Abstract
Folding to a well-defined conformation is essential for the function of structured ribonucleic acids (RNAs) like the ribosome and tRNA. Structured elements in the untranslated regions (UTRs) of specific messenger RNAs (mRNAs) are known to control expression. The importance of unstructured regions adopting multiple conformations, however, is still poorly understood. High-resolution SHAPE-directed Boltzmann suboptimal sampling of the Homo sapiens Retinoblastoma 1 (RB1) 5′ UTR yields three distinct conformations compatible with the experimental data. Private single nucleotide variants (SNVs) identified in two patients with retinoblastoma each collapse the structural ensemble to a single but distinct well-defined conformation. The RB1 5′ UTRs from Bos taurus (cow) and Trichechus manatus latirostris (manatee) are divergent in sequence from H. sapiens (human) yet maintain structural compatibility with high-probability base pairs. SHAPE chemical probing of the cow and manatee RB1 5′ UTRs reveals that they also adopt multiple conformations. Luciferase reporter assays reveal that 5′ UTR mutations alter RB1 expression. In a traditional model of disease, causative SNVs disrupt a key structural element in the RNA. For the subset of patients with heritable retinoblastoma-associated SNVs in the RB1 5′ UTR, the absence of multiple structures is likely causative of the cancer. Our data therefore suggest that selective pressure will favor multiple conformations in eukaryotic UTRs to regulate expression.
Keywords: SHAPE, UTR, covariation, retinoblastoma, riboSNitch
INTRODUCTION
The process of RNA transcription from DNA dictates a direct sequence relationship between the two nucleic acid polymers (Crick 1970). As a result, any sequence variants in the genome will necessarily exist in the transcriptome as well (Naruse et al. 2002; Macias et al. 2008; Cancer Genome Atlas Research Network 2012). Unlike DNA, the nucleotides in RNA are free to interact in an intramolecular fashion resulting in folding of the polymer chain (Celander and Cech 1991; Zarrinkar and Williamson 1994; Thirumalai and Woodson 2000; Woodson 2002; Schroeder et al. 2004). Stretches of RNA that are complementary in sequence have a propensity to pair, forming elements of RNA secondary structure (Zuker and Sankoff 1984; Agius et al. 2010). The functional consequences of these structural elements depend on their molecular context (Bartel 2009; Ulitsky and Bartel 2013). Since the secondary structure of an RNA transcript is dependent on its sequence, variants occurring in transcripts have the potential to disrupt this structure resulting in an altered phenotype.
A riboSNitch is broadly defined as an element in a noncoding RNA or an untranslated region (UTR) of an mRNA where a single nucleotide variant (SNV) results in a functionally important structural rearrangement (Halvorsen et al. 2010; Martin et al. 2012; Ritz et al. 2012; Lokody 2014; Wan et al. 2014). It is similar to a bacterial riboswitch, where binding of a small molecule results in a conformational rearrangement and gene regulation (Mandal et al. 2003; Tucker and Breaker 2005; Weinberg et al. 2007, 2011). RiboSNitches exist because of RNA's propensity to adopt multiple conformations (Sanchez et al. 2006; Taft et al. 2010; Lee and Tarn 2013; Rogler et al. 2014). A single point mutation has the potential to alter the thermodynamic folding landscape, favoring alternative conformations (Russell et al. 2002; Laederach et al. 2007; Solomatin et al. 2010; Ritz et al. 2013). We first described riboSNitches when we analyzed the structural consequences of human disease-associated mutations on UTRs and noncoding RNAs (ncRNAs) (Halvorsen et al. 2010). We identified six human diseases (Hyperferritinemia Cataract Syndrome, β-Thalassemia, Cartilage-Hair Hypoplasia, Retinoblastoma, Chronic Obstructive Pulmonary Disease, and Hypertension) where more than one associated SNV was predicted to alter the structure of a UTR or ncRNA using the SNPfold algorithm (Halvorsen et al. 2010). Retinoblastoma, or cancer of the retina, is frequently caused by SNVs in Retinoblastoma 1 (RB1), a tumor suppressor gene (Lee et al. 1987; Jacks et al. 1992; Valverde et al. 2005). We report here a detailed structural and functional analysis of the RB1 5′ UTR including two SNVs observed in individuals diagnosed with retinoblastoma (Cowell et al. 1996; Macias et al. 2008).
We use here high-resolution SHAPE (selective 2′-hydroxyl acylation analyzed by primer extension) to experimentally probe the structures of the wild-type and mutant RB1 5′ UTRs, revealing that the 5′ UTR of RB1 is indeed a riboSNitch. We used the SHAPE reactivities that we obtained to direct Boltzmann suboptimal sampling and predict the structural ensemble of each sequence. These different structural ensembles provide a structural framework for understanding the etiology of retinoblastoma in these individuals. To further validate our structural model of disease, we probed other eukaryotic RB1 5′ UTRs whose sequences were different from the human sequence but maintained structural compatibility with our model. By identifying regions in the UTR that are both disrupted by disease-associated mutations and conserved phylogenetically, we reveal the important regulatory structural features of the RB1 5′ UTR and propose a mechanism, supported by changes in expression observed in luciferase assays, for how these mutations lead to the retinoblastoma phenotype.
RESULTS
The RB1 5′ UTR is a riboSNitch associated with retinoblastoma
We previously predicted with the SNPfold algorithm that two retinoblastoma-associated SNVs mapping to the RB1 5′ UTR alter its structure (Halvorsen et al. 2010). The two SNVs, G17C and G18U (with respect to the transcription start site), were identified in a clinical genetics panel of patients with retinoblastoma (Cowell et al. 1996; Macias et al. 2008), suggesting that the G17C and G18U SNVs were causative of retinoblastoma (Cowell et al. 1996; Macias et al. 2008). Importantly, the patients with these SNVs had no other mutations near their RB1 gene that could explain the phenotype. Subsequent analysis of RB1 protein expression concluded that the G17C SNV altered the levels of the tumor suppressor, but no further characterization was carried out (Cowell et al. 1996).
The RB1 5′ UTR is particularly barren in terms of functional or experimental genomic annotations (Fig. 1A,B). Indeed, only a single Argonaute RNA Binding Protein (RBP, Fig. 1B) site was identified in genome-wide PAR-CLIP (Photoactivatable-Ribonucleoside-Enhanced Crosslinking and Immunoprecipitation) experiments (Anders et al. 2012). No Rfam motifs scored significantly on the sequence, indicating there are no known structural motifs (Griffiths-Jones et al. 2003, 2005; Gardner et al. 2009; Hafner et al. 2010a,b). This contrasts with the RB1 3′ UTR and coding sequence where a high density of RBP binding was suggested by PAR-CLIP data (Fig. 1A; Anders et al. 2012). The vast majority of known retinoblastoma-associated SNVs lie in the RB1 coding sequence (vertical lines, Fig. 1A,B), indicating that mutations and/or alterations to the protein are the cause of the disease etiology in most patients. Nonetheless, the 5′ UTR of RB1 and the two associated SNVs we identified present a unique system to study the specifics of SNV-induced RNA structure change, where structural mechanisms potentially cause human disease in the absence of known RBP-binding motifs or structural elements.
FIGURE 1.
