Abstract
Plasmodium simium is a parasite from New World monkeys that is most closely related to the human malaria parasite Plasmodium vivax; it also naturally infects humans. The blood-stage infection of P. vivax depends on Duffy binding protein II (PvDBPII) and its cognate receptor on erythrocytes, the Duffy antigen receptor for chemokines (hDARC), but there is no information on the P. simium erythrocytic invasion pathway. The genes encoding P. simium DBP (PsDBPII) and simian DARC (sDARC) were sequenced from Southern brown howler monkeys (Alouatta guariba clamitans) naturally infected with P. simium because P. simium may also depend on the DBPII/DARC interaction. The sequences of DBP binding domains from P. vivax and P. simium were highly similar. However, the genetic variability of PsDBPII was lower than that of PvDBPII. Phylogenetic analyses demonstrated that these genes were strictly related and clustered in the same clade of the evolutionary tree. DARC from A. clamitans was also sequenced and contained three new non-synonymous substitutions. None of these substitutions were located in the N-terminal domain of DARC, which interacts directly with DBPII. The interaction between sDARC and PvDBPII was evaluated using a cytoadherence assay of COS7 cells expressing PvDBPII on their surfaces. Inhibitory binding assays in vitro demonstrated that antibodies from monkey sera blocked the interaction between COS-7 cells expressing PvDBPII and hDARC-positive erythrocytes. Taken together, phylogenetic analyses reinforced the hypothesis that the host switch from humans to monkeys may have occurred very recently in evolution, which sheds light on the evolutionary history of new world plasmodia. Further invasion studies would confirm whether P. simium depends on DBP/DARC to trigger internalization into red blood cells.
Introduction
Invasion of erythrocytes by Plasmodium vivax merozoites is highly dependent on the interaction between the Duffy Antigen Receptor for Chemokines (DARC) and its ligand in the parasite, the Duffy binding protein (DBP) [1,2]. Individuals whose erythrocytes do not express DARC are highly resistant to invasion by P. vivax and Plasmodium knowlesi, a primate malaria parasite commonly found in Southeast Asia [3,4].
Plasmodium vivax and P. knowlesi merozoites exploit the DARC/DBP interaction to undergo internalization into several non-human primate erythrocytes, and erythrocyte susceptibility is partially dependent on the N-terminal tail of the DARC protein [2,5]. The interaction domain of DBP lies in region II of the protein, which is highly polymorphic [6–8]. This variability is associated with parasite evasion from the host immune system [9,10]. Therefore, antibodies that block DARC/DBPII interactions confer some variant-specific protection, which hampers vaccine development based on DBP [10,11]. Consequently, an understanding of erythrocyte invasion pathways of simian malaria is essential to comprehend the zoonotic potential of malaria in some regions of the world, and it might aid the development of a suitable model for the testing of drugs and vaccines against vivax malaria.
In addition to the hundreds of malaria cases that occur annually in the Amazon region (North Region of Brazil), autochthonous cases of malaria were described in the Atlantic Forest (South and Southeast of Brazil) [12–14]. Many of these cases were associated with a simian malaria parasite, Plasmodium simium, which is morphologically, immunologically and genetically indistinguishable from P. vivax [15–18]. This parasite was described previously as naturally infecting only two genera of monkeys, Alouatta (howler monkeys) and Brachyteles (spider monkey). Humans are susceptible to P. simium, but the natural infections described so far were accidentally acquired [14,19–23].
Our research group recently demonstrated the prevalence of P. simium infection and high levels of seropositivity against P. vivax antigens (MSP-1, DBPII and AMA-1) in wild Alouatta guariba clamitans from the Atlantic Forest in the Santa Catarina state in southern Brazil [24]. These findings suggested that some wild monkeys act as a Plasmodium reservoir for humans. P. simium infects humans, but the pathway used by this parasite to invade host erythrocytes has not been identified. We investigated the genetic variability of binding domains from P. simium DBPII (PsDBPII) and simian DARC on erythrocytes from Alouatta g. clamitans (sDARC) to gain insight into the evolution of Plasmodium species in the New World. We also aimed to gain insight into P. simium DBP and simian DARC interactions.
Materials and Methods
Animal information
Captive and wild Alouatta g. clamitans (Southern brown howler monkeys) were studied. Captive monkeys were obtained from the Conservationist Center of Biological Research at Indaial—CEPESBI (IBAMA Registration No. 1/42/98/000708-90) in the Indaial municipally, and wild monkeys were obtained from the Geisler Mountain in the surroundings areas of Indaial in the Santa Catarina state in southern Brazil. CEPESBI is a scientific captivity center of the University of Blumenau for the maintenance and conservation of A. clamitans. It is part of the Southern Brown Howler Project, which also has an observatory of non-human primates at Geisler Mountain connected to the National Park of Serra do Itajaí, part of the Atlantic Forest. CEPESBI researchers follow approximately 60 red howler monkeys, and 20 monkeys were caught for sample collection. Free-living animals were captured using anesthetic darts (tiletamine and zolazepam 3.9 mg/kg) with the aid of a compressed air gun (Dist-Inject moldel 70). The captivity area has 45 red howler monkeys that were obtained from different places around the municipality of Indaial, and these monkeys have been maintained in captivity from one year to more than 10 years. Animals were housed in couples in 3 m x 5 m x 2.6 m rooms containing ropes, logs and feeding platforms, and they were fed six times daily with a diet consisting of a variety of fruits and vegetables, a commercial ration (Nuvilab CR- 1, Sogorb, São Paulo, Brazil), leaves of Cecropia sp. and Sechium edule and water ad libitum. No animals were sacrificed in this study, and all free-ranging animals returned to their natural habitat after sample collection. The Brazilian government authorized this study and the access to and transport of biological samples through SISBIO no. 28953-1/2011 and 28953-2/2012. The Ethics Committee of the Regional University of Blumenau approved this study (no.003/12 and no. 516/2012).
Sample collection, diagnosis and DNA extraction
Captive animals were contained using dip nets and sedated via intramuscular administration of a combination of tiletamine and zolazepam. A veterinarian from CEPESBI obtained blood samples (5 mL) with and without anticoagulant (EDTA) using femoral venipuncture from each animal. The heart rate, breathing, eye reflexes and temperature were monitored during sedation. Only one animal showed symptoms related to malaria as previously described by us [24]. The other animals were healthy at the time of blood collection. DNA extraction from blood samples was performed using the Gentra Puregene Blood kit (Qiagen, Venlo, Netherlands) according to the manufacturer’s recommendations. Infection was identified in 9 animals (2 captive and 7 wild) using microscopy of panoptic-stained thin blood smear and confirmed using molecular techniques including nested PCR and real-time PCR as P. vivax/P. simium [24].
Plasmodium simium dbpII amplification
Extracted DNA samples from infected monkeys were used as a template in PCR to amplify a fragment of the P. simium DBPII-encoding gene as previously described [8]. Platinum high fidelity Taq DNA polymerase (1 U) (Invitrogen Life Technologies, Thermo Fisher Technologies, Grand Island, NY, USA) was used in a 10 μL PCR reaction with 100–200 ng DNA, 0.125 mM dNTPs, 0.5 μM primers (forward and reverse for P. vivax dbpII), 0.75 mM MgCl2, and 1 μL enzyme buffer. The following amplification parameters were used: 94°C for 3 min, 35 cycles of 94°C for 30 sec, 61°C for 30 sec and 72°C for 2 min, followed by a last extension cycle of 72°C for 5 min.
