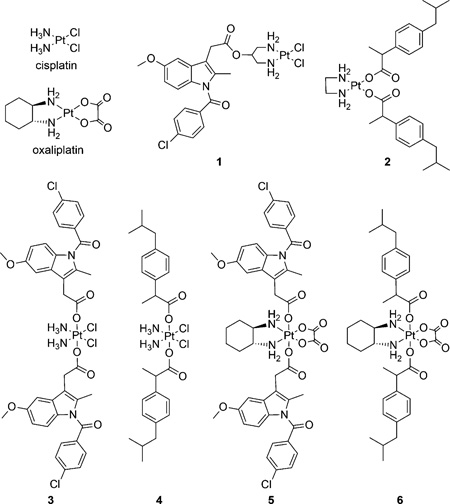
Abstract
Cyclooxygenase (COX) is an enzyme involved in tumorigenesis and is associated with tumor cell resistance against platinum-based antitumor drugs. Cisplatin analogues were conjugated with COX inhibitors (indomethacin, ibuprofen) to study the synergistic effects that were previously observed in combination treatments. The conjugates ensure concerted transport of both drugs into cells, and subsequent intracellular cleavage enables a dual-action mode. Whereas the platinum(II) complexes showed cytotoxicities similar to those of cisplatin, the platinum(IV) conjugates revealed highly increased cytotoxic activities and were able to completely overcome cisplatin-related resistance. Although some of the complexes are potent COX inhibitors, the conjugates appear to execute their cytotoxic action via COX-independent mechanisms. Instead, the increased lipophilicity and kinetic inertness of the conjugates seem to facilitate cellular accumulation of the platinum drugs and thus improve the efficacy of the antitumor agents. These conjugates are important tools for the elucidation of the direct influence of COX inhibitors on platinum-based anticancer drugs in tumor cells.
Keywords: antitumor agents, cyclooxygenase inhibitors, drug design, platinum, prodrugs
1. Introduction
Cisplatin and its analogues are among the most widely used chemotherapeutic agents in the treatment of various types of cancer.[1] However, platinum-based antitumor therapy is complicated by severe side effects and resistance of tumor cells. While the side effects mainly result from the low selectivity of these drugs for tumor cells, many cells exhibit an intrinsic resistance against the agents or acquire one upon treatment. Resistance mechanisms include decreased influx, increased efflux, detoxification of the drugs, repair or tolerance of DNA lesions, and interference with apoptotic pathways, which are usually activated by these drugs.[2] Implicated in cisplatin resistance is cyclooxygenase (COX), an enzyme that catalyzes key steps in the biosynthesis of prostanoids. These mediators play an important role in generating inflammatory responses, which are also associated with tumorigenesis. The inducible isoform, COX-2, is overexpressed in many tumor tissues and is involved in tumor initiation and progression.[3] COX-2 is also reported to cause poor outcome and low overall survival in several types of cisplatin-treated cancers.[2d, 4] Therefore, COX inhibitors, including non-steroidal anti-inflammatory drugs (NSAIDs) and COX-2-selective inhibitors (COXIBs), are increasingly used as cancer-preventive and adjuvant chemotherapeutic agents. Clinical studies have shown synergistic effects if various antitumor agents (e.g., cisplatin, paclitaxel, doxorubicin) were administered in combination with COX inhibitors, either improving the cytotoxic potency or decreasing the severity of cisplatin-induced side effects.[5] However, the mechanisms by which COX is involved in tumorigenesis are still largely unknown, and conflicting results have been reported. Whereas several studies revealed the positive effects of COX inhibitors to be COX-2 independent,[6] even antagonistic effects have been observed.[7] Furthermore, increased COX-2 expression in tumor cells upon treatment with antitumor agents such as cisplatin was shown in preclinical studies.[8]
Prior studies to investigate the influence of COX inhibitors on the efficacy of antitumor agents have used combination treatments resulting in potential discrepancies between clinical and tissue culture studies. Due to differential pharmacokinetic profiles and biodistribution parameters, delivery of the drugs to a tumor in vivo may fail to recapitulate administration of the drugs to cells in culture when administered individually. Hence, to study the direct influence of COX inhibitors on antitumor agents, tools that ensure concerted transport into tumor cells and that enable intracellular release of both drugs are necessary. To address this issue, we prepared several conjugates of cisplatin analogues and NSAIDs to be used as single prodrugs. Our first conjugates of cisplatin with indomethacin and ibuprofen (complexes 3 and 4, respectively) already showed much higher cytotoxicities than cisplatin and were even able to completely overcome cisplatin-related resistance.[9] Despite potently inhibiting COX-2 as well, the conjugates seemed to execute their cytotoxic action via COX-2-independent mechanisms. Although less potent, the conjugate between cisplatin and aspirin also revealed an improved efficacy of the platinum drug.[10]
Herein we present further conjugates, including platinum(II) and platinum(IV) complexes, with various intracellularly cleavable linkers and investigations into their mode of action. Only conjugation strategies that allow release of the NSAIDs (indomethacin, ibuprofen) without derivatization upon intracellular cleavage were used, thus enabling a direct comparison with combination treatments. Conjugation at the backbone of the non-leaving amine ligand of a cisplatin analogue was achieved via an ester bond. Ester bonds are stable under neutral conditions, but are cleaved in the acidic environment of the lysosome or endosome after uptake into the cells. Furthermore, they are hydrolyzed by esterases.[11] While the high level of esterases in the blood plasma should have a rather low impact on the stability of the conjugate in cell-culture studies, esterase inhibitors could help prevent undesired hydrolysis in blood during in vivo studies. Conjugate 1 was designed to release one molecule of the NSAID along with an antitumor-active complex containing two chloro ligands as leaving ligands. Besides conjugation at the non-leaving ligand, NSAIDs were coordinated at platinum(II) as leaving ligands via their carboxylic group (conjugate 2). Dissociation of the ligands from the metal center inside the tumor cell, similar to the activation of cisplatin, leads to release of two NSAID molecules per antitumor agent. Whereas platinum(II) compounds are rather labile due to dissociation, platinum(IV) complexes are much more inert against ligand exchange. This high kinetic inertness even enables oral administration, whereas platinum(II) compounds have to be injected intravenously.[12] As the activity is not affected, oxidation allows stabilization of platinum(II) complexes, which reveal a promising spectrum of activity, but are hampered by poor pharmacokinetics.[13] Intracellular reduction of platinum(IV) complexes by redox-active biomolecules such as glutathione (GSH) or ascorbate leads to release of the respective antitumor-active platinum(II) species.[14] Furthermore, the higher lipophilicity of platinum(IV) complexes facilitates the transport of the agents across cell membranes, thereby increasing cellular accumulation and the efficacy of the drugs.[14a, 15] Exerting activity in environments often associated with resistance to chemotherapeutic agents, platinum(IV) complexes are also generally not subject to multicellular resistance.[13] While high levels of GSH in cisplatin-resistant cells lead to deactivation of platinum(II) complexes, the reducing potency of GSH may activate platinum(IV) complexes, thereby presenting a mechanism of circumventing cisplatin resistance. Additionally, the axial ligands enable specific targeting of established platinum(II) drugs or the introduction of a dual mode of action, as derivatization of the equatorial ligands is not necessary. In this study, NSAIDs served as axial ligands of oxidized cisplatin (conjugates 3 and 4)[9] and oxaliplatin (conjugates 5 and 6), enabling release of the platinum-based agents along with two molecules of the respective COX inhibitor upon intracellular reduction.
2. Results and Discussion
2.1. Synthesis of conjugates
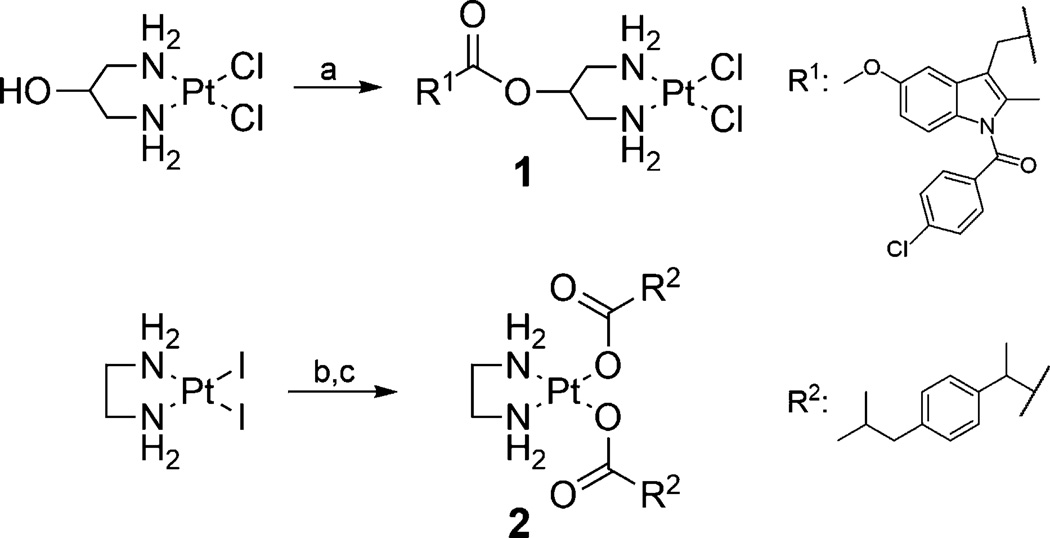
Indomethacin, an unselective COX inhibitor, was chosen for an ester-linked conjugate 1. As indomethacin esters have been reported to be COX-2 selective,[16] and COXIBs are selectively accumulated in tumors,[17] this NSAID could help enrich the conjugate in tumor cells. Formation of the ester bond was carried out after coordination of diaminopropanol at platinum(II) by using BOP-Cl (N,N’-bis(2-oxo-3-oxazolidinyl)phosphinic chloride) as activating agent (Scheme 1). Thereby, the coordination at platinum was also used as a protecting strategy for the amine groups during esterification to prevent the formation of amide bonds. Attempts to coordinate the respective diaminopropanyl ester of indomethacin at platinum(II) resulted in rearrangement of the ester to an amide upon the required usage of a base.
