Abstract
Micelleplexes are a class of nucleic acid carriers that have gained acceptance due to their size, stability, and ability to synergistically carry small molecules. MicroRNAs (miRNAs) are small non-coding RNA gene regulator that is consists of 19–22 nucleotides. Altered expression of miRNAs plays an important role in many human diseases. Using a model 22-nucleotide miRNA sequence, we investigated the interaction between charged groups on the micelle surface and miRNA. The model micelle system was formed from methoxy-poly(ethylene glycol)-b-poly(lactide) (mPEG-PLA) mixed with methoxy-poly(ethylene glycol)-b-poly(lactide)-b-oligoarginine (mPEG-PLA-Rx, x = 8 or 15). Surface properties of the micelles were varied by controlling the oligoarginine block length and conjugation density. Micelles were observed to have a core-shell conformation in the aqueous environment where the PLA block constituted the hydrophobic core, mPEG and oligoarginine formed a hydrophilic corona. Significantly different thermodynamic behaviors were observed during the interaction of single stranded miRNA with micelles of different surface properties, and the resulting micelleplexes mediated substantial cellular association. Depending upon the oligoarginine length and density, micelles exhibited miRNA loading capacity directly related to the presentation of charged groups on the surface. The effect of charged group accessibility of cationic micelle on micelleplex properties provides guidance on future miRNA delivery system design.
In this article, we investigated charged group presentation on the micelle surfaces and their interaction with miRNA using thermodynamics, biochemistry, and molecular dynamics simulation. Charged group accessibility on cationic micelle surfaces was shown as a mechanism to alter micelleplex formation and stability.
Keywords: miRNA, micelle, micelleplex, isothermal titration calorimetry, nanoparticle
Over the last decade, miRNA has attracted significant attention due to their critical gene regulatory function in both normal biological process as well as various human diseases.1 Micelleplexes, complexes formed by binding miRNA to micelles, have been designed for miRNA delivery. To form micelleplexes, miRNA is loaded onto the corona of the pre-formed micelles through multivalent ionic interactions.2–8 Micelleplexes provide control over particle size through the hydrophobic interactions of the core and hydrophilic stability through the corona. To further develop and optimize micelleplexes for miRNA delivery, we sought to better understand the physical events driving micelleplex formation, structure, and stability. Despite the fact that block copolymer micelles properties have been relatively well studied, little has been described for the processes driving micelleplexes formation and stabilization. For the first time, the effects of charged group surface presentation on miRNA binding are investigated from a thermodynamic standpoint to improve the understanding of the interactions of miRNA with micelles.
Modification of the surface of nanoparticles has significant effect on the stability and biologic interactions of the nanoparticles.9–11 To promote miRNA-micelle interactions, surface-exposed charged groups have been placed on micelles.2 Unfortunately, the hydrophilic corona-forming components of micelles also tend to interfere with the binding of miRNA. The hydrophilic corona-forming components, generally poly(ethylene glycol) (PEG), form a ‘hydration shell’ on nanoparticles excluding the adsorption of proteins resulting in steric stabilization of the micelle.12 Even with this shell, it is clear that proteins interact with the surface of micelles and other nanoparticles.13–19 However, PEG on the surface also impairs cationic polymer interactions with nucleic acids due to decreased positive charge density, hydrogen bonding, and steric hindrance.20, 21 Moreover, once the micelleplexes arrive at targeted site, PEG is expected to diminish micelleplex-cell interactions, reducing cellular uptake of the micelleplexes resulting in a significant decrease in miRNA activity.22, 23 Design criteria that aid in the balance between binding of miRNA and steric stabilization would greatly improve the efficiency of micelleplex design.
