Abstract
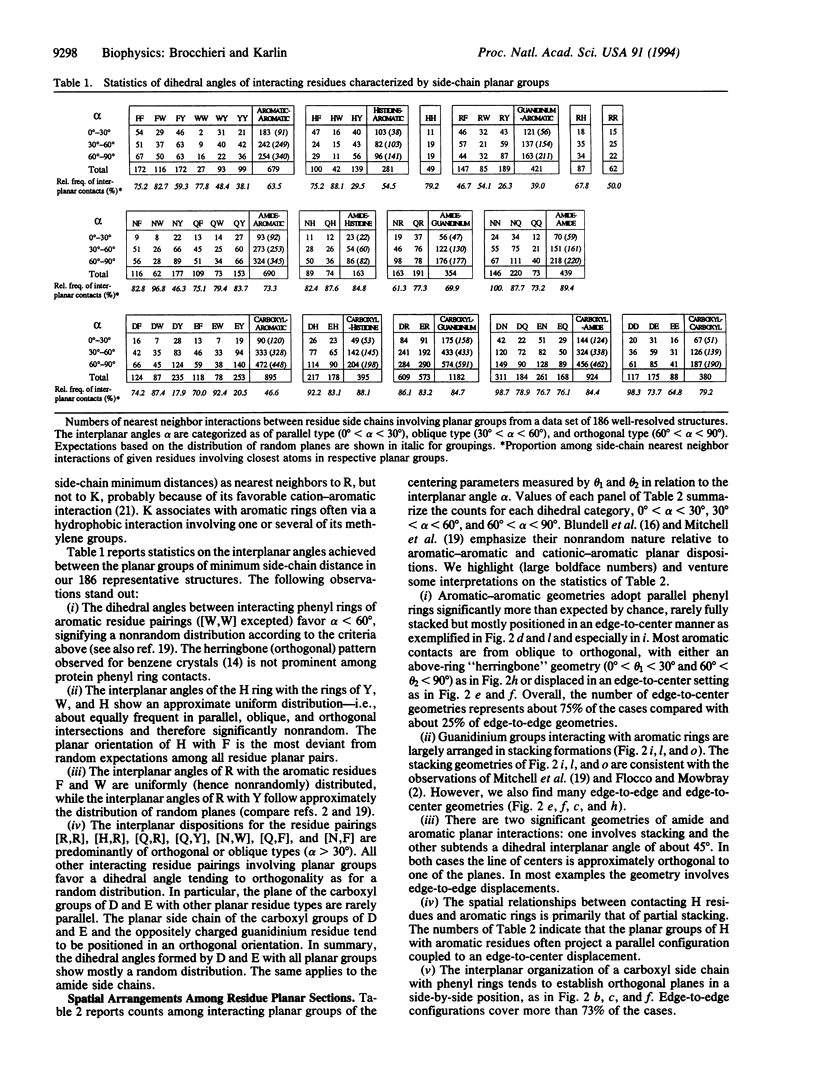
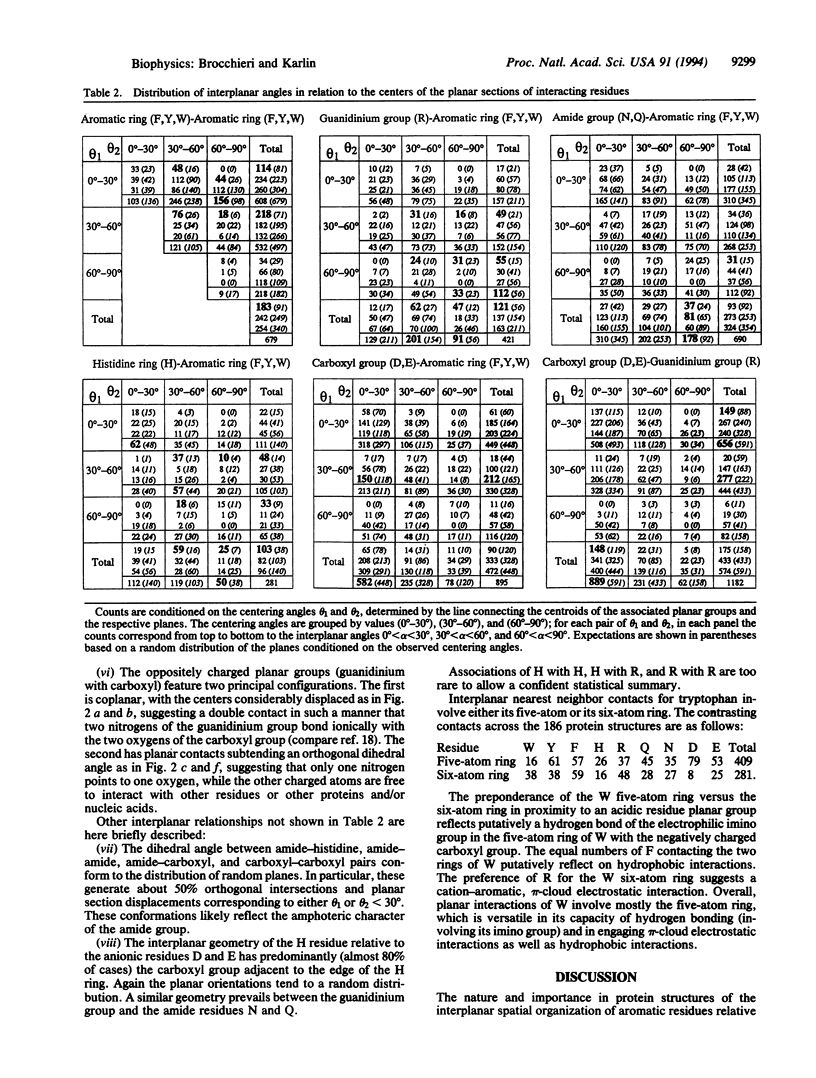
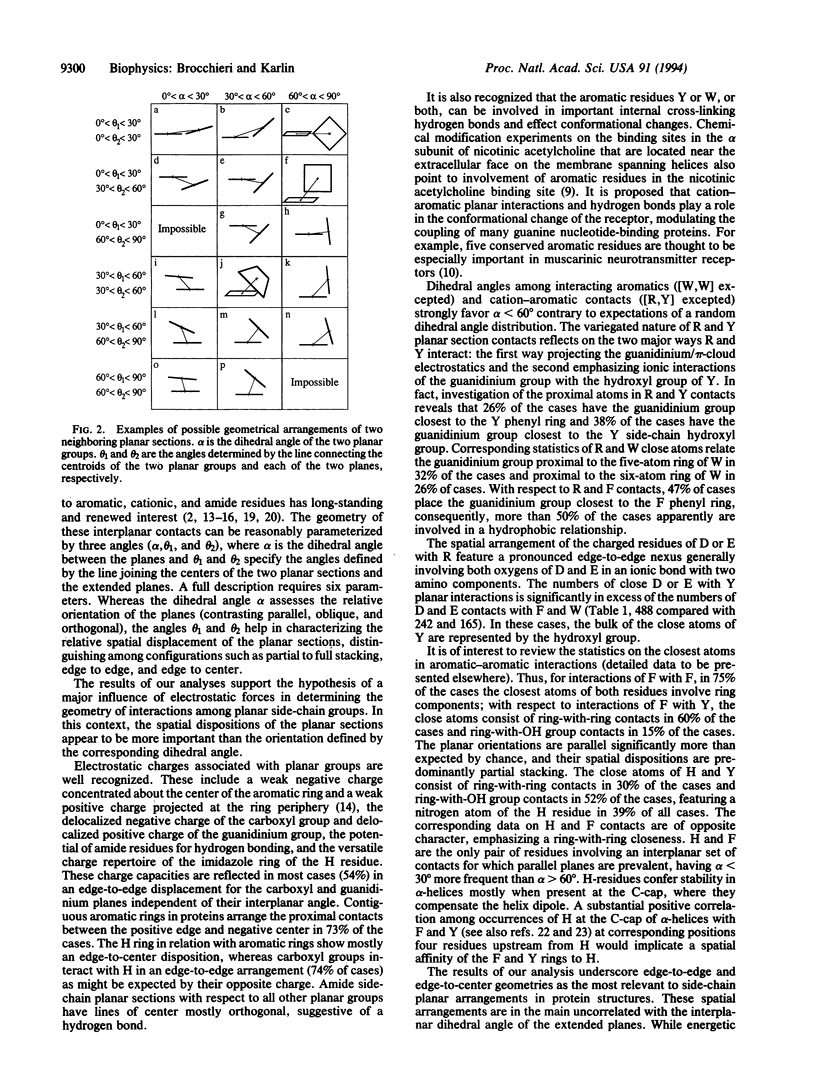
The relative spatial disposition of interacting side-chain planar groups (aromatic, guanidinium, amide, carboxyl, imidazole) is analyzed for 186 non-homologous well-resolved protein structures. The dihedral angle of amide or carboxyl planar groups with other planar groups accords with a random distribution of planes. By contrast, the dihedral angle of the planes between close aromatic rings or of the histidine ring interacting with aromatic residues is significantly nonrandom, showing an approximately uniform distribution. Our results indicate that edge-to-edge and edge-to-center spatial dispositions of residue planar sections are prevalent, while complete stacking configurations are uncommon. The hypothesis that electrostatic forces are a major determinant of the geometry of interactions between side-chain planar groups is discussed.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Armstrong K. M., Fairman R., Baldwin R. L. The (i, i + 4) Phe-His interaction studied in an alanine-based alpha-helix. J Mol Biol. 1993 Mar 5;230(1):284–291. doi: 10.1006/jmbi.1993.1142. [DOI] [PubMed] [Google Scholar]
- Bradley E. K., Thomason J. F., Cohen F. E., Kosen P. A., Kuntz I. D. Studies of synthetic helical peptides using circular dichroism and nuclear magnetic resonance. J Mol Biol. 1990 Oct 20;215(4):607–622. doi: 10.1016/S0022-2836(05)80172-X. [DOI] [PubMed] [Google Scholar]
- Burley S. K., Petsko G. A. Amino-aromatic interactions in proteins. FEBS Lett. 1986 Jul 28;203(2):139–143. doi: 10.1016/0014-5793(86)80730-x. [DOI] [PubMed] [Google Scholar]
- Burley S. K., Petsko G. A. Aromatic-aromatic interaction: a mechanism of protein structure stabilization. Science. 1985 Jul 5;229(4708):23–28. doi: 10.1126/science.3892686. [DOI] [PubMed] [Google Scholar]