Scheme 1.
Synthetic routes for conjugates 1 and 2. Reagents and conditions: a) R1COOH, N,N’-bis(2-oxo-3-oxazolidinyl)phosphinic chloride (BOP-Cl), Et3N, CH2Cl2, RT, 3 d, 37%; b) Ag2CO3, H2O, RT, 2.5 d, (not isolated); c) R2COOH, H2O, 55°C, 14 h, 10%.
In contrast to the use of an ester linkage, the NSAID ibuprofen in conjugate 2 was coordinated directly at the metal center via its carboxylic group. Several approaches for the synthesis of platinum(II) carboxylato complexes have been described. The synthetic procedures usually start from the respective diamine complex containing chloro or iodo ligands and involve the use of a silver salt to achieve coordination of the carboxylic acid at the metal center. If the product complex is water soluble, it is most convenient to precipitate the by-products from the reaction mixture (e.g., AgCl).[18] In the case of conjugate 2, however, the water solubility of the product complex is extremely low, so that any by-product should remain in the aqueous reaction solution or evolve as gas to ensure separation from the precipitating product. Therefore, [PtCl2(en)] (en: ethylenediamine) was treated with AgNO3 to form the respective diaqua complex [Pt(OH2)2(en)](NO3)2, which was then further reacted with the sodium salt of ibuprofen.[18a, c, 19] The reaction, however, led to a mixture of the desired product 2 and side products, which probably resulted from salt formation between the starting materials. Therefore, conjugate 2 was instead prepared by reaction of ibuprofen with a platinum(II) carbonato complex [Pt(CO3)(en)], which was formed by reacting [PtCl2(en)] with Ag2CO3.[20] With the only by-products being water and carbon dioxide, complex 2 could be obtained in high purity. Attempts to synthesize the respective indomethacin analogue were not successful. Although this conjugate was formed, it seemed to decompose, as it turned black upon isolation and could therefore not be prepared as a pure compound. The sterically more demanding indomethacin ligands might lead to destabilization of the conjugate. Furthermore, due to the more hydrophobic ligands, the indomethacin conjugate revealed even lower water solubility than conjugate 2.
The platinum(IV) conjugates 3, 4, 5, and 6 were synthesized by reacting the dihydroxoplatinum(IV) analogue of cisplatin or oxaliplatin with the acyl chloride of the respective NSAID (indomethacin, ibuprofen) in the presence of a base.[9, 21] All conjugates were identified by various analytical methods, including NMR and IR spectroscopy as well as mass spectrometry.
2.2. Release of antitumor-active species
The platinum(II) conjugates 1 and 2 are expected to execute their antitumor activity similar to cisplatin via dissociation of their leaving ligands. Analogous to cisplatin, both conjugates undergo rapid exchange of one ligand upon dissolution in DMSO, resulting in the formation of the respective mono-cationic derivatives (Scheme S1, Supporting Information).[22] As a soft metal, platinum prefers coordination of soft S-donors to hard O-donors, resulting in fast ligand exchange at platinum(II) complexes and strong coordination of the nucleophilic sulfur atom of DMSO at the metal center. While in conjugate 1 one chloro ligand is exchanged, conjugate 2 releases one ibuprofen molecule, enabling a dual action mode of both drugs. The time courses of the dissociation of 1 and 2 were followed by 1H NMR spectroscopy, and the formed species were further proved by mass spectrometry (Figure S1, Supporting Information). The ligand exchange started immediately upon dissolution of the conjugates in DMSO and was completed within less than one day.[22d] Similar to solvolysis in DMSO, aquation of the conjugates is expected, enabling intracellular activation prior to binding to DNA.
In contrast to platinum(II) complexes, platinum(IV) complexes are kinetically inert to ligand exchange, thus enabling oral administration.[12] However, these complexes are intracellularly reduced by redox-active biomolecules, such as GSH and ascorbate, to more labile platinum(II) complexes.[14] Thus, their pharmacological profile is significantly influenced by the reduction potential, and this in turn depends on the nature of the axial ligands. Reduction of platinum(IV) complexes most readily occurs with axial chloro ligands, least readily for axial hydroxo ligands and is intermediate for axial carboxylato ligands.[23] Reduction potentials that are too high lead to reduction in the blood, resulting in severe side effects (e.g., tetraplatin, systemic toxicity),[24] while excessively low potentials result in excretion of the intact compounds (e.g., iproplatin, weak activity).[25] The platinum(IV) conjugates 3, 4, 5, and 6 showed irreversible reduction, resulting in only one peak in the cyclic voltammograms (Figure S2, Supporting Information), which is caused by the loss of the axial ligands and change of the coordination geometry.[9] All conjugates exhibited similar reduction potentials (Table 1) which are similar to those of other reported platinum(IV) carboxylato complexes.[15d, 26] The intermediate reduction potentials should ensure in vivo stability of the complexes in blood during transport but enable intracellular reduction. In addition to the reduction potential, the biological properties of platinum(IV) compounds are also influenced by the reduction rate of the complexes.[23] Reduction of the platinum(IV) conjugates by redox-active biomolecules was demonstrated by incubation of 3 and 4 with ascorbic acid at 37°C and monitoring by 1H NMR spectroscopy.[9] About 40% of the conjugates were reduced within three days, a reduction rate which should be compatible with the clearance rate of such drugs.[27]
Table 1.
Reduction potentials of platinum(IV) conjugates 3–6.
Normalized vs. normal hydrogen electrode (NHE) in DMF; values are the mean of triplicates with standard deviation <5 %.
As determined previously.[9]
2.3. COX inhibition
Although the conjugates are expected to be cleaved intracellularly, they might also be capable of binding at the COX enzyme and could thereby contribute to targeted delivery of the antitumor agents.[17] The COX potency and selectivity of the conjugates were tested on the purified COX-1 and COX-2 enzymes (Table 2). Neither of the isoforms was inhibited by any of the ibuprofen conjugates (2, 4, or 6). Ibuprofen itself is a weak, rapidly reversible inhibitor of COX (IC50(COX-2): >25 µm),[28] and this probably results in only weak binding of the conjugates as well.[29] In contrast, the indomethacin conjugates (1, 3, and 5) represent potent COX inhibitors with different activities and selectivities. Conjugate 1 inhibited the enzymes unselectively with a rather low potency. Although the IC50 values of this compound resemble those of indomethacin (IC50(COX-1): 0.05 µm, IC50(COX-2): 0.75 µm),[16] the ester bond of the conjugate should be stable under the applied assay conditions and not lead to a release of indomethacin. Conjugation with the platinum complex via the short linker, however, might hinder a strong interaction with the enzymes. The platinum(IV) conjugates 3 and 5 revealed markedly higher COX inhibitory activity, but showed different selectivities for the isozymes. Whereas 3 is highly COX-2 selective,[9] conjugate 5 was COX-1 selective, although the complexes only differ in their equatorial coordination spheres. Esterification of indomethacin often generates inhibitor selectivity for COX-2.[16] Analogously, conjugate 3 represents a highly potent inhibitor with 100-fold selectivity for COX-2. The different equatorial ligands in 5, however, seem to result in a different binding mode at the enzyme leading to COX-1 selectivity. To get insight into the binding modes, docking studies were carried out for the two conjugates. The predicted trends are in agreement with the results of the inhibition studies. While 3 favors COX-2 over COX-1, 5 might only bind at COX-1 but not at COX-2 (Table S3, Supporting Information). With one indomethacin unit bound in the active site, the equatorial ligands of the complexes interact with side chains at the entrance of the catalytic domain while the second indomethacin unit protrudes into the membrane-binding domain of the enzymes (Figures S5–S7, Supporting Information). The different orientations and interactions of the equatorial ligands, cisplatin and oxaliplatin, with the isoforms seem to lead to the COX selectivities of the conjugates. The high COX-2 selectivity of 3 might result from an additional interaction of the outer indomethacin unit with the active site (Figure S4, Supporting Information). In contrast, the constricted entrance in COX-2 might hinder 5 with the large equatorial oxaliplatin from reaching the active site (Figure S6, Supporting Information). Upon entry of tumor cells, either the released NSAIDs or even the intact conjugates could lead to COX inhibition and thus decrease tumor-associated inflammation, as shown for a similar conjugate between cisplatin and aspirin.[10a]
Table 2.
Inhibitory activities of conjugates 1–6: COX inhibition as determined for COX-1 and COX-2; cytotoxicity as determined in proliferation assays for tumor cell lines HCT 116 and MDA-MB-231 for an incubation time of 72 h.
| Compd | IC50 [µm] | |||
|---|---|---|---|---|
| oCOX-1[a] | mCOX-2[b] | HCT 116[c] | MDA-MB-231[d] | |
| cisplatin | – | – | 12.0[e] | 20.0[e] |
| 1 | 1.033 | 0.563 | 53.0 | 39.8 |
| 1[f] | – | – | 56.0 | 47.4 |
| 2 | >25 | >25 | 66.4 | 45.5 |
| 3 | 4.1[e] | 0.045[e] | 1.1[e] | 1.65[e] |
| 4 | >25[e] | >25[e] | 0.065[e] | 0.05[e] |
| 5 | 0.745 | >25 | 1.3 | 0.55 |
| 6 | >25 | >25 | 0.31 | 0.33 |
Ovine COX-1.
Murine COX-2.
Colorectal carcinoma (no COX-2 expression).
Breast adenocarcinoma (COX-2 expression).