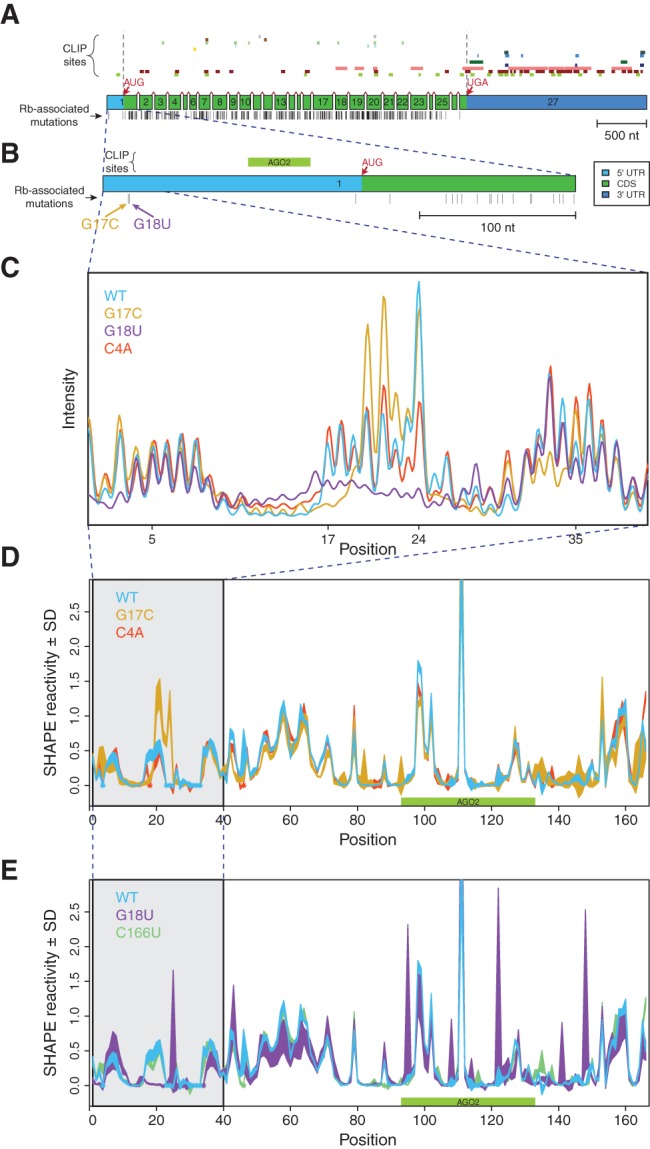
Disease-associated mutations in the 5′ UTR of RB1 change its SHAPE profile. (A) RB1 gene structure, protein-binding sites, and locations of retinoblastoma-associated mutations with reference to the 27 exons of the gene. (Top) Experimentally determined PAR-CLIP (Photoactivatable-Ribonucleoside-Enhanced Crosslinking and Immunoprecipitation) RNA binding protein (RBP) sites obtained from doRiNA database, from top to bottom: R521H, FUS, EWSR1, FMR1 isoform 1, FMR1 isoform 7, C17ORF85, PUM2, TIAL1, FXR2, ZC3H7B, TIA1, IGF2BP1-3, AGO1-4, ELAV1 (Anders et al. 2012). We observed that a majority of RBP binding sites are in the 3′ UTR and coding sequence. (Middle) Exons of the RB1 gene, to scale, including splice junctions. Light blue: 5′ UTR, green: coding sequence (CDS), dark blue: 3′ UTR. (Bottom) Positions of known retinoblastoma-associated point mutations, insertions, and deletions, from the Human Gene Mutation Database (HGMD), indicated as vertical black bars (Stenson et al. 2003; George et al. 2008). (B) Close-up schematic of exon 1 with a single PAR-CLIP site (Argonaute 2) mapping to the 5′ UTR. Corresponding retinoblastoma-associated mutations, G17C and G18U, which were previously predicted to alter the UTR structure (Halvorsen et al. 2010). (C) Representative raw SHAPE (selective 2′-hydroxyl acylation analyzed by primer extension) capillary electrophoresis traces for the WT (blue), G17C (gold), G18U (purple), and C4A (red) UTRs before normalization and averaging. Differences between the sequences across positions 17–24 show that the two disease-associated mutations result in large structural changes as predicted. (D,E) Normalized SHAPE profiles for wild-type, mutant, and structural control UTRs; area represents mean ± SD normalized SHAPE values over five repeats. The region containing nucleotides with mutation-induced structure change are highlighted in gray. Asterisks (in color) indicate positions where the background control peak was too high to accurately determine SHAPE reactivity for the nucleotide. (D) WT (blue), G17C (gold), C4A (structural control; red). (E) WT (blue), G18U (purple), C166U (structural control; green).
Representative raw SHAPE capillary electrophoresis (SHAPE-CE) (Wilkinson et al. 2005; Mitra et al. 2008; Karabiber et al. 2013) traces illustrated in Figure 1C clearly demonstrate that both the G17C (gold) and G18U (purple) SNVs significantly alter the UTR transcript structure, while the C4A control mutation (red) does not. When the data are averaged over five repeats and compared with two control mutations (C4A and C166U, red and green, respectively), distinct patterns of structural disruption occur with both retinoblastoma-associated SNVs (Fig. 1D,E; Supplemental Fig. S1A) consistent with our SNPfold predictions (Halvorsen et al. 2010). These data suggest that the human RB1 5′ UTR is a structural riboSNitch, as previously predicted.
We examined recent ChIP-seq data from the ENCODE project to locate transcription factor binding sites (TFBSs) near the RB1 gene locus in cell lines GM12878 (B-lymphocyte) and K562 (erythromyeloblastoid leukemia) (The ENCODE Project Consortium 2012). Although there are no publicly available ChIP-seq data sets for human retinal cells, hereditary mutations in RB1 also lead to cancer in other tissues (Marees et al. 2008). We found 61 ChIP-seq point-source peaks, corresponding to 32 unique proteins, within 200 nt of the annotated transcription start site (TSS) (Supplemental Fig. S2A). A majority of these peaks (74%) map 5′ of the TSS, indicating that the RB1 gene is conventionally regulated (Supplemental Fig. S2B). Out of these potential regulators, only 11 (18%) of the point-source peaks map to within 20 nt of the G17C and G18U SNVs (Supplemental Fig. S2C). We then investigated the sequence motifs for the transcription factors belonging to these 11 nearby peaks to see if any matched the beginning of the RB1 gene. We found that only MAZ and EGR1 have binding motifs that overlap G17C or G18U (Supplemental Fig. S2D). The mutations are located in the transcribed portion of the gene and sufficiently distant from a majority of known transcriptional regulators. Therefore, further investigation into the structure of the RB1 riboSNitch is warranted as a possible mechanism of disease.
Retinoblastoma SNVs and the 5′ UTR structural ensemble
No clear patterns of common structural disruption appear in the G17C and G18U mutant transcript SHAPE data (Fig. 1). The large peaks visible in the G18U trace (Fig. 1E, purple) that appear to differ from WT near nucleotide 100 are the result of our plotting standard error as line width. The resulting mean SHAPE reactivity at these sites does not suggest a significant structural disruption far downstream from the G18U mutation. To visualize potential common structural features of the two disease-associated transcripts, we performed suboptimal Boltzmann sampling to generate a representative ensemble of structures each sequence adopts (Ding et al. 2004, 2005). To generate an ensemble consistent with experimental observation, we used the SHAPE reactivities to direct the sampling as a pseudo-free energy term with the program RNAstructure (Mathews 2004; Deigan et al. 2009; Hajdin et al. 2013). Thus, the Boltzmann suboptimal sampling (Fig. 2) is consistent with the experimental SHAPE data shown in Figure 1D,E.
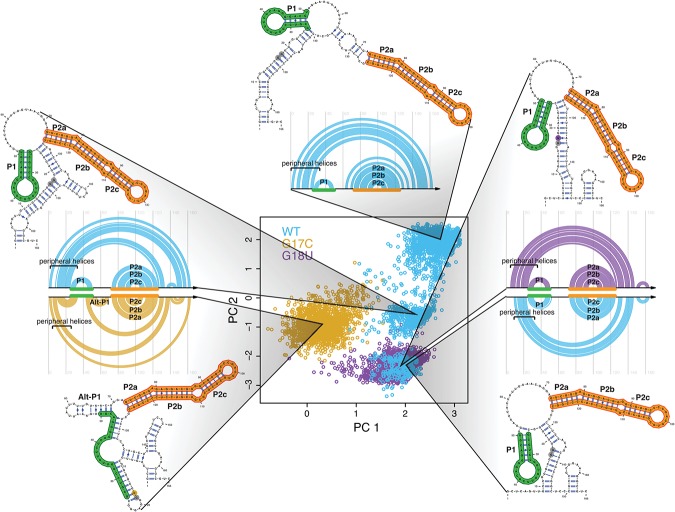
FIGURE 2.
Mutations collapse the structural space of the RB1 5′ UTR. (Center) Principal component decomposition of Boltzmann sampled suboptimal structures using SHAPE-directed free energy calculations (Deigan et al. 2009; Wilkinson et al. 2009). Representative structures are plotted next to the principal component space with their corresponding arc diagram. WT (blue) adopts three distinct conformations, while G17C (gold) adopts one cluster distinctly different from any WT structure. G18U (purple) adopts one major conformation that overlaps with one of the three WT structures, indicating that both sequences contain the same class of structures, seen in the arc diagrams (right). Positions 17 and 18 are denoted in gray when not mutated and in color when mutated. All structures include a major paired region (P2abc; orange). The WT and G18U conformations all contain another paired region (P1; green), while G17C favors an alternative P1 (Alt-P1) helix.