DARC amplification
Two pairs of primers were used to amplify two fragments of the gene encoding DARC from A. g. clamitans (sDARC1 and sDARC2). sDARC1 corresponding to the 5’ end of the gene, which includes the promoter, exon 1 (codons 1 to 7) and part of the intron (positions -154 to 157 of Homo sapiens DARC—hDARC, accession number JN251915.1), was amplified using primers KAT031 and KAT035 described by Demogines et al. [5]. The 20 μL reaction contained 100–200 ng of DNA, each primer at 0.5 μM, 0.125 mM dNTPs, 2 mM MgCl2, 1 U Taq DNA polymerase and 2 μL of enzyme buffer. PCR cycling was 94°C for 3 min, 35 cycles of 94°C for 40 sec, 64°C for 40 sec and 72°C for 1 min, followed by a final cycle of 72°C for 5 min. DARC2 includes part of exon 2 (nucleotides 1254 to 2072 of Saimiri sciureus DARC, accession number HQ285857.1), and it was amplified using primers DARC2F 5’CCCCTCCCACCTGCCCC3’ and DARC2R 5’GCCACCAGAAAATAAACCAG3’ in the same conditions described above, except for the annealing temperature of 60°C.
Sequencing of PCR-amplified DNA
PCR products were purified using the Purification Kit (Qiagen Inc., Valencia, CA, USA) following the manufacturer’s procedure. Approximately 1–10 ng of purified PCR products was amplified using each primer at 3.3 μM (forward or reverse) and 1 μL of Big Dye terminator kit (Life Technologies) in a program of 96°C for 1 min, 35 cycles of 96°C for 15 sec, the temperature of primer annealing for 15 sec and 60°C for 15 sec. The fragments were precipitated using ammonium acetate, resuspended in formamide HI-DI (Applied Biosystems, Thermo Fisher Scientific, Inc., Waltham, MA, USA) and analyzed in an ABI 3730 DNA (Applied Biosystems) automatic sequencer.
Serological assay
An ELISA (enzyme-linked immunosorbent assay) to detect IgG antibodies in monkey sera was performed using recombinant P. vivax Duffy binding protein (rDBP—regions II to IV) as previously described [24]. rDBP was used at a final concentration of 5 μg/mL with serum samples assayed at a 1:100 dilution, and Macaca mulatta anti-IgG was used as the secondary antibody (Sigma-Aldrich, St. Louis, MO). The cut-off was based on the mean plus three standard deviations of sera reactivity from 8 non-exposed monkeys, and the results are expressed as a reactivity index (IR = OD492nm values of test sample divided by the value of the cut-off).
COS-7 cell transfection and erythrocyte-binding assays
Sal-1 PvDBP-pEGFP recombinant plasmid [25,26] was transfected into COS-7 cells (American Type Culture Collection, Manassas, VA, USA) using lipofectamine and PLUS-reagent (Invitrogen Life Technologies, Carlsbad, CA, USA) according to the manufacturers’ protocols. Briefly, COS-7 cells in six-well culture plates (1.5 × 105 cells/well) were transfected with plasmid (0.5 μg/well)-liposome (5% Plus-reagent and 3% lipofectamine) complexes in Dulbecco’s Modified-Eagle Medium (DMEM, Sigma, St. Louis, MO, USA) without serum. Transfection medium was replaced by DMEM with 10% of fetal bovine serum (Gibco-BRL, Gaithersburg, MD, USA) after 6 h of cell exposure to the DNA liposome complexes (37°C, 5% CO2). Culture medium was replaced again 24 h after transfection, and the efficiency of transfection was assessed using fluorescence. Erythrocyte-binding assays were performed 48 h after transfection as previously described [27]. Briefly, erythrocytes from two monkeys (BL10 and BL34) were added to the cell cultures, and plates were incubated for 2 h at room temperature. Unbound erythrocytes were removed by washing the wells three times with phosphate-buffered saline (PBS). Binding was quantified by counting the number of rosettes observed over 10–20 fields of view (×200). Positive rosettes were defined as adherent erythrocytes that covered more than 50% of the COS cell surface.
Inhibition of erythrocyte binding assays
Blockade of erythrocyte binding was performed using sera of infected and non-infected monkeys added at 1:30, and the plates were incubated for 1 h at 37°C in 5% CO2. The 1:30 dilution was chosen because it provided a wide range of inhibitory activity in previous experiments (data not shown). Human O+ erythrocytes from a DARC-positive individual in a 10% suspension were added to each well (200 μL/well), and the plates were incubated for 2 h at room temperature. Unbound erythrocytes were removed by washing the wells three times with PBS. Binding was quantified by counting the number of rosettes observed over 10–20 fields of view (×200). Positive rosettes were defined as adherent erythrocytes that covered more than 50% of the COS cell surface. DARC-negative human serum was used as a negative control for each assay. The percent inhibition was calculated as 100 × (R c—R t)/R c, where R c is the average number of rosettes in the control wells, and R t is the average number of rosettes in the test wells.
Analyses
Identity of DNA sequences was confirmed using the BLAST program (www.ncbi.nlm.nih.gov/BLAST). DNA sequences were analyzed using BioEdit sequence alignment editor (www.mbio.ncsu.edu/BioEdit/bioedit.html) to align the sequences and identify polymorphisms. Genetic diversity was analyzed using DnaSP version 5.10 [28]. The three-D structure of the PvDBPII dimer (PDB– 3RRC) [29] was visualized using PyMol v1.1 [30], and it was used to identify polymorphic residues. Phylogenetic analyses were performed using the Maximum likelihood method with Tamura—Nei model and 5,000 bootstrap replicates in MEGA 5.2 [31]. Statistical analyses were performed using STATA software v12.
