Abstract
C-type lectins (CTLs) are a large family of Ca2+-dependent carbohydrate-binding proteins recognizing various glycoconjugates and functioning primarily in immunity and cell adhesion. We have identified 34 CTLDP (for CTL-domain protein) genes in the Manduca sexta genome, which encode proteins with one to three CTL domains. CTL-S1 through S9 (S for simple) have one or three CTL domains; immulectin-1 through 19 have two CTL domains; CTL-X1 through X6 (X for complex) have one or two CTL domains along with other structural modules. Nine simple CTLs and seventeen immulectins have a signal peptide and are likely extracellular. Five complex CTLs have both an N-terminal signal peptide and a C-terminal transmembrane region, indicating that they are membrane anchored. Immulectins exist broadly in Lepidoptera and lineage-specific gene duplications have generated three clusters of fourteen genes in the M. sexta genome, thirteen of which have similar expression patterns. In contrast to the family expansion, CTL-S1~S6, S8, and X1~X6 have 1:1 orthologs in at least four lepidopteran/dipteran/coleopteran species, suggestive of conserved functions in a wide range of holometabolous insects. Structural modeling suggests the key residues for Ca2+-dependent or independent binding of certain carbohydrates by CTL domains. Promoter analysis identified putative κB motifs in eighteen of the CTL genes, which did not have a strong correlation with immune inducibility in the mRNA or protein levels. Together, the gene identification, sequence comparisons, structure modeling, phylogenetic analysis, and expression profiling establish a solid foundation for future studies of M. sexta CTL-domain proteins.
Keywords: insect immunity, pattern recognition, carbohydrate recognition domain, expression profiling, comparative genomics
1. Introduction
Insect innate immune systems utilize soluble and membrane-bound receptors to recognize pathogen-associated molecular patterns (Kanost et al., 2004; Lemaitre and Hoffmann, 2007). Peptidoglycan recognition proteins, β-1,3-glucanase-related proteins, and an array of lectins bind to polysaccharides, glycoproteins and glycolipids on pathogen surface to induce defense responses (Charroux et al., 2009; Jiang et al., 2010; Weis et al., 1998). Lectins are classified based on the domain architectures and action mechanisms (Gallagher et al., 1984). C-type lectins (CTLs) constitute one of the largest and most diverse families of lectins in animals. They require Ca2+ for maintaining carbohydrate-binding activities and structures (Cambi et al., 2005). Each CTL contains one or more carbohydrate recognition domains (CRDs), known as CTL domains. A CTL domain is composed of β-sheets, α-helices, and loops (Weis et al., 1991). CTL domains may participate in protein interaction and binding to lipids and inorganic surfaces, which does not always require Ca2+ (Zelensky and Gready, 2005).
Specificity of CTLs is governed by key residues in the CRDs, which interact with the cognate oligosaccharides through Ca2+ coordination and a network of hydrogen bonds. In the Ca2+ binding site-2 of rat mannose-binding lectin A, Glu185, Asn187, Glu193, Asn205 and Asp206 are implicated in specific interactions (Weis and Drickamer, 1994; Weis et al., 1992). CTLs containing a Glu-Pro-Asn (EPN) motif in the CRD are characteristic of mannose-binding and thus called mannose-type CTLs. CTLs with a Gln-Pro-Asp (QPD) motif are generally galactose-type CTLs (Zelensky and Gready, 2005). Most CTLs contain a single CTL domain. Immulectins (IMLs) from lepidopteran insects have two. CTLs with dual CTLDs are also found in Tribolium castaneum (Zou et al., 2007) and crustaceans (Yu and Kanost, 2001), but it is unknown whether they share a common ancestor or arose independently.
Genome-wide analyses in insects revealed a number of genes encoding proteins with one or more CTL domains (Dodd and Drickamer, 2001; Christophides et al., 2002; Waterhouse et al., 2007; Zou et al., 2007; Tanaka et al., 2008). Drosophila melanogaster, Anopheles gambiae, Aedes aegypti, T. castaneum, and Bombyx mori have 34, 25, 39, 17 and 21 such genes, respectively. Since it is unclear whether these proteins bind carbohydrates or not in the presence or absence of Ca2+, we suggest that they be named CTL-domain proteins (CTLDPs) instead of CTLs. Individual immulectins have been identified and characterized in lepidopteran species (Yu and Kanost, 2008) (Table 1). In Manduca sexta, functions of IML-1~4 have been characterized biochemically. IML-1 can induce agglutination of Gram-positive and -negative bacteria and yeast in a Ca2+-dependent manner (Yu et al., 1999). Injection of IML-2 antiserum into M. sexta larvae inhibits clearance of a Gram-negative bacterial pathogen, Serratia marcescens, and decreases larval survival after bacterial infection (Yu and Kanost, 2003). Recombinant CRD2 of IML-2 directly binds to Caenorhabditis elegans and a human filarial nematode Brugia malayi and enhances encapsulation and melanization of C. elegans in vivo (Yu and Kanost, 2004). CTLD2 of IML-2 interacts with proPO, and the extended loop of CTLD2 is important for ligand binding and proPO activation (Shi and Yu, 2012). IML-3 is translocated into hemocytes in response to microbial stimulation (Ling et al., 2008). IML-4 can bind to immobilized LPS and lipoteichoic acid (LTA) in the absence of Ca2+, but agglutinate Escherichia coli, Staphylococcus aureus and Saccharomyces cerevisiae in a Ca2+-dependent manner (Yu et al., 2006).
Table 1.
Functions of some immulectins in lepidopteran insects
| Name | inducibility | tissue | agglutination | Ca2+-depen dence |
binding | proPO activation |
melanization | encapsulation | reference | |
|---|---|---|---|---|---|---|---|---|---|---|
| H | F | |||||||||
|
M.
sexta IML-1 |
Ec, Ml, Sc | no | yes | Ec, Sa, Sc | yes | - | yes | no | yes | Yu et al., 1999 |
|
M.
sexta IML-2 |
Ec, Ml, Sc | no | yes | Ec | yes/no* | lipid A, LPS, PG, LTA, mannan, laminarin |
yes | yes | yes |
Shi and Yu,
2012; Yu and Kanost, 2000, 2003, 2004; |
|
M.
sexta IML-3 |
Ec, Ml, Sc | no | yes | Ec | yes | LPS, LTA, laminarin |
- | no | yes |
Yu et al.,
2005; Ling et al., 2008; |
|
M.
sexta IML-4 |
Ec, Ml, Sc | no | yes | Ec, Sa, Sc | yes/no* | LPS, LTA, laminarin |
yes | yes | yes | Yu et al., 2006 |
|
H.
armigera CTL1~8 |
Bt, Sa, Pp, NPV, 20E |
yes | yes | Ec, Sa (1,3); Ca (1); - (2, 3~8) |
- | - | - | - | - |
Chai et al.,
2008; Wang et al., 2012 |
|
H.
cunea Hdd15 |
Ec, Ml | - | yes | - | - | LPS | - | - | - | Shin et al., 2000; |
|
B. mori LBP, MBP, etc. |
Ec, Ml, Sc | yes | yes | Ec, Smi (LBP); Sma, Ml, Sc (MBP) |
- (LBP); + (MBP) |
lipid A (LBP); TA, mannan, PG (MBP) |
- | yes (LBP) | - |
Koizumi et
al., 1997, 1999; Watanabe et al., 2006; Tanaka et al., 2008; |
Bt, Bacilus thuringiensis Ml, Micrococcus luteus; Sa, Staphylococcus aureus; Ec, Escherichia coli; Sma, Serratia marcescens; Smi, Salmonella minnesota, Sc, Saccharomyces cerevisiae; Ca, Candida albicans; Pp, Pichia pastoris; NPV, nucleopolyhedrovirus; 20E, 20-hydroxyecdysone; H: hemocytes; F: fat body; PG, peptidoglycan; LPS, lipopolysaccharide; LTA, lipoteichoic acid; TA, teichoic acid;
: Ca2+ required for agglutination of E. coli, but not for binding to microbial components.
To acquire an overview of M. sexta CTLDPs, we annotated CTLDP genes in the M. sexta genome based on the RNA-Seq data. Multiple sequence alignment and phylogenetic analysis revealed orthologs in other insects and lineage-specific expansion of the immulectin genes in lepidopterans. Analysis of the RNA-Seq reads provided expression patterns of the CTLDP genes in different tissues and stages. Putative immune responsive elements in the promoter regions were identified, and we examined whether presence of these elements correlates with mRNA and protein level changes in larval hemolymph before and after the immune challenge (Zhang et al., 2011 and 2014). We also studied sequence conservation and structure-function relationships via molecular modeling and discuss their potential roles insect physiological processes.
2. Materials and methods
2.1 Gene identification, sequence improvement, and feature prediction
Manduca Genome Assembly 1.0 and gene models in Manduca Official Gene Set 1.0 and Cufflinks Assembly 1.0 (X et al., 2014) were downloaded from Manduca Base (ftp://ftp.bioinformatics.ksu.edu/pub/Manduca/). CTL sequences from M. sexta and other insects were used as queries to search Cufflinks 1.0 using the TBLASTN algorithm with default settings. Hits with aligned regions longer than 30 residues and identity over 40% were retained for retrieving corresponding cDNA sequences. Correct open reading frames (ORFs) in the retrieved sequences were identified using ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). Errors resulting from problematic regions (e.g. NNN…) in the genome assembly were corrected after BLASTN search of Manduca Oases and Trinity Assemblies 3.0 of the RNA-Seq data (http://darwin.biochem.okstate.edu/blast/blast_links.html). The two genome-independent RNA-Seq assemblies (X et al., 2014) were developed to cross gaps between genome scaffolds/contigs and detect errors in the gene models. The manually improved sequences were incorporated into OGS 2.0. To uncover all genes in a cluster, which were often too similar to distinguish by Cufflinks 1.0, the relevant genome contigs were manually examined to identify exons based on the GT-AG rule and sequence alignment. All improved sequences were further validated by BLASTP homolog search of GenBank (http://www.ncbi.nlm.nih.gov/). Conserved domains and transmembrane regions were identified using SMART (http://smart.embl-heidelberg.de/smart/set_mode.cgi) and InterProScan (http://www.ebi.ac.uk/Tools/pfa/iprscan/). The domain architectures were plotted using DOG 2.0 (http://dog.biocuckoo.org/). Signal peptides were predicted using SignalP4.1 (http://www.cbs.dtu.dk/services/).
