Abstract
During sexual development, filamentous ascomycetes form complex, three-dimensional fruiting bodies for the generation and dispersal of spores. In previous studies, we identified genes with evolutionary conserved expression patterns during fruiting body formation in several fungal species. Here, we present the functional analysis of two developmentally up-regulated genes, chs7 and sec22, in the ascomycete Sordaria macrospora. The genes encode a class VII (division III) chitin synthase and a soluble N-ethylmaleimide-sensitive-factor attachment protein receptor (SNARE) protein, respectively. Deletion mutants of chs7 had normal vegetative growth and were fully fertile but showed sensitivity toward cell wall stress. Deletion of sec22 resulted in a reduced number of ascospores and in defects in ascospore pigmentation and germination, whereas vegetative growth was normal in the mutant. A SEC22-EGFP fusion construct under control of the native sec22 promoter and terminator regions was expressed during different stages of sexual development. Expression of several development-related genes was deregulated in the sec22 mutant, including three genes involved in melanin biosynthesis. Our data indicate that chs7 is dispensable for fruiting body formation in S. macrospora, whereas sec22 is required for ascospore maturation and germination and thus involved in late stages of sexual development.
Keywords: sexual development, fruiting body, chitin synthase, SNARE protein, Sordaria macrospora
Sexual development in filamentous ascomycetes involves the differentiation of fruiting bodies containing a number of specialized cell types that are not present in the vegetative mycelium (Bistis et al. 2003; Lord and Read 2011). During the last decades, much progress has been made in identifying the mechanisms that control this differentiation process at the molecular level, but a unified model has yet to emerge (Pöggeler et al. 2006a). The morphologic changes that occur during fruiting body formation are accompanied by drastic changes in gene expression that are thought to orchestrate the spatiotemporal sequence of events that finally lead to the differentiation and dispersal of ascospores. Not all genes that are differentially expressed during fruiting body development, however, are actually essential for this process, and one way to identify genes that might play an important role in differentiation is the analysis of evolutionary conserved expression patterns, because conserved expression can be a strong indicator of functional significance (Brawand et al. 2011; Lehr et al. 2014; Nowrousian 2014; Romero et al. 2012; Stuart et al. 2003). In previous studies, we have compared gene expression during fruiting body development in Sordaria macrospora and several other ascomycetes, namely Neurospora crassa (Gesing et al. 2013; Nowrousian 2009), Fusarium graminearum (Gesing et al. 2012), and Pyronema confluens (Nowrousian and Kück 2006; Traeger et al. 2013). In subsequent functional studies, a number of genes with conserved up-regulated expression during fruiting body formation were found to be involved in this process, indicating that conservation of expression is a suitable criterion to select candidate genes for downstream analyses (Gesing et al. 2013; Nowrousian 2009; Schindler and Nowrousian 2014).
Here, we present the functional analysis of two additional genes, chs7 and sec22, that were found previously to be up-regulated during sexual development in several fungi (Gesing et al. 2012, 2013). The role of both genes during fruiting body differentiation was analyzed in the ascomycete S. macrospora, a model organism for the study of sexual development (Engh et al. 2010; Teichert et al. 2014). chs7 encodes a chitin synthase (CHS), and sec22 encodes a SNARE (soluble N-ethylmaleimide-sensitive-factor attachment protein receptor) protein. CHS are important proteins for cell-wall biosynthesis and hyphal morphogenesis in fungi. They mediate polymerization of chitin, a major structural component of the fungal cell wall, from the monomer N-acetyl-glucosamine (Riquelme and Bartnicki-García 2008; Roncero 2002). Ascomycetes encode up to nine chitin synthase (chs) genes in their genomes, with Schizosaccharomyces pombe containing only one, the Saccharomycetes three to seven, and filamentous Ascomycetes four to nine genes (Pacheco-Arjona and Ramirez-Prado 2014; Riquelme and Bartnicki-García 2008). Studies of chs genes in several ascomycetes have indicated different roles for individual CHS; thus, the expansion of chs genes in filamentous ascomycetes might reflect specific needs for chitin biosynthesis capacities during different phases of the life cycle of the respective species (Kong et al. 2012; Martín-Udíroz et al. 2004; Roncero 2002; Sheng et al. 2013). The S. macrospora chs7 (SMAC_01722) gene is orthologous to the N. crassa chs-6 (NCU05268) gene, and both are strongly up-regulated during sexual development in these fungi; in addition, the S. macrospora chs7 gene is down-regulated in the sterile developmental mutant pro1 (Gesing et al. 2013; Teichert et al. 2012). Furthermore, a N. crassa chs-6 deletion mutant showed delayed sexual development and reduced sporulation as a female partner in crosses (Gesing et al. 2013); therefore, we chose chs7 as a candidate for functional analysis in S. macrospora.
The second gene that was analyzed is the putative SNARE protein-encoding sec22 (SMAC_06625). SNARE proteins are membrane-associated proteins that are required for intracellular membrane fusions that occur during vesicle trafficking between organelles or to and from the cell surface (Lee et al. 2004; Rossi et al. 2004; Ungar and Hughson 2003). Sec22p in the yeast Saccharomyces cerevisiae is involved in anterograde and retrograde trafficking of vesicles between the endoplasmic reticulum (ER) and the Golgi (Lewis et al. 1997; Liu and Barlowe 2002; Spang and Shekman 1998) and was shown to be important in autophagy and the selective accumulation of caesium ions (Dräxl et al. 2012; Nair et al. 2011). The role of sec22 homologs during fruiting body development has not been studied yet; however, formation of fruiting bodies requires transport of large amounts of nutrients from the vegetative mycelium to the developing fruiting bodies (Pöggeler et al. 2006a); therefore, a role for genes involved in vesicle trafficking might be envisioned. sec22 was found to be transcriptionally up-regulated during sexual development in S. macrospora and F. graminearum (Gesing et al. 2012; Qi et al. 2006) and was thus included in our study.
Materials and Methods
Strains and culture conditions
S. macrospora strains used in this study are given in Table 1. Unless stated otherwise, standard growth conditions and transformation protocols for S. macrospora were as described (Dirschnabel et al. 2014; Esser 1982; Nowrousian et al. 1999). For the analysis of sexual development, S. macrospora was grown on complete medium biomalt maize medium (BMM) (Esser 1982) or minimal medium Sordaria Westergaards medium (SWG) (Nowrousian et al. 2005). For stress-tolerance tests, S. macrospora was grown on SWG with stated stressors that were supplemented to the medium after sterilization. Growth radius was measured after 2 d under standard growth conditions or different temperatures. Mean and SDs were calculated from three technical replicates for each independent biological replicate. For RNA from cultures developing fruiting bodies, S. macrospora was grown at 25° in SWG in surface cultures as described (Nowrousian et al. 2005).
Table 1. Sordaria macrospora strains used in this study.
| Strain | Genotype | Reference |
|---|---|---|
| Wild type (S91327) | Wild type | Strain collectiona |
| fus1-1 (S84595) | Spore color mutant | (Nowrousian et al. 2012) |
| ∆ku70 (S96888) | ∆ku70 | (Pöggeler and Kück 2006) |
| ∆sec22 (S121397) | ∆sec22 | This study |
| SEC22-GFP-NA (ST13.1) | ∆sec22::Psec22::sec22-gfp | This study |
| SEC22-GFP-OE (S119860) | ∆sec22::Pgpd::sec22-gfp | This study |
| ∆chs7-1 (S123091) | ∆chs7-1 | This study |
| ∆chs7-2 (S123107) | ∆chs7-2 | This study |
| ∆chs7-3 (S123114) | ∆chs7-3 | This study |
| ∆chs7-4 (S123147) | ∆chs7-4 | This study |
All strains are single ascospore isolates.
Strain collection at the Department of General and Molecular Botany, Bochum.
Preparation of nucleic acids and reverse-transcription quantitative polymerase chain reaction (RT-qPCR)
DNA and RNA were extracted using standard protocols (Pöggeler et al. 1997; Yarden et al. 1992). RT-qPCR was performed as described previously (Schindler and Nowrousian 2014), oligonucleotides used as primers are given in Supporting Information, Table S1.