To elucidate the effect of charged group surface presentation of micelles on miRNA loading, stability and cellular uptake, we engineered polymeric micelles that are composed of hydrophilic oligoarginine (Rx, x=8 or 15) conjugated directly to the hydrophobic block of the mPEG-b-PLA block copolymer forming a triblock copolymer, mPEG-PLA-Rx8 based upon our previous experience with the R8 oligoarginine,24 the expected extended lengths of the oligoarginine and poly(ethylene glycol), and previously published research.8, 25 We hypothesized that the mPEG-PLA-Rx micelles would present the oligoarginine block exposed on the surface, dependent upon the length of the oligoarginine block and the poly(ethylene glycol) block. An alternative structure where the oligoarginine block were placed on the poly(ethylene glycol) block, PLA-PEG-Rx, could also be examined and will be in the future to determine if these structures are presented as described for poly(lysine)26, 27 or oriented toward the interior of the structure.10 Further, we hypothesized that the surface presentation of the oligoarginine would influence the miRNA-oligoarginine interactions and cellular uptake. In this way, the charged group was accessible for loading after formation, but the presentation at the surface could be controlled, unlike systems that bury the charged groups during the formation of the micelle and limit the loading to the point of formation of the particles.28
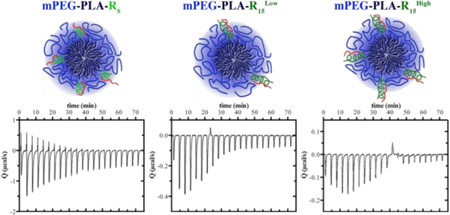
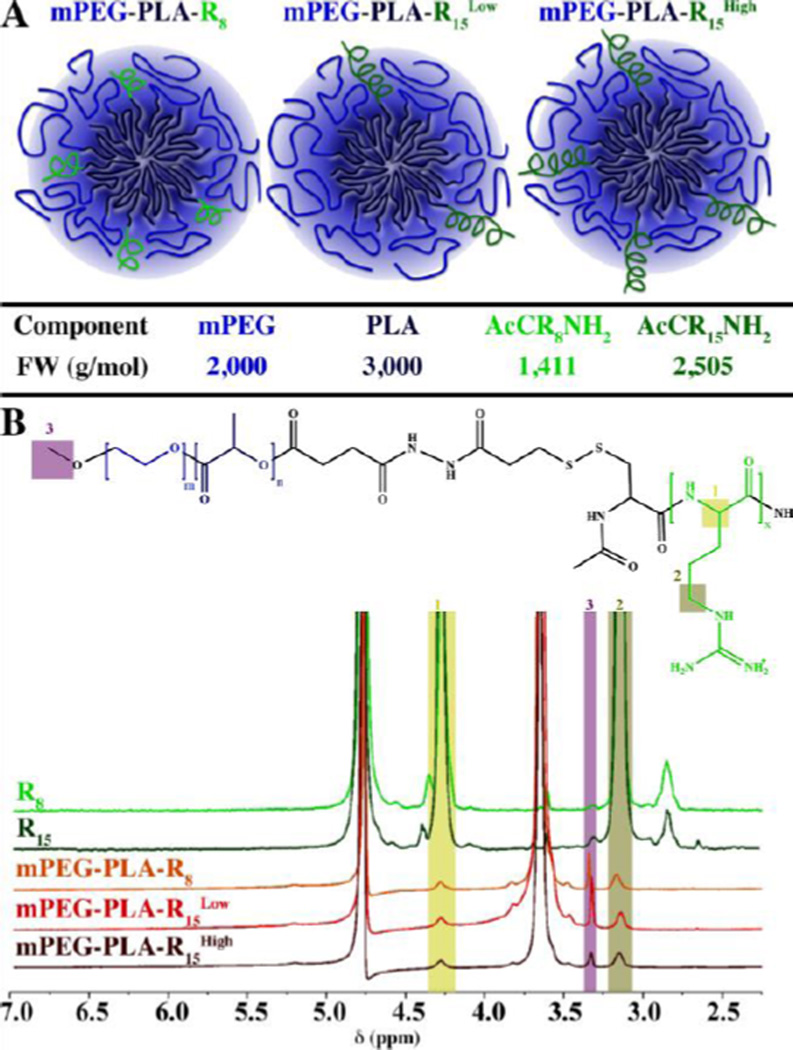
Micelles, mPEG-PLA-R8, mPEG-PLA-R15Low, and mPEG-PLA-R15High, with arginine block conjugation density (proportion of oligoarginine modified triblock copolymer to total mPEG-PLA polymer) of 27%, 14% and 28% (mol%), respectively (Figure 1A), were synthesized by coupling mPEG-PLA with CRx through disulfide bond exchange reaction and purified with a Sep-Pak C18 column and characterized with1H NMR (Figure S1). For the purpose of easily distinguishing 14% and 28% R15 substituted mPEG-PLA-R15 micelles, we denote them as mPEG-PLA-R15Low and mPEG-PLA-R15High, respectively. All of the mPEG-PLA-Rx copolymers have similar critical micelle concentration (CMC) to the mPEG-PLA polymer (Figure S2). The micelles from each group had similar diameter (Table S1). The ζ-potential of mPEG-PLA-R8 micelles (30.76±1.16) was similar to mPEG-PLA-R15High (30.46±1.88), suggesting that only a portion of the oligoarginine contributed directly to the surface charge of the micelles. This was further supported by the ζ-potential of mPEG-PLA-R8 and mPEG-PLA-R15High micelles being higher than mPEG-PLA-R15Low micelles (19.65±4.7). In this way, we produced micelles with similar size, oligoarginine content (mPEG-PLA-R8 and mPEG-PLA-R15Low) and charge (mPEG-PLA-R8 and mPEG-PLA-R15High). The morphology of mPEG-PLA-R8, mPEG-PLA-R15Low and mPEG-PLA-R15High micelles and micelleplexes were observed with transmission electron microscope, all of which demonstrated spherical shape and size similar to DLS measurements (Figure S3).
Figure 1. Micelles properties and surface oligoarginine presence.
(A) Schematic representation of the micelles formed and the relevant molecular weights of the components. (B) 1H NMR analysis of the peptides (CR8 and CR15) and micelles (mPEG-PLA-R8, mPEG-PLA-R15Low, and mPEG-PLA-R15High) showing the arginine α proton (1; δ = 4.40 ppm, 1H), δ protons (2; δ = 3.20 ppm, 2H), or ω-terminal methoxyl protons (arrow; δ = 3.40 ppm, 3H).
