts to explain the rapid evolution of CenH3 led to the hypothesis of centromere drive, which posits that CenH3 evolves to compensate for changes in centromeric DNA sequences that cause unequal binding to spindle microtubules, which results in skewed transmission frequencies for homologous chromosomes during asymmetric meiosis in females (Malik 2009; Roach et al. 2012). The strongest evidence in favor of this hypothesis is the finding that certain changes in CenH3 genes appear to have been fixed by recurrent episodes of positive selection (Malik and Henikoff 2001; Malik et al. 2002; Cooper and Henikoff 2004; Hirsch et al. 2009; Zedek and Bureš 2012). However, it should be noted that the functional relationship between CenH3 histones and centromeric DNA remains poorly understood and that the centromere drive hypothesis has yet to be tested experimentally. Moreover, the recruitment of CenH3 appears to be primarily an epigenetic process that is largely independent of underlying DNA sequence (Torras-Llort et al. 2009; Mehta et al. 2010). The few known exceptions are in budding yeast (Saccharomyces cerevisiae) and certain closely related species, whose point centromeres are determined genetically by a specific DNA sequence (Cole et al. 2011; Sanyal et al. 2014). In contrast, centromeres in most other species are composed of a single or few centromere-specific families of satellite DNA and/or retrotransposons (Zhong et al. 2002; Nagaki et al. 2003; Nagaki and Murata 2005; Houben et al. 2007; Nagaki, Kashihara, et al. 2009; Nagaki, Walling, et al. 2009; Tek et al. 2010, 2011; Neumann et al. 2012; Fukagawa and Earnshaw 2014; Plohl et al. 2014), although examples of repeat-less centromeres have also been documented (Piras et al. 2010; Shang et al. 2010; Gong et al. 2012). These centromeric DNA sequences evolve rapidly, resulting in a high level of sequence divergence between closely related species and the complete lack of DNA sequence conservation over long periods of evolutionary time (Lee et al. 2005; Nagaki, Walling, et al. 2009).
The size of centromere domains can also vary enormously. The smallest and simplest are the 125 bp point centromeres found in budding yeast, which encompass only a single nucleosome. Most species, however, have so called regional centromeres, which are much larger and more complex. The CenH3-binding region in fission yeast (Schizosaccharomyces pombe) encompasses 4–7 kb (Pidoux and Allshire 2004), but the corresponding regions in most multicellular eukaryotes are much larger, spanning tens of kilobases up to several megabases (Yan et al. 2008; Wolfgruber et al. 2009; Burrack and Berman 2012). In general, the CenH3-containing chromatin of regional centromeres forms a single compact domain localized within the primary constriction zone of monocentric chromosomes (Sullivan and Karpen 2004; Marshall et al. 2008; Ribeiro et al. 2010). Much larger diffuse centromeres are found in holocentric (polycentric) chromosomes. Although holocentric chromosomes lack primary constriction, they do possess CenH3-containing domains, which are distributed as contiguous loci in a linear axis over nearly the entire length of the chromosome (Maddox et al. 2004; Nagaki et al. 2005; Heckmann et al. 2011). Chromosome size in Caenorhabditis elegans, the best studied holocentric species, is 14–21 Mb, but it can exceed even 100 Mb in some species, such as Luzula nivea (Barlow and Nevin 1976; C. elegans Sequencing Consortium 1998).
It has been estimated that polycentric chromosomes independently evolved from monocentric ones at least 13 times (Melters et al. 2012), although the mechanisms behind the transition remain unknown. Because no intermediate state between monocentric and holocentric chromosomes had been documented, the separation between the two chromosome types appeared clear. However, we recently found that the seemingly monocentric chromosomes of the pea possess very large centromeres made up of three to five distinct CenH3-containing domains containing two substantially diverged yet fully colocalized CenH3 variants (hereafter referred to as CenH3-1_PSat and CenH3-2_PSat; Neumann et al. 2012). As the organization of CenH3-containing domains in the pea fits neither the monocentric nor holocentric centromere type, it was termed “metapolycentric.” The discovery of metapolycentric chromosomes in the pea provided evidence that chromosomes can possess multiple CenH3-containing domains without noticeably affecting their stability during cell division. This finding also suggests that centromeres can increase in size not only through the simple elongation of preexisting centromere domains but also through the formation of new centromere domains involving diverged sequences at relatively distant loci.
The discovery of metapolycentric chromosomes has dramatically changed our view of centromere plasticity and raises important questions concerning the mechanisms and limits of centromere dispersal, the evolutionary origin of metapolycentric organization, and its link to CenH3 evolution. To address these questions, we investigated variations in centromere size and CenH3 protein distribution in the chromosomes of 24 legume species, including the closest relatives to Pisum sativum that form the tribe Fabeae (P. fulvum and representatives of the genera Lathyrus, Vicia, and Lens), as well as non-Fabeae outgroup species from the inverted repeat lacking clade (IRLC) of the Fabaceae family. This analysis revealed profound changes in centromere structure that occurred in a subset of the studied species since the split of Fabeae from the other taxa of the IRLC clade approximately 16–23 Ma (Schaefer et al. 2012). Possible mechanisms for these changes are discussed in the context of CenH3-coding gene evolution in Fabeae.
Results
Metapolycentric Chromosomes Are Limited to Pisum and Lathyrus Species
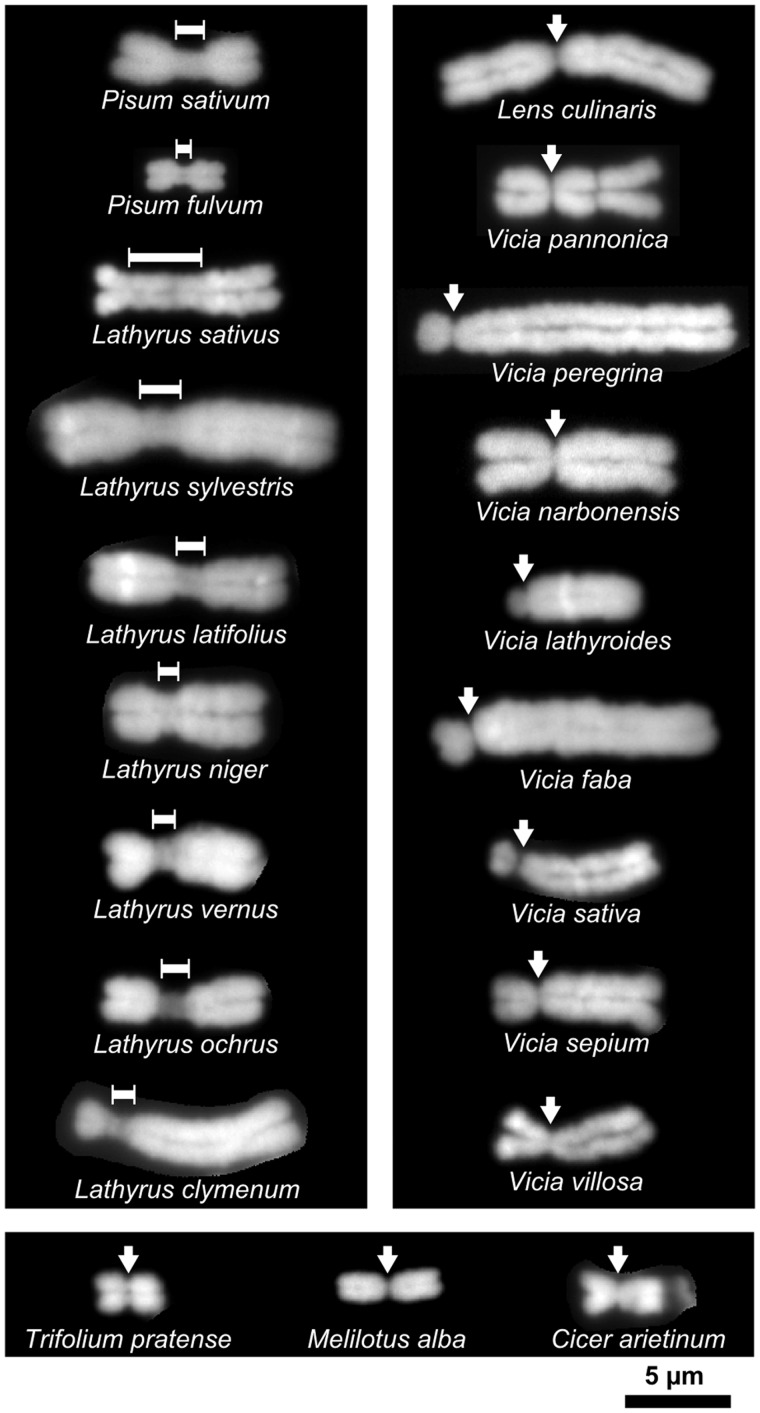
Investigation of centromere morphology in Fabeae species uncovered substantial variation in centromere size and shape. Elongated primary constrictions similar to those observed in P. sativum (Neumann et al. 2012) were only found in P. fulvum and the Lathyrus species, whereas Vicia, Lens, and non-Fabeae outgroup species displayed simple primary constrictions typical of the monocentric chromosomes found in most plants (fig. 1 and supplementary fig. S1, Supplementary Material online). Centromere morphology was reflected in the diversity of CenH3 domain organizations visualized using a set of antibodies raised against CenH3 variants in the investigated species (figs. 2 and 3). As expected, the short primary constrictions in Vicia and Lens species showed single dot-like signals for CenH3 (figs. 2D–F, I–K, P–S, and U and 3G–J). Single uninterrupted CenH3 signals, which were partially extended in some cases, were also detected at the early stages of mitotic chromosome condensation, indicating that there were no interspersed CenH3-negative chromatin blocks large enough to be detected at this level of resolution (fig. 2D–F). In contrast, the elongated primary constrictions in Pisum and Lathyrus species displayed complex patterns of CenH3 signals, which could be classified into two basic types. The “beads on a string” pattern, which is characteristic of metapolycentric organization, consisted of multiple dot-like signals clearly separated by CenH3-negative chromatin domains (figs. 2G, H, N, and O and 3A–E). Alternatively, CenH3 could be localized in a ribbon-like pattern along the entire length of the primary constriction (figs. 2G, H, L, M, N, and O and 3A–F). Some chromosomes displayed a beads on a string pattern at low level of chromosome condensation but showed a ribbon-like structure at higher chromatin condensation that caused merging of individual CenH3-containing domains (figs. 2C, G, H, and L–O and 3A–F). The most pronounced metapolycentric structure was observed in Lathyrus sativus, showing three to five clearly distinguished CenH3 domains which remained separate even on highly condensed chromosomes (figs. 2O, V, and W and 3A). Lathyrus sativus was also found to have the largest primary constrictions of all the investigated species, accounting for about one-third of chromosomal length and estimated to span 166–263 Mb (supplementary fig. S1 and table S1, Supplementary Material online). In contrast, chromosomes of the closely related species La. latifolius and La. sylvestris had considerably shorter primary constrictions, only some of which displayed a metapolycentric structure, with the remainder showing a ribbon-like pattern of CenH3 localization (figs. 2N and 3D and E). The smallest centromeres of the Lathyrus species were found in La. clymenum and La. ochrus, in which only 1–2 chromosomes had slightly elongated primary constrictions, whereas the remainder had very short constrictions similar to those found in Vicia and Lens spp. (figs. 1 and 3F, and supplementary fig. S1, Supplementary Material online). Differences in centromere size and CenH3 domain organization were also observed between P. sativum and P. fulvum, with shorter constrictions and ribbon-like patterns occurring more frequently in the latter species (figs. 2L and M and 3B and C and data not shown).
Fig. 1.
Examples of chromosomes with two types of centromere morphology. Elongated primary constrictions were observed only in Pisum and Lathyrus species (left panel), whereas all chromosomes in Vicia, Lens (right panel), and non-Fabeae species (bottom panel) showed the small primary constrictions typical of monocentric chromosomes found in most plants. Positions of primary constrictions are indicated by dimension lines and arrows, respectively. Chromosomes ofall species were fixed in 3:1 fixative (methanol:acetic acid) and prepared using the same technique. The staining was done with DAPI. Bar = 5 µm .