Generation of ∆chs7 mutants
For the generation of plasmids and homologous recombination in S. cerevisiae PJ69-4a, standard procedures were used (Bloemendal et al. 2012; Colot et al. 2006; James et al. 1996; Sambrooke and Russell 2001). For the chs7 knockout construct, the flanking regions of chs7 (SMAC_01722) were amplified with specific oligonucleotides [5′ region 1722-5-fw/rv (965 bp), 3′ region 1722-3-fw/rv (967 bp), Table S1] using PCR based on wild-type genomic DNA; the Hygromycin resistance cassette was obtained from vector pDrivehph (pSF27-34) (Nowrousian and Cebula 2005) by EcoRI digestion; and all fragments were inserted into pRS426 (Christianson et al. 1992) by homologous recombination in yeast. The resulting plasmid p∆chs7 was used as template to amplify the 3.4-kb knockout cassette with primers 1722-5-fw/-3-rv in a Phusion DNA Polymerase (Thermo Scientific)-PCR. The knockout cassette was used to transform S. macrospora strain Δku70 (Pöggeler and Kück 2006). Primary transformants were crossed against the fus1-1 strain to generate ∆chs7 strains without the Δku70 background, and transformants were screened for homologous integration by PCR and Southern blot analysis as described previously (Nowrousian and Cebula 2005) (Figure S1). For further analyses four strains (∆chs7-1: S123091; ∆chs7-2: S123107; ∆chs7-3: S123114; ∆chs7-4: S123147) were chosen.
Generation of ∆sec22 mutants and complementation
Generation of the sec22 deletion construct p∆sec22 was performed as described previously by the use of oligonucleotides 6625-5-fw/rv and 6625-3-fw/rv (Table S1) to amplify fragments of 1 kb upstream and 0.5 kb downstream of the sec22 open reading frame. The 3.4-kb knockout cassette was amplified from the resulting plasmid p∆sec22 with oligonucleotides 6625-5-fw/-3-rv and used to transform S. macrospora strain Δku70. Transformants were screened for homologous integration by PCR and Southern blot analysis as described previously (Nowrousian and Cebula 2005). Primary transformants were crossed against the fus1-1 strain to generate ∆sec22 strains without the Δku70 background, and transformants were screened for homologous integration by PCR and Southern blot analysis as described previously (Nowrousian and Cebula 2005).We were not able to isolate a single spore without the sec22 gene still detectable via PCR. Therefore we decided to follow a sheltered rescue plan to investigate if sec22 is an essential gene for S. macrospora. Overexpression construct pSEC22-GFP-OE with sec22::egfp under the control of the Aspergillus nidulans gpd promotor was cloned and ectopically integrated into the wild-type strain. An ascospore isolate with one copy of the overexpression plasmid was used for further crosses with transformants carrying the knockout construct at the homologous locus to establish a transformant carrying both the deletion background of sec22 and an ectopically integrated overexpression construct. One of these strains (S119860) was crossed against the fus1-1 strain to obtain three ∆sec22 strains, which were confirmed as deletion strains via PCR and Southern blot analysis (Figure S2). For complementation, plasmid pSEC22-GFP-NA was cloned with sec22::egfp under the native sec22 promotor and terminator. Plasmid pSEC22-GFP-NA was transformed in ∆sec22 (S121397), and single spore isolate SEC22-GFP-NA (ST13.1) was used for further analyses.
Microscopic analyses
To investigate differentiation, S. macrospora strains were grown on slides with 1 mL of BMM medium at 27° in continuous light (Engh et al. 2007b). Hyphal fusion assays were performed as described previously (Bloemendal et al. 2012). Light and fluorescence microscopy were performed as described previously (Dirschnabel et al. 2014; Engh et al. 2007b).
Ascospore quantification analyses
Germination analyses of ascospores were performed as described previously (ascospore germination assay of selfing strains) (Dirschnabel et al. 2014). For quantification of released ascospores and their pigmentation, Petri dish lids of three technical replicates per independent biological replicate grown on BMM were analyzed. Ascospores were washed off with distilled water, counted by hematocytometer, and analyzed for pigmentation.
Results
CHS7 belongs to division III of CHS
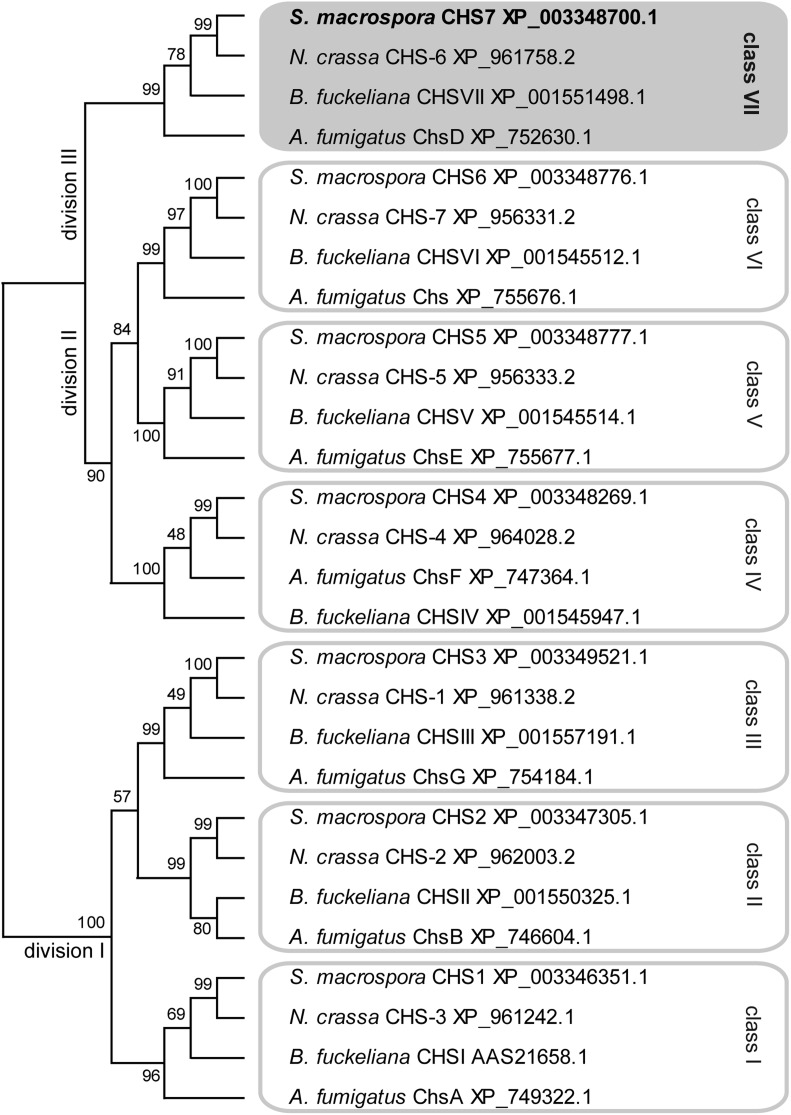
To characterize the predicted CHS SMAC_01722 and other CHS in S. macrospora, we used BLAST searches (Altschul et al. 1997) in the S. macrospora predicted peptides and identified seven putative CHS that are orthologous to the seven N. crassa CHS (Borkovich et al. 2004; Riquelme and Bartnicki-García 2008) (Figure 1). CHS in ascomycetes can be divided into seven classes according to sequence similarity and domain structure. Two nomenclature systems are in use that are similar for classes I to V but differ in that classes VI and VII are exchanged (Choquer et al. 2004; Mandel et al. 2006), going back to the assignment of class VI to two different types of CHS at almost the same time (Chigira et al. 2002; Roncero 2002). In this study, we use the nomenclature by Choquer et al. (2004), where class VI consists of CHS with a myosin-like domain that is not present in class VII CHS. Classes of CHS can be grouped into divisions by phylogenetic analysis, with division I consisting of classes I, II, and III, which are characterized by an additional catalytic subdomain pfam08407 (Fajardo-Somera et al. 2015; Riquelme and Bartnicki-García 2008). Division II contains the classes IV, V, and VI, which carry a cytochrome b5-like domain, with classes V and VI carrying an additional myosin-head-like domain (Choquer et al. 2004). Division III consists of class VII CHS that have the simplest domain structure and carry no other domains besides the conserved CHS catalytic domain (pfam03142) present in all CHS (Choquer et al. 2004; Riquelme and Bartnicki-García 2008). A phylogenetic analysis showed that the seven S. macrospora CHS represent the seven CHS classes, similar to N. crassa and several other filamentous ascomycetes (Figure 1). The developmentally upregulated SMAC_01722 (XP_003348700.1) encodes a CHS that belongs to class VII, division III; therefore, the gene was named chs7.
Figure 1.
