Abstract
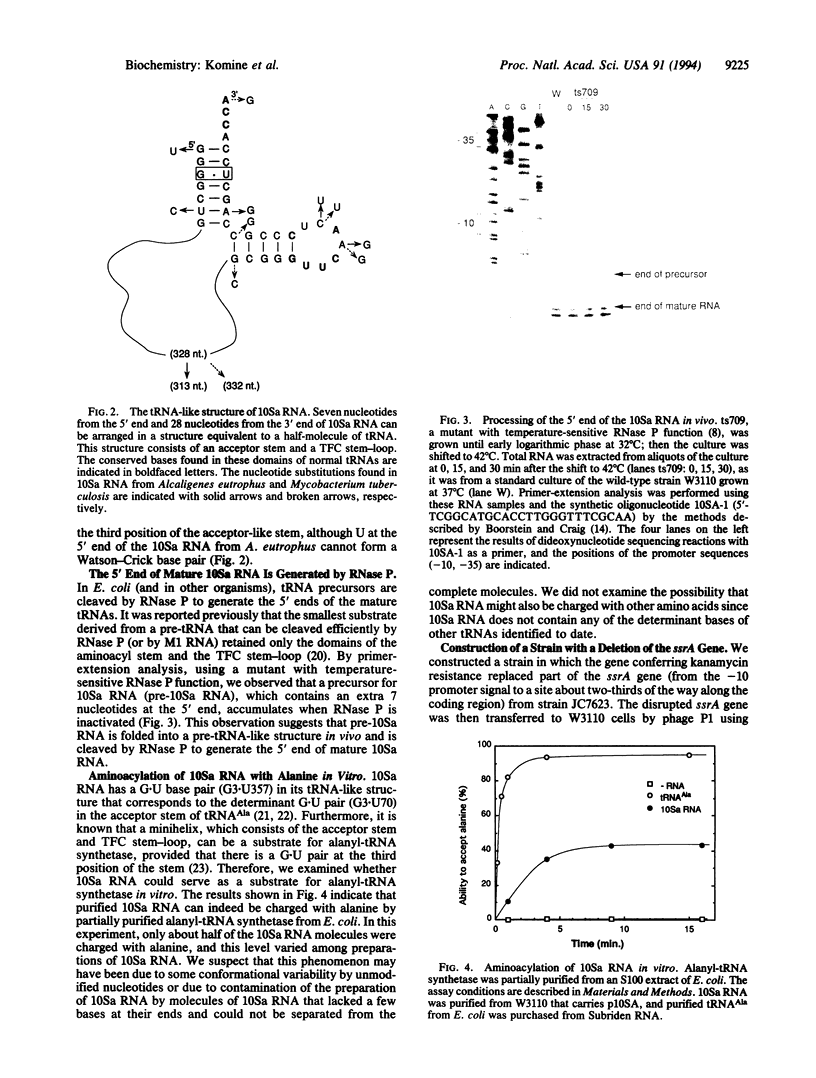
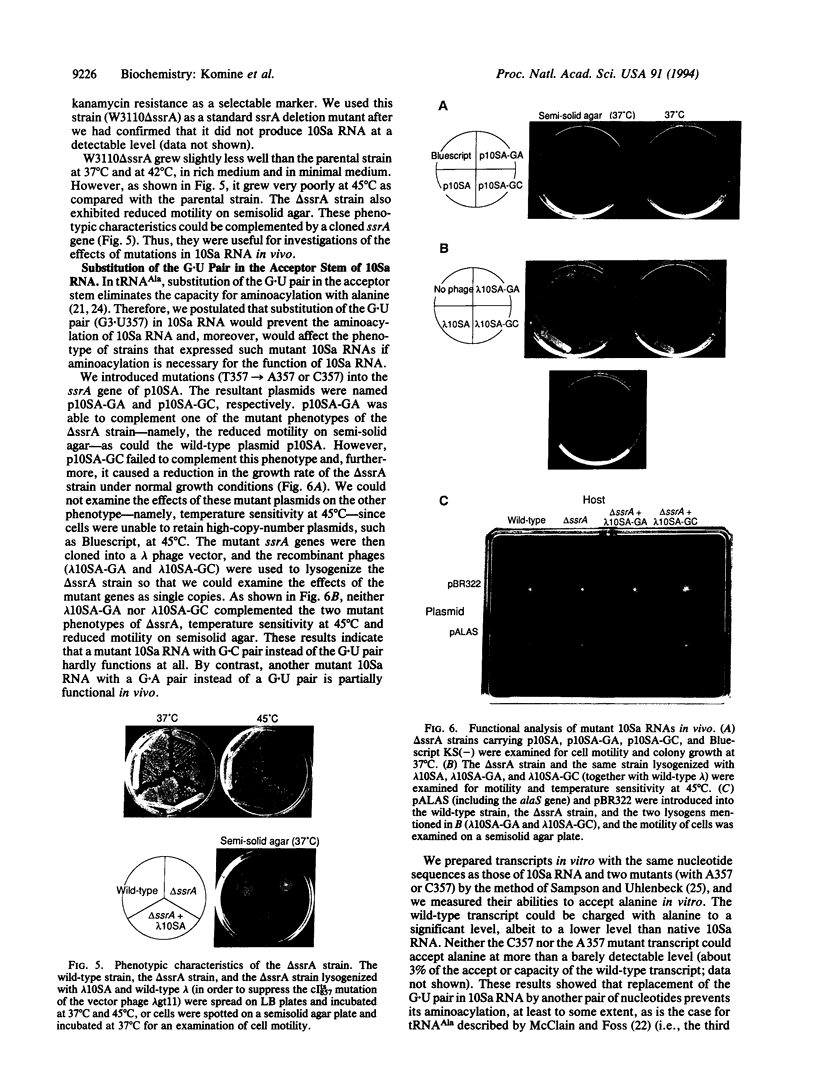
We have determined that 10Sa RNA (one of the small stable RNAs found in Escherichia coli) has an interesting structural feature: the 5' end and the 3' end of 10Sa RNA can be arranged in a structure that is equivalent to a half-molecule (acceptor stem and TFC stem-loop) of alanine tRNA of E. coli. Primer-extension analysis of 10Sa RNA extracted from a bacterial mutant with temperature-sensitive RNase P function revealed that the precursor to 10Sa RNA (pre-10Sa RNA) is folded into a pre-tRNA-like structure in vivo such that it can be cleaved by RNase P to generate the 5' end of the mature 10Sa RNA. The purified 10Sa RNA can be charged with alanine in vitro. Disruption of the gene encoding 10Sa RNA (ssrA) caused a reduction in the rate of cell growth, which was especially apparent at 45 degrees C, and a reduction in motility on semisolid agar. These phenotypic characteristics of the deletion strain (delta ssrA) allowed us to investigate the effects of some mutations in 10Sa RNA in vivo, although the exact function of 10Sa RNA still remains unclear. When the G.U pair (G3.U357) in 10Sa RNA, which may be equivalent to the determinant G.U pair of alanine tRNA, was changed to a G.A or G.C pair, the ability to complement the phenotypic mutations of the delta ssrA strain was lost. Furthermore, this inability to complement the mutant phenotypes that was caused by the substitution of the determinant bases by a G.A pair could be overcome by the introduction of a gene encoding alanyl-tRNA synthetase (alaS) on a multicopy plasmid. The evidence suggests that the proposed structural features of 10Sa RNA are indeed manifested in vivo.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altman S. Ribonuclease P: an enzyme with a catalytic RNA subunit. Adv Enzymol Relat Areas Mol Biol. 1989;62:1–36. doi: 10.1002/9780470123089.ch1. [DOI] [PubMed] [Google Scholar]
- Andachi Y., Yamao F., Muto A., Osawa S. Codon recognition patterns as deduced from sequences of the complete set of transfer RNA species in Mycoplasma capricolum. Resemblance to mitochondria. J Mol Biol. 1989 Sep 5;209(1):37–54. doi: 10.1016/0022-2836(89)90168-x. [DOI] [PubMed] [Google Scholar]