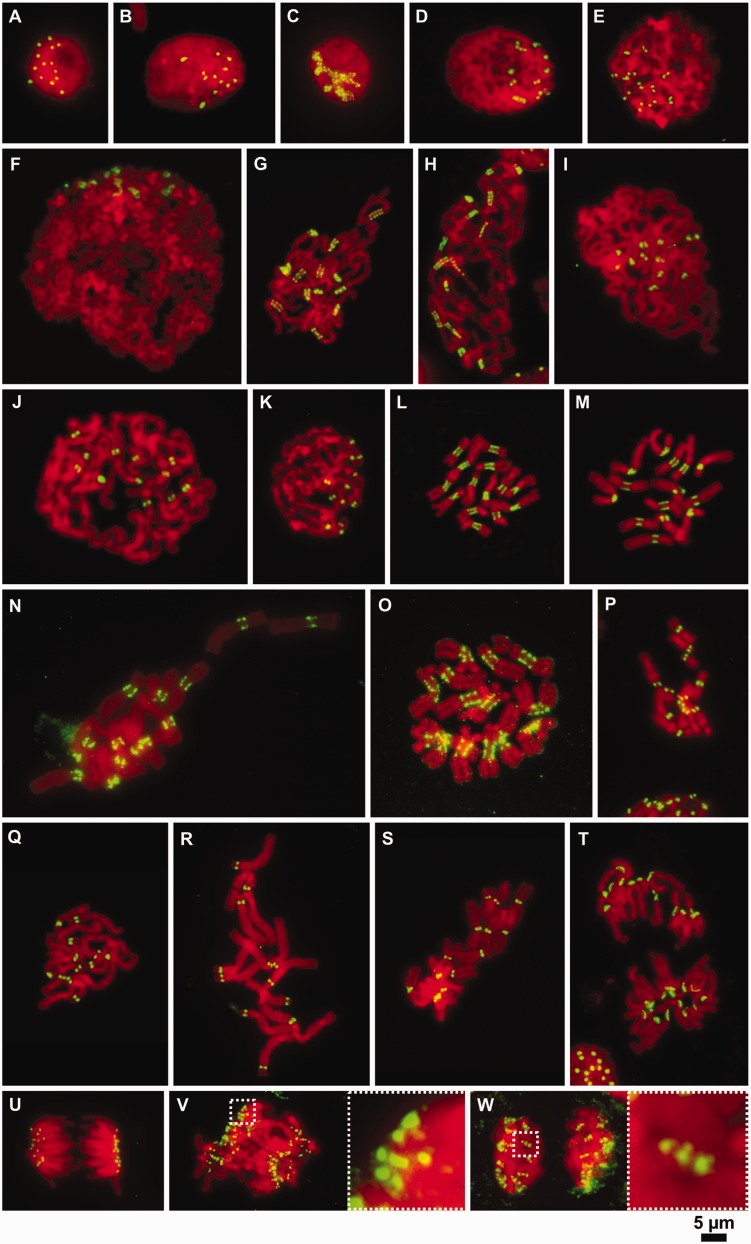
Fig. 2.
Organization of CenH3-containing domains on mitotic chromosomes at different levels of chromatin compaction. (A and B) Immunodetection of CenH3 in the nuclei of Pisum sativum (2n = 14) (A) and Vicia faba (2n = 12) (B). Note that the number of CenH3 signals corresponds to the number of chromosomes. (C–W) Immunodetection of CenH3 at different levels of chromatin compaction during mitosis. (C–F) Chromosomes at the early stages of chromatin condensation in P. sativum (C), V. pannonica (D), V. sativa (E), V. faba (F), P. sativum (G), P. fulvum (H), V. pannonica (I), V. faba (J), and V. sativa (K). Note that the low-condensed chromosomes in Pisum display the beads on a string pattern while those in Vicia possess a single uninterrupted CenH3 signal. (L and M) Metaphase chromosomes in P. sativum (L) and P. fulvum (M) displaying mostly ribbon-like patterns of CenH3 distribution. (N) Metaphase chromosomes in La. sylvestris showing either beads on a string or ribbon-like patterns of CenH3 distribution. (O) Metaphase chromosomes in La. sativus displaying the beads on a string pattern. (P and S) Metaphase chromosomes in V. sativa (P), V. pannonica (Q), V. faba (R), and Lens culinaris (S) showing single dot-like CenH3 signals. (T) Anaphase of chromosomes in P. fulvum, some of which show a ribbon-like pattern of CenH3 signal. (U) Anaphase chromosomes in Le. culinaris with single dot-like signals of CenH3 at each centromere. (V and W) anaphase (V) and telophase (W) chromosomes in La. sativus showing the beads on a string pattern. Insets show 5 × magnifications of the boxed regions. The immunodetection experiments were performed using squash preparations of synchronized root tip meristems fixed in 4% formaldehyde. CenH3 (green) was immunodetected with antibodies primarily raised to CenH3-1_PSat (P. sativum and P. fulvum), CenH3-2_PSat (La. sativus and La. sylvestris), CenH3-2_VF (V. faba, V. sativa and V. pannonica), and CenH3-2_LCul (Le. culinaris). Chromosomes were counterstained with DAPI (red). Bar = 5 µm.
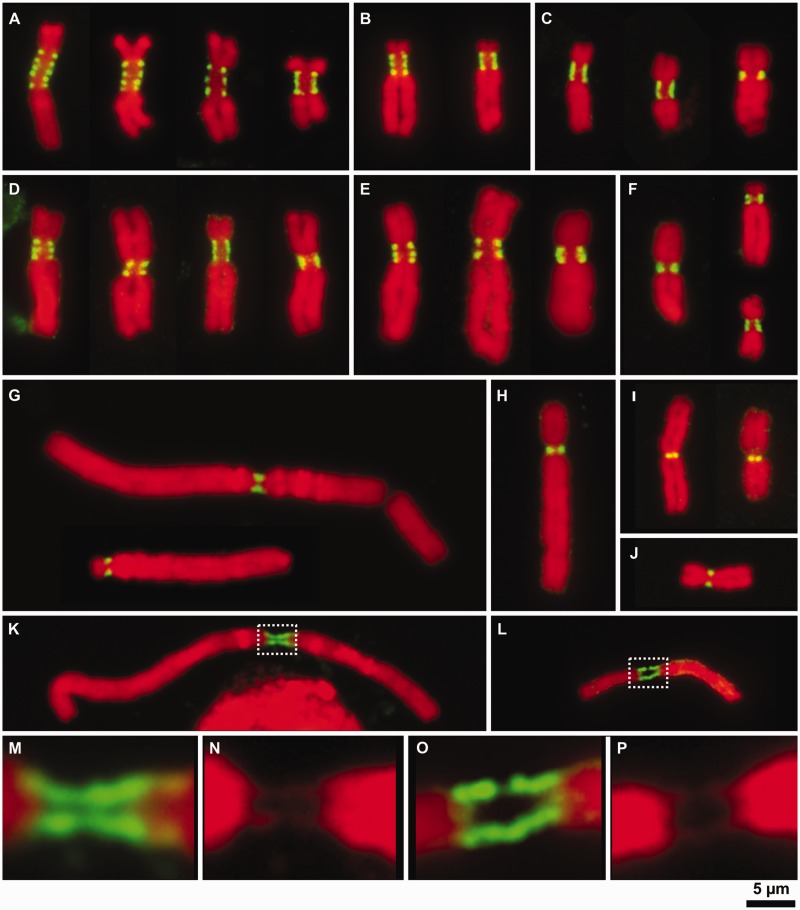
Fig. 3.
Patterns of CenH3 organization on metaphase chromosomes in selected Fabeae species. The organization of CenH3-containing domains was investigated in detail using isolated metaphase chromosomes; the gentle fixation process allowed for maximum sensitivity with the CenH3 antibodies while maintaining good preservation of chromosome morphology. (A) Chromosomes in Lathyrus sativus with the largest primary constrictions displayed 3–5 clearly separated CenH3-containing domains. The beads on a string pattern was clearly more common than the ribbon-like pattern, and we found no chromosomes with single dot-like signals at centromeres. (B) The type of CenH3 pattern in Pisum sativum was highly dependent on the level of chromatin condensation at the centromere, as shown for chromosome 3, which has three CenH3-containing domains that can merge into one contiguous signal. (C) The pattern of CenH3 signal on isolated chromosomes in P. fulvum depends mostly on a chromosome type. Chromosomes with elongated primary constrictions displayed ribbon-like patterns, whereas those with small primary constrictions showed a single dot-like signal. (D and E) Organization of CenH3-containing domains in La. latifolius (D) and La. sylvestris (E). Chromosomes displaying the beads on a string pattern possessed 2–3 CenH3-containing domains, whereas the remainder displayed ribbon-like signals of variable length. (F) Most chromosomes in La. clymenum possessed small primary constrictions displaying a single dot-like signal of CenH3, although two chromosome types showed ribbon-like patterns that appeared to consist of two or three closely adjacent domains. (G–J) All chromosomes in Vicia faba (G), V. pannonica (H), Lens culinaris (I), and V. sativa (J) displayed single dot-like CenH3 signals. (K–P) Stretched centromeres in V. faba (K; details of centromere are shown in M and N) and V. sativa (L; details of centromere are shown in O and P) displaying long, contiguous CenH3 signals along the entire length of the fibers connecting the chromosome arms, suggesting there are no blocks of CenH3-negative chromatin large enough to be detected at this level of resolution. CenH3 was immunodetected with antibodies to CenH3-1_PSat (P. sativum and P. fulvum), CenH3-2_PSat (La. sativus, La. latifolius, La. sylvestris, and La. clymenum), CenH3-2_VF (V. faba, V. sativa and V. pannonica), and CenH3-2_LCul (Le. culinaris). CenH3 signals are shown in green. Chromosomes were counterstained with DAPI (red). Bar = 5 µm.
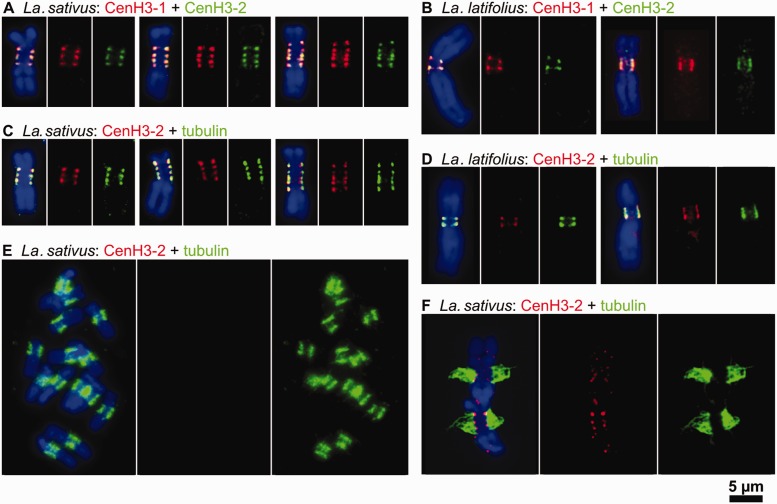
Regardless of pattern type, the CenH3 signals were always present at the poleward surface of the primary constriction. To investigate whether all CenH3 loci of multidomain centromeres were involved in chromosome segregation, simultaneous immunodetection of CenH3 and mitotic spindle protein tubulin (Pepper and Brinkley 1977) was performed on La. latifolius and La. sativus chromosomes. When applied to isolated chromosomes, the antibody against tubulin yielded signals at the surface of the primary constrictions adjacent to the CenH3 domains (fig. 4C and D). Mitotic spindle attachment to all CenH3 domains was evident in squash preparations (fig. 4E and F). Taken together, these results indicate that all CenH3 domains from ribbon-like and metapolycentric centromeres, even in the largest examples, function as sites for kinetochore formation.
Fig. 4.
Two variants of CenH3 in Lathyrus spp. fully colocalize with one another as well as with tubulin. (A and B) Simultaneous detection of CenH3-1 (red) and CenH3-2 (green) in Lathyrus sativus (A) and La. latifolius (B) revealed fully overlapping signals. (C and D) Simultaneous detection of CenH3-2 (red) and tubulin (green) in La. sativus (C) and La. latifolius (D). Note that tubulin signals are found at all CenH3-containing domains. (E and F) Detection of CenH3-2 (red) and tubulin (green) on La. sativus chromosomes prepared using the squash technique. This technique allowed some chromosomes to remain attached to larger microtubule fragments, although CenH3 signals were often missing due to the poor sensitivity of the CenH3 antibody in squash preparations of Lathyrus spp. (E). However, whenever both signals were detected, microtubules could be seen emanating from primary constrictions at all CenH3-containing domains (F). Chromosomes were counterstained with DAPI (blue). Bar = 5 µm.