2.2 Sequence alignment and phylogenetic analysis
Multiple sequence alignments of CTLs from M. sexta and other insects (http://www.ncbi.nlm.nih.gov/) were performed using MUSCLE, a module of MEGA 6.0 (http://www.megasoftware.net), at the following settings: refining alignment, gap opening penalty = −2.9, gap extension penalty = 0, hydrophobicity multiplier = 1.2, maximum iterations = 100, clustering method (for iterations 1 and 2) = UPGMB, and maximum diagonal length = 24. The aligned sequences were used to construct neighbor-joining trees with bootstrap method for the phylogeny test (1000 replications, Poisson model, uniform rates, and complete deletion of gaps or missing data).
2.3. Protein structure modeling
Amino acid sequences of the M. sexta CTL domains were submitted to the I-TASSER server (http://zhanglab.ccmb.med.umich.edu/I-TASSER/) for protein 3D–structure prediction (Zhang, 2008). Models were built based on multiple-threading alignments by LOMETS and iterative TASSER simulations (Roy et al., 2010). A representative model was chosen for the production of molecular graphics using PyMol (DeLano Scientific, Palo Alto, CA).
2.4. Gene expression profiling and promoter analysis
The 52 cDNA libraries, representing mRNA samples from whole insects, organs or tissues at various life stages, were constructed and sequenced by Illumina technology (Manduca sexta genome and transcriptome project; http://www.ncbi.nlm.nih.gov/bioproject/PRJNA81039). Reads from the individual RNA-Seq datasets were trimmed to 50 bp and mapped to the updated OGS 1.0 (Section 2.1) using Bowtie (0.12.8) (Langmead et al,, 2009). Numbers of the mapped reads were used to calculate FPKM (fragments per kilobase of exon per million fragments mapped) by RSEM (1.2.12) (Li and Dewey, 2011) for interlibrary comparisons. Hierarchical clustering of the log2(FPKM+1) values was performed using MultiExperiment Viewer (v4.9) (http://www.tm4.org/mev.html) with the Pearson correlation-based metric and average linkage clustering method. To study transcript changes after immune challenge, the entire set of CTL sequences were used as queries to search for corresponding contigs in the CIFH09 database (http://darwin.biochem.okstate.edu/blast/blast_links.html) (Zhang et al., 2011) by TBLASTN. The numbers of CF, CH, IF, and IH reads (C for control, I for induced after injection of bacteria, F for fat body, H for hemocytes) assembled into these contigs were retrieved for normalization and calculation of IF/CF and IH/CH ratios. When a polypeptide sequence corresponded to two or more contigs, sums of the normalized read numbers were used to calculate its relative mRNA abundances in fat body and hemocytes (Gunaratna and Jiang, 2013). Potential transcription factor binding sites in the 1000 bp region before the translation initiation site were searched using MacVector Sequence Analysis Software (Oxford Molecular Ltd.). Sequences, positions, and strand polarities of the perfectly matched GATA (WGATAR), R1 (KKGNNCTTTY), and CATTW boxes were documented. NF-κB motifs (GGGRAYYYYY) with 0, 1 or 2 mismatches were also identified.
3. Results
3.1. Occurrence and general properties of M. sexta CTL domain proteins
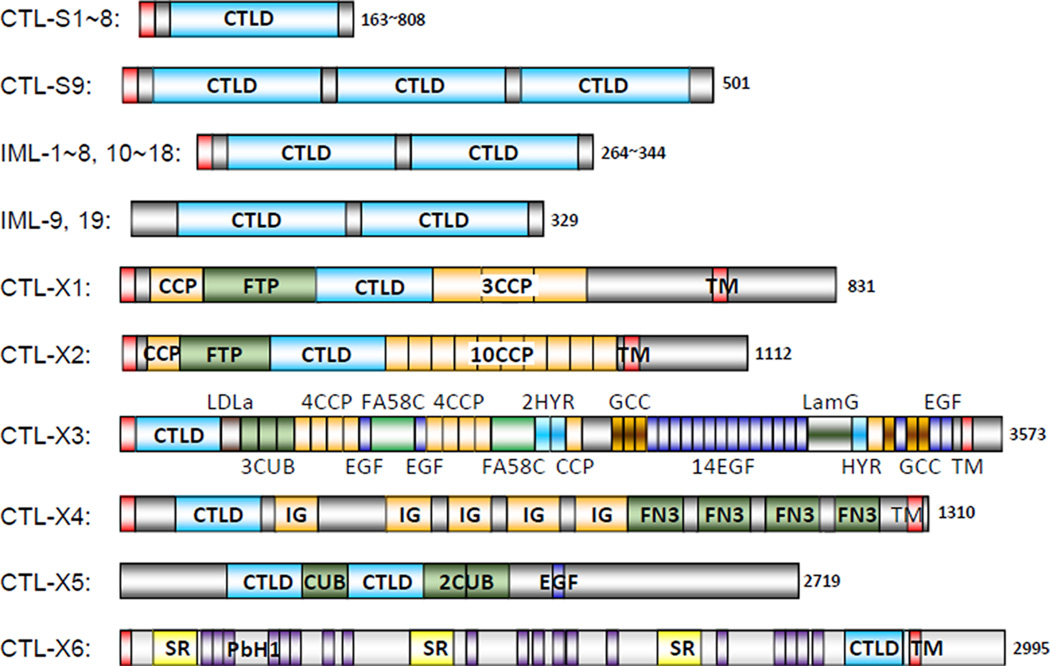
We identified 34 CTLDP genes in Manduca Genome Assembly 1.0, which encode proteins with 1~3 CTL domains (Table S1). Nine of the protein products (CTL-S1~S9) have a simple structure, nineteen (IML-1~19) belong to the immulectin (IML) family with two tandem CTL domains, and the remaining six (CTL-X1~X6) contain one or two CTL domains along with other structural units (Fig. 1). M. sexta CTL-S1~S9, IML-1~8, and IML-10~18 contain an N-terminal signal peptide and are likely secreted into plasma, as previously demonstrated for IML-1~4 (Table 1). In addition to the secretion signal, CTL-X1~X4 and X6 have a transmembrane (TM) region near the carboxyl-terminus, which may anchor them to cell membrane. The extracellular portion of these complex CTLs may interact with ligands or protein partners. In contrast, CTL-X5 and IML-19 lack a signal peptide and are probably cytosolic proteins. The IML-9 gene, lacking a translation start codon preceding the CTL domain-1, could be a pseudogene.
Fig. 1.
Domain architectures of the 34 M. sexta CTLDPs. Signal peptide and transmembrane (TM) region are in red. C-type lectin domains (CTLDs) are in cyan. Low complexity (LC) regions within the grey areas are not shown. Other domains including CCP/Sushi, FTP, LDLa, CUB, EGF, FA58C, HYR, GCC, LamG, IG, FN3, SR and PbH1 are in different colors. Protein sizes or size ranges are indicated at the end of each bar. The domain and protein sizes are not in proportion.
M. sexta CTL-S1 through S8 contain a single CTL domain while CTL-S9 has three. The CRD in CTL-S1, S2 and S3, containing the QPD motif, may be galactose-type, and so is the 2nd CRD in CTL-S9 (Table S1). CTL-S6 may bind mannose since its CRD has the EPN motif. It is unclear if the other putative CRDs associate with carbohydrates. In addition to their CTL domains, CTL-S3 and S5 contain an N-terminal extension of 77 (12 Arg) and 192 (36 Pro) residues whereas CTL-S4 and S8 have a C-terminal extension of 166 (20 Pro, 19 Ser, 19 Ala) and 657 (75 Thr, 56 Ser, 53 Asn, 53 Leu) residues, respectively. These extensions, including low complexity (LC) regions that are rich in certain amino acid residues, may have significant functions, since their orthologs from other insects share similar features (data not shown).
CTLDPs with the dual-CTLD architecture were first reported in lepidopteran insects and named immulectins in M. sexta (Table 1). We identified nineteen IML genes in the M. sexta genome (Table S1), each encoding two CTL domains. As an exception, IML-6 gene has a stop codon in the 2nd domain. The EPN motif is found in the first CRDs in IML-10 and IML-12 and in the second CRDs in IML-1, 2, 4, and 5. The QPD motif (which normally binds galactose) is found in the second CRDs of IML-3, 9~12, 16, and 17.
The CTL domains of M. sexta CTL-X1~X6 (Table S1), which are a small domain within a large complex protein, do not contain a QPD or EPN motif for galactose or mannose binding. Instead, they may be involved in protein-protein interactions. Like some of the CTL-Ss, CTL-Xs contains long extensions of low complexity sequence (Fig. 1). For instance, the C-terminal 292-residue segment (excluding the 23-residue TM region) of CTL-X1 is rich in Pro, Arg, Thr, and is highly basic (pI: 10.7). In contrast, the same region (106 residues) in the CTL-X2 C-terminus is rich in Thr, Glu, Ala, and is acidic (pI: 4.82). The N- and C-terminal extensions of CTL-X5 are basic (pI: 9.42 and 9.01); those of CTL-X6 are acidic (pI: 4.89 and 5.58). The functional relevance of these features is unclear.