Results
Single polymorphic nucleotides in Duffy binding protein II of Plasmodium simium (PsDBPII)
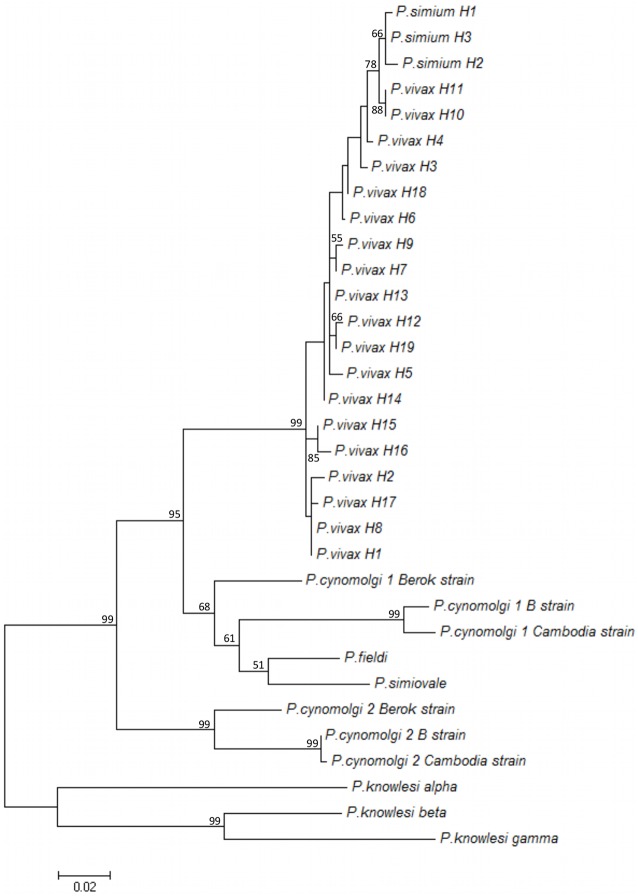
Plasmodium simium infection was detected in 9 of 65 monkeys (2 of 45 captive and 7 of 20 wild monkeys) using molecular diagnosis [24]. The DNA samples were further used to amplify PsDBPII. Good quality sequences of the PsDBPII-encoding gene (positions 1010–1447 of P. vivax Sal-1, accession number M61095.1) were obtained in 7 of 9 infected monkeys. Four polymorphic sites were identified in the P. simium sequences (nucleotide positions 1016, 1113, 1153 and 1154 of PvDBP sequence), and these sites all contained non-synonymous substitutions (S1 Fig). These polymorphisms were combined in three distinct haplotypes of PvDBPII (Table 1). Comparisons to the most prevalent P. vivax dbpII sequences available in GenBank identified 22 SNPs in P. simium dbp sequences (S1 Fig). The substitution 1233 A > C was present in all P. simium isolates, and it was never identified in P. vivax isolates worldwide. Comparisons of the genetic diversity of dbpII sequences from P. simium and all P. vivax sequences available in GenBank identified 84 polymorphic sites with a nucleotide diversity (π) of 0.0124 (Table 2). The comparison of P. simium sequences with a P. cynomolgi dbpII sequence allowed the identification of 56 polymorphic sites (π = 0.0348), and comparison with a P. knowlesi dbp alpha sequence identified 86 polymorphic sites (π = 0.0526). The nucleotide diversity of dbp sequences from P. vivax/P. simium (π = 0.0124) was slightly lower than those of P. vivax/P. cynomolgi (π = 0.0127) and P. vivax/P. knowlesi (π = 0.0130) (Table 2). Haplotype diversity was quite high for all species, but this result must be analyzed with caution because of the high dependency of the experimental sample size. Phylogenetic analyses were performed to identify relationships between dbpII sequences from different Plasmodium species. The three dbpII haplotypes from P. simium clustered together, and this clade exhibited a close relationship with dbpII from P. vivax predominant haplotypes (Fig 1). Polymorphic sites of P. simium and P. vivax were mapped on the PvDBPII 3D-structure (Fig 2). Polymorphisms of P. simium and P. vivax were concentrated in DBPII sub-domain 2. However, P. simium SNPs were not located near the DARC binding site, unlike the most important P. vivax polymorphisms.
Table 1. Single nucleotide polymorphisms (SNPs) in the ligand domain of the Duffy binding protein (DBPII)-encoding gene in seven isolates of Plasmodium simium.
| Nucleotide* (AA) | ||||
|---|---|---|---|---|
| Monkey code | Haplotype | 1016 (338) | 1113 (371) | 1153–1154 (385) |
| BL3, BL10 | 1 | AGA (R) | AAA (K) | ATA (I) |
| BL4, BL5, BL61, BL64 | 2 | AAA (K) | AAT (N) | CAA (Q) |
| BL6 | 3 | AGA (R) | AAA (K) | AAA (K) |
* Positions corresponding to P. vivax Sal-1 dbp, accession number M61095.1.
Underlined residues are polymorphic. Dots indicate the same residue in the sequences compared to the reference sequence.
Table 2. Genetic diversity of P. simium DBPII compared to other Plasmodium DBPII.
| Group | Plasmodium species (N) | S | Singleton variable sites | Parsimony informative sites | π ± SD | Synonymous changes | Non-synonymous changes | H | Hd ± SD |
|---|---|---|---|---|---|---|---|---|---|
| 1 | P. simium (7) | 3 | 0 | 3 | 0.0038 ± 0.0008 | 0 | 3 | 3 | 0.6670 ± 0.1600 |
| 2 | P. simium + P. vivax a (8) | 10 | 7 | 3 | 0.0080 ± 0.0031 | 1 | 7 | 4 | 0.7500 ± 0.1390 |
| 3 | P. simium + P. cynomolgi b (8) | 56 | 53 | 3 | 0.0348 ± 0.0224 | 17 | 39 | 4 | 0.7500 ± 0.1390 |
| 4 | P. simium + P. knowlesi c (8) | 86 | 83 | 3 | 0.0526 ± 0.0352 | 32 | 52 | 4 | 0.7500 ± 0.1390 |
| 5 | P. vivax (511) | 81 | 42 | 39 | 0.0122 ± 0.0003 | 14 | 64 | 118 | 0.9196 ± 0.0078 |
| 6 | P. vivax + P. simium (518) | 84 | 42 | 42 | 0.0124 ± 0.0003 | 14 | 65 | 121 | 0.9217 ± 0.0077 |
| 7 | P. vivax + P. cynomolgi b (512) | 122 | 81 | 41 | 0.0127 ± 0.0005 | 20 | 87 | 119 | 0.9199 ± 0.0078 |
| 8 | P. vivax + P. knowlesi c (512) | 145 | 101 | 44 | 0.0130 ± 0.0008 | 34 | 91 | 119 | 0.9199 ± 0.0078 |
Fig 1. Phylogenetic tree of dbpII from the 19 most predominant P. vivax haplotypes.
Haplotypes with > 1% frequency were used here: accession numbers (haplotype). P. vivax dbp: EU812840.1 (H1), EU812841.1 (H2), EU812842.1 (H3), EU812844.1 (H4), EU812845.1 (H5), EU812849.1 (H6), EU812861.1 (H7), EU812869.1 (H8), EU812874.1 (H9), EU812898.1 (H10), EU812915.1 (H11), EU812927.1 (H12), EU812954.1 (H13), AF220662 (H14), AF289650 (H15), AF289649 (H16), GU143965 (H17), GU143986 (H18), EF379128 (H19); P. cynomolgi dbp1: AB617788.1 (Cambodia strain), JQ422035.1 (Berok strain), XM_004221494.1 (B strain); P. cynomolgi dbp2: AB617789.1 (Cambodia strain), JQ422036.1 (Berok strain), XM004220981.1 (B strain); P. knowlesi dbp: M90466.1 (alpha), M90694.1 (beta) and (M90695.1 gamma); P. fieldi dbp: AB617790.1; P. simiovale dbp: AB617791.1; and three haplotypes of P. simium isolates herein (H1, H2, H3). The tree was build using Maximum likelihood method with the Tamura and Nei model (TN93 model) in Mega 5.2. Numbers in the nodes indicate bootstrap values greater than 50.
Fig 2. Three-dimensional structure of the PvDBPII dimer.
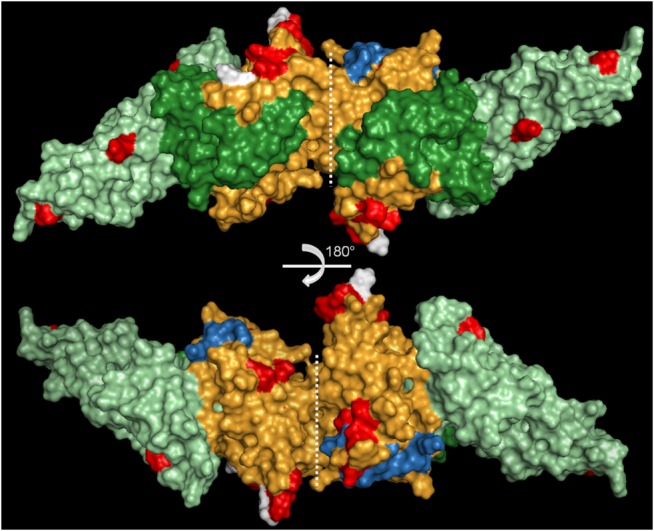
PvDBPII dimer (3RRC) showing the polymorphic sites in P. vivax (red) and P. simium (white). The three subdomains of the protein are shown in green (subdomain 1), orange (subdomain 2) and light green (subdomain 3). Residues important for the DARC interaction are shown in blue. 3-D structure visualized using PyMol. The structures correspond to a 180° rotation in the horizontal plan.
