Abstract
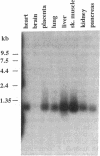
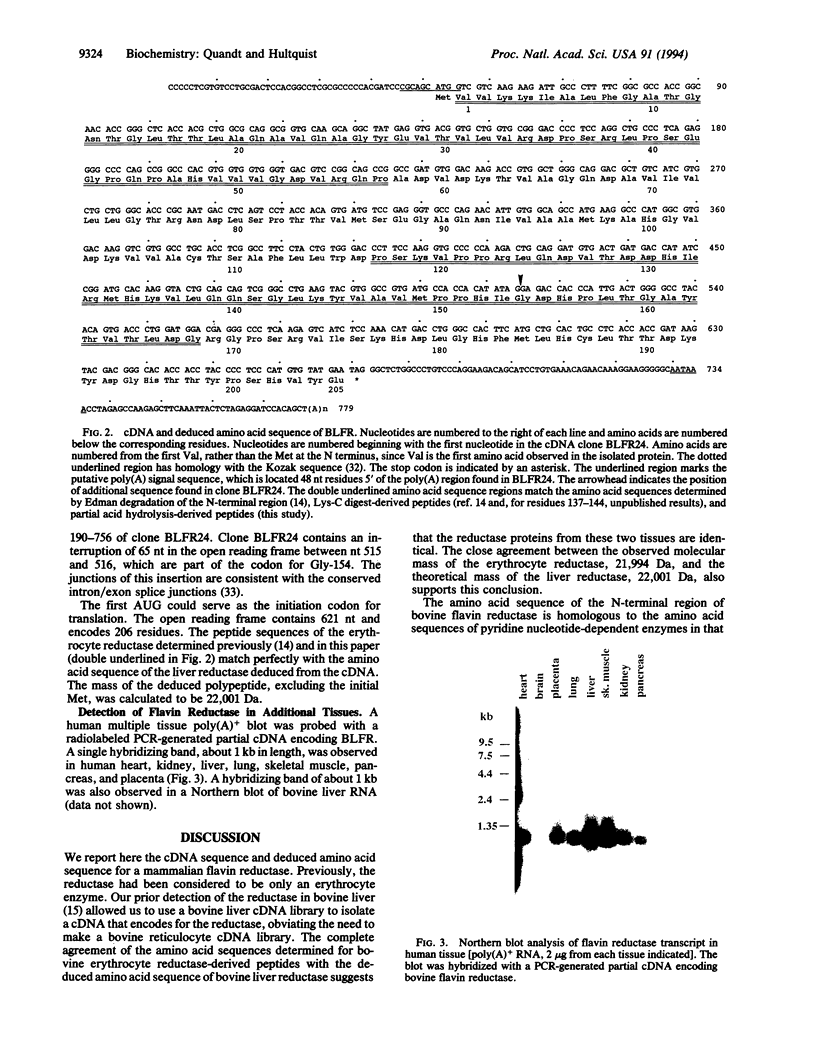
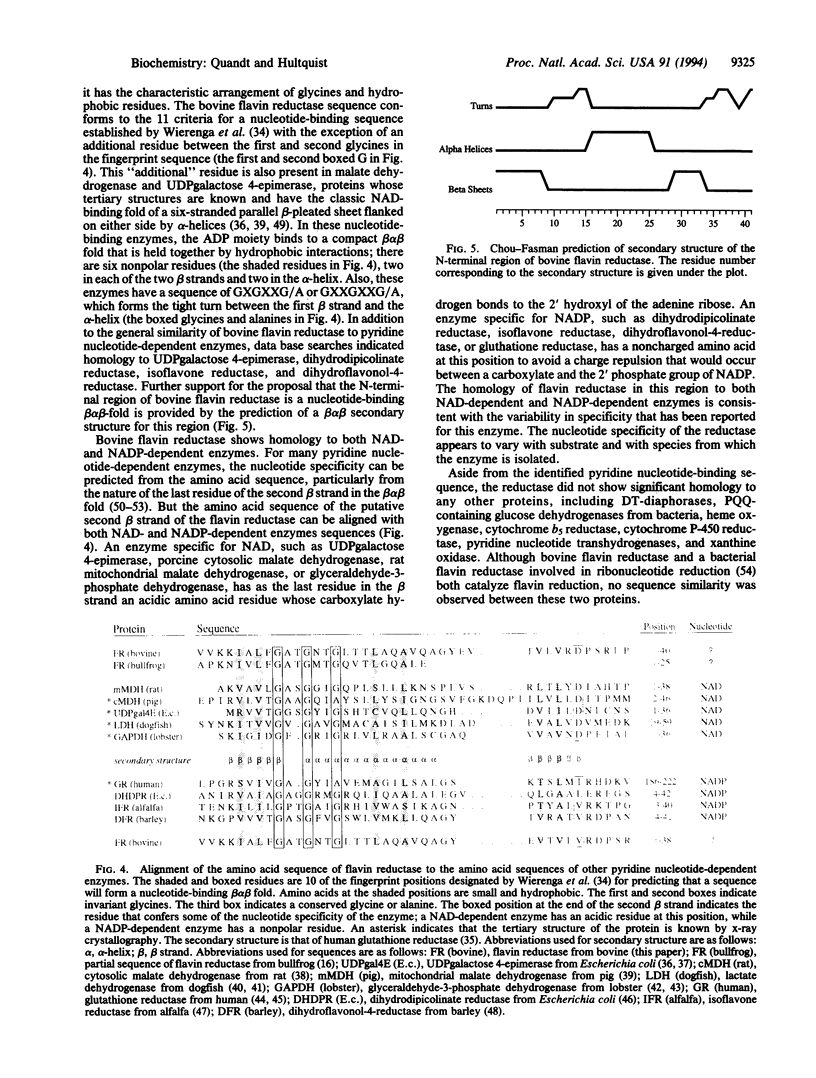
Flavin reductase catalyzes electron transfer from reduced pyridine nucleotides to methylene blue or riboflavin, and this catalysis is the basis of the therapeutic use of methylene blue or riboflavin in the treatment of methemoglobinemia. A cDNA for a mammalian flavin reductase has been isolated and sequenced. Degenerate oligonucleotides, with sequences based on amino acid sequences of peptides derived from bovine erythrocyte flavin reductase, were used as primers in PCR to selectively amplify a partial cDNA that encodes the bovine reductase. The template used in the PCR was first strand cDNA synthesized from bovine liver total RNA using oligo(dT) primers. A PCR product was used as a specific probe to screen a bovine liver cDNA library. The sequence determined from two overlapping clones contains an open reading frame of 621 nucleotides and encodes 206 amino acids. The amino acid sequence deduced from the bovine liver flavin reductase cDNA matches the amino acid sequences determined for erythrocyte reductase-derived peptides, and the predicted molecular mass of 22,001 Da for the liver reductase agrees well with the molecular mass of 21,994 Da determined for the erythrocyte reductase by electrospray mass spectrometry. The amino acid sequence at the N terminus of the reductase has homology to sequences of pyridine nucleotide-dependent enzymes, and the predicted secondary structure, beta alpha beta, resembles the common nucleotide-binding structural motif. RNA blot analysis indicates a single 1-kilobase reductase transcript in human heart, kidney, liver, lung, pancreas, placenta, and skeletal muscle.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abad-Zapatero C., Griffith J. P., Sussman J. L., Rossmann M. G. Refined crystal structure of dogfish M4 apo-lactate dehydrogenase. J Mol Biol. 1987 Dec 5;198(3):445–467. doi: 10.1016/0022-2836(87)90293-2. [DOI] [PubMed] [Google Scholar]
- Abe Y., Ito T., Okazaki T. Purification and characterization of NADPH-dependent methemoglobin reductase from the nucleated erythrocytes of bullfrog, Rana catesbeiana. J Biochem. 1990 Aug;108(2):255–260. doi: 10.1093/oxfordjournals.jbchem.a123190. [DOI] [PubMed] [Google Scholar]
- Adachi K., Okuyama T. Study on the reduced pyridine nucleotide dehydrogenase of bovine erythrocytes. I. Crystallization and properties of the reduced pyridine nucleotide dehydrogenase of bovine erythrocytes. Biochim Biophys Acta. 1972 Jun 16;268(3):629–637. doi: 10.1016/0005-2744(72)90266-5. [DOI] [PubMed] [Google Scholar]