To understand the conformation of the micelles, micelles were prepared in D2O for solution-phase NMR analysis.29–31 In this technique, the constituents in the aqueous phase are readily detected but the hydrophobic-phase constituents have repressed signature. In aqueous solution, the triblock mPEG-b-PLA-Rx polymers formed micelles where the PLA hydrophobic chains collapse, with a disappearance of the PLA proton peaks, to form a hydrophobic core while the PEG chains and the Rx block constitute a hydrophilic corona. While the PLA protons were not detected in D2O, the terminal methoxyl protons (δ = 3.40, 3H) from mPEG chains (Figure 1B), the δ methylene protons from arginine side chain (δ = 3.20, 2H) and the α proton (δ = 4.40, 1H, Figure 1B)8 in the peptide backbone of Rx blocks remained detectable in the 1H NMR spectrum. The prominent peaks (δ = 4.7 ppm and δ = 3.7 ppm) correspond to the solvent and methylene proton of poly(ethylene glycol), respectively. Based upon atomistic molecular dynamics simulation (Figure 2), the number of total copolymer monomers to produce a micelle with a size observed by DLS and TEM is between 60 and 100 monomers. Using this estimate and the neutral to slightly positive ζ–potenital of the micelles, it would be expected that the number of charged phosphate groups would match those of the charged arginine groups, or be slightly below unity. Therefore, the number of oligoarginines and miRNA loading of the micelles was estimated (Table 1).These observations combined suggest the presence of oligoarginine block on the micelles surface, i.e. the aqueous phase. Based upon this, each of the micelles presented a portion of the oligoarginine on the surface with access to the aqueous environment.
Figure 2. Molecular dynamics simulation of mPEG-PLA-R8 micelle.
Each monomer consisted of a methoxy-poly(ethylene-glycol)(mPEG; MW ~ 2,000 g/mol;  ) block coupled to the α-hydroxide of poly(lactide) (PLA; MW ~ 3,000 g/mol;
) block coupled to the α-hydroxide of poly(lactide) (PLA; MW ~ 3,000 g/mol;  ). As in the experiments, 28% of total monomers were modified with oligoarginine (R8;
). As in the experiments, 28% of total monomers were modified with oligoarginine (R8;  ) on the ω-carboxylate of the PLA block. The micelles were prepared with total 60 monomers.
) on the ω-carboxylate of the PLA block. The micelles were prepared with total 60 monomers.
Table 1.
Estimates of the number of oligo-arginine peptides and miRNA associated with micelles.
| Nagg (copolymer§/micelle) | 60 | 100 | ||
|---|---|---|---|---|
| NZ (oligoargine/micelle or miRNA/micelle) | NRx | NmiRNA | NRx | NmiRNA |
| mPEG-PLA-R8 | 16 | 6 | 27 | 10 |
| mPEG-PLA-R15Low | 8 | 6 | 14 | 10 |
| mPEG-PLA-R15High | 17 | 11 | 28 | 20 |
copolymers include both mPEG-PLA and mPEG-PLA-Rx. As presented in Table S1, 14, 27, and 28% of copolymers contain the arginine group for the mPEG-PLA-R8, mPEG-PLA-R15Low, mPEG-PLA-R15High micelles, respectively.
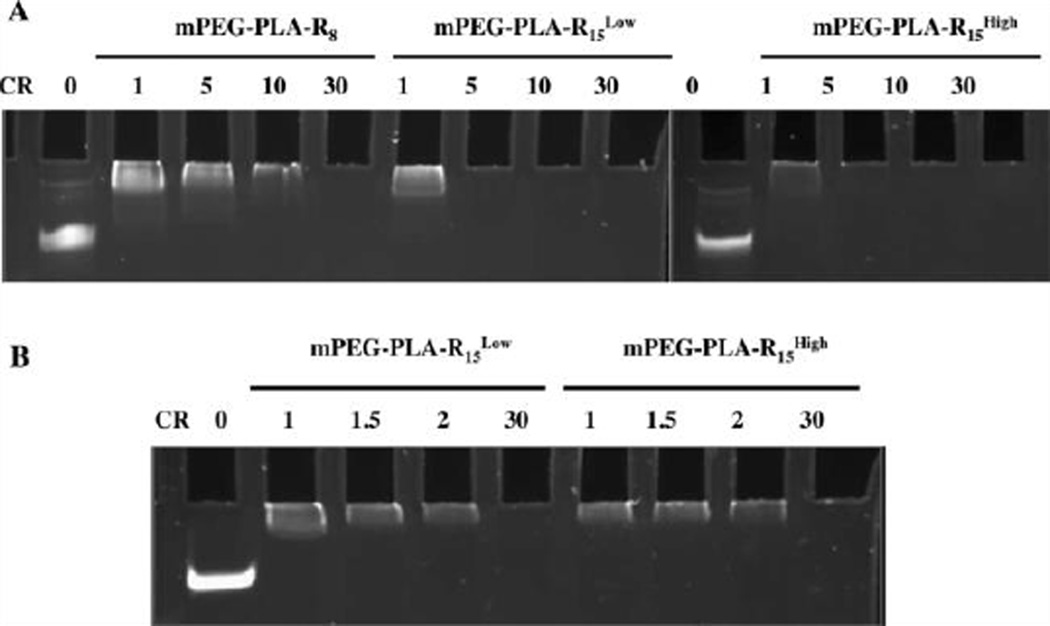
Being present in the aqueous phase, however, does not indicate that the oligoarginine is free to interact with biomolecules in the presence of the PEG corona. To determine the ability of the micelles to interact with miRNA, the gel shift assay was conducted (Figure 3A). mPEG-PLA-R8 micelles were able to fully retard miRNA mobility only at or above a positive to negative (+/−) charge ratio (arginine to nucleotide) of 30, while mPEG-PLA-R15Low and mPEG-PLA-R15High fully retarded miRNA mobility at a charge ratio of 5. The lower charge ratio needed to fully bind miRNA further suggested that the mPEG-PLA-R15 micelles contain arginine that is more accessible to the surface for biomolecular interactions. This is further supported by the fact that mPEG-PLA-R15Low micelles interact with the miRNA as efficiently as the mPEG-PLA-R15High despite the fact that there is approximately half the total arginine present. No significant difference was observed for miRNA interactions with mPEG-PLA-R15Low and mPEG-PLA-R15High at low charge ratios (Figure 3B). Based upon the interactions with miRNA, the longer oligoarginine chains are able to more readily interact with biomolecules at the surface of the micelles regardless of overall charge, i.e. ζ-potential, of the micelle. This is consistent with observation that PEGylation of cationic polymers generally deteriorates a polymer’s ability to interact with DNA and RNA.21
Figure 3. MiRNA complexation with micelles occurs at lower charge ratios for longer oligoarginine regardless of oligoarginine density.