3.2. Phylogenetic relationships and evolution of insect CTLDPs
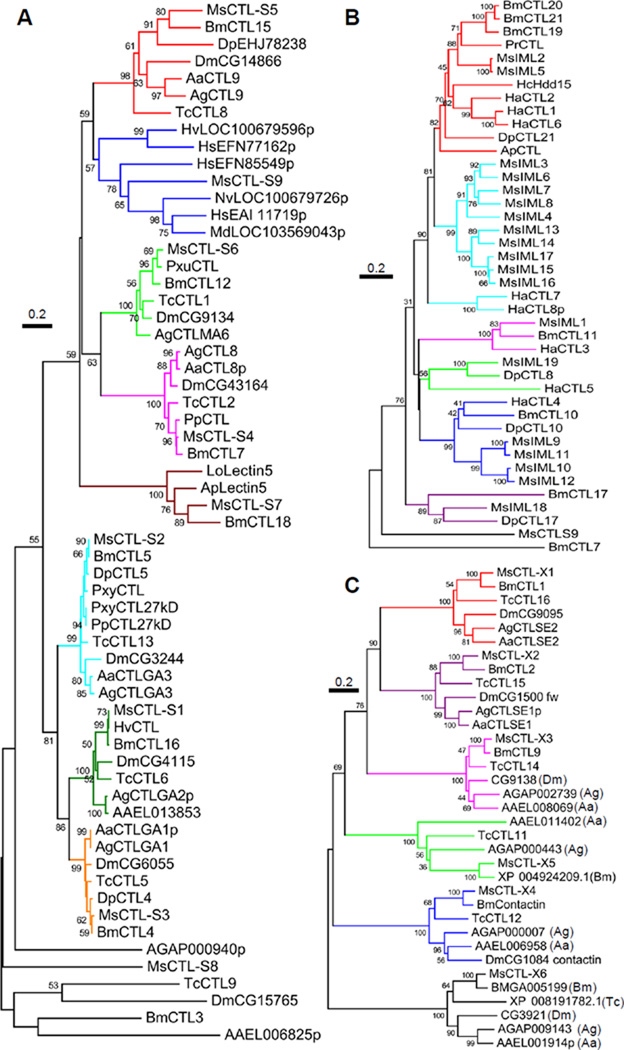
Alignment of 173 CTLDP sequences from different insects supported their classification into the CTL-S, IML and CTL-X groups (data not shown). Based on further BLASTP searches of GenBank, VectorBase and other databases, we retrieved more CTLDPs for group-specific sequence alignments. M. sexta CTL-S1 through S6 formed tight, monophyletic groups with their respective orthologs from the other species in Lepidoptera, Diptera, and Coleoptera (Fig. 2A). The CTL-S7 orthologs were only found in lepidopteran species. The relationships among M. sexta CTL-S8, B. mori CTL3, T. castaneum CTL9, and their dipteran orthologs are not strong, with relatively low sequence identity. The CTL-S9 and its hymenopteran homologs all have three CTL domains. However, the hymenopteran proteins are much longer at the C-terminus, including CCP or von Willebrand A domain and LC/TM region(s) (data not shown).
Fig. 2.
Phylogenetic relationships of M. sexta CTL-S1~S9 (A), IML-1~19 (B), and CTL-X1~X6 (C). Based on the preliminary analysis, M. sexta (Ms) CTL-S, IML and CTL-X groups were separately aligned with their homologs with similar domain architectures from other insects. Branches in each closely related group (bootstrap value >500 in 1000 trials) are shown in the same color. A. aegypti (Aa) AAEL011402 and AAEL011403, corresponding to the amino- and carboxyl-terminal regions of A. gambiae (Ag) AGAP000443, were combined prior to sequence alignment. The “intergenic" sequence in A. aegypti encodes most of the domains found in the center of AgAP000443 (data not shown). Ap, Antheraea pernyi; Bm, B. mori; Dm, D. melanogaster; Dp, Danaus plexippus; Ha, Helicoverpa armigera; Hc, Hyphantria cunea, Hs, Harpegnathos saltator; Hv, Heliothis virescens; Lo, Lonomia obliqua; Md, Microplitis demolitor; Nv, Nasonia vitripennis, Pp, Papilio polytes; Pr, Pieris rapae; Pxu, Papilio xuthus; Pxy, Plutella xylostella; Tc, T. castaneum; p for partial sequence.
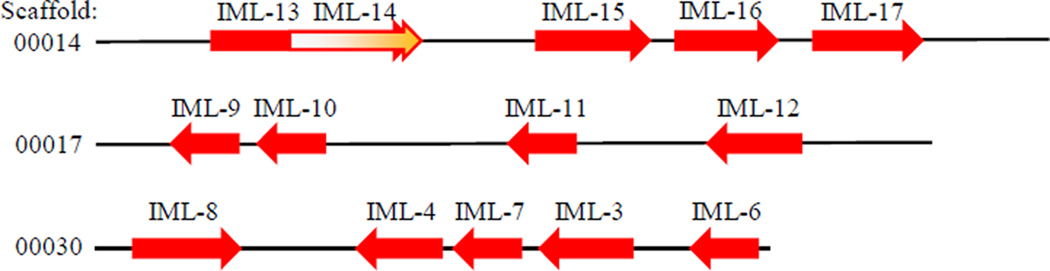
IMLs exist widely in lepidopteran insects (Fig. 2B). We identified 1:1 orthologs of IML-1, 18, and 19 in other lepidopteran species and found lineage-specific expansions in B. mori, H. armigera and M. sexta. For instance, M. sexta IML-3, 4, 6~8, IML-9~12, and IML-13~17 are three groups of related IMLs that evolved by multiple gene duplications. Identification of the gene clusters on Scaffolds 00014, 00017, and 00030 (Fig. 3) provides good support for the phylogenetic relationships based on the sequence comparison. M. sexta CTL-S9, which contains three CTL domains, is quite different in sequence from the IMLs (Fig. 2B), and its CTL domains did not align well with the IMLs (data not shown). Neither did the two in CTL3 of T. castaneum. Thus, emergence of the dual-CTLD genes in the lepidopteran insects seems independent from that in the beetle.
Fig. 3.
The M. sexta IML gene clusters. Exons 1~5 of IML-14 gene are located between exons 5 and 6 of IML-13 gene; whereas exon 6 of IML-14 closely follows exon 6 of IML-13.
The alignment of M. sexta CTL-X1~X6 with their homologs in B. mori, D. melanogaster, A. gambiae, A. aegypti, and T. castaneum established orthologous relationships within the six groups (Fig. 2C), suggesting their ancestral genes existed before divergence of the Lepidoptera, Diptera and Coleoptera. In the lineage of D. melanogaster, the ortholog of M. sexta CTL-X5 was apparently lost during evolution. Based on the sequence similarity and domain organization, we also found A. aegypti AAEL011402 and AAEL011403 are the N- and C-terminal fragments of the mosquito CTL-X5, respectively.
3.3. Evolutionary relationships and structural features of the CTL domains
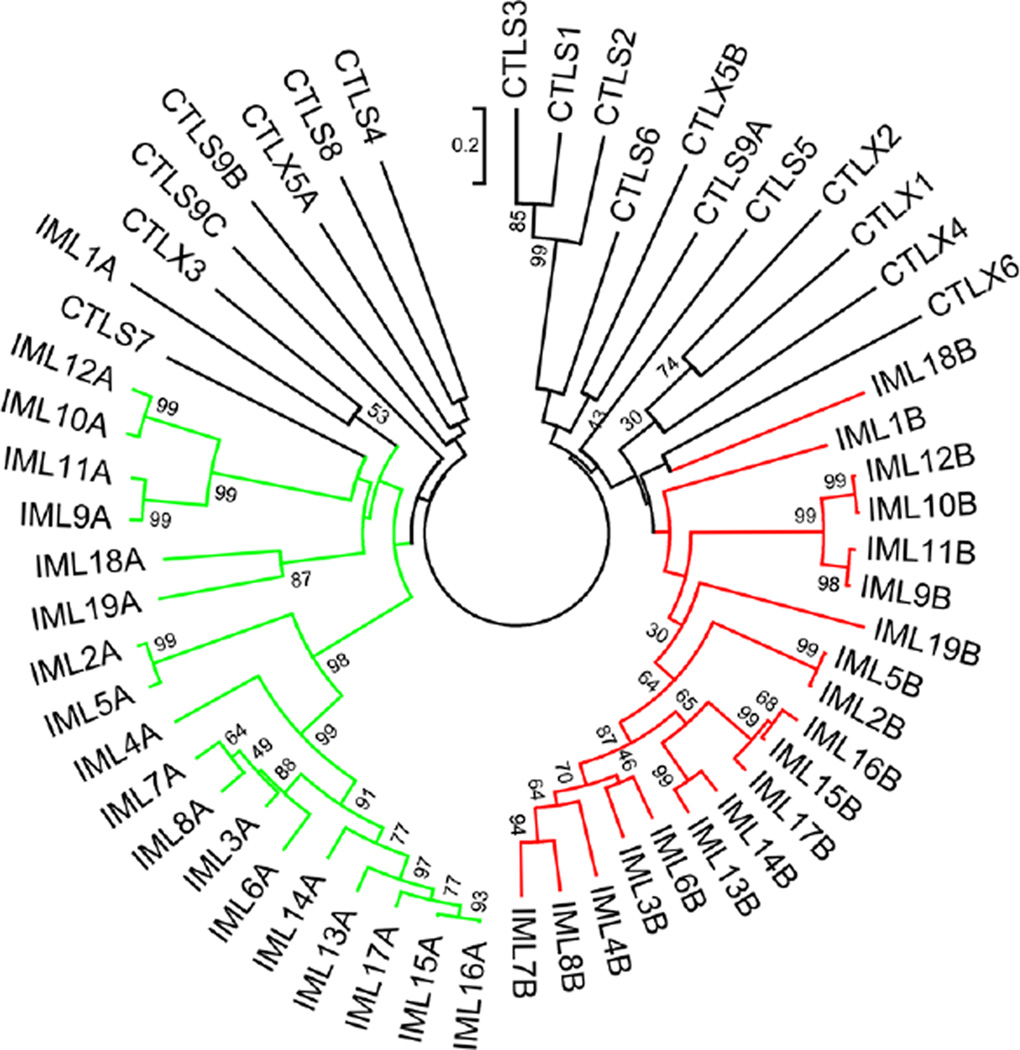
Can the phylogenetic relationships based on alignment of the entire CTLDPs (Fig. 2) be recreated by aligning their CTL domains alone? To test the possibility, we compared the 56 CTL domain sequences and constructed a tree based on the sequence alignment. Eighteen CTL domains formed branch A, and eighteen CTL domains formed branch B (Fig. 4). IML-1A and IML-18B are more diverged from the two major branches, as they arose in the early evolution of IMLs (Fig. 2B). These results suggest, after the ancestor IML gene came into being, entire gene duplications and sequence divergence gave rise to multiple tandem CTL domain IMLs in moths and butterflies before the radiation of Lepidoptera. Lineage-specific family expansions occurred more recently. There is no evidence for domain shuffling in the evolution of these genes. The three domains of CTL-S9 are not closely related to those in the IMLs (Fig. 4). The evolution of CTL-S and X groups mostly occurred before the emergence of IMLs. Nonetheless, the close relationships between CTL-X1 and X2 and among CTL-S1~S3 are also reflected at the domain level.
Fig. 4.
Phylogenetic relationships of the 56 M. sexta CTL domains. A neighbor-joining tree was constructed based on the sequence alignment, with the branches corresponding to the CTL domains A and B in the 19 IMLs colored green and red, respectively. Branches for the domains in CTL-S1~S9 and X1~X6 are shown in black.