- Adachi K. Studies on reduced pyridine nucleotide dehydrogenase in bovine erythrocytes. II. Electron acceptor specificity of two types of reduced pyridine nucleotide dehydrogenase in bovine erythrocytes. Biochim Biophys Acta. 1972 Dec 7;289(2):262–268. doi: 10.1016/0005-2744(72)90076-9. [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Baker P. J., Britton K. L., Rice D. W., Rob A., Stillman T. J. Structural consequences of sequence patterns in the fingerprint region of the nucleotide binding fold. Implications for nucleotide specificity. J Mol Biol. 1992 Nov 20;228(2):662–671. doi: 10.1016/0022-2836(92)90848-e. [DOI] [PubMed] [Google Scholar]
- Bauer A. J., Rayment I., Frey P. A., Holden H. M. The molecular structure of UDP-galactose 4-epimerase from Escherichia coli determined at 2.5 A resolution. Proteins. 1992 Apr;12(4):372–381. doi: 10.1002/prot.340120409. [DOI] [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Birktoft J. J., Rhodes G., Banaszak L. J. Refined crystal structure of cytoplasmic malate dehydrogenase at 2.5-A resolution. Biochemistry. 1989 Jul 11;28(14):6065–6081. doi: 10.1021/bi00440a051. [DOI] [PubMed] [Google Scholar]
- Bouvier J., Richaud C., Richaud F., Patte J. C., Stragier P. Nucleotide sequence and expression of the Escherichia coli dapB gene. J Biol Chem. 1984 Dec 10;259(23):14829–14834. [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Brillantes A. M., Allen P., Takahashi T., Izumo S., Marks A. R. Differences in cardiac calcium release channel (ryanodine receptor) expression in myocardium from patients with end-stage heart failure caused by ischemic versus dilated cardiomyopathy. Circ Res. 1992 Jul;71(1):18–26. doi: 10.1161/01.res.71.1.18. [DOI] [PubMed] [Google Scholar]
- Chikuba K., Yubisui T., Shirabe K., Takeshita M. Cloning and nucleotide sequence of a cDNA of the human erythrocyte NADPH-flavin reductase. Biochem Biophys Res Commun. 1994 Feb 15;198(3):1170–1176. doi: 10.1006/bbrc.1994.1165. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- Davidson B. E., Sajgò M., Noller H. F., Harris J. I. Amino-acid sequence of glyceraldehyde 3-phosphate dehydrogenase from lobster muscle. Nature. 1967 Dec 23;216(5121):1181–1185. doi: 10.1038/2161181a0. [DOI] [PubMed] [Google Scholar]
- Fan F., Lorenzen J. A., Plapp B. V. An aspartate residue in yeast alcohol dehydrogenase I determines the specificity for coenzyme. Biochemistry. 1991 Jul 2;30(26):6397–6401. doi: 10.1021/bi00240a008. [DOI] [PubMed] [Google Scholar]