Phylogenetic analysis of chitin synthases from four ascomycetes. A multiple alignments was created with CLUSTALW (Thompson et al. 2002) and used for maximum likelihood analysis with MEGA5.10 (Tamura et al. 2011). Numbers at branches indicate bootstrap support in % for 1000 bootstrap replications. Classes and divisions are labeled according to Choquer et al. (2004) and Riquelme and Bartnicki-García (2008), respectively. The predicted S. macrospora Chitin synthases (CHS) proteins are encoded by genes with the following locus tag numbers in the genome sequence (Nowrousian et al. 2010): CHS1, SMAC_07828; CHS2, SMAC_07162; CHS3, SMAC_03109; CHS4, SMAC_02767; CHS5, SMAC_01800; CHS6, SMAC_01799; CHS7, SMAC_01722.
A chs7 deletion mutant is fertile but sensitive to cell-wall stress
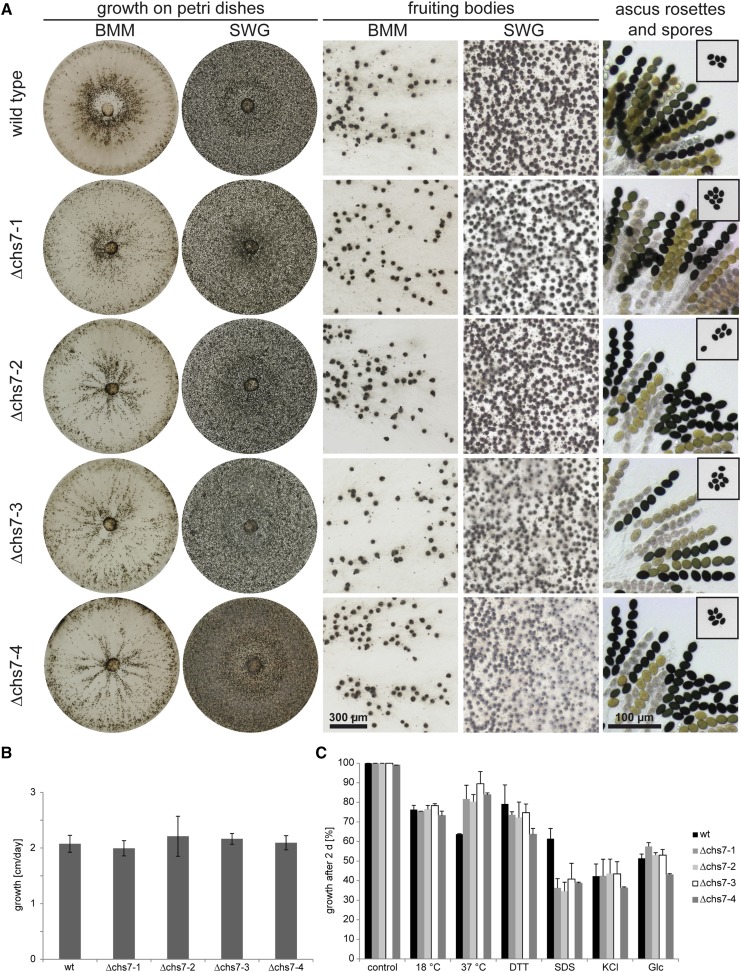
To test whether chs7 is involved in fruiting body formation in S. macrospora, we generated deletion mutants by homologous recombination (Figure S1). Four Δchs7 strains (Δchs7-1 to Δchs7-4) were analyzed for phenotypes related to sexual development, but all were fully fertile and produced perithecia, asci and ascospores in similar numbers as the wild type (Figure 2A). The growth rate of the vegetative mycelium also was unaffected in the Δchs7 mutants (Figure 2B). Chitin is an important structural component of the fungal cell wall, and CHS mutants in several fungi were reported to be sensitive to stress conditions, including cell wall stressors or osmotic stress (Kong et al. 2012; Martín-Udíroz et al. 2004; Sheng et al. 2013). Therefore, we tested growth of the Δchs7 strains under different stress conditions (Figure 2C). At 18°, growth of all strains was reduced compared to the standard growth temperature of 25°, but the reduction was similar to that of the wild type. Somewhat surprisingly, growth at 37° is less affected in the Δchs7 strains than in the wild type, indicating a greater heat stress tolerance in the mutant strains. Osmotic stress induced by KCl or glucose led to a similar growth reduction in the wild-type and the mutant strains, and the same was observed for the ER stress-inducing agent dithiothreitol (DTT); however, cell-wall stress induced by sodium dodecyl sulfate (SDS) in the medium led to much stronger growth reduction in the four Δchs7 deletion strains than in the wild type (Figure 2C). This finding might indicate that the Δchs7 strains have a reduced chitin content or modified cell-wall composition, as was described for mutants in the Aspergillus fumigatus homolog chsD (Mellado et al. 1996).
Figure 2.
Analysis of sexual development, vegetative growth, and stress resistance in Δchs7 strains. (A) Sexual development of the wild-type and four Δchs7 strains after 7 d on complete medium (BMM) and minimal medium (SWG). All strains form perithecia and ascus rosettes (right column), and eject ascospores toward the lids of the Petri dishes (spores on lids shown in small boxes in right column). (B) Vegetative growth was analyzed on BMM in race tubes over 10 d, error bars give SDs of three independent biological replicates. (C) Stress resistance was analyzed on SWG in Petri dishes with the indicated stress-inducing agents at the following concentrations: dithiothreitol (DTT) 0.04%, sodium dodecyl sulfate (SDS) 0.1%, KCl 0.6 M, glucose 0.6 M. The wild type was used as reference for the control. For each stress-inducing condition, the growth on the control plates for the corresponding strain was used as reference. Error bars indicate SDs for two independent biological replicates, each with three technical replicates.
A sec22 deletion mutant has defects in ascospore maturation and germination
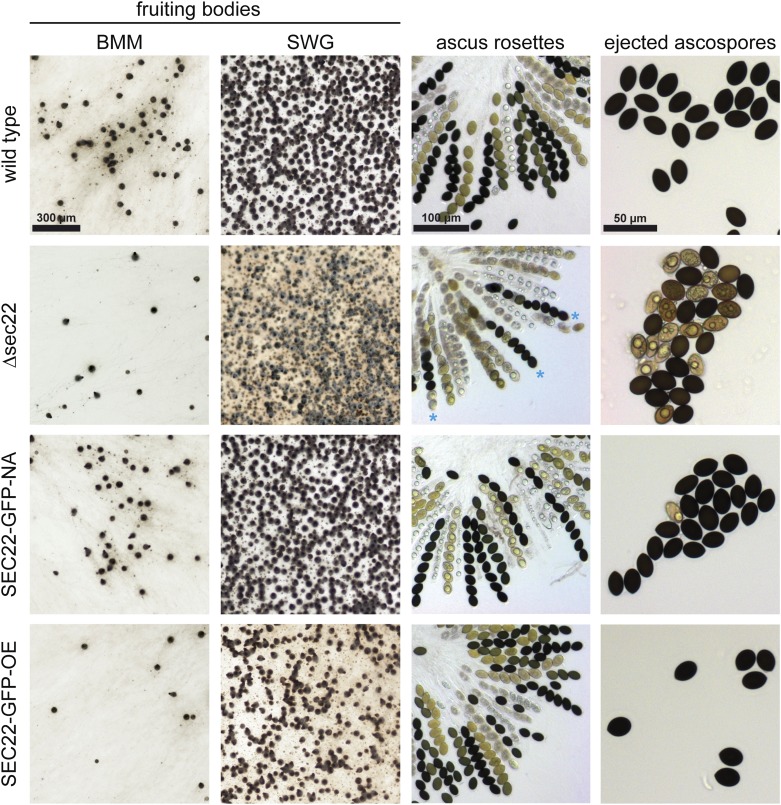
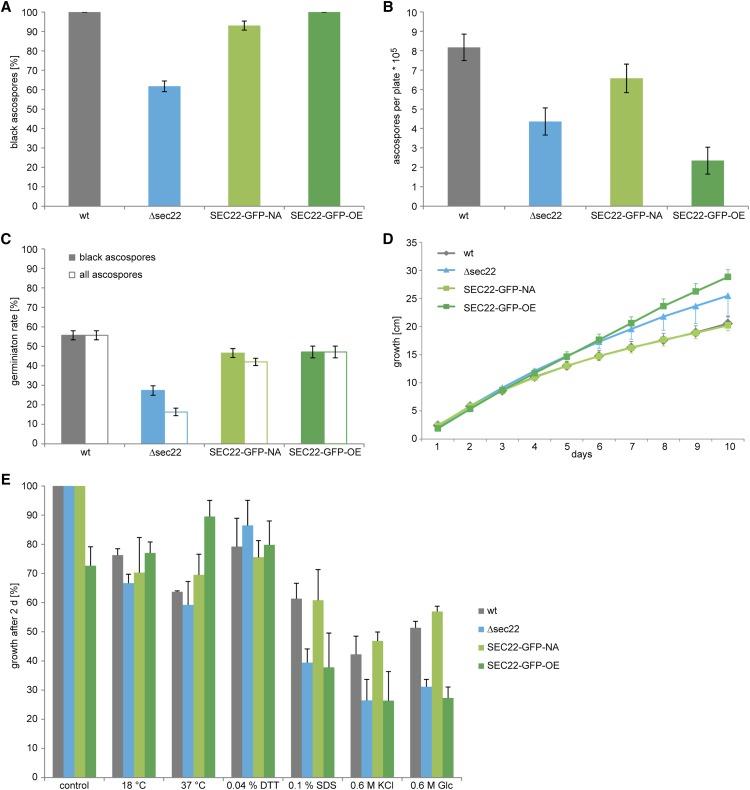
The developmentally up-regulated S. macrospora sec22 (SMAC_06625, XP_003346158.1) encodes a conserved protein with the typical domain structure described for this class of SNARE proteins (Rossi et al. 2004; Ungar and Hughson 2003) (Figure S3). To test whether sec22 plays a role in sexual development, we generated a sec22 deletion mutant (Figure S2). The mutant forms normal fruiting bodies and eight-spored asci; however, ascospore pigmentation is different from the wild type (Figure 3). In the wild type, the eight ascospores within an ascus usually display the same degree of maturation, which can be seen in the degree of pigmentation from immature, nonpigmented to mature, completely melanized spores. In the mutant strain, spores within an ascus frequently show different degrees of pigmentation, i.e., light-brown and fully melanized spores reside within one ascus (Figure 3). Furthermore, the mutant ejects ascospores that are not fully mature, in contrast to the wild type, where only fully melanized ascospores are released from the perithecia (Figure 3). Quantification of ascospores showed that the mutant ejects only about 60% of black ascospores compared to the wild type (Figure 4A), and that the overall number of ejected ascospores is also reduced by nearly 50% (Figure 4B). Furthermore, the germination rate of the ascospores from the Δsec22 mutant is reduced. Only the fully melanized ascospores from the mutant were able to germinate, whereas none of the not fully pigmented spores germinated. However, even the fully melanized ascospores from the mutant showed a lower germination rate than those of the wild type (Figure 4C), indicating other spore maturation or germination defects besides pigmentation.