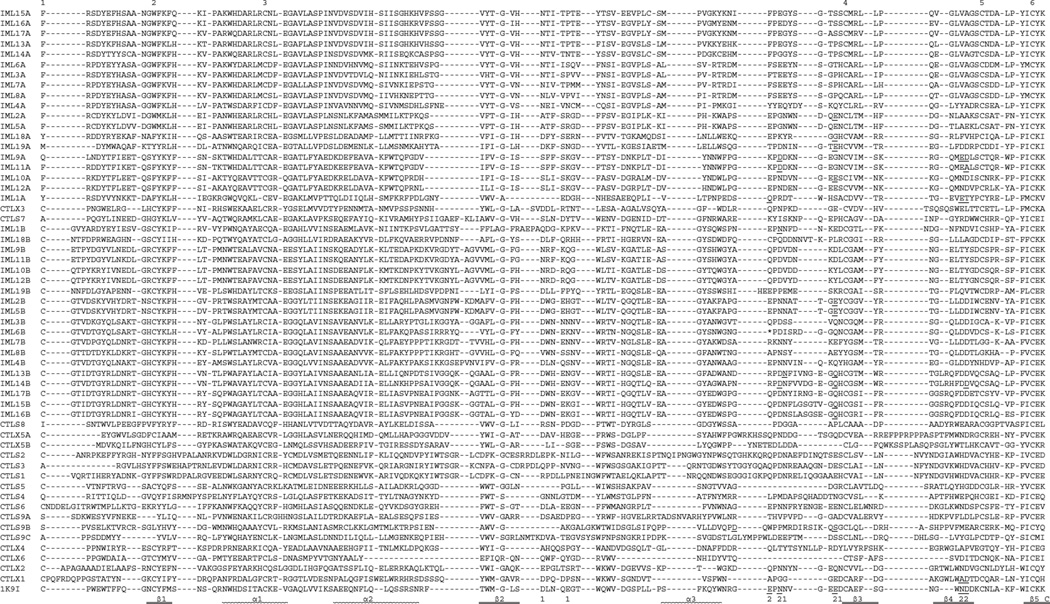
Alignment of the 56 CTL domain sequences with the domain in rat mannose binding lectin (MBL) allows a close examination of their sequence features (Fig. 5). The typical CTL domain is stabilized by three disulfide bonds between Cys-1 and −2, Cys-3 and −6, and Cys-4 and −5. These Cys residues are absolutely conserved in 23 CTL domains [IML-1~3, 5, 6, 9~19 B domains, CTL-S5, X1~X3, X5 (A, B), X6]. Both Cys-1 and −2 are absent in 26 other CTL domains [IML-1~19 A domains, CTL-S3, S4, S7, S8, and S9 (A, B, C domains), whereas Cys-4 and −5 are both absent in 4 CTL domains (IML-4, 7, 8 B domains and CTL-X4). The 3–6 and 4–5 linkages are predicted to form in CTL-S1, S2, and S6, but Cys-1 in CTL-S1 and S2 may pair with a Cys at the fourth position after Cys-3 based on their structure models (see below). The Cys residues in C(D/N)F(K/A)GC of CTL-S1, S2 and S3 may form a unique disulfide bond. We have also identified the residues corresponding to those in MBL involved in Ca2+ sugar binding (Fig. 5). Most of the IML A domains lack these residues whereas their B domains possess them. We suggest that CTL-S1, S3, S6, S9B, and X1 have the potential for carbohydrate binding.
Fig. 5.
Sequence alignment of the M. sexta CTL domains. Based on the domain predictions using SMART, the 56 CTL domain sequences are aligned with that of the rat mannose binding protein (IK9I). For IML-6, the sequence after the stop codon (*, shaded red) also included. In the aligned sequences, ‘EPN’ and ‘QPD’ motifs are highlighted at ‘P’ gray and cyan, respectively. Residues involved in Ca2+ binding in site-1 and −2 (labeled “1” and “2”, respectively) are shaded yellow. Residues involved in carbohydrate binding (based on the models in Fig. 6 and data not shown) are highlighted green, with those also binding Ca2+ underlined. Of the six conserved Cys residues, Cys-1 and −2, −3 and −6, −4 and −5 are predicted to form three disulfide bonds in 23 CTL domains. The other 30 CTL domains may lack the bond between Cys-1 and −2 or Cys-4 and −5. In other parts of the sequences, Cys residues (in red font) may form disulfide bridges with other linkage patterns. The SIDPLDVPQGSTAPQRTARHGTEHVM between D and T (shaded pink) in CTL-S4 and LDILRSEG between A and D (shaded pink) in CTL-S9 domain A are removed along with the gaps in the other sequences.
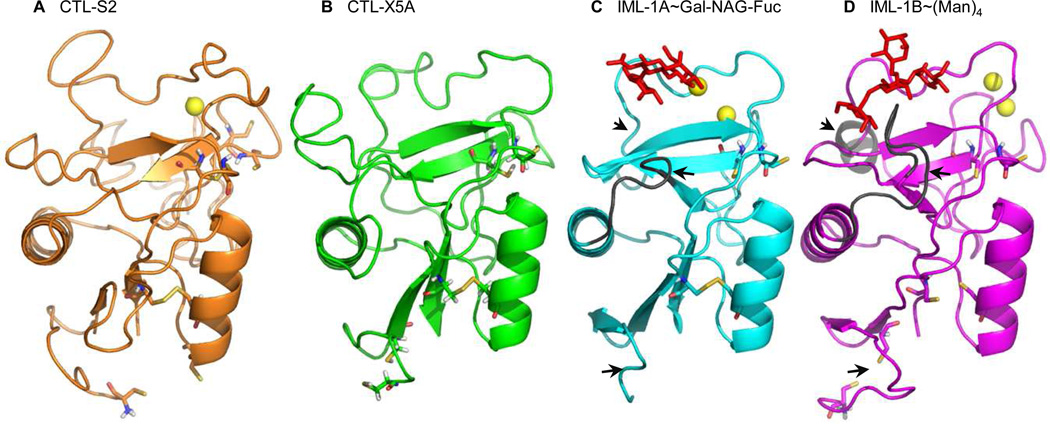
To further explore the binding sites, we performed structural modeling of 56 CTL domains and found their overall folds are predicted to be closely similar (Fig. 6, Table 2). Eighteen of the models may contain one or two Ca2+ ions for calcium-dependent sugar binding, and twenty-two may bind carbohydrates in a Ca2+-independent manner. Among the remaining sixteen CTL domains, one may bind Ca2+ but not sugar and fifteen may bind neither. Future analyses are needed to determine whether they bind specific carbohydrates and if Ca2+ enhances binding strength or specificity. While sequence alignment of the CTL domains in M. sexta CTLDPs and rat MBL allows us to predict residues important for Ca2+ coordination and sugar binding (Fig. 5), molecular modeling of the protein-Ca2+/sugar complexes provides similar information based on the three-dimensional structures (Fig. 6). Therefore, we have compared the results and found the predictions are mostly consistent. CTL-S1, S3, S9B, IML-9B, 10B, 11B, 12B, 16B, and 17B, containing the “QPD” motif, may form stable complexes with galactose, as suggested by the high C-scores of the models. CTL-S6, IML-1B, 4B, and 5B have the “EPN” motif and may bind mannose tightly. However, IML-2B, 10A and 12A (“EPN”) may bind mannose, but the C-scores are lower than those of the predicted protein-galactose complexes. In contrast, IML-3B (“QPD”) may form a more stable complex with mannose than galactose. IMB-15B (“QPD”) may not bind galactose at all (Table 2). Consequently, carbohydrate binding specificity of CTL domains may not be predicted with 100% accuracy based on the “EPN” or “QPD” signature. Experimental evidence will be required to validate the predicted specificities.
Fig. 6.
Structural models of the CTL domains of M. sexta CTL-S2 (A), CTL-X5A (B), IML-1A (C), and IML-1B (D). Ca2+ ions, yellow spheres; galactose (panel C) and mannose (panel D) containing carbohydrates, red stick; disulfide bonds, yellow sticks linking the side chains of paired Cys residues. Key residues involved in Ca2+ coordination and carbohydrate binding (Table 2) are indicated in Fig. 5. Regions that differ in IML-1A and −1B are marked by black arrows.
Table 2.