CenH3 Proteins Are Encoded by Two Different Genes in Pisum and Lathyrus and Colocalize along Extended Centromeres
As two variants of the CenH3 gene were previously discovered in P. sativum (Neumann et al. 2012), we carried out a more thorough survey of CenH3 gene duplications and sequence variability in Fabeae using a combination of experimental and bioinformatics approaches. Using reverse transcription-polymerase chain reaction (RT-PCR) and rapid amplification of cDNA ends (RACE) with several combinations of primers targeted to conserved regions of the CenH3 genes, we identified CenH3-2 orthologs in all Fabeae species, whereas CenH3-1 sequences were detected only in Pisum and Lathyrus species (supplementary table S2, Supplementary Material online). Nonexistence of the CenH3-1 variant in Vicia and Lens was confirmed by absence of CenH3-1 transcripts in the next-generation RNA-sequencing data from Vicia faba, V. sativa, and Lens culinaris (supplementary table S3, Supplementary Material online).
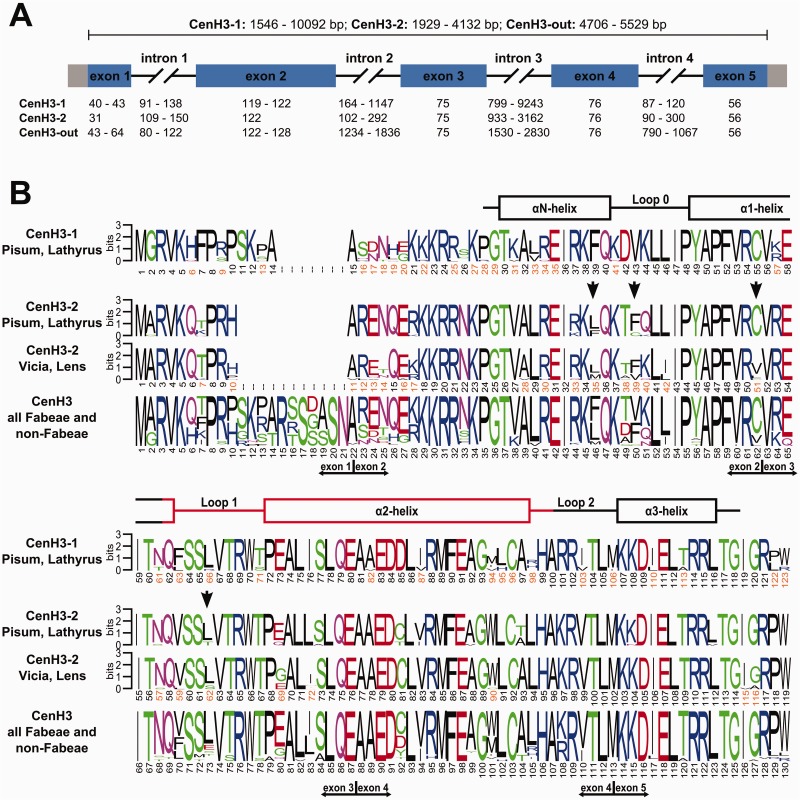
Cloned cDNA sequences from CenH3 transcripts were used to design primers for the retrieval and reconstruction of corresponding genomic loci. A total of 27 gene sequences originating from 18 species were compared, revealing a conserved structure for Fabeae species CenH3 genes. These genes contain five exons encoding proteins of 122–123 (CenH3-1) or 119 (CenH3-2) residues, separated by four introns with positions that are conserved between CenH3 paralogs (fig. 5 and supplementary table S4, Supplementary Material online). However, the overall size of the CenH3 genes varied considerably due to differences in intron sequences, which accumulated numerous indel mutations and ranged in total length from 1,174 to 9,720 bp (supplementary tables S4 and S5, Supplementary Material online).
Fig. 5.
Structure and divergence of CenH3 genes and histones. (A) Schematic of CenH3 genes. The basic structure of the CenH3-1 and -2 genes is highly conserved, consisting of 5 exons (rectangles; coding regions are in blue) of conserved size and four introns of highly variable size (black lines). (B) Sequence logos calculated separately for CenH3-1 and CenH3-2 as well as for all CenH3 sequences used in this study. The secondary structure of the HFD is shown above the logos, as adopted from Tachiwana et al. (2011). The putative centromere targeting domain (CATD) is shown in red. Sites in CenH3-2 that were predicted to evolve under positive selection are marked with vertical black arrows.
The presence and simultaneous transcription of two different CenH3 genes in Pisum and Lathyrus raises questions concerning the role of these proteins in centromere function and localization. Therefore, CenH3-1 and CenH3-2 proteins were analyzed in P. fulvum, La. sativus, and La. latifolius metaphase chromosomes using specific antibodies. Colocalization of the signals from these two antibodies was observed in all three species, indicating that CenH3-1 and CenH3-2 occupy the same domains and that the regions between these domains lack CenH3 of any type (fig. 4A and B and data not shown).
Duplication of the CenH3 Gene Predated the Split of Fabeae Species and Both Genes Evolved under Purifying Selection
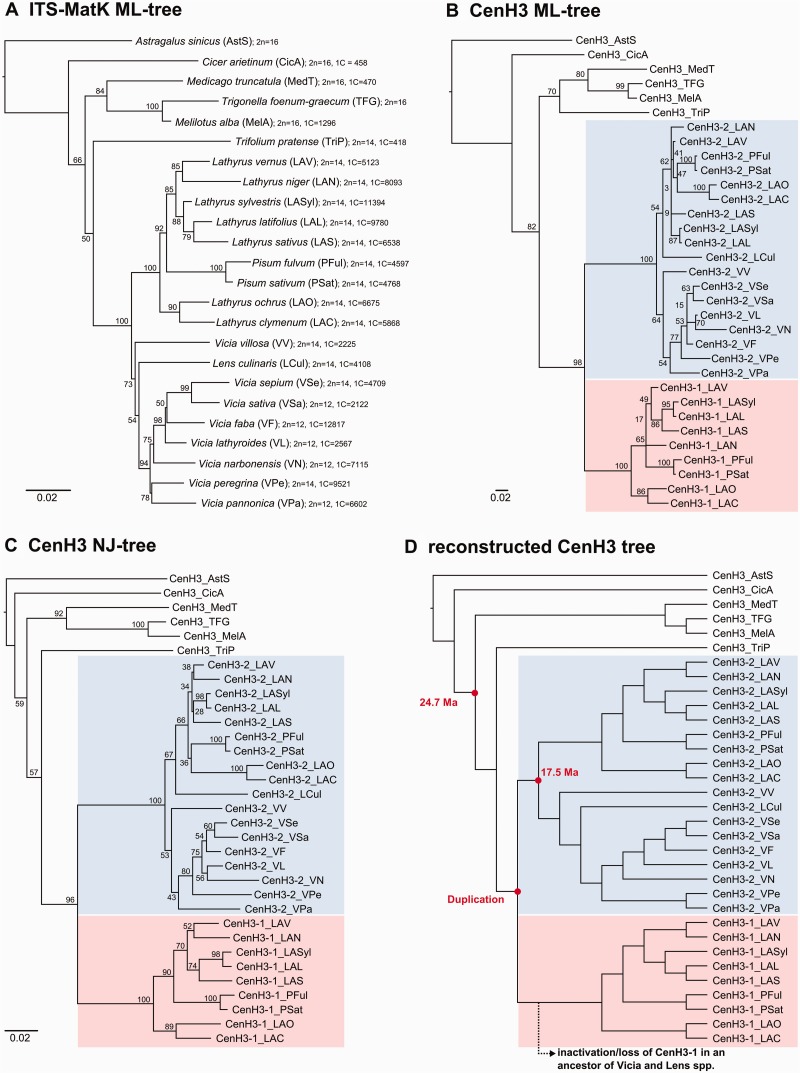
A phylogenetic analysis of CenH3-coding sequences from Fabeae and six outgroup species was performed to elucidate the history of the CenH3 duplication. The average pair-wise similarity of the orthologous proteins (calculated at 117 sites shared by all sequences) was 90.1% in CenH3-1 and 91.7% in CenH3-2, whereas the similarity was 75% between the two paralogous groups. The same calculation carried out for only species possessing both paralogs resulted in an average similarity of 95.0% for CenH3-2, indicating a slower mutation rate compared with CenH3-1. This observation was also reflected in the higher frequency of variable sites in CenH3-1 (32) compared with CenH3-2 (17), which were primarily frequent within the N-terminal regions of both gene variants (fig. 5). Phylogenetic trees inferred from a multiple sequence alignment of the CenH3-coding sequences using neighbor-joining (NJ) and maximum likelihood (ML) algorithms indicated that the two CenH3 variants identified in Pisum and Lathyrus originated from a single duplication event that occurred very early in the evolution of Fabeae, most likely in an ancestor common to all Fabeae species (fig. 6 and supplementary file S1, Supplementary Material online). These phylogenies also indicated that the duplicated genes also existed in the ancestor(s) of Lens and Vicia spp., although the CenH3-1 ortholog was most likely lost before species diversification in the two genera.
Fig. 6.
Phylogenetic trees. (A) A “species tree” based on a comparison of ITS-MatK sequences and inferred using the ML method. The topology of this tree is similar to one published previously (Schaefer et al. 2012) showing Pisum species nested in Lathyrus and Lens culinaris nested in Vicia. The species names are followed by codes that we used to distinguish CenH3 sequences from different species, chromosome number, and haploid genome size (1C) in megabases. The genome sizes were taken from the plant C-value database (Bennett et al. 2000; Bennett and Leitch 2005). (B and C) Phylogenetic trees inferred from a comparison of CenH3 sequences using ML (B) and NJ (C) methods. Note that the placement of CenH3-2_LCul does not fit the genus grouping shown in the species tree. (D) CenH3 tree that was reconstructed to reflect the species phylogeny shown in the tree “A”. Estimates of ages (million years ago) for the two nodes were taken from Lavin et al. (2005).
A variety of approaches and statistical models were employed to evaluate the observed ratios of nonsynonymous (Ka) to synonymous (Ks) nucleotide substitution rates (Ka/Ks = ω) to reveal the type of selective pressure acting on CenH3 genes during their diversification. All-to-all pair-wise, phylogeny-independent comparisons of full-length CenH3-coding sequences using the Ka-Ks calculator estimated ω to be less than 1 for the vast majority of sequence pairs, suggesting purifying selective pressure (supplementary table S6, Supplementary Material online). However, the same analyses carried out separately using sequences coding the C- and N-termini indicated purifying selective pressure for the former and relaxed selective pressure for the latter regions (supplementary tables S7 and S8, Supplementary Material online). Purifying selective pressure was also estimated by ML analysis using the codeml program of PAML, which takes into account the phylogenetic relationships among CenH3-coding sequences. Estimates of ω, calculated as an average over all branches in a tree, varied between 0.283 and 0.290, depending on tree topology (table 1). Further comparisons among the different branches revealed that CenH3-2 orthologs evolved under stronger purifying selective pressure than either the CenH3-1 orthologs or the non-Fabeae CenH3 genes (table 1). To identify potential episodes of positive selection, we estimated ω independently for each branch in the tree. Although some branches were estimated to have ω of greater than 1, none of the estimates was statistically significant (data not shown). These results suggest that positive selection either did not occur during the evolution of CenH3 genes in Fabeae or was limited to only a few sites that could not be detected using the branch models of codeml. To identify potential sites under positive selection, we carried out codeml analyses employing site models of codon substitution. These analyses predicted four sites that may have evolved under positive selection in CenH3-2, whereas none was found in CenH3-1 (table 2). All four sites occurred in the HFD, within the αN-helix, loop0, α1-helix, and loop1 in particular (fig. 5 and supplementary table S9, Supplementary Material online). Importantly, two and one of the sites corresponded to codons that were previously identified as showing adaptive evolution in Brasicaceae (site 51 and 62 [Cooper and Henikoff 2004]) and Caenorhabditis spp. (site 62 [Zedek and Bureš 2012]), respectively (supplementary table S9, Supplementary Material online). In addition, loop1 showed adaptive evolution in Drosophila, which contains a number of nonsynonymous changes in the codon corresponding to site 62 in CenH3-2 (Malik and Henikoff 2001; Malik et al. 2002). In contrast, these sites were invariant among CenH3 sequences from Oryza species (Hirsch et al. 2009) and were highly conserved in CenH3-1 sequences (supplementary table S9, Supplementary Material online). To test whether any of the amino acid replacement sites were positively selected for during the period immediately following the duplication event, we analyzed the particular branches of CenH3-1 and -2 using codeml employing the branch site model. However, results indicated that the early divergence of CenH3-1 and -2 from the ancestral sequence was not due to positive selection (supplementary table S10, Supplementary Material online).