- Feeney R., Clarke A. R., Holbrook J. J. A single amino acid substitution in lactate dehydrogenase improves the catalytic efficiency with an alternative coenzyme. Biochem Biophys Res Commun. 1990 Jan 30;166(2):667–672. doi: 10.1016/0006-291x(90)90861-g. [DOI] [PubMed] [Google Scholar]
- Galaris D., Eddy L., Arduini A., Cadenas E., Hochstein P. Mechanisms of reoxygenation injury in myocardial infarction: implications of a myoglobin redox cycle. Biochem Biophys Res Commun. 1989 May 15;160(3):1162–1168. doi: 10.1016/s0006-291x(89)80125-1. [DOI] [PubMed] [Google Scholar]
- Gibson Q. H. The reduction of methaemoglobin in red blood cells and studies on the cause of idiopathic methaemoglobinaemia. Biochem J. 1948;42(1):13–23. doi: 10.1042/bj0420013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant P. M., Tellam J., May V. L., Strauss A. W. Isolation and nucleotide sequence of a cDNA clone encoding rat mitochondrial malate dehydrogenase. Nucleic Acids Res. 1986 Aug 11;14(15):6053–6066. doi: 10.1093/nar/14.15.6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultquist D. E., Xu F., Quandt K. S., Shlafer M., Mack C. P., Till G. O., Seekamp A., Betz A. L., Ennis S. R. Evidence that NADPH-dependent methemoglobin reductase and administered riboflavin protect tissues from oxidative injury. Am J Hematol. 1993 Jan;42(1):13–18. doi: 10.1002/ajh.2830420105. [DOI] [PubMed] [Google Scholar]
- Kajita A., Kerwar G. K., Huennekens F. M. Multiple forms of methemoglobin reductase. Arch Biochem Biophys. 1969 Mar;130(1):662–673. doi: 10.1016/0003-9861(69)90085-x. [DOI] [PubMed] [Google Scholar]
- Karplus P. A., Schulz G. E. Refined structure of glutathione reductase at 1.54 A resolution. J Mol Biol. 1987 Jun 5;195(3):701–729. doi: 10.1016/0022-2836(87)90191-4. [DOI] [PubMed] [Google Scholar]
- Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J Biol Chem. 1991 Oct 25;266(30):19867–19870. [PubMed] [Google Scholar]
- Krauth-Siegel R. L., Blatterspiel R., Saleh M., Schiltz E., Schirmer R. H., Untucht-Grau R. Glutathione reductase from human erythrocytes. The sequences of the NADPH domain and of the interface domain. Eur J Biochem. 1982 Jan;121(2):259–267. doi: 10.1111/j.1432-1033.1982.tb05780.x. [DOI] [PubMed] [Google Scholar]
- Kristiansen K. N., Rohde W. Structure of the Hordeum vulgare gene encoding dihydroflavonol-4-reductase and molecular analysis of ant18 mutants blocked in flavonoid synthesis. Mol Gen Genet. 1991 Nov;230(1-2):49–59. doi: 10.1007/BF00290650. [DOI] [PubMed] [Google Scholar]
- Landon Cleavage at aspartyl-prolyl bonds. Methods Enzymol. 1977;47:145–149. doi: 10.1016/0076-6879(77)47017-4. [DOI] [PubMed] [Google Scholar]