Structural features of the 56 M. sexta CTL domain models
| domain | Ca2+ | putative Ca2+ coordinators | motif | sugar * | putative sugar binding residues | template |
|---|---|---|---|---|---|---|
| CTL-S1 | 0 | QPD | G+ | 116,125,127,129,138,155~6 | 2OX9 | |
| S2 | 1 | 75,79,95,132,142~3 | QPD | no | no | |
| S3 | 0 | QPD | G+ | 114,117,119,121,129,146~7 | 2OX9 | |
| S4 | 0 | LPM | no | no (0.16) | ||
| S5 | 0 | --- | G | 62,91,94,111~3,117 | 1SL5 | |
| S6 | 1 | 79,83,86,107,116~7 | EPN | M+ | 104,106,132~4 | 3G83 |
| S7 | 0 | KYI | M > G | 70,104,106,108,113,126~8 /105,113,117,119,126~8,132 |
2IT6 / 3P5I | |
| S8 | 0 | -PD | MLR | 94,101,115~7 | 2GGU | |
| S9A | 0 | THV | no | no | ||
| S9B | 1 | 97,111,128 | QPD | G+ | 95,97,99,111 | 2OX9 |
| S9C | 0 | LDE | no | no | ||
| CTL-X1 | 2 | 79,83,86,104,106~7 / 106,120~1 |
APG | M | 70,71,103,105,112~3,120~1 | 1K9I |
| X2 | 0 | QPN | M > G | 104,106,108,112,118~9,126~7, 129 / 101~4,106,112,126~7 |
1K9I / 3P5G | |
| X3 | 0 | QPN | G > M | 95,97,101,105,107,116~8, 122 / 95,101,116~7,119 |
3P5I / 1SL4 | |
| ×4 | 0 | QLT | no | no (0.13) | ||
| X5A | 0 | QPN | no | no | ||
| X5B | 0 | EDL | G > M | 101,107,123~4 / 123~5 | 3P5G / 1K9I | |
| X6 | 0 | --- | no | no | ||
| IML-1A | 2 |
92,95,99,111~2 / 69,95,99,100 |
QPR | G | 89,92,94,96,99,111~2 | 2OX9 |
| 1B | 2 | 112,116,118 / 111,115,118 |
EPN | M+ | 71,109,111,113,117, 123~4,130~1,133 |
1SL4 |
| 2A | 1 | 69,73,98,103,104 | EPG | Glc > M | 95,97,103,115~7 /
58,61~2, 95,97,99,103,109~10,115~6 |
1PWB / 1K9I |
| 2B | 1 | 80,84,87,110,115,116 | EPN | G > M± | 104~7,109,115,127~8 /
73, 107,109,111,115,121~2,127~8 |
3P5G / 1K9I |
| 3A | 0 | FTE | M | 63,105,108~9,116~8 | 2IT6 | |
| 3B | 0 | QPD | M > G± | 72,106,108,110,112,118~9,124~ 5,127 / 103,106,108,112,124~5 |
1SL4 / 2OX9 | |
| 4A | 0 | YYE | M | 64,99,101,106,112~3,118~20 | 1K9I | |
| 4B | 0 | EPN | M+ | 108,110,129~31 | 2IT6 | |
| 5A | 0 | EPG | M | 95,97,103,115~7 | 1K9I | |
| 5B | 2 | 80,84,87,110,115 116 / 83,110,114,116 |
EPN | M+ = G | 109,115,127~9/107,109, 111,115,121~2,126 |
1SL6 / 1K9I |
| 6A | 0 | FSE | no | no | ||
| 6B | 0 | *PD | no | no | ||
| 7A | 0 | FSE | no | no (0.11) | ||
| 7B | 0 | RKN | G | 72,107~8,110,112,123~5,129 | 1SL5 | |
| 8A | 0 | FSE | no | 2IT6 (0.13) | ||
| 8B | 0 | APN | M | 106,108,123~5 /
106,108, 112,116,123,125,131 |
2ORJ/3G84 | |
| 9A | 1 | 95,100,113~4 | KPD | M > G | 60,93,95,97,101,113~4,116 / 90~3,95,100,106,114~5 |
2IT5/3P5G |
| 9B | 0 | QPD | G+ | 103,106,108,110,113,125~6 | 2OX9 | |
| 10A | 1 | 67,71,74,96,101,102 | EPN | NPJ > M± | 93,95,97,101,107~9,113~5
/ 93,95,101,105,113,115 |
2GGX/3G84 |
| 10B | 0 | QPD | G+ | 103,106,108,113,125~6 | 2OX9 | |
| 11A | 1 | 95,100,114 | KPD | Glc = G | 90,93,95,97,101,113,114 | 2OX9 |
| 11B | 0 | QPD | G+ | 103,106,108,110,113,125~6 | 2OX9 | |
| 12A | 0 | EPN | G > M± | 90,93,95,97,101,113~4
/ 93,95,101,105,113,115 |
2OX9 / 3G84 | |
| 12B | 0 | QPD | G+ | 103,106,108,113,125,126 | 2OX9 | |
| 13A | 0 | FPE | no | no | ||
| 13B | 2 | 82,109,115,118 /
79, 83,86,109,117,118 |
RPD | M > G | 72,106,108,117,123~4,131~2, 134 / 72,109,117,131~3,137 |
1SL4 / 1SL5 |
| 14A | 0 | FPG | no | 1K9J | ||
| 14B | 2 | 108,117,132 /
79, 83,86,109,117,118 |
RPD | M | 72,106,108,117,131,132,134 | 2IT5 |
| 15A | 0 | FPE | no | no | ||
| 15B | 1 | 79,83,86,109,118,119 | QPD | M- | 72,106,108,110,118,132~3,135 | 2IT5 |
| 16A | 0 | FPE | no | no (0.12) | ||
| 16B | 2 | 79,82,109,118~9
/ 79,83,86,109,118~9 |
QPD | G+ | 103,106,108,110,118,132~3 | |
| 17A | 0 | FPE | no | |||
| 17B | 1 | 83,109,115,118 | QPD | G+ | 103,106,108,117,131~2 | |
| 18A | 2 | 70,74,77,104,105
/ 70,74,101,104,105 |
EPK | M | 98,100,104,108,116,118 | |
| 18B | 0 | CPQ | M | 69,103,105,107,114,126~7,129 | ||
| 19A | 1 | TPD | G | 93,96,98,100,104,116~7 | ||
| 19B | 0 | EPP | M | 112,118~9,124~5,127 |
G, galactose; M, mannose; MLR, maltotriose; NPJ, 4-nitrophenyl 4-O-α-D-glucopyranosyl-α-D-galactopyranoside; Glc: α-D-glucose; no, C-score ≤ 0.20; when X > Y> 0.20: if a sugar (X) has a higher C-score than the expected ( Y, marked ±) based on the EPN (mannose) or QPD (galactose) motif, both types are listed. Otherwise, only the sugar with highest C-score is listed and marked “+” for consistent predictions or “-” for contradictory ones. The residues involved in both Ca coordination and sugar binding are in bold.
3.4. Expression profiles and transcriptional regulation of M. sexta CTLDPs
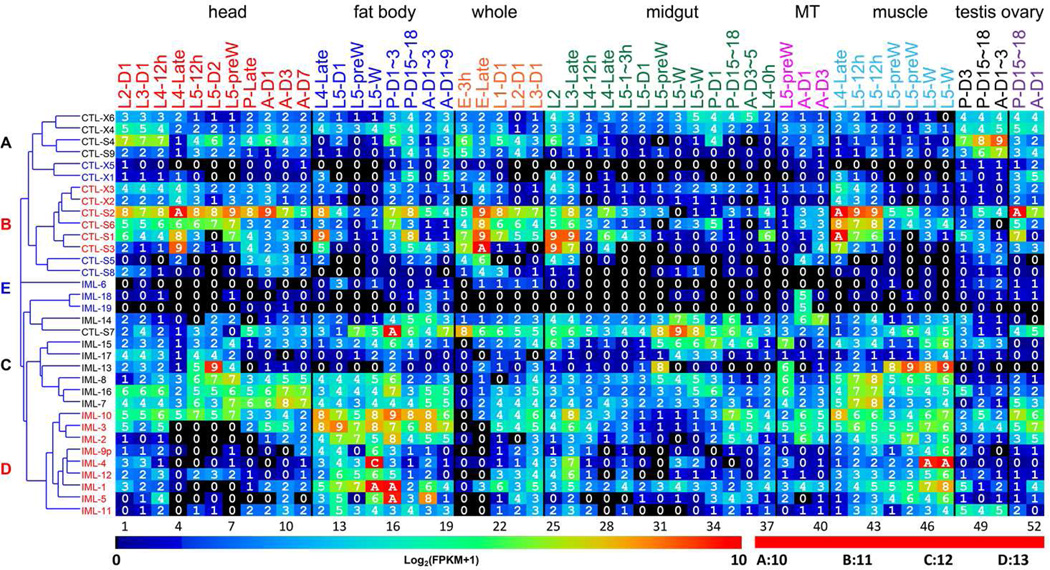
To analyze the expression of CTLDP genes, we examined their mRNA levels in 52 tissue samples from M. sexta at various developmental stages. Cluster analysis of the expression profiles revealed five groups (Fig. 7, Table S2). Group A includes CTL-S4, X4, X6, and IML-20 whose mRNA levels were low in muscle, Malpighian tubules and midgut, but moderate in head, fat body, ovary and testis. Group B consists of CTL-S1~S3, S6, X2 and X3. Although their mRNA levels are generally higher than those of the group A members, especially in muscle and Malpighian tubules, expression patterns of these two groups were similar. The CTL-S7, IML-7, 8 and 14~16 in group C are expressed at moderate levels in most of the samples. The mRNA levels of IML-1~5 and 9~12 (group D) are much higher in fat body and muscle, than in the other tissues. Group E is composed of CTL-X1, X5, S5, S8, IML-6, 18, and 19, and in this group transcript levels are low in most of the tissue samples.
Fig. 7. Transcript profiles of the M. sexta CTL genes in the fifty-two tissue samples.
The mRNA levels, as represented by log2(FPKM+1) values, are shown in the gradient heat map from blue (0) to red (≥10). The values of 0~0.49, 0.50~1.49, 1.50~2.49 … 8.50~9.49, 9.50~10.49 10.50~11.49, 11.50~12.49 and 12.50~13.49 are labeled as 0, 1, 2 … 9, A, B, C and D, respectively. The 52 cDNA libraries (1 through 52) are constructed from the following tissues and stages: head [1. 2nd (instar) L (larvae), d1 (day 1); 2. 3rd L, d1; 3. 4th L, d0.5; 4. 4th L, late; 5. 5th L, d0.5; 6. 5th L, d2; 7. 5th L, pre-W (pre-wandering); 8. P (pupae), late; 9. A (adults), d1; 10. A, d3; 11. A, d7], fat body (12. 4th L, late; 13. 5th L, d1; 14. 5th L, pre-W; 15. 5th L, W; 16. P, d1–3; 17. P, d15–18; 18. A, d1–3; 19. A, d7–9), whole animals [20. E (embryos), 3h; 21. E, late; 22. 1st L; 23. 2nd L; 24. 3rd L), midgut (25. 2nd L; 26. 3rd L; 27. 4th L, 12h; 28. 4th L, late; 29. 5th L, 1–3h; 30. 5th L, 24h; 31. 5th L, pre-W; 32–33. 5th L, W; 34. P, d1; 35. P, d15–18; 36. A, d3–5; 37. 4th L, 0h), Malpighian tubules (MT) (38. 5th L, pre-W; 39. A, d1; 40. A, d3), muscle (41. 4th L, late; 42–43. 5th L, 12h; 44–45. 5th L, pre-W; 46–47. 5th L, W), testis (48. P, d3; 49. P, d15–18; 50. A, d1–3), and ovary (51. P, d15–18; 52. A, d1). Cluster analysis has revealed four distinct groups (A~D), as shown on the left. The group E genes (in blue font) are expressed at low levels [log2(FPKM+1): 0~4] in nearly all the 52 samples. The data for this figure is included in Table S2.
We observed high mRNA levels of some CTLs in certain tissues and stages (Fig. 7, Table S2). For instance, the FPKM values of CTL-S1 in muscle of 4th instar larvae, CTL-S3 in eggs before hatching, CTL-S4 in adult testis, CTL-S7 in fat body of early pupae were 1378, 980, 484, and 1180, respectively. The FPKM values of CTL-S2 in ovary of late pupae, muscle and head of late 4th instar larvae were 926, 944, 820, 944, respectively. The IML-1, 5, 10, and 4 mRNA levels were high in fat body of early pupae (FPKM: 1268, 769, 414) and wandering larvae (3615), respectively. The FPKMs of IML-13 were 665, 515, and 637 in head of day 2, 5th instar larvae, muscle of pre-wandering and wandering 1arvae. In comparison, the FPKMs of CTL-S5, S6, S9, X1~X6, IML-2, 3, 6~9, 11, 12, and 14~19 were <400 (average: 12).