(A) mPEG-PLA-R8, mPEG-PLA-R15Low, and mPEG-PLA-R15High complexation with miRNA at +/− charge ratios of 1, 5, 10 and 30 (B) mPEG-PLA-R15Low and mPEG-PLA-R15High complexation with miRNA at +/− charge ratios 1, 1.5, 2 and 30.
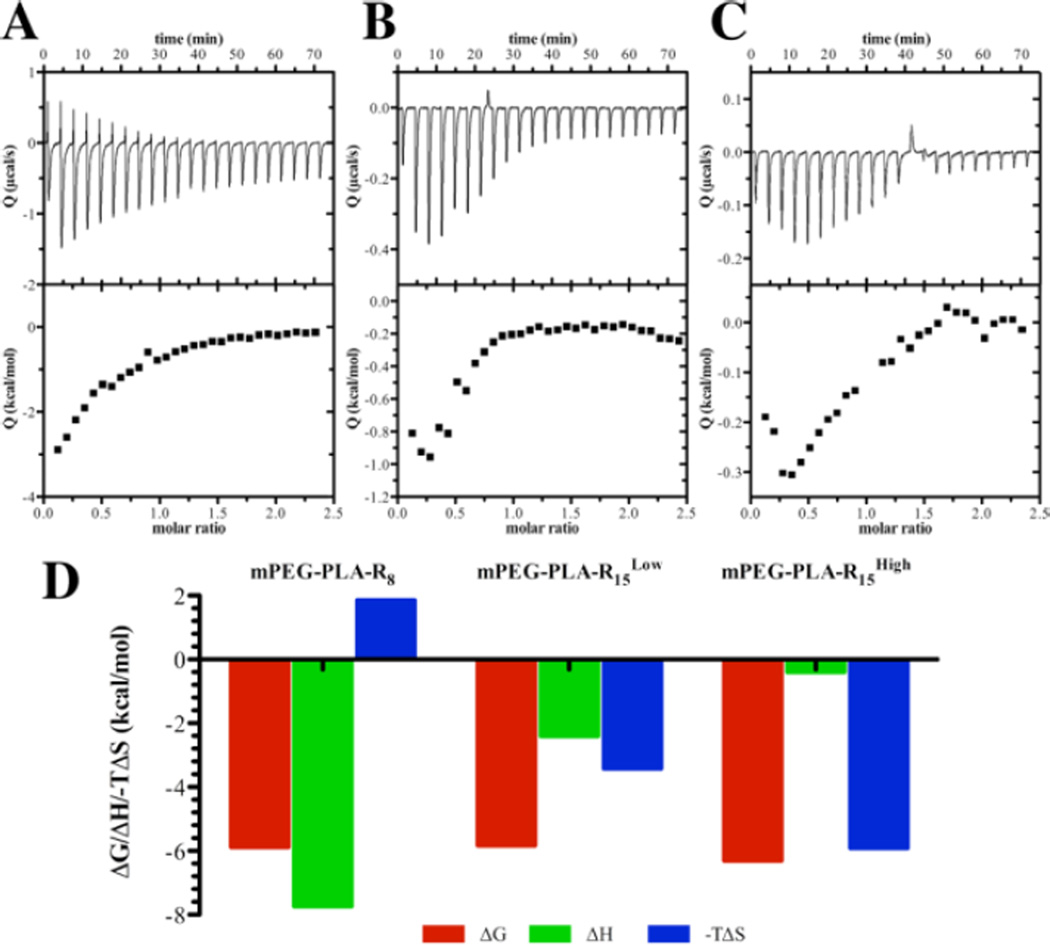
To better characterize the interactions and gain information about the events leading to miRNA-micelle interactions, thermodynamic analysis was utilized.32–35 An initial endothermic peak was observed for mPEG-PLA-R8, which suggests that a molecular rearrangement took place prior to the ionic interactions between the peptides and the miRNA (Figure 4). Unlike mPEG-PLA-R8, only exothermic processes were observed when miRNA interacts with mPEG-PLA-R15Low or mPEG-PLA-R15High. Only the shorter arginine-containing micelles, mPEG-PLA-R8, had an unfavorable entropic contribution while all micelles exhibited favorable enthalpy energy (Table S3). PEG chains are thought to develop a ‘hydration shell’ where water is structured in the local vicinity surrounding the micelles.12, 36 The entropic loss (−T∆ S > 0) during miRNA binding to mPEG-PLA-R8 process might result from the disturbance of the hydration shell surrounding mPEG-PLA-R8 in conjunction with unfavorable conformational changes necessary to accommodate the miRNA chain. Because the longer oligoarginine peptides are more accessible, the unfavorable rearrangements and the disruption of the ordered water layer are not necessary to accommodate the miRNA. The interactions of mPEG-PLA-R15Low and mPEG-PLA-R15High with miRNA are very similar to hyperbranched polyethylenimine (PEI) interactions with double strand siRNA,35 suggesting that arginine clusters on the micelle surface could interact with miRNA in similar manner to PEI.