- Lemaire H. G., Müller-Hill B. Nucleotide sequences of the gal E gene and the gal T gene of E. coli. Nucleic Acids Res. 1986 Oct 10;14(19):7705–7711. doi: 10.1093/nar/14.19.7705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moras D., Olsen K. W., Sabesan M. N., Buehner M., Ford G. C., Rossmann M. G. Studies of asymmetry in the three-dimensional structure of lobster D-glyceraldehyde-3-phosphate dehydrogenase. J Biol Chem. 1975 Dec 10;250(23):9137–9162. doi: 10.2210/pdb1gpd/pdb. [DOI] [PubMed] [Google Scholar]
- Niethammer D., Huennekens F. M. Electrophoretic separation and characterization of the multiple forms of methemoglobin reductase. Arch Biochem Biophys. 1971 Oct;146(2):564–573. doi: 10.1016/0003-9861(71)90162-7. [DOI] [PubMed] [Google Scholar]
- Nishiyama M., Birktoft J. J., Beppu T. Alteration of coenzyme specificity of malate dehydrogenase from Thermus flavus by site-directed mutagenesis. J Biol Chem. 1993 Mar 5;268(7):4656–4660. [PubMed] [Google Scholar]
- Paiva N. L., Edwards R., Sun Y. J., Hrazdina G., Dixon R. A. Stress responses in alfalfa (Medicago sativa L.) 11. Molecular cloning and expression of alfalfa isoflavone reductase, a key enzyme of isoflavonoid phytoalexin biosynthesis. Plant Mol Biol. 1991 Oct;17(4):653–667. doi: 10.1007/BF00037051. [DOI] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quandt K. S., Xu F., Chen P., Hultquist D. E. Evidence that the protein components of bovine erythrocyte green heme binding protein and flavin reductase are identical. Biochem Biophys Res Commun. 1991 Jul 15;178(1):315–321. doi: 10.1016/0006-291x(91)91816-u. [DOI] [PubMed] [Google Scholar]
- SCOTT E. M., DUNCAN I. W., EKSTRAND V. THE REDUCED PYRIDINE NUCLEOTIDE DEHYDROGENASES OF HUMAN ERYTHROCYTES. J Biol Chem. 1965 Jan;240:481–485. [PubMed] [Google Scholar]
- SHRAGO E., FALCONE A. B. Human-erythrocyte reduced triphosphopyridine nucleotide oxidase. Biochim Biophys Acta. 1963 Jan 8;67:147–149. doi: 10.1016/0006-3002(63)91805-5. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrutton N. S., Berry A., Perham R. N. Redesign of the coenzyme specificity of a dehydrogenase by protein engineering. Nature. 1990 Jan 4;343(6253):38–43. doi: 10.1038/343038a0. [DOI] [PubMed] [Google Scholar]
- Spyrou G., Haggård-Ljungquist E., Krook M., Jörnvall H., Nilsson E., Reichard P. Characterization of the flavin reductase gene (fre) of Escherichia coli and construction of a plasmid for overproduction of the enzyme. J Bacteriol. 1991 Jun;173(12):3673–3679. doi: 10.1128/jb.173.12.3673-3679.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S. S. Amino acid sequence of dogfish muscle lactate dehydrogenase. J Biol Chem. 1977 Mar 10;252(5):1799–1806. [PubMed] [Google Scholar]