Our studies on mRNA and polypeptide level changes (Zhang et al., 2011 and 2014; Gunaratna and Jiang, 2013) provided an overview of the immune system in response to an immune challenge. In light of the genome sequence, we reanalyzed the results on CTLDPs (Table 2) in conjunction with a search for potential regulatory elements in the 1000 bp region upstream of each gene. We identified one R1 binding motif in IML-8, 93 GATA boxes, and 247 LPS responsive elements. The R1 binding site is required for Rel protein-mediated up-regulation of cecropin A1 transcription in D. melanogaster (Uvell and Engström, 2003). The search for putative NF-κB binding sites (GGGRAYYYYY) in the thirty CTLDP genes uncovered 1, 22 and 290 motifs with 0, 1 and 2 mismatches, respectively. M. sexta IML-7 contains all three forms, CTL-S1, S2, S8, X1, X3, X4, IML-1, 3, 7, 10, 14~16, and 19 have the motifs with 1 and 2 mismatches, and 15 other genes have 2 mismatches. Five genes (IML-1, 2, 4, 12 and 15) showed increase in mRNA (>4.0 fold) and/or protein (≥1.6 fold) levels after immune challenge. However, the dataset is too limited to analyze whether the presence of κB elements correlates with inducibility.
4. Discussion
Annotation of the 34 CTLDP genes in M. sexta genome provide insights into structures of this diverse family of proteins in insects. Based on their domain organization, we divide them into three groups. Simple and complex CTLDPs exist widely in insects and other animals including human. Immulectins represent a unique group of C-type lectins with two tandem CTLDs (Fig. 1), which independently evolved in lepidopteran insects (Fig. 2). As a common structural module of CTLDPs, the CTL domain adopts a double-loop fold. The closely located N- and C-termini make the entire domain a “loop”, whereas the region between β2 and β3 strands forms another “loop” on the other side of the domain (Fig. 6) (Zelensky and Gready, 2005). The “domain loop” consists of β helices, β sheets, and loops; the other “interstrand loop” includes most of the residues interacting with Ca + and carbohydrates (Fig. 5). While these residues display certain levels of conservation, gaps and inserts are common in this part of the domain in CTLPDs (e.g. CTL-S4, S5, S9A, and X6), reminiscent of the complementarity determining regions of antibodies. Such variable sequences, may be important for its specific recognition of glycoconjugates. To explore the possibility, we constructed three-dimensional models of all the 56 CTL domains and observed a similar overall fold (Fig. 6; data not shown). Multiple-threading alignments and iterative template assembly simulations allowed us to predict key residues in the structures, which may participate in Ca and sugar binding (Table 2). Based on these predictions, eighteen CTL domains may bind Ca2+ ion(s) and carbohydrates; 22 may bind sugars in a Ca2+-independent manner; 16 may not bind any carbohydrate. In comparison, sequence alignment-based predictions are vague (Fig. 5). It would be interesting to determine experimentally the calcium-dependence and carbohydrate-binding specificity of these domains, especially the ones yielding inconsistent predictions (IML-2B, 3B, 10A, 12A, 15B) (Table 2). While mannose-binding was suggested by the EPN motif in A. aegypti CTLMA15, Anguilla japonica lectin-2, and Spirinchus lanceolatus asialofetunin-binding CTL, experimental data supported galactose binding (Cheng et al., 2010, Tasumi et al., 2002, Hosono et al., 2005), which is consistent with our predictions based on their 3D structural models (data not shown).
The comparative genomic analysis of insect CTLDPs provides leads for their functional studies in the future. The orthologous relationships of CTL-X and S proteins (Fig. 2) suggest the orthologs may perform similar functions in diverse groups of insects. For instance, mutations in four CTL-X genes lead to phenotype changes in D. melanogaster. The product of Drosophila furrowed gene (an ortholog of M. sexta CTL-X2) plays a role in homophilic cell adhesion that affects planar cell polarity. Flies deficient in furrowed exhibited disrupted development of compound eyes and bristles (Chin and Mlodzik, 2013; Leshko-Lindsay and Corces, 1997). Functions of the gene uninflatable (uif, CTL-X3 ortholog) were implied in two manners: uif deficient flies displayed deficiencies in tracheal growth, tracheal cuticle molting and trachea inflation during embryogenesis; uif antagonizes the Notch signaling pathway by modulating ligand accessibility to the extracellular domain of Notch (Xie et al., 2012; Zhang and Ward, 2009). Acting with neuroglian and neurexin IV, contactin (CTL-X4 ortholog) is essential for septate junction organization in epithelial and neuronal cells, normal blood-nerve barrier in peripheral nervous system, and blood-brain barrier functions (Banerjee et al., 2006; Faivre-Sarrailh et al., 2004; Stork et al., 2008). D. melanogaster CG3921 product (CTL-X6 ortholog) interacts with the claudin protein megatrachea (Jaspers et al., 2012). Due to the three scavenger receptor (SR) Cys-rich domains, CG3921 and its orthologs in the mosquitoes may have SR activity. We have also identified orthologs of CTL-S1~S6 (Fig. 2A), but little is known about their supposedly conserved functions.
In contrast to these CTLPDs, others have evolved to perform unique functions in diverse groups of insects. A 26 kDa regenectin (CTL-S) assists organization or stabilization of epidermis during leg regeneration in the American cockroach (Kubo et al., 1993). The heterodimer of A. gambiae CTL4 (CTL-S) and CTLMA2 (CTL-S), defending the mosquito against Gram-negative bacteria (Schnitger et al., 2009), acts as an agonist of Plasmodium berghei development (Osta et al., 2004). A. aegypti CLSP2 (CTL-X), with a CTL domain following N-terminal serine protease domain, negatively regulates proPO activation (Shin et al., 2011). West Nile virus induces A. aegypti mosGCTL-1 (formerly CTLMA15, a CTL-S), which enables the viral attachment to cells and enhances entry (Cheng et al., 2010). A CTL-S on the surface of the parasitic wasp embryonic cells may recognize N-linked carbohydrate chains with fucose residues on the host embryos for infiltration (Takahashi-Nakaguchi et al., 2011). Since no ortholog is found in M. sexta, we consider these functions as results of evolution under specific conditions by other insects.
Certain immulectins (e.g. M. sexta IML-1, B. mori CTL11, D. plexippus CTL17) form ortholog groups in lepidopteran insects. Others (e.g. H. armigera CTL1, 2 and 6; M. sexta IML-9~12) seem to be the result of lineage-specific gene duplications (Fig. 2B and Fig. 3). Although the functional implications of such evolutionary relationships are unclear, we suggest that most of these proteins participate in defense responses, based on the known IML functions (Table 1). These functions are pattern recognition, agglutination/nodulation, opsonization for phagocytosis, encapsulation, and proPO activation/melanization. As recognition molecules, gene duplications and sequence divergence in the loop regions facilitate specific binding of carbohydrates and other surface molecules of the invading pathogens. Having two CTLDs in one protein may assist dimerization, as observed in certain CTL-Ss (Schnitger et al., 2009; Haq et al., 1996), and further increase recognition spectra and binding affinity (Watanabe et al., 2006).
While the phylogenetic relationships of entire proteins (Fig. 2) and CTLDs (Fig. 4) revealed functional conservation of some CTLPDs and diversification of others, the emergence of CTLs with two CTLDs in Lepidoptera and their major expansion in B. mori, H. armigera, and M. sexta, are truly remarkable. We do not know the exact origins of domains A and B, but the CTLD tree (Fig. 4) suggests they are results of a merger of two ancient CTL-S genes similar to S7 and S5. Cys-1 and Cys-2, missing in the 19 immulectin A domains and CTL-S7 CTLD, are present in all the 19 B domains and CTL-S5 CTLD. There are three other differences between domains A and B. The gap between α2 and β2 (Fig. 5) results in the shortening of a loop connecting the two elements in IML-1 domain A (Fig. 6, C and D, arrows). The shorter linker between β2 and β3 causes the α-helix-3 to become a strand in the A domain. Substitutions of the key residues for carbohydrate interaction in domain A of IML-6~8 and 13~17, together with the two other changes, may lead to potential loss of sugar-binding capability (Table 2). Similarly, major expansions of the CTL-S family occurred in the lineages of D. melanogaster, A. gambiae and A. aegypti (Christophides et al., 2002; Waterhouse et al., 2007) to meet the need for pathogen recognition, perhaps.
Temporospatial expression of the CTLDP genes reflects functional importance of their protein products (Fig. 7, Table S2). For instance, the low levels of CTL-X1~X4 and X6 transcripts in the 52 tissue samples seem consistent with their attachment to cell membrane via the C-terminal TM region. Unlike most of the hemolymph CTLPDs, these cell adhesion molecules, limited in numbers and tissue locations, interact with specific protein partners. The IML-6 gene is hardly expressed in all the tissues, perhaps because the protein truncation rendered the gene nonfunctional. The increase in IML-1, 2 and 4 mRNA and protein levels (Table 3) after immune challenge is consistent with their known roles in defense responses (Table 1). It would be interesting to explore the functions of IML-12 and 15, which display similar patterns of the induced expression. As reported in Section 3.4, tissue or time-specific expression of CTL-S1~S4, S7, IML13 suggests functions, and so do high mRNA levels of IML-1, 4, 5 and 10. Based on its high transcript levels in fat body of early pupae, similar to those of IML-1 and 4, we predict CTL-S7 (Fig. 2A and Fig. 4) plays a key role in innate immunity of M. sexta.
Table 3.