As determined previously.[9]
Pre-incubation of stock in DMSO for 24 h before dilution with medium. For the proliferation assays, all compounds except cisplatin were dissolved in DMSO before dilution with culture medium. Cisplatin was directly dissolved in culture medium.[9]
Values are the mean of duplicates (COX assay) or six replicates (proliferation assay), with standard deviation <10% (and <5% for 95% of the values).
2.4. Antitumor activity
To evaluate their potential as antitumor agents, cell proliferation assays of the conjugates were carried out on two tumor cell lines with different sensitivities toward cisplatin and different levels of COX-2 expression (Table 2). While the cisplatinsensitive HCT 116 cells (colorectal carcinoma) do not express COX-2, MDA-MB-231 cells (breast adenocarcinoma) show high constitutive COX-2 expression and are cisplatin resistant.[9] Although bearing one or two NSAID molecules, conjugates 1 and 2 were less potent than cisplatin in the tested cell lines. The coupled COX inhibitors unfavorably led to a markedly decreased solubility of these complexes so that neither growth medium nor DMF could be used as solvents, but DMSO instead had to be used for the preparation of the stock solutions. Although the samples were immediately diluted with medium, the fast solvolysis in DMSO, as described above, probably led to partial ligand exchange at the complexes. Due to the strong coordination of sulfur at platinum, binding of DMSO at the metal center usually leads to a decrease of the cytotoxic activity of platinum(II) complexes.[22d, f, 30] Steric hindrance of the strongly bound DMSO results in a decreased ability of the DMSO adducts to bind at double-stranded DNA. Therefore, a sample of conjugate 1 was also pre-incubated in DMSO for 24 h prior to dilution with growth medium to form the respective mono-cationic DMSO complex with a chloride counter-ion. Indeed, the cytotoxicity of 1 decreased after preincubation. However, the IC50 values only slightly differed from those of the immediately diluted sample, thereby confirming that the determined values of 1 and 2 rather represent those of the solvolyzed and thus deactivated compounds. Thus, due to their low solubility as well as their rather low cytotoxicity and lability, these conjugates are less suitable as dual-acting antitumor agents.
In contrast, the platinum(IV) conjugates 3, 4, 5, and 6 revealed markedly higher cytotoxic activities than cisplatin.[9] Notably, the ibuprofen conjugates 4 and 6 showed the highest antitumor activities, with conjugate 4 being the most potent, exhibiting an IC50 value in the low-nanomolar range. The conjugates are among the most active compounds of the class of platinum(IV) carboxylato complexes, most of which exhibited potencies similar to that of cisplatin.[10, 15c, d, 26a, 31] Only few of the previously reported derivatives have shown highly increased cytotoxicities, with activities in the sub-micromolar range.[15d, 32] Especially potent, however, are the benzoate derivatives, which resemble the structure of the ibuprofen conjugates and showed activities similar to 4 and 6.[15b, e, 33] The NSAID conjugates were also able to completely overcome cisplatin-related resistance in MDA-MB-231 breast cancer cells. Although this cell line shows constitutive expression of COX-2, the expression of this isozyme does not necessarily cause resistance of tumor cells against platinum-based agents.[9] The highly resistant cell line, however, expresses a mutant p53 protein, which results in resistance to cisplatin-induced apoptosis.[34] Furthermore, overexpression of anti-apoptotic Bcl-2 proteins[35] and IκB kinase[36] are reported to promote chemoresistance in this cell line.
To evaluate whether the cytotoxicity of the platinum(IV) complexes is associated with apoptosis, the cleavage of poly-(ADP-ribose) polymerase (PARP) was determined after incubation of the tumor cells with conjugate 3 or 4. PARP is a key nuclear enzyme in DNA repair and plays an important role in programmed cell death. The cleavage of PARP1, analyzed by western blot, revealed the induction of apoptosis by the conjugates in both cell lines (Figure S3, Supporting Information) similar to cisplatin, with a faster induction in the more sensitive HCT 116 cells.[22f, 37]
The cytotoxicities of the conjugates were similar for a distinct NSAID, although the complexes differ in the released antitumor agent, cisplatin or oxaliplatin. Thus, differences in the cytotoxicity mainly depended on the conjugated NSAID, indomethacin or ibuprofen, whereas the reduction potentials were similar for all platinum(IV) conjugates. However, while the indomethacin conjugates (3 and 5) represent potent COX inhibitors, the non-inhibiting ibuprofen conjugates (4 and 6) revealed the highest potencies in tumor cells. Furthermore, the cytotoxic activities per conjugate were similar in both cell lines. Thus, the cytotoxicity of the complexes is unrelated to the COX inhibitory activity of the conjugates, the potency of the coordinated NSAID or the COX-2 expression of the tumor cell lines. Consistently, combined treatment of the cancer cells with cisplatin and the respective NSAID (ratio 1:2) did not reveal any increase in cytotoxicity over that of cisplatin alone,[9] which was similarly observed for the combination of cisplatin with aspirin.[10a] Furthermore, the NSAIDs themselves did not reveal antitumor activity at pharmacological concentrations.[5c, 9] These findings are consistent with previous studies suggesting that the positive effects of COX inhibitors on the potency of antitumor agents are COX-2-independent.[6] Notably, indomethacin and ibuprofen differ in their lipophilicity. While indomethacin represents a highly lipophilic molecule (logP 4.27),[38] ibuprofen exhibits intermediate lipophilicity (logP 3.50).[38] Consequently, the indomethacin conjugates should be more lipophilic than the ibuprofen analogues. In general, the lipophilicity of platinum(IV) complexes is strongly correlated with drug accumulation,[39] and the increased uptake usually improves the efficacy of drugs.[14a, 15] However, the beneficial effect of increased lipophilicity reaches an upper limit when the corresponding drop in water solubility limits the bioavailability of the complex.[40] Thus, the high lipophilicity of the indomethacin conjugates might impede passing of cell membranes and accumulation in cells.
2.5. Cellular platinum accumulation
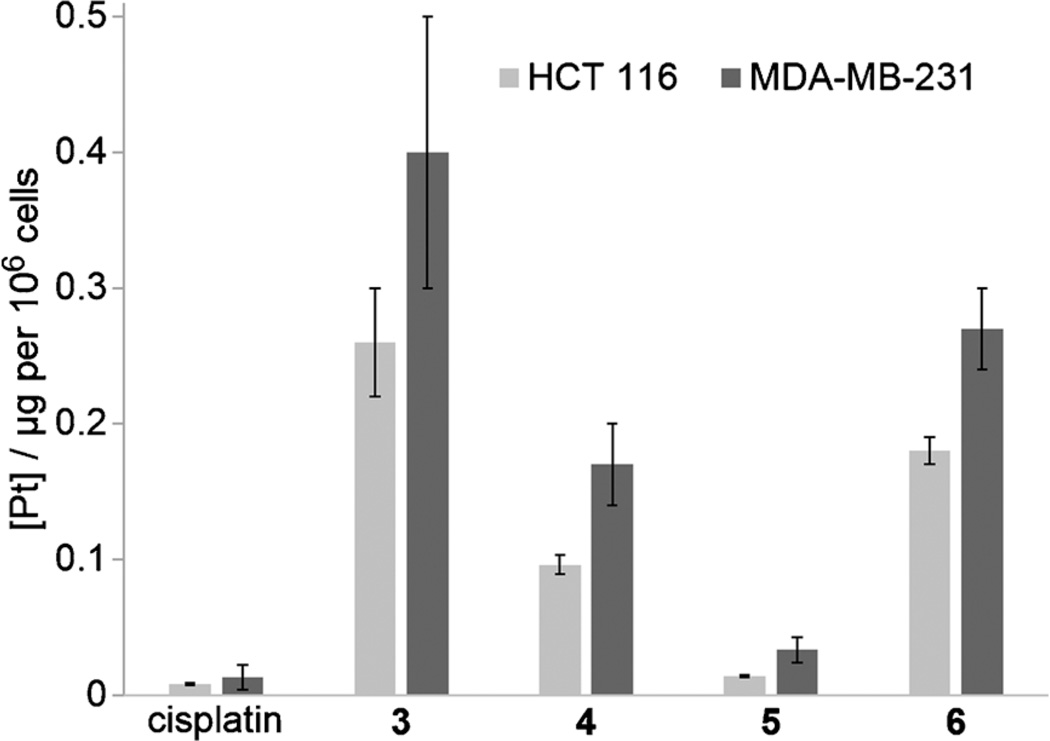
To investigate if conjugation of COX inhibitors facilitates the uptake of the antitumor drugs into cells, the cellular platinum accumulation for conjugates 3, 4, 5, and 6 was determined for both tumor cell lines and compared with that of cisplatin (Figure 1 and Supporting Information Table S2). After incubation of the tumor cells for a short duration — sufficient for drug uptake, but insufficient for post-exposure modification or cell death — the total platinum accumulation (net effect of influx and efflux) was determined by ICP-MS. All platinum(IV) conjugates revealed a higher platinum accumulation than cisplatin and the determined values were similar to those of other accumulation studies.[10b, 15d] Although the most potent conjugates 4 and 6 showed some of the highest accumulation values, the platinum uptake did not directly correlate with cytotoxicity. While the uptake of indomethacin conjugate 5 was only slightly higher than that of cisplatin, the uptake of its congener 3 was even higher than that of the ibuprofen derivatives. Despite the overall correlation between complex lipophilicity and accumulation, the lipophilicity does not fully rationalize the uptake trend, implying that the uptake is not based solely on passive diffusion and that there could be more complex drug–membrane interactions.[15b] Active transport processes might be involved, such as the copper transporter CTR1, reported to participate in the uptake of cisplatin.[41] Otherwise, efflux mechanisms, such as the multidrug resistance transporter (MDR1) P-glycoprotein (Pgp), could also play a role. Although cisplatin itself is not a substrate of Pgp,[42] lipophilicity is a main descriptor for Pgp substrates.[43] In contrast to other transporters, Pgp recognizes its substrates when dissolved in the lipid membrane,[44] implying that the membrane concentration of the substrate determines activation.[45] Furthermore, it has to be taken into account that the cellular accumulation was measured after a short incubation time (2 h), whereas the IC50 values were determined after a long exposure time (72 h), so that the equilibrium of uptake and efflux might not have been reached for every conjugate by then.[15d] In general, the cytotoxic activity of the conjugates seems to be mainly associated with the cellular uptake of the drugs. Congruently, the drug accumulation per compound was similar for both cell lines, as was cytotoxic activity. Although COX-2 inhibitors are selectively accumulated in COX-2-expressing tumors,[17] the COX-2 expression in MDA-MB-231 cells seemed not to enhance the uptake of COX-2-selective 3. The trend of accumulation was similar to that of the other conjugates, all revealing a slightly higher accumulation in MDA-MB-231 than in HCT 116 cells. As COX is an integral membrane protein, localized in the endoplasmic reticulum and the nuclear envelope, intracellular reduction of the conjugate probably impedes selective accumulation of the platinum complex. However, combination treatments revealed that indomethacin can lead to an increased intracellular uptake of cisplatin, even when administered individually. This was explained by changes in the fatty acid composition in tumor cells, which altered membrane fluidity and permeability and thus affected the entry of agents.[5c]
Figure 1.
