Abstract
The early stages of carcinogenesis are linked to defects in the cell cycle. A series of cell cycle checkpoints are involved in this process. The G1/S checkpoint that serves to integrate the control of cell proliferation and differentiation is linked to carcinogenesis and the mitotic spindle checkpoint with the development of chromosomal instability. This paper presents the outcome of systems biology studies designed to evaluate if networks of covariate cell cycle gene transcripts exist in proliferative mammalian tissues including mice, rats and humans. The GeneNetwork website that contains numerous gene expression datasets from different species, sexes and tissues represents the foundational resource for these studies (www.genenetwork.org). In addition, WebGestalt, a gene ontology tool, facilitated the identification of expression networks of genes that co-vary with key cell cycle targets, especially Cdc20 and Plk1 (www.bioinfo.vanderbilt.edu/webgestalt). Cell cycle expression networks of such covariate mRNAs exist in multiple proliferative tissues including liver, lung, pituitary, adipose and lymphoid tissues among others but not in brain or retina that have low proliferative potential. Sixty-three covariate cell cycle gene transcripts (mRNAs) compose the average cell cycle network with p = e−13 to e−36. Cell cycle expression networks show species, sex and tissue variability and they are enriched in mRNA transcripts associated with mitosis many of which are associated with chromosomal instability.
Keywords: cell cycle, systems biology, covariate mRNA expression, GeneNetwork
Introduction
The cell cycle serves critically important roles in the control of cell proliferation and cell differentiation (Kasperbauer et al., 1990; Krawisz and Scott, 1982; Scott et al., 1982a, 1982b; Wilke et al., 1988; Wille et al., 1985, 1984). Advances in addressing the integration of regulatory information through systems biology strategies can provide valuable insight into both the biology and pathology of cell cycle control (de Oliveira Dal’Molin et al., 2015; Hartmann and Schreiber, 2014; Mason and Watt, 2015; Oberhardt and Gianchandani, 2014; Petrey et al., 2015; Roy and Singer, 2015; Tortolina et al., 2014; Yang et al., 2015; Zhu et al., 2015). Integration of regulatory activities that mediate cell cycle and cell growth control, as well as proliferation with competency for expression of phenotype, are fundamentally important for biological processes and cancer (Aguilar et al., 2014; Asghar et al., 2015; Datla et al., 2014; Dudakovic et al., 2014; Faulknor et al., 2015; Fu et al., 2015; Gordon et al., 2014; Jongsma et al., 2014; Kapinas et al., 2015; Lamerton, 1974; Lopez-Camacho et al., 2014; Lopez-Mejia and Fajas, 2014; Meadows and Millar, 2015; Mundade et al., 2014; Schwanbeck, 2015; Sharma et al., 2014; Stachowiak et al., 2015; Stumpff et al., 2014; Sun et al., 2015; Vadia and Levin, 2015; Voskas et al., 2014; Weber and Ryan, 2014). Dysfunction in the cell cycle linked integrated control of both cell proliferation and cell differentiation is involved in the early stages of carcinogenesis (Scott and Florine, 1982; Scott and Maercklein, 1985; Scott et al., 1988; Sparks et al., 1986; Weinstein, 2000; Wier and Scott, 1985; Wille and Scott, 1986; Wille et al., 1982). This conclusion is strongly supported by Hanahan and Weinberg in their classic hallmarks of cancer review (Hanahan and Weinberg, 2000). In addition, a signature of 70 mRNAs for chromosomal instability (CIN) that is involved in the early stages of carcinogenesis was defined in 2006 by Carter et al.; they functionally linked 29 signature CIN mRNAs to the cell cycle, specifically mitosis (Carter et al., 2006).
Cell cycle gene products commonly function as complexes; over 60 cell cycle complexes have been characterized (de Lichtenberg et al., 2005). An important mechanism that serves to influence the function of such cell cycle complexes is the relative expression of various cell cycle proteins. Protein stoichiometry control involving changes in the expression of groups of proteins by as little as two fold have been reported to influence cellular behavior by modifying the normal stoichiometry of protein complex subunits, such as those required for competency to proliferate (Blagosklonny and Pardee, 2002; Ford and Pardee, 1999; Pardee and Qiao, 2008; Pardee, 2002; Pardee et al., 2004), initiation of S-Phase and histone gene expression (Aziz et al., 1998; Ghule et al., 2014; Liu et al., 2011; Miele et al., 2005; Mitra et al., 2007; Stein et al., 2006; Vaughan et al., 1998, 1995; Xie et al., 2009, 2001), cell cycle progression and mitosis (Bertoli et al., 2013; Ferrell, 2013; Hanahan and Weinberg, 2011; Herrup, 2013; Lim and Kaldis, 2013; London and Biggins, 2014; Torres et al., 2008; Williams and Fuchs, 2013; Yang and Ferrell, 2013; Zaidi et al., 2013).
This perspective is affirmed by the fact that differences in the dosage of cell cycle mRNAs and thereby in protein stoichiometry have been reported to result in aneuploidy that is linked to CIN. Microarray-based expression analyses indicate that gene copy number variants of aneuploid cells correlate with gene transcript levels. In addition, diploid yeast strains that contain different chromosome numbers show a corresponding change in protein transcript levels (Pollack et al., 2002; Torres et al., 2007; Tsafrir et al., 2006). The disruption of normal cell cycle control mechanisms associated with aneuploidy causally leads to carcinogenesis (Ricke and van Deursen, 2013). In addition, the transcript levels of genes encoded on chromosome 21 parallel the increase in gene copy number in patients with Down’s syndrome (Mao et al., 2003).
Systems biology analysis performed using microarray databases and GeneNetwork represents a very useful approach to evaluate the expression of covariate cell cycle mRNAs that form networks. In the proliferative tissues and cells studied, distinct expression networks of cell cycle mRNAs that are covariate with Cdc20 or Plk1 have been discovered and such cell cycle expression networks show species, sex, and tissue variability. The composition of the average cell cycle expression network includes 63 cell cycle transcripts and the observed cell cycle expression networks are enriched in cell cycle genes associated with mitosis. These cell cycle expression networks are envisioned to have the potential to facilitate stabilization of the control of normal cell proliferation and differentiation and to antagonize development of CIN and cancer. Thereby such networks have the potential to be of importance in the emergence of precision medicine (Curtis, 2015; Desautels et al., 2014; Glade Bender et al., 2015; Jorgensen, 2015; Kummar et al., 2015; Marquet et al., 2015; Rubin, 2014) wherein therapeutic strategies need to be aligned with specific properties of tumors.
Methods
GeneNetwork and WebGestalt
GeneNetwork is an open access, online data analysis resource for systems biology and systems genetics. It contains a large number of microarray datasets from multiple tissues of male and female rats, and mice of multiple strains and strain combinations in addition to human datasets. These dataset have been deposited in GeneNetwork from investigators around the world using a vast spectrum of microarray platforms as described (www.genenetwork.org).
Using GeneNetwork it is possible to extract sets of transcripts that co-vary tightly with a target transcript(s) across genetically diverse populations. This was specifically achieved by searching for covariates that showed correlation coefficients >0.5 with the expression of Cdc20 or Plk1 across the genetically diverse population studied in each database. Then multiple types of analysis were performed including: 1) correlation matrix / principal components analysis and 2) network analysis and graphing that enables the examination and depiction of the network of associations among large groups of traits.
To facilitate the identification of transcripts that co-vary with cell cycle probes, such as, Cdc20 and Plk1, the WebGestalt gene ontology system was used (www.bioinfo.vanderbilt.edu/webgestalt). WebGestalt is a "WEB-based GEne SeT AnaLysis Toolkit" designed for functional genomic, proteomic and large-scale genetic studies in which large number of gene lists are to be evaluated. In these studies sets of transcripts covariate with Cdc20 or Plk1 were sent to WebGestalt for analysis wherein WebGestalt defined those members of the submitted transcript set that had functions in the cell cycle. WebGestalt also designated the probability of occurrence of the observed set of cell cycle covariates relative to predicted occurrence levels. Probability (p) values for the various cell cycle expression networks presented in this paper represent these values. Thereby cell cycle expression networks were identified.
We have used the following GeneNetwork datasets in the present study:
Human lymphoblast panel [UTHSC CEPH B-cells Illumina (Sep09) Both Sexes] - 180 individuals; Cdc20 probe: ILMN_1663390; normalization system is RankInv.
BXD female mouse liver panel [UNC Agilent G4121A Liver LOWESS Stanford (Jan06) Females] - 41 strains including 38 BXD strains plus BXD parent and an F1 progeny (C57BL/6J, DBA/2J, B6D2F1, and BXDs 01, 02, 05, 06, 08, 09, 11,12, 13, 14, 15, 16, 19, 21, 23, 24, 28, 29, 31, 32, 34, 36, 38, 39, 40, 42, 43, 44, 45, 48, 51, 60, 62, 69, 73, 77, 85, and 86); Cdc20 probe: A_51_P361022; normalization system is Lowess.
BXD male mouse liver panel [UNC Agilent G4121A Liver LOWESS Stanford (Jan06) Males] - 41 strains including 38 BXD strains plus BXD parents and an F1 progeny (C57BL/6J, DBA/2J, B6D2F1, and BXDs 01, 02, 05, 06, 08, 09, 11,12, 13, 14, 15, 16, 19, 21, 23, 24, 28, 29, 31, 32, 34, 36, 38, 39, 40, 42, 43, 44, 45, 48, 51, 60, 62, 69, 73, 77, 85, and 86); Cdc20 probe: A_51__361022; normalization system is Lowess.
BHHBF2 mouse liver panel [UCLA BHHBF2 Liver (2005) Mlratio, Male] - 141 strains; Cdc20 probe: 10024401435; normalization system is Miratio.
Mouse diversity panel [Harrill-Rusyn MDP Liver Acetaminophen Tox Study (G4121A, 2009)] - 36 strains; Plk1 probe: A_51_P344566; normalization system is Lowess.
HXBBXH Rat liver panel [MDC/CAS/UCL Liver 230v2(Dec08) RMA] - 31 strains; Cdc20 probe: 1370294_a_at; normalization system is RMA.
BXD mouse lung [HZI Lung M430v2 (Apr08) RMA] - 57 strains: 47 BXD stains plus 9 additional strains including the BXD parents; Cdc20 probe: 1439377_x_at; normalization system is RMA
BXD mouse pituitary [INIA Pituitary Affy MoGene 1.0ST (Jun12) RMA] - 52 stains - 48 BXD strains plus 4 strains including the BXD parents and F1 progeny; Cdc20 probe: 10515744; normalization system is RMA.
HBXXBH male mouse adipose tissue [UCLA BHHBF2 Adipose Male Only] - 152 mouse groups, Cdc20 probe: 10023301435; normalization system is Miratio.
HBXXBH female mouse adipose tissue [UCLA BHHBF2 Adipose Female Only] - 141 mouse groups, Cdc20 probe: 10024401435; normalization system is Miratio.
Using GeneNetwork combined with WebGestalt, covariate expression sets of cell cycle transcripts were found in proliferative tissues and cells. Such enriched sets of genes with common functions (cell cycle) are designated cell cycle expression networks. When these networks are illustrated in graphic form in various figures of this paper, the color of the lines connecting the genes indicates the degree to which their expression co-varies among the strains that were evaluated (red = positive correlation coefficient >0.7, orange = positive correlation coefficient >0.5, blue = negative correlation coefficient >0.7, green = negative correlation coefficient >0.5).
Results
Initial studies searched for the presence of cell cycle expression networks in a BXD mouse liver database by looking for covariate networks of the following cell cycle mRNAs using GeneNetwork and WebGestalt to evaluate the following probes: Cdc20, Aurka, Prc1, Plk1, Bric5, Mki67, Nuf2, Aurkb, Rb1, Cenpe and Nek2. Of these Rb1, Cenpe and Nek2 showed no network characteristics. In contrast, the other eight probes identified networks consisting of between 70 to 95 cell cycle mRNAs. Follow-up studies in other tissues established Cdc20 as the most robust probe to detect cell cycle expression networks. In this regard, all but one set of data presented herein was derived using Cdc20 as the key cell cycle mRNA covariance probe.
I. What are the characteristics of liver cell cycle expression networks in rodents?
The first GeneNetwork dataset that was investigated was that of BXD female mouse liver (UNC Agilent G4121A Liver LOWESS Stanford (Jan06) Females). All the mRNAs that were covariate with Cdc20 that had correlation coefficients >0.5 were exported to WebGestalt to secure gene ontology classification data. Thereby it was established that in the liver of BXD female mice, 75 cell cycle mRNAs showed significant covariant expression with Cdc20 with p = 8.68e−21. Figure 1a and its legend present the characteristics and composition of that cell cycle expression network.
Figure 1. BXD female mouse liver.
Cell cycle expression network for Cdc20 covariates with correlation coefficients >0.50. The 75 cell cycle mRNAs that comprise this network are: Cdc20, Lig1, Ube2c, Mapre3, Cdca8, Casc5, 9930021A08Rik (Ubr2), Fancm, Sgol2, Jtb, Ptp4a1, 2410004L17Rik (Esco2), Pkmyt1, Sgol1, Mcm5, Cryaa, Psrc1, Ccng2, Chtf18, 5830426I05Rik (Ncapg2), Ska3, 1500003O22Rik (Rrp8), Melk, Plk1, Mastl, Birc5, Cdca5, Nek2, Rec8, Sept9, 4930563F16Rik (Dixdc1), Ncapg, Trip13, Aurkb, Ncapd2, Cenpe, Mcm2, Sept5, Kif20b, Npm1, BC027246 (Rfwd3), 1700022L09Rik (Dsn1), Cit, Ccnb2, Incenp, Spag5, Cdc25c, Cdkn1c, BC025462 (Fanci), Zwilch, Ccna2, Mcm7, Mki67, Stmn1, Nuf2, Ect2, Cdk1, Anapc2, Uhrf1, Ckap2, Aurka, Cenpf, Nusap1, Zfp318, Prc1, Plk4, Cenph, Ccnb1, Tpx2, Cdca2, Fignl1, Racgap1, Ptpn6, Pml, and 7330406P05Rik (Appl1). In the following network graph and in subsequent graphs, the various mRNA transcripts of the network can show an either positive (red and orange) or negative (blue and green) covariance. Correlation coefficients for these cell cycle gene transcript interactions are: red + >0.7, orange + >0.5 and blue - >0.7, green - >0.5.
Next, a comparison was made concerning the cell cycle expression network to determine if sexual dimorphism exists in the liver of female and male BXD mice derived from identical strain populations. Figure 2 presents such results for BXD male mouse liver (UNC Agilent G4121A Liver LOWESS Stanford (Jan06) males). All the mRNAs that were covariate with Cdc20 that had correlation coefficients >0.5 were exported to WebGestalt to secure gene ontology classification data. Thereby it was established that in the liver of BXD male mice, 44 cell cycle mRNAs showed significant covariant expression with Cdc20; p = 4.07e−19. Figure 2 shows that the cell cycle expression network of these mRNAs and the legend to this figure presents lists of genes that comprise this network.
Figure 2. BXD male mouse liver.
Cell cycle expression network for Cdc20 covariates with correlation coefficients >0.50. The 44 cell cycle mRNAs that comprise this network are: Cdc20, Ube2c, Ccnb2, Spag5, 120008O12Rik (Cep55), Cdca8, Cdc25c, D1Ertd8e (Sgol2), Ccna2, Cul4a, Mki67, Stmn1, Nuf2, Sgol1, Mcm5, Ect2, Cdc2a (Cdk1), Psrc1, Uhrf1, Asz1, Aurka, Cenpf, Nusap1, Plk1, Prc1, Rbl1, Dscc1, Plk4, Birc5, Cdca5, Nek2, Tpx2, Cenph, Ncapg, Fignl1, Aurkb, Mcm4, Cenpe, Racgap1, Mcm2, Fem1b, Kif20b (Mphosh1), Cyp26b1, and Sept11.
The third GeneNetwork database that was investigated with respect to the cell cycle expression network was BHHBF2 mouse liver (UCLA BHHBF2 liver (2005) Miratio - male). Cdc20 was again used to search for cell cycle mRNA covariates and its use again identified an impressive cell cycle expression network. Figure 3 illustrates and lists the 70 cell cycle mRNAs comprising the cell cycle expression network for which WebGestalt gave a p = 1.41e−23.
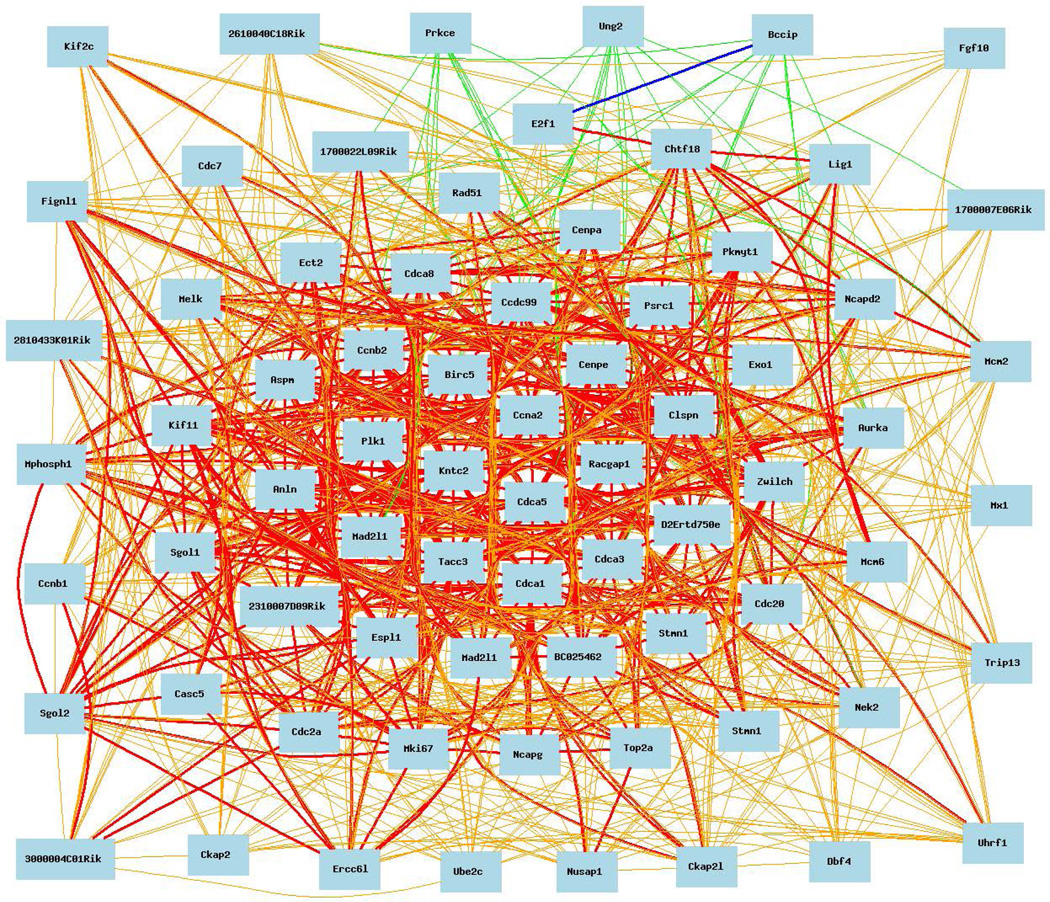
Figure 3. BHHBF2 male mouse liver.
Cell cycle expression network for Cdc20 covariates with correlation coefficients >0.50. The 70 cell cycle mRNAs that comprise this network are: Cdc20, Lig1, Exo1, Cep192, Ube2c, Cdca8, Casc5, Sgol2, Pkmyt1, Sgol1, Psrc1, Mcm6, Chtf18, Tacc3, Kntc2 (Ndc80), Melk, Plk1, Anin, Rad51, Aspm, Kif2c, Cdca5, Birc5, Nek2, Ncapg, Trip13, Ncapd2, 2810433K01Rik (Ska1), Cenpe, Mcm2, Mphosph1 (Kif20b), Cdcc99, 3000004O01Rik (Kif18b), 1700022L09Rik (Dsn1), Clspn, E2f1, Ccnb2, Kif11, BC025462 (Fanci), Ccna2, Zwilch, Mki67, Stmn1, Nuf2, Ect2, Cdc2a (Cdk1), 2310007D09Rik (Fam83d), Uhrf1, Ckap2, Aurka, Prkce, D3Ertd750e, 2610040C18Rik (Apitd1), Erc6l, Nusap1, Cdc7, Mad2l1, Bccip, UNg2 (Ccno), Mx1, Cdca3, Ccnb1, Fignl1, Racgap1, Anapc5, Cenpa, 1700007E06Rik (Syce3), Dbf4, Espl1, and Fgf10.
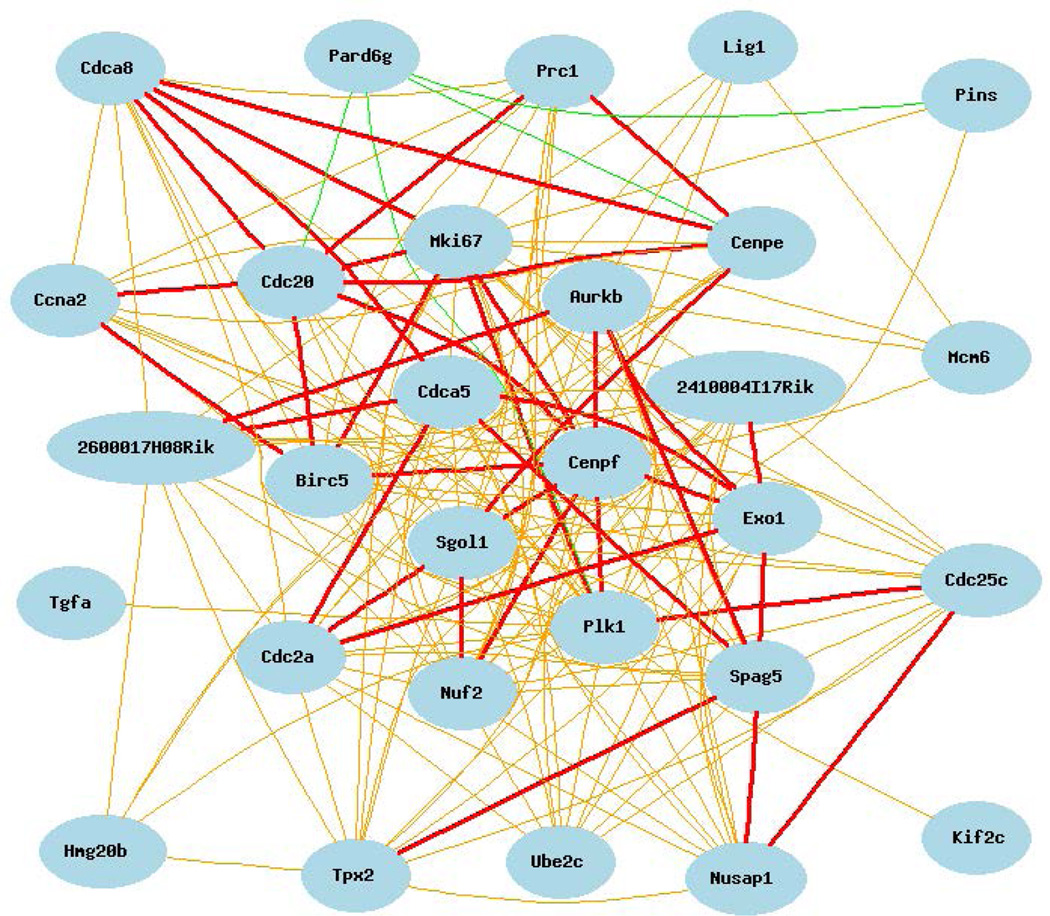
The fourth GeneNetwork database investigated for the existence of a cell cycle expression network was a mouse diversity panel [Harrill-Rusyn MDP Liver Acetaminophen Tox Study (G4121A, 2009)]. The search for a cell cycle expression network in this dataset was challenging because a robust network could not be identified using Cdc20, Cenpe or Tpx2 as covariate bait. However, when Plk1 was used Figure 4 shows cell cycle network of 29 cell cycle genes was evident with a p = 8.90e−12.
Figure 4. Mouse Diversity Panel Liver.
Cell cycle expression network for Plk1 covariates with correlation coefficients >0.50. The 29 cell cycle mRNAs that comprise this network are: Plk1, Lig1, Pard6g, Nusap1, Cenpf, Exo1, Cdc20, 2600017H08Rik (Spc25), Prc1, Ube2c, Spag5, Kif2c, Cdca8, Cdc25c, Cdca5, Birc5, Tgfa, Ccna2, Tpx2, Mki67, Gpsm2, 241994I17Rik (Esco2), Nuf2, Sgol1, Aurkb, Cenpe, Cdk1, Hmg20b, and Mcm6.
Further analyses showed that all four murine liver cell cycle expression networks are enriched in mitosis genes by 2.1 fold relative to the ratio of mitosis genes to cell cycle mRNAs in the WebGestalt murine gene ontology resource (Table 1).
Table 1.
Mitosis mRNA enrichment in murine liver cell cycle expression networks detected using Cdc20 covariates with correlation coefficients >0.50.
| All Covariates |
Cell Cycle | Mitosis | |||
|---|---|---|---|---|---|
| Tissues | Total | Observed | Total | Observed | |
| BXD Mouse Liver F | 350 | 1067 | 77 | 293 (27.5%) | 39 (50.6%) |
| BXD Mouse Liver M | 134 | 1067 | 43 | 293 (27.5%) | 26 (60.5%) |
| BHHBF2 Mouse Liver M | 308 | 1111 | 70 | 306 (27.5%) | 41 (58.6%) |
| MDP Mouse Liver M/F | 103 | 1067 | 29 | 293 (27.5%) | 19 (65.5%) |
| Averages | 27.5% | 58.8% | |||
| Enrichment | 2.14 | ||||
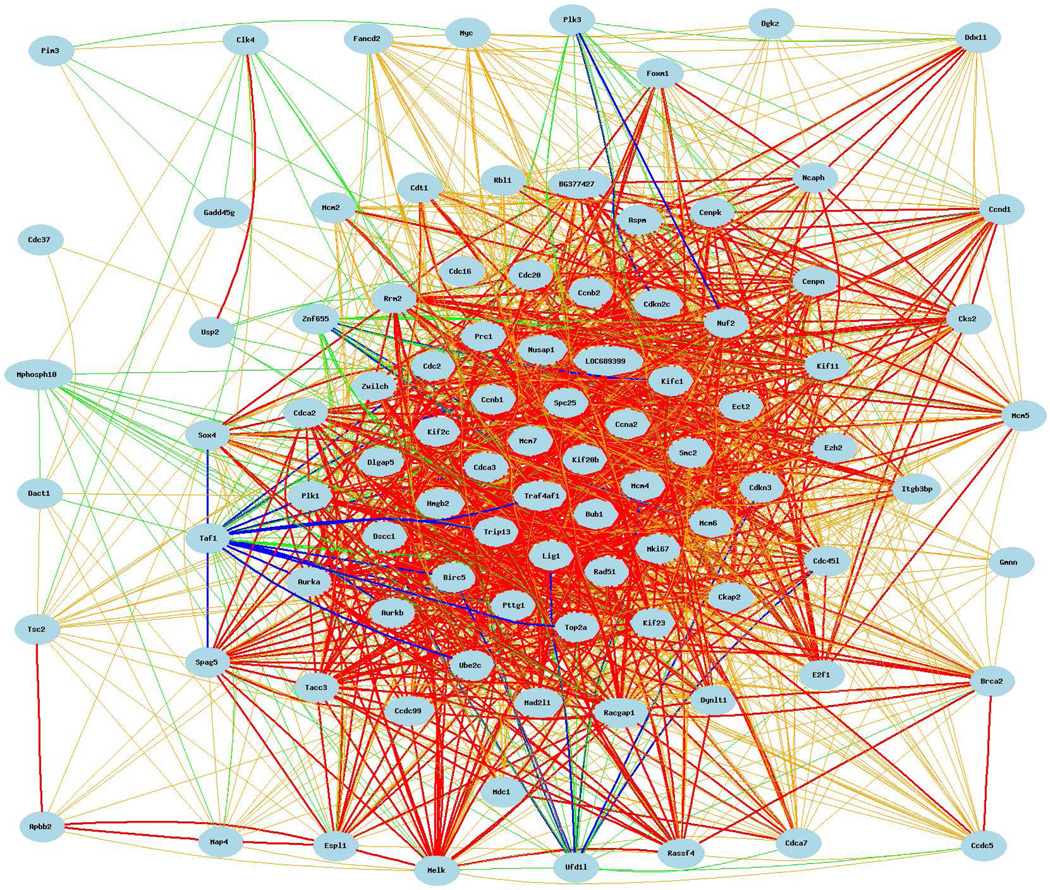
To confirm that the cell cycle expression network is evident in the liver of a different rodent, studies evaluated HXBBXH rat liver database that includes specimens of both sexes [MDC/CAS/UCL Liver 230v2 (Dec08) RMA]. The cell cycle expression network in rat liver is composed of 77 cell cycle mRNAs that were detected to be covariate with Cdc20 with a p = 2.95e−14 (Figure 5). The composition of the cell cycle network in rat liver is distinct from mouse liver and from lung, adipose tissue and pituitary shown below. Distinct rat liver mRNAs include: Apbb2, Cdkn2c, Cenpk, Cenpm, Cenpf, Pim3, Plk3, Brca2, Mdc1, Ccnd1, Cdc16, Csk2, CLk4, Dact1, Rrm2, Dynlt1, E2f1, Fancd2, Map4, Gadd45g, Hmgb2, Itgb3bp, Kifc1, Cenpw, Mphosph10, Myc, Pttg1, Ddx11, Dgkz, Rassf4, Taf1, Top2a, Traf4af1, Tsc2, Cdc37, Cdca7, Usp2, and Znf655. This analysis is important because it documents that cell cycle networks show variability not only on the basis of mouse strain and sex characteristics but also species variability.
Figure 5. Rat HXBBXH Liver.
Cell cycle expression network for Cdc20 covariates with correlation coefficients >0.50. The 93 cell cycle mRNAs that comprise this network are: Cdc20, Apbb2, Aspm, Cdkn2c, Cdkn3, Cdt1, Cenpk, Cenpm, Aurka, Aurkb, BG377427 (Cenpf), Pim3, Plk1, Plk3, Bric5, Brca2, Bub1, Ccdc5, Ccdc99, Ccnb2, Mcm7, Mdc1, Melk, Mki67, Ccnd1, Cdc16, Cdc2, Ckap2, Cks2, Clk4, Dact1, Digap5, Dscc1, Rrm2, Smc2, Sox4, Spag5, Dynlt1, E2f1, Ect2, Espl1, Ezh2, Fancd2, Foxm1, Lig1, Mad2l1, Map4, Mcm2, Gadd45g, Gmnn, Hmgb2, Itgb3bp, Kif11, Kif20b, Kif23, Kif2c, Kifc1, LOC689399 (Cenpw), Mcm4, Mcm5, Mcm6, Mphosph10, Myc, Ncaph, Cdc45l, Cdca2, Nuf2, Nusap1, Prc1, Pttg1, Racgap1, Rad51, Ddx11, Dgkz, Rassf4, Rbl1, Spc25, Tacc3, Taf1, Top2a, Ccna2, Ccnb1, Traf4af1, Trip13, Tsc2, Ube2c, Ufd1l, Cdc37, Cdca3, Cdca7, Usp2, Znf655, and Zwilch.
II. Can cell cycle expression networks be identified in other mouse tissues?
a. Lung cell cycle expression network
The GeneNetwork lung database includes microarray results based on analysis of 45 BXD mouse strains and 12 additional strains that include the BXD parents. Analysis of this database revealed a cell cycle expression network of 102 cell cycle mRNAs that are Cdc20 covariates with correlation coefficients >0.5 and a p = 4.75e−36. Figure 6 illustrates this 102 cell cycle network and its composition. This network of 102 cell cycle mRNAs show a 2.1 fold enrichment in mitosis mRNAs and contains a subset of 20 CIN signatures. These include: Cdc20, Ube2c, Cep55, Melk, Cdc45l, Trip13, Aurkb, Ncaph, Ccnb2, Mcm7, Ect2, Ezh2, Mad2l1, Prc1, Cdca3, Ccnb1, Tpx2, H2afx, Espl1 and Foxm1.
Figure 6. BXD lung.
Cell cycle expression network for Cdc20 covariates with correlation coefficients >0.50. The 102 cell cycle mRNAs that comprise this network are: Cdc20, C12orf32 (Rhno1), Cdk2, Kif23, Spc25, Ube2c, Cdca8, Cep55, C79407 (Mis18bp1), Sgol2, Gpsm2, Esco2, Sgol1, Mcm5, Chek1, 5830416I05Rik (Ncapg2), Klhl13, Tacc3, Ndc80 (Kntc2), Melk, Slbp, Casp3, E2f7, Plk1, Anin, Mastl, 8430435B07Rik (Gas213), Rad51, Aspm, Kif2c, Gsg2, Birc5, Cdca5, D030034H08 (Iqgap3), Cdc45, Mapre2, 5730507H05Rik (Ncapg), Trip13, Aurkb, 2810433K01Rik (Ska1), Cenpe, Kif20b, Cpeb1, Cdkn3 2600001J17Rik (Ccdc99), 2810047L02Rik (Dtl), Usp2, 3000004C01Rik (Kif18b), Rfwd3, Adam17, Clspn, Ncaph (Brrn1) Ccnb2, Incenp, Spag5, Fbxo5, Cks1b, Kif11, Ccna2, Mki67, Mcm7, Stmn1, Abl1, Cdca1 (Nuf2), Sox4, Ect2, Cdc2a (Cdk1), Smc2, Uhrf1, Rb1cc1, Marveld1, Ckap2, Aurka, D2Ertd750e, Brca1, 4432406C08Rik (E2f8), Ezh2, BC004701 (Ercc61), Mad2l1, Nusap1, Cenpf, Prc1, 2610036L11Rik (Cenpw), Plk4, Wee1, 2410030K01Rik (Spc24), Cdca3, Ccnb1, Tpx2, E2f3, H2afx, Pole, Cdca2, Fignl1, Racgap1, Bub1, Ccnf, Cenpa, Hells, Espl1, Mapk12, and Foxm1.
b. Pituitary cell cycle expression network
Analysis of the GeneNetwork pituitary database consisting of the results based on 45 BXD mouse strains and 7 additional strains that include the 2 parental BXD strains documents an expression network of 32 cell cycle mRNAs that are Cdc20 covariates with correlation coefficients >0.5. This network shows 2.0 fold enrichment in mitosis mRNAs and within this group of 32 network genes are 6 genes that are members of the CIN signature (Fig. 7). These include Ube2c, Cep55, Cdca3, Ncaph, Prc1, Cdc45l and Cdc20. The significance of the existence of this network has a p = 4.08e−13.
Figure 7. BXD pituitary.
Cell cycle expression network for Cdc20 covariates with correlation coefficients >0.50. The 32 cell cycle mRNAs that comprise this network are: Cdc20, Kif18b, Kif23, Spc25, Ube2c, Ncaph (Brnn1), Ube2s, Cep55, C79407 (Mis18bp1), Sgol2, Mki67, Kntc1, Mcm5, Ect2, Cdk1, Anapc2(Affy_10342641), Aurka, Pmf1, Plk1, Wfs1, Prc1, Katnb1, Digap5, Cdc45, Cdca3, Cenpe, Cdt1, Mcm2, Bub1, Cenpa, Ddb1, and Dtl.
c. Adipose cell cycle expression network
Analysis of the male and female BHHBF2 mouse adipose tissue microarray database provides important additional evidence that the cell cycle expression network shows sex and tissue specificity. To simplify this analysis Cdc20 covariates with correlation coefficients >0.7 were studied instead of covariates with >0.5. Figure 8 shows the female adipose network that consist of 42 cell cycle mRNAs that are Cdc20 covariates with a p = 6.18e−36.
Figure 8. BHHBF2 mouse adipose tissue.
a. Cell cycle expression network in females for Cdc20 covariates with correlation coefficients >0.70. The 42 cell cycle mRNAs that comprise this network are: Cdc20, 300004C01Rik (Kif18b), Exo1, Clspn, Cit, Ncaph, Ccnb2, Cdca8, Casc5, Csk2, Sgol2, BC025462 (Facni), Kif11, Ccna2, Mki67, Kntc1, Cdca1 (Nuf2), Sgol1, Cdk1, Psrc1, Uhrf1, Aurka, D2Ertd750e, Tacc3, E2f8, Melk, Plk1, Rad51, Aspm, Cdca5, Birc5, Cdca3, Cenph, Trip13, Fignl1, 2810443K01Rik (Ska1), Cenpe, Racgap1, Kif20b (Mphosph1), Ccdc99, Dbf4, and Espl1.
b. Cell cycle expression network in males for Cdc20 covariates with correlation coefficients >0.70. The 42 cell cycle mRNAs that comprise this network are: Cdc20, Exo1, Clspn, Cit, Ccnb2, Cdca8, Csk2, Kif11, BC025462 (Fanci), Ccna2, Sgol1, Aif1, Nuf2 (Cdca1), Mcm5, Psrc1, Cdk1, Mcm6, Uhrf1, Aurka, Tacc3, D2Ertd750e, E2f8, Melk, Plk1, Rad51, Aspm, Cdca5, Birc5, Gmnn, Cdca3, Cenph, Trip13, Fignl1, Mcm4, Cenpe, Racgap1, Mcm2, Cenpa, Ptpn6, Ccdc99, Dbf4 and Espl1.
Female and male adipose cell cycle gene expression networks share 34 mRNAs in common and each have an additional 8 mRNAs that shows sex specificity. The common mRNAs are: Cdc20, Exo1, Clspn, Cit, Ccnb2, Cdca8, Csk2, Kif11, Fanci, Ccna2, Sgol1, Nuf2, Psrc1, Cdk1, Utfr1, Aurka, Tacc3, D2Ertd750e, E2f8, Melk, Plk1, Rad51, Cdca5, Birc5, Cdca3, Cenpe, Rcgap1, Dbf4, Expl1, Ccdc99, Trip13, Aspm, Fignl1 and Cenpa. Female specific network mRNAs are: Kif18, Ncaph, Casc5, Sgol2, Mki67, Kntc1, Ska1, and Kif20b. Male specific network mRNAs are Aif1, Mcm5, Mcm6, Gmnn, Mcm4, Mcm2, Ptpn6, and Cenph.
These results show that female and male adipose tissues cell cycle expression networks contain different compositions of mRNAs. This further establishes the sex variability of cell cycle expression networks.
Adipose cell cycle expression networks from tissues of both sexes also contain CIN signatures. There are ten female adipose CIN signature mRNAs that include: Cdc20, Cit, Ncaph, Ccnb2, Cdca8, Csk2, Melk, Cdca3, Trip13, and Expl1. There are also nine male adipose CIN signatures that include: Cdc20, Ccnb2, Cdca8, Csk2, Melk, Cdca3, Trip13, Mcm2, and Espl1.
III. Does the existence of cell cycle networks translate to humans?
To establish that a cell cycle expression network exists in humans, lymphoblasts from 180 individuals (UTHSC CEPH B-cells Illumina (Sep09) RankInv) database were evaluated for covariates with Cdc20. Figure 9 shows a network of 59 such cell cycle mRNAs with correlation coefficients >0.50. This network showed p = 1.19e−19 according to WebGestalt. This human lymphoblast network contains a subset of cell cycle mRNAs not typically observed in the various rodent networks described previously in this paper. There include: Hjurp, Tubb4b, Dynlt3, Smarca4, Rcc2, Setd8, Tnfaip3, Rps27l, Pole, Suv39h1 and others. This observation establishes not only that the discovery of cell cycle expression networks translates to humans but also that human networks can show variability.
Figure 9. Human lymphoblasts.
Cell cycle expression network of 57 Cdc20 covariates with correlation coefficients >0.50. These mRNAs include: Cdc20, Casp3, Pdcd6ip, Hjurp, Tubb4b, Dynlt3, Cenpf, Cdca8, Tpx2, Smarca4, Kif23, Gsg2, Gpsm2, