Abstract
Genome-wide dissection of the heat stress response (HSR) is necessary to overcome problems in crop production caused by global warming. To identify HSR genes, we profiled gene expression in two Chinese cabbage inbred lines with different thermotolerances, Chiifu and Kenshin. Many genes exhibited >2-fold changes in expression upon exposure to 0.5– 4 h at 45°C (high temperature, HT): 5.2% (2,142 genes) in Chiifu and 3.7% (1,535 genes) in Kenshin. The most enriched GO (Gene Ontology) items included ‘response to heat’, ‘response to reactive oxygen species (ROS)’, ‘response to temperature stimulus’, ‘response to abiotic stimulus’, and ‘MAPKKK cascade’. In both lines, the genes most highly induced by HT encoded small heat shock proteins (Hsps) and heat shock factor (Hsf)-like proteins such as HsfB2A (Bra029292), whereas high-molecular weight Hsps were constitutively expressed. Other upstream HSR components were also up-regulated: ROS-scavenging genes like glutathione peroxidase 2 (BrGPX2, Bra022853), protein kinases, and phosphatases. Among heat stress (HS) marker genes in Arabidopsis, only exportin 1A (XPO1A) (Bra008580, Bra006382) can be applied to B. rapa for basal thermotolerance (BT) and short-term acquired thermotolerance (SAT) gene. CYP707A3 (Bra025083, Bra021965), which is involved in the dehydration response in Arabidopsis, was associated with membrane leakage in both lines following HS. Although many transcription factors (TF) genes, including DREB2A (Bra005852), were involved in HS tolerance in both lines, Bra024224 (MYB41) and Bra021735 (a bZIP/AIR1 [Anthocyanin-Impaired-Response-1]) were specific to Kenshin. Several candidate TFs involved in thermotolerance were confirmed as HSR genes by real-time PCR, and these assignments were further supported by promoter analysis. Although some of our findings are similar to those obtained using other plant species, clear differences in Brassica rapa reveal a distinct HSR in this species. Our data could also provide a springboard for developing molecular markers of HS and for engineering HS tolerant B. rapa.
Introduction
Heat stress (HS) and high temperature (HT) disturb cellular homeostasis and can interfere with various plant metabolic and physiological processes, including photosynthesis, growth, development, pollen fertility, and productivity in both crop species and model plants [1–3]. Therefore, global warming will threaten food safety by reducing crop production [4]. Understanding the molecular mechanisms underlying the heat stress response (HSR) will be necessary for the development of heat-tolerant crop plants capable of sustainable productivity in a changing environment.
Plants can perceive HS in various ways, and they tolerate or acclimate to heat via activation of signaling pathways, expression of stress proteins, and production of defensive materials. In particular, the activation of HSR genes, such as heat stress transcription factors (Hsfs), heat shock proteins (Hsps), and some other transcription factors (TFs), are very important for heat tolerance [5–7]. The activation of HSR genes begins with the perception of HS by a plasma membrane channel and various secondary messengers, such as calcium ions (Ca2+), nitric oxide (NO), and hydrogen peroxide (H2O2). In addition, a histone sensor in the nucleus and unfolded protein sensors present in the ER (endoplasmic reticulum) and cytosol play important roles in the perception process. These signals are then transmitted to downstream components, such as calmodulins, mitogen-activated protein kinases, Hsp90, and Hsfs. Finally the signals induce expression of HSR transcription factors (TFs) like WRKY, Dehydration-Responsive-Element Binding Factor (DREB), and bZIP [3,6–9]. Chaperones for protein homeostasis, osmolytes, and secondary metabolites responsible for ROS detoxification and osmoprotection are also important for thermotolerance [9].
Many Hsfs and Hsps play key roles in plant heat tolerance. The rapid accumulation of Hsps, which is required to protect the cell against HS conditions, is primarily controlled by Hsfs [10,11]. HsfA1s are major regulators in the HSR of tomato and Arabidopsis [12,13], HSfA2 is another major HS regulator in plants [10,14], and HsfA4 acts as a sensor of the H2O2 signal [15], whereas HsfA5 is a negative component of this pathway [16]. Hsps have been implicated in numerous cellular processes including protein folding, assembly, translocation, and degradation [17]. Hsps are classified according to their molecular weights into six major families: Hsp100 (Clp), Hsp90, Hsp70 (DnaK), Hsp60 (chaperonin and GroEL), Hsp40 (DnaJ), and Hsp20 (small HSP; sHsp). Chaperone functions prevent protein aggregation and misfolding, and are mainly mediated by Hsp70s, Hsp90s, and Hsp60s, whereas Hsp20s and Hsp100s are involved, respectively, in protection against protein aggregation and resolubilization of protein aggregates [18,19].
Identification of HSR genes from suitable genotypes can provide a springboard to the development of HS tolerance mechanisms. Omics approaches (genomics, transcriptomics, epigenomics, proteomics, and metabolomics) have been widely applied to investigations of plant HSR, and these analyses have identified many HSR genes [9]. Genome-wide identification of genes required for acquired thermotolerance have shown that expression of genes involved in protection of proteins, translation, and limiting oxidative stress is elevated, whereas expression of genes involved in programed cell death, basic metabolism, and biotic stress responses is reduced [20].
Chinese cabbage (Brassica rapa ssp. pekinensis) is one of the most important leafy vegetable crops in Korea and other eastern Asian countries, including China. Two inbred lines of Chinese cabbage, Chiifu and Kenshin, originated in different geographic regions: Chiifu originated in temperate regions, and Kenshin in subtropical and tropical regions [21]. Therefore, Kenshin has been traditionally used as breeding stock to develop heat-tolerant plants [22,23]. So far, most studies of HS have focused on cereals and model plants. Transcriptomics studies of stress responses have been performed in B. rapa in recent years, but these have primarily focused on cold, salt, and drought stresses [24–30]. Previously, the expression pattern of HSR genes in Chinese cabbage had been analyzed using 24K microarray, which does not include enough genes to cover the entire B. rapa genome [31]. In this study, we used version 3 microarrays (Br135K) to analyze gene expression in the Chiifu and Kenshin lines. We identified differentially expressed genes (DEGs), specifically expressed genes (SEGs), HSR genes, HS marker genes, and membrane leakage-related (MLR) genes, and we discuss these genes in the context of HS.
Materials and Methods
Plant materials and heat treatment
Seeds of two Chinese cabbage (Brassica rapa ssp. pekinensis) inbred lines, Chiifu and Kenshin, were obtained from the Korea Brassica rapa Genome Resource Bank, and the plants were grown for 4 weeks in a growth chamber at 22°C under a 16 h light/8 h dark photoperiod with a photon flux density of 140 μmol m-2 s-1. For heat shock treatments, leaf discs (1 cm in diameter) were incubated at 45°C for 0.5, 1, 2, 3, or 4 h by floating on a water bath. For control samples, the leaf discs were incubated at 22°C for 0.5h by floating on distilled water. Then, leaf discs were blotted, frozen immediately in liquid nitrogen, and stored at -70°C.
Electrolyte leakage test
Immediately after treatment, electrolyte leakage from HS-treated and control plants was measured as previously described [32,33] with some modifications [28]. Briefly, five leaf discs were randomly selected from mixed discs after excision from fully expanded leaves of six plants (10 discs/plant and mixed), and placed in a glass tube with 10 ml distilled water. The samples were then incubated on an orbital shaker at 150 rpm for 30 min at room temperature, after which the initial conductivity (I) was measured using a CON110 conductivity meter (Oakton Ins. USA). The leaf discs were then kept in a boiling water bath for 10 min, after which they were cooled to room temperature, and the final conductivity (F) was measured. The relative electrolyte leakage was calculated using the formula I/F ×100.
Construction of Br135K chips
The Br135K microarray (Brapa_V3_microarray, 3′-Tiling microarray) is a high-density DNA array prepared by NimbleGen (http://www.nimblegen.com/) using Maskless Array Synthesizer (MAS) technology as described in Jung et al. [28]. Labeling, data processing, and background correction were performed as described previously [28]. To assess the reproducibility of the microarray analysis, we repeated the experiment using independently prepared total RNA from two or three biological replicates. The complete raw microarray data have been deposited in the Omics database of NABIC (http://nabic.rda.go.kr) with accession numbers NC-0023-000001–NC-0023-000024.
Gene chip data analysis
Genes with adjusted P-value or false discovery rate (FDR) below 0.05 were collected and further selected for those genes with expression greater than or less than in at least one treatment. Multivariate statistical tests such as principal component analysis and multidimensional scaling were performed with Acuity 3.1 (Molecular Devices, USA). Clustering analysis was carried out using MultiExperiment Viewer version 4.9 (MeV4.9, http://www.tm4.org/mev.html). To obtain insights regarding the putative biological functions and biochemical pathways of DEGs, we carried out enrichment analyses by searching Gene Ontology (GO) [34], agriGO [35], and the Kyoto Encyclopedia of Genes and Genomes [36].
RNA extraction, RT-PCR, and qRT-PCR analysis
Total RNA was treated with RNase-free DNase I (Promega, USA) to remove genomic DNA contamination. First-strand cDNA synthesis was carried out using the Ace-α kit (Toyobo, Osaka, Japan). RT-PCR was performed using 25 ng of cDNA from plants exposed to HS treatments. The gene-specific primers for the stress-responsive genes are listed in S1 Table. Reactions were performed in 0.2 mL PCR tubes containing 10 pmol of each primer, 150 μM of each dNTP, 1.2 U of Taq polymerase, and 1X Taq polymerase buffer; double-distilled water was added to a total volume of 20 μL. The PCR cycle consisted of pre-denaturation at 94°C for 5 min, followed by 25 cycles of denaturation at 94°C for 30 s, annealing at 58°C for 30 s, and extension at 72°C for 45 s, followed by an additional extension step for 5 min at 72°C. PCR products were electrophoresed on 1.5% agarose gels. For qRT-PCR, pre-denaturation was performed at 95°C for 30 seconds, followed by a 40-cycle three-step reaction (5 seconds at 95°C, 20 seconds at 60°C, and 15 seconds at 72°C) with the primer sets shown in S2 Table.
Analysis of enriched conserved cis-elements
HSR genes were subjected to the Short Time-series Expression Miner (STEM) [37], and 1,000 bp upstream sequences of DEG start codons were analyzed for the presence of plant cis-acting elements (PLACE) [38]. The occurrences of these motifs were compared with their frequencies among all promoters represented in the 135K microarray. A P-value was then calculated for each motif and profile combination, based on the hypergeometric distribution [39]. Motifs with P-values below 10-4 were considered to be significantly enriched.