Cellular platinum accumulation in tumor cell lines upon incubation with cisplatin or conjugates 3, 4, 5 or 6 (20 µm, 2 h). Values are the mean±-standard deviation of triplicates.
3. Conclusions
The NSAIDs indomethacin and ibuprofen were conjugated with cisplatin analogues to explain the synergistic effects that were observed in combination treatments.[5] Different intracellularly cleavable linking strategies were used for the platinum(II) and platinum(IV) conjugates, ensuring a concerted transport of both drugs into tumor cells as well as enabling release and simultaneous action of the agents inside the cells. The platinum(II) complexes revealed cytotoxic activities similar to that of cisplatin. In contrast, the platinum(IV) complexes, which are activated upon intracellular reduction, showed remarkably higher cytotoxicities in all tested tumor cell lines and were able to completely overcome cisplatin-related resistance. Although the indomethacin conjugates were potent COX inhibitors, they appear to execute their cytotoxic action via COX-independent mechanisms. Their cytotoxic activities were similar in all cell lines independent of COX-2 expression and the COX potency of the conjugate or the respective NSAID. However, the potent platinum(IV) conjugates exhibited an increased accumulation in the cells relative to that of cisplatin, probably resulting from their higher lipophilicity and kinetic inertness and thus leading to a clearly enhanced cytotoxicity.
The conjugates and the combination treatments of the cells carried out in this study revealed no synergistic effects based on COX inhibition. In contrast, cell culture studies revealed direct chemosensitizing effects of indomethacin on cisplatin, either using a large excess of the NSAID,[6d] or even if the inhibitor was only used as low concentrated additive.[5c] However, disparities in the effects of indomethacin between in vitro and in vivo studies were also reported.[5c, 46] The NSAIDs used in in vivo treatments possibly did not directly inhibit tumor growth, but led to its suppression by modulating the cytokine production associated with inflammation and by regulating the formation of immunosuppressive prostaglandin E2, thus modifying cellular immune responses.[5c, 47] The conjugates reported herein might also decrease tumor-associated inflammation by the released NSAIDs and might have the potential to reduce the severity of cisplatin-induced side effects similar to aspirin.[10a, 48] However, due to the different pharmacokinetics and biodistribution parameters, the drugs might act at different compartments of the body, leading to a synergy when administered successively. Thus, our investigations using conjugates demonstrate that the observations of combination treatments are not directly transferable onto dual-acting prodrugs and that the conjugates are important tools for the elucidation of the direct influence of COX inhibitors on cisplatin analogues in cells.
Experimental Section
Synthesis of conjugates
General
Syntheses of conjugates 1, 5 and 6 were carried out under a nitrogen atmosphere. Chemicals were used as purchased. Anhydrous dichloromethane was purified using an MBRAUN Solvent Purification System MB SPS-800; acetone was dried over molecular sieves (3 Å); pyridine was dried over CaH2 and distilled under nitrogen atmosphere. Indomethacin and ibuprofen acyl chloride,[49] (2-hydroxy-1,3-diaminopropane-κ2N,N’)-dichloro platinum(II) [PtCl2(HO-pda)],[50] (ethane-1,2-diamine)-dichloro platinum(II) [PtCl2(en)],[51] (SP-4-2)-(trans-R,R-cyclohexane-1,2-diamine)-oxalato platinum(II) [Pt(ox)(DACH)],[52] (SP-4-2)-(trans-R,R-cyclohexane-1,2-diamine)-dihydroxo(oxalato) platinum(IV) [Pt(OH)2(ox)(DACH)],[53] and conjugates 3 and 4[9] were prepared according to the respective published procedures. NMR spectra were recorded on a Bruker AVANCE DRX 400 (400 MHz), a Bruker AVANCE III HD (400 MHz) or a VARIAN Mercury 300 plus (300 MHz) spectrometer; 1H and 13C NMR spectra were referenced to tetramethylsilane (TMS) as internal standard; 195Pt NMR spectra were referenced to Na2[PtCl6] as external standard. FTIR spectra were recorded on a PerkinElmer System 2000 FTIR spectrometer, scanning between 400 and 4000 cm−1. Mass spectra were recorded on an ESQUIRE 3000 Plus Bruker-Daltonics ESI spectrometer; HR mass spectra were recorded on a 7 Tesla Apex II Bruker-Daltonics ESI-FT-ICR spectrometer. Melting points were measured in sealed tubes.
[PtCl2(indomethacin 1,3-diaminopropan-2-yl ester)] (1)
Triethylamine (0.34 mL, 2.4 mmol, 4 equiv) was added to a suspension of indomethacin (0.43 g, 1.2 mmol, 2 equiv) and N,N’-bis(2-oxo-3-oxazolidinyl) phosphinic chloride (BOP-Cl) (0.31 g, 1.2 mmol, 2 equiv) in CH2Cl2 (30 mL) and the reaction mixture was stirred for 10 min. [PtCl2(HO-pda)] (0.21 g, 0.6 mmol, 1 equiv) was added to the then clear solution and the resulting suspension was stirred at room temperature for 3 days in the dark. The solvent was evaporated and after addition of water (30 mL), the excess indomethacin was extracted with Et2O. The solid was filtered off and washed with water, EtOH and Et2O and dried in vacuo to give compound 1 as a beige powder (0.16 g, 37 %): mp: decomposition above 229°C; 1H NMR (400 MHz, [D7]DMF): δ = 2.33 (s, 3 H, CH3), 2.95 (m, 2 H, CH2), 2.99 (m, 2 H, CH2), 3.85 (s, 3 H, OCH3), 4.00 (s, 2H, OOC-CH2), 5.09 (m, 2H, NH2), 5.25 (m, 1H, CH), 5.43 (m, 2H, NH2), 6.77 (dd, 1H, 3JH,H = 12 Hz, 4JH,H = 4 Hz, CHind), 7.06 (d, 1 H, 3JH,H = 12 Hz, CHind), 7.22 (d, 1 H, 4JH,H = 4 Hz, CHind), 7.72 (d, 2 H, 3JH,H = 8 Hz, CHPhe), 7.81 ppm (d, 2 H, 3JH,H = 8 Hz, CHPhe); 13C{1H} (75 MHz, [D7]DMF): δ = 170.5 (Cq), 168.8 (Cq), 156.8 (Cq), 138.7 (Cq), 136.5 (Cq), 135.3 (Cq), 132.0 (CHPhe), 131.7 (Cq), 131.4 (Cq), 129.8 (CHPhe), 115.4 (CHind), 113.6 (Cq), 112.1 (CHind), 102.3 (CHind), 70.2 (CHpda), 55.9 (O-CH3), 45.2 (CH2), 30.4 (CH2,pda), 29.8 (CH2,pda), 13.5 ppm (CH3); IR (KBr): ν̃ = 3440 (br), 3256 (s), 3214 (s), 3123 (m), 2961 (m), 2835 (w), 1744 (s), 1683 (s), 1591 (s), 1509 (w), 1506 (w), 1477 (s), 1456 (s), 1438 (m), 1401 (m), 1359 (s), 1322 (s), 1291 (m), 1261 (s), 1224 (s), 1153 (s), 1089 (s), 1069 (s), 1033 (s), 1015 (s), 956 (w), 926 (m), 911 (w), 853 (m), 833 (m), 805 (m), 755 (m), 739 (w), 711 (w), 690 (w), 673 (w), 660 (w), 631 (w), 601 (w), 566 (w), 549 (w), 482 (w), 468 (w), 434 (w), 414 cm−1 (w); ESI-MS (positive mode, DMSO/CH3OH) m/z (%): 718 (100) [M+Na]+, 738 (72) [M−Cl+DMSO]+; HR-ESI-MS (positive mode, DMSO/CH3OH) m/z [M−Cl+DMSO]+: calcd for C24H30N3O5SPtCl2 : 738.0912, found: 738.0911; the observed isotopic patterns were in agreement with the calculated ones.