Table 1.
PAML Branch-Specific Models.
| Model | NP | ML | NJ | Reconstructed |
|---|---|---|---|---|
| One-ratio (R1) | 1 | ωCenH3-1 = 0.283 | ωCenH3-1 = 0.290 | ωCenH3-1 = 0.285 |
| ωCenH3-2 = ωCenH3-1 | ωCenH3-2 = ωCenH3-1 | ωCenH3-2 = ωCenH3-1 | ||
| ωCenH3-out = ωCenH3-1 | ωCenH3-out = ωCenH3-1 | ωCenH3-out = ωCenH3-1 | ||
| ln L =−3011.832 | ln L = −3025.45 | ln L = −3089.698 | ||
| Two-ratios (R2) | 2 | ωCenH3-1 = 0.328 | ωCenH3-1 = 0.337 | ωCenH3-1 = 0.335 |
| ωCenH3-2 = 0.227 | ωCenH3-2 = 0.231 | ωCenH3-2 = 0.224 | ||
| ωCenH3-out = ωCenH3-1 | ωCenH3-out = ωCenH3-1 | ωCenH3-out = ωCenH3-1 | ||
| ln L =−3010.042 | ln L = −3023.51 | ln L = −3087.444 | ||
| 2ΔlR2:R1 = 3.579 | 2ΔlR2:R1 = 3.872 | 2ΔlR2:R1 = 4.509 | ||
| P = 0.059 | P = 0.0491 | P = 0.034 | ||
| Three-ratios (R3) | 3 | ωCenH3-1 = 0.321 | ωCenH3-1 = 0.339 | ωCenH3-1 = 0.330 |
| ωCenH3-2 = 0.227 | ωCenH3-2 = 0.231 | ωCenH3-2 = 0.224 | ||
| ωCenH3-out = 0.331 | ωCenH3-out = 0.337 | ωCenH3-out = 0.339 | ||
| ln L =−3010.034 | ln L = −3023.51 | ln L = −3087.438 | ||
| 2ΔlR3:R2 = 0.015 | 2Δl R3:R2 = 0.001 | 2Δl R3:R2 = 0.01 | ||
| P = 0.902 | P = 0.982 | P = 0.915 |
Note.—The ω values were estimated separately for all branches preceding the CenH3 duplication event (CenH3-out) and for both CenH3-1 and CenH3-2 branches following the duplication event. The analysis was performed using three phylogenetic trees that differed partially in topology (see fig. 6B–D). Below the ω estimates are logarithm likelihood values (ln L) and results of LRT (2Δl) with P-values (P) for comparing model R2 with R1 and R3 with R2. Note that the R2 model provided a significantly better fit (P < 0.05) of the data than the R1 model for two trees (marked in underline), indicating stronger purifying selection at the CenH3-2 branch. The R3 model did not provide better fit of the data than R2, suggesting that there is no significant difference between the CenH3-1 and CenH3-out branches. NP is the number of freely estimated ω ratios.
Table 2.
Results of PAML Site Model Comparisons.
| Tree | Gene | Likelihood Ratio Tests (2Δl) of Model Comparisons |
Parameters Estimates |
Positively Selected Sites |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M0/M3 | M1a/M2a | M7/M8 | M0 | M3 | M1a | M2a | M7 | M8 | M2a | M8 | ||
| ML | CenH3-1 | 2Δl = 15.482 | 2Δl = 0.801 | 2Δl = 1.38 | ω = 0.31 | ω0 = 0.000 | ω0 = 0.069 | ω0 = 0.092 | ω ≤ 1 | ω ≤1 | ||
| P = 0.0038 | P = 0.670 | P = 0.502 | ω1 = 0.545 | ω1 = 1.000 | ω1 = 1.000 | ß_p = 0.117 | ωs = 3.897 | |||||
| ω2 = 3.715 | p0 = 0.714 | ω2 = 4.392 | ß_q = 0.234 | p0 = 0.976 | ||||||||
| p0 = 0.493 | p1 = 0.286 | p0 = 0.742 | ps = 0.024 | |||||||||
| p1 = 0.477 | p1 = 0.242 | ß_p = 0.276 | ||||||||||
| p2 = 0.03 | p2 = 0.016 | ß_q = 0.692 | ||||||||||
| NJ | CenH3-1 | 2Δl = 22.461 | 2Δl = 3.431 | 2Δl = 4.125 | ω = 0.335 | ω0 = 0.000 | ω0 = 0.065 | ω0 = 0.089 | ω ≤ 1 | ω ≤ 1 | ||
| P = 0.0002 | P = 0.1798 | P = 0.1271 | ω1 = 0.635 | ω1 = 1.000 | ω1 = 1.000 | ß_p = 0.080 | ωs = 5.309 | |||||
| ω2 = 5.187 | p0 = 0.712 | ω2 = 5.537 | ß_q = 0.153 | p0 = 0.977 | ||||||||
| p0 = 0.525 | p1 = 0.288 | p0 = 0.731 | ps = 0.023 | |||||||||
| p1 = 0.451 | p1 = 0.248 | ß_p = 0.218 | ||||||||||
| p2 = 0.025 | p2 = 0.021 | ß_q = 0.503 | ||||||||||
| Rec. | CenH3-1 | 2Δl = 28.468 | 2Δl = 3.822 | 2Δl = 4.862 | ω = 0.285 | ω0 = 0.132 | ω0 = 0.068 | ω0 = 0.124 | ω ≤ 1 | ω ≤ 1 | ||
| P = 0.0000 | P = 0.1479 | P = 0.0879 | ω1 = 1.430 | ω1 = 1.000 | ω1 = 1.000 | ß_p = 0.064 | ωs = 2.453 | |||||
| ω2 = 3.419 | p0 = 0.738 | ω2 = 2.843 | ß_q = 0.128 | p0 = 0.900 | ||||||||
| p0 = 0.847 | p1 = 0.262 | p0 = 0.822 | ps = 0.1 | |||||||||
| p1 = 0.115 | p1 = 0.110 | ß_p = 1.135 | ||||||||||
| p2 = 0.038 | p2 = 0.068 | ß_q = 5.262 | ||||||||||
| ML | CenH3-2 | 2Δl = 80.124 | 2Δl = 10.663 | 2Δl = 12.021 | ω = 0.245 | ω0 = 0.051 | ω0 = 0.05 | ω0 = 0.052 | ω ≤ 1 | ω ≤ 1 | 35 | 35, 39, 51, 62 |
| P = 0.0000 | P = 0.0048 | P = 0.0025 | ω1 = 0.978 | ω1 = 1.000 | ω1 = 1.000 | ß_p = 0.111 | ωs = 5.330 | |||||
| ω2 = 5.757 | p0 = 0.806 | ω2 = 5.807 | ß_q = 0.374 | p0 = 0.987 | ||||||||
| p0 = 0.797 | p1 = 0.194 | p0 = 0.800 | ps = 0.013 | |||||||||
| p1 = 0.192 | p1 = 0.189 | ß_p = 0.130 | ||||||||||
| p2 = 0.011 | p2 = 0.011 | ß_q = 0.464 | ||||||||||
| NJ | CenH3-2 | 2Δl = 80.666 | 2Δl = 10.816 | 2Δl = 11.912 | ω = 0.249 | ω0 = 0.04 | ω0 = 0.045 | ω0 = 0.046 | ω ≤ 1 | ω ≤ 1 | 35 | 35, 39, 51, 62 |
| P = 0.0000 | P = 0.0045 | P = 0.0026 | ω1 = 0.899 | ω1 = 1.000 | ω1 = 1.000 | ß_p = 0.102 | ωs = 5.634 | |||||
| ω2 = 5.868 | p0 = 0.790 | ω2 = 6.131 | ß_q = 0.330 | p0 = 0.988 | ||||||||
| p0 = 0.766 | p1 = 0.21 | p0 = 0.783 | ps = 0.012 | |||||||||
| p1 = 0.223 | p1 = 0.207 | ß_p = 0.116 | ||||||||||
| p2 = 0.011 | p2 = 0.011 | ß_q = 0.396 | ||||||||||
| Rec. | CenH3-2 | 2Δl = 95.756 | 2Δl = 14.758 | 2Δl = 15.579 | ω = 0.229 | ω0 = 0.035 | ω0 = 0.036 | ω0 = 0.037 | ω ≤ 1 | ω ≤ 1 | 35 | 35, 39, 51, 62 |
| P = 0.0000 | P = 0.0006 | P = 0.0004 | ω1 = 0.95 | ω1 = 1.000 | ω1 = 1.000 | ß_p = 0.09 | ωs = 6.722 | |||||
| ω2 = 6.912 | p0 = 0.786 | ω2 = 7.023 | ß_q = 0.301 | p0 = 0.991 | ||||||||
| p0 = 0.772 | p1 = 0.214 | p0 = 0.779 | ps = 0.009 | |||||||||
| p1 = 0.219 | p1 = 0.212 | ß_p = 0.095 | ||||||||||
| p2 = 0.009 | p2 = 0.009 | ß_q = 0.322 | ||||||||||
note.—Likelihood ratio statistics for comparing models representing different hypothesis concerning the variability of selective pressure among sites: M0 (uniform selective pressure among sites; one ω ratio), M1a (variable selective pressure but no positive selection; ω0 < 1, ω1 = 1), M2a (variable selective pressure with positive selection; ω0 < 1, ω1 = 1, ω2 > 1), M3 (variable selective pressure among sites; discrete distribution (three classes): ω0, ω1, ω2), M7 (beta distributed variable selective pressure; 0 ≤ ω ≤ 1), and M8 (beta distributed variable selective pressure plus positive selection; 0 ≤ ω ≤ 1 plus one discrete class of ωs > 1). Proportions of individual classes of ω values described above are marked p0, p1, p2, and ps. β_p and β_q in models M7 and M8 are parameters of the beta distribution. The analyses were carried out separately for CenH3-1 and CenH3-2 using three alternative topologies of the phylogenetic trees. An LRT comparing M0 with M3 was used to test for variable selective pressure among sites. The M3 model provided a significantly better fit (P < 0.05) of the data than the M0 model in both CenH3 paralogs regardless of tree topology, suggesting that type of selection pressure differed among the sites. Two LRTs were conducted to test for sites evolving under purifying selection, comparing model M2a against M1a, and M8 against M7. For CenH3-2 but not for CenH3-1, the M2a and M8 models fitted (P < 0.05) the data significantly better (P < 0.05) than the M1a and M7 models, respectively. Sites evolving under positive selection in CenH3-2 were predicted using Bayes empirical Bayes method. Only site 35 was predicted with high posterior probability (0.997–1.0) regardless of tree topology and model. Three additional sites were predicted only using the M8 model and had low posterior probabilities (site 39: 0.51–0.74; 51: 0.5–0.64, 62: 0.59–0.75).
Despite Sequence Diversification, CenH3 Paralogs Target to Centromeres in Heterologous Fabeae Species
To study functionality of the P. sativum CenH3-1 gene in species lacking this gene variant, a fusion protein constructed using the CenH3-1_PSat fragment with reporter yellow fluorescent protein (YFP) sequence (CenH3-1_PSat-YFP) was expressed in transgenic hairy root cultures of V. faba. Regardless of the type of the fusion (N-terminal or C-terminal), 12 spots of YFP fluorescence most likely corresponding to centromeres of V. faba were visible in interphase nuclei of live cells (supplementary fig. S2, Supplementary Material online). Similar results were obtained when a fusion protein constructed using V. faba CenH3-2 gene (CenH3-2_VF-YFP) was expressed in transgenic roots of P. sativum (supplementary fig. S2, Supplementary Material online). In contrast, diffuse fluorescence with no distinct spots was observed in control variants transformed with fusion constructs containing the CenH3 gene from Arabidopsis thaliana (data not shown).
