Abstract
Microorganisms responsible for the degradation of phenanthrene in a clean forest soil sample were identified by DNA-based stable isotope probing (SIP). The soil was artificially amended with either 12C- or 13C-labeled phenanthrene, and soil DNA was extracted on days 3, 6 and 9. Terminal restriction fragment length polymorphism (TRFLP) results revealed that the fragments of 219- and 241-bp in HaeIII digests were distributed throughout the gradient profile at three different sampling time points, and both fragments were more dominant in the heavy fractions of the samples exposed to the 13C-labeled contaminant. 16S rRNA sequencing of the 13C-enriched fraction suggested that Acidobacterium spp. within the class Acidobacteria, and Collimonas spp. within the class Betaproteobacteria, were directly involved in the uptake and degradation of phenanthrene at different times. To our knowledge, this is the first report that the genus Collimonas has the ability to degrade PAHs. Two PAH-RHDα genes were identified in 13C-labeled DNA. However, isolation of pure cultures indicated that strains of Staphylococcus sp. PHE-3, Pseudomonas sp. PHE-1, and Pseudomonas sp. PHE-2 in the soil had high phenanthrene-degrading ability. This emphasizes the role of a culture-independent method in the functional understanding of microbial communities in situ.
Introduction
Polycyclic aromatic hydrocarbons (PAHs) can enter the environment from both anthropogenic and natural sources. They are primarily derived from the incomplete combustion of organic matter at high temperatures. In addition, PAHs can be released into the environment through natural processes such as forest fires, and direct biosynthesis by microbes and plants [1]. Their presence in the environment poses a considerable threat to public health and ecosystems because of their acute toxicity, potential mutagenicity, and carcinogenicity. This has led to their classification as priority pollutants by the U.S. Environmental Protection Agency [2].
The contamination resulting from PAHs can be removed by microorganisms, as has been shown for other pollutants. The role of bacteria in the degradation of organic pollutants has attracted much attention. A successful bioremediation strategy involves either encouraging the native microbial community to rapidly degrade the pollutants, or the addition of more-effective degradative microbes, in the absence of a native degrading biota [3]. The identification of pollutant-degrading populations within microbial communities is therefore essential for the design, control, and optimization of the bioremediation of PAHs-contaminated aquifers. Traditional isolation/cultivation studies have provided most of our knowledge regarding how pollutants are biodegraded under artificial conditions and the PAHs degradation pathways. The genes associated with the degradation process have been identified, with particular emphasis given to the PAHs ring hydroxylating dioxygenase (PAHs-RHD) [4, 5]. However, it has proven difficult to obtain a pure culture of target bacteria in the laboratory, with only a small percentage of the organisms found in soil communities capable to be cultured, which does not reveal the real picture within a specific site. Furthermore, inoculation cannot always achieve an ideal remediation, because of the lack of understanding of where the target microorganisms are located within the overall biota [6]. In recent years, culture-independent methods, such as stable-isotope probing (SIP), have been used to link the growth of microbial populations to specific metabolic processes. This has been achieved by feeding microorganisms a substrate containing an isotope to label the microbial DNA, allowing the identification and characterization of novel organisms that previously escaped detection. The 13C-labeled DNA is subsequently separated from 12C-DNA on density gradients, and analyzed by molecular techniques such as high-throughput sequencing, terminal restriction fragment length polymorphism (TRFLP) and denaturing gradient gel electrophoresis (DGGE), for phylogenetic characterization. Microbial populations within complex communities that are responsible for the degradation of targeted contaminants can then be identified [7–9]. Besides DNA, RNA and phospholipid fatty acid (PLFA) can also be used as biomarkers in a fingerprint analysis [10, 11]. To date, SIP has been successfully used to identify a variety of bacteria that assimilate diverse compounds [9, 12–18].
Phenanthrene was used as a model PAHs in the present study due to its ubiquity in nature and the fused-ring structure resembling higher-molecular-weight PAHs that are known to be carcinogenic. To date, SIP studies undertaken on PAHs biodegradation have been limited to previously contaminated media such as coal-tar polluted sediments [19], road runoff polluted soils [20, 21], and polluted soils from a former gas-manufacturing plant [22–24], former wood preserving facility [25], former coking plant site [5], and a tarmacadam-producing plant [18]. However, SIP has not yet been used to investigate potential PAHs degraders in non-contaminated soils. It is well-known that bacterial diversity can usually be repressed by the existence of toxic organic pollutants [26–28]. Hence, unpolluted forest soils are likely to have a more varied bacterial community than contaminated soils, which could be quite different from the community in contaminated sites. Identification of the functional community responsible for the degradation of phenanthrene is important for understanding the biogeochemistry of PAHs in natural soils, and the clean-up ability of the soil itself. In this study, 13C-DNA targeted SIP was applied to a clean forest soil to investigate phenanthrene degradation therein, and to determine the organisms responsible for this in situ degradation. Traditional isolation and pure culture techniques were also used for comparison with the effectiveness of SIP. The aim was to achieve a full understanding of phenanthrene degradation in this ‘clean’ medium.
Materials and Methods
Development of phenanthrene-degrading microcosms
Soil samples were collected from a forest in Luoji Mountain (27°34′N, 102°25′E, altitude 2200 m), Sichuan Province, China. The research site is owned by Guangzhou Institute of Geochemistry. The field studies did not involve endangered or protected species and no specific permits were required for the described field studies.The soil (pH = 5.5) had a high organic matter content (6.2%), and was classified as a sandy loam soil based on the texture analysis. In laboratory, the samples were homogenized, sieved through a 4 mm screen and stored at 4°C until use.
A sample of 6 g soil (dry weight) was placed in a 150 mL serum bottle containing 20 mL phosphate-buffered mineral medium [29]. The bottles were sealed with rubber stoppers and compressed with an aluminum seal. After sealing, 12C-labeled phenanthrene (99%, Cambridge Isotope Laboratories, Inc., Tewksbury, MA, USA) or 13C-labeled phenanthrene (13C6-PHE, 99%, Cambridge Isotope Laboratories, Inc.) was added to the bottles via a gas-tight syringe, to a final phenanthrene concentration of 1 mg/kg. The serum bottles were opened each day, kept in the air for 1 hour to ensure that the oxygen in the microcosms remained close to environment and resealed. The treatments included: no phenanthrene controls, autoclaved controls, and 12C- and 13C-labeled phenanthrene samples. Six samples were prepared for each treatment. The cultures were incubated at 20°C, with reciprocal shaking. On days 3, 6, and 9 after incubation, two samples from each treatment were sacrificed for phenanthrene analysis and DNA extraction.