[Pt(ibuprofen-H)2(en)] (2)
[PtCl2(en)] (0.50 g, 1.5 mmol, 1 equiv) and Ag2CO3 (1.27 g, 4.6 mmol, 3 equiv) were suspended in water (125 mL) and after short sonication the reaction mixture was stirred at room temperature for 2.5 days in the dark. The formed AgCl was removed by filtration through Celite. Ibuprofen (1.27 g, 6.2 mmol, 4 equiv) was added to the solution and after short sonication the reaction mixture was stirred at 55°C for 14 h in the dark. The solvent was evaporated and the solid was suspended in Et2O and left to settle overnight. The precipitated solid was filtered off and washed with Et2O to remove the excess ibuprofen. The solid was further washed with small amounts of water, EtOH and Et2O and dried in vacuo to give compound 2 as a beige powder (0.11 g, 10%): mp: decomposition above 197°C; 1H NMR (400 MHz, [D7]DMF): δ = 0.87 (d, 12H, 3JH,H = 8 Hz, CH3), 1.29 (d, 6 H, 3JH,H = 8 Hz, CH3), 1.82 (m, 2H, CH), 2.33 (m, 4 H, N-CH2), 2.42 (d, 4 H, 3JH,H = 8 Hz, CH2), 3.51 (m, 2 H, CH), 5.78 (br s, 2 H, NH2), 5.82 (br s, 2H, NH2), 7.05 (d, 4H, 3JH,H = 8 Hz, CHPhe), 7.26 ppm (d, 4 H, 3JH,H = 8 Hz, CHPhe); 13C{1H} (100 MHz, [D7]DMF): δ = 181.3 (Cq), 141.7 (Cq), 139.1 (Cq), 128.8 (CHPhe), 127.5 (CHPhe), 48.6 (CH2), 47.2 (CH), 44.9 (CH2), 30.3 (CH3), 22.1 (CH3), 20.0 ppm (CH); IR (KBr): ν̃ = 3427 (br), 3268 (s), 3214 (s), 3022 (m), 2956 (s), 2928 (s), 2869 (m), 2848 (m), 1618 (s), 1513 (m), 1463 (m), 1456 (m), 1420 (w), 1377 (s), 1349 (s), 1261 (m), 1245 (m), 1191 (w), 1165 (w), 1115 (w), 1094 (w), 1058 (m), 1022 (w), 922 (w), 888 (w), 849 (w), 802 (m), 757 (w), 737 (w), 705 (w), 677 (w), 661 (w), 638 (w), 590 (w), 569 (w), 550 (w), 502 (w), 484 (w), 438 (w), 417 (w), 409 cm−1 (w); ESI-MS (positive mode, DMSO/CH3OH) m/z (%): 460 (38) [M−ibuprofen]+, 538 (100) [M−ibuprofen+DMSO]+, 688 (57) [M+Na]+, ESI-MS (negative mode, DMSO/CH3OH) m/z (%): 701 (100) [M+Cl]−; HR-ESI-MS (positive mode, DMSO/CH3OH) m/z [M+Na]+: calcd for C28H42N2O4PtNa: 688.2687, found: 688.2692; the observed isotopic patterns were in agreement with the calculated ones.
[Pt(indomethacin-H)2(ox)(DACH)] (5)
Pyridine (0.2 mL, 2.5 mmol, 10 equiv) was added to a suspension of [Pt(OH)2(ox)(DACH)] (0.10 g, 0.2 mmol, 1 equiv) in acetone (6 mL). A solution of indomethacin acyl chloride (0.44 g, 1.1 mmol, 5 equiv) in acetone (12 mL) was added and the reaction mixture was held at reflux (75 °C) for 8 h and stirred at room temperature for another 1.5 days. The reaction was quenched with water (10 mL) and the solvent was evaporated. The excess indomethacin was extracted with Et2O and the remaining solid was filtered off, washed with water, EtOH and Et2O to remove the formed pyridinium salt and dried in vacuo to give compound 5 as a pale yellow powder (0.13 g, 48%): mp: decomposition above 212°C; 1H NMR (400 MHz, [D6]DMSO): δ = 0.94 (m, 2H, CHDACH), 1.31 (m, 2 H, CHDACH), 1.45 (m, 2H, CHDACH), 2.04 (m, 2H, CHDACH), 2.17 (s, 6 H, CH3), 2.45 (m, 2 H, CHDACH), 3.68 (m, 4 H, CH2), 3.76 (s, 6H, OCH3), 6.69 (dd, 2H, 3JH,H = 8 Hz, 4JH,H = 2 Hz, CHind), 6.89 (d, 2H, 3JH,H = 8 Hz, CHind), 7.02 (d, 2H, 4JH,H = 2 Hz, CHind), 7.64 (d, 2H, 3JH,H = 8 Hz, CHPhe), 7.69 (d, 2 H, 3JH,H = 8 Hz, CHPhe), 8.06 (m, 2H, NH2), 8.34 ppm (m, 2H, NH2); 13C{1H} (100 MHz, [D6]DMSO): δ=178.5 (Cq), 168.3 (Cq), 163.8 (Cq), 156.0 (Cq), 138.0 (Cq), 135.4 (Cq), 134.6 (Cq), 131.7 (CHPhe), 131.2 (Cq), 130.7 (Cq), 129.4 (CHPhe), 115.0 (CHind), 114.6 (Cq), 111.6 (CHind), 102.4 (CHind), 61.6 (CH), 55.9 (O-CH3), 31.9 (CH2), 31.4 (CH2), 24.9 (CH2), 13.8 (CH3) ppm; 195Pt{1H} (86 MHz, [D6]DMSO): δ = 1605 ppm (br s); IR (KBr): ν̃ = 3436 (br), 3246 (w), 3098 (w), 2937 (m), 1733 (s), 1708 (w), 1679 (s), 1648 (m), 1595 (m), 1554 (w), 1515 (w), 1506 (w), 1477 (s), 1456 (m), 1400 (w), 1372 (m), 1355 (s), 1323 (s), 1287 (m), 1261 (m), 1225 (s), 1179 (m), 1145 (m), 1089 (m), 1068 (m), 1034 (m), 1015 (m), 927 (w), 882 (w), 834 (w), 805 (m), 755 (m), 738 (w), 725 (w), 689 (w), 629 (w), 580 (w), 550 (w), 517 (w), 502 (w), 482 (w), 441 (w), 417 cm−1 (w); ESI-MS (positive mode, DMSO/CH3OH) m/z (%): 1133 (100) [M+Na]+, ESI-MS (negative mode, DMSO/CH3OH) m/z (%): 1109 (100) [M−H]−; HR-ESI-MS (positive mode, DMSO/CH3OH) m/z [M+Na]+: calcd for C46H44N4O12PtCl2Na: 1133.1869, found: 1133.1860; the observed isotopic patterns were in agreement with the calculated ones.
[Pt(ibuprofen-H)2(ox)(DACH)] (6)
The synthesis was carried out as described for 5, with [Pt(OH)2(ox)(DACH)] (0.20 g, 0.5 mmol, 1 equiv) and ibuprofen acyl chloride (0.44 g, 2.0 mmol, 5 equiv). After extraction of excess of ibuprofen with Et2O, the remaining solid was washed with water and Et2O to give compound 6 as a white powder (0.21 g, 55%): mp: decomposition above 190°C; 1H NMR (400 MHz, [D6]DMSO): δ = 0.85 (d, 12H„ 3JH,H = 6 Hz, CH3), 0.94 (m, 2 H, CH), 1.24 (m, 2H, CHDACH), 1.29 (d, 6 H, 3JH,H = 8 Hz, CH3), 1.42 (m, 2 H, CHDACH), 1.78 (m, 2 H, CHDACH), 2.00 (m, 2 H, CHDACH), 2.24 (m, 2H, CHDACH), 2.38 (d, 4 H, 3JH,H = 8 Hz, CH2), 3.69 (q, 2H, 3JH,H = 8 Hz, CH), 7.03 (d, 4H, 3JH,H = 8 Hz, CHPhe), 7.15 (d, 4 H, 3JH,H = 8 Hz, CHPhe), 8.09 (m, 2H, NH2), 8.33 ppm (m, 2 H, NH2); 13C{1H} (100 MHz, [D6]DMSO): δ = 182.3 (Cq), 163.5 (Cq), 139.8 (Cq), 139.4 (Cq), 129.3 (CHPhe), 127.6 (CHPhe), 61.4 (CH), 46.7 (CH), 44.7 (CH2), 31.4 (CH2), 30.1 (CH3), 23.9 (CH2), 22.7 (CH3), 19.7 (CH), 19.2 ppm (CH); 195Pt{1H} (86 MHz, [D6]DMSO): δ = 1596 ppm (br s); IR (KBr): ν̃ = 3452 (br), 3180 (m), 3091 (m), 3054 (m), 3025 (m), 2954 (s), 2868 (m), 1732 (s), 1695 (m), 1646 (m), 1575 (m), 1554 (m), 1536 (w), 1513 (m), 1456 (m), 1420 (w), 1345 (s), 1302 (m), 1261 (m), 1224 (m), 1180 (m), 1137 (w), 1094 (w), 1065 (m), 1023 (m), 921 (w), 891 (w), 850 (w), 808 (m), 758 (w), 732 (w), 703 (w), 672 (w), 644 (w), 578 (w), 551 (w), 509 (w), 462 (w), 444 (w), 426 (w), 417 cm−1 (w); ESI-MS (positive mode, DMSO/CH3OH) m/z (%): 830 (100) [M+ Na]+, ESI-MS (negative mode, DMSO/CH3OH) m/z (%): 806 (100) [M−H]−; HR-ESI-MS (positive mode, DMSO/CH3OH) m/z [M+Na]+: calcd for C34H48N2O8PtNa: 830.2954, found: 830.2958; the observed isotopic patterns were in agreement with the calculated ones.
Solvolysis experiments
Conjugate 1 or 2 was dissolved in [D6]DMSO at room temperature. Solvolysis was monitored by recording 1H NMR spectra at different time intervals. Solvolysis products were further characterized by ESI-MS (positive mode, [D6]DMSO/CH3OH; see Supporting Information).