The suboptimal structures projected onto the common principal component analysis (PCA) space for the three transcripts (wild-type blue, G17C gold, and G18U purple) reveal important similarities in the type of structural change observed for the RB1 5′ UTR (Fig. 2). The wild-type (WT) RB1 5′ UTR (blue dots) forms three structural clusters while both disease-associated SNVs collapse the ensemble to a single cluster (gold and purple). This analysis suggests that retinoblastoma-associated SNVs “decrease” the structural diversity of the UTR, favoring a structurally homogenous ensemble compared with that of WT. Representative structures for each cluster of conformations, near each cluster's centroid, find a P1 stem (green) present in each WT structure. In addition, the G18U SNV forms a single structure similar to one of the three WT structures. G17C also collapses the ensemble, but favors an entirely different structure with an alternative P1 (Alt-P1) stem. The representative structures are both informed by and compatible with the SHAPE profile for that sequence (Supplemental Fig. S1B). All four structural conformations contain a conserved P2 stem (orange) not disrupted by either SNV.
Two sequential high-probability hairpins in the human RB1 5′ UTR
The principal component projection in Figure 2 is ideal for visualizing the entire Boltzmann ensemble, but it does not reveal specifics of most common structural features in the mRNA. The SHAPE reactivities were used to direct estimation of the partition function for each sequence using the program suite RNAstructure, from which we obtained the base-pairing probabilities (McCaskill 1990; Mathews 2004; Bernhart et al. 2006; Deigan et al. 2009). As can be seen in Figure 3A, two hairpin loops (one small and one large, denoted P1 and P2) occur with over 90% frequency. Thus, although the WT RB1 5′ UTR adopts three classes of conformations (Fig. 2), these two hairpins occur in a large majority of the sampled structures and are a common feature of all three conformations.
FIGURE 3.
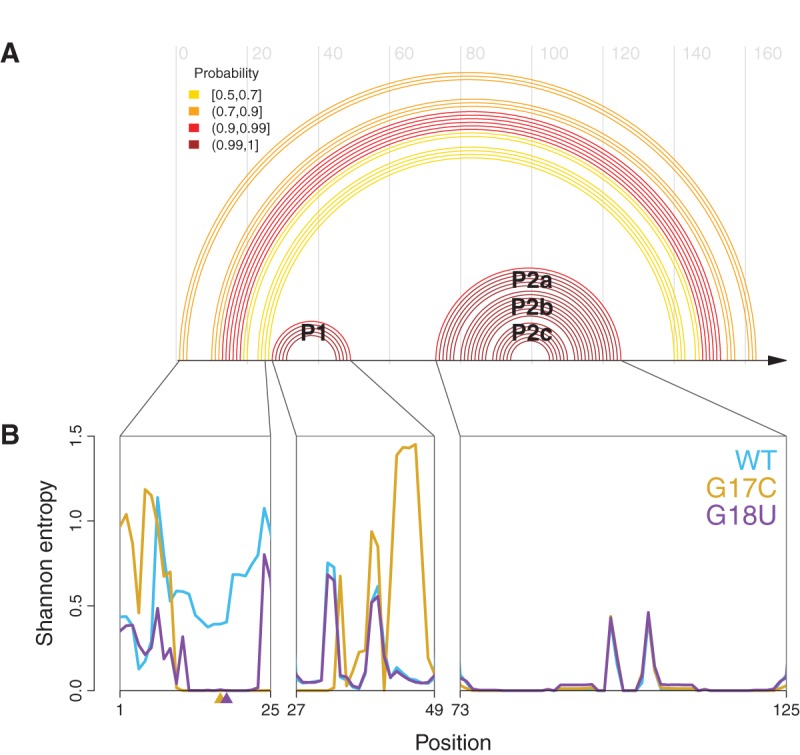
Elements of the core structure in the 5′ UTR can be disrupted by mutation. (A) WT base-pair probabilities >50% computed using a SHAPE-directed free energy function (Deigan et al. 2009; Wilkinson et al. 2009), plotted as an arc diagram; they reveal a core structure containing two helices, P1 and P2. The P2 helix has two bulges yielding the P2a, P2b, and P2c paired regions. Peripheral helices are lower probability and thus more variable. (B) Shannon entropies for individual nucleotides in WT (blue), G17C (gold), and G18U (purple). The two retinoblastoma-associated mutations drastically reduce the entropy of nucleotides 13–23 relative to WT, and G17C changes the pattern of entropy over the P1 helix. The P2 helix has low entropy in all three constructs, indicating a well-defined structure and suggesting the mutations do not alter this region of the mRNA.
The Shannon entropy of the base-pair probabilities for the WT and two mutant constructs are consistent with the changes observed in PCA space. In this context, Shannon entropy is a measure of structural homogeneity at a particular nucleotide; a low value indicates that the nucleotide always forms the same base pair (or lack thereof), and a high value indicates that the nucleotide exists in a variety of base-pairing contexts (Mantegna et al. 1994; Huynen et al. 1997; Kennedy et al. 2008). We calculated the Shannon entropy of each nucleotide using the SHAPE-directed base-pair probabilities (Equation 1). As can be seen in Figure 3B, both disease-associated SNVs lower the local entropy of the bases near the site of mutation. The G17C mutation drastically increases the entropy of bases in the P1 helix (nucleotides 44–49) while the entropy of the P2 helices remain unaffected by mutation.
Phylogenetically related RB1 5′ UTRs
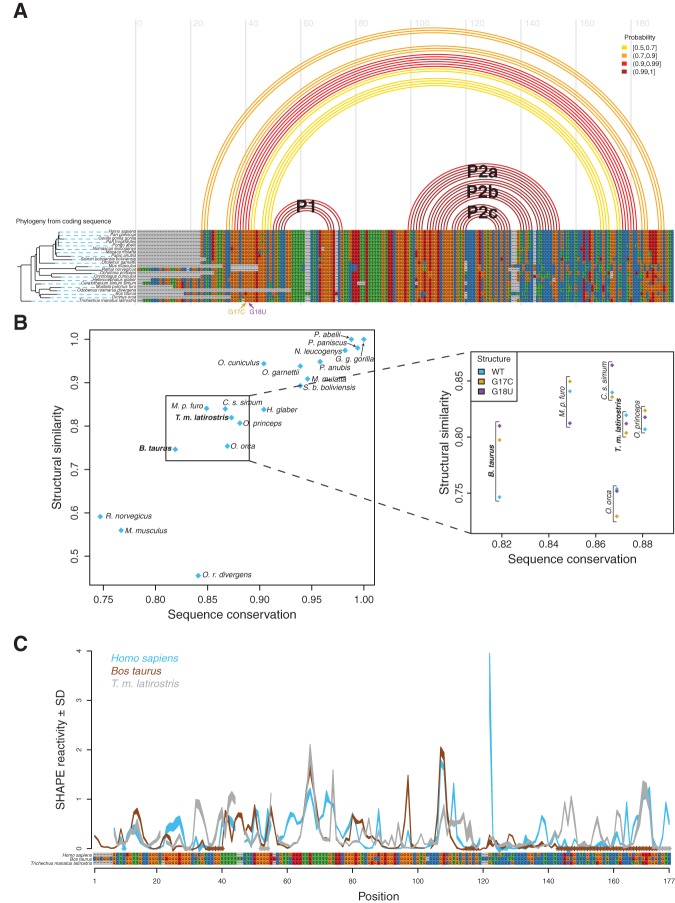
The high-probability hairpins in the human RB1 5′ UTR provide a starting point for structurally informed phylogenetic comparisons to other eukaryotic sequences. Structural alignment based solely on sequence comparisons is challenging in eukaryotic UTRs, as these tend to be either too highly conserved or too divergent for traditional covariation analysis (Griffiths-Jones et al. 2003; Eddy 2006; Nawrocki et al. 2009). As such, traditional covariation approaches do not produce strong or useful models, explaining the dearth of structural annotations in the RB1 5′ UTR. Nonetheless, our data on the WT and mutant human RB1 5′ UTRs suggest that an important structural element is present (two high-probability hairpins, P1 and P2). Furthermore, based on the data presented here, we propose that multiple conformations are critical for the proper regulation of this transcript.