Figure 3.
Sexual development in the Δsec22 mutant and complemented strains. Strain SEC22-GFP-NA expresses sec22 under its own promoter and terminator regions, whereas in SEC22-GFP-OE sec22 is under control of the strong, constitutive A. nidulans gpd promoter. Fruiting body formation was analyzed after 7 d on complete medium (BMM) and minimal medium (SWG). Ascus rosettes from perithecia grown on BMM medium were analyzed after 7−9 d. Ascospores from the Δsec22 strain show different degrees of melanization within one ascus (blue asterisks). Ascospores that were ejected toward the lid of the plate from strains grown on BMM were analyzed after 7−9 d (right column).
Figure 4.
Analysis of ascospore production and germination, and stress resistance in the Δsec22 mutant and complemented strains. Strains are the same as in Figure 3. (A) Proportion of black ascospores that are ejected from perithecia. Error bars give standard deviations for three independent biological replicates. (B) Number of ejected ascospores per Petri dish. Error bars give SDs for three independent biological replicates. (C) Germination rate of ascospores. The wild type and strain SEC22-GFP-OE eject only black spores; therefore, the germination rate is identical for black and all ascospores. Strains Δsec22 and SEC-22-NA eject a certain proportion of nonmelanized ascospores (A), none of which were observed to germinate, thereby reducing the germination rate of all ascospores. Error bars give standard deviations of three independent biological replicates. (D) Vegetative growth was analyzed on race tubes over 10 d. Error bars give SDs of three independent biological replicates, for clarity, only one direction (plus or minus) is shown for each error bar. (E) Stress resistance was analyzed on SWG in petri dishes with the indicated stress-inducing agents at the following concentrations: dithiothreitol (DTT) 0.04%, sodium dodecyl sulfate (SDS) 0.1%, KCl 0.6 M, glucose 0.6 M. The wild type was used as reference for the control. For each stress-inducing condition, the growth on the control plates for the corresponding strain was used as reference. Error bars indicate SDs for two independent biological replicates, each with three technical replicates.
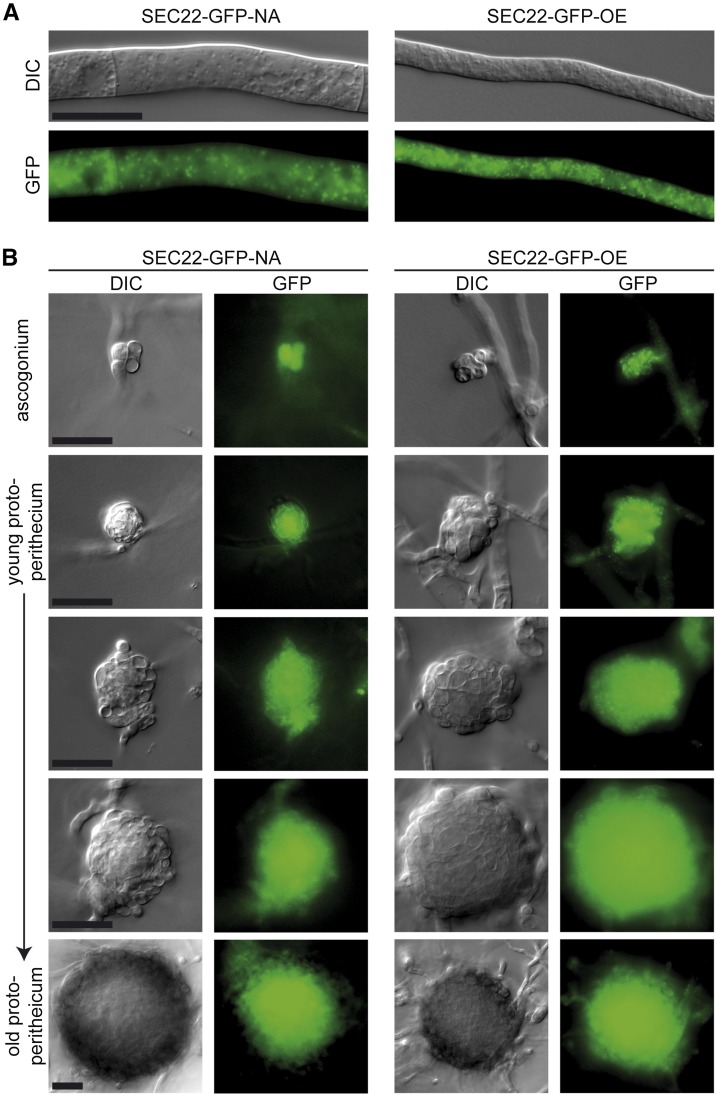
Two complementation plasmids were constructed for sec22, one expressing a sec22-egfp fusion gene under its own promoter and terminator, and the other an overexpression construct with sec22-egfp under control of a constitutive promoter. Both constructs led to EGFP fluorescence in dots within the cytoplasm of hyphae of complemented strains (Figure 5A), and fluorescence in developing fruiting bodies was strong with both constructs (Figure 5B). Expression of sec22 was detectable at transcript level in both strains, with the expression from the overexpression construct stronger, and from the native construct somewhat lower than in the wild type (Figure 6). The spore pigmentation and germination defects of the Δsec22 mutant were complemented in the mutant strain carrying the construct that expresses sec22 under its own promoter and terminator regions (strain SEC22-GFP-NA, Figure 3 and Figure 4). This strain produces mostly fully pigmented ascospores, and germination of these spores is in the same range as for the wild type. Interestingly, a mutant strain carrying the construct overexpressing sec22 (strain SEC22-GFP-OE) shows full restoration of ascospore pigmentation and germination, but the number of ascospores that are produced is more similar to the mutant than to the wild type (Figure 3 and Figure 4). One reason for this finding might be a reduced number of fruiting bodies that are produced in BMM medium by the Δsec22 mutant and the overexpressing strain, which is the medium that was used for the ascospore quantification and germination assays (Figure 3).
Figure 5.
Microscopic analysis of SEC22-EGFP fusion proteins shows that sec22 is expressed throughout the developmental cycle. Strain SEC22-GFP-NA expresses sec22 under its own promoter and terminator regions, whereas in SEC22-GFP-OE sec22 is under control of the strong, constitutive A. nidulans gpd promoter. EGFP fluorescence was observed in vegetative hyphae (A) and developing ascogonia and protoperithecia (B). Scale bars indicate 20 µm.
Figure 6.
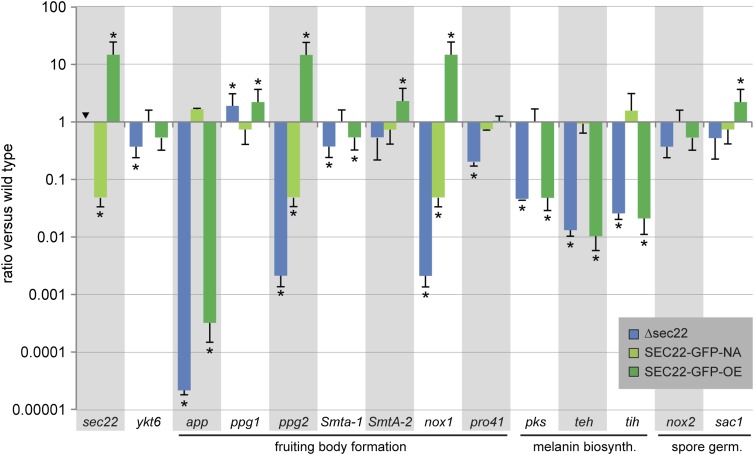
Expression of developmentally regulated genes in the Δsec22 mutant and complemented strains. Strains are the same as in Figure 3. Transcript levels determined by RT-qPCR are given relative to the wild type; the mean of two independent biological replicates is shown. Error bars indicate SD (positive values; for better visualization standard deviations for negative expression ratios are shown in the negative direction). For sec22, no expression was detected in the sec22 deletion mutant as expected (indicated by an inverted triangle). Asterisks indicate significantly differential expression (P ≤ 0.001, calculated with REST) (Pfaffl et al. 2002).
We also investigated vegetative growth and stress tolerance of Δsec22 and complemented strains. Vegetative growth was similar to the wild type for all strains, with the deletion and overexpression strains showing slightly but not significantly faster growth (Figure 4D). When grown under stress-inducing conditions, the mutant as well as the overexpression strain showed growth reduction under cell-wall stress induced by sodium dodecyl sulfate, and osmotic stress induced by glucose, and to a lesser degree by KCl, whereas the complemented strain SEC22-GFP-NA had wild type-like growth (Figure 4E).
sec22 is required for correct expression of several development-associated genes
Because sec22 is necessary for full fertility, we wondered whether the deletion of sec22 has an effect on the expression of other genes known to be involved in or associated with sexual development in S. macrospora. RT-qPCR was performed for 12 development-associated genes from S. macrospora as well as for ykt6 (SMAC_01930), because ykt6 in S. cerevisiae was shown to be upregulated and able to functionally replace sec22 in a sec22 mutant strain (Liu and Barlowe 2002). In contrast to S. cerevisiae, however, ykt6 is down-regulated in the Δsec22 mutant of S. macrospora (Figure 6), and the ascospore maturation and germination phenotype of the Δsec22 mutant shows that ykt6 cannot functionally replace sec22 in S. macrospora.
Several genes that are required or associated with fruiting body formation were tested for expression in Δsec22 and the complemented strains (Figure 6). The two mating-type genes Smta-1 and SmtA-2 that are essential for fruiting body formation (Klix et al. 2010; Pöggeler et al. 2006b) are slightly down-regulated in the sec22 mutant, although only expression of Smta-1 is significantly different from the wild type. In addition to the mating-type genes, the two pheromone genes ppg1 and ppg2 are involved in sexual development (Mayrhofer et al. 2006). Interestingly, while ppg1 is slightly up-regulated, ppg2 is strongly down-regulated in Δsec22. One might speculate that the different expression patterns of the pheromone genes in Δsec22 are related to the different modes of secretion of the corresponding pheromones. While the ppg1-derived pheromone is similar to the S. cerevisiae α-factor, and therefore most likely secreted via the classical ER-based secretion pathway, the ppg2-derived pheromone is similar to the S. cerevisiae a-factor, which is secreted by an ATP-binding cassette transporter (Mayrhofer and Pöggeler 2005). Thus, secretion of the PPG1 pheromone might involve a SEC22-dependent pathway, and it might be hypothesized that autoregulatory feedback leads to upregulation of ppg1 expression when this pathway is not fully functional. Secretion of the PPG2 pheromone is unlikely to involve SEC22, and therefore the down-regulation of ppg2 is probably a more indirect effect.
The pro41 gene that encodes an ER membrane protein essential for fruiting body formation (Nowrousian et al. 2007a) is also down-regulated in the sec22 mutant. Interestingly, the PRO41 orthologs BcNoxD and PaNoxD from Botrytis cinerea and Podospora anserina, respectively, colocalize and/or interact with an NADPH oxidase (NOX) at the ER and vacuolar membranes as part of a NOX complex (Lacaze et al. 2015; Siegmund et al. 2015). The NOX proteins that are part of this complex are PaNox1 in P. anserina, and BcNoxA in B. cinerea, which are homologs of NOX1 in S. macrospora (Dirschnabel et al. 2014; Malagnac et al. 2004; Siegmund et al. 2013), and nox1 is also strongly down-regulated in the Δsec22 mutant (Figure 6). Thus, one might hypothesize that genes for ER proteins or proteins that require ER-dependent export are misregulated in Δsec22, probably due to feedback reactions caused by insufficient or de-regulated transport events.
This hypothesis would also explain the down-regulation of the three melanin biosynthesis genes pks, teh, and tih (Figure 6). Melanin is present in the cell wall of many fungal structures, and its intermediates and/or biosynthetic enzymes are thought to require vesicle-based transport (Eisenman and Casadevall 2012). In S. macrospora, melanin is required for the black pigmentation of perithecia and ascospores, and the melanin genes are up-regulated during sexual development (Engh et al. 2007a; Nowrousian et al. 2012). The down-regulation of the melanin biosynthesis genes in Δsec22 correlates well with the ascospore pigmentation defects in the mutant; however, in the overexpression strain SEC22-GFP-OE, the genes are also down-regulated (Figure 6), despite the normal pigmentation in this strain (Figure 3). One possible explanation for this finding might be that the bottleneck of melanin biosynthesis is not the level of transcription of the biosynthesis genes, but ER-related transport processes. If this were the case, transcriptional down-regulation in the Δsec22 strain might be caused by feedback due to ER stress, whereas in the overexpression strain, it might be possible that more efficient transport leads to similar transcriptional down-regulation through product-based inhibition.
One other gene that was tested is the development-associated app gene, which encodes a protein that accumulates in fruiting bodies in S. macrospora and N. crassa. Previously, app was found to be down-regulated in mutants with a block at the stage of young fruiting bodies (protoperithecia), but normal in a developmental mutant that produced fruiting bodies, but no mature spores (Nowrousian et al. 2007b). Therefore, it was surprising that app is strongly down-regulated in the sec22 mutant, even though the mutant produces fruiting bodies (Figure 6). However, in a recent analysis of a polyketide synthase mutant that only produces protoperithecia, it was found that app was expressed normally in this mutant and down-regulated in overexpressing strains, indicating that app expression is regulated by at least two different developmental pathways (Schindler and Nowrousian 2014), which could also explain down-regulation of app in the fertile sec22 mutant.
We also analyzed transcript levels of nox2 and sac1, both of which are involved in ascospore germination in S. macrospora (Dirschnabel et al. 2014; Kamerewerd et al. 2008); however, both genes are expressed normally in Δsec22 (Figure 6).