Figure 4. Thermodynamic profiles of single stranded miRNA binding to micelles indicate different interactions occur dependent upon oligoarginine length and density.
(A) mPEG-PLA-R8 (B) mPEG-PLA-R15Low (C) mPEG-PLA-R15High (D) Thermodynamic parameters, derived from ITC, of miRNA binding to mPEG-PLA-R8, mPEG-PLA-R15Low, and mPEG-PLA-R15High micelles.
The entropic energy that was needed to form the complexes was coupled with a lower number of arginines that could interact with the miRNA. The mPEG-PLA-R8 micelles only had about 2.6 arginines per peptide available to interact while the mPEG-PLA-R15Low and mPEG-PLA-R15High micelles had 6.2 or 11.0 arginines per peptide available to interact with miRNA, respectively. The differences in available arginines do not mirror the charge on the micelles (Table S2), which were all positive, and the density of peptide being the only factor influencing the ζ-potential. This suggests that the available content of arginine increased on the surface as the amount of peptide in the micelles increased.
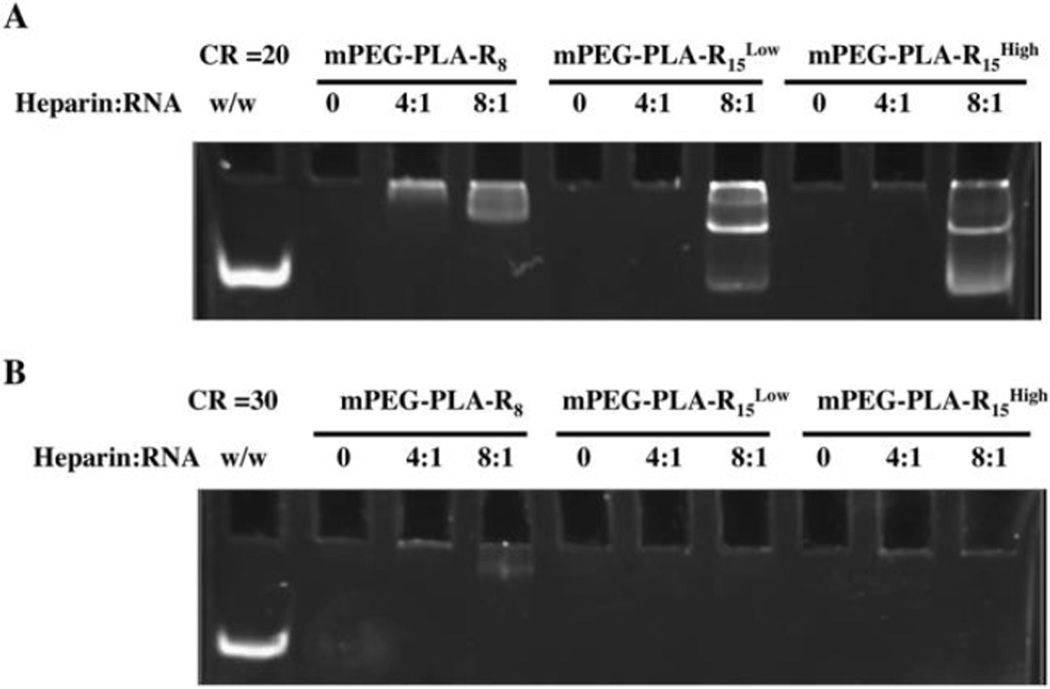
In addition to the role of the PEG corona shielding the charged groups during complexation, the PEG corona is also thought to influence the stability of the complexes by altering the surface accessibility of charged biomolecules, particularly anionic biomacromolecules that compete with negatively charged nucleic acids on the micelle surface. In all micelles, the micelleplex ζ-potentials were neutral compared to the original highly positive ζ-potential. Using the heparin sulfate competition assay to mimic the natural anionic macromolecules, shorter oligoarginine-containing micelles, mPEG-PLA-R8, were not able to condense miRNA as tightly as longer oligoarginine containing micelles, mPEG-PLA-R15Low and mPEG-PLA-R15High. mPEG-PLA-Rx micelleplexes migrate in the polyacrylamide gel, but to a lesser extent than miRNA control due to the charge neutralization and the size of the micelleplexes (Figure 3). At a charge ratio of 20, heparin did not dissociate miRNA from mPEG-PLA-R8 micelles at a heparin to miRNA weight ratio of 4:1 (Figure 5A), but based upon the diminished dye exclusion the miRNA appears to be less tightly bound to the micelleplex. The association between miRNAs and micelles was similarly loosened in mPEG-PLA-R15Low and mPEG-PLA-R15High micelleplexes in the presence of 8:1 heparin to miRNA. At a charge ratio of 30, heparin was unable to compete with miRNA or loosen the interactions. This suggests that the poorly accessible arginine content in mPEG-PLA-R8 was less available for interactions, but once the interaction was formed the interaction was stable to heparin competition. mPEG-PLA-R15Low and mPEG-PLA-R15High have longer peptide sequence and appear to present a greater proportion of miRNAs closer to the surface than in mPEG-PLA-R8 micelleplexes, and are thus less resistant to heparin competition. At a higher charge ratio of 30 to 1, all micelles were able to maintain the interaction with miRNA, (Figure 5B) suggesting that the interaction was stabilized with further ionic crosslinking. This observation agrees with others who observed that the macromolecules that freely interact with RNA are not necessarily the most stable.37
Figure 5. Micelleplexes are stable to heparin competition.