A multiple sequence alignment of the 20 known homologous eukaryotic RB1 5′ UTRs indicates a high level of sequence conservation in this transcript (Fig. 4A). This high level of conservation is paralleled in the coding sequence as well (Supplemental Fig. S3). From this alignment alone a covariance model cannot be derived as very few columns reveal significant covariation signal. When the SHAPE-derived human base-pairing probabilities (or partition function) are projected on the alignment, however, it is clear the P1 and P2 stem-loops occur in highly conserved regions of the alignment, suggesting structural conservation of this element. To quantify this conservation, we computed a structural similarity score by summing the base-pair probabilities for consistent base pairs for each aligned homologous sequence (Equation 2). We plotted these structural similarity scores against the corresponding sequence similarity scores, revealing the correlation between the two metrics (Fig. 4B).
FIGURE 4.
Comparative sequence analysis predicts manatee and domestic cow RB1 5′ UTRs have conserved structural features present in the human construct. (A) (Top) Ensemble of human structures for the RB1 WT 5′ UTR, represented by predicted arcs colored by base-pairing probability. (Bottom) Multiple sequence alignment of the RB1 5′ UTR, showing that the UTR is highly conserved in mammals. Phylogenetic tree was created from the RB1 protein sequence from each organism (Supplemental Fig. S3). The length of the black branches indicates evolutionary distance, and the dashed blue lines connect the leaves of the tree to their corresponding organism. (B) (x-axis) Sequence similarity score. (y-axis) Structural similarity score (consistency of each sequence from the alignment to the SHAPE-directed partition function for human WT). The sequences we are interested in, which are most divergent in sequence yet highly conserved in structure, are easily visualized by trending to the top left corner. We used this plot to identify candidate UTR sequences for further SHAPE structural characterization. We chose to study the domestic cow (B. taurus) as it diverges significantly from human in sequence but has a relatively high structural similarity. In addition, the transcription start site of the cow RB1 5′ UTR was recently verified experimentally (Zimin et al. 2009, 2012). The manatee RB1 5′ UTR was chosen for further experimental characterization since it is structurally similar to human WT (blue diamond, inset) and differs significantly from G17C and G18U (gold and purple diamonds, inset). (C) SHAPE structure probing for human (blue), domestic cow (brown), and manatee (gray) mapped onto the alignment of these sequences. Qualitative similarities in the protection patterns suggest similar properties of the RNA structural ensemble.
The purpose of this analysis was to identify RB1 5′ UTR sequences that are highly divergent from human in sequence but maintain consistency with the WT structure. In addition, because the G17C and G18U disease-associated constructs adopt different structural ensembles, we also computed structural similarity scores relative to those two ensembles (Fig. 4B, inset). Although the rat and mouse RB1 5′ UTR sequences are the most divergent from the human sequence, they are also missing the region containing the G17C and G18U mutations (Fig. 4A). As such, we did not consider these RNAs for further structural analysis.
Both the domestic cow (Bos taurus) and the manatee (Trichechus manatus latirostris) RB1 5′ UTR sequences diverge from human sequence. When the cow sequence is compared by alignment to the SHAPE-informed human structural model (Fig. 4A), it becomes apparent that the major structural features of the UTR (the P1 and P2 stems) are still compatible with both sequences. Furthermore, when we compute the structural similarity scores (Fig. 4B), we see that despite being relatively divergent in sequence, both the cow and manatee sequences remain compatible with the human SHAPE-directed structural model. In addition, the transcription start site for the cow RB1 transcript has been experimentally determined as part of the domestic cow whole-genome assembly (Zimin et al. 2009, 2012). Further inspection of structural similarity scores for WT, G17C, and G18U (blue, gold, and purple, Fig. 4B, inset) reveal that the manatee RB1 5′ UTR is most consistent (as measured by structural similarity score) with the human WT structure relative to the two disease-associated mutants. The manatee is also the organism most phylogenetically distant from human in the multiple sequence alignment (Fig. 4A; Sayers et al. 2009). We therefore chose to further characterize the structure of the manatee and cow RB1 5′ UTRs by SHAPE structural probing.
Next, we performed SHAPE for the cow and manatee RB1 5′ UTRs (Fig. 4C; Supplemental Fig. S4A). We aligned the SHAPE data for the cow (brown) and manatee (gray) UTRs to human (blue) according to the multiple sequence alignment in Figure 4A. Qualitatively, we observed similar patterns of reactivity in the most conserved regions of the sequence. However, only SHAPE-directed prediction of the Boltzmann suboptimal ensemble reveals the common features of these three RNAs.
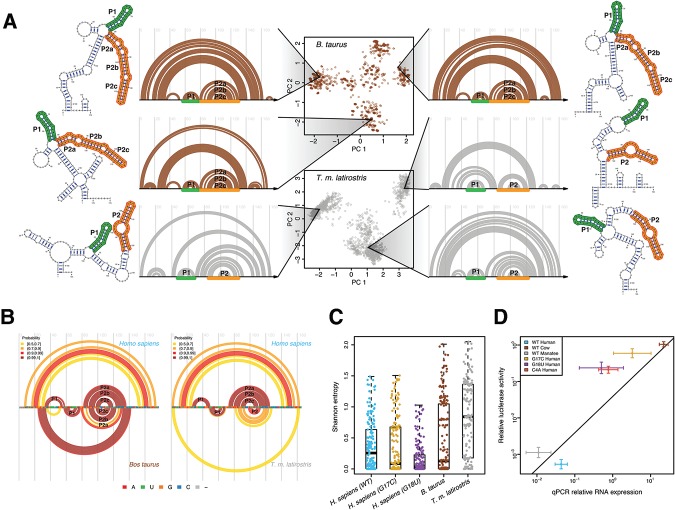
The conservation of multiple, populated alternative structures as a feature of RB1 5′ UTRs is further supported by SHAPE-directed Boltzmann suboptimal sampling for cow and manatee (Fig. 5A). Indeed, both sequences can adopt multiple conformations, with the manatee 5′ UTR even adopting three conformations like the human WT. Representative structures for each sequence demonstrate that the P1 and P2 helices are a common feature for each structural conformation. These structures correspond to the SHAPE reactivities for the cow and manatee sequences (Supplemental Fig. S4B). The conservation of the core P1 and P2 helices are confirmed when the high probability base pairs for cow and manatee are compared with human (Fig. 5B). Interestingly, in both cow and manatee, the P1 helix is shifted 3′, to a position analogous to the Alt-P1 helix observed in the G17C mutant. Thus, while the precise structure of the core helices is not perfectly conserved, the overall architecture of the ensemble is present in all three WT sequences. The main structural feature conserved in the human, cow, and manatee WT RB1 5′ UTRs is the presence of alternative conformations. Multiple conformations are also lost as a structural feature in the G17C and G18U SNVs (Fig. 2) suggesting multiple conformations are an important component of the disease etiology.
FIGURE 5.
Multiple divergent conformations are a conserved feature of the cow and manatee RB1 5′ UTRs. (A) (Center) Principal component decomposition of SHAPE-directed Boltzmann suboptimal sampling for the B. taurus (brown) and T.m. latirostris (gray) RB1 5′ UTRs. Both these RNAs display multiple well-populated conformations. Representative structures are plotted next to their corresponding arc diagrams. The P1 and P2 stem structures, analogous to those observed in human, are annotated with green and orange, respectively. (B) Arc diagrams of high-probability base pairs for the B. taurus (left) and T.m. latirostris (gray) compared with high-probability base pairs in H. sapiens. The P1 and P2 stem structures are consistent with the predicted structural similarity of these sequences. (C) Shannon entropy for each base using SHAPE-directed prediction of the partition function. The WT human, cow, and manatee UTRs have the highest median entropies, consistent with these UTRs forming multiple structures, while the two disease-associated UTRs have lower Shannon entropy. (D) Scatter plot of luciferase activity (y-axis) versus luciferase RNA abundance (x-axis) for the human, cow, and manatee WT constructs (blue, brown, and gray, respectively) and the three human mutant UTRs (C4A, G17C, G18U; red, gold, and purple, respectively). Values reported for both luciferase activity and RNA abundance are relative to an empty vector control. In general, higher RNA transcription yields higher luciferase activity, as expected. Also plotted is the linear regression through the three WT constructs (human, manatee, and cow). The three mutant constructs have slightly higher expression than WT as they all decrease above this line; however, the largest regulatory effects of UTR variation are at the level of RNA.