Characterization of sDARC
Fragments of the sDARC-encoding genes (promoter and nucleotides 1 to 768) from 10 A. g. clamitans individuals were sequenced, and no polymorphisms were identified (data not shown). Comparisons to the other primate DARC sequences available in GenBank identified 222 polymorphic sites (S2 Fig). Alouatta g. clamitans exhibited 7 SNPs that were not previously identified in any other primate species, and 3 of these SNPs were non-synonymous substitutions (at nucleotide positions 530, 745 and 746) (S2 Fig). N-terminal sDARC (aa 1–42) exhibited the highest genetic variability (Table 3). Polymorphic sites were not only concentrated in this region but were distributed throughout the entire molecule, including the transmembrane segments (Fig 3). The mutation indicative of a null allele (-67 T > C) (data not shown) and the substitution responsible for the FY*A allele (Asp42Gly) were not observed in our A. clamitans DARC sequences (S2 Fig).
Table 3. Genetic diversity of DARC from Southern brown howler monkeys (Alouatta g. clamitans).
| DARC (AA) | S | Singleton variable sites | Parsimony informative sites | π ± SD | Synonymous changes | Non-synonymous changes |
|---|---|---|---|---|---|---|
| 1–42 | 54 | 25 | 29 | 0.07499 ± 0.00642 | 17 | 14 |
| 43–256 | 168 | 75 | 93 | 0.04622 ± 0.00268 | 88 | 64 |
| 1–256 | 222 | 100 | 122 | 0.05088 ± 0.00265 | 105 | 78 |
AA: amino acid position; S: number of segregating sites; π: nucleotide diversity; H: haplotypes; Hd: haplotype diversity; SD: standard deviation.
Fig 3. Schematic model of DARC showing polymorphic sites.
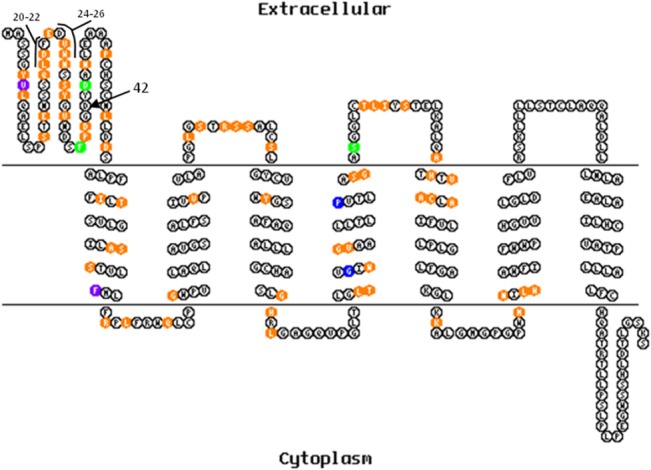
The 2-D model of DARC from Homo sapiens was constructed using TOPO2 software. Polymorphic sites in primates are shown in orange, purple (SNPs exclusive to the Cercopithecidae family), green (SNPs exclusive to the Platyrrhini parvorder) and blue (SNPs exclusive to the Hominoidea superfamily). Residues involved in direct binding to DARC are indicated (20–22 and 24–26) according to Batchelor et al. [53]. The arrow indicates Asp42Gly. Polymorphisms were annotated only until codon 255 (fragment available in our sequences).
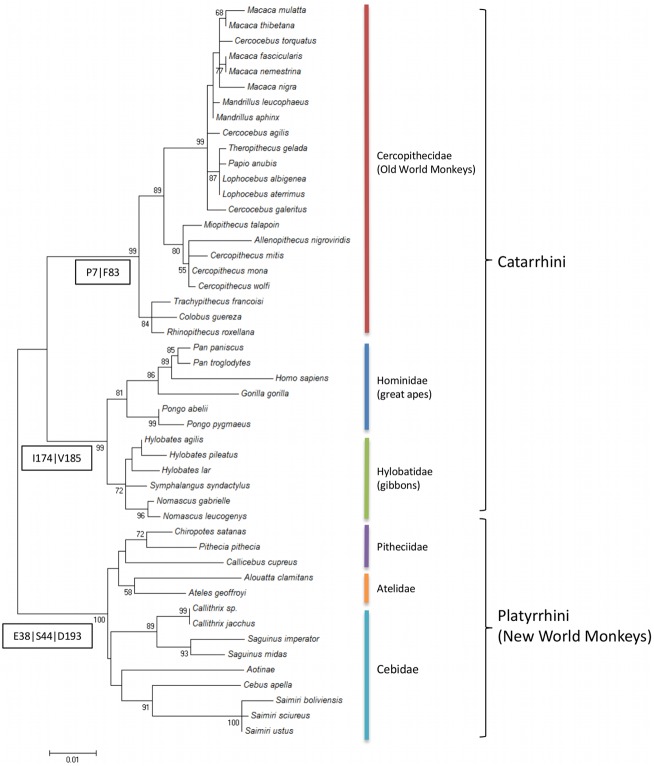
The reconstruction of the phylogenetic relationships of primates based on the N-terminal DARC sequence (responsible for DBP binding) clustered A. g. clamitans with Ateles geoffroyi in the clade of Platyrrhini parvorder (New World monkeys) (Fig 4). Notably, only a few SNPs were exclusive to each group: two from the Cercopithecidae family (P7 and F83), two from the Hominoidea superfamily (I174 and V185) and three from the Platyrrhini family (E38, S44 and D193) (Figs 3 and 4).
Fig 4. Phylogenetic tree of DARC from primates.
Sequences of DARC from 47 primate species available from GenBank were aligned with the Alouatta g. clamitans DARC sequences described here. Clusters of parvorder: Platyrrhini and Catarrhini, and Families: Cercopithecidae, Hominidae, Hylobatidae, Pitheciidae, Atelidae and Cebidae are indicated. The numbers in the boxes correspond to the exclusive codons from each group (one letter amino acid and position).
DARC/DBP in vitro interaction and inhibitory antibodies
We next investigated whether monkey sera blocked the interaction between human DARC (hDARC) and PvDBPII. A preliminary experiment investigated whether sDARC from A. clamitans bound DBPII from P. vivax. The results clearly demonstrated that simian erythrocytes (all DARC positive) recognized PvDBPII on the surface of COS-7 cells. Fig 5A illustrates one of two experiments in which monkey erythrocytes formed rosettes in COS-7 cells expressing PvDBPII. hDARC-negative erythrocytes exhibited no binding to DBPII expressed on COS-7 cells (Fig 5B).
Fig 5. Interaction of Alouatta g. clamitans DARC and PvDBPII.
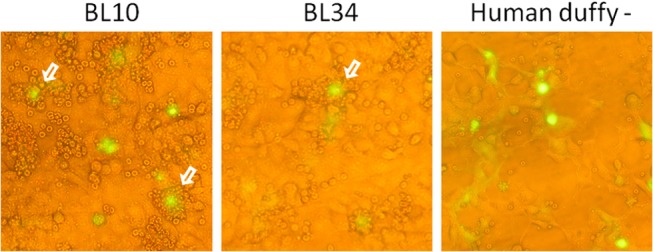
(A) Interaction between P. vivax DBPII expressed on the COS7 cell surface and DARC on the surface of A. clamitans erythrocytes (BL10 and BL34), representative of two experiments. Green fluorescence indicates transfected cells expressing GFP. (B) The negative control was performed using human DARC-negative erythrocytes. The cells were observed using conventional epifluorescence microscopy (×200). White arrows indicate the rosettes.