Cyclic voltammetry
Electrochemical measurements were conducted at ambient temperature (~20°C) using a potentiostat/galvanostat SP-50 (Bio-Logic SAS, France), equipped with a three-electrode cell. A glassy carbon electrode was used as working electrode, a platinum wire as counter electrode and an Ag wire as reference electrode. A 1 mm solution of conjugate 5 or 6 in 0.1m (nBu4N)BF4/DMF was purged with nitrogen before measurement of the potentials at a scan rate of 100 mVs−1. Ferrocene [Fe(η5-C5H5)2] was used as internal standard (E1/2 = +0.72 V vs. NHE in DMF),[54] and the potentials were quoted relative to the normal hydrogen electrode (NHE) (Figure S2 and Table S1, Supporting Information).
Biological methods
COX inhibition assays
COX inhibitory activity was assayed by a method that quantifies the COX-mediated conversion of [1-14C]arachidonic acid to [1-14C]prostaglandin products. Stock solutions of the compounds were prepared in DMF (1, 2) or DMSO (5, 6). The compounds were tested for inhibition of ovine COX-1 and murine COX-2 by a published procedure.[16] Briefly, hematin-reconstituted ovine COX-1 (44 nm) or murine COX-2 (66 nm) was pre-incubated in 100 mm Tris·HCl (pH 8.0) (Tris: tris(hydroxymethyl)aminomethane) containing 500 µm phenol for 20 min with 5 µL DMSO or the test compound at various concentrations followed by the addition of [1-14C]arachidonic acid (50 µm) for 30 s at 37°C. Total reaction volume was 200 µL and final DMF or DMSO concentration was 2.5 %. Reactions were terminated, processed and analyzed as stated in the above reference. Product conversion for control protein (DMF- or DMSO-treated) was limited to 30–35% of total substrate in the reaction. [1-14C]Arachidonic acid (≈55 mCi mmol−1) was purchased from PerkinElmer (Waltham MA, USA) and TLC plates (EMD Kieselgel 60, 20×20 cm with pre-concentration zone, EM-11798-7) were obtained from VWR (West Chester, PA, USA).
Cell lines and tissue culture conditions
The human cell lines HCT 116 (colorectal carcinoma, no COX-2 expression) and MDMBA-231 (breast adenocarcinoma, COX-2 expression) were purchased from ATCC. The cells were maintained in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS; Atlas Biologicals, Fort Collins, CO, USA) without antibiotics and were cultured in monolayer and maintained at 37 °C supplemented with 5% CO2.
Cell proliferation assays
Stock solutions of the conjugates were prepared by dissolving the compounds in DMSO (1 and 2: 0.006–12.5 mm; 5 and 6: 0.006–25 mm). The stocks were further diluted with RPMI-1640 medium (0.02–100 µm) before addition to the cells. The low solubility of 1 and 2 impeded preparation of stocks in PBS (phosphate-buffered saline) buffer or DMF and DMSO was used instead. Due to their fast solvolysis in DMSO, dilution with medium was carried out immediately. A further sample of 1 was pre-incubated in DMSO (12.5 mm) for 24 h before dilution to form the respective mono-cation by solvolysis.
Cells were trypsinized with 0.25% trypsin-EDTA (ATCC) and manually counted with 0.4% trypan blue. Cells were seeded in 96-well plates with 5 000–10000 cells per well. Once cells were attached (≈24 h post seeding), the medium was removed and replaced with drug-containing medium. A WST-1 cell proliferation assay was performed 72 h post treatment using WST-1 Cell Proliferation Reagent (Roche Diagnostics Corporation, Indianapolis/IN, USA) according to the manufacturer’s instructions. Briefly, 10 µL of the WST-1 reagent was added to each well and the cells were incubated at 37 °C until absorption of the vehicle-treated cells reached a value of ~0.7. Absorbance at 450 nm was measured using a VersaMax microplate spectrophotometer (Molecular Devices, Sunnyvale/CA, USA) and Prism (ver. 6d; 2013, GraphPad, USA) was used to calculate the half maximal inhibitory (IC50) values for each cell line.
Apoptosis assays
The cleavage of PARP1 was used as marker for apoptosis of the cells incubated with conjugates 3 or 4 (full-length PARP1: 116 kDa; cleaved PARP: 89 kDa). Stock solutions of conjugates 3 and 4 were prepared in DMSO (3: 2 mm; 4: 0.2 mm). The stocks were further diluted with RPMI-1640 medium (containing 1% penicillin/streptomycin) (3: 2 µm; 4: 0.2 µm; concentrations chosen based on IC50 values determined in cell proliferation assays) before addition to the cells.
HCT 116 or MDA-MB-231 cells were seeded in 150 mm dishes and incubated in RPMI-1640 medium (25 mL) for 24 h. The medium was replaced with drug-containing medium and the cells were incubated for 12 h, 24 h or 48 h (two plates per compound and cell line were prepared for the control and 12 h time point to obtain enough cells). Cells were harvested and washed with PBS (centrifugation: 1000 rpm, room temperature, 5 min). Cell pellets were lysed with M-PER mammalian protein extraction reagent (Thermo Scientific; with 1:100 protease inhibitor (Protease Inhibitor Cocktail, Sigma–Aldrich) and stored at −80 °C. Protein concentrations were determined by a BCA (bicinchoninic acid) assay. Aliquots of the cell lysates (20 µg protein) were resolved by SDS-PAGE, followed by electroblotting to nitrocellulose membranes (Bio-Rad). Membranes were blocked in 5% powdered milk in TBST (Tris-buffered saline with Tween 20) and probed with primary PARP antibody (#9542, Cell Signaling Technology), followed by horseradish peroxidase (HRP)-linked anti-rabbit IgG secondary antibody (Promega). Protein bands were visualized by a chemiluminescence detection kit (Pierce ECL, Thermo Scientific). Actin was used as loading control.
Platinum accumulation assays
The total cell accumulation (net effect of uptake and efflux) was determined. Stock solutions of conjugates 3, 4, 5, and 6 were prepared in DMSO (5 mm). A stock solution of cisplatin (~1 mm) was prepared in RPMI-1640 medium (containing 1% penicillin/streptomycin; without FBS), followed by filtration through a 0.2 µm syringe filter and the final concentration was determined by ICP-OES (Varian ICP model 720-ES; see Supporting Information). The stocks were further diluted with RPMI-1640 medium (final concentration: 20 µm) before addition to the cells.
HCT 116 or MDA-MB-231 cells were seeded in 100 mm dishes (~2×106 cells) and incubated in RPMI-1640 medium (10 mL) for 24 h. The medium was replaced with drug-containing medium and the cells were incubated for 2 h. The drug-containing medium was removed and the cells were washed thrice with warm PBS. Cells were trypsinized with 0.25% trypsin-EDTA (ATCC), harvested and centrifuged (1000 rpm, room temperature, 5 min). After removal of trypsin by washing with RPMI-1640 medium and addition of PBS, the cells were counted automatically (Bio-Rad TC10 automated cell counter), followed by centrifugation and removal of any remaining buffer. Cell pellets were stored at 4 °C. For digestion, cell pellets were treated with concentrated HNO3 (300 µL; 69%, TraceSELECT, Sigma–Aldrich, USA) at 100°C for 2 h in Wheaton glass vials with PTFE-faced rubber-lined caps. The digested samples were further diluted with deionized water to a final HNO3 concentration of 5% (v/v) and were filtered through 0.2 µm syringe filters. The platinum content of the samples was determined by ICP-MS (PerkinElmer model ELAN DRC II, see Supporting Information); if necessary, the samples were further diluted. The total cell accumulation of platinum was calculated as µg Pt/106 cells. All uptake assays were performed in triplicate.
Supplementary Material
Acknowledgements
This work was supported by the Fonds der Chemischen Industrie (doctoral grant for W.N.), the Graduate School “Building with Molecules and Nano-objects (BuildMoNa)” funded by the Deutsche Forschungsgemeinschaft, the Europäischer Sozialfond and the Freistaat Sachsen, the US National Institutes of Health (CA89450), and the German Academic Exchange Service (PPP USA). The authors thank Umicore AG & Co. KG for a generous donation of chemicals and are grateful to Ms. Rosanne DeLapp (Vanderbilt University, USA) for ICP-OES and ICP-MS measurements and Dr. Carol Rouzer (Vanderbilt University, USA) for editorial assistance.
Footnotes
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/cmdc.201402353.