Micelleplexes were prepared at (A) +/− charge ratio 20 or (B) 30 and incubated with heparin at a heparin to miRNA weight ratio (w/w) of 0:1, 4:1, or 8:1 prior to electrophoresis and staining.
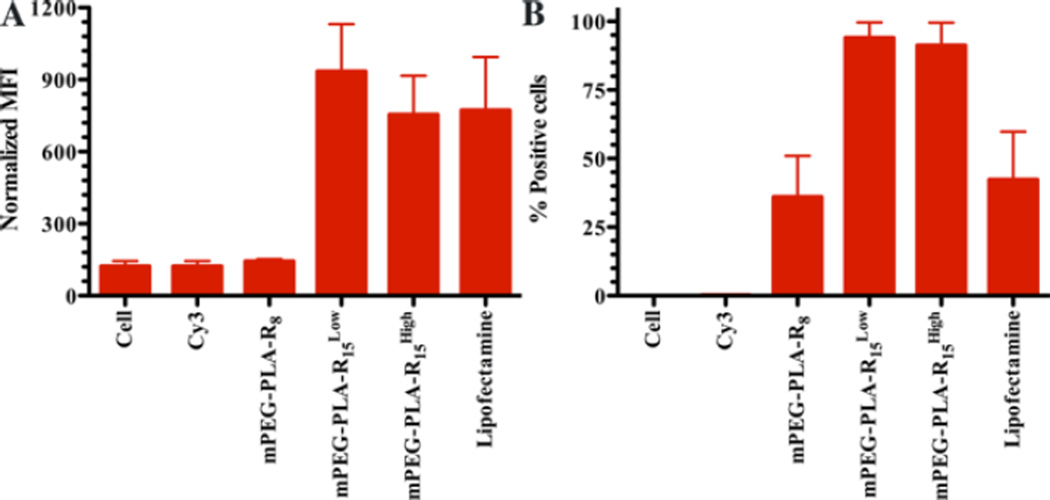
Finally, surface exposed charged groups on the micelles are believed to play key roles in their cellular interactions, with neutral particles (ζ-potential −10 to +10 mV) having limited non-specific cellular interactions.10, 38, 39 mPEG-PLA-R8 and mPEG-PLA-R15High micelles had higher ζ-potential than mPEG-PLA-R15Low, but the ζ-potential range of all three micelleplexes were generally neutral after miRNA complexation (Table S1 & S2). mPEG-PLA-R15Low and mPEG-PLA-R15High micelleplexes showed significantly more cellular association than mPEG-PLA-R8 micelleplexes (Figure 6). With the similar charge and size, the oligoarginine available on the micelle surface was expected to influence the ability of micelles to associate with or enter cells, but the total arginine and surface charge was not directly related to cellular interaction.24, 40, 41 Only the available arginine content on the micelle surface correlated with the ability of the micelles to interact with cells. This further supports the idea that the oligoarginine on the mPEG-PLA-R15 micelles was more available for interactions.
Figure 6. Cellular association mediated by different micelleplexes.
(A) The mean fluorescence intensity observed on U251 glioma cells following interaction with mPEG-PLA-R8, mPEG-PLA-R15Low, and mPEG-PLA-R15High micelleplexes or untreated cells or Cy3-labeled miRNA. (B) The relative population (%) of cells associated with the mPEG-PLA-R8, mPEG-PLA-R15Low, and mPEG-PLA-R15High micelleplexes or untreated cells or Cy3-labeled miRNA. N=3, mean ± standard error of the mean.
In summary, we have presented the synthesis of triblock copolymers that form micelles and expose different amounts of charged groups on their surface. We systematically studied the interactions of the micelles with miRNA and the miRNA-micelle complexes with biologic macromolecules and cells. By precise control of polymer structure, the accessibility of charged groups on micelle surface was used to control the interaction of miRNA with micelles and the eventual interaction with cells and the presence of oligoarginine near the hydrophobic core of the micelles was confirmed experimentally and through molecular modeling. The accessibility of the charged groups has direct impact on miRNA interaction with the micelles, micelleplexes stability and cellular interaction. These results guide the design of materials for RNA/DNA interactions by suggesting that the total charge of the groups used is not the primary factor for determining RNA binding or cellular association. Future work will further elucidate if the shielding of ionic groups can be controlled in alternate architectures. The PEG corona can shield the ionic groups thus diminishing RNA-micelle binding and minimizing cell binding; however, this PEG corona protects the RNA from competition yielding a more stable micelleplex.