Discussion
chs7 orthologs play different roles during growth and development in different species
In this study, we found that the S. macrospora chs7 is not required for sexual development but instead plays a role under stress conditions. Interestingly, a deletion mutant of the N. crassa ortholog chs-6 has a very different phenotype. It is strongly reduced in vegetative growth (Fajardo-Somera et al. 2015; Sánchez-León et al. 2011), in contrast to the S. macrospora Δchs7 strain, which grows normally (Figure 2). Furthermore, the N. crassa Δchs-6 is delayed in sexual development and sporulation when present as the female partner in a cross and forms fewer perithecia and no ascospores in homozygous crosses (Fajardo-Somera et al. 2015; Gesing et al. 2013), whereas the S. macrospora Δchs7 has wild type-like sexual development (Figure 2). Thus, these orthologous genes have evolved different functions in these relatively closely related species. However, even though orthologs from S. macrospora and N. crassa often have similar functions, different roles for ortholog pairs in these species have been observed previously. Examples are the orthologous transcription factor genes pro1 and adv-1 that are involved in sexual development, but not in vegetative growth, in S. macrospora, and in sexual development as well as vegetative growth in N. crassa (Colot et al. 2006; Masloff et al. 1999), and pro45 and its N. crassa ortholog ham-4, which are required for sexual development in S. macrospora but not N. crassa (Nordzieke et al. 2015; Simonin et al. 2010). In the case of CHS, the presence of multiple paralogs in the genomes of most ascomycetes might facilitate rapid evolution leading to functional differentiation (Pacheco-Arjona and Ramirez-Prado 2014; Riquelme and Bartnicki-García 2008). In Magnaporthe oryzae, which is a member of the Sordariomycetes, but only distantly related to S. macrospora and N. crassa, the chs7 ortholog is required for the formation of appressoria from germ tubes on hydrophobic surfaces, and for penetration and invasive growth during plant infection (Kong et al. 2012). However, sexual development in the corresponding mutant has not yet been studied in M. oryzae. In the Eurotiomycete Aspergillus nidulans, the chs7 ortholog chsG is not involved in vegetative growth or sexual development (Fajardo-Somera et al. 2015), whereas the A. fumigatus ortholog chsD is required for the full chitin content of the cell wall, but is dispensable for infection in a mouse model of aspergillosis (Mellado et al. 1996). Thus, chs7 orthologs have evolved to fulfill a number of species-specific roles in growth, sexual development, and pathogenicity in different ascomycetes. Nevertheless, several CHS paralogs can be partially redundant and can complement each other in case of deletion of one paralog. For example, N. crassa single mutants of chs-1 and chs-3 have normal vegetative growth, whereas the double mutant shows severe growth reduction (Fajardo-Somera et al. 2015; Sánchez-León et al. 2011), and in M. oryzae, chs5 can partially complement the function of chs6 (Kong et al. 2012). However, these functional redundancies occur for CHS that are members of the same division, with N. crassa chs-1 and chs-3 being members of division I, and M. oryzae chs5 and chs6 members of division II (Kong et al. 2012; Riquelme and Bartnicki-García 2008). The S. macrospora chs7 is the only member of division III; however, CHS from division III have the simplest domain structure without additional domains apart from the essential CHS domain (Fajardo-Somera et al. 2015), therefore functional replacement by other CHS might seem possible, which could explain the lack of a developmental phenotype despite a conserved expression pattern.
The SNARE protein SEC22 is involved in late stages of sexual development
The functional analysis of sec22 showed that this gene is required for normal ascospore maturation and germination in S. macrospora. To the best of our knowledge, this is the first time that an involvement of sec22 in sexual development in fungi is shown. In S. cerevisiae, sec22 mutants are sensitive to heat stress, but grow normally under standard laboratory conditions, because of functional redundancy with the SNARE-encoding ykt6 gene, whereas a double mutant of both genes is lethal (Dascher et al. 1991; Liu and Barlowe 2002). The S. macrospora sec22 mutant is not heat sensitive, but overexpression of sec22 leads to increased vegetative growth at higher temperature (Figure 4E). In filamentous fungi, sec22 was analyzed in M. oryzae, where it was found to be involved in conidiation, pathogenicity, and growth; furthermore, the corresponding mutant is more sensitive to oxidative stress (Song et al. 2010). Sexual development or the reaction to heat stress was not analyzed in the M. oryzae mutant yet. Interestingly, SEC22 in Arabidopsis thaliana is essential for male and female gametophyte development, and thus for the completion of the developmental cycle of this plant (El-Kasmi et al. 2011). The findings in the filamentous fungi M. oryzae and S. macrospora and the plant A. thaliana indicate that sec22 might have species-specific roles in multicellular development.
In S. cerevisiae, sec22 was shown to be involved in macroautophagy, a process that mediates intracellular degradation of proteins and organelles through the formation of autophagosome vesicles. Interestingly, this requirement for sec22 in yeast is not compensated by ykt6 as described for other functions of sec22 (Nair et al. 2011). Autophagy is conserved in all eukaryotes, and in S. macrospora, several autophagy genes are essential for sexual development (Teichert et al. 2014). Among these are the genes Smatg4 and Smatg8. Mutants for both genes are unable to form mature fruiting bodies, and display reduced ascospore germination in crosses against the wild type or each other (Voigt and Pöggeler 2013). The latter phenotype is similar to the reduced spore germination in Δsec22, and therefore one might hypothesize that sec22 is also involved in autophagy in S. macrospora. However, any role of sec22 in autophagy in S. macrospora is unlikely to be an essential one, because mutants in core autophagy genes that were generated so far are either nonviable or unable to generate fruiting bodies (Nolting et al. 2009; Voigt et al. 2013; Voigt et al. 2014; Voigt and Pöggeler 2013), whereas the Δsec22 phenotype concerns ascospore maturation and germination, and thus occurs during later stages of sexual development.
Our analysis revealed transcriptional deregulation of several developmental genes in the Δsec22 mutant (Figure 6). Several of these genes encode ER-associated proteins or proteins that are predicted to require vesicle-dependent transport steps, suggesting that their deregulation might be caused by feedback mechanisms that sense problems in the vesicle trafficking pathway. The hyphae of filamentous fungi are extremely polarized cells that require a continuous flow of vesicles to the hyphal tips to support polarized growth (Fischer et al. 2008; Sánchez-León et al. 2015), and the differentiation of fruiting bodies and spores in particular requires the transfer of large amounts of metabolites within the fungal mycelium (Pöggeler et al. 2006a; Wessels 1993). It is conceivable that sec22 might be involved in the trafficking processes required for ascospore maturation. Highly specific roles for sec22 in transport processes can also be found in other species, e.g., it is essential for accumulation of caesium ions (Cs+), but not for accumulation of the essential K+ ions in the vacuoles of S. cerevisiae and A. thaliana (Dräxl et al. 2012). Thus, the molecular function of the SNARE protein SEC22 might be conserved, while integration into cellular and organismic processes appears to be variable among eukaryotes.
Supplementary Material
Acknowledgments
We thank Swenja Ellßel, Silke Nimtz, and Susanne Schlewinski for excellent technical assistance and Prof. Dr. Ulrich Kück (Bochum) for continuing support. This work was supported by the German Research Foundation (DFG, grant NO407/4-1). We also acknowledge support for publication by the German Research Foundation and the Open Access Publication Funds of the Ruhr-Universität Bochum.
Footnotes
Supporting information is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.115.017681/-/DC1
Communicating editor: M. S. Sachs
Literature Cited
- Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., et al. , 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bistis G. N., Perkins D. D., Read N. D., 2003. Different cell types in Neurospora crassa. Fungal Genet. Newsl. 50: 17–19. [Google Scholar]
- Bloemendal S., Bernhards Y., Bartho K., Dettmann A., Voigt O., et al. , 2012. A homolog of the human STRIPAK complex controls sexual development in fungi. Mol. Microbiol. 84: 310–323. [DOI] [PubMed] [Google Scholar]
- Borkovich K. A., Alex L. A., Yarden O., Freitag M., Turner G. E., et al. , 2004. Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol. Mol. Biol. Rev. 68: 1–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawand D., Soumillon M., Necsulea A., Julien P., Csardi G., et al. , 2011. The evolution of gene expression levels in mammalian organs. Nature 478: 343–348. [DOI] [PubMed] [Google Scholar]
- Chigira Y., Abe K., Gomi K., Nakajima T., 2002. chsZ, a gene for a novel class of chitin synthase from Aspergillus oryzae. Curr. Genet. 41: 261–267. [DOI] [PubMed] [Google Scholar]
- Choquer M., Boccara M., Goncalves I. R., Soulié M. C., Vidal-Cros A., 2004. Survey of the Botrytis cinerea chitin synthase multigenic family through the analysis of six euascomycete genomes. Eur. J. Biochem. 271: 2153–2164. [DOI] [PubMed] [Google Scholar]
- Christianson T. W., Sikorski R. S., Dante M., Shero J. H., Hieter P., 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110: 119–122. [DOI] [PubMed] [Google Scholar]
- Colot H. V., Park G., Turner G. E., Ringelberg C., Crew C. M., et al. , 2006. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. USA 103: 10352–10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dascher C., Ossig R., Gallwitz D., Schmitt H. D., 1991. Identification and structure of four yeast genes (SLY) that are able to suppress the functional loss of YPT1, a member of the RAS superfamily. Mol. Cell. Biol. 11: 872–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirschnabel D. E., Nowrousian M., Cano-Domínguez N., Aguirre J., Teichert I., et al. , 2014. New insights into the roles of NADPH oxidases in sexual development and ascospore germination in Sordaria macrospora. Genetics 196: 729–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dräxl S., Müller J., Li W. B., Michalke B., Scherb H., et al. , 2012. Caesium accumulation in yeast and plants is selectively repressed by loss of the SNARE Sec22p/SEC22. Nat. Commun. 4: 2092. [DOI] [PubMed] [Google Scholar]
- Eisenman H. C., Casadevall A., 2012. Synthesis and assembly of fungal melanin. Appl. Microbiol. Biotechnol. 93: 931–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Kasmi F., Pacher T., Strompen G., Stierhof Y. D., Müller L. M., et al. , 2011. Arabidopsis SNARE protein SEC22 is essential for gametophyte development and maintenance of Golgi-stack integrity. Plant J. 66: 268–279. [DOI] [PubMed] [Google Scholar]
- Engh I., Nowrousian M., Kück U., 2007a Regulation of melanin biosynthesis via the dihydroxynaphtalene pathway is dependent on sexual development in the ascomycete Sordaria macrospora. FEMS Microbiol. Lett. 275: 62–70. [DOI] [PubMed] [Google Scholar]
- Engh I., Würtz C., Witzel-Schlömp K., Zhang H. Y., Hoff B., et al. , 2007b The WW domain protein PRO40 is required for fungal fertility and associates with Woronin bodies. Eukaryot. Cell 6: 831–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engh I., Nowrousian M., Kück U., 2010. Sordaria macrospora, a model organism to study fungal cellular development. Eur. J. Cell Biol. 89: 864–872. [DOI] [PubMed] [Google Scholar]
- Esser K., 1982. Cryptogams - Cyanobacteria, Algae, Fungi, Lichens. Cambridge University Press, London. [Google Scholar]
- Fajardo-Somera R. A., Jöhnk B., Bayram Ö., Valerius O., Braus G. H., et al. , 2015. Dissecting the function of the different chitin synthases in vegetative growth and sexual development in Neurospora crassa. Fungal Genet. Biol. 75: 30–45. [DOI] [PubMed] [Google Scholar]
- Fischer R., Zekert N., Takeshita N., 2008. Polarized growth in fungi - interplay between the cytoskeleton, positional markers and membrane domains. Mol. Microbiol. 68: 813–826. [DOI] [PubMed] [Google Scholar]
- Gesing S., Schindler D., Fränzel B., Wolters D., Nowrousian M., 2012. The histone chaperone ASF1 is essential for sexual development in the filamentous fungus Sordaria macrospora. Mol. Microbiol. 84: 748–765. [DOI] [PubMed] [Google Scholar]
- Gesing S., Schindler D., Nowrousian M., 2013. Suppression subtractive hybridization and comparative expression analysis to identify developmentally regulated genes in filamentous fungi. J. Basic Microbiol. 53: 742–751. [DOI] [PubMed] [Google Scholar]
- James P., Halladay J., Craig E. A., 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144: 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamerewerd J., Jansson M., Nowrousian M., Pöggeler S., Kück U., 2008. Three alpha subunits of heterotrimeric G proteins and an adenylyl cyclase have distinct roles in fruiting body development in the homothallic fungus Sordaria macrospora. Genetics 180: 191–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klix V., Nowrousian M., Ringelberg C., Loros J. J., Dunlap J. C., et al. , 2010. Functional characterization of MAT1–1-specific mating-type genes in the homothallic ascomycete Sordaria macrospora provides new insights into essential and non-essential sexual regulators. Eukaryot. Cell 9: 894–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L. A., Yang J., Li G. T., Qi L. L., Zhang Y. J., et al. , 2012. Different chitin synthase genes are required for various developmental and plant infection processes in the rice blast fungus Magnaporthe oryzae. PLoS. Path. 8: e1002526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacaze I., Lalucque H., Siegmund U., Silar P., Brun S., 2015. Identification of NoxD/Pro41 as the homologue of the p22phox NADPH oxidase subunit in fungi. Mol. Microbiol. 95: 1006–1024. [DOI] [PubMed] [Google Scholar]
- Lee M. C. S., Miller E. A., Goldberg J., Orci L., Shekman R., 2004. Bi-directional protein transport between the ER and Golgi. Annu. Rev. Cell Dev. Biol. 20: 87–123. [DOI] [PubMed] [Google Scholar]
- Lehr N. A., Wang Z., Li N., Hewitt D. A., López-Giráldez F., et al. , 2014. Gene expression differences among three Neurospora species reveal genes required for sexual reproduction in Neurospora crassa. PLoS ONE 9: e110398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis M. J., Rayner J. C., Pelham H. R. B., 1997. A novel SNARE complex implicated in vesicle fusion with the endoplasmic reticulum. EMBO J. 16: 3017–3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Barlowe C., 2002. Analysis of Sec22p in endoplasmic reticulum/Golgi transport reveals cellular redundancy in SNARE protein function. Mol. Biol. Cell 13: 3314–3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord K. M., Read N. D., 2011. Perithecium morphogenesis in Sordaria macrospora. Fungal Genet. Biol. 49: 388–399. [DOI] [PubMed] [Google Scholar]
- Malagnac F., Lalucque H., Lepere G., Silar P., 2004. Two NADPH oxidase isoforms are required for sexual reproduction and ascospore germination in the filamentous fungus Podospora anserina. Fungal Genet. Biol. 41: 982–997. [DOI] [PubMed] [Google Scholar]
- Mandel M. A., Galgiani J. N., Kroken S., Orbach M. J., 2006. Coccidioides posadasii contains single chitin synthase genes corresponding to classes I to VII. Fungal Genet. Biol. 43: 775–788. [DOI] [PubMed] [Google Scholar]
- Martín-Udíroz M., Madrid M. P., Roncero M. I. G., 2004. Role of chitin synthase genes in Fusarium oxysporum. Microbiology 150: 3175–3187. [DOI] [PubMed] [Google Scholar]
- Mayrhofer S., Pöggeler S., 2005. Functional characterization of an {alpha}-factor-like Sordaria macrospora peptide pheromone and analysis of its interaction with Its cognate receptor in Saccharomyces cerevisiae. Eukaryot. Cell 4: 661–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masloff S., Pöggeler S., Kück U., 1999. The pro1+ gene from Sordaria macrospora encodes a C6 zinc finger transcription factor required for fruiting body development. Genetics 152: 191–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayrhofer S., Weber J. M., Pöggeler S., 2006. Pheromones and pheromone receptors are required for proper sexual development in the homothallic ascomycete Sordaria macrospora. Genetics 172: 1521–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellado E., Specht C. A., Robbins P. W., Holden D. W., 1996. Cloning and characterization of chsD, a chitin synthase-like gene of Aspergillus fumigatus. FEMS Microbiol. Lett. 143: 69–76. [DOI] [PubMed] [Google Scholar]
- Nair U., Jotwani A., Geng J., Gammoh N., Richerson D., et al. , 2011. SNARE proteins are required for macroautophagy. Cell 146: 290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolting N., Bernhards Y., Pöggeler S., 2009. SmATG7 is required for viability in the homothallic ascomycete Sordaria macrospora. Fungal Genet. Biol. 46: 531–542. [DOI] [PubMed] [Google Scholar]