References
- 1.Wheate NJ, Walker S, Craig GE, Oun R. Dalton Trans. 2010;39:8113. doi: 10.1039/c0dt00292e. [DOI] [PubMed] [Google Scholar]
- 2.a) Kartalou M, Essigmann JM. Mutat. Res. 2001;478:23. doi: 10.1016/s0027-5107(01)00141-5. [DOI] [PubMed] [Google Scholar]; b) Ohmichi M, Hayakawa J, Tasaka K, Kurachi H, Murata Y. Trends Pharmacol. Sci. 2005;26:113. doi: 10.1016/j.tips.2005.01.002. [DOI] [PubMed] [Google Scholar]; c) Rabik CA, Dolan ME. Cancer Treat. Rev. 2007;33:9. doi: 10.1016/j.ctrv.2006.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Stewart DJ. Crit. Rev. Oncol. Hematol. 2007;63:12. doi: 10.1016/j.critrevonc.2007.02.001. [DOI] [PubMed] [Google Scholar]; e) Galluzzi L, Senovilla L, Vitale I, Michels J, Martins I, Kepp O, Castedo M, Kroemer G. Oncogene. 2012;31:1869. doi: 10.1038/onc.2011.384. [DOI] [PubMed] [Google Scholar]; f) Shen D-W, Pouliot LM, Hall MD, Gottesman MM. Pharmacol. Rev. 2012;64:706. doi: 10.1124/pr.111.005637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.a) Simmons DL, Botting RM, Hla T. Pharmacol. Rev. 2004;56:387. doi: 10.1124/pr.56.3.3. [DOI] [PubMed] [Google Scholar]; b) Tsatsanis C, Androulidaki A, Venihaki M, Margioris AN. Int. J. Biochem. Cell Biol. 2006;38:1654. doi: 10.1016/j.biocel.2006.03.021. [DOI] [PubMed] [Google Scholar]; c) Sarkar FH, Adsule S, Li Y, Padhye S. Mini-Rev. Med. Chem. 2007;7:599. doi: 10.2174/138955707780859431. [DOI] [PubMed] [Google Scholar]; d) Marnett LJ. Annu. Rev. Pharmacol. Toxicol. 2009;49:265. doi: 10.1146/annurev.pharmtox.011008.145638. [DOI] [PubMed] [Google Scholar]; e) Greenhough A, M. Smartt HJ, Moore AE, Roberts HR, Williams AC, Paraskeva C, Kaidi A. Carcinogenesis. 2009;30:377. doi: 10.1093/carcin/bgp014. [DOI] [PubMed] [Google Scholar]; f) Ghosh N, Chaki R, Mandal V, Mandal SC. Pharmacol. Rep. 2010;62:233. doi: 10.1016/s1734-1140(10)70262-0. [DOI] [PubMed] [Google Scholar]
- 4.a) Ferrandina G, Lauriola L, Distefano MG, Zannoni GF, Gessi M, Legge F, Maggiano N, Mancuso S, Capelli A, Scambia G, Ranelletti FO. J. Clin. Oncol. 2002;20:973. doi: 10.1200/JCO.2002.20.4.973. [DOI] [PubMed] [Google Scholar]; b) Ferrandina G, Lauriola L, Zannoni GF, Fagotti A, Fanfani F, Legge F, Maggiano N, Gessi M, Mancuso S, Ranelletti FO, Scambia G. Ann. Oncol. 2002;13:1205. doi: 10.1093/annonc/mdf207. [DOI] [PubMed] [Google Scholar]; c) Ferrandina G, Ranelletti FO, Legge F, Gessi M, Salutari V, Distefano MG, Lauriola L, Zannoni GF, Martinelli E, Scambia G. Clin. Cancer Res. 2004;10:3117. doi: 10.1158/1078-0432.ccr-1090-3. [DOI] [PubMed] [Google Scholar]
- 5.a) Knapp DW, Glickman NW, Widmer WR, DeNicola DB, Adams LG, Kuczek T, Bonney PL, Amalia AE, Han C, Glickman LT. Cancer Chemother. Pharmacol. 2000;46:221. doi: 10.1007/s002800000147. [DOI] [PubMed] [Google Scholar]; b) Hattori K, Matsushita R, Kimura K, Abe Y, Nakashima E. Biol. Pharm. Bull. 2001;24:1214. doi: 10.1248/bpb.24.1214. [DOI] [PubMed] [Google Scholar]; c) Ogino M, Minoura S. Int. J. Clin. Oncol. 2001;6:84. doi: 10.1007/pl00012088. [DOI] [PubMed] [Google Scholar]; d) Barnes AP, Miller BE, Kucera GL. Gynecol. Oncol. 2007;104:443. doi: 10.1016/j.ygyno.2006.08.008. [DOI] [PubMed] [Google Scholar]; e) Li G, Sha SH, Zotova E, Arezzo J, Van de Water T, Schacht J. Lab. Invest. 2002;82:585. doi: 10.1038/labinvest.3780453. [DOI] [PubMed] [Google Scholar]
- 6.a) Grösch S, Tegeder I, Niederberger E, Bräutigam L, Geisslinger G. FASEB J. 2001;15:2742. doi: 10.1096/fj.01-0299fje. [DOI] [PubMed] [Google Scholar]; b) Minter HA, Eveson JW, Huntley S, Elder DJ, Hague A. Clin. Cancer Res. 2003;9:1885. [PubMed] [Google Scholar]; c) Jendrossek V, Handrick R, Belka C. FASEB J. 2003;17:1547. doi: 10.1096/fj.02-0947fje. [DOI] [PubMed] [Google Scholar]; d) Byun S-S, Kim SW, Choi H, Lee C, Lee E. BJU Int. 2005;95:1086. doi: 10.1111/j.1464-410X.2005.05472.x. [DOI] [PubMed] [Google Scholar]
- 7.a) Czembirek C, Eder-Czembirek C, Erovic BM, Turhani D, Selzer E, Thurnher D. Oncol. Rep. 2005;14:1523. [PubMed] [Google Scholar]; b) Bijman MN, Hermelink CA, van Berkel MP, Laan AC, Janmaat ML, Peters GJ, Boven E. Biochem. Pharmacol. 2008;75:427. doi: 10.1016/j.bcp.2007.09.005. [DOI] [PubMed] [Google Scholar]; c) Yu L, Chen M, Li Z, Wen J, Fu J, Guo D, Jiang Y, Wu S, Cho C-H, Liu S. Mol. Pharmacol. 2011;79:608. doi: 10.1124/mol.110.069393. [DOI] [PubMed] [Google Scholar]
- 8.Mercer SJ, Di Nicolantonio F, Knight LA, Gabriel FG, White-house PA, Sharma S, Fernando A, Bhandari P, Somers SS, Toh SK, Cree IA. Anti-Cancer Drugs. 2005;16:495. doi: 10.1097/00001813-200506000-00004. [DOI] [PubMed] [Google Scholar]
- 9.Neumann W, Crews BC, Marnett LJ, Hey-Hawkins E. Chem Med Chem. 2014;9:1150. doi: 10.1002/cmdc.201402074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.a) Pathak RK, Marrache S, Choi JH, Berding TB, Dhar S. Angew. Chem. Int. Ed. 2014;53:1963. doi: 10.1002/anie.201308899. Angew. Chem. 2014, 126, 1994. [DOI] [PubMed] [Google Scholar]; b) Cheng Q, Shi H, Wang H, Min Y, Wang J, Liu Y. Chem. Commun. 2014;50:7427. doi: 10.1039/c4cc00419a. [DOI] [PubMed] [Google Scholar]
- 11.Williams FM. Clin. Pharmacokinet. 1985;10:392. doi: 10.2165/00003088-198510050-00002. [DOI] [PubMed] [Google Scholar]
- 12.a) Kelland LR, Abel G, McKeage MJ, Jones M, Goddard PM, Valenti M, Murrer BA, Harrap KR. Cancer Res. 1993;53:2581. [PubMed] [Google Scholar]; b) Lemma K, Sargeson AM, Elding LI. J. Chem. Soc. Dalton Trans. 2000:1167. [Google Scholar]
- 13.Hall MD, Mellor HR, Callaghan R, Hambley TW. J. Med. Chem. 2007;50:3403. doi: 10.1021/jm070280u. [DOI] [PubMed] [Google Scholar]
- 14.a) Hall MD, Hambley TW. Coord. Chem. Rev. 2002;232:49. [Google Scholar]; b) Graf N, Lippard SJ. Adv. Drug Delivery Rev. 2012;64:993. doi: 10.1016/j.addr.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Wexselblatt E, Gibson D. J. Inorg. Biochem. 2012;117:220. doi: 10.1016/j.jinorgbio.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 15.a) Mistry P, Kelland LR, Loh SY, Abel G, Murrer BA, Harrap KR. Cancer Res. 1992;52:6188. [PubMed] [Google Scholar]; b) Ang WH, Pilet S, Scopelliti R, Bussy F, Juillerat-Jeanneret L, Dyson PJ. J. Med. Chem. 2005;48:8060. doi: 10.1021/jm0506468. [DOI] [PubMed] [Google Scholar]; c) Ang WH, Khalaila I, Allardyce CS, Juillerat-Jeanneret L, Dyson PJ. J. Am. Chem. Soc. 2005;127:1382. doi: 10.1021/ja0432618. [DOI] [PubMed] [Google Scholar]; d) Reithofer MR, Bytzek AK, Valiahdi SM, Kowol CR, Groessl M, Hartinger CG, Jakupec MA, Galanski M, Keppler BK. J. Inorg. Biochem. 2011;105:46. doi: 10.1016/j.jinorgbio.2010.09.006. [DOI] [PubMed] [Google Scholar]; e) Chin CF, Tian Q, Setyawati MI, Fang W, Tan ESQ, Leong DT, Ang WH. J. Med. Chem. 2012;55:7571. doi: 10.1021/jm300580y. [DOI] [PubMed] [Google Scholar]
- 16.Kalgutkar AS, Marnett AB, Crews BC, Remmel RP, Marnett LJ. J. Med. Chem. 2000;43:2860. doi: 10.1021/jm000004e. [DOI] [PubMed] [Google Scholar]
- 17.Uddin MJ, Crews BC, Blobaum AL, Kingsley PJ, Gorden DL, McIntyre JO, Matrisian LM, Subbaramaiah K, Dannenberg AJ, Piston DW, Marnett LJ. Cancer Res. 2010;70:3618. doi: 10.1158/0008-5472.CAN-09-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.a) Harrison RC, McAuliffe CA, Zaki AM. Inorg. Chim. Acta. 1980;46:L15. [Google Scholar]; b) Khokhar AR, Krakoff IH, Hacker MP, McCormack JJ. Inorg. Chim. Acta. 1985;108:63. [Google Scholar]; c) Rochon FD, Gruia LM. Inorg. Chim. Acta. 2000;306:193. [Google Scholar]
