Abstract
The Gram-negative bacterium Escherichia coli is routinely used as the chassis for a variety of biotechnology and synthetic biology applications. Identification and analysis of reliable chromosomal integration and expression target loci is crucial for E. coli engineering. Chromosomal loci differ significantly in their ability to support integration and expression of the integrated genetic circuits. In this study, we investigate E. coli K12 MG1655 flagellar regions 2 and 3b. Integration of the genetic circuit into seven and nine highly conserved genes of the flagellar regions 2 (motA, motB, flhD, flhE, cheW, cheY and cheZ) and 3b (fliE, F, G, J, K, L, M, P, R), respectively, showed significant variation in their ability to support chromosomal integration and expression of the integrated genetic circuit. While not reducing the growth of the engineered strains, the integrations into all 16 target sites led to the loss of motility. In addition to high expression, the flagellar region 3b supports the highest efficiency of integration of all E. coli K12 MG1655 flagellar regions and is therefore potentially the most suitable for the integration of synthetic genetic circuits.
Introduction
The Gram-negative model bacterium Escherichia coli is capable of thriving in a wide variety of environments (Juhas et al., 2014a). Easily amenable to genetic manipulations, E. coli strain K-12 is among the most frequently used hosts for cloning and the intermediate and the final destination chassis for engineering large DNA fragments. Escherichia coli K-12 is also important for a number of industrial applications, biomanufacturing and metabolic engineering (Ajikumar et al., 2010; Zhang et al., 2010; Clomburg and Gonzalez, 2011; Yim et al., 2011; Zhou et al., 2012). With the advent of synthetic biology, E. coli K-12 has become one of the most frequently used synthetic biology host organisms (Juhas et al., 2013; 2014a; Juhas, 2015).
Introduction of the synthetic DNA fragments into the E. coli genome by chromosomal integration has many advantages over the plasmid-borne transformation (Cunningham et al., 2009; Marcellin et al., 2010). Furthermore, integration into the chromosome could be exploited for heterologous protein expression, particularly for expression of toxic proteins in E. coli. Work on plasmids has shown that regulation of expression is tighter when the copy number is low (Anthony et al., 2004; Guan et al., 2013). The frequently used methods of the E. coli chromosomal integration include the integrase-mediated recombination between the phage attachment sites (att) (St-Pierre et al., 2013) and the λ bacteriophage Red recombinase-mediated recombination employing knock-in/knock-out (KIKO) vectors (Sabri et al., 2013), plasmid pSB1K3(FRTK) (Juhas et al., 2014b) and the yeast mitochondrial homing endonuclease I-SceI (Ublinskaya et al., 2012). Chromosomal integration target sites differ significantly in their ability to support integration and expression of the integrated genetic circuits (Juhas et al., 2014b). As the traditionally used att sites are missing in a number of industrially important E. coli strains, identification and validation of the reliable chromosomal integration target sites is crucial for E. coli engineering. Ideally, integration target sites should be well-characterized, non-essential, conserved and highly expressed (Fraser et al., 1999; Baba et al., 2006; Vora et al., 2009; Kahramanoglou et al., 2011; Juhas et al., 2014b). Genes encoding flagellar functions meet all these prerequisites (Juhas et al., 2014b). Previous analyses of the E. coli K12 MG1655 flagellar regions 3a and 1 led to the identification of only three potential integration target sites (Juhas et al., 2014b; Juhas and Ajioka, 2015). The identification and validation of alternative integration sites is crucial for the development of a robust synthetic biology toolkit (Juhas and Ajioka, 2015). This is critical particularly for applications that require integrations of multiple genetic circuits into the chromosome. Here, we investigate the E. coli K12 MG1655 flagellar regions 2 and 3b. Analysis of the seven and nine highly conserved genes of the flagellar regions 2 and 3b, respectively, revealed significant variability in their suitability for integration and expression of genetic circuits. Furthermore, we show that in addition to high expression, the E. coli K12 MG1655 flagellar region 3b supports highest efficiency of chromosomal integration of all E. coli flagellar regions.
Results and discussion
Integration target loci in the E. coli flagellar regions 2 and 3b
Identification of the reliable chromosomal integration target loci is crucial for engineering E. coli cells (Sabri et al., 2013; Juhas et al., 2014b). Chromosomal integration target sites should be well-characterized, conserved, non-essential and highly expressed (Fraser et al., 1999; Baba et al., 2006; Vora et al., 2009; Kahramanoglou et al., 2011; Juhas et al., 2014b; Juhas, 2015). Genes encoding flagellar functions are considered to be among the best targets for integration of genetic circuits into the E. coli chromosome (Juhas et al., 2014b). Previous studies investigating E. coli K12 MG1655 flagellar regions 3a (Juhas et al., 2014b) and 1 (Juhas and Ajioka, 2015) led to the identification of three putative chromosomal integration target sites. Identification and validation of the alternative loci is important particularly for those biotechnology and synthetic biology applications that require integrations of multiple genetic circuits into E. coli chromosome.
Here, we investigate E. coli K12 MG1655 flagellar regions 2 and 3b. Escherichia coli K12 MG1655 flagellar regions 2 (Fig. 1A) and 3b (Fig. 1B) show high probability of the RNA polymerase binding. This suggests that genetic circuits integrated into these regions will be strongly transcribed. Escherichia coli K12 MG1655 flagellar regions 2 and 3b harbour 28 open reading frames (flhA, flhB, flhC, flhD, flhE, motA, motB, cheA, cheB, cheR, cheW, cheY, cheZ, tar, fliE, fliF, fliG, fliH, fliI, fliJ, fliK, fliL, fliM, fliN, fliO, fliP, fliQ, fliR) (Fig. 1). Our investigation revealed that seven and five genes of the E. coli K12 MG1655 flagellar regions 2 and 3b, respectively, are not fitting for integration of genetic circuits because of low conservation or lack of suitable integration target sequences. The other seven genes of the flagellar region 2 (motA, motB, flhD, flhE, cheW, cheY, cheZ) (Table 1) and nine genes of the flagellar region 3b (fliE, fliF, fliG, fliJ, fliK, fliL, fliM, fliP, fliR) (Table 2) are highly conserved among E. coli strains, including industrially relevant strains, such as BL21-DE3, W3110, DH10B and MG1655. The function and location of the analysed chromosomal integration target loci in the E. coli K12 MG1655 flagellar regions 2 and 3b are shown in Fig. 1 and Tables 1 and 2. We have integrated the genetic circuit Repr-ts-1 (Fig. 2) harbouring thermosensitive lambda repressor into these loci using the modified lambda Red recombinase integration method (Juhas et al., 2014b).
Fig 1.
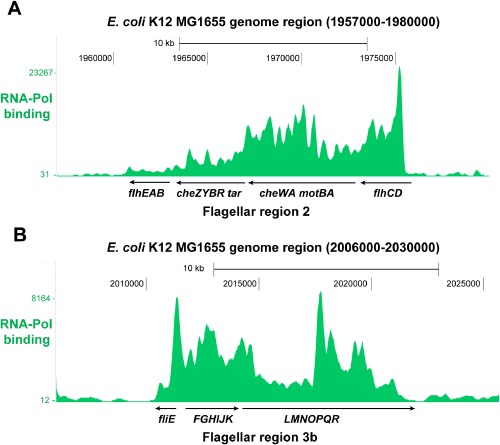
RNA polymerase binding to E. coli flagellar regions 2 and 3b. Figure depicts the probability of the RNA polymerase (RNA-Pol) binding (green peaks) to the E. coli K12 MG1655 genome regions 1957000-1980000 (A) and 2006000-2030000 (B). The investigated E. coli K12 MG1655 flagellar regions 2 (1962580-1978197) and 3b (2011038-2021702) show high probability of being occupied by RNA polymerase. Figure was created by uploading the ChIP-seq RNA-Pol data (Kahramanoglou et al., 2011) to the UCSC genome browser for E. coli K12 MG1655.