- Wierenga R. K., Drenth J., Schulz G. E. Comparison of the three-dimensional protein and nucleotide structure of the FAD-binding domain of p-hydroxybenzoate hydroxylase with the FAD- as well as NADPH-binding domains of glutathione reductase. J Mol Biol. 1983 Jul 5;167(3):725–739. doi: 10.1016/s0022-2836(83)80106-5. [DOI] [PubMed] [Google Scholar]
- Wierenga R. K., Terpstra P., Hol W. G. Prediction of the occurrence of the ADP-binding beta alpha beta-fold in proteins, using an amino acid sequence fingerprint. J Mol Biol. 1986 Jan 5;187(1):101–107. doi: 10.1016/0022-2836(86)90409-2. [DOI] [PubMed] [Google Scholar]
- Wilbur W. J., Lipman D. J. Rapid similarity searches of nucleic acid and protein data banks. Proc Natl Acad Sci U S A. 1983 Feb;80(3):726–730. doi: 10.1073/pnas.80.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F., Hultquist D. E. Coupling of dihydroriboflavin oxidation to the formation of the higher valence states of hemeproteins. Biochem Biophys Res Commun. 1991 Nov 27;181(1):197–203. doi: 10.1016/s0006-291x(05)81401-9. [DOI] [PubMed] [Google Scholar]
- Xu F., Mack C. P., Quandt K. S., Shlafer M., Massey V., Hultquist D. E. Pyrroloquinoline quinone acts with flavin reductase to reduce ferryl myoglobin in vitro and protects isolated heart from re-oxygenation injury. Biochem Biophys Res Commun. 1993 May 28;193(1):434–439. doi: 10.1006/bbrc.1993.1642. [DOI] [PubMed] [Google Scholar]
- Xu F., Quandt K. S., Hultquist D. E. Characterization of NADPH-dependent methemoglobin reductase as a heme-binding protein present in erythrocytes and liver. Proc Natl Acad Sci U S A. 1992 Mar 15;89(6):2130–2134. doi: 10.1073/pnas.89.6.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yubisui T., Matsuki T., Takeshita M., Yoneyama Y. Characterization of the purified NADPH-flavin reductase of human erythrocytes. J Biochem. 1979 Mar;85(3):719–728. [PubMed] [Google Scholar]
- Yubisui T., Matsuki T., Tanishima K., Takeshita M., Yoneyama Y. NADPH-flavin reductase in human erythrocytes and the reduction of methemoglobin through flavin by the enzyme. Biochem Biophys Res Commun. 1977 May 9;76(1):174–182. doi: 10.1016/0006-291x(77)91683-7. [DOI] [PubMed] [Google Scholar]
- Yubisui T., Tamura M., Takeshita M. Characterization of a second form of NADPH-flavin reductase purified from human erythrocytes. Biochem Int. 1987 Jul;15(1):1–8. [PubMed] [Google Scholar]