- Burley S. K., Petsko G. A. Weakly polar interactions in proteins. Adv Protein Chem. 1988;39:125–189. doi: 10.1016/s0065-3233(08)60376-9. [DOI] [PubMed] [Google Scholar]
- Changeux J. P., Devillers-Thiéry A., Galzi J. L., Bertrand D. New mutants to explore nicotinic receptor functions. Trends Pharmacol Sci. 1992 Aug;13(8):299–301. doi: 10.1016/0165-6147(92)90094-m. [DOI] [PubMed] [Google Scholar]
- Dougherty D. A., Stauffer D. A. Acetylcholine binding by a synthetic receptor: implications for biological recognition. Science. 1990 Dec 14;250(4987):1558–1560. doi: 10.1126/science.2274786. [DOI] [PubMed] [Google Scholar]
- Flocco M. M., Mowbray S. L. Planar stacking interactions of arginine and aromatic side-chains in proteins. J Mol Biol. 1994 Jan 14;235(2):709–717. doi: 10.1006/jmbi.1994.1022. [DOI] [PubMed] [Google Scholar]
- Hunter C. A., Singh J., Thornton J. M. Pi-pi interactions: the geometry and energetics of phenylalanine-phenylalanine interactions in proteins. J Mol Biol. 1991 Apr 20;218(4):837–846. doi: 10.1016/0022-2836(91)90271-7. [DOI] [PubMed] [Google Scholar]
- Karlin S., Zuker M., Brocchieri L. Measuring residue associations in protein structures. Possible implications for protein folding. J Mol Biol. 1994 Jun 3;239(2):227–248. doi: 10.1006/jmbi.1994.1365. [DOI] [PubMed] [Google Scholar]
- Kumpf R. A., Dougherty D. A. A mechanism for ion selectivity in potassium channels: computational studies of cation-pi interactions. Science. 1993 Sep 24;261(5129):1708–1710. doi: 10.1126/science.8378771. [DOI] [PubMed] [Google Scholar]
- Mitchell J. B., Nandi C. L., McDonald I. K., Thornton J. M., Price S. L. Amino/aromatic interactions in proteins: is the evidence stacked against hydrogen bonding? J Mol Biol. 1994 Jun 3;239(2):315–331. doi: 10.1006/jmbi.1994.1370. [DOI] [PubMed] [Google Scholar]