Phenanthrene measurement
On days 3, 6, and 9 after incubation, two samples from each treatment were used for phenanthrene analysis as follows. A freeze-dried soil sample was homogenized, pulverized, spiked with 1,000 ng of deuterated PAHs as a surrogate standard and extracted with dichloromethane (DCM) in a Soxhlet apparatus for 48 h with activated copper added to remove the sulfur. The extract was concentrated to about 0.5 mL after a solvent exchange with hexane. The soil extracts were purified with a silica gel/alumina column (8 mm i.d.) filled with anhydrous Na2SO4 (1 cm), neutral silica gel (3 cm, 3% deactivated) and neutral alumina (3 cm, 3% deactivated) from top to bottom, with an eluent of 15 mL hexane/DCM (1:1, v/v). After being concentrated to approximately 50 μL using a gentle stream of N2, 1,000 ng of hexamethylbenzene as an internal standard was added to all samples prior to instrumental analysis.
Phenanthrene was analyzed by gas chromatography (model 7890, Agilent, Santa Clara, CA, USA), using a capillary column (DB-5MS, 30 m, 0.25 mm, 0.25 μm) and a mass spectrometric detector (MSD, model 5975, Agilent). Samples (1 μL) were injected under splitless mode with a 10 min solvent delay time. High-purity helium was used as a carrier gas with a flow velocity of 1.83 mL/min. The temperature of the injector and transfer line were 290°C and 300°C, respectively. The initial oven temperature was set at 60°C for 1 min and raised to 290°C at a rate of 3°C/min, and then held for 20 min.
DNA extraction and ultracentrifugation
DNA extraction was conducted on two soil samples from each treatment that were incubated for 3, 6, and 9 days, using a Powersoil DNA extraction kit (MO BIO Laboratories, Inc. Carlsbad, CA, USA) following the manufacturer’s instructions. The DNA content was quantified with an ND-2,000 UV-Vis spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). Thereafter, about 10,000-ng DNA was added to Quick-Seal polyallomer tubes (13 × 51 mm, 5.1 mL, Beckman Coulter, Pasadena, CA, USA) and spun in Tris-EDTA (TE, pH 8.0)/CsCl solution. Prior to sealing the tubes with one cordless quick-seal tube topper (Beckman Coulter), the average buoyant density (BD) of all prepared gradients was confirmed with a digital refractometer (model AR200, Leica Microsystems Inc., Buffalo Grove, IL, USA), and adjusted by adding small volumes of CsCl solution or Tris-EDTA buffer. The tubes were transferred to an ultracentrifuge (Optima L-100XP, Beckman Coulter). Centrifugation was performed at 45,000 g (20°C) for 48 h. Subsequently, the centrifuge tubes were placed onto a fraction recovery system (Beckman Coulter) and fractions (150 μL for each) were collected. The BD of each fraction was then measured, and CsCl was removed by introducing concentrated ethanol [30].
PCR and TRFLP
The ultracentrifugation fractions were subjected to a polymerase chain reaction (PCR). The 16S rRNA PCR primers (Operon Biotechnologies, Bangalore, India) were 27F-FAM (5’-AGAGTTTGATCMTGGCTCAG, 5’ end-labeled with carboxyfluorescine) and 1492R (5’-GGTTACCTTGTTACGACTT), and the PCR amplifications were conducted in a Peltier thermal cycler (BIO-RAD, Hercules, CA, USA) using a protocol adopted previously: 94°C (5 min); 94°C (30 s), 55°C (30 s), 72°C (1.5 min) (30 cycles); and 72°C (5 min) [8]. The presence of PCR products was confirmed by 1% agarose gel electrophoresis and the subsequent staining of the gels with ethidium bromide. PCR products were further purified with an Omega ENZA Cycle-Pure kit (Omega Bio-Tek, Inc., Norcross, GA, USA). The purified PCR products from the total DNA and fractions were digested with HaeIII, following the manufacturer’s instructions (New England Biolabs, Ipswich, MA, USA). One nanogram of labeled PCR product was run on a genetic analyzer (ABI 3730, Applied Biosystems, Foster City, CA, USA) with Peak Scanner Software v. 1.0 (Applied Biosystems), and using ROX500 as the internal standard to separate the DNA fragments. Data was analyzed with Peak Scanner Software v. 1.0. The percentage abundance of each fragment was determined as described previously [30].
Detection of the PAH-RHDα gene in microcosms
The presence of the PAH-RHDα gene was investigated using both the PAH-RHDα GP and GN primer pairs on the heavy DNA fractions. A gradient PCR was performed with annealing temperatures ranging from 52 to 62°C [31]. However, only the PAH-RHDα GP primers 641F (5’-CGGCGCCGACAAYTTYGTNGG) and 933R (5’-GGGGAACACGGTGCCRTGDATRAA) produced a strong and specific amplicon and were therefore selected for the study. Further amplification reactions were conducted in a volume of 50 μL, as reported in a previous study: 95°C (5 min); 95°C (30 s), 54°C (30 s), 72°C (30 s) (30 cycles); and 72°C (7 min) [31].
Sequencing of 16S rRNA genes and the partial PAH-RHDα gene
Clone libraries of the 16S rRNA genes were constructed with the 13C fraction of 13C-labeled DNA, amplified using a previously adopted protocol, where 27F-FAM was replaced by 27F [8]. In addition, amplicons generated with PAH-RHDα primer pairs were also prepared for cloning and sequencing. Purified PCR products were cloned into Escherichia coli JM109 using a TA cloning kit (TaKaRa Biotechnology, Inc., Shiga, Japan). E. coli clones were grown on Luria-Bertani (LB) medium, solidified with 15 g agar/L in the presence of 50 μg/L ampicillin for 16 h at 37°C. The positive single colonies were isolated and cultivated in the LB medium containing 50 μg/L ampicillin overnight. Colonies with inserts were verified by PCR with primers M13 forward (5’-TGTAAAACGACGGCCAGT-3’) and M13 reverse (5’-AACAGCTATGACCATG-3’), and the plasmids were extracted with an EZNA Plasmid Mini kit (Omega Bio-Tek, Inc.). The insertions were sequenced with a genetic analyzer (ABI 3730, Applied Biosystems), using the pair of M13 universal primers. The sequences obtained were compared with the GenBank database, using the Basic Local Alignment Search Tool (BLAST) algorithm (National Center for Biotechnology Information, Bethesda, MD, USA). Phylogenic trees for the partial PAH-RHDα sequences along with the closest matches in GenBank were obtained by the neighbor-joining method using Molecular Evolutionary Genetics Analysis, version 5.0 (MEGA 5.0).