Discussion
Two distinct types of centromere organization are generally recognized in higher plants (Malik 2009; Melters et al. 2012). The regional centromeres of monocentric chromosomes are characterized by CenH3 deposition at a specific region which constitutes narrow primary constriction on metaphase chromosomes. To date, the sizes of these regions have only been estimated in rice and maize, which were between 0.4 and 3.2 Mb (Yan et al. 2008; Wolfgruber et al. 2009). Holocentric chromosomes represent a second type of centromere morphology, with CenH3 protein and kinetochore formation spread along the entire chromosome length. CenH3 is located in longitudinal grooves formed along the chromatids, and thus no primary constrictions occur on holocentric chromosomes (Heckmann et al. 2011). In higher plants, the former centromere type is much more common than the latter, which has only been reported in species from five families (Cyperaceae, Juncaceae, Melanthiaceae, Convolvulaceae, and Droseraceae) (Melters et al. 2012). It is thought that holocentric chromosomes originated from monocentric ones independently in several taxa, although the causes and mechanisms of these transitions are unknown (Melters et al. 2012). Moreover, no intermediate centromere type had been reported and neither of these basic types display significant structural variability. Therefore, the extensive variation in centromere size and morphology reported here for the tribe Fabeae is unlike that in any other group of plants studied to date, and thus may provide a new model system for studying centromere evolution.
Although it has been shown that the regional centromeres typical of most eukaryotes are composed of intermingled arrays of CenH3-containing and CenH3-negative chromatin (Sullivan and Karpen 2004; Black and Bassett 2008; Jin et al. 2008), they always form a single compact CenH3-containing domain on metaphase chromosomes (Jin et al. 2004; Nagaki et al. 2004; Nagaki and Murata 2005; Houben et al. 2007; Liu et al. 2008; Nagaki, Kashihara et al. 2009; Tek et al. 2010, 2011; Gong et al. 2012). An investigation of centromere total size variation revealed a strong correlation with the genome size and moderate but significant correlation with average chromosome size (Zhang and Dawe 2012). Intriguingly, when maize chromosomes were transferred into the oat genome, which is approximately four times larger than maize, their centromeres expanded dramatically, suggesting the flexible regulation of centromere size and a tendency toward uniformity regardless of chromosome size (Wang et al. 2014). Although big differences in reactivity of our antibodies to divergent CenH3 histones made it impossible to accurately quantify and compare amounts of CenH3 among species, our data clearly show that the enormous differences in the size of primary constrictions and number of CenH3-containing domains in the Fabeae spp. do not correlate with either genome or chromosome size. For instance, La. sativus (haploid genome size [1C] = 6.5 Gb), which has enormous primary constrictions with three to five CenH3-containing domains, has a smaller genome size than La. sylvestris (1C = 11.4 Gb) and La. latifolius (1C = 9.8 Gb), which have significantly shorter primary constrictions with one to three CenH3-containing domains (figs. 1, 2N, O, 3A, D, and E, and 6A and supplementary fig. S1, Supplementary Material online). In addition, P. sativum (1C = 4.8 Gb) and Le. culinaris (1C = 4.1 Gb) differ markedly in the length of primary constrictions as well as number of CenH3-containing domains even though they have similar genome sizes (fig. 2L and S and supplementary fig. S1, Supplementary Material online). Finally, Vicia species vary greatly in genome size, including V. faba (1C = 12.8 Gb), V. pannonica (1C = 6.6 Gb), and V. sativa (1C = 2.1 Gb), without significant differences in the length of primary constrictions or the number of CenH3 signals (fig. 2I–K and P–R and supplementary fig. S1, Supplementary Material online). These findings indicate that centromere expansion and genome expansion occurred independently in Fabeae and that large chromosomes do not require longer primary constrictions and more CenH3-containing domains or vice versa.
It should be noted that vast majority of chromatin within the primary constrictions of Pisum and Lathyrus species apparently lacks CenH3 (figs. 2 and 3). The association of individual CenH3-containing domains with specific families of satellite repeats, inferred from their precise colocalization (Neumann et al. 2012), allowed us to estimate the proportion of CenH3 chromatin in the centromeres of several metapolycentric chromosomes from P. sativum. Using previously published data concerning the genomic abundance of P. sativum satellites (Macas et al. 2007; Neumann et al. 2012), it was estimated that CenH3-containing domains associated with TR18, TR20, and TR21/22/23 repeats on chromosomes 3, 7, and 6 span 0.57, 0.19, and 0.5 Mb, respectively. Using the same approach, the average size of two CenH3-associated clusters of TR7 on chromosome 1 was 2.9 Mb. Assuming that the sizes of the other CenH3-binding loci are similar and that there are three to five of these in the primary constrictions, comprising 69 and 107 Mb (Neumann et al. 2012), then we predict that the total proportion of CenH3-associated DNA in the primary constrictions of P. sativum is on the order of a few percentage points, and most likely does not exceed 15%.
According to the centromere drive model, it has been proposed that the evolution of centromere size is driven by a conflict between centromeric DNA and CenH3 (Malik 2009; Roach et al. 2012). The model predicts that centromeric repeats change and/or expand to increase their segregation properties while CenH3 changes adaptively to restore equal segregation frequencies. Thus, the adaptive evolution of CenH3 genes should be strongest in the evolutionary history of species that differ largely in centromere size, such the Fabeae species. Contrary to this prediction, only four of 33 variable sites in the protein sequences of CenH3-2 showed adaptive evolution and none was predicted among the 55 CenH3-1 variable sites (table 2 and fig. 5), indicating that the majority of the variability among Fabeae CenH3 sequences is not due to positive selection. CenH3 histones in Brassicaceae and Caenorhabditis spp. were predicted to evolve adaptively at 12 and 11 sites, respectively (Cooper and Henikoff 2004; Zedek and Bureš 2012). Although two of the sites predicted in CenH3-2 matched those found in Brassicaceae and Caenorhabditis spp. (supplementary table S9, Supplementary Material online), we did not find any correlation between centromere expansion and the changes at these adaptively evolving sites in CenH3-2. For example, the CenH3-2 sequences in P. sativum and P. fulvum are identical, yet some of the centromeres in the former species are larger. In addition, centromeres in La. sativus are much larger than in La. vernus, although both species share the same amino acid residues at the adaptively evolving sites. Together, these data suggest that the enormous expansion of centromeres in Pisum and Lathyrus evoked relatively little to no response in the adaptive selection of CenH3 genes. These findings are not necessarily in conflict with centromere drive model because there are other proteins beside CenH3 which may be positively selected to suppress centromere drive. Indeed, adaptive evolution has already been demonstrated for kinetochore protein Cenp-C in both plants and animals, providing an explanation for absence of adaptive evolution in CenH3 from mammals and grasses (Talbert et al. 2004). We should also mention that it remains possible, that we did not detect all adaptively evolving sites because the power and accuracy of PAML to detect such sites depends strongly on the data set being analyzed as well as on the strength of positive selection (Anisimova et al. 2001). Therefore, further research is still needed to unravel the entire extent of positive selection in CenH3 evolution in Fabeae species and to finally prove or refute a causal relationship between changes in the size or DNA sequence composition of centromeres and changes in CenH3. One line of research that might shed light on this relationship is investigation of localization of CenH3 expressed in heterologous species that differ considerably in the properties of their centromeres. Centromeric localization of CenH3-1_PSat-YFP and CenH3-2_VF-YFP fusion proteins in V. faba and P. sativum (supplementary fig. S2, Supplementary Material online), respectively, indicated that the sequence differences between the CenH3 histones of these two species, some of which occurred at the adaptively evolving sites, did not abolish centromere targeting in the heterologous species. This is in agreement with a recently published study which demonstrated that even CenH3 from phylogenetically very distant species can functionally complement A. thaliana CenH3, indicating that essential functions of CenH3 are conserved across a broad evolutionary landscape (Maheshwari et al. 2015). Importantly, although the plants possessing the variant CenH3 were viable and fertile when selfed, they showed dramatic segregation errors in zygotic mitosis when crossed to a wild-type (Maheshwari et al. 2015). This suggests that naturally occurring differences in CenH3 may be an adaptation to lineage-specific cellular, most likely centromeric, environment. However, the molecular basis of this adaptation is unclear.
Metapolycentric organization of CenH3-containing domains is yet another puzzling feature of Pisum and Lathyrus centromeres. In general, di- or multicentric chromosomes are unstable unless the distance between the centromeres allows them to function together as a single centromere or because there is an unknown mechanism ensuring that kinetochores on the same chromatid attach to the same spindle pole (Higgins et al. 2005; Zhang et al. 2010). Observed transitions from beads on a string to ribbon-like patterns of CenH3 distribution during metaphase chromosome condensation suggests that the former mechanism ensures chromosome stability in most Pisum and Lathyrus species. However, the preservation of the beads on a string pattern throughout metaphase and anaphase in La. sativus suggests that distant CenH3-containing domains do not necessarily have to merge to ensure attachment to the same spindle pole. Stable dicentric chromosome was found in maize, although the two CenH3-containing domains are only separated by approximately 2.8 Mb (Wolfgruber et al. 2009). The largest documented distance between two functional centromeres was in engineered human dicentric chromosomes, being located approximately 20 Mb apart (Higgins et al. 2005). Considering that the primary constrictions in La. sativus, which contain 3–5 CenH3-containing domains, were estimated to span 166–263 Mb, the average distance between these domains is likely 2–3 times larger.
Considering the lack of an obvious correlation between centromere and genome/chromosome size and the existence of intraspecific variation in centromere morphology, it is possible that the metapolycentric centromeres of Pisum and Lathyrus represent a dynamic intermediate state between monocentric and polycentric chromosomes. Although the functions and mechanisms of centromere expansion are not clear from the available data, it is striking that this phenomenon occurred only in those Fabeae species with two CenH3 genes. The presence of two CenH3 genes has been demonstrated in only a very few diploid species, including A. lyrata, Hordeum bulbosum, H. vulgare, Lu. nivea, and C. elegans (Monen et al. 2005; Kawabe et al. 2006; Moraes et al. 2011; Sanei et al. 2011). It is notable that Lu. nivea and C. elegans possess holocentric chromosomes and that the regional centromeres of H. vulgare are significantly larger compared with maize, which only has a single CenH3 gene (Zhong et al. 2002; Zhang and Dawe 2012). In contrast, most eukaryotic species, including some that have undergone whole-genome duplications, possess just a single CenH3 gene (Hirsch et al. 2009; Malik 2009). This suggests that one of duplicated copies of CenH3 gene is subsequently degenerated or eliminated in most cases. Indeed, we found evidence for such an event in the legume species Glycine max (soybean), which possesses one functional CenH3-coding gene on chromosome 7 and truncated fragments of at least two others on chromosomes 16 and 17 (data not shown).
Phylogenetic analyses of the CenH3 genes presented in this study suggest that CenH3-1 and CenH3-2 paralogs originated from a single duplication event that either predated the emergence of Fabeae or occurred very early in the evolution of this tribe more than 17.5 Ma (fig. 6). This period was long enough for noncoding intron sequences to evolve considerably among the Fabeae species due to frequent substitutions and indel mutations (supplementary tables S4 and S5, Supplementary Material online). However, the coding sequences of both CenH3 variants were relatively conserved and were found to evolve under purifying selection. This strongly suggests that both CenH3 paralogs are still required for proper centromere function in Pisum and Lathyrus spp. To explain the preservation of such duplicated genes, three major outcomes of duplication that maintain the new copy can be considered: neofunctionalization, subfunctionalization, and conservation of function (Hahn 2009). Neofunctionalization, or the development of a completely new function, appears unlikely, as both CenH3 variants fully colocalize, which is more consistent with conserved CenH3 function as a constitutive epigenetic mark of centromeric chromatin (Torras-Llort et al. 2009). Subfunctionalization, or the division of functions between paralogous genes, can occur to varying degrees. At the protein level, we consider this rather unlikely due to the relatively high sequence similarity of the CenH3-2 proteins in Pisum and Lathyrus spp. to the CenH3-2 proteins of Vicia and Lens spp., suggesting the maintenance of all ancestral functions. The conservation of function model assumes that functions of the duplicated genes have remained unchanged over the course of evolution. If true, then the positive impact of the presence of two genes is usually due to a dosage effect. Intriguingly, there has been a number of reports documenting the relationship between increased expression and centromere spreading or CenH3 mislocalization to ectopic sites (Van Hooser et al. 2001; Tomonaga et al. 2003; Heun et al. 2006; Burrack et al. 2011; Roy et al. 2011). On the other hand, the potential dosage effect of CenH3 duplication alone cannot account for the dramatic increase in centromere size and variability among certain Pisum and Lathyrus species.
Although no final conclusions can be made to explain the dramatic centromere expansion observed in Pisum and Lathyrus, or its relationship to the presence of two CenH3 paralogs, these observations point the way toward new investigations into centromere function and evolution in plants. Various strategies can be envisioned for future work on Fabeae centromeres, including surveys of DNA sequence composition in CenH3-containing domains to uncover potential roles for repetitive sequences in centromere expansion, inactivation or modification of the CenH3 paralogs to study their roles in centromere function and evolution, and investigations into the meiotic behavior of chromosomes with large centromeres.
Materials and Methods
Plant Material
Seeds of the plants used in this study were obtained from Selgen, Stupice, Czech Republic (P. sativum cv. Terno), Osiva, Boršov nad Vltavou, Czech Republic (Trifolium pratense cv. Tábor, V. faba cv. Merkur and V. pannonica cv. “Dětenická panonská”), Research Institute for Fodder Crops, Troubsko, Czech Republic (Cicer arietinum cv. Irenka, Melilotus alba, cv. Adéla, and Trigonella foenum-graecum cv. Hanka), Nohel Garden, Dobříš, Czech Republic (Le. culinaris cv. Eston), IPK Gaterslaben, Germany (V. lathyroides [VIC 874], V. peregrina [VIC 765], V. sepium [VIC 55]), Central Institute for Supervising and Testing in Agriculture, Brno, Czech Republic (V. villosa cv. Modra), Agriculture Research Institute, Kromeříž, Czech Republic (V. sativa cv. Ebena), Crop Research Institute, Prague-Ruzyně, Czech Republic (Medicago truncatula cv. Jemalong), Fratelli Ingegnoli, Milano, Italy (La. sativus), and SEMO, Smržice, Czech Republic (La. latifolius). Mature La. vernus and La. niger plants were purchased from Arboretum Paseka Makču Pikču, Paseka, Czech Republic. Seeds of La. sylvestris were harvested from natural populations at Vidov (Czech Republic; GPS: 48°54′51.238″N, 14°29′50.680″E). Seeds of P. fulvum (NS-15 IG 64207), La. ochrus (ATC 80432), and La. clymenum (RBGE 541) were a gift from Dr Petr Smýkal (Palacký University, Olomouc, Czech Republic). Seeds of V. narbonensis (ICARDA 14) were provided by Dr Anna Torres (IFAPA, Córdoba, Spain).
Antibodies
Antibodies raised previously to CenH3-1_PSat and CenH3-2_PSat (Neumann et al. 2012) were used to test Pisum and Lathyrus species. As these two antibodies failed to detect any signals in squash preparations of mitotic chromosomes and interphase nuclei in Vicia and Lens spp. (data not shown), we raised another two antibodies to the CenH3-2 histones of Le. culinaris and V. faba. These antibodies were custom raised (Genscript, Piscataway, NJ) in rabbits using peptides from CenH3-2_VF (CQT PRH ARE TQE KKK RRN KPG) and CenH3-2_LCul (PRP VLQ NQE RKK RRN KPG C). The mouse antibody to α-tubulin was purchased from Sigma-Aldrich (St. Louis, MO; catalog number T6199).
Chromosome Preparation and CenH3 Immunodetection
The immunostaining experiments were carried out using chromosomes prepared from root tip meristem cells synchronized with 1.25–2.5 mM hydroxyurea and blocked at metaphase with 15 µM oryzalin or 2.5 µM amiprophos-methyl (APM), as described previously (Neumann et al. 2002). Modifications to the protocol are shown in supplementary table S11, supplementary Material online.
Centromere morphology was assessed in mitotic chromosome preparations prepared according to Dong et al. (2000), with a few modifications. Synchronized root tip meristems were fixed in 3:1 fixative (3:1 solution of methanol and glacial acetic acid) for 2 days at 4°C. After washing twice in ice-cold distilled water (5 min each), the meristems were cut off and incubated in a solution of 4% cellulase ONOZUKA R10 (SERVA Electrophoresis, Heidelberg, Germany), 2% pectinase, and 0.4% pectolyase Y23 (both MP Biomedicals, Santa Ana, CA) in 0.01 M citrate buffer (pH 4.5) at 37°C for 1–2 h. The root tips were then washed carefully with ice-cold distilled water three times for 5 min each and then postfixed in the 3:1 fixative solution for 1 day at 4°C. One to five meristems were transferred to a glass slide, macerated in a drop of 3:1 fixative using a fine-pointed forceps and a scalpel, and then warmed over an alcohol flame. After air-drying, the slides were immediately stained with 4′,6-diamidino-2-phenylindole (DAPI) and mounted in Vectashield mounting medium (Vector Laboratories, Burlingame, CA).
Chromosome isolation was performed as described previously (Neumann et al. 2002). Isolated chromosomes were spun on slides using cytospin columns, and the slides were washed in 1 × phosphate buffered saline (PBS) for 5 min. The squash preparations for CenH3 immunodetection were performed in LB01 buffer (Doležel et al. 1989) by squashing synchronized root tip meristems fixed in 4% formaldehyde for 25 min at 20°C and digested with 2% cellulase ONOZUKA R10 and 2% pectinase in 1 × PBS for 30-120 min at 28°C. The squash preparations were washed in 1 × PBS for 5 min, 1 × PBS-T1 buffer (1 × PBS and 0.5% Triton, pH 7,4) for 25 min, and twice in 1 × PBS for 5 min. Prior to incubation with the primary antibody, the slides were incubated in PBS-T2 buffer (1 × PBS and 0.1% Tween 20, pH 7,4) for 30 min at room temperature (RT). The slides were incubated with primary antibodies diluted in PBS-T2 overnight at 4°C. Dilution ratios were 1:1,000–5,000 for the CenH3 antibodies and 1:100 for the α-tubulin antibody. Following two washes in 1 × PBS for 5 min and one wash in 1 × PBS-T2 for 5 min, the antibodies were detected with anti-rabbit-Rhodamine Red-X-AffiniPure (1:500, Jackson ImmunoResearch, Suffolk, UK; catalog number 111-295-144), anti-chicken-DyLight488 (1:500, Jackson ImmunoResearch; catalog number 103-485-155) or anti-mouse-FITC (1:100, Abcam, Cambridge, UK; catalog number ab6785) in PBS-T2 buffer for 1 h at RT. After two washes in 1 × PBS for 5 min and one wash in 1 × PBS-T2 for 5 min, the slides were counterstained with DAPI and mounted in Vectashield mounting medium. The chromosomes were examined using a Nikon Eclipse 600 microscope. Images were captured using a DS-Qi1Mc cooled camera and analyzed using the NIS Elements 3.0 software program (Laboratory Imaging, Praha, Czech Republic).
Estimation of Centromere Size in La. sativus
Chromosome sizes in megabases were estimated from the relative chromosome lengths of individual chromosomes and a haploid genome size of 6,538 Mb (Bennett and Leitch 2005) using the following formula: genome size × relative chromosome length. Centromere size was estimated using chromosomes stained with the DNA-binding fluorescent dye DAPI as the proportion of integrated fluorescence intensity at the primary constriction site compared with that of the whole chromosome. Images from nine complete sets of metaphase chromosomes were used to measure both relative chromosome lengths and fluorescent intensities. Image analyses were performed using the NIS Elements 3.0 software program.
Identification and Cloning of CenH3 Genes
Internal fragments of CenH3-coding sequences were amplified using RT–PCR with degenerate primers based on the most conserved regions of previously characterized sequences from Viciaceae species, including P. sativum, M. truncatula, Lotus japonicus, and G. max, as well as from sequences obtained during the course of this work. Total RNA was isolated from leaves using Trizol reagent (Invitrogen, Carlsbad, CA) and treated with DNase I (Ambion, Austin, TX). First strand synthesis was performed using the SuperScript III First-Strand Synthesis System for RT-PCR kit (Invitrogen, Carlsbad, CA) or Transcriptor High Fidelity cDNA Synthesis kit (Roche, Basel, Switzerland), according to the manufacturer’s recommendations with random hexamers or oligo dT as primers. The obtained partial CenH3-coding sequences were used to design species-specific primers which were then used for 5′- and 3′-RACE to obtain sequences at 5' and 3' ends of the CenH3 transcripts. Both types of RACE were carried out using the GeneRacer kit (Invitrogen, Carlsbad, CA, USA), as described previously (Neumann et al. 2007). Full-length CenH3-coding sequences were obtained using RT-PCR with species-specific primers. Lists of successful primers and their combinations are available in supplementary file S2 and table S2, Supplementary Material online, respectively. All amplification products were cloned into the pCR4-TOPO or pCR8/GW/TOPO vectors (Invitrogen, Carlsbad, CA).
Transcriptome sequence data for P. sativum (SOLiD, total RNA) and V. faba (Illumina, mRNA) were generated in our lab and deposited in the short read archives (accession numbers are shown in supplementary table S3, Supplementary Material online). Additional transcriptome data for P. sativum (Study accession: SRP006313), V. sativa (SRP010681), and Le. culinaris (SRP007008), were downloaded from http://www.ebi.ac.uk/ena/ (last accessed March 23, 2015). Assemblies of the reads from the transcriptome sequencing were constructed using the Trinity software program (Grabherr et al. 2011). Searches for CenH3 sequences were carried out using tBLASTn and BLASTn with all CenH3 sequences identified in this study used as queries.
Long fragments of CenH3 genes were amplified by PCR from genomic DNA using LA DNA polymerase (Top-Bio, Prague, Czech Republic). Genomic DNA was extracted from young leaves as described by Dellaporta et al. (1983). PCR primers are shown in supplementary table S2, Supplementary Material online, and the composition of reaction mixture was as described previously (Neumann et al. 2012). As reverse transcription and PCR can introduce sequence errors, all final CenH3-coding sequences used in this analysis were validated by comparing all sequences obtained from the respective species. The validation process allowed us to create a highly reliable sequence data set suitable for further analysis. Accession numbers for the CenH3 sequences obtained experimentally in this study, as well as those downloaded from the GenBank, are provided in supplementary table S2, Supplementary Material online.
Sequence Analysis
Sequence analysis was conducted using the EMBOSS, Staden and MEGA5.2 (Staden 1996; Rice et al. 2000; Tamura et al. 2011), multiple alignments were performed using Muscle (Edgar 2004), and pair-wise alignments were performed using the Stretcher software program (Myers and Miller 1988). Similarity searches were performed using BLAST software (Altschul et al. 1990). Sequence logos were generated using Weblogo (Crooks et al. 2004). Phylogenetic analyses were performed using NJ and ML algorithms implemented in the SeaView (Galtier et al. 1996) and PhyML 3.0 (Guindon et al. 2010) programs, respectively. Bootstrap values were calculated from at least 1,000 replications. Phylogenetic trees were drawn and edited using the FigTree program (http://tree.bio.ed.ac.uk/software/figtree/, last accessed March 23, 2015). The reconstructed tree was created using TreeGraph (Stover and Muller 2010). The combined alignment of ITS and MatK sequences used to infer the species tree is provided in supplementary file S3, Supplementary Material online.
Tests for Selective Pressure
The type of selective pressure acting on the CenH3 genes was assessed by estimating the ratio between synonymous (Ks) and nonsynonymous (Ka) nucleotide substitution rate. An omega ratio (ω = Ka/Ks) = 1 indicates neutral evolution, whereas ω < 1 and ω > 1 indicates purifying (i.e., stabilizing) selection and positive selection (i.e., adaptive evolution), respectively. Estimation of ω from all-to-all pair-wise sequence comparisons was performed using the KaKs_Calculator (Zhang et al. 2006). P-values were computed using the Fischer exact test, and only estimates with P-values < 0.05 were considered to be statistically significant.
More sophisticated tests of selective pressure were carried out using the codeml software by employing branch, site and branch-site models, as implemented in PAML4.7 (Yang 2007). This program allowed us both to estimate the selective pressure at different times in the phylogenetic history of the CenH3 genes and to statistically test hypotheses using the likelihood ratio test (LRT). LRT was performed by taking twice the log likelihood difference between two nested models (2Δl = 2 × (l1–l0); where l1 and l0 are ML estimates for the test and null model, respectively) representing null and alternative hypotheses concerning selective pressure, and then comparing this value to the chi-square distribution, with degrees of freedom equal to the difference in a number of parameters for the two models being compared. The test was considered statistically significant if the P-value < 0.05. Whenever applicable, analyses using codeml were performed multiple times with different initial values for omega (0.001, 0.01, 0.1, 0.25, 0.5, 0.75, 1, 2, 3, 4, 5, or 10) to obtain a globally optimum likelihood score. To minimize bias due to tree topology, all PAML analyses were carried out using both CenH3-inferred trees (fig. 6B and C), as well as the tree reconstructed from the species tree inferred from ITS-MatK sequences (fig. 6D).
Branch models were employed to determine whether selection pressure differed among branches. A free ratio model was used to estimate independent ω values for each branch in a tree. The statistical significance of ω estimates was determined for each individual branch with ω > 1 using the strict branch test. In this test, a two ratio model (R2) was used to estimate one ω value for the branch of interest and another for all other branches. The LRT was conducted by comparing a test model allowing ω > 1 (positive selection) with a null model having ω fixed to 1 (neutral evolution) at the specified branch. The R2 and three ratio (R3) models were employed to estimate independent ω ratios for two and three specified sets of branches, respectively. To test for statistical significance for R2, an LRT was performed by comparing this model with a one ratio model (R1), assuming no variation in selection pressure among branches (i.e., same ω for all branches). In addition, an LRT comparing R3 with R2 was performed to test the statistical significance of the R3 model.
To test for differences in selection type among the sites and to identify potential sites that evolved under positive selection, we employed the following site models of codon substitution representing various hypotheses about selective pressure: M0 (uniform selective pressure among sites; one ω ratio), M1a (variable selective pressure but no positive selection; ω0 < 1, ω1 = 1), M2a (variable selective pressure with positive selection; ω0 < 1, ω1 = 1, ω2 > 1), M3 (variable selective pressure among sites; discrete distribution (three classes): ω0, ω1, ω2), M7 (beta distributed variable selective pressure; 0 ≤ ω ≤ 1) and M8 (beta distributed variable selective pressure plus positive selection; 0 ≤ ω ≤ 1 plus one discrete class of ωs > 1). An LRT comparing M0 with M3 was used to test for variable selective pressure among sites. Two LRTs were conducted to test for sites undergoing positive selection, comparing model M1a against M2a, and M7 against M8. Posterior probabilities of the sites predicted to evolve under positive selection were calculated using Bayes empirical Bayes method.
A branch-site test was employed to identify potential sites evolving under positive selection at either of two branches immediately following the duplication event. A codeml analysis was carried out using the branch-site model A, and the results were evaluated using branch-site test 2, as described previously (Zhang et al. 2005).
Fusion Constructs and Transformation
RT-PCR-amplified fragments encoding CenH3-1_PSat and CenH3-2_VF were cloned into the pCR8/GW/TOPO entry vectors using the pCR8/GW/TOPO TA Cloning Kit (Invitrogen, Carlsbad, CA). Fragments in the appropriate orientation were subsequently recombined into the destination vectors pEarleyGate104 and pEarleyGate101 (obtained from TAIR; http://www.arabidopsis.org/, last accessed March 23, 2015), allowing for C- and N-terminal fusions with YFP, respectively. The recombination reactions were carried out using Gateway LR Clonase II Enzyme Mix (Invitrogen, Carlsbad, CA), according to the manufacturer's instructions. Nucleotide sequences of all constructs were verified by sequencing. Transgenic hairy root cultures expressing the fusion constructs were obtained as described previously (Neumann et al. 2012). Images of transgenic cells expressing the fusion constructs were captured using an Olympus FV1000 confocal microscope and processed using FW10-ASW software.
Supplementary Material
Supplementary files S1–S3, tables S1–S11, and figures S1–S2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
This research was financially supported by grants from the Czech Science Foundation of the Czech Republic (GAP501/11/1843 to P.Ne.); the Ministry of Education, Youth, and Sport of the Czech Republic (KONTAKT II LH11058 to J.M.); and the Academy of Sciences of the Czech Republic (RVO:60077344). Access to computing and storage facilities owned by parties and projects contributing to the Czech National Grid Infrastructure MetaCentrum, provided under the program “Projects of Large Infrastructure for Research, Development, and Innovations” (LM2010005), is greatly appreciated. The authors thank J. Látalová and V. Tetourová for technical assistance. Institution at which research was done: Biology Centre ASCR.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Anisimova M, Bielawski JP, Yang ZH. Accuracy and power of the likelihood ratio test in detecting adaptive molecular evolution. Mol Biol Evol. 2001;18:1585–1592. doi: 10.1093/oxfordjournals.molbev.a003945. [DOI] [PubMed] [Google Scholar]
- Barlow PW, Nevin D. Quantitative karyology of some species of Luzula. Plant Syst Evol. 1976;125:77–86. [Google Scholar]
- Bennett MD, Bhandol P, Leitch IJ. Nuclear DNA amounts in angiosperms and their modern uses—807 new estimates. Ann Bot. 2000;86:859–909. [Google Scholar]
- Bennett MD, Leitch IJ. Nuclear DNA amounts in angiosperms: progress, problems and prospects. Ann Bot. 2005;95:45–90. doi: 10.1093/aob/mci003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, Bassett EA. The histone variant CENP-A and centromere specification. Curr Opin Cell Biol. 2008;20:91–100. doi: 10.1016/j.ceb.2007.11.007. [DOI] [PubMed] [Google Scholar]
- Black BE, Foltz DR, Chakravarthy S, Luger K, Woods VL, Cleveland DW. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- Burrack LS, Applen SE, Berman J. The requirement for the Dam1 complex is dependent upon the number of kinetochore proteins and microtubules. Curr Biol. 2011;21:889–896. doi: 10.1016/j.cub.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrack LS, Berman J. Flexibility of centromere and kinetochore structures. Trends Genet. 2012;28:204–212. doi: 10.1016/j.tig.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Cole HA, Howard BH, Clark DJ. The centromeric nucleosome of budding yeast is perfectly positioned and covers the entire centromere. Proc Natl Acad Sci U S A. 2011;108:12687–12692. doi: 10.1073/pnas.1104978108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JL, Henikoff S. Adaptive evolution of the histone fold domain in centromeric histones. Mol Biol Evol. 2004;21:1712–1718. doi: 10.1093/molbev/msh179. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB. A plant DNA mini-preparation: version II. Plant Mol Biol Report. 1983;1:19–21. [Google Scholar]
- Doležel J, Binarová P, Lucretti S. Analysis of nuclear-DNA content in plant-cells by flow-cytometry. Biol Plant. 1989;31:113–120. [Google Scholar]
- Dong F, Song J, Naess SK, Helgeson JP, Gebhardt C, Jiang J. Development and applications of a set of chromosome-specific cytogenetic DNA markers in potato. Theor Appl Genet. 2000;101:1001–1007. [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa T, Earnshaw WC. The centromere: chromatin foundation for the kinetochore machinery. Dev Cell. 2014;30:496–508. doi: 10.1016/j.devcel.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N, Gouy M, Gautier C. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput Appl Biosci. 1996;12:543–548. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- Gong ZY, Wu YF, Koblížková A, Torres GA, Wang K, Iovene M, Neumann P, Zhang WL, Novák P, Buell CR, et al. Repeatless and repeat-based centromeres in potato: implications for centromere evolution. Plant Cell. 2012;24:3559–3574. doi: 10.1105/tpc.112.100511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng QD, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Hahn MW. Distinguishing among evolutionary models for the maintenance of gene duplicates. J Hered. 2009;100:605–617. doi: 10.1093/jhered/esp047. [DOI] [PubMed] [Google Scholar]
- Heckmann S, Schroeder-Reiter E, Kumke K, Ma L, Nagaki K, Murata M, Wanner G, Houben A. Holocentric chromosomes of Luzula elegans are characterized by a longitudinal centromere groove, chromosome bending, and a terminal nucleolus organizer region. Cytogenet Genome Res. 2011;134:220–228. doi: 10.1159/000327713. [DOI] [PubMed] [Google Scholar]
- Heun P, Erhardt S, Blower MD, Weiss S, Skora AD, Karpen GH. Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev Cell. 2006;10:303–315. doi: 10.1016/j.devcel.2006.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins AW, Gustashaw KM, Willard HF. Engineered human dicentric chromosomes show centromere plasticity. Chromosome Res. 2005;13:745–762. doi: 10.1007/s10577-005-1009-2. [DOI] [PubMed] [Google Scholar]
- Hirsch CD, Wu YF, Yan HH, Jiang JM. Lineage-specific adaptive evolution of the centromeric protein CENH3 in diploid and allotetraploid Oryza species. Mol Biol Evol. 2009;26:2877–2885. doi: 10.1093/molbev/msp208. [DOI] [PubMed] [Google Scholar]
- Houben A, Schroeder-Reiter E, Nagaki K, Nasuda S, Wanner G, Murata M, Endo TR. CENH3 interacts with the centromeric retrotransposon cereba and GC-rich satellites and locates to centromeric substructures in barley. Chromosoma. 2007;116:275–283. doi: 10.1007/s00412-007-0102-z. [DOI] [PubMed] [Google Scholar]
- Jin WW, Lamb JC, Zhang WL, Kolano B, Birchler JA, Jiang JM. Histone modifications associated with both A and B chromosomes of maize. Chromosome Res. 2008;16:1203–1214. doi: 10.1007/s10577-008-1269-8. [DOI] [PubMed] [Google Scholar]
- Jin WW, Melo JR, Nagaki K, Talbert PB, Henikoff S, Dawe RK, Jiang JM. Maize centromeres: organization and functional adaptation in the genetic background of oat. Plant Cell. 2004;16:571–581. doi: 10.1105/tpc.018937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabe A, Nasuda S, Charlesworth D. Duplication of centromeric histone H3 (HTR12) gene in Arabidopsis halleri and A. lyrata, plant species with multiple centromeric satellite sequences. Genetics. 2006;174:2021–2032. doi: 10.1534/genetics.106.063628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin M, Herendeen PS, Wojciechowski MF. Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the tertiary. Syst Biol. 2005;54:575–594. doi: 10.1080/10635150590947131. [DOI] [PubMed] [Google Scholar]
- Lee HR, Zhang W, Langdon T, Jin W, Yan H, Cheng Z, Jiang J. Chromatin immunoprecipitation cloning reveals rapid evolutionary patterns of centromeric DNA in Oryza species. Proc Natl Acad Sci U S A. 2005;102:11793–11798. doi: 10.1073/pnas.0503863102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Yue W, Li DY, Wang RRC, Kong XY, Lu K, Wang GX, Dong YS, Jin WW, Zhang XY. Structure and dynamics of retrotransposons at wheat centromeres and pericentromeres. Chromosoma. 2008;117:445–456. doi: 10.1007/s00412-008-0161-9. [DOI] [PubMed] [Google Scholar]