Results and Discussion
Membrane leakage test
Along with survival percentage, lipid peroxidation, and peroxidase (POD) activity, electrolyte leakage was considered to be a good indicator of thermotolerance in Brassica species [40]. Therefore, we measured leakage during HT exposure of two inbred Chinese cabbage lines, Chiifu and Kenshin. As shown in Fig 1, the two lines exhibited similar electrolyte leakage after 1 h exposure to 45°C, but a significant difference was observed after 2 h. On the basis of this result, we concluded that Kenshin is more resistant to > 2 h exposure to HT at 45°C, exactly the reverse of the two strains’ relative tolerance to freezing temperatures [28]. The major parameters used to phenotype thermotolerance of crop plants include viability, pollen development and fertility, photosynthetic rate, and germination [41]. However, breeders and farmers of the leafy vegetable B. rapa pay much more attention to leaf defense against pathogens, growth, and the heading process, complicating the choice of good marker for thermotolerance. Our results indicate that electrolyte leakage could be a useful marker for temperature sensitivity in B. rapa.
Fig 1. Electrolyte leakage of two DH lines, Chiifu and Kenshin, subjected to treatment at 45°C.
Leakage is expressed as the ratio (%) of the conductivity of the initial (I) and final (F) solution. Error bars represent SD of six replicates.
Microarray analysis to identify HSR genes
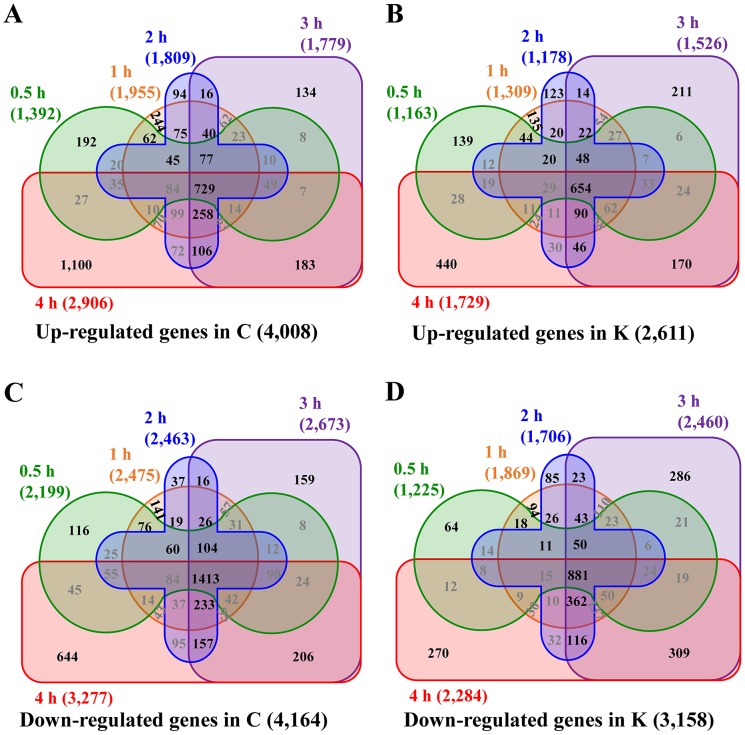
To identify HSR genes, we identified DEGs under HS from among 41,173 genes represented on Br135K microarrays (S3 Table). After removing genes with PI (Probe Intensity) values below 500 for all samples (usually it’s hard to detect the gene expression level using RT-PCR under 25 cycles condition), DEGs were defined as genes with > 2-fold changes in at least one time point. A large number of genes, 4,008 for Chiifu and 2,611 for Kenshin, were up-regulated (Fig 2A and 2B, S4 and S5 Tables). Among these, 729 and 654 genes in Chiifu and Kenshin, respectively, were up-regulated > 2-fold during HS treatments of 0.5–4 h, suggesting that about 20% of HS up-regulated genes (18.2% in Chiifu; 25% in Kenshin) were expressed more strongly during HSR (Fig 2A and 2B, S4 and S5 Tables). In addition, many genes were down-regulated by HS (4,164 in Chiifu and 3,158 in Kenshin) (Fig 2C and 2D, S6 and S7 Tables). Among these, 1,413 and 881 genes in Chiifu and Kenshin, respectively, maintained > 2-fold down-regulation during HS treatments of 0.5–4 h; thus, HS suppressed about 30% of HS-down-regulated genes at all-time points used (Fig 2C and 2D, S6 and S7 Tables). The proportion of DEGs under HS was similar to that obtained in an analysis of the tomato transcriptome (2,203 genes, 9.6%) [13], but higher than the 2% reported by other studies [42–44].
Fig 2. Distribution of genes up- or down-regulated by heat.
A, Distribution of genes up-regulated by heat in Chiifu; B, Distribution of genes up-regulated by heat in Kenshin. Gray number means the stepwise priming up-regulated genes. C, Distribution of heat down-regulated genes in Chiifu; D, Distribution of heat down-regulated genes in Kenshin. Black numbers indicate the genes down-regulated at a single time point or continuously.
Next, we functionally analyzed HSR genes by GO enrichment using agriGO [35], based on information about Arabidopsis homologs. All Arabidopsis counterparts of our microarray probes were used as background references, and significantly represented GO items were defined as FDR values below 0.05. At a significance of < 0.05 FDR, 63 and 106 GO items were significantly enriched among the up-regulated genes in Chiifu and Kenshin, respectively (S8 Table). On the other hand, 126 and 145 GO items in Chiifu and Kenshin, respectively, were significantly enriched among the down-regulated genes (S9 Table). The most enriched GO items among the up-regulated genes in both genotypes were associated with HS: response to heat (GO:0009408), response to ROS (GO:0000302), response to temperature stimulus (GO:0009266), response to abiotic stimulus (GO:0009628), MAPKKK cascade (GO:0000165), etc. (S8 Table). Some GO terms exhibited genotype-specific up-regulation: for Chiifu, response to water deprivation (GO:0009414), and for Kenshin, calcium-mediated signaling (GO:0019722), protein targeting to membrane (GO:0006612), and respiratory burst (GO:0045730) (S8 Table).
To determine the association of HSR genes with specific pathways, we placed the Arabidopsis counterparts of the identified DEGs on KEGG pathway maps. This analysis identified 126 pathways (S10 Table), including plant hormone signal transduction, starch and sucrose metabolism, plant–pathogen interaction, glutathione metabolism, and so on. Most genes related to the RNA degradation pathway were up-regulated by HS in both Chiifu and Kenshin. Most of genes related to cyanoamino acid metabolism and the N-glycan biosynthesis pathways were down-regulated by HS in both Chiifu and Kenshin. We identified no pathway specific for the HS response in Chiifu or Kenshin.
Intrinsic transcriptome differences between Chiifu and Kenshin prior to HT treatment
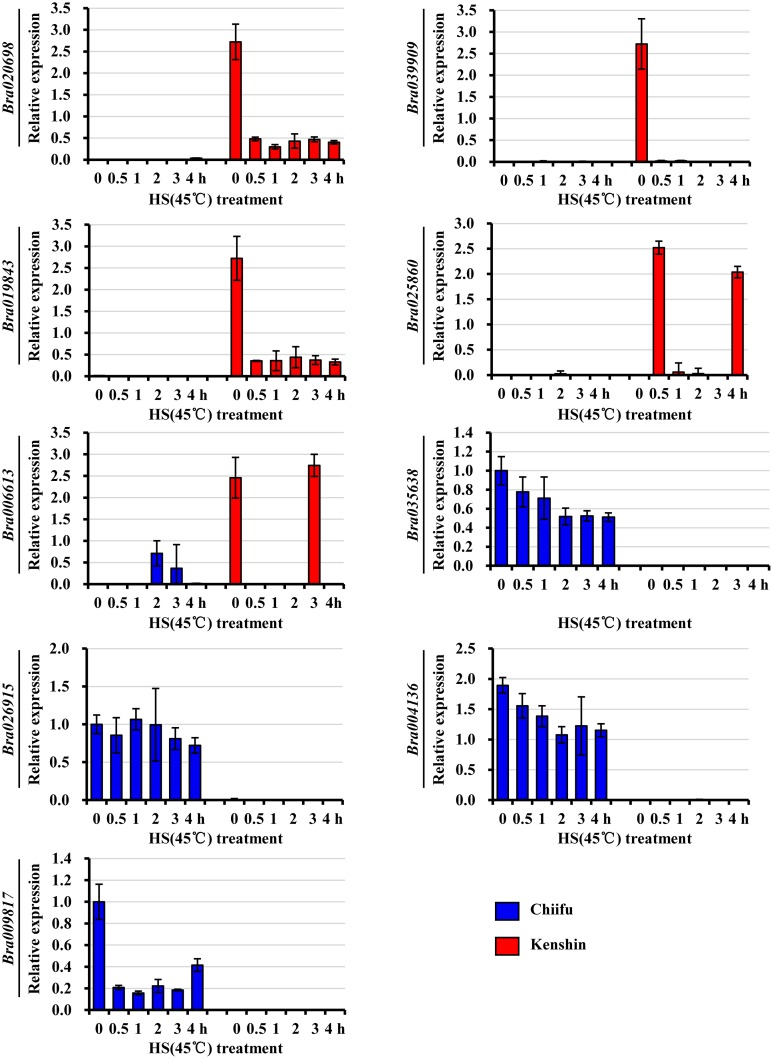
Because previous reports suggested that plant HS tolerance was largely due to constitutive expression of many genes prior to stress treatment [45,46], we compared the intrinsic differences between two Chinese cabbage inbred lines. Intrinsic genes were selected by fold change on the basis of PI values. A large number of genes, 522 genes for Chiifu and 205 genes for Kenshin (S11 Table), exhibited a > 4-fold change compared to the other, but the functions of most genes exhibiting high levels of expression could not be associated with HS. Many unknown genes (including orphan genes) were in this category, suggesting that they play roles in the HSR. To analyze the expression patterns of SEGs in each inbred line, we selected some of the top-ranking genes from S11 Table and subjected them to qRT-PCR analysis (Fig 3). None of these genes have yet been functionally characterized, but expression of all of them except Bra026915 was reduced upon HS treatment. To isolate the candidate genes which may work for the heat tolerant in Kenshin, we isolated genes with intrinsically expressed in Kenshin and showed up-regulation in Keshin after heat treatment at the same time. In the end, 14 genes, including one Hsf gene (Bra029292, HSFB2A), one methionine sulfoxide reductase (Bra019187, PMSR4), three orphan genes (Bra039796, Bra016685, and Bra028606), were identified.
Fig 3. qRT-PCR analysis of several SEGs from either Kenshin or Chiifu.
Relative expression was normalized against BrActin. Two biological repeats were performed, and error bars represent SD. HS, heat shock.
To obtain functional clues about the genes in this category, we carried out GO annotation on the basis of the Arabidopsis homolog data (Table 1). Genotypic SEGs included those encoding kinases, TFs, and factors involved in response to stress, response to abiotic or biotic stimulus, transferase activity, hydrolase activity, and signal transduction. Noteworthy categories include cellular respiration, unsaturated fatty acid biosynthesis, and Hsf genes, which were over-represented in Kenshin relative to Chiifu. For instance, 5 Hsf genes, Bra013409, Bra029292 (HSFB2A), Bra029291 (HSFB2A), Bra004272 (HSFA8), and Bra000557 (HSFA2) were highly intrinsic expressed in Kenshin. Otherwise, only one Hsf gene, Bra012828 (HSFA7A), showed highly intrinsic expressed in Chiifu. We also carried out KEGG pathway analysis to reveal differences between the two genotypes (S12 Table). In total, 100 and 86 pathways were represented by the up-regulated genes in Chiifu and Kenshin, respectively. Metabolic pathways were most prominent in both genotypes, suggesting that a metabolomics approach will be required in future studies.