For another measure of conformational flexibility, we computed the Shannon entropy of each nucleotide in the ensemble for human, cow, and manatee WT sequences as well as the sequences with the disease-associated mutations (Fig. 5C). The distributions of the human WT and mutant entropies show that the two disease-associated mutations reduced the median entropy, corresponding to the collapse of the structural ensemble observed with the principal component decomposition (Fig. 2). The cow and manatee ensembles (brown and gray dots, respectively) both have higher median entropies than the disease-associated constructs, consistent with our sequence/structure analysis.
Structure/function relationships with luciferase reporter assays
To understand the functional consequences of the observed structural changes in the RB1 5′ UTR, we performed quantitative luciferase reporter assays in transiently transfected cells. We measured both Firefly luciferase activity (Supplemental Fig. S5A) and RNA levels (Supplemental Fig. S5B) for each construct relative to an empty vector control (Fig. 5D). We also measured transfection efficiency with a Renilla luciferase control and found no difference between the different constructs (Supplemental Fig. S5C). The x-axis in Figure 5D represents luciferase transcript levels, while the y-axis shows luciferase activity relative to the control. The line in Figure 5D is a regression through the three WT constructs (human, cow, and manatee) and represents mean relative luciferase expression. We observe a qualitative inverse correlation between expression and the entropy of each sequence (Fig. 5C), consistent with the hypothesis that multiple structures are important to WT function. Surprisingly, the C4A mutation also increases expression relative to WT, even though it does not significantly affect structure. This mutation was computationally predicted as not disruptive of the structural ensemble, but its close proximity to the 5′ cap may affect translation initiation. It is important to note that the C4A mutation has not been observed in any individual (healthy or diseased); in fact, the RB1 5′ UTR is highly conserved in mammalian genomes (Fig. 4A).
DISCUSSION
The role of structure in eukaryotic UTR evolution is still poorly understood since traditional approaches, such as covariation analysis, have not revealed conserved features in a majority of mRNAs (Eddy 2006; Nawrocki et al. 2009; Gardner et al. 2011). Indeed, UTRs and a majority of noncoding RNAs produced in eukaryotes are either not conserved or so highly conserved (as is the case for the RB1 5′ UTR) that strong covariation signals cannot be derived (Gardner and Giegerich 2004; Stevens et al. 2011; Widmann et al. 2012). In contrast, alignment-based structure prediction has identified thousands of conserved secondary structure motifs in prokaryotic genomes (Weinberg et al. 2007, 2009, 2011; Weinberg and Breaker 2011).
The cellular milieu is quite different in prokaryotes and eukaryotes; multiple RNA helicases in the latter likely reduce the importance of RNA structure in many regulatory processes (Burckin et al. 2005; Coller and Parker 2005; Russell et al. 2012). Nonetheless, mutations that affect RNA structure and consequently alter human phenotypes are not limited to the RB1 5′ UTR (Halvorsen et al. 2010; Martin et al. 2012; Lokody 2014; Wan et al. 2014). Interestingly, when genome-wide association studies include genetic variation in noncoding regions of the genome, a majority of highly associated SNPs (Single Nucleotide Polymorphisms) map outside of the coding region (Benjamin et al. 2007; Martin et al. 2012; Bulik-Sullivan et al. 2013). The lack of retinoblastoma-associated mutations mapping to noncoding regions of the gene (Fig. 1A) is likely the result of clinical genomics sequencing bias; until recently such studies focused almost exclusively on coding regions of the genome (Naruse et al. 2002; Macias et al. 2008; Cancer Genome Atlas Research Network 2012). It is therefore likely that other SNVs in the RB1 5′ UTR will cause retinoblastoma, but have yet to be reported in publicly available databases. In fact, a third private SNV was identified in a patient with retinoblastoma (Fig. 1B), but it was not predicted to alter the mRNA's structure (Halvorsen et al. 2010). This mutation is near the start codon and likely affects expression through a different mechanism than structure change.
A recent genome-wide characterization of human transcriptome secondary structure in three individuals identified almost 2000 riboSNitches in a family trio (Wan et al. 2014). These data suggest that SNV-induced structure change is quite common and in most cases phenotypically benign; the study was carried out on three healthy individuals. The propensity of SNVs to affect RNA structure thus appears to be a general phenomenon. What makes the RB1 5′ UTR system particularly interesting as a novel riboSNitch is the nature of the observed structure changes. For both disease-associated SNVs, the mutations collapse the ensemble into a single structure, and for the case of G18U, the single structure is very similar to a wild-type conformation, with no disruption of important structural motifs. Our data suggest that the formation of a single structure is deleterious to the regulation of RB1. This conclusion is supported by the fact that the two other wild-type RB1 UTRs we investigated (cow and manatee) also adopt multistructure ensembles.
Our transient transfection assays (Fig. 5D; Supplemental Fig. S5) reveal the complexity of structure/function relationships in eukaryotic gene regulation. We observe that the three mutant RB1 5′ UTRs have higher expression compared with WT (regression line, Fig. 5D). Renilla cotransfection controls (Supplemental Fig. S5C) suggest the important difference in RNA expression of the WT and mutant constructs (as measured by qRT-PCR) is not the result of differences in transfection efficiency. Our assay is nonetheless primarily designed to measure translation efficiency, but the possibility that a riboSNitch could alter RNA expression is intriguing and warrants further study. There are many transcriptional riboswitches in bacteria, and changes in UTR structure in eukaryotes may affect transcription too (Batey et al. 2004; Stoddard et al. 2008, 2010). One other aspect of expression we could not directly measure in these assays is RNA degradation. It is possible that the large changes in expression we observe are due to differential stabilities of the mutant mRNAs in the cell. One final consideration with these assays is that they were performed in HeLa cells, which are known to have altered transcriptional and post-transcriptional programs (Murray et al. 2004; Landry et al. 2013).
Phylogenetic comparison of RNAs remains a powerful tool for determining structure, especially in prokaryotic systems where covariation signals are sufficient to unambiguously determine secondary structure (Michel and Westhof 1990; Gutell et al. 2002; Mertz et al. 2009). Our comparative analysis of the human, cow, and manatee RB1 5′ UTRs suggests that structural elements are conserved in noncoding regions of messages as a result of selective pressure. For this UTR, a single structure is not selected for; instead, diversity of the structural landscape is conserved. This observation may explain the high degree of evolutionary sequence conservation (Fig. 4A) and lack of covariation signal observed in the UTR. Selection for a single structure favors canonical covariation, but little is known about how tolerant specific structural ensembles are to mutation and covariation. With the advent of high-throughput techniques for obtaining high-resolution structural probing data (Siegfried et al. 2014), along with new methods of profiling RNA structural ensembles (Rogers and Heitsch 2014), it will become feasible to determine the role of specific structural ensembles in regulating eukaryotic expression through genomic analysis.
MATERIALS AND METHODS
SHAPE data collection
The human WT RB1 5′ UTR sequence with hairpin adapters for SHAPE (GGCCTTCGGGCCAAGCTCAGTTGCCGGGCGGGGGAGGGCGCGTCCGGTTTTTCTCAGGGGACGTTGAAATTATTTTTGTAACGGGAGTCGGGAGAGGACGGGGCGTGCCCCGACGTGCGCGCGCGTCGTCCTCCCCGGCGCTCCTCCACAGCTCGCTGGCTCCCGCCGCGGAAAGGCGTCATGCCGTCGATCCGGTTCGCCGGATCCAAATCGGGCTTCGGTCCGGTTC) was inserted between the SgfI and MluI sites of the pCMV6-AC nontagged precision shuttle vector (Origene). Hairpin adapters are indicated in bold. The mutant sequences, which varied only by point mutations, were inserted into pUC57 by Genscript. The Bos taurus RB1 (GGCCTTCGGGCCAACGCGGCGCTCGGTTGCCGGGCGAGGGAGGGCCGGCCCGGTTTTTTTCTCAGGGGAACGTTCAAATTATTTTTGTAACGGGAGTCGGCCGAGGACGGGGCGTGCCCGAGGTGCGCGCGTCCTCTCCCTTCCCCGGCCCTCCTCCAGCGCCCGCCGGCGCCTGCCCAGCGAGCGCGTCATGCCGTCGATCCGGTTCGCCGGATCCAAATCGGGCTTCGGTCCGGTTC) and T.m. latirostris RB1 (GGCCTTCGGGCCAAGCTCGGTTGCCGGGTGGGGAGGGCTTGTCCGGTTTTTCTCAGGGGACGTTCAAATTATTTTTGTAACGGGAGTCGAGAGAGGACGGGGCGTGCCCCGACGTGTGCGCGCGTCCCCCGCCCCCGCCCTCCTCCACAGCTCTCTAGCTCCTACCCTGTAAGGGCGTCATGCCGTCGATCCGGTTCGCCGGATCCAAATCGGGCTTCGGTCCGGTTC) sequences were also cloned by Genscript into pUC57. A T7 promoter (TAATACGACTCACTATAGGG) was introduced to the 5′ end of the 5′ UTR during PCR amplification followed by transcription with the T7 high-yield RNA synthesis kit (New England Biolabs) and cleanup by MegaClear (Ambion).
Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) experiments were performed as previously described (Martin et al. 2012) with a few modifications. Of note, 2 pmol RNA were used for each reaction and, after denaturation as previously described, were folded in a final concentration of 100 mM HEPES, pH 8.0, 10 mM MgCl2, 100 mM KCl at 37°C for 15 min. Primer extension was performed as previously described, but with 2 pmol of Vic or Ned-labeled primer without RNase inhibitor. The samples with and without NMIA were reverse transcribed with the Vic-labeled primer; the Ned-labeled primer was used to make sequencing ladders using unreacted RNA and 1 µl 5 mM ddGTP, ddCTP, ddATP, or ddTTP. The base and neutralization steps used to degrade remaining RNA in the cDNA samples were found to be unnecessary and were eliminated in later experiments. The cDNA pellets were dried, resuspended in Hi-Di formamide (Applied Biosystems/Life Technologies), and run on an Applied Biosystems 3500 Genetic Analyzer. The resulting data were analyzed using QuSHAPE (Karabiber et al. 2013).
SHAPE data averaging and visualization
A minimum of five experimental repeats were collected for each construct, and the data were filtered for quality control as previously described (Martin et al. 2012). SHAPE reactivities for each experiment were normalized as described in (Wilkinson et al. 2008). Briefly, the mean reactivity at each nucleotide was found for the background (-NMIA) conditions. The positions with the highest background signal (3.6%–7.0% of nucleotides) were manually identified for each construct, not used in the averaging, and indicated as no data (asterisks in Figs. 1D,E, 4C). For each SHAPE experiment, the background was scaled so that the mean of those positions in the background would equal the mean of those positions with the reagent. To find the SHAPE reactivities, the background reactivities were subtracted from the reagent reactivities, and these were normalized by the 2%–8% method described in Wilkinson et al. (2005, 2006, 2008). Nucleotides 142–176 in the B. taurus sequence were not considered in structure prediction because of difficulty aligning to the ladder.
The normalized SHAPE data is presented in Supplemental Data 1 as well as available for download in ISATab format at https://docs.google.com/spreadsheet/ccc?key=0AhfD_gVAiWMBdENTNEEwUDNZRm52T052SUVac1VCOXc&usp=sharing
Boltzmann suboptimal sampling and principal component visualization
The partition program in RNAstructure (v5.6) was used to compute the partition function used for all structure prediction and sampling calculations (Mathews 2004; Deigan et al. 2009; Hajdin et al. 2013). SHAPE data were used to direct all simulations as described in (Deigan et al. 2009) using standard parameters (SHAPE Intercept: −0.6 kcal/mol, SHAPE slope: 1.8 kcal/mol, and temperature: 310.15 K). For suboptimal sampling, we used RNAstructure's stochastic to generate 5000 structures for each sampled sequence. These structures were coded as binary vectors by whether each nucleotide was base-paired. Principal component visualization of suboptimal structures was carried out with the R Project for Statistical Computing (v3.1.0). The principal component space for human structures was created by first predicting which mutations in the human sequence would maximize overall entropy, sampling suboptimal structures from both entropy-maximizing mutations and the WT sequence (without SHAPE data), and finding the principal components of those sampled structures in aggregate. For cow and manatee, the native principal component space was used. Representative structures for each cluster were chosen from sampled structures near each cluster's centroid, as the centroid does not necessarily correspond to a sampled structure. Arc diagrams were created using R4RNA (Lai et al. 2012). Base-pairing probabilities used in arc diagrams were those reported by RNAstructure's ProbabilityPlot.
Shannon entropy
The SHAPE-directed base-pairing probabilities found by RNAstructure's ProbabilityPlot were used for calculations of Shannon entropy. The Shannon entropy of each nucleotide was calculated as described (Huynen et al. 1997; Siegfried et al. 2014):
| (1) |
where Si is the entropy of nucleotide i and Pi,j is the probability of nucleotides i and j base-pairing (which is the probability of nucleotide i being unpaired when i = j).
Sequence analysis of homologous RB1 transcripts
Homologous sequences to the human RB1 5′ UTR were identified with NCBI BLAST. A multiple sequence alignment was created with MAFFT (v6.850) (Katoh and Toh 2008) with the EMBL-EBI webserver (McWilliam et al. 2013) and then refined manually. Using each partition function for the human sequences (WT, G17C, G18U) a “structure similarity score” for each homologous sequence was computed according to Equation 2:
| (2) |
The structure similarity score for sequence q (Tq) quantifies the compatibility of a given sequence with the human, SHAPE-directed partition function. Phuman(i,j) is the probability of alignment positions i and j base based on the human, SHAPE-directed partition function. Each probability is only included in the sum if those alignment positions can form a valid Watson–Crick or wobble base pair in sequence q. The denominator contains the sum of all base-pairing probabilities for the human sequence. As such, structure similarity scores have a value between 0 and 1. Sequence similarity was computed using the alistat software, which is part of the HMMER package (Eddy 2009; Johnson et al. 2010; Finn et al. 2011). To visualize sequence and structural divergence, the structural similarity score to WT for each homologous sequence was plotted against the sequence conservation score.
The phylogeny of the RB1 gene was found through an NCBI BLAST search of the human RB1 coding DNA sequence. The translations of these coding sequences were aligned by MAFFT (Katoh and Toh 2008). From this multiple sequence alignment, a phylogenetic tree was created using PhyML (Guindon et al. 2010). The tree was rooted by the manatee sequence, the only non-Boreoeutherian mammal in the tree (Sayers et al. 2009).
Luciferase assays and qPCR to measure expression
For luciferase assays, each RB1 construct was cloned into the pGL3-control vector between the SV40 promoter and the Firefly luciferase CDS by Genscript. HeLa cells were transfected with 0.5 μg plasmid DNA and harvested 24 h later using Cell Culture Lysis Reagent (Promega # E153A). Luciferase activity was measured on a luminometer (Molecular Devices) using Luciferase Assay Substrate (Promega # E151C). The protein content of the samples was determined by Bradford assay. The luciferase readings were normalized to protein content in each lysate, as determined by the Bradford assay (n = 4).
To control for differences in transfection efficiency we repeated our transfections including a common Renilla luciferase construct (n = 2). Following measurement of Firefly luciferase we measured the exact same sample for Renilla abundance. The abundance of Renilla was normalized to sample protein content.
Total RNA was extracted from the same lysates used in the luciferase assays using TRIzol reagent. The RNA was DNase treated using Ambion Turbo DNA-free (AM1907). cDNA was generated using Ambion High Capacity cDNA Reverse Transcription Kit (#4368813). cDNA abundance was measured by quantitative real time PCR (qRT-PCR) on a BioRad CFX96 Real-Time System using the following primers: luciferase 5′-ACAAAGGCTATCAGGTGGCT-3′, 5′-CGTGCTCCAAAACAACAACG-3′; GAPDH 5′-CTGTTGCTGTAGCCAAATTCGT-3′, 5′-ACCCACTCCTCCACCTTTGAC-3′. The abundance of luciferase RNA was determined by the ▵▵Ct method using GAPDH as the reference transcript (n = 4). Values reported for both luciferase activity and RNA abundance are relative to an empty vector control.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This work is supported by US National Institutes of Health grants NHLBI R01 HL111527 and NIGMS R01 GM101237 to A.L., R01 GM064803 to K.M.W., and funds from NIAID R01 AI03311 and the North Carolina University Cancer Research Fund to N.M.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.049221.114.
Freely available online through the RNA Open Access option.
REFERENCES
- Agius P, Bennett KP, Zuker M. 2010. Comparing RNA secondary structures using a relaxed base-pair score. RNA 16: 865–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders G, Mackowiak SD, Jens M, Maaskola J, Kuntzagk A, Rajewsky N, Landthaler M, Dieterich C. 2012. doRiNA: a database of RNA interactions in post-transcriptional regulation. Nucleic Acids Res 40: D180–D186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batey RT, Gilbert SD, Montange RK. 2004. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature 432: 411–415. [DOI] [PubMed] [Google Scholar]
- Benjamin EJ, Dupuis J, Larson MG, Lunetta KL, Booth SL, Govindaraju DR, Kathiresan S, Keaney JF Jr, Keyes MJ, Lin JP, et al. 2007. Genome-wide association with select biomarker traits in the Framingham Heart Study. BMC Med Genet 8: S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhart SH, Tafer H, Muckstein U, Flamm C, Stadler PF, Hofacker IL. 2006. Partition function and base pairing probabilities of RNA heterodimers. Algorithms Mol Biol 1: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik-Sullivan B, Selitsky S, Sethupathy P. 2013. Prioritization of genetic variants in the microRNA regulome as functional candidates in genome-wide association studies. Hum Mutat 34: 1049–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burckin T, Nagel R, Mandel-Gutfreund Y, Shiue L, Clark TA, Chong JL, Chang TH, Squazzo S, Hartzog G, Ares M Jr. 2005. Exploring functional relationships between components of the gene expression machinery. Nat Struct Mol Biol 12: 175–182. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. 2012. Comprehensive genomic characterization of squamous cell lung cancers. Nature 489: 519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celander DW, Cech TR. 1991. Visualizing the higher order folding of a catalytic RNA molecule. Science 251: 401–407. [DOI] [PubMed] [Google Scholar]
- Coller J, Parker R. 2005. General translational repression by activators of mRNA decapping. Cell 122: 875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowell JK, Bia B, Akoulitchev A. 1996. A novel mutation in the promoter region in a family with a mild form of retinoblastoma indicates the location of a new regulatory domain for the RB1 gene. Oncogene 12: 431–436. [PubMed] [Google Scholar]
- Crick F. 1970. Central dogma of molecular biology. Nature 227: 561–563. [DOI] [PubMed] [Google Scholar]
- Deigan KE, Li TW, Mathews DH, Weeks KM. 2009. Accurate SHAPE-directed RNA structure determination. Proc Natl Acad Sci 106: 97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Chan CY, Lawrence CE. 2004. Sfold web server for statistical folding and rational design of nucleic acids. Nucleic Acids Res 32: W135–W141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Chan CY, Lawrence CE. 2005. RNA secondary structure prediction by centroids in a Boltzmann weighted ensemble. RNA 11: 1157–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. 2006. Computational analysis of RNAs. Cold Spring Harb Symp Quant Biol 71: 117–128. [DOI] [PubMed] [Google Scholar]
- Eddy SR. 2009. A new generation of homology search tools based on probabilistic inference. Genome Inform 23: 205–211. [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Clements J, Eddy SR. 2011. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 39: W29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner PP, Giegerich R. 2004. A comprehensive comparison of comparative RNA structure prediction approaches. BMC Bioinformatics 5: 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner PP, Daub J, Tate JG, Nawrocki EP, Kolbe DL, Lindgreen S, Wilkinson AC, Finn RD, Griffiths-Jones S, Eddy SR, et al. 2009. Rfam: updates to the RNA families database. Nucleic Acids Res 37: D136–D140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner PP, Daub J, Tate J, Moore BL, Osuch IH, Griffiths-Jones S, Finn RD, Nawrocki EP, Kolbe DL, Eddy SR, et al. 2011. Rfam: Wikipedia, clans and the “decimal” release. Nucleic Acids Res 39: D141–D145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George RA, Smith TD, Callaghan S, Hardman L, Pierides C, Horaitis O, Wouters MA, Cotton RG. 2008. General mutation databases: analysis and review. J Med Genet 45: 65–70. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy SR. 2003. Rfam: an RNA family database. Nucleic Acids Res 31: 439–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A. 2005. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res 33: D121–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59: 307–321. [DOI] [PubMed] [Google Scholar]
- Gutell RR, Lee JC, Cannone JJ. 2002. The accuracy of ribosomal RNA comparative structure models. Curr Opin Struct Biol 12: 301–310. [DOI] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M Jr, Jungkamp AC, Munschauer M, et al. 2010a. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141: 129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jungkamp AC, Munschauer M, et al. 2010b. PAR-CliP—a method to identify transcriptome-wide the binding sites of RNA binding proteins. J Vis Exp: e2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdin CE, Bellaousov S, Huggins W, Leonard CW, Mathews DH, Weeks KM. 2013. Accurate SHAPE-directed RNA secondary structure modeling, including pseudoknots. Proc Natl Acad Sci 110: 5498–5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halvorsen M, Martin JS, Broadaway S, Laederach A. 2010. Disease-associated mutations that alter the RNA structural ensemble. PLoS Genet 6: e1001074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynen M, Gutell R, Konings D. 1997. Assessing the reliability of RNA folding using statistical mechanics. J Mol Biol 267: 1104–1112. [DOI] [PubMed] [Google Scholar]
- Jacks T, Fazeli A, Schmitt EM, Bronson RT, Goodell MA, Weinberg RA. 1992. Effects of an Rb mutation in the mouse. Nature 359: 295–300. [DOI] [PubMed] [Google Scholar]
- Johnson LS, Eddy SR, Portugaly E. 2010. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinformatics 11: 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karabiber F, McGinnis JL, Favorov OV, Weeks KM. 2013. QuShape: rapid, accurate, and best-practices quantification of nucleic acid probing information, resolved by capillary electrophoresis. RNA 19: 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Toh H. 2008. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform 9: 286–298. [DOI] [PubMed] [Google Scholar]
- Kennedy R, Lladser ME, Yarus M, Knight R. 2008. Information, probability, and the abundance of the simplest RNA active sites. Front Biosci 13: 6060–6071. [DOI] [PubMed] [Google Scholar]
- Laederach A, Shcherbakova I, Jonikas MA, Altman RB, Brenowitz M. 2007. Distinct contribution of electrostatics, initial conformational ensemble, and macromolecular stability in RNA folding. Proc Natl Acad Sci 104: 7045–7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai D, Proctor JR, Zhu JY, Meyer IM. 2012. R-CHIE: a web server and R package for visualizing RNA secondary structures. Nucleic Acids Res 40: e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry JJ, Pyl PT, Rausch T, Zichner T, Tekkedil MM, Stütz AM, Jauch A, Aiyar RS, Pau G, Delhomme N, et al. 2013. The genomic and transcriptomic landscape of a HeLa cell line. G3 (Bethesda) 3: 1213–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KM, Tarn WY. 2013. Coupling pre-mRNA processing to transcription on the RNA factory assembly line. RNA Biol 10: 380–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WH, Bookstein R, Hong F, Young LJ, Shew JY, Lee EY. 1987. Human retinoblastoma susceptibility gene: cloning, identification, and sequence. Science 235: 1394–1399. [DOI] [PubMed] [Google Scholar]
- Lokody I. 2014. RNA: RiboSNitches reveal heredity in RNA secondary structure. Nat Rev Genet 15: 219. [DOI] [PubMed] [Google Scholar]
- Macias M, Dean M, Atkinson A, Jiménez-Morales S, Garcia-Vazquez FJ, Saldaña-Alvarez Y, Ramirez-Bello J, Chávez M, Orozco L. 2008. Spectrum of RB1 gene mutations and loss of heterozygosity in Mexican patients with retinoblastoma: identification of six novel mutations. Cancer Biomark 4: 93–99. [DOI] [PubMed] [Google Scholar]
- Mandal M, Boese B, Barrick JE, Winkler WC, Breaker RR. 2003. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell 113: 577–586. [DOI] [PubMed] [Google Scholar]
- Mantegna RN, Buldyrev SV, Goldberger AL, Havlin S, Peng CK, Simons M, Stanley HE. 1994. Linguistic features of noncoding DNA sequences. Phys Rev Lett 73: 3169–3172. [DOI] [PubMed] [Google Scholar]
- Marees T, Moll AC, Imhof SM, de Boer MR, Ringens PJ, van Leeuwen FE. 2008. Risk of second malignancies in survivors of retinoblastoma: more than 40 years of follow-up. J Natl Cancer Inst 100: 1771–1779. [DOI] [PubMed] [Google Scholar]
- Martin JS, Halvorsen M, Davis-Neulander L, Ritz J, Gopinath C, Beauregard A, Laederach A. 2012. Structural effects of linkage disequilibrium on the transcriptome. RNA 18: 77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews DH. 2004. Using an RNA secondary structure partition function to determine confidence in base pairs predicted by free energy minimization. RNA 10: 1178–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaskill JS. 1990. The equilibrium partition function and base pair binding probabilities for RNA secondary structure. Biopolymers 29: 1105–1119. [DOI] [PubMed] [Google Scholar]
- McWilliam H, Li W, Uludag M, Squizzato S, Park YM, Buso N, Cowley AP, Lopez R. 2013. Analysis tool web services from the EMBL-EBI. Nucleic Acids Res 41: W597–W600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertz JA, Chadee AB, Byun H, Russell R, Dudley JP. 2009. Mapping of the functional boundaries and secondary structure of the mouse mammary tumor virus Rem-responsive element. J Biol Chem 284: 25642–25652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F, Westhof E. 1990. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J Mol Biol 216: 585–610. [DOI] [PubMed] [Google Scholar]
- Mitra S, Shcherbakova IV, Altman RB, Brenowitz M, Laederach A. 2008. High-throughput single-nucleotide structural mapping by capillary automated footprinting analysis. Nucleic Acids Res 36: e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JI, Whitfield ML, Trinklein ND, Myers RM, Brown PO, Botstein D. 2004. Diverse and specific gene expression responses to stresses in cultured human cells. Mol Biol Cell 15: 2361–2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naruse TK, Kawata H, Inoko H, Isshiki G, Yamano K, Hino M, Tatsumi N. 2002. The HLA-DOB gene displays limited polymorphism with only one amino acid substitution. Tissue Antigens 59: 512–519. [DOI] [PubMed] [Google Scholar]
- Nawrocki EP, Kolbe DL, Eddy SR. 2009. Infernal 1.0: inference of RNA alignments. Bioinformatics 25: 1335–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritz J, Martin JS, Laederach A. 2012. Evaluating our ability to predict the structural disruption of RNA by SNPs. BMC Genomics 13: S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritz J, Martin JS, Laederach A. 2013. Evolutionary evidence for alternative structure in RNA sequence co-variation. PLoS Comput Biol 9: e1003152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers E, Heitsch CE. 2014. Profiling small RNA reveals multimodal substructural signals in a Boltzmann ensemble. Nucleic Acids Res 42: e171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogler LE, Kosmyna B, Moskowitz D, Bebawee R, Rahimzadeh J, Kutchko K, Laederach A, Notarangelo LD, Giliani S, Bouhassira E, et al. 2014. Small RNAs derived from lncRNA RNase MRP have gene-silencing activity relevant to human cartilage-hair hypoplasia. Hum Mol Genet 23: 368–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell R, Zhuang X, Babcock HP, Millett IS, Doniach S, Chu S, Herschlag D. 2002. Exploring the folding landscape of a structured RNA. Proc Natl Acad Sci 99: 155–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell R, Jarmoskaite I, Lambowitz AM. 2012. Toward a molecular understanding of RNA remodeling by DEAD-box proteins. RNA Biol 10: 44–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez M, Galy B, Dandekar T, Bengert P, Vainshtein Y, Stolte J, Muckenthaler MU, Hentze MW. 2006. Iron regulation and the cell cycle: identification of an iron-responsive element in the 3′-untranslated region of human cell division cycle 14A mRNA by a refined microarray-based screening strategy. J Biol Chem 281: 22865–22874. [DOI] [PubMed] [Google Scholar]
- Sayers EW, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, et al. 2009. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 37: D5–D15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder R, Barta A, Semrad K. 2004. Strategies for RNA folding and assembly. Nat Rev Mol Cell Biol 5: 908–919. [DOI] [PubMed] [Google Scholar]
- Siegfried NA, Busan S, Rice GM, Nelson JA, Weeks KM. 2014. RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP). Nat Methods 11: 959–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomatin SV, Greenfeld M, Chu S, Herschlag D. 2010. Multiple native states reveal persistent ruggedness of an RNA folding landscape. Nature 463: 681–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenson PD, Ball EV, Mort M, Phillips AD, Shiel JA, Thomas NS, Abeysinghe S, Krawczak M, Cooper DN. 2003. Human Gene Mutation Database (HGMD): 2003 update. Hum Mutat 21: 577–581. [DOI] [PubMed] [Google Scholar]
- Stevens SG, Gardner PP, Brown C. 2011. Two covariance models for iron-responsive elements. RNA Biol 8: 792–801. [DOI] [PubMed] [Google Scholar]
- Stoddard CD, Gilbert SD, Batey RT. 2008. Ligand-dependent folding of the three-way junction in the purine riboswitch. RNA 14: 675–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddard CD, Montange RK, Hennelly SP, Rambo RP, Sanbonmatsu KY, Batey RT. 2010. Free state conformational sampling of the SAM-I riboswitch aptamer domain. Structure 18: 787–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. 2010. Non-coding RNAs: regulators of disease. J Pathol 220: 126–139. [DOI] [PubMed] [Google Scholar]
- Thirumalai D, Woodson SA. 2000. Maximizing RNA folding rates: a balancing act. RNA 6: 790–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker BJ, Breaker RR. 2005. Riboswitches as versatile gene control elements. Curr Opin Struct Biol 15: 342–348. [DOI] [PubMed] [Google Scholar]
- Ulitsky I, Bartel DP. 2013. lincRNAs: genomics, evolution, and mechanisms. Cell 154: 26–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde JR, Alonso J, Palacios I, Pestana A. 2005. RB1 gene mutation up-date, a meta-analysis based on 932 reported mutations available in a searchable database. BMC Genet 6: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Qu K, Zhang QC, Flynn RA, Manor O, Ouyang Z, Zhang J, Spitale RC, Snyder MP, Segal E, et al. 2014. Landscape and variation of RNA secondary structure across the human transcriptome. Nature 505: 706–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z, Breaker RR. 2011. R2R—software to speed the depiction of aesthetic consensus RNA secondary structures. BMC Bioinformatics 12: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z, Barrick JE, Yao Z, Roth A, Kim JN, Gore J, Wang JX, Lee ER, Block KF, Sudarsan N, et al. 2007. Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline. Nucleic Acids Res 35: 4809–4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z, Perreault J, Meyer MM, Breaker RR. 2009. Exceptional structured noncoding RNAs revealed by bacterial metagenome analysis. Nature 462: 656–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z, Wang JX, Bogue J, Yang J, Corbino K, Moy RH, Breaker RR. 2011. Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea, and their metagenomes. Genome Biol 11: R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widmann J, Stombaugh J, McDonald D, Chocholousova J, Gardner P, Iyer MK, Liu Z, Lozupone CA, Quinn J, Smit S, et al. 2012. RNASTAR: an RNA STructural Alignment Repository that provides insight into the evolution of natural and artificial RNAs. RNA 18: 1319–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson KA, Merino EJ, Weeks KM. 2005. RNA SHAPE chemistry reveals nonhierarchical interactions dominate equilibrium structural transitions in tRNAAsp transcripts. J Am Chem Soc 127: 4659–4667. [DOI] [PubMed] [Google Scholar]
- Wilkinson KA, Merino EJ, Weeks KM. 2006. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc 1: 1610–1616. [DOI] [PubMed] [Google Scholar]
- Wilkinson KA, Gorelick RJ, Vasa SM, Guex N, Rein A, Mathews DH, Giddings MC, Weeks KM. 2008. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol 6: e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson KA, Vasa SM, Deigan KE, Mortimer SA, Giddings MC, Weeks KM. 2009. Influence of nucleotide identity on ribose 2′-hydroxyl reactivity in RNA. RNA 15: 1314–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson SA. 2002. Folding mechanisms of group I ribozymes: role of stability and contact order. Biochem Soc Trans 30: 1166–1169. [DOI] [PubMed] [Google Scholar]
- Zarrinkar PP, Williamson JR. 1994. Kinetic intermediates in RNA folding. Science 265: 918–924. [DOI] [PubMed] [Google Scholar]
- Zimin AV, Delcher AL, Florea L, Kelley DR, Schatz MC, Puiu D, Hanrahan F, Pertea G, Van Tassell CP, Sonstegard TS, et al. 2009. A whole-genome assembly of the domestic cow, Bos taurus. Genome Biol 10: R42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimin AV, Kelley DR, Roberts M, Marcais G, Salzberg SL, Yorke JA. 2012. Mis-assembled “segmental duplications” in two versions of the Bos taurus genome. PLoS One 7: e42680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M, Sankoff D. 1984. RNA secondary structures and their prediction. Bull Math Bio 46: 591–621. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.