- Nordzieke S., Zobel T., Fränzel B., Wolters D. A., Kück U., et al. , 2015. A fungal SLMAP homolog plays a fundamental role in development and localizes to the nuclear envelope, ER, and mitochondria. Eukaryot. Cell 14: 345–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowrousian M., 2009. A novel polyketide biosynthesis gene cluster is involved in fruiting body morphogenesis in the filamentous fungi Sordaria macrospora and Neurospora crassa. Curr. Genet. 55: 185–198. [DOI] [PubMed] [Google Scholar]
- Nowrousian M., 2014. Genomics and transcriptomics to analyze fruiting body development, pp. 149–172 in The Mycota XIII. Fungal Genomics, ed. 2, edited by Nowrousian M. Springer, Berlin, Heidelberg. [Google Scholar]
- Nowrousian M., Cebula P., 2005. The gene for a lectin-like protein is transcriptionally activated during sexual development, but is not essential for fruiting body formation in the filamentous fungus Sordaria macrospora. BMC Microbiol. 5: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowrousian M., Kück U., 2006. Comparative gene expression analysis of fruiting body development in two filamentous fungi. FEMS Microbiol. Lett. 257: 328–335. [DOI] [PubMed] [Google Scholar]
- Nowrousian M., Masloff S., Pöggeler S., Kück U., 1999. Cell differentiation during sexual development of the fungus Sordaria macrospora requires ATP citrate lyase activity. Mol. Cell. Biol. 19: 450–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowrousian M., Ringelberg C., Dunlap J. C., Loros J. J., Kück U., 2005. Cross-species microarray hybridization to identify developmentally regulated genes in the filamentous fungus Sordaria macrospora. Mol. Genet. Genomics 273: 137–149. [DOI] [PubMed] [Google Scholar]
- Nowrousian M., Frank S., Koers S., Strauch P., Weitner T., et al. , 2007a The novel ER membrane protein PRO41 is essential for sexual development in the filamentous fungus Sordaria macrospora. Mol. Microbiol. 64: 923–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowrousian M., Piotrowski M., Kück U., 2007b Multiple layers of temporal and spatial control regulate accumulation of the fruiting body-specific protein APP in Sordaria macrospora and Neurospora crassa. Fungal Genet. Biol. 44: 602–614. [DOI] [PubMed] [Google Scholar]
- Nowrousian M., Stajich J. E., Chu M., Engh I., Espagne E., et al. , 2010. De novo assembly of a 40 Mb eukaryotic genome from short sequence reads: Sordaria macrospora, a model organism for fungal morphogenesis. PLoS Genet. 6: e1000891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowrousian M., Teichert I., Masloff S., Kück U., 2012. Whole-genome sequencing of Sordaria macrospora mutants identifies developmental genes. G3 (Bethesda) 2: 261–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco-Arjona J. R., Ramirez-Prado J. H., 2014. Large-scale phylogenetic classification of fungal chitin synthases and identification of a putative cell-wall metabolism gene cluster in Aspergillus genomes. PLoS One 9: e104920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M. W., Horgan G. W., Dempfle L., 2002. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöggeler S., Kück U., 2006. Highly efficient generation of signal transduction knockout mutants using a fungal strain deficient in the mammalian ku70 ortholog. Gene 378: 1–10. [DOI] [PubMed] [Google Scholar]
- Pöggeler S., Nowrousian M., Jacobsen S., Kück U., 1997. An efficient procedure to isolate fungal genes from an indexed cosmid library. J. Microbiol. Methods 29: 49–61. [Google Scholar]
- Pöggeler S., Nowrousian M., Kück U., 2006a Fruiting-body development in ascomycetes, pp. 325–355 in The Mycota I, edited by Kües U., Fischer R. Springer, Berlin, Heidelberg. [Google Scholar]
- Pöggeler S., Nowrousian M., Ringelberg C., Loros J. J., Dunlap J. C., et al. , 2006b Microarray and real time PCR analyses reveal mating type-dependent gene expression in a homothallic fungus. Mol. Genet. Genomics 275: 492–503. [DOI] [PubMed] [Google Scholar]
- Qi W., Kwon C., Trail F., 2006. Microarray analysis of transcript accumulation during perithecium development in the filamentous fungus Gibberella zeae (anamorph Fusarium graminearum). Mol. Genet. Genomics 276: 87–100. [DOI] [PubMed] [Google Scholar]
- Riquelme M., Bartnicki-García S., 2008. Advances in understanding hyphal morphogenesis: Ontogeny, phylogeny and cellular localization of chitin synthases. Fungal Biol. Rev. 22: 56–70. [Google Scholar]
- Romero I. G., Ruvinsky I., Gilad Y., 2012. Comparative studies of gene expression and the evolution of gene regulation. Nat. Rev. Genet. 13: 505–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roncero C., 2002. The genetic complexity of chitin synthesis in fungi. Curr. Genet. 41: 367–378. [DOI] [PubMed] [Google Scholar]
- Rossi V., Banfield D. K., Vacca M., Dietrich L. E. P., Ungermann C., et al. , 2004. Longins and their longin domains: regulated SNAREs and multifunctional SNARE regulators. Trends Biochem. Sci. 29: 682–688. [DOI] [PubMed] [Google Scholar]
- Sambrooke J., Russell D. W., 2001. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. [Google Scholar]
- Sánchez-León E., Verdín J., Freitag M., Roberson R. W., Bartnicki-García S., et al. , 2011. Traffic of chitin synthase 1 (CHS-1) to the Spitzenkörper and developing septa in hyphae of Neurospora crassa: Actin dependence and evidence of distinct microvesicle populations. Eukaryot. Cell 10: 683–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-León E., Bowman B., Seidel C., Fischer R., Novick P., et al. , 2015. The Rab GTPase YPT-1 associates with Golgi cisternae and Spitzenkörper microvesicles in Neurospora crassa. Mol. Microbiol. 95: 472–490. [DOI] [PubMed] [Google Scholar]
- Schindler D., Nowrousian M., 2014. The polyketide synthase gene pks4 is essential for sexual development and regulates fruiting body morphology in Sordaria macrospora. Fungal Genet. Biol. 68: 48–59. [DOI] [PubMed] [Google Scholar]
- Sheng W., Yamashita S., Ohta A., Horiuchi H., 2013. Functional differentiation of chitin synthases in Yarrowia lipolytica. Biosci. Biotechnol. Biochem. 77: 1275–1281. [DOI] [PubMed] [Google Scholar]
- Siegmund U., Heller J., Van Kan J. A., Tudzynski P., 2013. The NADPH oxidase complexes in Botrytis cinerea: evidence for a close association with the ER and the tetraspanin Pls1. PLoS One 8: e55879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegmund U., Marschall R., Tudzynski P., 2015. BcNoxD, a putative ER protein, is a new component of the NADPH oxidase complex in Botrytis cinerea. Mol. Microbiol. 95: 988–1005. [DOI] [PubMed] [Google Scholar]
- Simonin A. R., Rasmussen C. G., Yang M., Glass N. L., 2010. Genes encoding a striatin-like protein (ham-3) and a forkhead associated protein (ham-4) are required for hyphal fusion in Neurospora crassa. Fungal Genet. Biol. 47: 855–868. [DOI] [PubMed] [Google Scholar]
- Song W., Dou X., Qi Z., Wang Q., Zhang X., et al. , 2010. R-SNARE homolog MoSec22 is required for conidiogenesis, cell wall integrity, and pathogenesis of Magnaporthe oryzae. PLoS One 5: e13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A., Shekman R., 1998. Reconstitution of retrograde transport from the Golgi to the ER in vitro. J. Cell Biol. 143: 589–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart J. M., Segal E., Koller D., Kim S. K., 2003. A gene-coexpression network for global discovery of conserved genetic modules. Science 302: 249–255. [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., et al. , 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teichert I., Wolff G., Kück U., Nowrousian M., 2012. Combining laser microdissection and RNA-seq to chart the transcriptional landscape of fungal development. BMC Genomics 13: 511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teichert I., Nowrousian M., Pöggeler S., Kück U., 2014. The filamentous fungus Sordaria macrospora as a genetic model to study fruiting body development. Adv. Genet. 87: 199–244. [DOI] [PubMed] [Google Scholar]
- Thompson J. D., Gibson T. J., Higgins D. G., 2002. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinformatics 2: Unit 2.3. [DOI] [PubMed] [Google Scholar]
- Traeger S., Altegoer F., Freitag M., Gabaldon T., Kempken F., et al. , 2013. The genome and development-dependent transcriptomes of Pyronema confluens: a window into fungal evolution. PLoS Genet. 9: e1003820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungar D., Hughson F. M., 2003. SNARE protein struture and function. Annu. Rev. Cell Dev. Biol. 19: 493–517. [DOI] [PubMed] [Google Scholar]
- Voigt O., Pöggeler S., 2013. Autophagy genes Smatg8 and Smatg4 are required for fruiting-body development, vegetative growth and ascospore germination in the filamentous ascomycete Sordaria macrospora. Autophagy 9: 33–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt O., Herzog B., Jakobshagen A., Pöggeler S., 2013. bZIP transcription factor SmJLB1 regulates autophagy-related genes Smatg8 and Smatg4 and is required for fruiting-body development and vegetative growth in Sordaria macrospora. Fungal Genet. Biol. 61: 50–60. [DOI] [PubMed] [Google Scholar]
- Voigt O., Herzog B., Jakobshagen A., Pöggeler S., 2014. Autophagic kinases SmVPS34 and SmVPS15 are required for viability in the filamentous ascomycete Sordaria macrospora. Microbiol. Res. 169: 128–138. [DOI] [PubMed] [Google Scholar]
- Wessels J. G. H., 1993. Fruiting in the higher fungi. Adv. Microb. Physiol. 34: 147–202. [DOI] [PubMed] [Google Scholar]
- Yarden O., Plamann M., Ebbole D., Yanofsky C., 1992. cot-1, a gene required for hyphal elongation in Neurospora crassa encodes a protein kinase. EMBO J. 11: 2159–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.