The 16S rRNA gene clone libraries and the partial PAH-RHDα gene sequences determined in this study were deposited under accession numbers KF035825 and KF035826 for partial 16S rRNA gene sequences, and KF656718 and KF656719 for partial PAH-RHDα gene sequences.
Isolation and identification of phenanthrene-degrading bacteria
The isolation of the phenanthrene degrading microorganisms that were capable of degrading phenanthrene in soils, and their pure cultivation, was attempted. Soil samples (10 g) were suspended in 100 mL mineral salts medium (MSM) (NaCl 0.5 g/L, (NH4)2SO4 1.0 g/L, K2HPO4 1.5 g/L, KH2PO4 0.5 g/L, MgSO4·7H2O 0.2 g/L, pH = 7.0), with 1- and 10-mg/kg (MSM-C1 and MSM-C10) phenanthrene as the sole carbon source, which corresponded to 1- and 10-times the phenanthrene used in the SIP study, respectively. The inoculum was incubated at 30°C at 180 rpm for 4 days. Subsequently, 100 μL of the culture was subcultured in 100 mL fresh MSM-C1 and MSM-C10 respectively, and then incubated under the same conditions for another 4 days. After 3 sequential rounds of enrichment, the cultured medium was diluted 10–105-fold with sterilized water, and then daubed on a solid mineral salts medium supplemented with 1 and 10 mg/L phenanthrene, respectively. Single colonies were selected and cultured in MSM-C1 or MSM-C10 for 4 days, and the degrading capacities of the isolates were tested in liquid culture by adding 3- or 30-μg phenanthrene to 3-mL cell suspensions. Cultures were incubated in the dark at 180 rpm at 30°C. The degradation efficiency was determined after 3 days using the method described above. Genomic DNA was extracted and 16S rRNA was amplified by PCR using the primers 27F and 1492R as above. Cultivable phenanthrene degraders were identified by 16S rRNA sequencing, and the sequences of the isolates were deposited in the GenBank database under accession numbers KF035827 to KF035829.
Results and Discussion
Previous SIP studies regarding the biodegradation of PAHs focused primarily on samples from formerly contaminated sites. To expand on this, this study applied SIP to a clean forest soil, and therefore fresh insights on the biogeochemistry of PAHs in natural soils were anticipated.
Biodegradation of phenanthrene in soil
As shown in Table 1, the phenanthrene concentration remained constant from day 3 to 9 in the sterile soils. In contrast, significant phenanthrene biodegradation was observed after 3 days of incubation in the unsterilized microcosms. Phenanthrene in the unsterilized soils degraded quickly, with 70% degradation achieved after 9 days. Throughout the process, no significant difference (p >0.05) was found between soils amended with 13C- and 12C-labeled phenanthrene.
Table 1. Ranges for percentage of phenanthrene remaining in soil-liquid slurries over time.
| Time (days) | Sterile controls | 12C- phenanthrene amended samples | 13C- phenanthrene amended samples |
|---|---|---|---|
| 3 | 85–90 | 66–72 | 64–75 |
| 6 | 83–87 | 40–45 | 42–48 |
| 9 | 86–89 | 30–36 | 26–35 |
Phenanthrene is a widespread pollutant that biodegrades more rapidly than many other organic pollutants [32]. The degradation rate observed in this study was comparable with results reported previously [5, 21, 22], but considerably higher than the rates reported by Renzt et al. and Powell et al [33, 34]. This may be attributed to the soil properties, experimental conditions such as the laboratory microcosm versus field scale conditions, and the concentration and composition of contaminants in the system tested. It was found that when phenanthrene co-existed with other PAHs, such as naphthalene and pyrene, it degraded at a slower rate than when present alone, which was explained by potential interactive effects, the toxicity, and both non-competitive and competitive inhibition [34]. In addition, soils used in laboratory experiments are usually homogeneous and the growth conditions tend to be optimized, which enhances the degradation capacity compared with the heterogeneous natural soil in the field.
TRFLP results for SIP
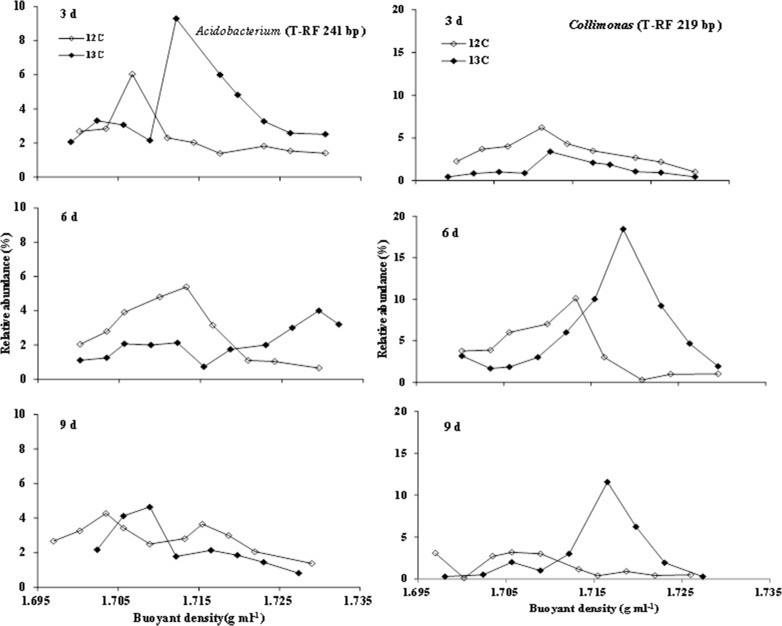
The organisms responsible for 13C assimilation were identified by a comparison of the relative abundances of specific terminal restriction fragments between the control (amended with 12C-labeled phenanthrene) and the sample (amended with 13C-labeled phenanthrene), at selected time points for each fraction. Based on the TRFLP results, the fragment 241-bp HaeIII TRF displayed a clear trend of increased relative abundance at higher buoyant densities in samples amended with 13C-phenanthrene compared to the 12C-phenanthrene treatment on days 3 and 6. This indicated that 13C was incorporated by the microorganisms represented by these fragments (Fig 1). However, this trend was not evident on day 9. For fragment 219-bp HaeIII TRF, an increased relative abundance at higher buoyant densities in samples amended with 13C-phenanthrene compared to the 12C-phenanthrene control was observed on days 6 and 9 (Fig 1), but not on day 3. These results may indicate that organisms represented by fragments 241- and 219-bp played a role at different stages of phenanthrene degradation, whereas in the first 3 days the organisms represented by 241-bp played a major role, and by day 9, their role was replaced by organisms represented by the 219-bp band.
Fig 1. The shift trendency of 241-bp and 219-bp fragments.
The relative abundance of the dominant 241-bp and 219-bp fragments over a range ofbuoyant density (BD) from DNA extracted after 3, 6, 9 days from the soil added with either 13C or 12C labeled phenanthrene.
Phylogenetic identification of 13C-labeled phenanthrene degraders
Fragment lengths from enzyme digestion were compared to those obtained from in silico digests of 16S rRNA sequence data from 13C fractions, to determine the clones they represented. Of the 50 clones sequenced, 21 and 17 clones had restriction enzyme cut sites that matched the TRFLP results for fragments 241- and 219-bp, respectively. The slight difference (2–3 bases) between the measured fragment lengths and those predicted using sequence data has been noted by others [8, 10, 30, 35, 36]. On the basis of the comparative analyses of 16S rRNA genes in this study, the phenanthrene degrading microorganisms represented by 241- and 219-bp were classified as Acidobacterium sp. within the class Acidobacteria, and Collimonas sp. within the class Betaproteobacteria, respectively.
The phylum Acidobacteria was first identified in 1997 [37], and accounts for around 20% of the soil bacterial community, with large genetic and metabolic diversity in environmental samples [38–41]. Acidobacterium species have previously been linked with the degradation of both natural and anthropogenic organic compounds, including petroleum in a coastal stream and beach sediment [42, 43], PAHs in coal-tar contaminated soil [44], landfill soil [45] the rhizosphere [46], and polychlorinated biphenyls (PCBs) from contaminated sediment and soil [45, 47, 48]. Some species in this genus were shown via SIP to be actively involved in the oxidation of propionate [49], and the biodegradation of biocides [50], herbicides [51] and benzene [17]. However, this genus has not been previously linked with phenanthrene degradation; thus uncertainty remains as to whether these microbes are directly involved in the degradation of PAHs, either in situ or in a mixed culture. Our results provide strong evidence that microorganisms in this genus are the primary organisms responsible for the degradation of phenanthrene in the mixed community of a forest soil.
The genus Collimonas was first described in 2004, with the remarkable characteristic of growing at the expense of living fungal hyphae [52, 53]. An important role of the bacterial genus Collimonas in mineral weathering for forest soils has been reported [54, 55]. However, studies reported its ability to degrade organic pollutants [56], that the isolated Collimonas sp. is an alkane degrader, and its 16S rRNA sequence is similar to a naphthalene-degrading strain originally classified as Herbaspirillum sp.
To our knowledge, this is the first study to use a DNA-based SIP technique to link the phenanthrene-degrading ability of these two genera in a real environment, although a number of studies have been undertaken to identify phenanthrene degrading microbes using SIP [9, 18–25, 34]. This result improves our understanding of the ability of Acidobacterium and Collimonas species to degrade PAHs pollutants.
Occurrence of PAH-RHDα functional genes in the microcosm
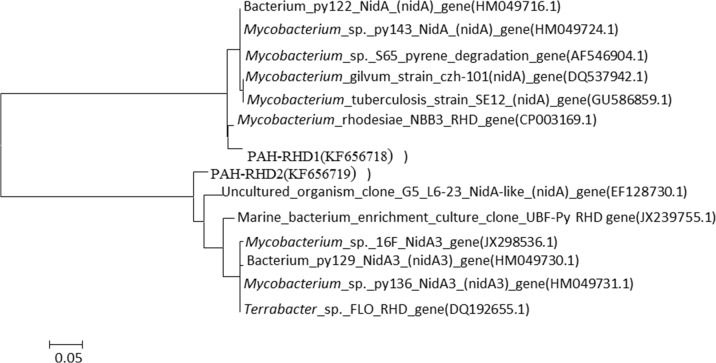
PAH-RHDα genes from Gram-positive (GP) and Gram-negative (GN) bacteria were analyzed in heavy DNA fractions. On the global scale, the proportion of PAH-RHDα GN tends to be higher than that of the PAH-RHDα GP [31]. However, in the present study only two types of PAH-RHDα GP gene were detected in the heavy DNA, and were affiliated to PAH-RHDα genes from Mycobacterium rhodesiae NBB3 (CP003169) and an uncultured organism (EF128730.1) (Fig 2). These may be the functional genes associated with the phenanthrene-degrading strains of Acidobacterium and Collimonas identified by SIP. It should be noted that the active phenanthrene degrading bacteria might contain other functional genes that could not be targeted by the primer used in the present study.
Fig 2. Phylogenetic tree of PAH-RHDα gene.
Phylogenetic relationship of PAH-RHDα gene cloned from soil treated by 1 mg/kg phenanthrene. PAH-RHD1 and PAH-RHD2 showed 97% and 92% similarity with genes from Mycobacterium_rhodesiaee and uncultured organism, respectively.
Phenanthrene-degrading bacteria derived from isolation and pure culture
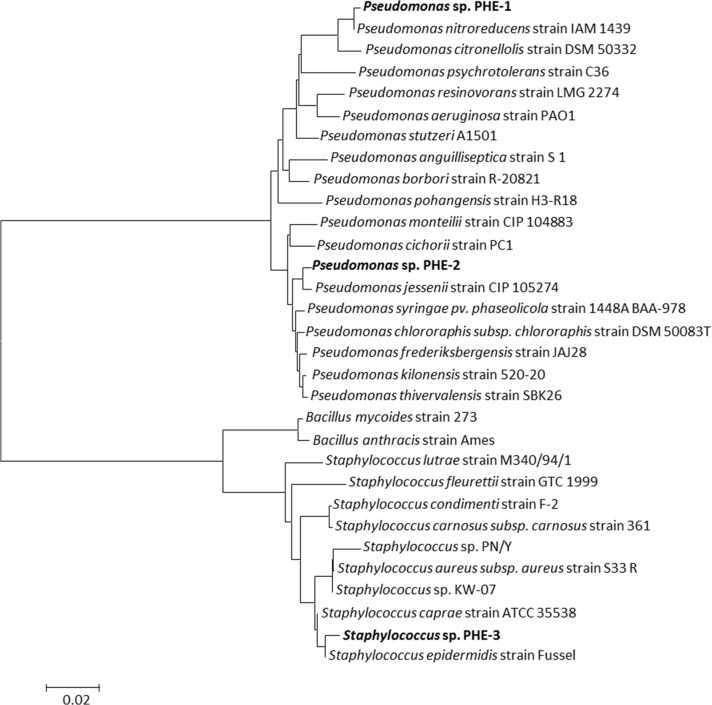
Among the phenanthrene-degrading bacteria using MSM with 1 mg/L and 10 mg/L (w/v) phenanthrene as the sole carbon source, three colonies were selected based on the size of the clear zone in the phenanthrene layer, respectively. DNA sequence analysis identified these three isolates as Pseudomonas sp. PHE-1, Pseudomonas sp. PHE-2, and Staphylococcus sp. PHE-3 (Fig 3). As shown in S1 Table, for the degradation of 1 mg/kg phenanthrene, the most efficient strain was Pseudomonas sp. PHE-1 (98.5% degradation within three days), followed by Pseudomonas sp. PHE-2 and Staphylococcus sp. PHE-3, with 82.3% and 75.6% degradation in the same time period, respectively. At 10 mg/kg phenanthrene, a 51.2% degradation performance was evident in the presence of Pseudomonas sp. PHE-1.