- Mitchell J. B., Thornton J. M., Singh J., Price S. L. Towards an understanding of the arginine-aspartate interaction. J Mol Biol. 1992 Jul 5;226(1):251–262. doi: 10.1016/0022-2836(92)90137-9. [DOI] [PubMed] [Google Scholar]
- Nandi C. L., Singh J., Thornton J. M. Atomic environments of arginine side chains in proteins. Protein Eng. 1993 Apr;6(3):247–259. doi: 10.1093/protein/6.3.247. [DOI] [PubMed] [Google Scholar]
- Padlan E. A. On the nature of antibody combining sites: unusual structural features that may confer on these sites an enhanced capacity for binding ligands. Proteins. 1990;7(2):112–124. doi: 10.1002/prot.340070203. [DOI] [PubMed] [Google Scholar]
- Prochnicka-Chalufour A., Casanova J. L., Avrameas S., Claverie J. M., Kourilsky P. Biased amino acid distributions in regions of the T cell receptors and MHC molecules potentially involved in their association. Int Immunol. 1991 Sep;3(9):853–864. doi: 10.1093/intimm/3.9.853. [DOI] [PubMed] [Google Scholar]
- Ripoll D. R., Faerman C. H., Axelsen P. H., Silman I., Sussman J. L. An electrostatic mechanism for substrate guidance down the aromatic gorge of acetylcholinesterase. Proc Natl Acad Sci U S A. 1993 Jun 1;90(11):5128–5132. doi: 10.1073/pnas.90.11.5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman J. L., Harel M., Frolow F., Oefner C., Goldman A., Toker L., Silman I. Atomic structure of acetylcholinesterase from Torpedo californica: a prototypic acetylcholine-binding protein. Science. 1991 Aug 23;253(5022):872–879. doi: 10.1126/science.1678899. [DOI] [PubMed] [Google Scholar]
- Trumpp-Kallmeyer S., Hoflack J., Bruinvels A., Hibert M. Modeling of G-protein-coupled receptors: application to dopamine, adrenaline, serotonin, acetylcholine, and mammalian opsin receptors. J Med Chem. 1992 Sep 18;35(19):3448–3462. doi: 10.1021/jm00097a002. [DOI] [PubMed] [Google Scholar]