Features of the M. sexta CTLDP genes, transcripts, and polypeptides
| name | length a (bp) | κB b (2, 1, 0) |
GATA | R1 | CATTW | mRNA c | protein d | ||
|---|---|---|---|---|---|---|---|---|---|
| IF/CF | IH/CH | I/C | p | ||||||
| CTL-S1 | 10, 1, 0 | 3 | 6 | ||||||
| CTL-S2 | 13, 4, 0 | 3 | 8 | ||||||
| CTL-S3 | 11, 0, 0 | 3 | 6 | ||||||
| CTL-S4 | 8, 0, 0 | 2 | 9 | ||||||
| CTL-S5 | 6, 0, 0 | 5 | 7 | ||||||
| CTL-S6 | 16, 0, 0 | 3 | 12 | ||||||
| CTL-S7 | 4, 0, 0 | 2 | 9 | ||||||
| CTL-S8 | 6, 1, 0 | 5 | 4 | ||||||
| CTL-S9 | 15, 0, 0 | 3 | 6 | ||||||
| IML-1 | 13, 2, 0 | 2 | 10 | 4.9 | 0.9 | 7.3 | 0.00 | ||
| IML-2 | 5, 0, 0 | 4 | 5 | 45.4 | nd | 1.6 | 0.01 | ||
| IML-3 | 10, 1, 0 | 3 | 10 | 1.7 | 3.0 | 1.1 | 0.72 | ||
| IML-4 | 13, 0, 0 | 2 | 13 | 2573.6 | 5.8 | 6.2 | 0.16 | ||
| IML-5 | 289 | 4, 0, 0 | 1 | 2 | |||||
| IML-6 | ? | ? | ? | ? | ? | ||||
| IML-7 | 13, 1, 1 | 5 | 9 | 0.1 | 0.8 | 0.8 | 0.06 | ||
| IML-8 | 12, 4, 0 | 5 | 1 | 7 | 0.6 | 1.5 | 1.1 | 0.89 | |
| IML-9 | ? | ? | ? | ? | ? | 1.9 | 1.5 | 0.1 | 0.00 |
| IML-10 | 6, 1, 0 | 5 | 8 | 1.8 | nd | 0.8 | 0.08 | ||
| IML-11 | ? | ? | ? | ? | ? | 1.9 | 1.5 | 0.6 | 0.30 |
| IML-12 | ? | ? | ? | ? | ? | 332.3 | nd | 101.4 | 0.03 |
| IML-13 | 7, 0, 0 | 2 | 10 | ||||||
| IML-14 | 10, 1, 0 | 3 | 7 | ||||||
| IML-15 | 19, 1, 0 | 2 | 6 | 15.0 | 2.4 | ||||
| IML-16 | 8, 1, 0 | 4 | 10 | 0.3 | 0.3 | 1.3 | 0.27 | ||
| IML-17 | 15, 0, 0 | 4 | 8 | 0 | 0.37 | ||||
| IML-18 | 7, 0, 0 | 1 | 17 | ||||||
| IML-19 | 12, 1, 0 | 4 | 6 | ||||||
| CTL-X1 | 6, 1, 0 | 1 | 8 | ||||||
| CTL-X2 | 8, 0, 0 | 2 | 6 | ||||||
| CTL-X3 | 10, 1, 0 | 6 | 4 | ||||||
| CTL-X4 | 775 | 4, 1, 0 | 4 | 9 | |||||
| CTL-X5 | 9, 0, 0 | 1 | 11 | ||||||
| CTL-X6 | 10, 0, 0 | 3 | 14 | ||||||
length of the analyzed region before the longest transcript in Cufflink1.0: 1000 bp if unspecified. ?: no sequence available for search.
numbers of the κB motif (GGGRAYYYYY) with 2, 1, and 0 mismatch. R1 site, KKGNNTTTY; GATA box, WGATAR.
relative abundances of the mRNA in fat body (IF/CF) and hemocytes (IH/CH) (Zhang et al., 2011). nd, not detected.
I/C ratio from the peptidome data (Zhang et al., 2014). p-value from the Student’s t-test of normalized spectral counts.
In summary, we performed an integrated investigation of the 34 CTLDP genes in the M. sexta genome. Sequence comparisons and phylogenetic analysis revealed evolution dynamics of these genes. We found changes were relatively few in CTL-X and some -S genes and, on the other hand, gene merging and duplications gave rise to 19 IMLs in M. sexta in a short period of evolutionary time. Structural variations such as indels in IML B domains caused conformation changes in the loops that interact with carbohydrates to cover a broader range of pathogens, perhaps. Molecular modeling of 3D structures is useful for predicting sugar binding specificity and visualizing structural differences between IML A and B domains. Gene expression profiling provides new information for functional exploration of CTLDPs in this biochemical model species.
Supplementary Material
Identified 9 simple and 6 complex CTL-domain proteins and 19 immulectins;
Analyzed their structural features, evolution dynamics, and expression profiles;
Modeled 56 CTL domains to predict their carbohydrate binding capability and specificity.
Acknowledgements
This work was supported by NIH grants GM58634 (to H. Jiang) and GM041247 (to M. Kanost), National Natural Science Foundation of China grant 31402017 (to X. Rao), and a DARPA grant (to G. Blissard). Computation for this project was performed at OSU High Performance Computing Center at Oklahoma State University supported in part through the National Science Foundation grant OCI–1126330. This work was approved for publication by the Director of Oklahoma Agricultural Experimental Station, and supported in part under project OKLO2450.
Abbreviations
- CTL
C-type lectin
- CRD
carbohydrate recognition domain
- CTLDP
CTL-domain protein
- IML
immulectin
- LC
low complexity
- proPO
prophenoloxidase
- MBL
rat mannose binding lectin
- FPKM
fragments per kilobase of exon per million fragments mapped
- C
control
- I
induced
- F
fat body
- H
hemocytes
- TM
transmembrane
- SR
scavenger receptor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Banerjee S, Pillai AM, Paik R, Li J, Bhat MA. Axonal ensheathment and septate junction formation in the peripheral nervous system of Drosophila. J. Neurosci. 2006;26:3319–3329. doi: 10.1523/JNEUROSCI.5383-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambi A, Koopman M, Figdor CG. How C-type lectins detect pathogens. Cell. Microbiol. 2005;7:481–488. doi: 10.1111/j.1462-5822.2005.00506.x. [DOI] [PubMed] [Google Scholar]
- Chai LQ, Tian YY, Yang DT, Wang JX, Zhao XF. Molecular cloning and characterization of a C-type lectin from the cotton bollworm, Helicoverpa armigera. Dev. Comp. Immunol. 2008;32:71–83. doi: 10.1016/j.dci.2007.04.006. [DOI] [PubMed] [Google Scholar]
- Charroux B, Rival T, Narbonne-Reveau K, Royet J. Bacterial detection by Drosophila peptidoglycan recognition proteins. Microbes Infect. 2009;11:631–636. doi: 10.1016/j.micinf.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Cheng G, Cox J, Wang P, Krishnan MN, Dai J, Qian F, Anderson JF, Fikrig E. A C-type lectin collaborates with a CD45 phosphatase homolog to facilitate West Nile virus infection of mosquitoes. Cell. 2010;142:714–725. doi: 10.1016/j.cell.2010.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin ML, Mlodzik M. The Drosophila selectin furrowed mediates intercellular planar cell polarity interactions via frizzled stabilization. Dev. Cell. 2013;26:455–468. doi: 10.1016/j.devcel.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophides GK, Zdobnov E, Barillas-Mury C, Birney E, Blandin S, Blass C, Brey PT, Collins FH, Danielli A, Dimopoulos G, Hetru C, Hoa NT, Hoffmann JA, Kanzok SM, Letunic I, Levashina EA, Loukeris TG, Lycett G, Meister S, Michel K, Moita LF, Muller HM, Osta MA, Paskewitz SM, Reichhart JM, Rzhetsky A, Troxler L, Vernick KD, Vlachou D, Volz J, von Mering C, Xu J, Zheng L, Bork P, Kafatos FC. Immunity-related genes and gene families in Anopheles gambiae. Science. 2002;298:159–165. doi: 10.1126/science.1077136. [DOI] [PubMed] [Google Scholar]
- Dodd RB, Drickamer K. Lectin-like proteins in model organisms: implications for evolution of carbohydrate-binding activity. Glycobiol. 2001;11:71R–79R. doi: 10.1093/glycob/11.5.71r. [DOI] [PubMed] [Google Scholar]
- Faivre-Sarrailh C, Banerjee S, Li JJ, Hortsch M, Laval M, Bhat MA. Drosophila contactin, a homolog of vertebrate contactin, is required for septate junction organization and paracellular barrier function. Development. 2004;131:4931–4942. doi: 10.1242/dev.01372. [DOI] [PubMed] [Google Scholar]
- Gallagher JT. Carbohydrate-binding properties of lectins: a possible approach to lectin nomenclature and classification. Biosci Rep. 1984;4:621–632. doi: 10.1007/BF01121015. [DOI] [PubMed] [Google Scholar]
- Gunaratna RT, Jiang H. A comprehensive analysis of the Manduca sexta immunotranscriptome. Dev. Comp. Immunol. 2013;39:388–398. doi: 10.1016/j.dci.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haq S, Kubo T, Kurata S, Kobayashi A, Natori S. Purification, characterization, and cDNA cloning of a galactose-specific C-type lectin from Drosophila melanogaster. J. Biol. Chem. 1996;271:20213–20218. doi: 10.1074/jbc.271.33.20213. [DOI] [PubMed] [Google Scholar]
- Hosono M, Sugawara S, Ogawa Y, Kohno T, Takayanagi M, Nitta K. Purification, characterization, cDNA cloning, and expression of asialofetuin-binding C-type lectin from eggs of shishamo smelt (Osmerus [Spirinchus] lanceolatus) Biochim. Biophys. Acta. 2005;1725:160–173. doi: 10.1016/j.bbagen.2005.07.009. [DOI] [PubMed] [Google Scholar]
- Jaspers MH, Nolde K, Behr M, Joo SH, Plessmann U, Nikolov M, Urlaub H, Schuh R. The claudin megatrachea protein complex. J. Biol. Chem. 2012;287:36756–36765. doi: 10.1074/jbc.M112.399410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Vilcinskas A, Kanost MR. Immunity in lepidopteran insects. Adv. Exp. Med. Biol. 2010;708:181–204. doi: 10.1007/978-1-4419-8059-5_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanost MR, Jiang H, Yu XQ. Innate immune responses of a lepidopteran insect, Manduca sexta . Immunol. Rev. 2004;198:97–105. doi: 10.1111/j.0105-2896.2004.0121.x. [DOI] [PubMed] [Google Scholar]
- Koizumi N, Imai Y, Morozumi A, Imamura M, Kadotani T, Yaoi K, Iwahana H, Sato R. Lipopolysaccharide-binding protein of Bombyx mori participates in a hemocyte-mediated defense reaction against gram-negative bacteria. J. Insect Physiol. 1999;45:853–859. doi: 10.1016/s0022-1910(99)00069-4. [DOI] [PubMed] [Google Scholar]
- Koizumi N, Morozumi A, Imamura M, Tanaka E, Iwahana H, Sato R. Lipopolysaccharide-binding proteins and their involvement in the bacterial clearance from the hemolymph of the silkworm Bombyx mori . Eur. J. Biochem. 1997;248:217–224. doi: 10.1111/j.1432-1033.1997.t01-1-00217.x. [DOI] [PubMed] [Google Scholar]
- Kubo T, Kawasaki K, Natori S. Transient appearance and localization of a 26-kDa lectin, a novel member of the Periplaneta lectin family, in regenerating cockroach leg. Dev. Biol. 1993;156:381–390. doi: 10.1006/dbio.1993.1085. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Ann. Rev. Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- Leshko-Lindsay LA, Corces VG. The role of selectins in Drosophila eye and bristle development. Development. 1997;124:169–180. doi: 10.1242/dev.124.1.169. [DOI] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling E, Ao J, Yu X-Q. Nuclear translocation of immulectin-3 stimulates hemocyte proliferation. Mol. Immunol. 2008;45:2598–2606. doi: 10.1016/j.molimm.2007.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osta MA, Christophides GK, Kafatos FC. Effects of mosquito genes on Plasmodium development. Science. 2004;303:2030–2032. doi: 10.1126/science.1091789. [DOI] [PubMed] [Google Scholar]
- Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protocols. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnitger AK, Yassine H, Kafatos FC, Osta MA. Two C-type lectins cooperate to defend Anopheles gambiae against Gram-negative bacteria. J. Biol. Chem. 2009;284:17616–17624. doi: 10.1074/jbc.M808298200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X-Z, Yu X-Q. The extended loop of the C-terminal carbohydrate-recognition domain of Manduca sexta immulectin-2 is important for ligand binding and functions. Amino Acids. 2012;42:2383–2391. doi: 10.1007/s00726-011-0980-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin SW, Park DS, Kim SC, Park HY. Two carbohydrate recognition domains of Hyphantria cunea lectin bind to bacterial lipopolysaccharides through O-specific chain. FEBS Lett. 2000;467:70–74. doi: 10.1016/s0014-5793(00)01127-3. [DOI] [PubMed] [Google Scholar]
- Shin SW, Zou Z, Raikhel AS. A new factor in the Aedes aegypti immune response: CLSP2 modulates melanization. EMBO Rep. 2011;12:938–943. doi: 10.1038/embor.2011.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stork T, Engelen D, Krudewig A, Silies M, Bainton RJ, Klambt C. Organization and function of the blood-brain barrier in Drosophila . J. Neurosci. 2008;28:587–597. doi: 10.1523/JNEUROSCI.4367-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi-Nakaguchi A, Hiraoka T, Iwabuchi K. The carbohydrate ligands on the host embryo mediate intercellular migration of the parasitic wasp embryo. FEBS Lett. 2011;585:2295–2299. doi: 10.1016/j.febslet.2011.05.055. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Ishibashi J, Fujita K, Nakajima Y, Sagisaka A, Tomimoto K, Suzuki N, Yoshiyama M, Kaneko Y, Iwasaki T, Sunagawa T, Yamaji K, Asaoka A, Mita K, Yamakawa M. A genome-wide analysis of genes and gene families involved in innate immunity of Bombyx mori . Insect Biochem. Mol. Biol. 2008;38:1087–1110. doi: 10.1016/j.ibmb.2008.09.001. [DOI] [PubMed] [Google Scholar]
- Tasumi S, Ohira T, Kawazoe I, Suetake H, Suzuki Y, Aida K. Primary structure and characteristics of a lectin from skin mucus of the Japanese eel Anguilla japonica . J. Biol. Chem. 2002;277:27305–27311. doi: 10.1074/jbc.M202648200. [DOI] [PubMed] [Google Scholar]
- Uvell H, Engström Y. Functional characterization of a novel promoter element required for an innate immune response in Drosophila . Mol. Cell Biol. 2003;23:8272–8281. doi: 10.1128/MCB.23.22.8272-8281.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JL, Liu XS, Zhang Q, Zhao HB, Wang YF. Expression profiles of six novel C-type lectins in response to bacterial and 20E injection in the cotton bollworm (Helicoverpa armigera) Dev. Comp. Immunol. 2012;37:221–232. doi: 10.1016/j.dci.2012.04.004. [DOI] [PubMed] [Google Scholar]
- Watanabe A, Miyazawa S, Kitami M, Tabunoki H, Ueda K, Sato R. Characterization of a novel C-type lectin, Bombyx mori multibinding protein, from the B. mori hemolymph: mechanism of wide-range microorganism recognition and role in immunity. J. Immunol. 2006;177:4594–4604. doi: 10.4049/jimmunol.177.7.4594. [DOI] [PubMed] [Google Scholar]
- Waterhouse RM, Kriventseva EV, Meister S, Xi Z, Alvarez KS, Bartholomay LC, Barillas-Mury C, Bian G, Blandin S, Christensen BM, Dong Y, Jiang H, Kanost MR, Koutsos AC, Levashina EA, Li J, Ligoxygakis P, Maccallum RM, Mayhew GF, Mendes A, Michel K, Osta MA, Paskewitz S, Shin SW, Vlachou D, Wang L, Wei W, Zheng L, Zou Z, Severson DW, Raikhel AS, Kafatos FC, Dimopoulos G, Zdobnov EM, Christophides GK. Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science. 2007;316:1738–1743. doi: 10.1126/science.1139862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis WI, Drickamer K. Trimeric structure of a C-type mannose-binding protein. Structure. 1994;2:1227–1240. doi: 10.1016/S0969-2126(94)00124-3. [DOI] [PubMed] [Google Scholar]
- Weis WI, Drickamer K, Hendrickson WA. Structure of a C-type mannose-binding protein complexed with an oligosaccharide. Nature. 1992;360:127–134. doi: 10.1038/360127a0. [DOI] [PubMed] [Google Scholar]
- Weis WI, Kahn R, Fourme R, Drickamer K, Hendrickson WA. Structure of the calcium-dependent lectin domain from a rat mannose-binding protein determined by MAD phasing. Science. 1991;254:1608–1615. doi: 10.1126/science.1721241. [DOI] [PubMed] [Google Scholar]
- Weis WI, Taylor ME, Drickamer K. The C-type lectin superfamily in the immune system. Immunol. Rev. 1998;163:19–34. doi: 10.1111/j.1600-065x.1998.tb01185.x. [DOI] [PubMed] [Google Scholar]
- X, et al. M. sexta genome paper. 2014 [Google Scholar]
- Xie GQ, Zhang HT, Du GP, Huang QL, Liang XH, Ma J, Jiao RJ. Uif, a large transmembrane protein with EGF-Like repeats, can antagonize notch signaling in Drosophila . PLoS One. 2012;7:e36362. doi: 10.1371/journal.pone.0036362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X-Q, Kanost MR. Manduca sexta lipopolysaccharide-specific immulectin-2 protects larvae from bacterial infection. Dev. Comp. Immunol. 2003;27:189–196. doi: 10.1016/s0145-305x(02)00099-x. [DOI] [PubMed] [Google Scholar]
- Yu X-Q, Kanost MR. Immulectin-2, a pattern recognition receptor that stimulates hemocyte encapsulation and melanization in the tobacco hornworm, Manduca sexta . Dev. Comp. Immunol. 2004;28:891–900. doi: 10.1016/j.dci.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Yu X-Q, Kanost MR. Activation of lepidopteran insect innate immune responses by C-type immulectins. In: Ahmed HA, Vasta GR, editors. Animal lectins: a functional view. Taylor and Francis: CRC Press; 2008. pp. 383–395. [Google Scholar]
- Yu X-Q, Tracy ME, Ling E, Scholz FR, Trenczek T. A novel C-type immulectin-3 from Manduca sexta is translocated from hemolymph into the cytoplasm of hemocytes. Insect Biochem. Mol. Biol. 2005;35:285–295. doi: 10.1016/j.ibmb.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Yu X-Q, Gan H, Kanost MR. Immulectin, an inducible C-type lectin from an insect, Manduca sexta, stimulates activation of plasma prophenol oxidase. Insect Biochem. Mol. Biol. 1999;29:585–597. doi: 10.1016/s0965-1748(99)00036-3. [DOI] [PubMed] [Google Scholar]
- Yu X-Q, Kanost MR. Immulectin-2, a lipopolysaccharide-specific lectin from an insect, Manduca sexta, is induced in response to gram-negative bacteria. J. Biol. Chem. 2000;275:37373–37381. doi: 10.1074/jbc.M003021200. [DOI] [PubMed] [Google Scholar]
- Yu X-Q, Kanost MR. A family of C-type lectins in Manduca sexta . Adv. Exp. Med. Biol. 2001;484:191–194. doi: 10.1007/978-1-4615-1291-2_17. [DOI] [PubMed] [Google Scholar]
- Yu X-Q, Ling E, Tracy ME, Zhu Y. Immulectin-4 from the tobacco hornworm Manduca sexta binds to lipopolysaccharide and lipoteichoic acid. Insect Mol. Biol. 2006;15:119–128. doi: 10.1111/j.1365-2583.2006.00618.x. [DOI] [PubMed] [Google Scholar]
- Zelensky AN, Gready JE. The C-type lectin-like domain superfamily. FEBS J. 2005;272:6179–6217. doi: 10.1111/j.1742-4658.2005.05031.x. [DOI] [PubMed] [Google Scholar]
- Zhang L, Ward RE. uninflatable encodes a novel ectodermal apical surface protein required for tracheal inflation in Drosophila . Dev. Biol. 2009;336:201–212. doi: 10.1016/j.ydbio.2009.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Gunaratna RT, Zhang X, Najar F, Wang Y, Roe B, Jiang H. Pyrosequencing-based expression profiling and identification of differentially regulated genes from Manduca sexta, a lepidopteran model insect. Insect Biochem. Mol. Biol. 2011;41:733–746. doi: 10.1016/j.ibmb.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Cao X, He Y, Hartson S, Jiang H. Semi-quantitative analysis of changes in the plasma peptidome of Manduca sexta larvae and their correlation with the transcriptome variations upon immune challenge. Insect Biochem. Mol. Biol. 2014;47:46–51. doi: 10.1016/j.ibmb.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Z, Evans JD, Lu Z, Zhao P, Williams M, Sumathipala N, Hetru C, Hultmark D, Jiang H. Comparative genomic analysis of the Tribolium immune system. Genome Biol. 2007;8:R177. doi: 10.1186/gb-2007-8-8-r177. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.