Integrin αβ transmembrane heterodimers play central roles in metazoan development and physiology by mediating adhesion and by transmitting forces and biochemical signals across the plasma membrane (Hynes, 2002; Kim et al., 2011). Integrin α and β cytoplasmic domains (tails) have many reported binding partners; however, one β-tail-binding protein, talin, is indispensable for inducing the high affinity form of integrins (“activation”) and most integrin-mediated adhesive functions require talin (Calderwood et al., 2013; Kim et al., 2011). Thus talin can be viewed as a central player in integrin functions.
A large and complex repertoire of integrins and their associated proteins forms the “integrin adhesome” (Geiger and Zaidel-Bar, 2012). The critical role of talin in integrin functions suggests that a scheme to classify the building blocks (modules) that form the adhesome could center around the interactions of talin with integrin β-tails. One set of modules are formed by proteins that displace talin to create alternative integrin cytoskeletal linkages or signaling complexes (competitive). When modules are formed by talin, non-competitive β-tail interactors can cooperate with talin to stabilize adhesion complexes or generate signals. In addition, the talin-integrin interaction can be induced by a group of talin-binding regulatory proteins (direct regulators) that can specify functional outputs (Lee et al., 2013).
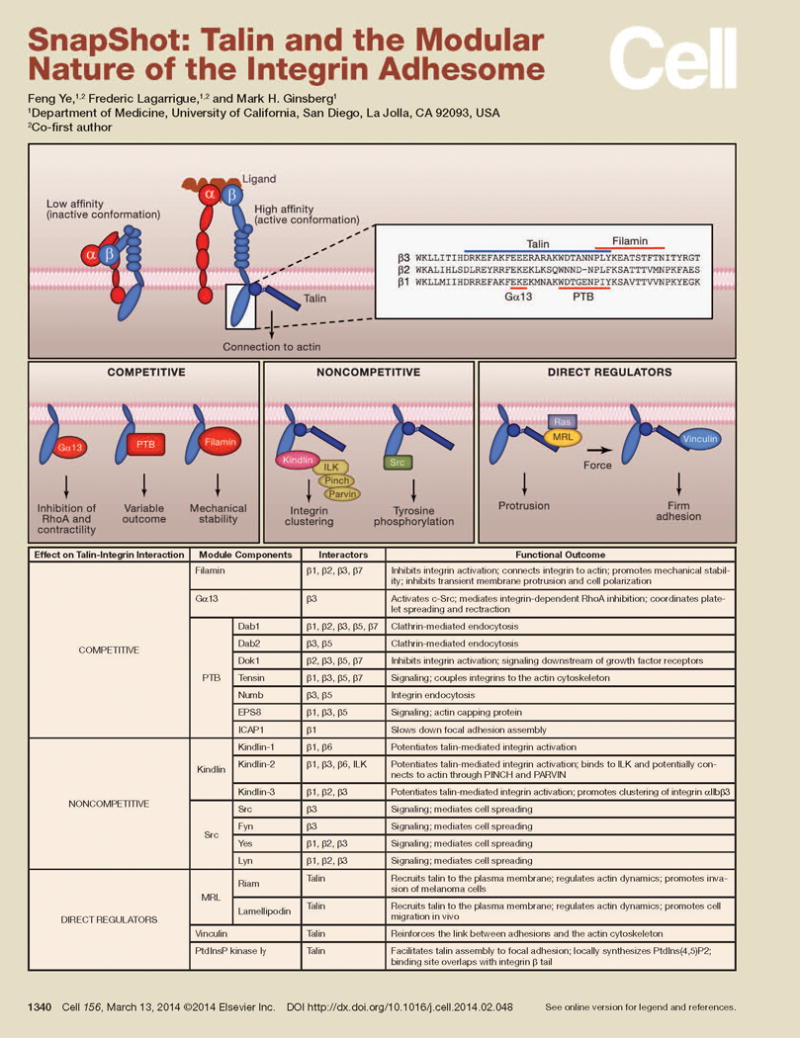
In this Snapshot, we present a simplified “modular” view of the integrin adhesome, centered on the talin-integrin interaction, and provide examples of how it can help unravel the adhesome’s remarkable functional diversity and plasticity. Furthermore, we suggest that studies to isolate, characterize, and visualize individual modules are promising approaches to understand the many functions of integrins.
The figure and accompanying table depict a talin-centric view of some of the components that comprise integrin modules that form the adhesome. The upper panels illustrate the essential role of talin in inducing a high affinity form of the integrin that can form stable, force bearing bonds with the extracellular matrix (Calderwood et al., 2013). To the right are illustrated an alignment of the β1, β2 and β3 tails with the approximate location of binding sites for talin and overlapping sites for three competitive interactors indicated by lines (Gα13, phosphotyrosine-binding (PTB) domain containing proteins, filamin). Filamin inhibits integrin activation and cell migration by blocking talin binding to β tails (Kiema et al., 2006) and establishes an alternative integrin-cytoskeleton linkage. In platelets, Gα13-β3 transiently replaces talin and inhibits RhoA (Shen et al., 2013). Many proteins containing PTB-domains compete with talin (Calderwood et al., 2003) with diverse biological consequences indicated in the table. Importantly, tyrosine phosphorylation of the integrin β-tail can favor binding of specific PTB domains relative to talin (Anthis et al., 2009). Thus, these competitors can form talin-independent modules with a wide variety of functions; importantly they can coexist in the same macro-adhesion with the talin-containing modules.