Table 1.
Integration target loci in the E. coli flagellar region 2
| Gene | Function | References |
|---|---|---|
| motA | Flagellar motor component | Mohawk et al., 2014; Takahashi and Ito, 2014 |
| motB | Flagellar motor component | Reboul et al., 2011; Takahashi et al., 2014 |
| flhD | Master regulator of flagellar genes | Chatterjee et al., 2009; Mitra et al., 2013 |
| flhE | Proton influx regulator via T3SS | Lee and Harshey, 2012 |
| cheW | Chemotaxis signal transduction | Cashman et al., 2013 |
| cheY | Chemotaxis response regulator, clockwise flagellar rotation | Fraiberg et al., 2015 |
| cheZ | Phosphatase, cheY dephosphorylation | Freeman et al., 2011 |
Table 2.
Integration target loci in the E. coli flagellar region 3b
| Gene | Function | References |
|---|---|---|
| fliE | Flagellar basal body component | Dyszel et al., 2010 |
| fliF | Membrane and supramembrane (MS)-ring collar protein, flagellar basal body | Ogawa et al., 2015 |
| fliG | Flagellar motor switching | Lam et al., 2012 |
| fliJ | Flagellar protein export apparatus, rotor like function | Kishikawa et al., 2013 |
| fliK | Flagellar hook-length control | Aizawa, 2012 |
| fliL | Flagellar motor output control | Partridge et al., 2015 |
| fliM | Flagellar motor energizing | Delalez et al., 2014 |
| fliP | Flagellar export apparatus | Boyd and Gober, 2001 |
| fliR | Flagellar export apparatus | Minamino and Macnab, 1999 |
Fig 2.
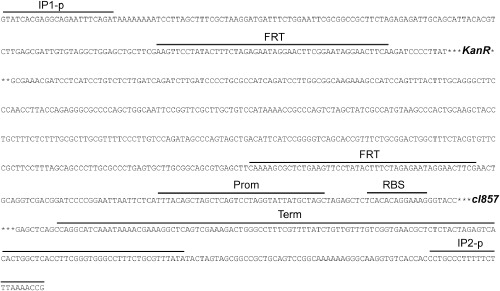
Genetic circuit Repr-ts-1. Figure shows sequence of the genetic circuit Repr-ts-1 integrated into the chromosome. IP1-p and IP2-p (integration primer parts); FRT (Flp recombinase target site); KanR (kanamycin resistance gene); Prom (promoter); RBS (ribosome binding site); cI857 (temperature-sensitive lambda repressor); Term (terminator).
High efficiency integration into E. coli flagellar region 3b
As E. coli chromosomal loci differ in their ability to support integration of genetic circuits (Juhas et al., 2014b), we investigated the integration efficiency for each of the 16 target loci. Genetic circuit was integrated into the investigated target sites [motA (motAi), motB (motBi), flhD (flhDi), flhE (fhlEi), cheW (cheWi), cheY (cheYi), cheZ (cheZi)], fliE (fliEi), fliF (fliFi), fliG (fliGi), fliJ (fliJi), fliK (fliKi), fliL (fliLi), fliM (fliMi), fliP (fliPi) and fliR (fliRi)] (Figs 3 and 4) and the integration efficiency was determined from the number of colony-forming units per microgram of electroporated DNA. Chromosomal integrations were confirmed with polymerase chain reaction (PCR) using flanking primers (Fig. S1) and sequencing. The primers used for the integration of the genetic circuit into the chromosome and the flanking primers are listed in Table 3. The investigated target loci differed significantly in their suitability to support integration of the genetic circuit. From the analysed genes of the E. coli K12 MG1655 flagellar region 2, the integration efficiency into motA (motAi) was highest (Fig. 5). From the E. coli K12 MG1655 flagellar region 3b, fliK (fliKi) supports the highest integration efficiency (Fig. 5). Notably, integrations into one and four loci of the flagellar regions 2 (motAi) and 3b (fliEi, fliJi, fliKi, fliRi), respectively, occurred with the higher efficiency than integrations into the previously examined flagellar regions 3a (Juhas et al., 2014b) and 1 (Juhas and Ajioka, 2015). Furthermore, integration efficiency into fliK (fliKi) was significantly higher than that of motA (motAi) (Fig. 5). Hence, the E. coli K12 MG1655 flagellar region 3b supports the highest efficiency of integration of all E. coli flagellar regions.
Fig 3.

Escherichia coli flagellar region 2 integrations. Figure shows the sequences of the integration target sites in the E. coli K12 MG1655 flagellar region 2 (motA (motAi), motB (motBi), flhD (flhDi), flhE (flhEi), cheW (cheWi), cheY (cheYi). The exact positions within the target genes where the integrations occurred are highlighted with stars.
Fig 4.

Escherichia coli flagellar region 3b integrations. Figure shows the sequences of the integration target sites in the E. coli K12 MG1655 flagellar region 3b (fliE (fliEi), fliF (fliFi), fliG (fliGi), fliJ (fliJi), fliK (fliKi), fliL (fliLi), fliM (fliMi), fliP (fliPi). The exact positions within the target genes where the integrations occurred are highlighted with stars.
Table 3.