We performed in vitro erythrocyte binding inhibition assays, in which sera of infected-monkeys were used to block the interaction between hDARC/PvDBPII, to further investigate the inhibitory activity of immune plasma samples. We selected 8 monkeys (acute or chronically infected) with different levels of anti-PvDBP ELISA-detected IgG antibodies (Table 4). Binding inhibition ranged from 0 to 94%, with one monkey (BL37) showing the strongest DARC/DBP inhibition. Titration of the inhibitory antibody responses of the two highest inhibitory sera exhibited a dose-response curve (S3 Fig). A positive correlation was identified between the reactivity index in ELISA and the percentage of DARC/DBP interaction blockade (Spearman’s correlation coefficient = 0.843, P = 0.011).
Table 4. Characteristics of immune sera from monkeys used for the inhibition of the hDARC/PvDBPII interaction.
| Monkey Code | Acute infection b | Anti-PvDBPII IgG, RI c | Percentage of inhibition |
|---|---|---|---|
| BL69 | Pos | 22 | 44% |
| BL64 | Pos | 18 | 40% |
| BL10 a | Pos | 12 | 36% |
| BL34 | Neg | 22 | 43% |
| BL37 | Neg | 14 | 94% |
| BL40 | Neg | 7 | 10% |
| BL41 | Neg | 6 | 0 |
| BL44 | Neg | 1 | 0 |
aBL10 was the only symptomatic animal [24]
bPeripheral blood infection was detected using conventional microscopy, and P. vivax/P. simium identification was confirmed using amplification of the P. vivax 18SSU rRNA gene with two different PCR-based protocols (Nested and Real-time) [24]
cAnti-PvDBPII IgG antibodies were detected by ELISA using recombinant PvDBP (aa 132–771)[24], and the results are expressed as Reactivity Index (RI), which corresponds to OD492nm values of test samples divided by the cut-off value.
Values of RI > 1 were considered positive.
Discussion
The major pathway for erythrocyte invasion by P. vivax merozoites is dependent on the interaction between parasite DBPII and DARC on the host cell surface [32]. This dependency and the capacity of DBP to elicit a protective immune response rendered DBP one of the most prominent vaccine candidates against malaria caused by P. vivax [33–36]. The elicited immunity is related to the genetic diversity of the DBP ligand domain, and blockade of the DBPII/DARC interaction appears to be variant-specific. The close proximity between P. vivax and P. simium suggests that the DBPII/DARC pathway is also involved in P. simium erythrocyte invasion. We analyzed the DBPII sequences from isolates of P. simium from seven naturally infected monkeys to gain insight into the genetic variability of P. simium DBPII. The results demonstrated much less variability in PvDBPII than its orthologous protein in P. vivax available in GenBank. Numerous segregating sites and a high overall nucleotide diversity in P. vivax DBPII [37] were not observed for P. simium DBPII. This result corroborates the hypothesis of a recent transfer of Plasmodium species from humans to New World monkeys (Atelidae family), as previously suggested [38–44]. Tazi and Ayala [43] and Lim et al. [44] noted biological and historical reasons in favor of the transfer from humans to New World monkeys, such as the worldwide distribution of P. vivax compared to a very limited geographic description of P. simium and the restricted number of natural hosts of P. simium. We cannot disregard the possibility that this low variability does not mean that P. simium is evolutionarily more recent than P. vivax, but it could be a consequence of a recent bottleneck in the P. simium population, which is consistent with the absence of synonymous substitutions observed in PsDBPII. However, the high genetic identity found here between P. simium and P. vivax DBPII reinforces their close phylogenetic relationship and strengthens the hypothesis that the host switch arose in very recent evolutionary times. This switch may have occurred only after the latest five centuries of European colonization [44,45]. Accordingly, the genetic identity previously reported for P. vivax and P. simium at rapidly evolving loci, such as microsatellites and tandem repeats, confirms that the host transfer must have occurred recently in the evolutionary scale [40].
Similar to P. vivax, P. simium exhibited an excessive number of non-synonymous substitutions in DBPII, which suggests diversification due to selective pressure by the host immune response [37,46]. The spatial localization of polymorphic residues of P. vivax DBPII near the erythrocyte-binding domain, in which substitutions divert recognition by inhibitory antibodies, likely contributes to the variant specificity of the immune response [8,47]. Notably, polymorphic residues of PsDBPII were distinctly localized from the P. vivax polymorphic residues in the DBPII 3-D structure. Despite this localization difference, we identified at least one A. clamitans antibody in sera that strongly inhibited (94%) the in vitro interaction between PvDBPII and human DARC.
The P. simium erythrocyte invasion pathway was not the focus of the current work, but the availability of transfected COS cells expressing PvDBPII [48] offered an opportunity to gain insight into the biology of invasion. The in vitro experiments demonstrated that A. clamitans erythrocytes bound to PvDBPII, and this in vitro interaction, as evaluated by the number of rosettes, was similar to that with human erythrocytes (data not shown). Most interestingly, the results demonstrated that simian immune sera inhibited the interaction between hDARC and PvDBPII. ELISA reactivity positively correlated with the inhibition levels, but great variability in the profile of inhibitory response was observed, and most of the monkeys generated weak or non-neutralizing antibodies to DBPII. We cannot exclude the possibility that the use of a homologous system (PsDBP/sDARC) could increase this response. However, these results are consistent with P. vivax-exposed populations, which showed that PvDBPII was weakly immunogenic and induced strain-specific immunity [10,49]. Consequently, it was not surprising that a single animal produced high levels of binding inhibitory antibodies (BIAbs) that cross-reacted with PvDBPII. These findings are interesting because few infected people developed a significant BIAbs response (defined as “elite responders”) and produced broadly reactive anti-DBPII binding inhibitory antibodies that were directed against more conserved B cell epitopes ([48], revised in [50]). Therefore, these results reinforce the possibility of using P. simium/New World monkeys as a promising tool to guide the development of DBPII as a vaccine against P. vivax.
Although more definitive invasion/inhibitory assays are needed to confirm whether P. simium infection depends on the DBP/DARC interaction, this invasion pathway is already well-established in another simian malaria parasite, P. knowlesi [51], which is also able to infect humans [52] but is evolutionarily less closely related to P. vivax than P. simium [53]. This parasite has two paralogous erythrocyte binding proteins, EBP β and γ, in addition to the orthologous DBP PkEBP α. These two P. knowlesi EBP proteins, which do not bind DARC positive-erythrocytes, exhibited much higher diversity than PvDBP than PsDBP [37], which corroborates the potential to use P. simium as a model for P. vivax infection.
Phylogenetic analysis of the sDARC fragment (aa 1 to 255) showed very good correspondence with highly resolved primate phylogeny [54]. DARC exhibited a high level of variability among the 48 primates, and the N-terminal domain (aa 8 to 42), which was identified as the binding domain for P. vivax erythrocyte invasion, was the most polymorphic protein region [55]. This finding reinforces the hypothesis of selective pressure acting in this domain, as previously demonstrated [5,56]. Recently, the structural analysis of PvDBP/hDARC binding defined amino acids 19 to 30 of DARC as a critical interaction site, in which L20/D21/F22 and D24/V25/W26 residues make direct contact with DBPII [57]. Different combinations of amino acids in these positions according to the group of primates are found (S2 Fig), and the most polymorphic residue was at position 25. This residue was implicated as an inter-species barrier to P. vivax invasion because Gorilla gorilla has mutation V25A, which disrupts an essential hydrophobic interaction with PvDBP that stabilizes DARC/DBP binding [5,57,58]. All New World monkeys exhibited a V25L mutation, which is expected to maintain the stabilization role of this residue, which suggests that all of these monkeys are susceptible to P. vivax invasion. This result indicates that there is not a clear signature in DARC for the high invasion dependency observed in some Plasmodium species, such as P. vivax and likely P. simium. Therefore, this feature does not have a monophyletic origin.
Beyond the evolutionary importance of simian malaria, the existence of a sylvatic P. simium reservoir of malaria infection might have public health implications. The presence of P. vivax-related parasites in monkeys was also described in Central Africa, where human P. vivax infection is not common because of the high levels of Duffy negativity in the human population [59]. This result suggests that humans living in close proximity to monkeys in specific geographic areas, such as the Atlantic Forest in Brazil and Central Africa, may harbor P. vivax-related simian malaria. Antibodies against P. vivax pre-erythrocytic stages were described in DARC- negative individuals in the Republic of Congo (West Central Africa), and P. vivax autochthonous cases may be common in travelers returning from this region [60]. Autochthon cases of malaria may have also originated in the Atlantic Forest area in Brazil [11–13]. Therefore, the potential of simian Plasmodium infections as a malaria reservoir should be considered in the international effort to reduce the global incidence of malaria.
Conclusions
The close phylogenetic relationship between P. simium and P. vivax and the lower DBPII variability in infected monkeys suggest a recent host switch from New World monkeys to humans. The possibility of a sylvatic reservoir of Plasmodium species in Atlantic Forests should be considered in the current efforts to control and eliminate human malaria. Furthermore, the possibility that P. vivax and P. simium share a similar erythrocyte invasion pathway, which was reinforced by the presence of inhibitory antibodies in wild A. clamitans, makes the P. simium/New World monkeys an attractive model for vaccine and drug development. Towards that goal, it will be important to validate this model using in vitro and in vivo assays, particularly using small New World monkeys, such as capuchin monkeys (subfamily Cebinae), which we found to be naturally infected with P. simium [61].
Supporting Information
dbpII sequences (positions 1021 to 1300 of P. vivax Sal-1 dbp, accession number M61095.1) from the 19 most prevalent haplotypes of P. vivax (sequences with > 1% frequency) in 511 available sequences, accession numbers (haplotype): EU812840.1 (H1), EU812841.1 (H2), EU812842.1 (H3), EU812844.1 (H4), EU812845.1 (H5), EU812849.1 (H6), EU812861.1 (H7), EU812869.1 (H8), EU812874.1 (H9), EU812898.1 (H10), EU812915.1 (H11), EU812927.1 (H12), EU812954.1 (H13), AF220662 (H14), AF289650 (H15), AF289649 (H16), GU143965 (H17), GU143986 (H18), EF379128 (H19); and the seven P. simium dbp sequences (herein). White head arrows indicate polymorphic sites, and black head arrows indicate polymorphisms among P. simium sequences. Identical residues are represented by dots. An asterisk shows the species-specific polymorphism (1233A>C).
(PDF)
darc sequences of Alouatta g. clamitans were aligned with other primate darc sequences available in GenBank (nucleotides 1 to 768, from Homo sapiens darc sequence, accession number JN251915.1). The numbered bars above the sequences indicate transmembrane domains. The grey box shows the N-terminal minimum-binding domain (19–30 aa), and residues responsible for direct interaction with DBPII are underlined (20,21,22 and 24,25,26). Colored bars on the left side of the alignment represent the phylogenetic group families: red (Cercopithecidae), blue (Hominidae), green (Hylobatidae), purple (Pitheciidae), orange (Atelidae) and aqua (Cebidae). The arrow indicates the polymorphism Asp42Gly, which is responsible for the FY*A and FY*B alleles.
(PDF)
The sera from two monkeys (BL37 and BL69), which showed the highest levels of blockade, were used in dilutions of 1:30, 1:90 and 1:270 in the inhibition assays of human DARC and PvDBPII. Inhibition was calculated based on the reduction in the rosette numbers observed in the presence of monkey serum compared to the rosette numbers in the absence of serum.
(TIF)
Acknowledgments
The authors thank Dr. Rosely Malfronte for providing serum from negative monkeys. We also thank the PDTIS sequencing facilities of FIOCRUZ.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors received funding from FAPEMIG for this work. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Haynes JD, Dalton JP, Klotz FW, McGinniss MH, Hadley TJ, Hudson DE, et al. Receptor-like specificity of a Plasmodium knowlesi malarial protein that binds to Duffy antigen ligands on erythrocytes. J Exp Med. 1988;167: 1873–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wertheimer SP, Barnwell JW. Plasmodium vivax interaction with the human Duffy blood group glycoprotein: identification of a parasite receptor-like protein. Exp Parasitol. 1989;69: 340–350. [DOI] [PubMed] [Google Scholar]
- 3. Miller LH, Mason SJ, Dvorak JA, McGinniss MH, Rothman IK. Erythrocyte receptors for (Plasmodium knowlesi) malaria: Duffy blood group determinants. Science. 1975;189: 561–563. [DOI] [PubMed] [Google Scholar]
- 4. Miller L, Mason S, Clyde D, McGinniss M. The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype, FyFy. N Engl J Med. 1976;295: 302–304. [DOI] [PubMed] [Google Scholar]
- 5. Demogines A, Truong KA, Sawyer SL. Species-specific features of DARC, the primate receptor for Plasmodium vivax and Plasmodium knowlesi . Mol Bio Evol. 2012;29: 445–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Adams J, Sim BKL, Dolan SA, Fang XD, Kaslow DC, Miller LH. A family of erythrocyte binding-proteins of malaria parasites. Proc Natl Acad Sci USA. 1992;89: 7085–7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Singh S, Hora R, Belrhali H, Chitnis C, Sharma A. Structural basis for Duffy recognition by the malaria parasite Duffy-binding-like domain. Nature. 2006;439: 741–744. [DOI] [PubMed] [Google Scholar]