Fig 3. Phylogenetic tree of the isolated phenanthrene degraders.
Phylogenetic tree for the taxonomic location of Pseudomonas sp. PHE-1, Pseudomonas sp. PHE-2 and Staphylococcus sp. PHE-3. This tree based on 16S rRNA sequence was produced by MEGA 5.0.
Pseudomonas sequences are common in PAHs-contaminated soils, and many PAHs degrading isolates are found in this genus. The pathways used by this genus in the degradation of low-molecular-biomass PAHs, including naphthalene and phenanthrene, have been investigated extensively [57]. In addition, the genetic control of the pathways involved in naphthalene degradation by this genus has been studied in detail, and the nucleotide sequences of the genes encoding the upper pathway enzymes in some Pseudomonas strains have been clarified [58]. In this study Pseudomonas sp. PHE-1 and Pseudomonas sp. PHE-2 displayed 99% similarity with Pseudomonas nitroreducens and Pseudomonas jessenii. However, no enrichment of the appropriate Pseudomonas TRF was seen in the 13C-labeled fractions, indicating that these microbes were not likely to be the major degraders in situ.
Although the genus Staphylococcus is not as common as Pseudomonas in PAHs-contaminated soils, a few species in this genus have been found to degrade phenanthrene [59, 60]. The species, Staphylococcus warneri, has been successfully used to build a consortium with Bacillus pumilus capable of efficient degradation of phenanthrene, pyrene, and benzoanthracene [61]. A novel phenanthrene assimilation pathway involving 2-hydroxy-1-naphthoic acid was found in Staphylococcus sp. PN/Y, which provided new insight into the microbial degradation of PAHs [59]. By analyzing the 16S rRNA sequence, it was found that Staphylococcus sp. PHE-3 displayed 96% similarity to Staphylococcus sp. PN/Y and 97% similarity to Staphylococcus sp. KW-07. To our knowledge, this is the third species of the Staphylococcus genus displaying an ability to degrade phenanthrene.
Implications for the Understanding of PAHS Biogeochemistry in Natural Soils
These results illustrate the importance of culture-independent approaches compared to culture-dependent approaches for understanding the functions of mixed cultures. Biodegradation has been extensively studied for many decades, and has become a popular option for the elimination of toxic organic pollutants in air, water, and soil media because of its efficacy, and environmentally sustainable and economical characteristics. Phenanthrene can be degraded by some strains under aerobic conditions, and many successful field remediation cases have been reported for PAHs-contaminated sediments and soils. With the aid of advanced methods, if these bacteria can be isolated and cultured under normal laboratory conditions, the pathway adopted by Acidobacterium and Collimonas in phenanthrene degradation could be clarified. This would provide a new perspective on the biogeochemistry of phenanthrene in natural soils, and a suitable method for the biodegradation of phenanthrene in contaminated environments.
Strain inoculation as a superior method is used in the remediation of sites contaminated with organic pollutants. In this study, the dramatically different results obtained from the isolation and SIP provides some insight into why some artificial constructed systems in the field are not as efficient as expected. This could be a consequence of the intensive competition between native organisms and the inoculated strains. With the aid of the culture-independent SIP technique, the ecological roles of the functional microbes in a specific biota can be identified and characterized. Further artificial manipulation and optimization could stimulate the functional strains; hence, the maximum degradation efficiency would be achieved.
Supporting Information
Phylogenetic tree for the taxonomic location of the bacteria corresponding 219- and 241-bp TRFs. The tree is based on 16S rDNA sequence and produced by MEGA 5.0. 2.
(PDF)
The density distribution of total DNA in the micocosms amended with unlabled (12C) or labled (13C) PHE after centrifugation on days 3, 6 and 9.
(PDF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
This study was supported by the Joint Funds of the National Natural Science Foundation of China and the Natural Science Foundation of Guangdong Province, China (No. U1133004), and the National Natural Science Foundation of China (Nos. 41173082 and 41322008). This is contribution No. IS-2082 from GIGCAS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data Availability
Data relevant to Table 1, S1 Table, S2 Fig, and Fig 1 are within the paper and its Supporting Information files. The information about the DNA sequences are available from the GenBank database (KF035825 and KF035826 for partial 16S rRNA gene sequences, and KF656718 and KF656719 for partial RHD gene sequences, KF035827 to KF035829 for sequences of the isolates).