Non-competitive interactors
When talin is bound to the integrin, additional proteins can bind to the integrin β tail, thereby creating modules with new functional capabilities. For example, the c-Src SH3 domain interacts equally well with the C-terminus of the integrin β tail in the presence or absence of an integrin binding fragment of talin and C-Src can phosphorylate numerous substrates including integrins. Similarly, kindlins can bind to integrin β tails simultaneously with talin, resulting in increased integrin clustering that stabilizes adhesions(Ye et al., 2013) and recruitment of cytoskeletal adaptors such as integrin-linked kinase (Calderwood et al., 2013).
Direct regulators of talin function
Talin-binding proteins can enable talin to bind to integrin β-tails. Recent studies identified two such talin-binding proteins that form a binary switch that specify the function of the resulting module. MRL proteins, lamellipodin and RIAM, and vinculin bind talin mutually exclusively and both can drive talin to interact with and activate integrins. Furthermore, the transition from MRL-talin-integrin modules in the lamellipodium to vinculin-talin-integrin modules at the lamellum-lamellipodium border, which occurs during force-induced adhesion maturation, results in evolution of dynamic protrusion-promoting attachments into stable force-bearing adhesions (Lee et al., 2013).
Implications of a Modular Approach to the Adhesome
The integrin adhesions, classically defined by size and shape, are a heterogeneous mixture of integrin modules that are the basic units of adhesions. The admittedly incomplete scheme proposed here suggests that we may define specific modules based on a simple combinatorial scheme that divides them into those in which talin is bound and the integrin is in a high affinity state and those in which one of the competitive interactors is present in place of talin. The talin-containing modules may be further subdivided depending on which talin regulator is driving the talin-integrin interaction and the presence or absence of non-competitive interactors such as kindlins or Src kinases. Since the individual modules can differ in assembly mechanism and functional output, the changing modular composition of adhesions can help explain the changing assembly dynamics and functional output of adhesions. The challenge will be to isolate, characterize, and visualize such modules to parse the complexity and plasticity of the integrin adhesome.
References
- Anthis NJ, Haling JR, Oxley CL, Memo M, Wegener KL, Lim CJ, Ginsberg MH, Campbell ID. beta integrin tyrosine phosphorylation is a conserved mechanism for regulating talin-induced integrin activation. J Biol Chem. 2009;284:36700–36710. doi: 10.1074/jbc.M109.061275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood DA, Campbell ID, Critchley DR. Talins and kindlins: partners in integrin-mediated adhesion. Nat Rev Mol Cell Biol. 2013;14:503–517. doi: 10.1038/nrm3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood DA, Fujioka Y, de Pereda JM, Garcia-Alvarez B, Nakamoto T, Margolis B, McGlade CJ, Liddington RC, Ginsberg MH. Integrin beta cytoplasmic domain interactions with phosphotyrosine-binding domains: a structural prototype for diversity in integrin signaling. Proc Natl Acad Sci U S A. 2003;100:2272–2277. doi: 10.1073/pnas.262791999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger T, Zaidel-Bar R. Opening the floodgates: proteomics and the integrin adhesome. Curr Opin Cell Biol. 2012;24:562–568. doi: 10.1016/j.ceb.2012.05.004. [DOI] [PubMed] [Google Scholar]
- Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- Kiema T, Lad Y, Jiang P, Oxley CL, Baldassarre M, Wegener KL, Campbell ID, Ylanne J, Calderwood DA. The molecular basis of filamin binding to integrins and competition with talin. Mol Cell. 2006;21:337–347. doi: 10.1016/j.molcel.2006.01.011. [DOI] [PubMed] [Google Scholar]
- Kim C, Ye F, Ginsberg MH. Regulation of integrin activation. Annu Rev Cell Dev Biol. 2011;27:321–345. doi: 10.1146/annurev-cellbio-100109-104104. [DOI] [PubMed] [Google Scholar]
- Lee HS, Anekal P, Lim CJ, Liu CC, Ginsberg MH. Two modes of integrin activation form a binary molecular switch in adhesion maturation. Mol Biol Cell. 2013;24:1354–1362. doi: 10.1091/mbc.E12-09-0695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Zhao X, O’Brien KA, Stojanovic-Terpo A, Delaney MK, Kim K, Cho J, Lam SC, Du X. A directional switch of integrin signalling and a new anti-thrombotic strategy. Nature. 2013;503:131–135. doi: 10.1038/nature12613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F, Petrich BG, Anekal P, Lefort CT, Kasirer-Friede A, Shattil SJ, Ruppert R, Moser M, Fassler R, Ginsberg MH. The mechanism of kindlin-mediated activation of integrin alphaIIbbeta3. Curr Biol. 2013;23:2288–2295. doi: 10.1016/j.cub.2013.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]