- 8. Sousa TN, Tarazona-Santos EM, Wilson DJ, Madureira AP, Kuser PR, Falcão PR, et al. Genetic variability and natural selection at the ligand domain of the Duffy binding protein in brazilian Plasmodium vivax populations. Malar J. 2010;9: 334 10.1186/1475-2875-9-334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. VanBuskirk KM, Cole-Tobian JL, Baisor M, Sevova ES, Bockarie M, King CL, et al. Antigenic drift in the ligand domain of Plasmodium vivax duffy binding protein confers resistance to inhibitory antibodies. J Infect Dis. 2004;190: 1556–1562. [DOI] [PubMed] [Google Scholar]
- 10. Ceravolo IP, Sanchez BAM, Sousa TN, Guerra BM, Soares IS, Braga EM, et al. Naturally acquired inhibitory antibodies to Plasmodium vivax Duffy binding protein are short-lived and allele-specific following a single malaria infection. Clin Exp Immun. 2009;156: 502–510. 10.1111/j.1365-2249.2009.03931.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. King CL, Michon P, Shakri AR, Marcotty A, Stanisic D, Zimmerman PA, et al. Naturally acquired Duffy-binding protein-specific binding inhibitory antibodies confer protection from blood-stage Plasmodium vivax infection. Proc Natl Acad Sci USA. 2008;105: 8363–8368. 10.1073/pnas.0800371105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Curado I, Duarte AM, Lal AA, Oliveira SG, Kloetzel JK. Antibodies anti bloodstream and circumsporozoite antigens (Plasmodium vivax and Plasmodium malariae/P. brasilianum) in areas of very low malaria endemicity in Brazil. Mem Inst Oswaldo Cruz. 1997;92: 235–243. [DOI] [PubMed] [Google Scholar]
- 13. Cerutti C Jr, Boulos M, Coutinho AF, Hatab Mdo C, Falqueto A, Rezende HR, et al. Epidemiologic aspects of the malaria transmission cycle in an area of very low incidence in Brazil. Malar J. 2007;6: 33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wanderley DM, da Silva RA, de Andrade JC. Epidemiological aspects of malaria in the State of São Paulo, Brazil, 1983 to 1992. Rev Saude Publica. 1994;28: 192–197. [DOI] [PubMed] [Google Scholar]
- 15. Fonseca F. Plasmodium of a primate of Brazil. Mem Inst Oswaldo Cruz. 1951;49: 543–553. [DOI] [PubMed] [Google Scholar]
- 16. Collins WE, Contacos PG, Guinn EG. Observations on the sporogonic cycle and transmission of Plasmodium simium Da Fonseca. J Parasitol. 1969;55: 814–816. [PubMed] [Google Scholar]
- 17. Coatney GR, Collins WE, Warren McW, Contacos PG. The Primate Malarias. Washington: U.S. Government Printing Office; 1971. [Google Scholar]
- 18. Goldman IF, Qari SH, Millet PG, Collins WE, Lal AA. Circumsporozoite protein gene of Plasmodium simium, a Plasmodium vivax-like monkey malaria parasite. Mol Biochem Parasitol. 1993;57: 177–180. [DOI] [PubMed] [Google Scholar]
- 19. Deane M, Neto JF, Sitônio JG. New host of natural Plasmodium simium and Plasmodium brasilianum: mono, Brachyteles arachnoides . J Inst Med Trop São Paulo. 1968;10: 287–288. [PubMed] [Google Scholar]
- 20. Deane LM. Simian malaria in Brazil. Mem Inst Oswaldo Cruz. 1992;87: 1–20. [DOI] [PubMed] [Google Scholar]
- 21. Yamasaki T, Duarte AMRC, Curado I, Summa MEL, Neves DV, Wunderlich G, et al. Detection of etiological agents of malaria in howler monkeys from Atlantic Forests, rescued in regions of São Paulo city, Brazil. J Med Primatol. 2011;40: 392–400. 10.1111/j.1600-0684.2011.00498.x [DOI] [PubMed] [Google Scholar]
- 22. Deane LM, Deane MP, Ferreira Neto J. Studies on transmission of simian malaria and on the natural infection of man with Plasmodium simium in Brazil. Bull World Health Organ. 1966;35: 805–808. [PMC free article] [PubMed] [Google Scholar]
- 23. Carréri-Bruno GC, Ciaravolo RM, Pereira M. Malaria acquired during entomological research in the Serra do Mar, southeastern region of Brazil. Rev Saude Publica. 1995;29: 142–143. [DOI] [PubMed] [Google Scholar]
- 24. Costa DC, Cunha VP, Assis GMP, Souza JCS Jr, Hirano ZMB, de Arruda ME, et al. Plasmodium simium/P. vivax infections in southern brown howler monkeys from Atlantic Forest. Mem Inst Oswaldo Cruz. 2014;109: 641–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chitnis CE, Miller LH. Identification of the erythrocyte binding domains of Plasmodium vivax and Plasmodium knowlesi proteins involved in erythrocyte invasion. J Exp Med. 1994;180: 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Michon P, Fraser T, Adams JH. Naturally acquired and vaccine-elicited antibodies block erythrocyte cytoadherence of the Plasmodium vivax Duffy binding protein. Infect Immun. 2000;68: 3164–3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ceravolo IP, Souza-Silva FA, Fontes CJ, Braga EM, Madureira AP, Krettli AU, et al. Inhibitory properties of the antibody response to Plasmodium vivax Duffy binding protein in an area with unstable malaria transmission. Scand J Immunol. 2008;67: 270–278. 10.1111/j.1365-3083.2007.02059.x [DOI] [PubMed] [Google Scholar]
- 28. Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25: 1451–1452. 10.1093/bioinformatics/btp187 [DOI] [PubMed] [Google Scholar]
- 29. Batchelor JD, Zahm JA, Tolia NH. Dimerization of Plasmodium vivax DBP is induced upon receptor binding and drives recognition of DARC. Nat Struct Mol Biol. 2011;18: 908–914. 10.1038/nsmb.2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wijeyesakere S, Richardson R, Stuckey J. Modeling the tertiary structure of the patatin domain of neuropathy target esterase. Protein J. 2007;26: 165–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol. 2011;28: 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Adams JH, Blair PL, Kaneko O, Peterson DS. An expanding ebl family of Plasmodium falciparum . Trends Parasitol. 2001;17: 297–299. [DOI] [PubMed] [Google Scholar]
- 33. Chitnis CE, Sharma A. Targeting the Plasmodium vivax Duffy-binding protein. Trends Parasitol. 2008;24: 29–34. [DOI] [PubMed] [Google Scholar]
- 34. Xainli J, Baisor M, Kastens W, Bockarie M, Adams JH, King CL. Age-dependent cellular immune responses to Plasmodium vivax Duffy binding protein in humans. J Immunol. 2002;169: 3200–3207. [DOI] [PubMed] [Google Scholar]
- 35. Michon PA, Arevalo-Herrera M, Fraser T, Herrera S, Adams JH. Serologic responses to recombinant Plasmodium vivax Duffy binding protein in a Colombian village. Am J Trop Med Hyg. 1998;59: 597–599. [DOI] [PubMed] [Google Scholar]
- 36. Fraser T, Michon P, Barnwell JW, Noe AR, Al-Yaman F, Kaslow DC, et al. Expression and serologic activity of a soluble recombinant Plasmodium vivax Duffy binding protein. Infect Immun. 1997;65: 2772–2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cole-Tobian J, King CL. Diversity and natural selection in Plasmodium vivax Duffy binding protein gene. Mol Biochem Parasitol. 2003;127: 121–132. [DOI] [PubMed] [Google Scholar]
- 38. Escalante AA, Ayala FJ. Phylogeny of the malarial genus Plasmodium, derived from rRNA gene sequences. Proc Natl Acad Sci USA. 1994;91: 11373–11377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ayala FJ, Escalante AA, Rich SM. Evolution of Plasmodium and the recent origin of the world populations of Plasmodium falciparum . Parassitologia. 1999;41: 55–68. [PubMed] [Google Scholar]
- 40. Leclerc MC, Hugot JP, Durand P, Renaud F. Evolutionary relationships between 15 Plasmodium species from new and old world primates (including humans): an 18S rDNA cladistic analysis. Parasitology. 2004;129: 677–684. [DOI] [PubMed] [Google Scholar]