- Macas J, Neumann P, Navrátilová A. Repetitive DNA in the pea (Pisum sativum L.) genome: comprehensive characterization using 454 sequencing and comparison to soybean and Medicago truncatula. BMC Genomics. 2007;8:427. doi: 10.1186/1471-2164-8-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox PS, Oegema K, Desai A, Cheeseman IM. “Holo"er than thou: chromosome segregation and kinetochore function in C. elegans. Chromosome Res. 2004;12:641–653. doi: 10.1023/B:CHRO.0000036588.42225.2f. [DOI] [PubMed] [Google Scholar]
- Maheshwari S, Tan EH, West A, Franklin FC, Comai L, Chan SW. Naturally occurring differences in CENH3 affect chromosome segregation in zygotic mitosis of hybrids. PLoS Genet. 2015;11:e1004970. doi: 10.1371/journal.pgen.1004970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik HS. The centromere-drive hypothesis: a simple basis for centromere complexity. Prog Mol Subcell Biol. 2009;48:33–52. doi: 10.1007/978-3-642-00182-6_2. [DOI] [PubMed] [Google Scholar]
- Malik HS, Henikoff S. Adaptive evolution of Cid, a centromere-specific histone in Drosophila. Genetics. 2001;157:1293–1298. doi: 10.1093/genetics/157.3.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik HS, Vermaak D, Henikoff S. Recurrent evolution of DNA-binding motifs in the Drosophila centromeric histone. Proc Natl Acad Sci U S A. 2002;99:1449–1454. doi: 10.1073/pnas.032664299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall OJ, Marshall AT, Choo KH. Three-dimensional localization of CENP-A suggests a complex higher order structure of centromeric chromatin. J Cell Biol. 2008;183:1193–1202. doi: 10.1083/jcb.200804078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta GD, Agarwal MP, Ghosh SK. Centromere identity: a challenge to be faced. Mol Genet Genomics. 2010;284:75–94. doi: 10.1007/s00438-010-0553-4. [DOI] [PubMed] [Google Scholar]
- Melters DP, Paliulis LV, Korf IF, Chan SWL. Holocentric chromosomes: convergent evolution, meiotic adaptations, and genomic analysis. Chromosome Res. 2012;20:579–593. doi: 10.1007/s10577-012-9292-1. [DOI] [PubMed] [Google Scholar]
- Monen J, Maddox PS, Hyndman F, Oegema K, Desai A. Differential role of CENP-A in the segregation of holocentric C. elegans chromosomes during meiosis and mitosis. Nat Cell Biol. 2005;7:1248–1255. doi: 10.1038/ncb1331. [DOI] [PubMed] [Google Scholar]
- Moraes ICR, Lermontova I, Schubert I. Recognition of A. thaliana centromeres by heterologous CENH3 requires high similarity to the endogenous protein. Plant Mol Biol. 2011;75:253–261. doi: 10.1007/s11103-010-9723-3. [DOI] [PubMed] [Google Scholar]
- Myers EW, Miller W. Optimal alignments in linear space. Comput Appl Biosci. 1988;4:11–17. doi: 10.1093/bioinformatics/4.1.11. [DOI] [PubMed] [Google Scholar]
- Nagaki K, Cheng ZK, Ouyang S, Talbert PB, Kim M, Jones KM, Henikoff S, Buell CR, Jiang JM. Sequencing of a rice centromere uncovers active genes. Nat Genet. 2004;36:138–145. doi: 10.1038/ng1289. [DOI] [PubMed] [Google Scholar]
- Nagaki K, Kashihara K, Murata M. Visualization of diffuse centromeres with centromere-specific histone H3 in the holocentric plant Luzula nivea. Plant Cell. 2005;17:1886–1893. doi: 10.1105/tpc.105.032961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaki K, Kashihara K, Murata M. A centromeric DNA sequence colocalized with a centromere-specific histone H3 in tobacco. Chromosoma. 2009;118:249–257. doi: 10.1007/s00412-008-0193-1. [DOI] [PubMed] [Google Scholar]
- Nagaki K, Murata M. Characterization of CENH3 and centromere-associated DNA sequences in sugarcane. Chromosome Res. 2005;13:195–203. doi: 10.1007/s10577-005-0847-2. [DOI] [PubMed] [Google Scholar]
- Nagaki K, Talbert PB, Zhong CX, Dawe RK, Henikoff S, Jiang JM. Chromatin immunoprecipitation reveals that the 180-bp satellite repeat is the key functional DNA element of Arabidopsis thaliana centromeres. Genetics. 2003;163:1221–1225. doi: 10.1093/genetics/163.3.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaki K, Walling J, Hirsch C, Jiang J, Murata M. Structure and evolution of plant centromeres. Prog Mol Subcell Biol. 2009;48:153–179. doi: 10.1007/978-3-642-00182-6_6. [DOI] [PubMed] [Google Scholar]
- Neumann P, Navrátilová A, Schroeder-Reiter E, Koblížková A, Steinbauerová V, Chocholová E, Novák P, Wanner G, Macas J. Stretching the rules: monocentric chromosomes with multiple centromere domains. PLoS Genet. 2012;8:e1002777. doi: 10.1371/journal.pgen.1002777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann P, Požárková D, Vrána J, Doležel J, Macas J. Chromosome sorting and PCR-based physical mapping in pea (Pisum sativum L.) Chromosome Res. 2002;10:63–71. doi: 10.1023/a:1014274328269. [DOI] [PubMed] [Google Scholar]
- Neumann P, Yan HH, Jiang JM. The centromeric retrotransposons of rice are transcribed and differentially processed by RNA interference. Genetics. 2007;176:749–761. doi: 10.1534/genetics.107.071902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper DA, Brinkley BR. Localization of tubulin in the mitotic apparatus of mammalian cells by immunofluorescence and immunoelectron microscopy. Chromosoma. 1977;60:223–235. doi: 10.1007/BF00329772. [DOI] [PubMed] [Google Scholar]
- Pidoux AL, Allshire RC. Kinetochore and heterochromatin domains of the fission yeast centromere. Chromosome Res. 2004;12:521–534. doi: 10.1023/B:CHRO.0000036586.81775.8b. [DOI] [PubMed] [Google Scholar]
- Piras FM, Nergadze SG, Magnani E, Bertoni L, Attolini C, Khoriauli L, Raimondi E, Giulotto E. Uncoupling of satellite DNA and centromeric function in the genus Equus. PLoS Genet. 2010;6:e1000845. doi: 10.1371/journal.pgen.1000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plohl M, Mestrovic N, Mravinac B. Centromere identity from the DNA point of view. Chromosoma. 2014;123:313–325. doi: 10.1007/s00412-014-0462-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro SA, Vagnarelli P, Dong Y, Hori T, McEwen BF, Fukagawa T, Flors C, Earnshaw WC. A super-resolution map of the vertebrate kinetochore. Proc Natl Acad Sci U S A. 2010;107:10484–10489. doi: 10.1073/pnas.1002325107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Roach KC, Ross BD, Malik HS. Rapid evolution of centromeres and centromeric/kinetochore proteins. In: Singh RS, Xu J, Kulathinal RJ, editors. Rapidly evolving genes and genetic systems. Oxford: Oxford University Press; 2012. pp. 83–93. [Google Scholar]
- Roy B, Burrack LS, Lone MA, Berman J, Sanyal K. CaMtw1, a member of the evolutionarily conserved Mis12 kinetochore protein family, is required for efficient inner kinetochore assembly in the pathogenic yeast Candida albicans. Mol Microbiol. 2011;80:14–32. doi: 10.1111/j.1365-2958.2011.07558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanei M, Pickering R, Kumke K, Nasuda S, Houben A. Loss of centromeric histone H3 (CENH3) from centromeres precedes uniparental chromosome elimination in interspecific barley hybrids. Proc Natl Acad Sci U S A. 2011;108:498–505. doi: 10.1073/pnas.1103190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal K, Sridhar S, Yadav V, Heitman J, Kozubowski L. Structure-function analysis of the centromere-kinetochore and spindle assembly checkpoint machinery in Cryptococcus neoformans var grubii. Mycoses. 2014;57:71–72. [Google Scholar]
- Schaefer H, Hechenleitner P, Santos-Guerra A, de Sequeira MM, Pennington RT, Kenicer G, Carine MA. Systematics, biogeography, and character evolution of the legume tribe Fabeae with special focus on the middle-Atlantic island lineages. BMC Evol Biol. 2012;12:250. doi: 10.1186/1471-2148-12-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang WH, Hori T, Toyoda A, Kato J, Popendorf K, Sakakibara Y, Fujiyama A, Fukagawa T. Chickens possess centromeres with both extended tandem repeats and short non-tandem-repetitive sequences. Genome Res. 2010;20:1219–1228. doi: 10.1101/gr.106245.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. The Staden sequence analysis package. Mol Biotechnol. 1996;5:233–241. doi: 10.1007/BF02900361. [DOI] [PubMed] [Google Scholar]
- Stover BC, Muller KF. TreeGraph 2: combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformatics. 2010;11:7. doi: 10.1186/1471-2105-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan BA, Karpen GH. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat Struct Mol Biol. 2004;11:1076–1083. doi: 10.1038/nsmb845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachiwana H, Kagawa W, Shiga T, Osakabe A, Miya Y, Saito K, Hayashi-Takanaka Y, Oda T, Sato M, Park SY, et al. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature. 2011;476:232–235. doi: 10.1038/nature10258. [DOI] [PubMed] [Google Scholar]
- Talbert PB, Bryson TD, Henikoff S. Adaptive evolution of centromere proteins in plants and animals. J Biol. 2004;3:18. doi: 10.1186/jbiol11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tek AL, Kashihara K, Murata M, Nagaki K. Functional centromeres in soybean include two distinct tandem repeats and a retrotransposon. Chromosome Res. 2010;18:337–347. doi: 10.1007/s10577-010-9119-x. [DOI] [PubMed] [Google Scholar]
- Tek AL, Kashihara K, Murata M, Nagaki K. Functional centromeres in Astragalus sinicus include a compact centromere-specific histone H3 and a 20-bp tandem repeat. Chromosome Res. 2011;19:969–978. doi: 10.1007/s10577-011-9247-y. [DOI] [PubMed] [Google Scholar]
- Tomonaga T, Matsushita K, Yamaguchi S, Oohashi T, Shimada H, Ochiai T, Yoda K, Nomura F. Overexpression and mistargeting of centromere protein-A in human primary colorectal cancer. Cancer Res. 2003;63:3511–3516. [PubMed] [Google Scholar]
- Torras-Llort M, Moreno-Moreno O, Azorin F. Focus on the centre: the role of chromatin on the regulation of centromere identity and function. EMBO J. 2009;28:2337–2348. doi: 10.1038/emboj.2009.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hooser AA, Ouspenski II, Gregson HC, Starr DA, Yen TJ, Goldberg ML, Yokomori K, Earnshaw WC, Sullivan KF, Brinkley BR. Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J Cell Sci. 2001;114:3529–3542. doi: 10.1242/jcs.114.19.3529. [DOI] [PubMed] [Google Scholar]
- Wang K, Wu YF, Zhang WL, Dawe RK, Jiang JM. Maize centromeres expand and adopt a uniform size in the genetic background of oat. Genome Res. 2014;24:107–116. doi: 10.1101/gr.160887.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfgruber TK, Sharma A, Schneider KL, Albert PS, Koo DH, Shi JH, Gao Z, Han FP, Lee H, Xu RH, et al. Maize centromere structure and evolution: sequence analysis of centromeres 2 and 5 reveals dynamic loci shaped primarily by retrotransposons. PLoS Genet. 2009;5: e1000743. doi: 10.1371/journal.pgen.1000743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan HH, Talbert PB, Lee HR, Jett J, Henikoff S, Chen F, Jiang JM. Intergenic locations of rice centromeric chromatin. PLoS Biol. 2008;6:2563–2575. doi: 10.1371/journal.pbio.0060286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZH. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Zedek F, Bureš P. Evidence for centromere drive in the holocentric chromosomes of Caenorhabditis. PLoS One. 2012;7:e30496. doi: 10.1371/journal.pone.0030496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Dawe RK. Total centromere size and genome size are strongly correlated in ten grass species. Chromosome Res. 2012;20:403–412. doi: 10.1007/s10577-012-9284-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JZ, Nielsen R, Yang ZH. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
- Zhang WL, Friebe B, Gill BS, Jiang JM. Centromere inactivation and epigenetic modifications of a plant chromosome with three functional centromeres. Chromosoma. 2010;119:553–563. doi: 10.1007/s00412-010-0278-5. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Li J, Zhao XQ, Wang J, Wong GK, Yu J. KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genomics Proteomics Bioinformatics. 2006;4:259–263. doi: 10.1016/S1672-0229(07)60007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong CX, Marshall JB, Topp C, Mroczek R, Kato A, Nagaki K, Birchler JA, Jiang JM, Dawe RK. Centromeric retroelements and satellites interact with maize kinetochore protein CENH3. Plant Cell. 2002;14:2825–2836. doi: 10.1105/tpc.006106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.