- 19.a) Khokhar AR, Deng Y, Kido Y, Siddik ZH. J. Inorg. Biochem. 1993;50:79. doi: 10.1016/0162-0134(93)80015-2. [DOI] [PubMed] [Google Scholar]; b) Zhang JC, Zhao XJ. Eur. J. Med. Chem. 2007;42:286. doi: 10.1016/j.ejmech.2006.09.013. [DOI] [PubMed] [Google Scholar]; c) Gabano E, Ravera M, Cassino C, Bonetti S, Palmisano G, Osella D. Inorg. Chim. Acta. 2008;361:1447. [Google Scholar]
- 20.a) Khokhar AR, Lumetta G, Doran SL. Inorg. Chim. Acta. 1988;151:87. [Google Scholar]; b) Howell BA, Beholz LG, Sastry BBS. J. Therm. Anal. 1993;40:395. [Google Scholar]
- 21.Galanski M, Keppler BK. Inorg. Chem. 1996;35:1709. doi: 10.1021/ic9509490. [DOI] [PubMed] [Google Scholar]
- 22.a) Kerrison SJS, Sadler PJ. J. Chem. Soc. Chem. Commun. 1977:861. [Google Scholar]; b) Kerrison SJS, Sadler PJ. Inorg. Chim. Acta. 1985;104:197. [Google Scholar]; c) Sundquist WI, Ahmed KJ, Hollis LS, Lippard SJ. Inorg. Chem. 1987;26:1524. [Google Scholar]; d) Fischer SJ, Benson LM, Fauq A, Naylor S, Windebank AJ. NeuroToxicology. 2008;29:444. doi: 10.1016/j.neuro.2008.02.010. [DOI] [PubMed] [Google Scholar]; e) Platts JA, Ravera M, Gabano E, Sardi M, Bianco S, Osella D. Eur. J. Inorg. Chem. 2012:5625. [Google Scholar]; f) Hall MD, Telma KA, Chang K-E, Lee TD, Madigan JP, Lloyd JR, Goldlust IS, Hoeschele JD, Gottesman MM. Cancer Res. 2014;74:3913. doi: 10.1158/0008-5472.CAN-14-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.a) Choi S, Filotto C, Biszano M, Delaney S, Lagasee D, Whitworth JL, Jusko A, Li C, Wood NA, Willingham J, Schwenker A, Spaulding K. Inorg. Chem. 1998;37:2500. [Google Scholar]; b) Ellis LT, Er HM, Hambley TW. Aust. J. Chem. 1995;48:793. [Google Scholar]
- 24.Schilder RJ, LaCreta FP, Perez RP, Johnson SW, Brennan JM, Rogatko A, Nash S, McAleer C, Hamilton TC, Roby D, Young RC, Ozols RF, O’Dwyer PJ. Cancer Res. 1994;54:709. [PubMed] [Google Scholar]
- 25.Trask C, Silverstone A, Ash CM, Earl H, Irwin C, Bakker A, Tobias JS, Souhami RL. J. Clin. Oncol. 1991;9:1131. doi: 10.1200/JCO.1991.9.7.1131. [DOI] [PubMed] [Google Scholar]
- 26.a) Wilson JJ, Lippard S. Inorg. Chem. 2011;50:3103. doi: 10.1021/ic2000816. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Varbanov HP, Valiahdi SM, Kowol CR, Jakupec MA, Galanski M, Keppler BK. Dalton Trans. 2012;41:14404. doi: 10.1039/c2dt31366a. [DOI] [PubMed] [Google Scholar]
- 27.Siddik ZH, Jones M, Boxall FE, Harrap KR. Cancer Chemother. Pharmacol. 1988;21:19. doi: 10.1007/BF00262732. [DOI] [PubMed] [Google Scholar]
- 28. Ghebreselasie K, Crews BC, Marnett LJ. unpublished results. IC50(COX-1) has not been determined.
- 29.Prusakiewicz JJ, Duggan KC, Rouzer CA, Marnett LJ. Biochemistry. 2009;48:7353. doi: 10.1021/bi900999z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Massart C, Le Tellier C, Gibassier J, Leclech G, Nicol M. Toxicol. in Vitro. 1993;7:87. doi: 10.1016/0887-2333(93)90116-m. [DOI] [PubMed] [Google Scholar]
- 31.a) Reithofer MR, Valiahdi SM, Jakupec MA, Arion VB, Egger A, Galanski M, Keppler BK. J. Med. Chem. 2007;50:6692. doi: 10.1021/jm070897b. [DOI] [PubMed] [Google Scholar]; b) Song Y, Suntharalingam K, Yeung JS, Royzen M, Lippard SJ. Bioconjugate Chem. 2013;24:1733. doi: 10.1021/bc400281a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.a) Kwon Y-E, Whang K-J, Park Y-J, Kim KH. Bioorg. Med. Chem. 2003;11:1669. doi: 10.1016/s0968-0896(03)00044-0. [DOI] [PubMed] [Google Scholar]; b) Reithofer MR, Schwarzinger A, Valiahdi SM, Galanski M, Jakupec MA, Keppler BK. J. Inorg. Biochem. 2008;102:2072. doi: 10.1016/j.jinorgbio.2008.07.006. [DOI] [PubMed] [Google Scholar]; c) Varbanov H, Valiahdi SM, Legin AA, Jakupec MA, Roller A, Galanski M, Keppler BK. Eur. J. Med. Chem. 2011;46:5456. doi: 10.1016/j.ejmech.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gandin V, Marzano C, Pelosi G, Ravera M, Gabano E, Osella D. Chem-Med Chem. 2014;9:1299. doi: 10.1002/cmdc.201400061. [DOI] [PubMed] [Google Scholar]
- 34.Olivier M, Eeles R, Hollstein M, Khan MA, Harris CC, Hainaut P. Hum. Mutat. 2002;19:607. doi: 10.1002/humu.10081. [DOI] [PubMed] [Google Scholar]
- 35.Kutuk O, Arisan ED, Tezil T, Shoshan MC, Basaga H. Carcinogenesis. 2009;30:1517. doi: 10.1093/carcin/bgp165. [DOI] [PubMed] [Google Scholar]
- 36.Tezil T, Bodur C, Kutuk O, Basaga H. Cell. Signalling. 2012;24:1361. doi: 10.1016/j.cellsig.2012.01.012. [DOI] [PubMed] [Google Scholar]
- 37.a) Del Bello B, Valentini MA, Mangiavacchi P, Comporti M, Maellaro E. Exp. Cell Res. 2004;293:302. doi: 10.1016/j.yexcr.2003.10.024. [DOI] [PubMed] [Google Scholar]; b) Wu YJ, Muldoon LL, Neuwelt EA. J. Pharmacol. Exp. Ther. 2005;312:424. doi: 10.1124/jpet.104.075119. [DOI] [PubMed] [Google Scholar]
- 38.Lewis DFV, Lake BG, Ito Y, Dickins M. J. Enzyme Inhib. Med. Chem. 2006;21:385. doi: 10.1080/14756360600703313. [DOI] [PubMed] [Google Scholar]
- 39.Oldfield SP, Hall MD, Platts JA. J. Med. Chem. 2007;50:5227. doi: 10.1021/jm0708275. [DOI] [PubMed] [Google Scholar]
- 40.Song R, Kim KM, Sohn YS. Bull. Korean Chem. Soc. 2000;21:1000. [Google Scholar]
- 41.Ishida S, Lee J, Thiele DJ, Herskowitz I. Proc. Natl. Acad. Sci. USA. 2002;99:14298. doi: 10.1073/pnas.162491399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nooter K, Stoter G. Pathol. Res. Pract. 1996;192:768. doi: 10.1016/S0344-0338(96)80099-9. [DOI] [PubMed] [Google Scholar]
- 43.Chiba P, Ecker G, Schmid D, Tell B, Goldenberg S, Gekeler V. Mol. Pharmacol. 1996;49:1122. [PubMed] [Google Scholar]
- 44.Stein WD. Physiol. Rev. 1997;77:545. doi: 10.1152/physrev.1997.77.2.545. [DOI] [PubMed] [Google Scholar]
- 45.Seelig A, Landwojtowicz E. Eur. J. Pharm. Sci. 2000;12:31. doi: 10.1016/s0928-0987(00)00177-9. [DOI] [PubMed] [Google Scholar]
- 46.Maca RD. J. Biol. Response Mod. 1988;7:568. [PubMed] [Google Scholar]
- 47.a) Chouaib S, Chatenoud L, Klatzmann D, Fradelizi D. J. Immunol. 1984;132:1851. [PubMed] [Google Scholar]; b) Lala PK, Parhar RS, Singh P. Cell. Immunol. 1986;99:108. doi: 10.1016/0008-8749(86)90220-0. [DOI] [PubMed] [Google Scholar]; c) Strassmann G, Jacob CO, Evans R, Beall D, Fong M. J. Immunol. 1992;148:3674. [PubMed] [Google Scholar]; d) Ogino M, Hanazono M. Int. J. Clin. Oncol. 1998;3:176. [Google Scholar]
- 48.Mukherjea D, Rybak LP, Sheehan KE, Kaur T, Ramkumar V, Jajoo S, Sheth S. Expert Opin. Drug Discovery. 2011;6:491. doi: 10.1517/17460441.2011.562887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang Z, Velázquez CA, Abdellatif KRA, Chowdhury MA, Reisz JA, DuMond JF, King SB, Knaus EE. J. Med. Chem. 2011;54:1356. doi: 10.1021/jm101403g. [DOI] [PubMed] [Google Scholar]
- 50.Liu WP, Chen XZ, Xie MJ, Lou LG, Ye QS, Yu Y, Hou SQ SQ. J. Inorg. Biochem. 2008;102:1942. doi: 10.1016/j.jinorgbio.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 51.Failes TW, Hall MD, Hambley TW. Dalton Trans. 2003:1596. [Google Scholar]
- 52.Habala L, Galanski M, Yasemi A, Nazarov AA, Keyserlingk NGv, Keppler BK. Eur. J. Med. Chem. 2005;40:1149. doi: 10.1016/j.ejmech.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 53.Khokhar AR, Al-Baker S, Siddik ZH. J. Inorg. Biochem. 1994;54:39. doi: 10.1016/0162-0134(94)85122-0. [DOI] [PubMed] [Google Scholar]
- 54.Barrette WC, Jr, Johnson HW, Jr, Sawyer DT. Anal. Chem. 1984;56:1890. doi: 10.1021/ac00275a030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.