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge Dr. Hayat Onyuksel for generous sharing of instrumentation, Dr. Gerd Prehna in University of Illinois at Chicago (UIC) Center for Structural Biology for assistance with ITC experiments and Dr. Larry L. Klein for helpful discussion on polymer conjugation.
Funding Sources
This investigation was conducted in a facility constructed with support from Research Facilities Improvement Program Grant Number C06 RR15482 from the National Center for Research Resources, NIH and funded by the UIC Center for Clinical and Translational Science (CCTS) award UL1 TR000050 (RAG), Provost’s Award for Graduate Research from UIC Graduate College (YZ), and National Science Foundation award DMR 1309765 (PK). Yu Zhang was supported by University Fellowship from UIC.
Footnotes
ASSOCIATED CONTENT
Experimental procedures and additional supporting data, specifically the 1H NMR spectra, micelle particle size, ζ–potential, CMC, and thermodynamic parameters of ITC. This material is available free of charge via the Internet at http://http://pubs.rsc.org/.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
REFERENCES
- 1.Zhang Y, Wang ZJ, Gemeinhart RA. J. Control. Release. 2013;172:962–974. doi: 10.1016/j.jconrel.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gary DJ, Lee H, Sharma R, Lee JS, Kim Y, Cui ZY, Jia D, Bowman VD, Chipman PR, Wan L, Zou Y, Mao G, Park K, Herbert BS, Konieczny SF, Won YY. ACS Nano. 2011;5:3493–3505. doi: 10.1021/nn102540y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim YM, Song SC. Biomaterials. 2014;35:7970–7977. doi: 10.1016/j.biomaterials.2014.05.070. [DOI] [PubMed] [Google Scholar]
- 4.Mao CQ, Du JZ, Sun TM, Yao YD, Zhang PZ, Song EW, Wang J. Biomaterials. 2011;32:3124–3133. doi: 10.1016/j.biomaterials.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 5.Sharma R, Lee JS, Bettencourt RC, Xiao C, Konieczny SF, Won YY. Biomacromolecules. 2008;9:3294–3307. doi: 10.1021/bm800876v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun TM, Du JZ, Yao YD, Mao CQ, Dou S, Huang SY, Zhang PZ, Leong KW, Song EW, Wang J. ACS Nano. 2011;5:1483–1494. doi: 10.1021/nn103349h. [DOI] [PubMed] [Google Scholar]
- 7.Yu H, Zou Y, Wang Y, Huang X, Huang G, Sumer BD, Boothman DA, Gao J. ACS Nano. 2011;5:9246–9255. doi: 10.1021/nn203503h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao ZX, Gao SY, Wang JC, Chen CJ, Zhao EY, Hou WJ, Feng Q, Gao LY, Liu XY, Zhang LR, Zhang Q. Biomaterials. 2012;33:6793–6807. doi: 10.1016/j.biomaterials.2012.05.067. [DOI] [PubMed] [Google Scholar]
- 9.Pearson RM, Hsu H-j, Bugno J, Hong S. MRS Bull. 2014;39:227–237. [Google Scholar]
- 10.Pearson RM, Patra N, Hsu HJ, Uddin S, Kral P, Hong S. ACS Macro Lett. 2013;2:77–81. doi: 10.1021/mz300533w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vukovic L, Khatib FA, Drake SP, Madriaga A, Brandenburg KS, Kral P, Onyuksel H. J. Am. Chem. Soc. 2011;133:13481–13488. doi: 10.1021/ja204043b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allen C, Dos Santos N, Gallagher R, Chiu GN, Shu Y, Li WM, Johnstone SA, Janoff AS, Mayer LD, Webb MS, Bally MB. Biosci. Rep. 2002;22:225–250. doi: 10.1023/a:1020186505848. [DOI] [PubMed] [Google Scholar]
- 13.Pozzi D, Colapicchioni V, Caracciolo G, Piovesana S, Capriotti AL, Palchetti S, De Grossi S, Riccioli A, Amenitsch H, Lagana A. Nanoscale. 2014;6:2782–2792. doi: 10.1039/c3nr05559k. [DOI] [PubMed] [Google Scholar]
- 14.Casals E, Pfaller T, Duschl A, Oostingh GJ, Puntes V. ACS nano. 2010;4:3623–3632. doi: 10.1021/nn901372t. [DOI] [PubMed] [Google Scholar]
- 15.Cedervall T, Lynch I, Lindman S, Berggard T, Thulin E, Nilsson H, Dawson KA, Linse S. Proc Natl Acad Sci U S A. 2007;104:2050–2055. doi: 10.1073/pnas.0608582104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsu H-J, Sen S, Pearson RM, Uddin S, Kral P, Hong S. Macromolecules. 2014;47:6911–6918. doi: 10.1021/ma501258c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lundqvist M, Stigler J, Elia G, Lynch I, Cedervall T, Dawson KA. Proc. Natl. Acad. Sci. U. S. A. 2008;105:14265–14270. doi: 10.1073/pnas.0805135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monopoli MP, Walczyk D, Campbell A, Elia G, Lynch I, Bombelli FB, Dawson KA. J. Am. Chem. Soc. 2011;133:2525–2534. doi: 10.1021/ja107583h. [DOI] [PubMed] [Google Scholar]