Funding Statement
This study was supported by the Joint Funds of the National Natural Science Foundation of China and the Natural Science Foundation of Guangdong Province, China (No. U1133004), and the National Natural Science Foundation of China (Nos. 41173082 and 41322008). This is contribution No. IS-2082 from GIGCAS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Eisler R. Polycyclic aromatic hydrocarbon hazards to fish, wildlife, and invertebrates: A synoptic review report. Laurel, MD: Patuxent Wildlife Research Center, 1987. [Google Scholar]
- 2. Keith LH, Telliard WA. Priority Pollutants I-a Perspective View. Environ Sci Technol. 1979;13(4):416–23. 10.1021/Es60152a601 . [DOI] [Google Scholar]
- 3. Peng RH, Xiong AS, Xue Y, Fu XY, Gao F, Zhao W, et al. Microbial biodegradation of polyaromatic hydrocarbons. Fems Microbiol Rev. 2008;32(6):927–55. 10.1111/j.1574-6976.2008.00127.x . [DOI] [PubMed] [Google Scholar]
- 4. Moser R, Stahl U. Insights into the genetic diversity of initial dioxygenases from PAH-degrading bacteria. Appl Microbiol Biot. 2001;55(5):609–18. . [DOI] [PubMed] [Google Scholar]
- 5. Cebron A, Louvel B, Faure P, France-Lanord C, Chen Y, Murrell JC, et al. Root exudates modify bacterial diversity of phenanthrene degraders in PAH-polluted soil but not phenanthrene degradation rates. Environ Microbiol. 2011;13(3):722–36. 10.1111/j.1462-2920.2010.02376.x . [DOI] [PubMed] [Google Scholar]
- 6. Oren A. Prokaryote diversity and taxonomy: current status and future challenges. Philos T Roy Soc B. 2004;359(1444):623–38. 10.1098/rstb.2003.1458 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Radajewski S, Ineson P, Parekh NR, Murrell JC. Stable-isotope probing as a tool in microbial ecology. Nature. 2000;403(6770):646–9. . [DOI] [PubMed] [Google Scholar]
- 8. Luo CL, Xie SG, Sun WM, Li XD, Cupples AM. Identification of a Novel Toluene-Degrading Bacterium from the Candidate Phylum TM7, as Determined by DNA Stable Isotope Probing. Appl Environ Microb. 2009;75(13):4644–7. 10.1128/Aem.00283-09 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gutierrez T, Singleton DR, Aitken MD, Semple KT. Stable Isotope Probing of an Algal Bloom To Identify Uncultivated Members of the Rhodobacteraceae Associated with Low-Molecular-Weight Polycyclic Aromatic Hydrocarbon Degradation. Appl Environ Microb. 2011;77(21):7856–60. 10.1128/Aem.06200-11 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Clement BG, Kehl LE, DeBord KL, Kitts CL. Terminal restriction fragment patterns (TRFPs), a rapid, PCR-based method for the comparison of complex bacterial communities. J Microbiol Meth. 1998;31(3):135–42. 10.1016/S0167-7012(97)00105-X . [DOI] [Google Scholar]
- 11. Whiteley AS, Manefield M, Lueders T. Unlocking the 'microbial black box' using RNA-based stable isotope probing technologies. Curr Opin Biotech. 2006;17(1):67–71. 10.1016/j.copbio.2005.11.002 . [DOI] [PubMed] [Google Scholar]
- 12. Lueders T, Pommerenke B, Friedrich MW. Stable-isotope probing of microorganisms thriving at thermodynamic limits: Syntrophic propionate oxidation in flooded soil. Appl Environ Microb. 2004;70(10):5778–86. 10.1128/Aem.70.10.5778-5786.2004 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Borodina E, Cox MJ, McDonald IR, Murrell JC. Use of DNA-stable isotope probing and functional gene probes to investigate the diversity of methyl chloride-utilizing bacteria in soil. Environ Microbiol. 2005;7(9):1318–28. 10.1111/j.1462-5822.2005.00819.x . [DOI] [PubMed] [Google Scholar]
- 14. DeRito CM, Pumphrey GM, Madsen EL. Use of field-based stable isotope probing to identify adapted populations and track carbon flow through a phenol-degrading soil microbial community. Appl Environ Microb. 2005;71(12):7858–65. 10.1128/Aem.71.12.7858-7865.2005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mahmood S, Paton GI, Prosser JI. Cultivation-independent in situ molecular analysis of bacteria involved in degradation of pentachlorophenol in soil. Environ Microbiol. 2005;7(9):1349–60. 10.1111/j.1462-2920.2005.00822.x . [DOI] [PubMed] [Google Scholar]
- 16. Sul WJ, Park J, Quensen JF, Rodrigues JLM, Seliger L, Tsoi TV, et al. DNA-Stable Isotope Probing Integrated with Metagenomics for Retrieval of Biphenyl Dioxygenase Genes from Polychlorinated Biphenyl-Contaminated River Sediment. Appl Environ Microb. 2009;75(17):5501–6. 10.1128/Aem.00121-09 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xie SG, Sun WM, Luo CL, Cupples AM. Novel aerobic benzene degrading microorganisms identified in three soils by stable isotope probing. Biodegradation. 2011;22(1):71–81. 10.1007/s10532-010-9377-5 . [DOI] [PubMed] [Google Scholar]
- 18. Uhlik O, Wald J, Strejcek M, Musilova L, Ridl J, Hroudova M, et al. Identification of Bacteria Utilizing Biphenyl, Benzoate, and Naphthalene in Long-Term Contaminated Soil. Plos One. 2012;7(7). doi: ARTN e40653 10.1371/journal.pone.0040653 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jeon CO, Park W, Padmanabhan P, DeRito C, Snape JR, Madsen EL. Discovery of a bacterium, with distinctive dioxygenase, that is responsible for in situ biodegradation in contaminated sediment. P Natl Acad Sci USA. 2003;100(23):13591–6. 10.1073/pnas.1735529100 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Martin F, Torelli S, Le Paslier D, Barbance A, Martin-Laurent F, Bru D, et al. Betaproteobacteria dominance and diversity shifts in the bacterial community of a PAH-contaminated soil exposed to phenanthrene. Environ Pollut. 2012;162:345–53. 10.1016/j.envpol.2011.11.032 . [DOI] [PubMed] [Google Scholar]
- 21. Regonne RK, Martin F, Mbawala A, Ngassoum MB, Jouanneau Y. Identification of soil bacteria able to degrade phenanthrene bound to a hydrophobic sorbent in situ. Environ Pollut. 2013;180:145–51. 10.1016/j.envpol.2013.04.038 . [DOI] [PubMed] [Google Scholar]