Table 1. GO annotation of intrinsically expressed genes.
One gene can be associated with two or more GO terms. NA, no Arabidopsis homologs.
| GO Term | Kenshin-up | Chiifu-up |
|---|---|---|
| Kinase | 173 | 322 |
| Response to stress | 167 | 320 |
| Response to abiotic or biotic stimulus | 154 | 302 |
| Transferase activity | 110 | 220 |
| Hydrolase activity | 110 | 189 |
| Signal transduction | 66 | 161 |
| Transcription factors | 61 | 103 |
| Organelle membrane | 70 | 61 |
| Vacuole | 60 | 53 |
| Zinc ion binding | 39 | 71 |
| Oxidation-reduction process | 39 | 67 |
| Response to salicylic acid stimulus | 22 | 71 |
| Carbohydrate catabolic process | 38 | 31 |
| Response to water deprivation | 25 | 43 |
| Response to ethylene stimulus | 19 | 43 |
| Response to auxin stimulus | 17 | 30 |
| Response to heat | 13 | 29 |
| Photosynthetic membrane | 24 | 16 |
| Chlorophyll binding/metabolic process | 14 | 12 |
| Photosynthesis | 14 | 10 |
| Heat acclimation | 3 | 15 |
| Heat shock protein | 5 | 13 |
| Proline transport | 10 | 6 |
| Respiration | 9 | 7 |
| Water transport | 8 | 7 |
| Sugar transmembrane transporter activity | 5 | 8 |
| Cellular respiration | 8 | 2 |
| Unsaturated fatty acid biosynthetic process | 6 | 2 |
| Heat shock transcription factor | 5 | 1 |
| Unclassified | 292 | 465 |
| NA | 203 | 196 |
Analysis of some HSR genes
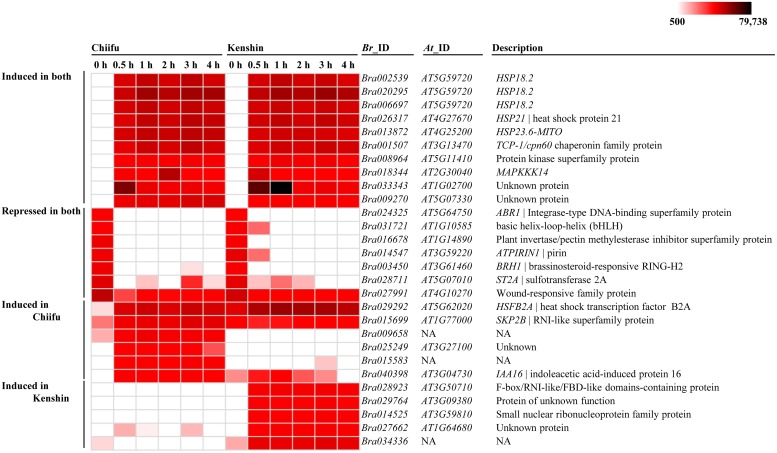
As a first step toward identifying putative regulatory genes for thermotolerance, we identified several genes exhibiting very high levels of expression or specific expression changes, and confirmed them by RT-PCR (Fig 4, S1 Fig, and S13 Table). The top ten genes induced by HS in both genotypes included genes encoding small Hsps, chaperones, kinases, and unknown proteins. Many of these are well-known genes involved in thermotolerance in plants [3,19,47]. Among these, Bra002538 (HSP18.2) exhibited the strongest induction in both genotypes, > 410-fold at 0.5 h and > 630-fold at 1 h, and maintained its level thereafter. An unknown gene (Bra033343, AT1G02700) exhibited a peculiar pattern of expression upon HS treatments: in Chiifu, 1,213-fold induction at 0.5 h and 217-fold induction at 1 h, but in Kenshin, 1,573-fold at 0.5 h, 2,462-fold at 1 h, and 176-fold at 2 h. These patterns are similar to Hsp90 and Hsp70 expression, as expected for the HSR. Several genes were specifically induced in either genotype. Among them, Kenshin SEGs (Bra028923, Bra029764, Bra014525, Bra027662, and Bra034336) could be good targets for thermotolerance studies in Chinese cabbage because Kenshin has been traditionally used as a breeding stock to develop heat-tolerant plants [22,23].
Fig 4. Heat map of genes induced/repressed by HS treatment.
Hsfs and Hsps
Many Hsfs and Hsps play important roles in plant heat tolerance. In the Br135K microarray, 38 Hsf- and 206 Hsp-encoding genes (Hsp100/ClpB, Hsp90, Hsp70, Hsp60, Hsp40, and sHsp) are represented (S14 Table). Among them, 49 genes (including 11 Hsf genes) exhibited very low levels of expression (PI values < 500 of in all samples). Upon 1 h HS treatment, however, 10 and 13 Hsf genes were up-regulated > 2-fold in Chiifu and Kenshin, respectively. Among these, nine Hsf genes were up-regulated in both genotypes: HsfA2 (Bra000557), HsfA4A (Bra020988), HsfA7A (Bra012828, Bra033468), HsfB2A (Bra029292, Bra029291, Bra010049), HsfB2B (Bra000749), and HsfB3 (Bra000235). Up-regulated expression of some genes was confirmed by RT-PCR (S2 Fig). In particular, one HsfA2 and two HsfA7As were up-regulated over 4-fold in both genotypes, whereas one HsfB2A (Bra029292) was induced over 20-fold only in Chiifu. The expression levels of three HsfB2As in Kenshin (Bra010049, Bra029291, & Bra029292) were relatively high under normal growth conditions and increased further upon HS treatment, whereas the levels of two HsfB2As in Chiifu (Bra029291, Bra010049) exhibited similar patterns upon HS treatment but were lower than those in Kenshin. This result suggests that BrHsfB2A (Bra029292) plays an important regulatory role in heat tolerance in Kenshin. In tomato, HsfA1a, HsfA3, HsfB1, and HsfB2a are most abundant and exhibit only minor changes in response to heat. On the other hand, expression of low-abundance genes, such as HsfA1b, HsfA1e, HsfA2, HsfA4b, and HsfA6a, increase upon HS treatment [13]. Some Chinese cabbage Hsfs (HsfB1, HsfB2A) exhibited expression profiles similar to those observed in tomato, whereas others did not, indicating the presence of distinct regulatory mechanisms in different plant species.
The Arabidopsis genome contains 21 Hsf genes that can be categorized into three classes (A, B, and C) [10,48]. HsfA1a is a master regulator of the HSR [10,12,13,49]. However, given the low expression level of HsfA1 following HS, our data imply that other Hsfs could serve as the major TFs in B. rapa. HsfA4a and HsfA8 are sensors of reactive oxygen species (ROS), which are produced as a secondary stress signal during the HSR [50,51]. In particular, HsfA4 acts as a sensor of the H2O2 signal [15], whereas HsfA5 acts as a negative regulator of this pathway [16]. In our microarrays, both genes were highly expressed. Although the expression of HsfA4 increased slightly upon HS, the expression of HsfA5 decreased slightly (S14 Table). All these results suggest that heat tolerance in B. rapa might be primarily regulated at the protein level, with only minor regulation at the transcriptional level. Alternatively, heat tolerance might involve key regulators that are different from those Arabidopsis, as proposed in Wang et al. [52].
Like Hsfs, Hsps also play important roles in heat tolerance in plants, especially in protein folding, assembly, translocation, and degradation [17]. In our microarrays (S14 Table), most Hsp70, Hsp90, and Hsp101 genes were constitutively and highly expressed in all samples. However, small Hsps encoded by genes such as Hsp18.2, Hsp20, Hsp22, and Hsp23.6 exhibited strong induction (15–656-fold) by HS treatment in both genotypes. We confirmed the expression of some genes by RT-PCR (S2 Fig). These results are similar to those in tomato, in which the majority of HS-induced chaperone genes belong to the sHsp family [13].
In Arabidopsis, HS-associated 32 kD protein (HSA32) (AT4G21320), an Hsp, is essential for acquired thermotolerance during long-term recovery after acclimation treatment [53] and in rice [54]. B. rapa does not contain ortholog of this gene, providing further evidence for the existence of different HSRs among plants. Recently, Wang et al. [52] reported that different key genes and regulatory mechanisms are involved in abiotic stress responses in B. rapa and Arabidopsis.
Upstream components of the HSR, including ROS
Upstream events of the HSR include perception and signal transduction of HS. Jia et al. [55] demonstrated that Hsf activity and Hsp production involved in thermotolerance are a result of cross-talk among H2O2, nitric oxide (NO), Ca2+ channels, and calmodulin (CaM). In particular, CaM activates Hsfs, protein kinase, phosphatase, and cyclic nucleotide-gated ion channels. Among these, ROS including H2O2 are considered to be the first signaling components produced by HS [15,56,57]. The ROS signal can then be recognized by histidine kinases and Hsfs (HsfA4 and HsfA8) [19,50,51]. Hsf sensing activates multiple TFs (Zat family, WRKY, and MBF1c) through the MAPK signal pathway [19].
Regarding the expression of ROS-scavenging genes upon initiation of the HSR, two possibilities exist: such genes could be induced in order to reduce levels of ROS produced by HS, or they could be suppressed in order to maintain the levels of ROS involved in downstream signaling. Among 170 PODs on the Br135K microarray, only seven were differentially expressed upon HS treatment (S15 Table). Three genes were up-regulated in both genotypes: ascorbate peroxidase 1 (APX1) (Bra031598), Fe-superoxide dismutase 3 (FSD3) (Bra026503), and Fe-superoxide dismutase 2 (FSD2) (Bra029190). Previous work revealed APX1 as a central component of the ROS gene network [58] and a key player in stress responses [59], whereas FSD2 and FSD3 confer protection against oxidative stress [60,61]. Glutathione peroxidase 2 (GPX2) (AT2G31570; Bra022853), which exhibits Kenshin-specific up-regulation, protects against stress through activation of cytosolic superoxide dismutase [61]. Thus, BrGPX2 is a promising candidate thermotolerance-related gene in Kenshin. In addition, one peroxidase superfamily protein (Bra016930), which is also likely to be involved in ROS signaling, was down-regulated upon HS in both genotypes.
Because calcium signaling can engage in cross-talk with ROS signaling to trigger thermotolerance, we investigated the expression profiles of several calcium-related genes represented on the Br135K chip: 24 calcium channels, 39 CaM, 31 calcium-dependent protein kinases (CDPKs), five CaM-binding protein phosphatases, and 33 cyclic nucleotide-gated ion channels (CNGCs) genes (S16 Table). Most calcium channel genes were highly expressed at all-time points, suggesting that they are regulated at the protein level, if at all. In contrast to the nine Arabidopsis CaM genes up-regulated by HS treatment [62], 14 BrCaM genes (with the exception of CaM8) exhibited high constitutive expression under most conditions. These data further support the idea that the HSR is differentially regulated among species. Expression of CDPK and CNGC genes exhibited no significant changes after HS treatment, with the exception of Bra001676, a homolog of Arabidopsis salt-induced CNG20 [63], which had Kenshin-specific expression. Taken together, these data suggest that regulation of calcium-related proteins in B. rapa HSR is exerted at the protein level.
ROS signaling recognized by histidine kinases must be transmitted to other kinases and phosphatases. To obtain further insight into the HSR signaling pathway, we analyzed expression profiles of 2,110 kinase genes represented on the Br135K microarray. Several genes exhibited differential expression in response to HS treatment: in total, we detected 13 up-regulated, three down-regulated, five Kenshin-specific, and seven Chiifu-specific genes (S17 Table). The 13 up-regulated genes included receptor kinase 3 (RK3) (Bra001630, Bra040589, Bra038767, Bra040589), mitogen-activated protein kinase kinase kinase 14 (MAPKKK14) (Bra018344), a protein kinase superfamily (AT2G28940; Bra040931) (AT5G11410; Bra008964) (AT5G13290; Bra008839), a U-box domain-containing protein kinase family (Bra024393), and somatic embryogenesis receptor-like kinase 5 (Bra032128). The three down-regulated genes included CBL-interacting protein kinase 13 (Bra021897), a receptor-like protein kinase-related family (AT5G48540; Bra037480), and a leucine-rich repeat protein kinase family (AT1G51800; Bra030416). The five Kenshin-specific genes included with-no-lysine (K) kinase 4 (Bra024518), a protein kinase superfamily (AT3G26700; Bra034245), a concanavalin A-like lectin protein kinase family (Bra010340), a cysteine-rich receptor-like protein kinase (RLK) 36 (Bra029496), and a leucine-rich repeat protein kinase family (AT5G59680; Bra020301). Finally, the seven Chiifu-specific genes included wall-associated kinase 5 (Bra025886), phosphoglycerate kinase 1 (Bra034729), receptor serine/threonine kinase (AT4G18250; Bra008687), a leucine-rich repeat protein kinase family protein (AT5G59670; Bra006700), a cysteine-rich RLK 39 (Bra017552), and a protein kinase superfamily (AT3G46930; Bra018183). High expression of most kinase genes under all conditions suggested that these factors are primarily regulated at the protein level.
Although most phosphatase genes were highly expressed in all samples (S18 Table), three were differentially expressed: two were up-regulated genes upon HS (AT3G51470; Bra036796, AT4G29690; Bra011129), and one was expressed specifically in Kenshin (probable apyrase 5, Bra019669). The functions of these genes have not yet been determined. As with the kinases, the expression patterns suggest that most phosphatases function at the post-transcriptional level.
Transcription factors (TFs)
TFs regulate many genes involved in plant growth and development at the transcriptional level. The Br135K microarray includes 3,354 TFs (8% of the total of 41,174 genes), of over 43 different classes (Table 2, S19 Table). Among them, 109 TFs exhibited significant changes in their expression levels following HS treatment (Table 2). Apart from the heat-induced Hsfs, the majority of these TFs belong to the Integrase-type, NAC, homeodomain, HB, bHLH, and DREB families. Members of these TF families are involved in the HSR and thermotolerance in different plant species [45,64,65], indicating the existence of complex transcriptional regulatory networks beyond the direct regulatory control of Hsfs.
Table 2. Summary of transcription factor genes represented on the Br135K microarray.
| Up-regulated by HS | Down-regulated by HS | Genotype specific | |||||||
|---|---|---|---|---|---|---|---|---|---|
| No | Chiifu | Kenshin | Both | Chiifu | Kenshin | Both | Chiifu | Kenshin | |
| MYB | 289 | 1 | - | 3 | - | - | 1 | - | - |
| bHLH | 234 | - | - | 1 | - | - | 4 | - | - |
| NAC | 191 | - | - | 7 | - | - | 3 | - | - |
| Homeodomain | 183 | - | - | 3 | 1 | - | 1 | 1 | - |
| C2H2 | 159 | - | - | 1 | - | - | 2 | 1 | - |
| Integrase-type | 153 | - | - | 9 | - | - | 6 | - | - |
| WRKY | 150 | 1 | - | - | - | - | 1 | - | - |
| AGL | 109 | 1 | - | - | - | - | 2 | - | - |
| bZIP | 108 | - | - | 1 | - | - | - | 1 | - |
| AP2 | 100 | - | 1 | - | - | - | - | - | - |
| HB | 93 | - | - | 2 | - | - | 5 | 3 | - |
| GATA | 69 | - | - | - | - | - | 1 | - | - |
| Dof | 66 | - | - | - | - | - | - | - | - |
| MADS | 62 | - | - | - | - | - | - | 1 | - |
| B3 family | 59 | - | - | - | - | - | - | 1 | - |
| NF-Y | 51 | - | - | 1 | - | - | - | - | - |
| C3HC4 | 50 | - | - | 2 | - | - | - | - | - |
| C-x8-C-x5-C-x3 | 43 | - | - | - | - | - | 3 | - | 1 |
| DHHC | 42 | - | - | - | - | - | 1 | 1 | - |
| CCCH | 41 | - | - | 2 | - | - | 1 | - | - |
| LOB | 41 | - | - | - | - | - | 1 | - | - |
| HS | 38 | - | - | 6 | - | - | - | - | - |
| ERF | 38 | - | - | 1 | - | - | - | - | - |
| ARF | 36 | - | - | 1 | - | - | 2 | - | - |
| GRAS | 34 | - | - | - | 1 | - | - | - | - |
| TCP | 33 | - | - | - | - | - | - | - | - |
| B-box | 32 | - | - | 1 | - | - | 2 | - | - |
| Squamosa | 29 | - | - | - | - | - | 2 | - | - |
| DREB (TINY) | 28 | - | - | 5 | - | - | 1 | - | - |
| JAZ | 26 | - | - | 1 | - | - | - | - | - |
| LSH | 26 | - | - | 4 | - | - | - | - | 1 |
| A20/AN1 | 24 | - | - | - | - | - | - | - | |
| PLATZ | 23 | - | - | - | - | - | - | - | - |
| RWP-RK | 23 | - | - | - | - | - | - | - | - |
| Jumonji | 22 | - | 1 | - | - | - | - | - | |
| CCHC | 22 | - | - | - | - | - | - | - | - |
| ZFP | 20 | - | - | - | - | - | - | - | - |
| GRF | 17 | - | - | - | - | - | - | - | - |
| Alfin-like | 17 | ||||||||
| KNAT | 16 | - | - | - | - | - | - | 1 | - |
| BZR1 family | 16 | - | - | - | - | - | - | - | - |
| Winged-helix | 12 | - | - | - | - | - | - | - | - |
| AUX/IAA | 8 | - | - | - | - | - | - | - | - |
| OTHER | 521 | 3 | - | 6 | 1 | - | 10 | - | - |
| Total | 3,354 | 3 | 1 | 52 | 2 | 0 | 39 | 10 | 2 |
Although only AP2/B3-like TF (AT5G60142) (Bra002509) exhibited Kenshin-specific up-regulation, Chiifu-specific up-regulation was detected for three genes: MYB-like 2 (AT1G71030) (Bra016164), WRKY33 (Bra000064), and AGL64 (Bra032347). Zinc-finger CCCH domain-containing-protein (Bra012068) and LSH6 (Bra031552) were Kenshin-specifically expressed without much change upon HS, and ten genes were specific to Chiifu. Except for WRKY33, which may play a role in defense signaling, the functions of these genes have not been determined.
According to a recent study, the ER membrane-associated basic leucine zipper (bZIP) transcription factor bZIP28 is activated by ER stress resulting from the accumulation of misfolded or unfolded proteins following HS; this factor stimulates expression of stress response genes [66]. Two BrbZIP28 genes were highly expressed in all Chinese cabbage samples; expression of one of them (Bra034147) increased slightly upon HS treatment, whereas the other gene (Bra023224) exhibited no significant change.
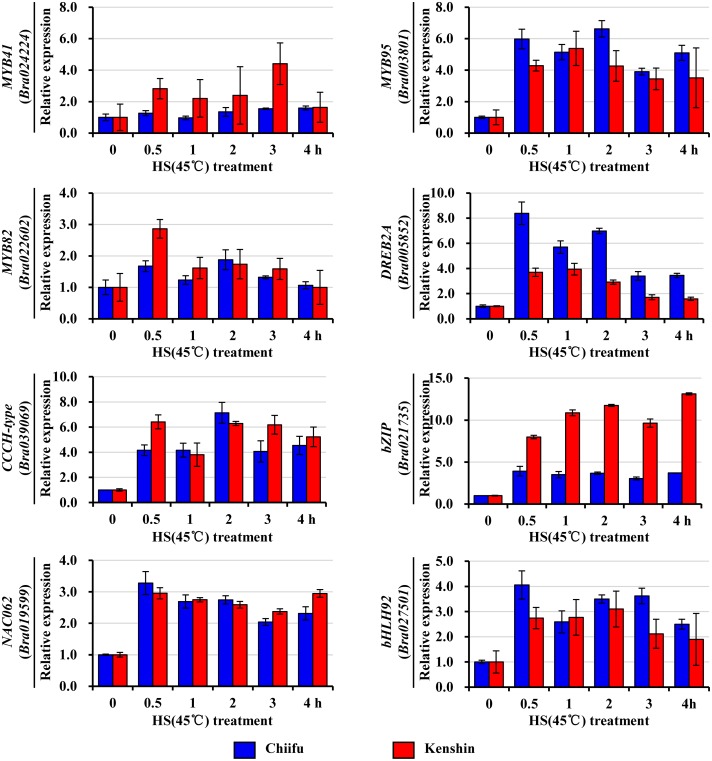
Several TFs were selected and subjected to qRT-PCR analysis. Three genes (Bra022602, Bra039069, Bra019599) were induced in both lines at similar levels upon HS, and three and two genes exhibited prominent expression in Chiifu (Bra003801, Bra005852, Bra027501) and Kenshin (Bra021735, Bra024224), respectively (Fig 5). Due to the large standard deviation resulting from normalization, the qRT-PCR data of several genes (MYB41, MYB95, MYB82, and bHLH92) were not consistent with the microarray values. Except for Bra022602 (MYB82), expression of most genes was highly increased by HS treatment. With the exception of three genes (CCH-type, MYB82, and MYB95) whose functions have not been identified, these genes are related to abiotic stress responses in Arabidopsis. Two genes with prominent expression in Chiifu, Bra005852 (DREB2A: dehydration-responsive element binding protein 2A) and Bra027501 (bHLH92), are responsive to cold stress [28] suggesting that they serve another role in HSR. In Arabidopsis, bHLH92 functions in response to NaCl, dehydration, and cold [67], but its homolog, Bra027501, would be expected to function in HS of B. rapa, based on its expression change. Arabidopsis DREB2A controls the levels of HsfA3, Hsp18.1-CI, Hsp26.5-MII, and Hsp70 during heat and drought stress [68–70], implying that it responds to both heat and cold stresses. B. rapa contains three DREB2A genes: two of these (Bra005852, Bra019162) were expressed at low basal levels under normal growth conditions and were greatly up-regulated by HS, whereas the other (Bra009112) was expressed at a relatively high level that increased further upon HS treatment (S19 Table). These results indicate that expression of Hsps in B. rapa might be also induced by DREB2A.
Fig 5. qRT-PCR analysis of transcription factor genes that were up-regulated by HS treatment.
Relative expression was normalized to BrActin and is presented relative to 0 h expression levels. Two biological repeats were performed, and error bars represent SD. HS, heat shock.
Two genes exhibiting a similar response to HS treatment in both inbred lines, Bra003801 (MYB95) and Bra019599 (NAC062/NTL6), are predicted to function in both cold and HT, even though only the cold response has been characterized in Arabidopsis [71,72]. In Arabidopsis, NAC062/NTL6 processing is stimulated by cold and ABA to activate the expression of defense response genes, but in B. rapa, its processing is reduced by HT [71]. Given that the expression of Bra019599 was greatly increased upon HS in both lines, it is possible that BrNAC62 functions in both cold and HT stress.
Two genes with a notable response to HS treatment in Kenshin, Bra024224 (MYB41) and Bra021735 (a bZIP/AIR1 [Anthocyanin-Impaired-Response-1]), may function in HS tolerance. Arabidopsis MYB41 activates suberin synthesis under abiotic stress conditions [73], implying a relationship to HS. Arabidopsis AIR1 regulates various steps in the flavonoid and anthocyanin accumulation pathway [74]. Its expression is regulated by salt stress, but not by HT or drought. It is possible that BrAIR1 could function under HS conditions, particularly for Kenshin. Therefore, BrAIR1 could be a useful molecular marker for HT tolerance in B. rapa.
HSR genes and proteins can be divided into two groups, signaling components (protein kinases and TFs) and functional genes (HSPs and catalase) [75,76]. Zat7 [77], Zat10 [15], and Zat12 [78] respond to HS. In particular, Zat12 is necessary for the expression of APX, Zat7, and WARKY 25. B. rapa contains Zat1, 6, 10, and 12, but not Zat7. The expression levels of Zat12 (four alleles) and Zat10 (two alleles) were very high and slightly increased by HS. Also, WRKY25 expression was very high. These results suggest the possible involvement of TFs through the MAPK signaling pathway.
HS marker genes
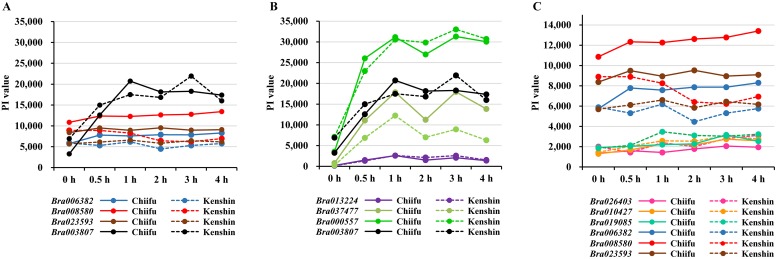
According to a previous report [41], Arabidopsis plants exhibit four classes of tolerance to HS regimes: basal thermotolerance (BT), short-term acquired thermotolerance (SAT), long-term acquired thermotolerance (LAT), and thermotolerance to moderately high temperature (TMHT). Because genes representing each regime have been identified in Arabidopsis, we examined expression of the B. rapa genes that correspond to the Arabidopsis genes involved in the tolerance phenotypes (S20 Table, Fig 6). Expression of most genes did not differ between Chiifu and Kenshin, but only expression of XPO1A (exportin 1A) (Bra008580, Bra006382), which is involved in BT and TMHT in Arabidopsis, seemed to be correlated with thermotolerance as well as membrane leakage. LAT-related genes could be up-regulated (induced or stimulated) even after 30 min HT treatments, but no difference was observed between the two genotypes. The Hsa32 (HS-associated 32 kD: AT4G21320) homolog, which is also a marker of LAT, was absent from the B. rapa gene database. These results suggest that (1) HSR markers differ between Arabidopsis [41] and B. rapa, and (2) BT and LAT from B. rapa are controlled by different genes.
Fig 6. Expression patterns of marker genes for thermotolerance in Chiifu and Kenshin.
PI value indicates probe intensity (see also S20 Table). A, Expression patterns of basal thermotolerance marker genes (BT) and short-term acquired thermotolerance (SAT) in Chiifu and Kenshin. B, Expression patterns of long-term acquired thermotolerance (LAT) marker genes in Chiifu and Kenshin. C, Expression patterns of thermotolerance to moderately high temperatures (TMHT) marker genes in Chiifu and Kenshin.
Membrane leakage-related (MLR) genes
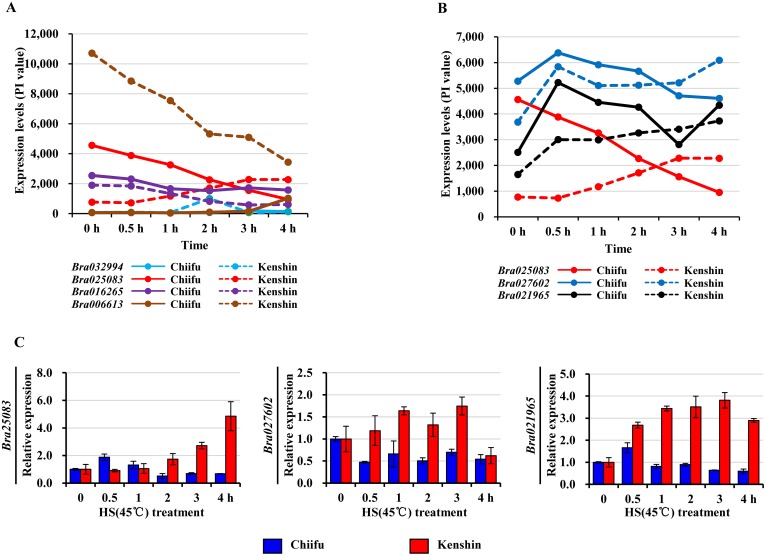
Even though HSR genes were immediately induced by HT exposure, membrane leakage of B. rapa remained constant until 1 h HT exposure (Fig 1). Regarding electrolyte leakage, Kenshin is less sensitive to prolonged HT exposure than Chiifu, implying that the HT responses of these lines are negatively correlated with leakage. Because electrolyte leakage reflects the traits of these two inbred lines, analysis of the MLR genes is important for an understanding of the B. rapa HT-response. MLR genes can be defined as those whose expression patterns correlate with the membrane leakage pattern either positively or negatively, i.e., expression of genes should not exhibit a significant change from no treatment to 1 h HS treatment, but should start to change upon 2 h HS treatment (over 2-fold, up or down) and increase thereafter. As shown in S21 Table, expression levels of 15 and 65 genes were changed over 2-fold in Chiifu and Kenshin, respectively. Among them, four genes specifically expressed in Kenshin were selected as putative MLR genes: two up-regulated genes [Bra032994 (NA), Bra025083 (cytochrome P450; CYP707A3)], and two down-regulated genes [Bra006613 (FAD/NAD(P)-binding oxidoreductase) and Bra016265 (protein kinase superfamily protein)](Fig 7A and 7B). In particular, the Bra025083 expression pattern was well matched to the membrane leakage pattern. B. rapa includes three CYP707A3 genes, and two of them, Bra025083 and Bra021965, exactly followed the pattern of membrane leakage (Fig 7C). The reported functions of CYP707A3, a major ABA 8’-hydroxylase, includes the dehydration response [79,80], stomata closure [81], and salt stress [80]. Our data suggests another function for this gene in HS conditions.
Fig 7. Examples of membrane leakage-related (MLR) genes.
A, Four genes: two up-regulated genes, Bra032994 (NA) and Bra025083 (cytochrome P450; CYP707A3), and two down-regulated genes, Bra006613 (FAD/NAD(P)-binding oxidoreductase) and Bra016265 (protein kinase superfamily protein). B, Three B. rapa genes for CYP707A3. C, qRT-PCR of three B. rapa cytochrome P450 genes (abscisic acid 8’-hydroxylase). Relative expression was normalized to BrActin and is presented relative to 0 h expression levels. Two biological repeats were performed and error bars represent SD. HS, heat shock.
Orphan genes
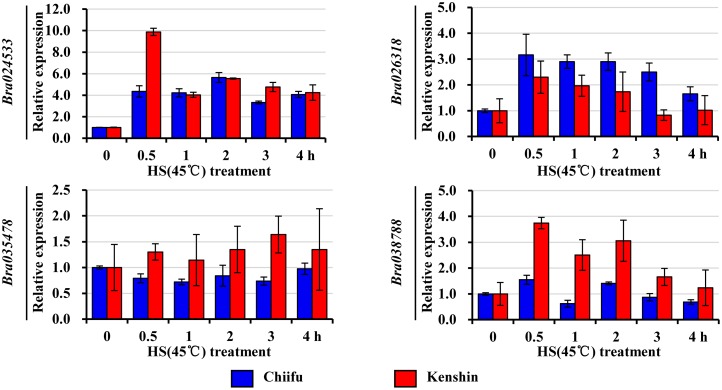
Orphan genes, which are protein-coding genes unique to a species, are widespread across all organisms [82]. Estimates of the percentage of orphan genes in various species range from 1% to 71%, with 5–15% being fairly typical [83]. Orphan genes are associated with abiotic stresses, including HS [83–85]. About 13% of genes on the Br135K microarray were orphan genes (5,349 out of 41,173 genes) (S22 Table). Among them, 34 were induced or up-regulated by HS treatment, and 16 and 13 genes were induced in a Chiifu- or Kenshin-specific manner (S22 Table, Fig 8). Bra026318 (15–37-fold induction) and Bra024533 (25–34-fold induction) exhibited the strongest induction in Chiifu and Kenshin, respectively. In addition, 54 and 14 genes were specific to Chiifu and Kenshin, respectively, suggesting that they were associated with specific traits of the two genotypes, ncluding their HS responses. Several genes that were up-regulated in both inbred lines were confirmed by qRT-PCR (Fig 9). In contrast to the PI value changes observed in the microarray experiment, the induction revealed by qRT-PCR was less obvious, even though the expression levels clearly increased upon HS.
Fig 8. Heat map of orphan genes induced by HS treatment.
Fig 9. qRT-PCR verifications of the expression of several orphan genes.
Relative expression was normalized to BrActin and is presented relative to 0 h expression levels. Two biological repeats were performed, and error bars represent SD. HS, heat shock.
Analysis of conserved cis-elements enriched for HSR genes
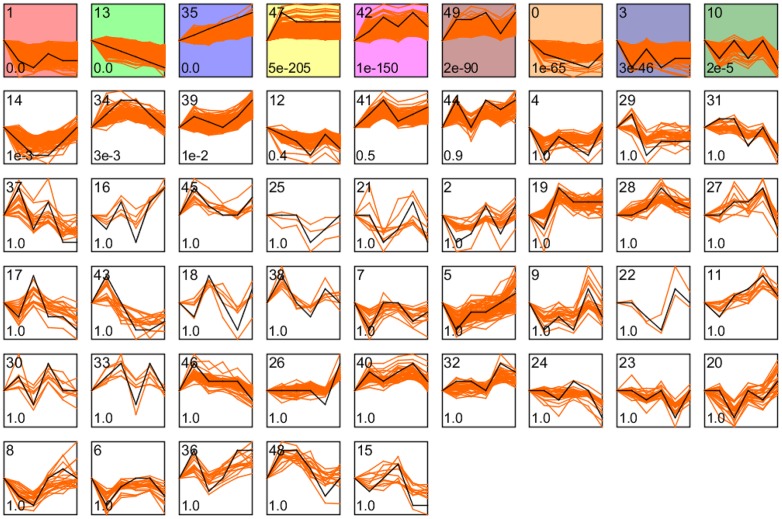
Because the full genome sequence has already been reported for Chiifu (http://brassicadb.org/brad/), we performed an analysis of cis-elements of the HSR genes in order to extend our knowledge of HSR mechanisms in B. rapa. Although the genome sequence of Kenshin has not yet been published, results obtained using the Chiifu genome sequence should be sufficient to provide information about Kenshin because expression of most TFs exhibited similar patterns in both inbred lines (Fig 5). We performed our clustering analysis with STEM [37] using HSR genes that were > 2-fold DEGs (8,160 genes) in Chiifu. Fifty profiles were obtained from STEM, and nine of them significant overrepresentation (p < 0.01) (S23 Table, Fig 10). We then subjected the 1,000 bp upstream sequences of the genes in the nine significantly overrepresented profiles to PLACE database for motif scanning [38]. The occurrences of these motifs were compared with their frequencies among all promoters represented in the 135K microarray. A P-value was then calculated for each motif and profile combination, based on the hypergeometric distribution [39]. Motifs with P-values below 10-4 were considered to be significantly enriched significantly enriched motifs (SEMs) (S24 Table). Among the SEMs in profiles P35, P42, P47, and P19, we identified several motifs related to the HSR: CCAATBOX1 (CCAAT) in P35 is required for heat shock promoter activity [86,87]; and ACGTABREMOTIFA2OSEM (ACGTGKC) in P42, ABRELATERD1 (ACGTG) in P47, and two calcium-responsive motifs in P47, ABRERATCAL (MACGYGB) [88], and CGCGBOXAT (VCGCGB) could be responsible for the HS response. These three motifs are also known as ABREs binding by a bZIP TF responding to multiple stresses, such as drought and cold [88–93]. Our motif and transcriptome analyses suggest that ABREs are also necessary for the HSR. SEMs found in down-regulated genes were also related to the HSR: the DRECRTCOREAT (RCCGAC) motif, represented in P1 and P3, is an AP2 TF-binding site involved in cold and ABA-responsive processes [94]; and SITEIIATCYTC (TGGGCY), in P1 and P13, is a TCP-domain TF-binding site important for mitochondrial oxidative phosphorylation. Furthermore, we subjected genes containing the same cis-elements to agriGO for GO enrichment analysis. Processes including response to heat, heat acclimation, MAPKKK cascade, calmodulin binding, and regulation of oxygen and ROS metabolic process were represented by up-regulated genes containing shared cis-elements (S24 Table). In addition, shoot system development, respiratory burst during defense response, and very-long-chain fatty acid metabolic processes were represented by down-regulated genes containing shared cis-elements (S24 Table). All of this information will be useful because combinatorial analysis of promoters, co-expression networks, and transcriptome can provide further insight into gene functions and signaling pathways [39,95].
Fig 10. Short Time-series Expression Miner (STEM) clusters of expression profiles with HSR genes in Chiifu.
The number of profiles in each cluster is at the top left corner of each STEM. Profiles are ordered based on the p-value significance of the number (at bottom-left corner) of assigned versus expected genes. Colored frame denotes significant profiles (P-value≤0.01). Each graph displays the mean expression pattern (black lines) of the profile genes.
Conclusion
Genome-wide analysis of the transcriptome following heat treatment in two inbred lines of Chinese cabbage provided important information regarding the HSR. First, even though Kenshin was more resistant to membrane leakage than Chiifu, the number of DEGs was higher in Chiifu. Second, expression of small Hsps was highly induced by HS treatment, whereas high-molecular weight Hsps exhibited constitutively high expression. Third, although expression of several upstream genes of the HSR was induced by HS treatment, most genes associated with the signaling pathway were constitutively expressed, suggesting that they are regulated at the protein level. Fourth, besides well-known TFs, many TFs and orphan genes seemed to be related to HSR in B. rapa. Fifth, most of the B. rapa HSR is likely to use mechanisms identified in other plants, but the critical or core genes could be different. To strengthen HSR mechanism in Chinese cabbage, it will be essential to perform combinatorial analyses of transcriptomes, proteomes, metabolomes, and small RNAs.
Supporting Information
(TIFF)
(TIFF)
(XLSX)
(XLSX)
Leaf discs were prepared from 4-week-old plants at 22°C (0) and subjected to 45°C for the indicated time (0.5–4 h). Probe intensity (PI) value are the means of two independent experiments. NA, no Arabidopsis counterpart.
(XLSX)
Leaf discs were prepared from 4-week-old plants at 22°C and subjected to 45°C for the indicated time. Probe values are the means of two independent experiments.
(XLSX)
Leaf discs were prepared from 4 week old plants at 22°C and subjected to 45°C for the indicated time. Probe values are the means of two independent experiments.
(XLSX)
Leaf discs were prepared from 4 week old plants at 22°C and subjected to 45°C for the indicated time. Probe values are the means of two independent experiments.
(XLSX)
Leaf discs were prepared from 4 week old plants at 22°C and subjected to 45°C for the indicated time. Probe values are the means of two independent experiments.
(XLSX)
Yellow shading indicates the FDR value below 0.05 (significantly represented items).
(XLSX)
Yellow shade indicates the FDR value below 0.05 (significantly represented items).
(XLSX)
Gene No. represents Arabidopsis counterparts involved in each pathways.
(XLSX)
Expression levels of all genes in this list changed by over 4-fold in both genotypes
(XLSX)
Gene No. represents Arabidopsis homologs involved in each pathway.
(DOCX)
(XLSX)
The total number of genes was 244 (38 for Hsfs and 206 for Hsps).
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
MLR genes were defined as those whose expression matters correlated negatively or positively with the membrane leakage pattern, i.e., genes should exhibit no significant change from no treatment to 1 h HS treatment, but should change after 2 h HS treatment (over 2-fold, up or down).
(XLSX)
(XLSX)
GO enrichment was carried out using agriGO. Significantly enriched GO items were represented (FDR≤0.05).
(XLSX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was supported by two research grants: (1) Golden Seed Project (Center for Horticultural Seed Development, nos. 213003-04-2-CG100 and 213003-04-2-SB230), Ministry of Agriculture, Food and Rural Affairs (MAFRA), Ministry of Oceans and Fisheries (MOF), Rural Development Administration (RDA), and Korea Forest Service (KFS), and (2) a grant from the Next-Generation BioGreen21 Program (PJ008118), RDA, Korea. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Mittler R (2006) Abiotic stress, the field environment and stress combination. Trends Plant Sci 11: 15–19. [DOI] [PubMed] [Google Scholar]
- 2. Mittler R, Blumwald E (2010) Genetic engineering for modern agriculture: challenges and perspectives. Annu Rev Plant Biol 61: 443–462. 10.1146/annurev-arplant-042809-112116 [DOI] [PubMed] [Google Scholar]
- 3. Hasanuzzaman M, Nahar K, Alam MM, Roychowdhury R, Fujita M (2013) Physiological, biochemical, and molecular mechanisms of heat stress tolerance in plants. Int J Mol Sci 14: 9643–9684. 10.3390/ijms14059643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bita CE, Gerats T (2013) Plant tolerance to high temperature in a changing environment: scientific fundamentals and production of heat stress-tolerant crops. Front Plant Sci 4: 273 10.3389/fpls.2013.00273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hua J (2009) From freezing to scorching, transcriptional responses to temperature variations in plants. Curr Opin Plant Biol 12: 568–573. 10.1016/j.pbi.2009.07.012 [DOI] [PubMed] [Google Scholar]
- 6. Saidi Y, Finka A, Goloubinoff P (2011) Heat perception and signalling in plants: a tortuous path to thermotolerance. New Phytol 190: 556–565. 10.1111/j.1469-8137.2010.03571.x [DOI] [PubMed] [Google Scholar]
- 7. Mittler R, Finka A, Goloubinoff P (2012) How do plants feel the heat? Trends Biochem Sci 37: 118–125. 10.1016/j.tibs.2011.11.007 [DOI] [PubMed] [Google Scholar]
- 8. Hu W, Hu G, Han B (2009) Genome-wide survey and expression profiling of heat shock proteins and heat shock factors revealed overlapped and stress specific response under abiotic stresses in rice. Plant Sci 176: 583–590. [DOI] [PubMed] [Google Scholar]
- 9. Bokszczanin KL, Solanaceae Pollen Thermotolerance Initial Training Network C, Fragkostefanakis S (2013) Perspectives on deciphering mechanisms underlying plant heat stress response and thermotolerance. Front Plant Sci 4: 315 10.3389/fpls.2013.00315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Baniwal SK, Bharti K, Chan KY, Fauth M, Ganguli A, Kotak S, et al. (2004) Heat stress response in plants: a complex game with chaperones and more than twenty heat stress transcription factors. J Biosci 29: 471–487. [DOI] [PubMed] [Google Scholar]
- 11. Scharf KD, Berberich T, Ebersberger I, Nover L (2012) The plant heat stress transcription factor (Hsf) family: structure, function and evolution. Biochim Biophys Acta 1819: 104–119. 10.1016/j.bbagrm.2011.10.002 [DOI] [PubMed] [Google Scholar]
- 12. Liu HC, Liao HT, Charng YY (2011) The role of class A1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis . Plant Cell Environ 34: 738–751. 10.1111/j.1365-3040.2011.02278.x [DOI] [PubMed] [Google Scholar]
- 13. Fragkostefanakis S, Roth S, Schleiff E, Scharf KD (2014) Prospects of engineering thermotolerance in crops through modulation of heat stress transcription factor and heat shock protein networks. Plant Cell Environ July 3 10.1111/pce.12396. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 14. Kotak S, Larkindale J, Lee U, von Koskull-Doring P, Vierling E, Scharf KD. (2007) Complexity of the heat stress response in plants. Curr Opin Plant Biol 10: 310–316. [DOI] [PubMed] [Google Scholar]
- 15. Miller G, Mittler R (2006) Could heat shock transcription factors function as hydrogen peroxide sensors in plants? Ann Bot 98: 279–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Baniwal SK, Chan KY, Scharf KD, Nover L (2007) Role of heat stress transcription factor HsfA5 as specific repressor of HsfA4 . J Biol Chem 282: 3605–3613. [DOI] [PubMed] [Google Scholar]
- 17. Vierling E (1991) The Roles of Heat Shock Proteins in Plants. Plant Mol Biol 42: 579–620. [Google Scholar]
- 18. Wang W, Vinocur B, Shoseyov O, Altman A (2004) Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci 9: 244–252. [DOI] [PubMed] [Google Scholar]
- 19. Qu AL, Ding YF, Jiang Q, Zhu C (2013) Molecular mechanisms of the plant heat stress response. Biochem Biophys Res Commun 432: 203–207. 10.1016/j.bbrc.2013.01.104 [DOI] [PubMed] [Google Scholar]
- 20. Larkindale J, Vierling E (2008) Core genome responses involved in acclimation to high temperature. Plant Physiol 146: 748–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee J, Song H, Han C-T, Lim Y, Chung S-M, Hur Y (2010) Expression characteristics of heat shock protein genes in two comparable inbred lines of Chinese cabbage, Chiifu and Kenshin. Genes Genom 32: 247–257. [Google Scholar]
- 22. Hossain MM, Inden H, Asahira T (1989) Interspecific hybrids between Brassica campestris L. and B. oleracea L. through embryo and ovary culture. Mem Coll Agric, Kyoto Univ 135: 21–30. [Google Scholar]
- 23. Yamagishi H, Hossain MM, Yonezawa K (1994) Morphology, fertility and cross-compatibility of somatic hybrids between Brassica oleracea L. and B. campestris L. Sci Hort 58: 283–288. [Google Scholar]
- 24. Lee SC, Lim MH, Kim JA, Lee SI, Kim JS, Jin M, et al. (2008) Transcriptome analysis in Brassica rapa under the abiotic stresses using Brassica 24K oligo microarray. Mol Cells 26: 595–605. [PubMed] [Google Scholar]
- 25. Lee SC, Lim MH, Yu JG, Park BS, Yang TJ (2012) Genome-wide characterization of the CBF/DREB1 gene family in Brassica rapa . Plant Physiol Biochem 61: 142–152. 10.1016/j.plaphy.2012.09.016 [DOI] [PubMed] [Google Scholar]
- 26. Song X, Li Y, Hou X (2013) Genome-wide analysis of the AP2/ERF transcription factor superfamily in Chinese cabbage (Brassica rapa ssp. pekinensis). BMC Genomics 14: 573 10.1186/1471-2164-14-573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hwang I, Jung HJ, Park JI, Yang TJ, Nou IS (2014) Transcriptome analysis of newly classified bZIP transcription factors of Brassica rapa in cold stress response. Genomics 104: 194–202. 10.1016/j.ygeno.2014.07.008 [DOI] [PubMed] [Google Scholar]
- 28. Jung HJ, Dong X, Park JI, Thamilarasan SK, Lee SS, Kim YK, et al. (2014) Genome-wide transcriptome analysis of two contrasting Brassica rapa doubled haploid lines under cold-stresses using Br135K oligomeric chip. PLoS One 9: e106069 10.1371/journal.pone.0106069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kayum MA, Jung HJ, Park JI, Ahmed NU, Saha G, Yang TJ, et al. (2015) Identification and expression analysis of WRKY family genes under biotic and abiotic stresses in Brassica rapa . Mol Genet Genomics 290: 79–95. 10.1007/s00438-014-0898-1 [DOI] [PubMed] [Google Scholar]
- 30. Song XM, Huang ZN, Duan WK, Ren J, Liu TK, Li Y, et al. (2014) Genome-wide analysis of the bHLH transcription factor family in Chinese cabbage (Brassica rapa ssp. pekinensis). Mol Genet Genomics 289: 77–91. 10.1007/s00438-013-0791-3 [DOI] [PubMed] [Google Scholar]
- 31. Lee J, Lim Y-P, Han C-T, Nou I-S, Hur Y (2013) Genome-wide expression profiles of contrasting inbred lines of Chinese cabbage, Chiifu and Kenshin, under temperature stress. Genes Genom. 2013;35(3):273–88. [Google Scholar]
- 32. Rapacz M (2002) Cold-deacclimation of oilseed rape (Brassica napus var. oleifera) in response to fluctuating temperatures and photoperiod. Ann Bot 89: 543–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Song L, Ding W, Zhao M, Sun B, Zhang L (2006) Nitric oxide protects against oxidative stress under heat stress in the calluses from two ecotypes of reed. Plant Sci 171: 449–458. 10.1016/j.plantsci.2006.05.002 [DOI] [PubMed] [Google Scholar]
- 34. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38: W64–70. 10.1093/nar/gkq310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, et al. (2008) KEGG for linking genomes to life and the environment. Nucleic Acids Res 36: D480–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ernst J, Bar-Joseph Z (2006) STEM: a tool for the analysis of short time series gene expression data. BMC Bioinformatics 7: 191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27: 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ma S, Bohnert HJ (2007) Integration of Arabidopsis thaliana stress-related transcript profiles, promoter structures, and cell-specific expression. Genome Biol 8: R49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wilson RA, Sangha MK, Banga SS, Atwal AK, Gupta S (2014) Heat stress tolerance in relation to oxidative stress and antioxidants in Brassica juncea . J Environ Biol 35: 383–387. [PubMed] [Google Scholar]
- 41. Yeh CH, Kaplinsky NJ, Hu C, Charng YY (2012) Some like it hot, some like it warm: phenotyping to explore thermotolerance diversity. Plant Sci 195: 10–23. 10.1016/j.plantsci.2012.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rizhsky L, Liang H, Shuman J, Shulaev V, Davletova S, Mittler R (2004) When defense pathways collide. The response of Arabidopsis to a combination of drought and heat stress. Plant Physiol 134: 1683–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rensink WA, Iobst S, Hart A, Stegalkina S, Liu J, Buell CR (2005) Gene expression profiling of potato responses to cold, heat, and salt stress. Funct Integr Genomics 5: 201–207. [DOI] [PubMed] [Google Scholar]
- 44. Lim CJ, Yang KA, Hong JK, Choi JS, Yun DJ, Hong JC, et al. (2006) Gene expression profiles during heat acclimation in Arabidopsis thaliana suspension-culture cells. J Plant Res 119: 373–383. [DOI] [PubMed] [Google Scholar]
- 45. Frank G, Pressman E, Ophir R, Althan L, Shaked R, Freedman M, et al. (2009) Transcriptional profiling of maturing tomato (Solanum lycopersicum L.) microspores reveals the involvement of heat shock proteins, ROS scavengers, hormones, and sugars in the heat stress response. J Exp Bot 60: 3891–3908. 10.1093/jxb/erp234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bita CE, Zenoni S, Vriezen WH, Mariani C, Pezzotti M, Gerats T (2011) Temperature stress differentially modulates transcription in meiotic anthers of heat-tolerant and heat-sensitive tomato plants. BMC Genomics 12: 384 10.1186/1471-2164-12-384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gilroy S, Suzuki N, Miller G, Choi WG, Toyota M, Devireddy AR, et al. (2014) A tidal wave of signals: calcium and ROS at the forefront of rapid systemic signaling. Trends Plant Sci 19: 623–630. 10.1016/j.tplants.2014.06.013 [DOI] [PubMed] [Google Scholar]
- 48. Nover L, Bharti K, Doring P, Mishra SK, Ganguli A, Scharf KD (2001) Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress Chaperones 6: 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mishra SK, Tripp J, Winkelhaus S, Tschiersch B, Theres K, Nover L, et al. (2002) In the complex family of heat stress transcription factors, HsfA1 has a unique role as master regulator of thermotolerance in tomato. Genes Dev 16: 1555–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Volkov RA, Panchuk II, Mullineaux PM, Schoffl F (2006) Heat stress-induced H(2)O (2) is required for effective expression of heat shock genes in Arabidopsis . Plant Mol Biol 61: 733–746. [DOI] [PubMed] [Google Scholar]
- 51. Charng YY, Liu HC, Liu NY, Chi WT, Wang CN, Chang SH, et al. (2007) A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis . Plant Physiol 143: 251–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang Z, Wang F, Tang J, Huang Z, Xiong A, Hou X (2014) C-repeat binding factor gene family identified in non-heading Chinese cabbage is functional in abiotic and biotic stress response but different from that in Arabidopsis . Acta Physiol Plant 36: 3217–3229. [Google Scholar]
- 53. Charng YY, Liu HC, Liu NY, Hsu FC, Ko SS (2006) Arabidopsis Hsa32, a novel heat shock protein, is essential for acquired thermotolerance during long recovery after acclimation. Plant Physiol 140: 1297–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lin MY, Chai KH, Ko SS, Kuang LY, Lur HS, Charng YY (2014) A positive feedback loop between HEAT SHOCK PROTEIN101 and HEAT STRESS-ASSOCIATED 32-KD PROTEIN modulates long-term acquired thermotolerance illustrating diverse heat stress responses in rice varieties. Plant Physiol 164: 2045–2053. 10.1104/pp.113.229609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jia L, Chu H, Wu D, Feng M, Zhao L (2014) Role of calmodulin in thermotolerance. Plant Signal Behav 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sgobba A, Paradiso A, Dipierro S, De Gara L, de Pinto MC (2015) Changes in antioxidants are critical in determining cell responses to short- and long-term heat stress. Physiol Plant 153: 68–78. 10.1111/ppl.12220 [DOI] [PubMed] [Google Scholar]
- 57. Wang L, Guo Y, Jia L, Chu H, Zhou S, Chen K, et al. (2014) Hydrogen peroxide acts upstream of nitric oxide in the heat shock pathway in Arabidopsis seedlings. Plant Physiol 164: 2184–2196. 10.1104/pp.113.229369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Davletova S, Rizhsky L, Liang H, Shengqiang Z, Oliver DJ, Coutu J, et al. (2005) Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis . Plant Cell 17: 268–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Koussevitzky S, Suzuki N, Huntington S, Armijo L, Sha W, Cortes D, et al. (2008) Ascorbate peroxidase 1 plays a key role in the response of Arabidopsis thaliana to stress combination. J Biol Chem 283: 34197–34203. 10.1074/jbc.M806337200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Myouga F, Hosoda C, Umezawa T, Iizumi H, Kuromori T, Motohashi R, et al. (2008) A heterocomplex of iron superoxide dismutases defends chloroplast nucleoids against oxidative stress and is essential for chloroplast development in Arabidopsis . Plant Cell 20: 3148–3162. 10.1105/tpc.108.061341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Xu XM, Lin H, Maple J, Bjorkblom B, Alves G, Larsen JP, et al. (2010) The Arabidopsis DJ-1a protein confers stress protection through cytosolic SOD activation. J Cell Sci 123: 1644–1651. 10.1242/jcs.063222 [DOI] [PubMed] [Google Scholar]
- 62. Al-Quraan NA, Locy RD, Singh NK (2010) Expression of calmodulin genes in wild type and calmodulin mutants of Arabidopsis thaliana under heat stress. Plant Physiol Biochem 48: 697–702. 10.1016/j.plaphy.2010.04.011 [DOI] [PubMed] [Google Scholar]
- 63. Kugler A, Kohler B, Palme K, Wolff P, Dietrich P (2009) Salt-dependent regulation of a CNG channel subfamily in Arabidopsis. BMC Plant Biol 9: 140 10.1186/1471-2229-9-140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. von Koskull-Doring P, Scharf KD, Nover L (2007) The diversity of plant heat stress transcription factors. Trends Plant Sci 12: 452–457. [DOI] [PubMed] [Google Scholar]
- 65. Li S, Zhou X, Chen L, Huang W, Yu D (2010) Functional characterization of Arabidopsis thaliana WRKY39 in heat stress. Mol Cells 29: 475–483. 10.1007/s10059-010-0059-2 [DOI] [PubMed] [Google Scholar]
- 66. Srivastava R, Deng Y, Howell SH (2014) Stress sensing in plants by an ER stress sensor/transducer, bZIP28. Front Plant Sci 5: 59 10.3389/fpls.2014.00059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Jiang Y, Yang B, Deyholos MK (2009) Functional characterization of the Arabidopsis bHLH92 transcription factor in abiotic stress. Mol Genet Genomics 282: 503–516. 10.1007/s00438-009-0481-3 [DOI] [PubMed] [Google Scholar]
- 68. Sakuma Y, Maruyama K, Osakabe Y, Qin F, Seki M, Shinozaki K, et al. (2006) Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 18: 1292–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sakuma Y, Maruyama K, Qin F, Osakabe Y, Shinozaki K, Yamaguchi-Shinozaki K (2006) Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc Natl Acad Sci U S A 103: 18822–18827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Schramm F, Larkindale J, Kiehlmann E, Ganguli A, Englich G, Vierling E, et al. (2008) A cascade of transcription factor DREB2A and heat stress transcription factor HsfA3 regulates the heat stress response of Arabidopsis . Plant J 53: 264–274. [DOI] [PubMed] [Google Scholar]
- 71. Seo PJ, Kim MJ, Song JS, Kim YS, Kim HJ, Park CM (2010) Proteolytic processing of an Arabidopsis membrane-bound NAC transcription factor is triggered by cold-induced changes in membrane fluidity. Biochem J 427: 359–367. 10.1042/BJ20091762 [DOI] [PubMed] [Google Scholar]
- 72. Seo PJ, Park CM (2010) A membrane-bound NAC transcription factor as an integrator of biotic and abiotic stress signals. Plant Signal Behav 5: 481–483. 10.4161/psb.11083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kosma DK, Murmu J, Razeq FM, Santos P, Bourgault R, Molina I, et al. (2014) AtMYB41 activates ectopic suberin synthesis and assembly in multiple plant species and cell types. Plant J 80: 216–229. 10.1111/tpj.12624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Van Oosten MJ, Sharkhuu A, Batelli G, Bressan RA, Maggio A (2013) The Arabidopsis thaliana mutant air1 implicates SOS3 in the regulation of anthocyanins under salt stress. Plant Mol Biol 83: 405–415. 10.1007/s11103-013-0099-z [DOI] [PubMed] [Google Scholar]
- 75. Gill SS, Tuteja N (2010) Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem 48: 909–930. 10.1016/j.plaphy.2010.08.016 [DOI] [PubMed] [Google Scholar]
- 76. Morito D, Nagata K (2012) ER Stress Proteins in Autoimmune and Inflammatory Diseases. Front Immunol 3: 48 10.3389/fimmu.2012.00048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ciftci-Yilmaz S, Morsy MR, Song L, Coutu A, Krizek BA, Lewis MW, et al. (2007) The EAR-motif of the Cys2/His2-type zinc finger protein Zat7 plays a key role in the defense response of Arabidopsis to salinity stress. J Biol Chem 282: 9260–9268. [DOI] [PubMed] [Google Scholar]
- 78. Davletova S, Schlauch K, Coutu J, Mittler R (2005) The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis . Plant Physiol 139: 847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Umezawa T, Okamoto M, Kushiro T, Nambara E, Oono Y, Seki M, et al. (2006) CYP707A3, a major ABA 8'-hydroxylase involved in dehydration and rehydration response in Arabidopsis thaliana . Plant J 46: 171–182. [DOI] [PubMed] [Google Scholar]
- 80. Zheng Y, Huang Y, Xian W, Wang J, Liao H (2012) Identification and expression analysis of the Glycine max CYP707A gene family in response to drought and salt stresses. Ann Bot 110: 743–756. 10.1093/aob/mcs133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Okamoto M, Tanaka Y, Abrams SR, Kamiya Y, Seki M, Nambara E (2009) High humidity induces abscisic acid 8'-hydroxylase in stomata and vasculature to regulate local and systemic abscisic acid responses in Arabidopsis . Plant Physiol 149: 825–834. 10.1104/pp.108.130823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Khalturin K, Hemmrich G, Fraune S, Augustin R, Bosch TC (2009) More than just orphans: are taxonomically-restricted genes important in evolution? Trends Genet 25: 404–413. 10.1016/j.tig.2009.07.006 [DOI] [PubMed] [Google Scholar]
- 83. Luhua S, Hegie A, Suzuki N, Shulaev E, Luo X, Cenariu D, et al. (2013) Linking genes of unknown function with abiotic stress responses by high-throughput phenotype screening. Physiol Plant 148: 322–333. 10.1111/ppl.12013 [DOI] [PubMed] [Google Scholar]
- 84. Gollery M, Harper J, Cushman J, Mittler T, Mittler R (2007) POFs: what we don't know can hurt us. Trends Plant Sci 12: 492–496. [DOI] [PubMed] [Google Scholar]
- 85. Luhua S, Ciftci-Yilmaz S, Harper J, Cushman J, Mittler R (2008) Enhanced tolerance to oxidative stress in transgenic Arabidopsis plants expressing proteins of unknown function. Plant Physiol 148: 280–292. 10.1104/pp.108.124875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Rieping M, Schoffl F (1992) Synergistic effect of upstream sequences, CCAAT box elements, and HSE sequences for enhanced expression of chimaeric heat shock genes in transgenic tobacco. Mol Gen Genet 231: 226–232. [DOI] [PubMed] [Google Scholar]
- 87. Haralampidis K, Milioni D, Rigas S, Hatzopoulos P (2002) Combinatorial interaction of cis elements specifies the expression of the Arabidopsis AtHsp90-1 gene. Plant Physiol 129: 1138–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kaplan B, Davydov O, Knight H, Galon Y, Knight MR, Fluhr R, et al. (2006) Rapid transcriptome changes induced by cytosolic Ca2+ transients reveal ABRE-related sequences as Ca2+-responsive cis elements in Arabidopsis . Plant Cell 18: 2733–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Hobo T, Kowyama Y, Hattori T (1999) A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription. Proc Natl Acad Sci U S A 96: 15348–15353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Narusaka Y, Nakashima K, Shinwari ZK, Sakuma Y, Furihata T, Abe H, et al. (2003) Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J 34: 137–148. [DOI] [PubMed] [Google Scholar]
- 91. Kim S, Kang JY, Cho DI, Park JH, Kim SY (2004) ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant J 40: 75–87. [DOI] [PubMed] [Google Scholar]
- 92. Nakashima K, Fujita Y, Katsura K, Maruyama K, Narusaka Y, Seki M, et al. (2006) Transcriptional regulation of ABI3- and ABA-responsive genes including RD29B and RD29A in seeds, germinating embryos, and seedlings of Arabidopsis . Plant Mol Biol 60: 51–68. [DOI] [PubMed] [Google Scholar]
- 93. Lindlof A, Brautigam M, Chawade A, Olsson O, Olsson B (2009) In silico analysis of promoter regions from cold-induced genes in rice (Oryza sativa L.) and Arabidopsis thaliana reveals the importance of combinatorial control. Bioinformatics 25: 1345–1348. 10.1093/bioinformatics/btp172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Suzuki M, Ketterling MG, McCarty DR (2005) Quantitative statistical analysis of cis-regulatory sequences in ABA/VP1- and CBF/DREB1-regulated genes of Arabidopsis . Plant Physiol 139: 437–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Ma S, Shah S, Bohnert HJ, Snyder M, Dinesh-Kumar SP (2013) Incorporating motif analysis into gene co-expression networks reveals novel modular expression pattern and new signaling pathways. PLoS Genet 9: e1003840 10.1371/journal.pgen.1003840 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIFF)
(TIFF)
(XLSX)
(XLSX)
Leaf discs were prepared from 4-week-old plants at 22°C (0) and subjected to 45°C for the indicated time (0.5–4 h). Probe intensity (PI) value are the means of two independent experiments. NA, no Arabidopsis counterpart.
(XLSX)
Leaf discs were prepared from 4-week-old plants at 22°C and subjected to 45°C for the indicated time. Probe values are the means of two independent experiments.
(XLSX)
Leaf discs were prepared from 4 week old plants at 22°C and subjected to 45°C for the indicated time. Probe values are the means of two independent experiments.
(XLSX)
Leaf discs were prepared from 4 week old plants at 22°C and subjected to 45°C for the indicated time. Probe values are the means of two independent experiments.
(XLSX)
Leaf discs were prepared from 4 week old plants at 22°C and subjected to 45°C for the indicated time. Probe values are the means of two independent experiments.
(XLSX)
Yellow shading indicates the FDR value below 0.05 (significantly represented items).
(XLSX)
Yellow shade indicates the FDR value below 0.05 (significantly represented items).
(XLSX)
Gene No. represents Arabidopsis counterparts involved in each pathways.
(XLSX)
Expression levels of all genes in this list changed by over 4-fold in both genotypes
(XLSX)
Gene No. represents Arabidopsis homologs involved in each pathway.
(DOCX)
(XLSX)
The total number of genes was 244 (38 for Hsfs and 206 for Hsps).
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
MLR genes were defined as those whose expression matters correlated negatively or positively with the membrane leakage pattern, i.e., genes should exhibit no significant change from no treatment to 1 h HS treatment, but should change after 2 h HS treatment (over 2-fold, up or down).
(XLSX)
(XLSX)
GO enrichment was carried out using agriGO. Significantly enriched GO items were represented (FDR≤0.05).
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.