- 41. Mu J, Joy DA, Duan J, Huang Y, Carlton J, Walker J, et al. Host switch leads to emergence of Plasmodium vivax malaria in humans. Mol Biol Evol. 2005;22: 1686–1693. [DOI] [PubMed] [Google Scholar]
- 42. Escalante AA, Cornejo OE, Freeland DE, Poe AC, Durrego E, Collins WE, et al. A monkey's tale: the origin of Plasmodium vivax as a human malaria parasite. Proc Natl Acad Sci USA. 2005;102: 1980–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tazi L, Ayala FJ. Unresolved direction of host transfer of Plasmodium vivax v. P. simium and P. malariae v. P. brasilianum . Infect Genet Evol. 2011;11: 209–221. 10.1016/j.meegid.2010.08.007 [DOI] [PubMed] [Google Scholar]
- 44. Lim CS, Tazi L, Ayala FJ. Plasmodium vivax: recent world expansion and genetic identity to Plasmodium simium . Proc Natl Acad Sci USA. 2005;102: 15523–15528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Culleton R, Coban C, Zeyrek FY, Cravo P, Kaneko A, Randrianarivelojosia M, et al. The origins of African Plasmodium vivax; insights from mitochondrial genome sequencing. PLoS One. 2011;6: e29137 10.1371/journal.pone.0029137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Baum J, Thomas AW, Conway DJ. Evidence for diversifying selection on erythrocyte-binding antigens of Plasmodium falciparum and P. vivax . Genetics. 2003;163: 1327–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. de Sousa TN, Kano FS, de Brito CF, Carvalho LH. The Duffy binding protein as a key target for a Plasmodium vivax vaccine: lessons from the Brazilian Amazon. Mem Inst Oswaldo Cruz. 2014;109: 608–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Souza-Silva FA, Torres LM, Santos-Alves JR, Tang ML, Sanchez BA, Sousa TN, et al. Duffy antigen receptor for chemokine (DARC) polymorphisms and its involvement in acquisition of inhibitory anti-duffy binding protein II (DBPII) immunity. PLoS One. 2014;9: e93782 10.1371/journal.pone.0093782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chootong P, Panichakul T, Permmongkol C, Barnes SJ, Udomsangpetch R, Adams JH. Characterization of inhibitory anti-Duffy binding protein II immunity: approach to Plasmodium vivax vaccine development in Thailand. PLoS One. 2012;7: e35769 10.1371/journal.pone.0035769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ntumngia FB, King CL, Adams JH. Finding the sweet spots of inhibition: understanding the targets of a functional antibody against Plasmodium vivax Duffy binding protein. Int J Parasitol. 2012;42: 1055–1062. 10.1016/j.ijpara.2012.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Haynes JD, Dalton JP, Klotz FW, McGinniss MH, Hadley TJ, Hudson DE, et al. Receptor-like specificity of a Plasmodium knowlesi malarial protein that binds to Duffy antigen ligands on erythrocytes. J Exp Med. 1988;167: 1873–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Singh B, Sung LK, Matusop A, Radhakrishnan A, Shamsul SS, Cox-Singh J, et al. A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet. 2004;363: 1017–1024. [DOI] [PubMed] [Google Scholar]
- 53. Waters AP, Higgins DG, McCutchan TF. Evolutionary relatedness of some primate models of Plasmodium. Mol Biol Evol. 1993;10: 914–923. [DOI] [PubMed] [Google Scholar]
- 54. Perelman P, Johnson WE, Roos C, Seuánez HN, Horvath JE, Moreira MA, et al. A molecular phylogeny of living primates. PLoS Genet. 2011;7: e1001342 10.1371/journal.pgen.1001342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chitnis CE, Chaudhuri A, Horuk R, Pogo AO, Miller LH. The domain on the Duffy blood group antigen for binding Plasmodium vivax and P. knowlesi malarial parasites to erythrocytes. J Exp Med. 1996;184: 1531–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Oliveira TY, Harris EE, Meyer D, Jue CK, Silva WA Jr. Molecular evolution of a malaria resistance gene (DARC) in primates. Immunogenetics. 2012;64: 497–505. 10.1007/s00251-012-0608-2 [DOI] [PubMed] [Google Scholar]
- 57. Tournamille C, Filipe A, Badaut C, Riottot MM, Longacre S, Cartron JP, et al. Fine mapping of the Duffy antigen binding site for the Plasmodium vivax Duffy-binding protein. Mol Biochem Parasitol. 2005;144: 100–103. [DOI] [PubMed] [Google Scholar]
- 58. Batchelor JD, Malpede BM, Omattage NS, DeKoster GT, Henzler-Wildman KA, Tolia NH. Red blood cell invasion by Plasmodium vivax: structural basis for DBP engagement of DARC. PLoS Pathog. 2014;10: e1003869 10.1371/journal.ppat.1003869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Liu W, Li Y, Shaw KS, Learn GH, Plenderleith LJ, Malenke JA, et al. African origin of the malaria parasite Plasmodium vivax . Nat Commun. 2014;5: 3346 10.1038/ncomms4346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Culleton R, Ndounga M, Zeyrek FY, Coban C, Casimiro PN, Takeo S, et al. Evidence for the transmission of Plasmodium vivax in the Republic of the Congo, West Central Africa. J Infect Dis. 2009;200: 1465–1469. 10.1086/644510 [DOI] [PubMed] [Google Scholar]
- 61. de Alvarenga DA, de Pina-Costa A, de Sousa TN, Pissinatti A, Zalis MG, Suaréz-Mutis MC, et al. Simian malaria in the Brazilian Atlantic forest: first description of natural infection of capuchin monkeys (Cebinae subfamily) by Plasmodium simium . Malar J. 2015;14: 81 10.1186/s12936-015-0606-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
dbpII sequences (positions 1021 to 1300 of P. vivax Sal-1 dbp, accession number M61095.1) from the 19 most prevalent haplotypes of P. vivax (sequences with > 1% frequency) in 511 available sequences, accession numbers (haplotype): EU812840.1 (H1), EU812841.1 (H2), EU812842.1 (H3), EU812844.1 (H4), EU812845.1 (H5), EU812849.1 (H6), EU812861.1 (H7), EU812869.1 (H8), EU812874.1 (H9), EU812898.1 (H10), EU812915.1 (H11), EU812927.1 (H12), EU812954.1 (H13), AF220662 (H14), AF289650 (H15), AF289649 (H16), GU143965 (H17), GU143986 (H18), EF379128 (H19); and the seven P. simium dbp sequences (herein). White head arrows indicate polymorphic sites, and black head arrows indicate polymorphisms among P. simium sequences. Identical residues are represented by dots. An asterisk shows the species-specific polymorphism (1233A>C).
(PDF)
darc sequences of Alouatta g. clamitans were aligned with other primate darc sequences available in GenBank (nucleotides 1 to 768, from Homo sapiens darc sequence, accession number JN251915.1). The numbered bars above the sequences indicate transmembrane domains. The grey box shows the N-terminal minimum-binding domain (19–30 aa), and residues responsible for direct interaction with DBPII are underlined (20,21,22 and 24,25,26). Colored bars on the left side of the alignment represent the phylogenetic group families: red (Cercopithecidae), blue (Hominidae), green (Hylobatidae), purple (Pitheciidae), orange (Atelidae) and aqua (Cebidae). The arrow indicates the polymorphism Asp42Gly, which is responsible for the FY*A and FY*B alleles.
(PDF)
The sera from two monkeys (BL37 and BL69), which showed the highest levels of blockade, were used in dilutions of 1:30, 1:90 and 1:270 in the inhibition assays of human DARC and PvDBPII. Inhibition was calculated based on the reduction in the rosette numbers observed in the presence of monkey serum compared to the rosette numbers in the absence of serum.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.