- 22. Singleton DR, Powell SN, Sangaiah R, Gold A, Ball LM, Aitken MD. Stable-isotope probing of bacteria capable of degrading salicylate, naphthalene, or phenanthrene in a Bioreactor treating contaminated soil. Appl Environ Microb. 2005;71(3):1202–9. 10.1128/Aem.71.3.1202-1209.2005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Singleton DR, Sangaiah R, Gold A, Ball LM, Aitken MD. Identification and quantification of uncultivated Proteobacteria associated with pyrene degradation in a bioreactor treating PAH-contaminated soil. Environ Microbiol. 2006;8(10):1736–45. 10.1111/j.1462-2920.2006.01112.x . [DOI] [PubMed] [Google Scholar]
- 24. Jones MD, Crandell DW, Singleton DR, Aitken MD. Stable-isotope probing of the polycyclic aromatic hydrocarbon-degrading bacterial guild in a contaminated soil. Environ Microbiol. 2011;13(10):2623–32. 10.1111/j.1462-2920.2011.02501.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jones MD, Singleton DR, Carstensen DP, Powell SN, Swanson JS, Pfaender FK, et al. Effect of incubation conditions on the enrichment of pyrene-degrading bacteria identified by stable-isotope probing in an aged, PAH-contaminated soil. Microb Ecol. 2008;56(2):341–9. 10.1007/s00248-007-9352-9 . [DOI] [PubMed] [Google Scholar]
- 26. Sawulski P, Clipson N, Doyle E. Effects of polycyclic aromatic hydrocarbons on microbial community structure and PAH ring hydroxylating dioxygenase gene abundance in soil. Biodegradation. 2014;25(6):835–47. 10.1007/s10532-014-9703-4 . [DOI] [PubMed] [Google Scholar]
- 27. Tian WJ, Zhao YG, Sun HM, Bai J, Wang YM, Wu CL. The effect of irrigation with oil-polluted water on microbial communities in estuarine reed rhizosphere soils. Ecol Eng. 2014;70:275–81. 10.1016/j.ecoleng.2014.06.003 . [DOI] [Google Scholar]
- 28. Zhang XZ, Xie JJ, Sun FL. Effects of Three Polycyclic Aromatic Hydrocarbons on Sediment Bacterial Community. Curr Microbiol. 2014;68(6):756–62. 10.1007/s00284-014-0535-6 . [DOI] [PubMed] [Google Scholar]
- 29. Mu DY, Scow KM. Effect of Trichloroethylene (Tce) and Toluene Concentrations on Tce and Toluene Biodegradation and the Population-Density of Tce and Toluene Degraders in Soil. Appl Environ Microb. 1994;60(7):2661–5. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sun WM, Xie SG, Luo CL, Cupples AM. Direct Link between Toluene Degradation in Contaminated-Site Microcosms and a Polaromonas Strain. Appl Environ Microb. 2010;76(3):956–9. 10.1128/Aem.01364-09 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cebron A, Norini MP, Beguiristain T, Leyval C. Real-Time PCR quantification of PAH-ring hydroxylating dioxygenase (PAH-RHD alpha) genes from Gram positive and Gram negative bacteria in soil and sediment samples. J Microbiol Meth. 2008;73(2):148–59. 10.1016/j.mimet.2008.01.009 . [DOI] [PubMed] [Google Scholar]
- 32. Andreoni V, Gianfreda L. Bioremediation and monitoring of aromatic-polluted habitats. Appl Microbiol Biot. 2007;76(2):287–308. 10.1007/s00253-007-1018-5 . [DOI] [PubMed] [Google Scholar]
- 33. Rentz JA, Alvarez PJJ, Schnoor JL. Repression of Pseudomonas putida phenanthrene-degrading activity by plant root extracts and exudates. Environ Microbiol. 2004;6(6):574–83. 10.1111/j.1462-2920.2004.00589.x . [DOI] [PubMed] [Google Scholar]
- 34. Powell SN, Singleton DR, Aitken MD. Effects of enrichment with salicylate on bacterial selection and PAH mineralization in a microbial community from a bioreactor treating contaminated soil. Environ Sci Technol. 2008;42(11):4099–105. 10.1021/Es703007n . [DOI] [PubMed] [Google Scholar]
- 35. Osborn AM, Moore ERB, Timmis KN. An evaluation of terminal-restriction fragment length polymorphism (T-RFLP) analysis for the study of microbial community structure and dynamics. Environ Microbiol. 2000;2(1):39–50. 10.1046/j.1462-2920.2000.00081.x . [DOI] [PubMed] [Google Scholar]
- 36. Cupples AM, Sims GK. Identification of in situ 2,4-dichlorophenoxyacetic acid-degrading soil microorganisms using DNA-stable isotope probing. Soil Biol Biochem. 2007;39(1):232–8. 10.1016/j.soilbio.2006.07.011 . [DOI] [Google Scholar]
- 37. Ludwig W, Bauer SH, Bauer M, Held I, Kirchhof G, Schulze R, et al. Detection and in situ identification of representatives of a widely distributed new bacterial phylum. Fems Microbiol Lett. 1997;153(1):181–90. 10.1016/S0378-1097(97)00256-5 . [DOI] [PubMed] [Google Scholar]
- 38. Dunbar J, Takala S, Barns SM, Davis JA, Kuske CR. Levels of bacterial community diversity in four arid soils compared by cultivation and 16S rRNA gene cloning. Appl Environ Microb. 1999;65(4):1662–9. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Janssen PH. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl Environ Microb. 2006;72(3):1719–28. 10.1128/Aem.72.3.1719-1728.2006 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kleinsteuber S, Muller FD, Chatzinotas A, Wendt-Potthoff K, Harms H. Diversity and in situ quantification of Acidobacteria subdivision 1 in an acidic mining lake. Fems Microbiol Ecol. 2008;63(1):107–17. 10.1111/j.1574-6941.2007.00402.x . [DOI] [PubMed] [Google Scholar]
- 41. Quaiser A, Lopez-Garcia P, Zivanovic Y, Henn MR, Rodriguez-Valera F, Moreira D. Comparative analysis of genome fragments of Acidobacteria from deep Mediterranean plankton. Environ Microbiol. 2008;10(10):2704–17. 10.1111/j.1462-2920.2008.01691.x . [DOI] [PubMed] [Google Scholar]
- 42. Abed RMM, Safi NMD, Koster J, de Beer D, El-Nahhal Y, Rullkotter J, et al. Microbial diversity of a heavily polluted microbial mat and its community changes following degradation of petroleum compounds. Appl Environ Microb. 2002;68(4):1674–83. 10.1128/Aem.68.4.1674-1683.2002 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rosano-Hernandez MC, Ramirez-Saad H, Fernandez-Linares L. Petroleum-influenced beach sediments of the Campeche Bank, Mexico: Diversity and bacterial community structure assessment. J Environ Manage. 2012;95:S325–S31. 10.1016/j.jenvman.2011.06.046 . [DOI] [PubMed] [Google Scholar]