- 19.Pearson RM, Juettner VV, Hong S. Front. Chem. 2014;2 doi: 10.3389/fchem.2014.00108. Article 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen CK, Jones CH, Mistriotis P, Yu Y, Ma X, Ravikrishnan A, Jiang M, Andreadis ST, Pfeifer BA, Cheng C. Biomaterials. 2013;34:9688–9699. doi: 10.1016/j.biomaterials.2013.08.063. [DOI] [PubMed] [Google Scholar]
- 21.Fitzsimmons RE, Uludag H. Acta Biomater. 2012;8:3941–3955. doi: 10.1016/j.actbio.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 22.Endo I, Nagamune T, Akita H. Nano/Micro Biotechnology. Springer; 2010. [Google Scholar]
- 23.Hatakeyama H, Akita H, Kogure K, Harashima H. Adv. Biochem. Eng. Biotechnol. 2010;119:197–230. doi: 10.1007/10_2008_40. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Köllmer M, Buhrman JS, Tang MY, Gemeinhart RA. Peptides. 2014;58:83–90. doi: 10.1016/j.peptides.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu P, Yu H, Sun Y, Zhu M, Duan Y. Biomaterials. 2012;33:4403–4412. doi: 10.1016/j.biomaterials.2012.02.041. [DOI] [PubMed] [Google Scholar]
- 26.Liu Y, Chen Z, Liu C, Yu D, Lu Z, Zhang N. Biomaterials. 2011;32:5167–5176. doi: 10.1016/j.biomaterials.2011.03.077. [DOI] [PubMed] [Google Scholar]
- 27.Liu Y, Liu C, Li M, Liu F, Feng L, Zhang L, Zhang N. J Biomed Nanotechnol. 2014;10:948–958. doi: 10.1166/jbn.2014.1796. [DOI] [PubMed] [Google Scholar]
- 28.Wang HX, Yang XZ, Sun CY, Mao CQ, Zhu YH, Wang J. Biomaterials. 2014;35:7622–7634. doi: 10.1016/j.biomaterials.2014.05.050. [DOI] [PubMed] [Google Scholar]
- 29.Jongpaiboonkit L, Zhou Z, Ni X, Wang YZ, Li J. J. Biomater. Sci. Polym. Ed. 2006;17:747–763. doi: 10.1163/156856206777656553. [DOI] [PubMed] [Google Scholar]
- 30.Darefsky AS, King JT, Jr, Dubrow R. Cancer. 2012;118:2163–2172. doi: 10.1002/cncr.26494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shin IG, Kim SY, Lee YM, Cho CS, Sung YK. J. Control. Release. 1998;51:1–11. doi: 10.1016/s0168-3659(97)00164-8. [DOI] [PubMed] [Google Scholar]
- 32.Holzerny P, Ajdini B, Heusermann W, Bruno K, Schuleit M, Meinel L, Keller M. J. Control. Release. 2012;157:297–304. doi: 10.1016/j.jconrel.2011.08.023. [DOI] [PubMed] [Google Scholar]
- 33.Gu L, Nusblat LM, Tishbi N, Noble SC, Pinson CM, Mintzer E, Roth CM, Uhrich KE. J. Control. Release. 2014;184:28–35. doi: 10.1016/j.jconrel.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jensen LB, Pavan GM, Kasimova MR, Rutherford S, Danani A, Nielsen HM, Foged C. Int. J. Pharm. 2011;416:410–418. doi: 10.1016/j.ijpharm.2011.03.015. [DOI] [PubMed] [Google Scholar]
- 35.Zheng M, Pavan GM, Neeb M, Schaper AK, Danani A, Klebe G, Merkel OM, Kissel T. ACS Nano. 2012;6:9447–9454. doi: 10.1021/nn301966r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kjellander R, Florin E. J Chem Soc Farad T 1. 1981;77 2053-&. [Google Scholar]
- 37.Mao S, Neu M, Germershaus O, Merkel O, Sitterberg J, Bakowsky U, Kissel T. Bioconjugate Chem. 2006;17:1209–1218. doi: 10.1021/bc060129j. [DOI] [PubMed] [Google Scholar]
- 38.Fortier C, Durocher Y, De Crescenzo G. Nanomedicine. 2014;9:135–151. doi: 10.2217/nnm.13.194. [DOI] [PubMed] [Google Scholar]
- 39.Xiao K, Li Y, Luo J, Lee JS, Xiao W, Gonik AM, Agarwal RG, Lam KS. Biomaterials. 2011;32:3435–3446. doi: 10.1016/j.biomaterials.2011.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu X, Jian Y, Li Y, Zhang X, Tu Z, Gu Z. ACS Nano. 2014;8:9255–9264. doi: 10.1021/nn503118f. [DOI] [PubMed] [Google Scholar]
- 41.Liu Y, Kim YJ, Ji M, Fang J, Siriwon N, Zhang LI, Wang P. Mol. Ther. Methods Clin. Dev. 2014;1 doi: 10.1038/mtm.2013.12. 10.1038/mtm.2013.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.