Primers used in this study
| Primer (Sequence 5′→ 3′) |
|---|
| motArepF: motA integration primer forward |
| CTCCAAATACACCAAAGCAATGTATATGGATCTGCTGGCTCTGCTTTATCGGTTGATGGCGAAATCGTATCACGAGGCAGAATTTCAGAT |
| motArepR: motA integration primer reverse |
| TTTCTGACGTAAAACAGTCGCTAATGGGGAAATAAATCCGTAAGCCAATAAAATGCCGAGGAAAGTCGGTTTTAAAGAAAAAGGGCAGG |
| motBrepF: motB integration primer forward |
| ACATGGATCGTGGAAGATTGCTTATGCCGACTTTATGACTGCGATGATGGCCTTTTTTCTGGTGATGTATCACGAGGCAGAATTTCAGAT |
| motBrepR: motB integration primer reverse |
| AATTTCCGCAATCGACTTTGCTCCATGCGTTTTTTCAGCTCTTCGATGTTCGGCTGCTTATTCACTCGGTTTTAAAGAAAAAGGGCAGG |
| flhDrepF: flhD integration primer forward |
| ATGCATACCTCCGAGTTGCTGAAACACATTTATGACATCAACTTGTCATATTTACTACTTGCACAGGTATCACGAGGCAGAATTTCAGAT |
| flhDrepR: flhD integration primer reverse |
| CTTCTTCAGGCTGATTAACATCATTCAGCAAGCGTGTTGAGAGCATGATGCCGGTATGAATTTGCTCGGTTTTAAAGAAAAAGGGCAGG |
| flhErepF: flhE integration primer forward |
| CAATTGGCGGCAAATAATGAGAACCTTATTAGCAATATTATTGTTTCCGCTGCTGGTGCAAGCCGGTATCACGAGGCAGAATTTCAGAT |
| flhErepR: flhE integration primer reverse |
| TTGTCCTTCAGCGATAATTCACAATCACTTCATTACGTTGTACCTTTAGCGGTGGAATTAACCGCCCGGTTTTAAAGAAAAAGGGCAGG |
| cheWrepF: cheW integration primer forward |
| TACCCTTGGTGATGAAGAGTACGGTATTGATATCCTGAAAGTGCAGGAGATCCGTGGCTACGATCAGTATCACGAGGCAGAATTTCAGAT |
| cheWrepR: cheW integration primer reverse |
| TATCTAACAGCGCCATCTCTTCGCTGTTCAGCAGTTTTTCGATGTTCACCAGAATCAACATCCGGTCGGTTTTAAAGAAAAAGGGCAGG |
| cheYrepF: cheY integration primer forward |
| AAAGAGCTGGGATTCAATAATGTTGAGGAAGCGGAAGATGGCGTCGACGCTCTCAATAAGTTGCAGGTATCACGAGGCAGAATTTCAGAT |
| cheYrepR: cheY integration primer reverse |
| AGTTTCTCAAAGATTTTGTTGAGTTTTTCCTCCAGCGTCGCGGCGGTAAATGGCTTCACCACATAGCGGTTTTAAAGAAAAAGGGCAGG |
| cheZrepF: cheZ integration primer forward |
| CAGGATTTTCAGGATCTCACCGGGCAGGTCATTAAGCGGATGATGGATGTCATTCAGGAGATCGAAGTATCACGAGGCAGAATTTCAGAT |
| cheZrepR: cheZ integration primer reverse |
| TCAAAATCCAAGACTATCCAACAAATCGTCCACCTGATCCTGACTGGCTACCACACCGGCTTTGCTCGGTTTTAAAGAAAAAGGGCAGG |
| motArepTF: motA integration test primer forward |
| GCGCTGCCGTTGCTGTTTCG |
| motArepTR: motA integration test primer reverse |
| GCAGAGTGACTTTGACGCACTGCA |
| motBrepTF: motB integration test primer forward |
| CGTCAAACGACGCAAAGCCAAAAGC |
| motBrepTR: motB integration test primer reverse |
| GACGTAACGCCCGCAGTTTCG |
| flhDrepTF: flhD integration test primer forward |
| GCTTCCCGGCGACATCACG |
| flhDrepTR: flhD integration test primer reverse |
| AGGCCCTTTTCTTGCGCAGC |
| flhErepTF: flhE integration test primer forward |
| CTGTCTGATAACCGACATATCCGCATGACG |
| flhErepTR: flhE integration test primer reverse |
| CACTGAGTTATTAAACATACTCGCGAGCGC |
| cheWrepTF: cheW integration test primer forward |
| ACCGCCGCCTGAATGAGTAAAAAGG |
| cheWrepTR: cheW integration test primer reverse |
| CGGGAGAATTACGCCACTTCTGACG |
| cheYrepTF: cheY integration test primer forward |
| CCACCATGCGACGCATAGTGC |
| cheYrepTR: cheY integration test primer reverse |
| GCATCATAGTCGCATCCTCACATGCC |
| cheZrepTF: cheZ integration test primer forward |
| CACGACAATTTCTGGCAGATGTACCCG |
| cheZrepTR: cheZ integration test primer reverse |
| TCAGACCGCCTGATATGACGTGGT |
| fliEreprF: fliE integration primer forward |
| GTTAAACGATGTGATGACCGATATGCAAAAAGCCTCAGTTTCTATGCAAATGGGGATTCAGGTGCGGTATCACGAGGCAGAATTTCAGAT |
| fliEreprR: fliE integration primer reverse |
| GGATCGTGGATGGCAGAAAACGTTCAGGATCAGGTATCCATTTTAAACCAGAAATTGAGTGAGTTTCGGTTTTAAAGAAAAAGGGCAGG |
| fliFreprF: fliF integration primer forward |
| ATTATTCAGCAATCTTTCCGATCAGGATGGTGGCGCAATTGTCAGCCAACTGACGCAAATGAATATGTATCACGAGGCAGAATTTCAGAT |
| fliFreprR: fliF integration primer reverse |
| CTTCAATACGCCGCTGAATACGGCCTTCGACATCGCTGGCATATTTCAACTGAGCGTCATTAAGATCGGTTTTAAAGAAAAAGGGCAGG |
| fliGreprF: fliG integration primer forward |
| GCCGCACTGAATATCAACGCCAACGATTATCTGCGCTCGGTATTGGTCAAAGCTCTGGGTGAAGAAGTATCACGAGGCAGAATTTCAGAT |
| fliGreprR: fliG integration primer reverse |
| CGCTGAATGCTGCGATCGTCGACATCCACCAGATTCTCGAACAGGAACATCTCGTCGATGATTTTCCGGTTTTAAAGAAAAAGGGCAGG |
| fliJreprF: fliJ integration primer forward |
| GAAATGCGTCGCGGATGTCAGCAGGCGGAAGAACAGCTCAAAATGCTGATTGATTATCAGAATGAAGTATCACGAGGCAGAATTTCAGAT |
| fliJreprR: fliJ integration primer reverse |
| TGCAAACGTTGTTTTTTTTCTCGCCAACTGTTCAGGGCAATGTCAACTTTCTGCGTCCACTGATTACGGTTTTAAAGAAAAAGGGCAGG |
| fliKreprF: fliK integration primer forward |
| TATTGTTTCCGACGCGCAACAAGCTAATTTACTGATCCCTGTGGATGAAACACCGCCTGTCATCAAGTATCACGAGGCAGAATTTCAGAT |
| fliKreprR: fliK integration primer reverse |
| GTGAAACCATCTGGATTTGCGCCTGGTTATCATCCACTTTGAGGGAGATTTGCACTTCACCTAAATCGGTTTTAAAGAAAAAGGGCAGG |
| fliLreprF: fliL integration primer forward |
| ATGACTGATTACGCGATAAGCAAGAAAAGCAAGCGATCGCTTTGGATCCCGATTCTGGTATTCATTGTATCACGAGGCAGAATTTCAGAT |
| fliLreprF: fliL integration primer reverse |
| GTGGTTTTAATCTCGGCAATCAGGTTTTTCTTGCCTTCTTCTGTCGCCAGTACGGCAGCATCCTGACGGTTTTAAAGAAAAAGGGCAGG |
| fliMreprF: fliM integration primer forward |
| AAATTTACCAATATCACCACCTCGCCGAACGACATTGTGGTTAACACGCCGTTCCATGTGGAGATTGTATCACGAGGCAGAATTTCAGAT |
| fliMreprF: fliM integration primer reverse |
| AAATCGGGTTAATCAAATGTTCTATCCGTAACGCATACTGACCGTTGAGGGTGCCATACTGACTGGCGGTTTTAAAGAAAAAGGGCAGG |
| fliPreprF: fliP integration primer forward |
| GGTATTGCTGGGGCTGGCACTGTTTTTGACCTTTTTTATTATGTCACCGGTGATCGACAAAATTTAGTATCACGAGGCAGAATTTCAGAT |
| fliPreprF: fliP integration primer reverse |
| AACACGCTGGCTATCACCAGGTCGATAATCAAAAAAGGGATGAAAATCGTGAAGCCTATCTGAAATCGGTTTTAAAGAAAAAGGGCAGG |
| fliRreprF: fliP integration primer forward |
| TATCATGGATATGCTGGCGTTACTGCTGTTCCTGACATTTAACGGTCATTTATGGTTGATTTCACTGTATCACGAGGCAGAATTTCAGAT |
| fliRreprF: fliP integration primer reverse |
| ATAATATCAGCCAGCAAATTAAAAATTTCACTGAATAAATGTTCGCAAAAAGGTGCAATTAACGGCCGGTTTTAAAGAAAAAGGGCAGG |
| fliErepTF: fliE integration test primer forward |
| GCCGCAACCGACCATTAGTTTTGC |
| fliErepTR: fliE integration test primer reverse |
| GCTCTGATGTTCAGGGGGGTTATCG |
| fliFrepTF: fliF integration test primer forward |
| GTCATGGTCGCACTGATCCTGTGG |
| fliFrepTR: fliF integration test primer reverse |
| CAGCTGCGCCGTAACCTGG |
| fliGrepTF: fliG integration test primer forward |
| CTGGCGGAGTTTGAGCAAGAAGC |
| fliGrepTR: fliG integration test primer reverse |
| CGGCGTCCAGGTTTTCCACG |
| fliJrepTF: fliJ integration test primer forward |
| GCTGGCGACCCTGAAAGATCTGG |
| fliJrepTR: fliJ integration test primer reverse |
| GCCGTTCCTGCAGTGTCTGC |
| fliKrepTF: fliK integration test primer forward |
| GCCCACGACAAAAGGCGAGC |
| fliKrepTR: fliK integration test primer reverse |
| GGTTCATGGTTTGCTGTGCGTTGG |
| fliLrepTF: fliL integration test primer forward |
| AGCAGTAGCGACACAGGAAGACC |
| fliLrepTR: fliL integration test primer reverse |
| CGGTGACATCCTGTTTCGGTTGC |
| fliMrepTF: fliM integration test primer forward |
| GTTGAGTACGTGCGTTCGGAAATGC |
| fliMrepTR: fliM integration test primer reverse |
| CACTCATTTGGGCTGTTCCTCGTTCAG |
| fliPrepTF: fliP integration test primer forward |
| GTGGACAAAGCTGGTCGCTCC |
| fliPrepTR: fliP integration test primer reverse |
| GGCAGAGCAATGGTGGCTGG |
| fliRrepTF: fliR integration test primer forward |
| ATGGGGCTGTCATTTGCGACG |
| fliRrepTR: fliR integration test primer reverse |
| CAGGCGGACTTACTATCCCGTAAAGTG |
Fig 5.
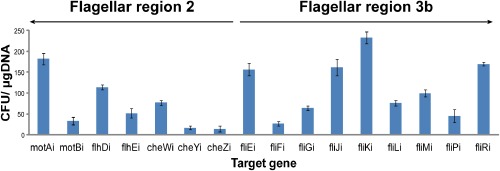
High integration efficiency into the E. coli flagellar region 3b. Figure showing the integration efficiency (CFU per microgram of electroporated DNA) into the investigated target loci of the E. coli K12 MG1655 flagellar regions 2 [motA (motAi), motB (motBi), flhD (flhDi), flhE (flhEi), cheW (cheWi), cheY (cheYi) and cheZ (cheZi)] and 3b [fliE (fliEi), fliF (fliFi), fliG (fliGi), fliJ (fliJi), fliK (fliKi), fliL (fliLi), fliM (fliMi), fliP (fliPi) and fliR (fliRi)]. The figure shows means and standard deviations from three experiments.
Integrations into flagellar regions 2 and 3b abolish motility
Flagellum is crucial for the motility of E. coli cells. Therefore, the disruptions of the flagellar functions-encoding genes usually have a negatively impact on motility (Juhas et al., 2014b). Integrations into two genes of the previously analysed flagellar region 3a only reduced motility of the engineered strains when compared with the wild type (Juhas et al., 2014b). We investigated the effect of the chromosomal integrations into the flagellar regions 2 and 3b by spotting 2 μl of the normalized overnight cultures of the engineered E. coli strains and E. coli K12 MG1655 wild type in the middle of the motility agar plates (Fig. 6). The motility of all strains harbouring integrations in the investigated genes of the flagellar regions 2 (Fig. 6A) and 3b (Fig. 6B) was completely abolished.
Fig 6.
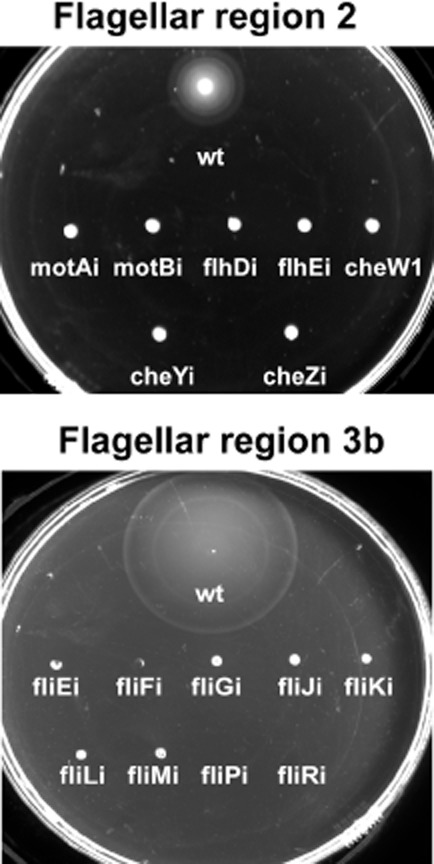
Integrations into the E. coli flagellar regions 2 and 3b abolish motility. The integrations into the investigated target sites of the flagellar region 2 [motA (motAi), motB (motBi), flhD (flhDi), flhE (flhEi), cheW (cheWi), cheY (cheYi) and cheZ (cheZi)] and 3b [fliE (fliEi), fliF (fliFi), fliG (fliGi), fliJ (fliJi), fliK (fliKi), fliL (fliLi), fliM (fliMi), fliP (fliPi)] completely abolished the motility of the engineered strains. Wt (E. coli K12 MG1655 wild type). E. coli cultures normalized to OD600 of 1 were inoculated in the motility agar plates and the picture was taken after 5 h of incubation at 37°C.
Integrations into flagellar regions do not have negative impact on the growth
As integrations of the synthetic genetic circuits into the E. coli chromosome should not negatively impact cell growth, target loci cannot be located within essential genes (Juhas et al., 2011; 2012a,b; 2014a). To assess the effect of chromosomal integrations into the seven investigated genes of the flagellar regions 2 (motA, motB, flhD, flhE, cheW, cheY and cheZ) and 3b (fliE, fliF, fliG, fliJ, fliK, fliL, fliM, fliP and fliR) on the growth rate, the absorbance of the engineered strains and K12 MG1655 wild type was measured with the microplate reader (Fluostar Omega). Integrations into all investigated genes of the flagellar regions 2 (Fig. S2) and 3b (Fig. S3) did not diminish growth rate when compared with the wild type at both 30°C and 37°C. This is consistent with previous results from flagellar regions 3a (Juhas et al., 2014b) and 1 (Juhas and Ajioka, 2015).
Transcription of the flagellar regions 2 and 3b
The relative transcription of the investigated genes of the flagellar regions 2 and 3b was measured by real-time polymerase chain reaction (RT-PCR) using arcA and rpoD as the reference housekeeping genes (Jandu et al., 2009; Minty et al., 2011). Real-time polymerase chain reaction (RT-PCR) showed that the relative expression of four genes from both analysed flagellar regions 2 (motA, motB, flhD, cheY) and 3b (fliJ, fliK, fliL, fliM) was higher (twofold to fivefold) than the average expression of the housekeeping genes (Fig. 7A). The relative transcription of fliG was not significantly different, whereas the transcription of the remaining genes was lower than the mean expression of the housekeeping genes (Fig. 7A). The transcription of the genetic circuit integrated into motA (motAi), motB (motBi), flhD (flhDi), flhE (fhlEi), cheW (cheWi), cheY (cheYi) and cheZ (cheZi) of the flagellar region 2 and fliE (fliEi), fliF (fliFi), fliG (fliGi), fliJ (fliJi), fliK (fliKi), fliL (fliLi), fliM (fliMi), fliP (fliPi) and fliR (fliRi) of the flagellar region 3b measured by RT-PCR was high at all analysed loci (Fig. 7B). From the flagellar region 2, highest expressed (8- to 11-fold higher than the housekeeping genes) was the genetic circuit integrated into motA (motAi), motB (motBi) and flhD (flhDi) (Fig. 7B). The expression at flhE (fhlEi), cheW (cheWi) and cheY (cheYi) was fourfold to sixfold higher than the mean expression of the housekeeping genes (Fig. 7B). From the flagellar region 3b, highest expressed (8- to 13-fold higher than the housekeeping genes) was the genetic circuit integrated into fliJ (fliJAi), fliL (fliLi) and fliR (fliRi) (Fig. 7B). The expression at the remaining loci of the flagellar region 3b was sixfold to eightfold higher than the mean expression of the housekeeping genes (Fig. 7B). Such strong expression of the genetic circuit integrated into this flagellar region is interesting, particularly when considering that the flagellar region 3b shows lowest probability of being occupied by RNA polymerase (Fig. 1B). This suggests that other factors might be also important for the expression of the integrated synthetic DNA and shows that empirical characterization is necessary for engineering into integration sites. Expression of the integrated genetic circuit was determined by the quantitative measurement of the green fluorescent protein (GFP) and the red fluorescent protein (mCherry) fluorescence over time with the microplate reader (FLUOstar Omega). For this, we have used plasmids pSB1A1(GFP) and pSB1A1(mCh) harbouring GFP and mCherry, respectively, regulated by the pR promoter. Both GFP and mCherry were not expressed at permissive conditions for the repressor (30°C), while the temperature shift to 42°C set off GFP and mCherry expression (Figs S4–S7).
Fig 7.
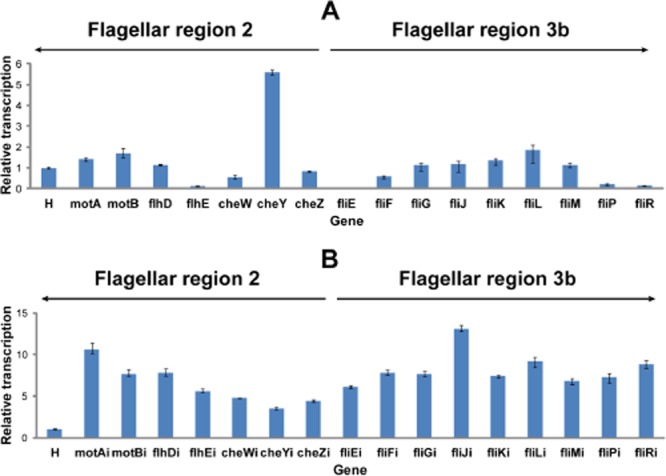
RT-PCR analysis.A. Relative transcription of the analysed target genes of the E. coli K12 MG1655 flagellar regions 2 (motA, motB, flhD, flhE, cheW, cheY and cheZ) and 3b (fliE, fliF, fliG, fliJ, fliK, fliL, fliM, fliP, fliR) compared with the housekeeping genes (H).B. Transcription of the genetic circuit integrated in the investigated integration target loci of the E. coli flagellar region 2 [motA (motAi), motB (motBi), flhD (flhDi), flhE (flhEi), cheW (cheWi), cheY (cheYi) and cheZ (cheZi)] and flagellar region 3b [(fliE (fliEi), fliF (fliFi), fliG (fliGi), fliJ (fliJi), fliK (fliKi), fliL (fliLi), fliM (fliMi), fliP (fliPi) and fliR (fliRi)] relative to the transcription of the housekeeping genes (H). Bars and errors represent averages and standard errors from three experiments. H (mean transcription of the reference housekeeping genes arcA and rpoD). Relative transcription was quantified with rest9 Software (Qiagen) employing Pfaffl method (Pfaffl et al., 2002).
Conclusions
Flagellar regions are good targets for integration of genetic circuits into the E. coli chromosome. The identification of reliable target loci is crucial for building a robust synthetic biology toolkit and for E. coli bioengineering. Furthermore, it can be exploited for tighter regulation of expression of toxic proteins in E. coli. In this study, we have integrated genetic circuit into 16 well-conserved open reading frames of the E. coli K12 MG1655 flagellar regions 2 (motA, motB, flhD, flhE, cheW, cheY and cheZ) and 3b (fliE, fliF, fliG, fliJ, fliK, fliL, fliM, fliP and fliR). The integrations into all target loci of these flagellar regions led to the loss of motility, but did not reduce the growth rate of the engineered strains. Escherichia coli K12 MG1655 flagellar region 3b supports highest efficiency of integration of all E. coli flagellar regions. Notably, the genetic circuit integrated into flagellar region 3b was also highly expressed although the probability of the RNA polymerase binding into this region is significantly lower than into other flagellar regions. This suggests that other factors might also play a role in the expression of the integrated synthetic DNA. There appears to be a weak inverse correlation between the probability of RNA polymerase binding to the target loci and their ability to support integration of the genetic circuit; however, this will require further investigation. Furthermore, as flagellar genes are closer to the terminal (TER) region of the E. coli chromosome than oriC, their copy number is approximately sixfold lower than those genes close to oriC during exponential growth. Therefore, genes nearer to oriC are also potentially interesting target loci for integration and expression of genetic circuits. Besides the modified lambda Red recombinase method used in our analysis, clustered regularly interspaced short palindromic repeats (CRISPR) and integrases could be exploited for E. coli engineering. A variety of high complexity integrase sites, such as phiC31, R4 and Bxb1, could be moved to the hotspot integration regions in the E. coli chromosome employing CRISPR for appending new functionalities. Overall, the E. coli K12 MG1655 flagellar region 3b is the most suitable of all E. coli flagellar regions for integration and expression of genetic circuits. However, there is a significant variation between individual target loci. For instance, motA of the E. coli K12 MG1655 flagellar region 2 supports the second highest integration and expression efficiency of all investigated target sites in this study (Figs 5 and 7). Therefore, when considered individually, fliJ and motA appear to be the most suitable integration target loci of the analysed flagellar regions 2 and 3b.
Experimental procedures
Bacterial strains, plasmids and growth conditions
All strains and plasmids used in this study are recorded in Table 4. Escherichia coli was routinely grown in Luria–Bertani (LB) medium supplemented with ampicillin (100 μg ml−1) or kanamycin (50 μg ml−1) when required. Liquid E. coli cultures were cultivated on a rotatory shaker at 200 r.p.m. at 30°C, 37°C or 42°C. Plate cultures were supplemented with 1% agar (w/v) and grown for about 24 h at 30°C, 37°C or 42°C.
Table 4.
Bacterial strains and plasmids
| Characteristics | Reference | |
|---|---|---|
| Strains | ||
| K12 MG1655 | E. coli wild type | Hayashi et al., 2006 |
| Ec:motAi | E. coli K12 MG1655, motA integration | This study |
| Ec:motBi | E. coli K12 MG1655, motB integration | This study |
| Ec:flhDi | E. coli K12 MG1655, flhD integration | This study |
| Ec:flhEi | E. coli K12 MG1655, flhE integration | This study |
| Ec:cheWi | E. coli K12 MG1655, cheW integration | This study |
| Ec:cheYi | E. coli K12 MG1655, cheY integration | This study |
| Ec:cheZi | E. coli K12 MG1655, cheZ integration | This study |
| Ec:fliEi | E. coli K12 MG1655 integration into fliE | This study |
| Ec:fliFi | E. coli K12 MG1655 integration into fliF | This study |
| Ec:fliGi | E. coli K12 MG1655 integration into fliG | This study |
| Ec:fliJi | E. coli K12 MG1655 integration into fliJ | This study |
| Ec:fliK | E. coli K12 MG1655 integration into fliK | This study |
| Ec:fliLi | E. coli K12 MG1655 integration into fliL | This study |
| Ec:fliMi | E. coli K12 MG1655 integration into fliM | This study |
| Ec:fliPi | E. coli K12 MG1655 integration into fliP | This study |
| Ec:fliRi | E. coli K12 MG1655 integration into fliR | This study |
| Plasmids | ||
| pCP20 | Plasmid encoding FLP recombinase | Datsenko and Wanner, 2000 |
| pKM208 | IPTG-induced Red recombinase system | Murphy and Campellone, 2003 |
| pSB1A1(GFP) | λ promoter-controlled GFP, AmpR | Juhas et al., 2014b |
| pSB1A1(mCh) | λ promoter-controlled mCherry, AmpR | This study |
| pSB1K3(FRTKr) | λ repressor, KanR | Juhas et al., 2014b |
DNA amplification and modification
DNA was amplified by PCR in 50 μl of reaction volumes employing Phusion DNA polymerase (Thermo Scientific) or Dream Taq master mix kit (Thermo Scientific) according to the supplier's instructions. Oligonucleotide primers for PCR amplifications were synthesized by Integrated DNA Technologies (IDT) and Sigma-Aldrich. DNA fragments were purified by gel electrophoresis, followed by gel extraction employing Qiaquick Gel Extraction kit (Qiagen), according to the manufacturer's instructions. Plasmid DNA was performed with the Qiaprep Spin Miniprep kit (Qiagen), according to the supplier's recommendations. Sequencing was performed by Source Bioscience (Cambridge, UK). A Gibson Isothermal Assembly method (Gibson et al., 2009; Merryman and Gibson, 2012) was employed to assemble DNA fragments. The original Gibson Isothermal Assembly method protocol was modified as described previously (Juhas et al., 2014b).
Integration of the genetic circuit into the chromosome
Altered Hannah (Hanahan et al., 1991) and Miller and Nickoloff (1995) protocols were used to prepare the chemically competent and electro-competent E. coli cells respectively. Integrations of the genetic circuit into target open reading frames of the analysed E. coli flagellar region were carried out using method described previously (Juhas et al., 2014b). Briefly, plasmid pKM208 was transformed into the wild-type E. coli K12 MG1655 and selected on plates with ampicillin at 30°C. Escherichia coli K12 MG1655 harbouring pKM208 was inoculated into LB with ampicillin and grown at 30°C. After reaching OD600 of 0.2, 1 mM IPTG was added and the bacterial culture was cultivated to the final OD600 of 0.4–0.6. Bacteria were subsequently washed and resuspended in 10% glycerol and transformed with the genetic circuit harbouring the flanking sequences of the target genes. Bacteria with chromosomal integrations were selected on plates with kanamycin at 37°C and subsequently grown at 42°C to cure out the temperature-sensitive plasmid pKM208. Chromosomal integrations were proved by PCR with flanking primers and sequencing.
GFP and mCherry fluorescence quantitation with the microplate reader
200 μl of the E. coli cultures (grown overnight and diluted to OD600 of 0.05) were transferred into flat-bottomed black 96 well plates (Greiner BioOne, UK). The plates with the E. coli cultures were placed into Fluostar Omega fluorimeter (BMG Labtech, UK) and incubated first at 30°C for 3 h and then at 42°C for 17 h or 7 h for GFP and mCherry fluorescence measurement respectively. GFP fluorescence was quantified with an automatically repeated protocol each 30 min using emission filter EM520, excitation filter 485-12, double orbital shaking at 200 r.p.m. and gain 1400. mCherry fluorescence was measured with an automatically repeated protocol each 30 min using emission filter EM620, excitation filter 584, double orbital shaking at 200 r.p.m. and gain 2800.
Absorbance measurement with the microplate reader
The diluted overnight E. coli cultures (OD600 of 0.05) were transferred into flat-bottomed clear 96 well plates (Sterilin Sero-Well, UK). The plates were then incubated in the microplate reader (Fluostar Omega, BMG Labtech, UK) at 37°C and 30°C for 24 h. Absorbance was measured each 30 min using 600 nm absorbance filter and double orbital shaking at 500 r.p.m.
RNA isolation and purification
Total RNA was isolated from 109 E. coli cells at mid-exponential phase with Isolate II RNA Mini Kit (Bioline) according to manufacturer's instructions. To elute RNA from the Isolate II RNA columns, 60 μl of RNAse-free H2O was used. To avoid contamination with genomic DNA, the isolated RNA was purified with TURBO DNA-free Kit (Applied Biosystems) according to supplier's instructions.
RT-PCR
Isolated and purified RNA (1 μg) was used to synthesize cDNA using SuperScript III Reverse Transcriptase (Invitrogen) according to supplier's instructions. Primers for RT-PCR designed with primer3 Software were prepared to generate 100–150 bp long DNA sequences. Expression levels were measured using QuantiTect SYBR Green PCR Kit (Qiagen). MicroAmp Fast Optical 96-Well Reaction Plates (Applied Biosystems) with RT-PCR reactions were incubated in the 7500 Fast Real-Time PCR System (Applied Biosystems) according to manufacturer's instructions. The relative expression was computed employing rest9 Software (Qiagen) with Pfaffl method (Pfaffl et al., 2002). The RT-PCR was performed in triplicate, and the means and standard errors were calculated.
Evaluation of motility
Motility agar plates for motility assay were made by transferring 100 ml of motility agar [composed of 0.25% Bacto-Agar (Difco), 5 g NaCl and 10 g tryptone] in the 13 cm plates and let to set overnight. Plates were then pre-warmed to 37°C and inoculated with the 2 μl of the overnight bacterial cultures normalized to OD600 of 1.0. Pictures were taken after incubation for 4–6 h at 37°C.
Sequence analyses
The annotated E. coli K-12 MG1655 genome from the E. coli K-12 project website (http://www.xbase.ac.uk/genome/escherichia-coli-str-k-12-substr-mg1655) was used to retrieve DNA sequences of the target loci. DNA sequencing was carried out by Source Bioscience (Cambridge, UK). The blastn (Altschul et al., 1990) and tblastx algorithms from the National Center for Biotechnology Information (NCBI) website (http://ncbi.nlm.nih.gov) and the position-specific iterated blast (PSI-BLAST) (Altschul et al., 1997) were used to compare DNA sequences.
Acknowledgments
We would like to thank to Dr. Kerstin Ewen for plasmid pKM208 and Dr. Gillian Fraser for constructive discussions.
Conflict of interest
There are no conflicts of interest associated with this manuscript.
Supporting Information
Additional Supporting Information may be found in the online version of this article at the publisher's web-site:
Fig. S1. Verification of chromosomal integrations into the E. coli flagellar region 2.
Fig. S2. Growth rates of the engineered E. coli strains with integrations in the flagellar region 2.
Fig. S3. Integrations into the E. coli flagellar region 3b do not negatively impact growth.
Fig. S4. Fluorescence measurement of the E. coli flagellar region 2 integrations.
Fig. S5. Fluorescence measurement of the E. coli flagellar region 3b integrations.
Fig. S6. mCherry fluorescence quantitation of the E. coli flagellar region 2 integrations.
Fig. S7. mCherry fluorescence quantitation of the E. coli flagellar region 3b integrations.
References
- Aizawa S. Mystery of fliK in length control of the flagellar hook. J Bacteriol. 2012;194:4798–4800. doi: 10.1128/JB.06239-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajikumar PK, Xiao WH, Tyo KE, Wang Y, Simeon F, Leonard E, et al. Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli. Science. 2010;330:70–74. doi: 10.1126/science.1191652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW. Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W. Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony LC, Suzuki H. Filutowicz M. Tightly regulated vectors for the cloning and expression of toxic genes. J Microbiol Methods. 2004;58:243–250. doi: 10.1016/j.mimet.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol Syst Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd CH. Gober JW. Temporal regulation of genes encoding the flagellar proximal rod in Caulobacter crescentus. J Bacteriol. 2001;183:725–735. doi: 10.1128/JB.183.2.725-735.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashman DJ, Ortega DR, Zhulin IB. Baudry J. Homology modeling of the CheW coupling protein of the chemotaxis signaling complex. PLoS ONE. 2013;8:e70705. doi: 10.1371/journal.pone.0070705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A, Cui Y. Chatterjee AK. RsmC of Erwinia carotovora subsp. carotovora negatively controls motility, extracellular protein production, and virulence by binding FlhD and modulating transcriptional activity of the master regulator, FlhDC. J Bacteriol. 2009;191:4582–4593. doi: 10.1128/JB.00154-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clomburg JM. Gonzalez R. Metabolic engineering of Escherichia coli for the production of 1,2-propanediol from glycerol. Biotechnol Bioeng. 2011;108:867–879. doi: 10.1002/bit.22993. [DOI] [PubMed] [Google Scholar]
- Cunningham DS, Koepsel RR, Ataai MM. Domach MM. Factors affecting plasmid production in Escherichia coli from a resource allocation standpoint. Microb Cell Fact. 2009;8:27. doi: 10.1186/1475-2859-8-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA. Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delalez NJ, Berry RM. Armitage JP. Stoichiometry and turnover of the bacterial flagellar switch protein FliN. MBio. 2014;5:e01216-01214. doi: 10.1128/mBio.01216-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyszel JL, Soares JA, Swearingen MC, Lindsay A, Smith JN. Ahmer BM. E. coli K-12 and EHEC genes regulated by SdiA. PLoS ONE. 2010;5:e8946. doi: 10.1371/journal.pone.0008946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraiberg M, Afanzar O, Cassidy CK, Gabashvili A, Schulten K, Levin Y. Eisenbach M. CheY's acetylation sites responsible for generating clockwise flagellar rotation in Escherichia coli. Mol Microbiol. 2015;95:231–244. doi: 10.1111/mmi.12858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser GM, Bennett JC. Hughes C. Substrate-specific binding of hook-associated proteins by FlgN and FliT, putative chaperones for flagellum assembly. Mol Microbiol. 1999;32:569–580. doi: 10.1046/j.1365-2958.1999.01372.x. [DOI] [PubMed] [Google Scholar]
- Freeman AM, Mole BM, Silversmith RE. Bourret RB. Action at a distance: Amino acid substitutions that affect binding of the phosphorylated CheY response regulator and catalysis of dephosphorylation can be far from the CheZ phosphatase active site. J Bacteriol. 2011;193:4709–4718. doi: 10.1128/JB.00070-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D, Young L, Chuang R, Venter J, Hutchison CR. Smith H. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Guan L, Liu Q, Li C. Zhang Y. Development of a Fur-dependent and tightly regulated expression system in Escherichia coli for toxic protein synthesis. BMC Biotechnol. 2013;13:25. doi: 10.1186/1472-6750-13-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Jessee J. Bloom FR. Plasmid transformation of Escherichia coli and other bacteria. Methods Enzymol. 1991;204:63–113. doi: 10.1016/0076-6879(91)04006-a. [DOI] [PubMed] [Google Scholar]
- Hayashi K, Morooka N, Yamamoto Y, Fujita K, Isono K, Choi S, et al. Highly accurate genome sequences of Escherichia coli K-12 strains MG1655 and W3110. Mol Syst Biol. 2006;2:2006.0007. doi: 10.1038/msb4100049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jandu N, Ho NK, Donato KA, Karmali MA, Mascarenhas M, Duffy SP, et al. Enterohemorrhagic Escherichia coli O157:H7 gene expression profiling in response to growth in the presence of host epithelia. PLoS ONE. 2009;4:e4889. doi: 10.1371/journal.pone.0004889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhas M. On the road to synthetic life: The minimal cell and genome-scale engineering. Crit Rev Biotechnol. 2015;12:1–8. doi: 10.3109/07388551.2014.989423. [DOI] [PubMed] [Google Scholar]
- Juhas M. Ajioka J. Identification and validation of novel chromosomal integration and expression loci in Escherichia coli flagellar region 1. PLoS ONE. 2015;10:e0123007. doi: 10.1371/journal.pone.0123007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhas M, Eberl L. Glass JI. Essence of life: Essential genes of minimal genomes. Trends Cell Biol. 2011;21:562–568. doi: 10.1016/j.tcb.2011.07.005. [DOI] [PubMed] [Google Scholar]
- Juhas M, Eberl L. Church GM. Essential genes as antimicrobial targets and cornerstones of synthetic biology. Trends Biotechnol. 2012a;30:601–607. doi: 10.1016/j.tibtech.2012.08.002. [DOI] [PubMed] [Google Scholar]
- Juhas M, Stark M, von Mering C, Lumjiaktase P, Crook DW, Valvano MA. Eberl L. High confidence prediction of essential genes in Burkholderia cenocepacia. PLoS ONE. 2012b;7:e40064. doi: 10.1371/journal.pone.0040064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhas M, Davenport PW, Brown JR, Yarkoni O. Ajioka JW. Meeting report: The Cambridge Biodesign Techevent – Synthetic Biology, a new ‘Age of Wonder’. Biotechnol J. 2013;8:761–763. [Google Scholar]
- Juhas M, Reuß DR, Zhu B. Commichau FM. Bacillus subtilis and Escherichia coli essential genes and minimal cell factories after one decade of genome engineering. Microbiology. 2014a;160:2341–2351. doi: 10.1099/mic.0.079376-0. [DOI] [PubMed] [Google Scholar]
- Juhas M, Evans LD, Frost J, Davenport PW, Yarkoni O, Fraser GM. Ajioka JW. Escherichia coli flagellar genes as target sites for integration and expression of genetic circuits. PLoS ONE. 2014b;9:e111451. doi: 10.1371/journal.pone.0111451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahramanoglou C, Seshasayee AS, Prieto AI, Ibberson D, Schmidt S, Zimmermann J, et al. Direct and indirect effects of H-NS and Fis on global gene expression control in Escherichia coli. Nucleic Acids Res. 2011;39:2073–2091. doi: 10.1093/nar/gkq934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishikawa J, Ibuki T, Nakamura S, Nakanishi A, Minamino T, Miyata T, et al. Common evolutionary origin for the rotor domain of rotary ATPases and flagellar protein export apparatus. PLoS ONE. 2013;8:e64695. doi: 10.1371/journal.pone.0064695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam KH, Ip WS, Lam YW, Chan SO, Ling TK. Au SW. Multiple conformations of the FliG C-terminal domain provide insight into flagellar motor switching. Structure. 2012;20:315–325. doi: 10.1016/j.str.2011.11.020. [DOI] [PubMed] [Google Scholar]
- Lee J. Harshey RM. Loss of FlhE in the flagellar Type III secretion system allows proton influx into Salmonella and Escherichia coli. Mol Microbiol. 2012;84:550–565. doi: 10.1111/j.1365-2958.2012.08043.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcellin E, Chen WY. Nielsen LK. Understanding plasmid effect on hyaluronic acid molecular weight produced by Streptococcus equi subsp. zooepidemicus. Metab Eng. 2010;12:62–69. doi: 10.1016/j.ymben.2009.09.001. [DOI] [PubMed] [Google Scholar]
- Merryman C. Gibson DG. Methods and applications for assembling large DNA constructs. Metab Eng. 2012;14:196–204. doi: 10.1016/j.ymben.2012.02.005. [DOI] [PubMed] [Google Scholar]
- Miller EM. Nickoloff JA. Escherichia coli electrotransformation. Methods Mol Biol. 1995;47:105–113. doi: 10.1385/0-89603-310-4:105. [DOI] [PubMed] [Google Scholar]
- Minamino T. Macnab RM. Components of the Salmonella flagellar export apparatus and classification of export substrates. J Bacteriol. 1999;181:1388–1394. doi: 10.1128/jb.181.5.1388-1394.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minty JJ, Lesnefsky AA, Lin F, Chen Y, Zaroff TA, Veloso AB, et al. Evolution combined with genomic study elucidates genetic bases of isobutanol tolerance in Escherichia coli. Microb Cell Fact. 2011;10:18. doi: 10.1186/1475-2859-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra A, Palaniyandi S, Herren CD, Zhu X. Mukhopadhyay S. Pleiotropic roles of uvrY on biofilm formation, motility and virulence in uropathogenic Escherichia coli CFT073. PLoS ONE. 2013;8:e55492. doi: 10.1371/journal.pone.0055492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohawk KL, Poly F, Sahl JW, Rasko DA. Guerry P. High frequency, spontaneous motA mutations in Campylobacter jejuni strain 81–176. PLoS ONE. 2014;9:e88043. doi: 10.1371/journal.pone.0088043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy KC. Campellone KG. Lambda Red-mediated recombinogenic engineering of enterohemorrhagic and enteropathogenic E. coli. BMC Mol Biol. 2003;4:11. doi: 10.1186/1471-2199-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa R, Abe-Yoshizumi R, Kishi T, Homma M. Kojima S. Interaction of the C-terminal tail of FliF with FliG from the Na+-driven flagellar motor of Vibrio alginolyticus. J Bacteriol. 2015;197:63–72. doi: 10.1128/JB.02271-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge JD, Nieto V. Harshey RM. A new player at the flagellar motor: Flil controls both motor output and bias. MBio. 2015;6:e02367–02314. doi: 10.1128/mBio.02367-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW. Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboul CF, Andrews DA, Nahar MF, Buckle AM. Roujeinikova A. Crystallographic and molecular dynamics analysis of loop motions unmasking the peptidoglycan-binding site in stator protein MotB of flagellar motor. PLoS ONE. 2011;6:e18981. doi: 10.1371/journal.pone.0018981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabri S, Steen JA, Bongers M, Nielsen LK. Vickers CE. Knock-in/knock-out (KIKO) vectors for rapid integration of large DNA sequences, including whole metabolic pathways, onto the Escherichia coli chromosome at well-characterised loci. Microb Cell Fact. 2013;12:60. doi: 10.1186/1475-2859-12-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre F, Cui L, Priest DG, Endy D, Dodd IB. Shearwin KE. One-step cloning and chromosomal integration of DNA. ACS Synth Biol. 2013;2:537–541. doi: 10.1021/sb400021j. [DOI] [PubMed] [Google Scholar]
- Takahashi Y. Ito M. Mutational analysis of charged residues in the cytoplasmic loops of MotA and MotP in the Bacillus subtilis flagellar motor. J Biochem. 2014;156:211–220. doi: 10.1093/jb/mvu030. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Koyama K. Ito M. Suppressor mutants from MotB-D24E and MotS-D30E in the flagellar stator complex of Bacillus subtilis. J Gen Appl Microbiol. 2014;60:131–139. doi: 10.2323/jgam.60.131. [DOI] [PubMed] [Google Scholar]
- Ublinskaya AA, Samsonov VV, Mashko SV. Stoynova NV. A PCR-free cloning method for the targeted φ80 Int-mediated integration of any long DNA fragment, bracketed with meganuclease recognition sites, into the Escherichia coli chromosome. J Microbiol Methods. 2012;89:167–173. doi: 10.1016/j.mimet.2012.03.013. [DOI] [PubMed] [Google Scholar]
- Vora T, Hottes AK. Tavazoie S. Protein occupancy landscape of a bacterial genome. Mol Cell. 2009;35:247–253. doi: 10.1016/j.molcel.2009.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim H, Haselbeck R, Niu W, Pujol-Baxley C, Burgard A, Boldt J, et al. Metabolic engineering of Escherichia coli for direct production of 1,4-butanediol. Nat Chem Biol. 2011;7:445–452. doi: 10.1038/nchembio.580. [DOI] [PubMed] [Google Scholar]
- Zhang X, Shanmugam KT. Ingram LO. Fermentation of glycerol to succinate by metabolically engineered strains of Escherichia coli. Appl Environ Microbiol. 2010;76:2397–2401. doi: 10.1128/AEM.02902-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Niu DD, Tian KM, Chen XZ, Prior BA, Shen W, et al. Genetically switched D-lactate production in Escherichia coli. Metab Eng. 2012;14:560–568. doi: 10.1016/j.ymben.2012.05.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Verification of chromosomal integrations into the E. coli flagellar region 2.
Fig. S2. Growth rates of the engineered E. coli strains with integrations in the flagellar region 2.
Fig. S3. Integrations into the E. coli flagellar region 3b do not negatively impact growth.
Fig. S4. Fluorescence measurement of the E. coli flagellar region 2 integrations.
Fig. S5. Fluorescence measurement of the E. coli flagellar region 3b integrations.
Fig. S6. mCherry fluorescence quantitation of the E. coli flagellar region 2 integrations.
Fig. S7. mCherry fluorescence quantitation of the E. coli flagellar region 3b integrations.