- 44. Kumar M, Khanna S. Diversity of 16S rRNA and dioxygenase genes detected in coal-tar-contaminated site undergoing active bioremediation. J Appl Microbiol. 2010;108(4):1252–62. 10.1111/j.1365-2672.2009.04523.x . [DOI] [PubMed] [Google Scholar]
- 45. Perez-Leblic MI, Turmero A, Hernandez M, Hernandez AJ, Pastor J, Ball AS, et al. Influence of xenobiotic contaminants on landfill soil microbial activity and diversity. J Environ Manage. 2012;95:S285–S90. 10.1016/j.jenvman.2010.07.017 . [DOI] [PubMed] [Google Scholar]
- 46. Tejeda-Agredano MC, Gallego S, Vila J, Grifoll M, Ortega-Calvo JJ, Cantos M. Influence of the sunflower rhizosphere on the biodegradation of PAHs in soil. Soil Biol Biochem. 2013;57:830–40. 10.1016/j.soilbio.2012.08.008 . [DOI] [Google Scholar]
- 47. Correa PA, Lin LS, Just CL, Hu DF, Hornbuckle KC, Schnoor JL, et al. The effects of individual PCB congeners on the soil bacterial community structure and the abundance of biphenyl dioxygenase genes. Environ Int. 2010;36(8):901–6. 10.1016/j.envint.2009.07.015 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Di Gregorio S, Azaizeh H, Lorenzi R. Biostimulation of the autochthonous microbial community for the depletion of polychlorinated biphenyls (PCBs) in contaminated sediments. Environ Sci Pollut R. 2013;20(6):3989–99. 10.1007/s11356-012-1350-x . [DOI] [PubMed] [Google Scholar]
- 49. Gan YL, Qiu QF, Liu PF, Rui JP, Lu YH. Syntrophic Oxidation of Propionate in Rice Field Soil at 15 and 30 degrees C under Methanogenic Conditions. Appl Environ Microb. 2012;78(14):4923–32. 10.1128/Aem.00688-12 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jakobs-Schonwandt D, Mathies H, Abraham WR, Pritzkow W, Stephan I, Noll M. Biodegradation of a Biocide (Cu-N-Cyclohexyldiazenium Dioxide) Component of a Wood Preservative by a Defined Soil Bacterial Community. Appl Environ Microb. 2010;76(24):8076–83. 10.1128/Aem.01092-10 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Liu YJ, Liu SJ, Drake HL, Horn MA. Alphaproteobacteria dominate active 2-methyl-4-chlorophenoxyacetic acid herbicide degraders in agricultural soil and drilosphere. Environ Microbiol. 2011;13(4):991–1009. 10.1111/j.1462-2920.2010.02405.x . [DOI] [PubMed] [Google Scholar]
- 52. Hoppener-Ogawa S, Leveau JHJ, Smant W, van Veen JA, de Boer W. Specific detection and real-time PCR quantification of potentially mycophagous bacteria belonging to the genus Collimonas in different soil ecosystems. Appl Environ Microb. 2007;73(13):4191–7. 10.1128/Aem.00387-07 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. de Boer W, Leveau JHJ, Kowalchuk GA, Gunnewiek PJAK, Abeln ECA, Figge MJ, et al. Collimonas fungivorans gen. nov., sp nov., a chitinolytic soil bacterium with the ability to grow on living fungal hyphae. Int J Syst Evol Micr. 2004;54:857–64. 10.1099/ijs.0.02920-0 . [DOI] [PubMed] [Google Scholar]
- 54. Uroz S, Calvaruso C, Turpault MP, Pierrat JC, Mustin C, Frey-Klett P. Effect of the mycorrhizosphere on the genotypic and metabolic diversity of the bacterial communities involved in mineral weathering in a forest soil. Appl Environ Microb. 2007;73(9):3019–27. 10.1128/Aem.00121-07 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Uroz S, Calvaruso C, Turpault MP, Sarniguet A, de Boer W, Leveau JHJ, et al. Efficient mineral weathering is a distinctive functional trait of the bacterial genus Collimonas. Soil Biol Biochem. 2009;41(10):2178–86. 10.1016/j.soilbio.2009.07.031 . [DOI] [Google Scholar]
- 56. Hamamura N, Olson SH, Ward DM, Inskeep WP. Microbial population dynamics associated with crude-oil biodegradation in diverse soils. Appl Environ Microb. 2006;72(9):6316–24. 10.1128/Aem.01015-06 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Haritash AK, Kaushik CP. Biodegradation aspects of Polycyclic Aromatic Hydrocarbons (PAHs): A review. J Hazard Mater. 2009;169(1–3):1–15. 10.1016/j.jhazmat.2009.03.137 . [DOI] [PubMed] [Google Scholar]
- 58. Bosch R, Garcia-Valdes E, Moore ERB. Genetic characterization and evolutionary implications of a chromosomally encoded naphthalene-degradation upper pathway from Pseudomonas stutzeri AN10. Gene. 1999;236(1):149–57. 10.1016/S0378-1119(99)00241-3 . [DOI] [PubMed] [Google Scholar]
- 59. Mallick S, Chatterjee S, Dutta TK. A novel degradation pathway in the assimilation of phenanthrene by Staphylococcus sp strain PN/Y via meta-cleavage of 2-hydroxy-1-naphthoic acid: formation of trans-2,3-dioxo-5-(2 '-hydroxyphenyl)pent-4-enoic acid. Microbiol-Sgm. 2007;153:2104–15. 10.1099/mic.0.2006/004218-0 . [DOI] [PubMed] [Google Scholar]
- 60. Chang CH, Lee J, Ko BG, Kim SK, Chang JS. Staphylococcus sp KW-07 contains nahH gene encoding catechol 2,3-dioxygenase for phenanthrene degradation and a test in soil microcosm. Int Biodeter Biodegr. 2011;65(1):198–203. 10.1016/j.ibiod.2010.11.003 . [DOI] [Google Scholar]
- 61. Moscoso F, Teijiz I, Sanroman MA, Deive FJ. On the Suitability of a Bacterial Consortium To Implement a Continuous PAHs Biodegradation Process in a Stirred Tank Bioreactor. Ind Eng Chem Res. 2012;51(49):15895–900. 10.1021/Ie3021736 . [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic tree for the taxonomic location of the bacteria corresponding 219- and 241-bp TRFs. The tree is based on 16S rDNA sequence and produced by MEGA 5.0. 2.
(PDF)
The density distribution of total DNA in the micocosms amended with unlabled (12C) or labled (13C) PHE after centrifugation on days 3, 6 and 9.
(PDF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
Data relevant to Table 1, S1 Table, S2 Fig, and Fig 1 are within the paper and its Supporting Information files. The information about the DNA sequences are available from the GenBank database (KF035825 and KF035826 for partial 16S rRNA gene sequences, and KF656718 and KF656719 for partial RHD gene sequences, KF035827 to KF035829 for sequences of the isolates).