- Boorstein W. R., Craig E. A. Primer extension analysis of RNA. Methods Enzymol. 1989;180:347–369. doi: 10.1016/0076-6879(89)80111-9. [DOI] [PubMed] [Google Scholar]
- Brown J. W., Hunt D. A., Pace N. R. Nucleotide sequence of the 10Sa RNA gene of the beta-purple eubacterium Alcaligenes eutrophus. Nucleic Acids Res. 1990 May 11;18(9):2820–2820. doi: 10.1093/nar/18.9.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan A. K., Apirion D. The gene for a small stable RNA (10Sa RNA) of Escherichia coli. Mol Microbiol. 1989 Nov;3(11):1481–1485. doi: 10.1111/j.1365-2958.1989.tb00133.x. [DOI] [PubMed] [Google Scholar]
- Francklyn C., Schimmel P. Aminoacylation of RNA minihelices with alanine. Nature. 1989 Feb 2;337(6206):478–481. doi: 10.1038/337478a0. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983 Dec;35(3 Pt 2):849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- Hou Y. M., Schimmel P. A simple structural feature is a major determinant of the identity of a transfer RNA. Nature. 1988 May 12;333(6169):140–145. doi: 10.1038/333140a0. [DOI] [PubMed] [Google Scholar]
- Inokuchi H., Yamao F., Sakano H., Ozeki H. Identification of transfer RNA suppressors in Escherichia coli. I. Amber suppressor su+2, an anticodon mutant of tRNA2Gln. J Mol Biol. 1979 Aug 25;132(4):649–662. doi: 10.1016/0022-2836(79)90380-2. [DOI] [PubMed] [Google Scholar]
- Jain S. K., Gurevitz M., Apirion D. A small RNA that complements mutants in the RNA processing enzyme ribonuclease P. J Mol Biol. 1982 Dec 15;162(3):515–533. doi: 10.1016/0022-2836(82)90386-2. [DOI] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Komine Y., Inokuchi H. Physical map locations of the genes that encode small stable RNAs in Escherichia coli. J Bacteriol. 1991 Sep;173(17):5252–5252. doi: 10.1128/jb.173.17.5252.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushner S. R., Nagaishi H., Templin A., Clark A. J. Genetic recombination in Escherichia coli: the role of exonuclease I. Proc Natl Acad Sci U S A. 1971 Apr;68(4):824–827. doi: 10.1073/pnas.68.4.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnab R. M. Proton-driven bacterial flagellar motor. Methods Enzymol. 1986;125:563–581. doi: 10.1016/s0076-6879(86)25046-6. [DOI] [PubMed] [Google Scholar]
- McClain W. H., Chen Y. M., Foss K., Schneider J. Association of transfer RNA acceptor identity with a helical irregularity. Science. 1988 Dec 23;242(4886):1681–1684. doi: 10.1126/science.2462282. [DOI] [PubMed] [Google Scholar]
- McClain W. H., Guerrier-Takada C., Altman S. Model substrates for an RNA enzyme. Science. 1987 Oct 23;238(4826):527–530. doi: 10.1126/science.2443980. [DOI] [PubMed] [Google Scholar]
- McClain W. H., Guerrier-Takada C., Altman S. Model substrates for an RNA enzyme. Science. 1987 Oct 23;238(4826):527–530. doi: 10.1126/science.2443980. [DOI] [PubMed] [Google Scholar]
- Miczak A., Chauhan A. K., Apirion D. Two new genes located between 2758 and 2761 kilobase pairs on the Escherichia coli genome. J Bacteriol. 1991 Jun;173(11):3271–3272. doi: 10.1128/jb.173.11.3271-3272.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh B. K., Apirion D. 10Sa RNA, a small stable RNA of Escherichia coli, is functional. Mol Gen Genet. 1991 Sep;229(1):52–56. doi: 10.1007/BF00264212. [DOI] [PubMed] [Google Scholar]
- Ray B. K., Apirion D. Characterization of 10S RNA: a new stable rna molecule from Escherichia coli. Mol Gen Genet. 1979 Jul 2;174(1):25–32. doi: 10.1007/BF00433301. [DOI] [PubMed] [Google Scholar]
- Retallack D. M., Johnson L. L., Friedman D. I. Role for 10Sa RNA in the growth of lambda-P22 hybrid phage. J Bacteriol. 1994 Apr;176(7):2082–2089. doi: 10.1128/jb.176.7.2082-2089.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakano H., Yamada S., Ikemura T., Shimura Y., Ozeki H. Temperature sensitive mutants of Escherichia coli for tRNA synthesis. Nucleic Acids Res. 1974 Mar;1(3):355–371. doi: 10.1093/nar/1.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson J. R., Uhlenbeck O. C. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc Natl Acad Sci U S A. 1988 Feb;85(4):1033–1037. doi: 10.1073/pnas.85.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M., Cameron J. R., Davis R. W. Viable molecular hybrids of bacteriophage lambda and eukaryotic DNA. Proc Natl Acad Sci U S A. 1974 Nov;71(11):4579–4583. doi: 10.1073/pnas.71.11.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi J. S., Kinger A. K. Identification of the 10Sa RNA structural gene of Mycobacterium tuberculosis. Nucleic Acids Res. 1992 Jan 11;20(1):138–138. doi: 10.1093/nar/20.1.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]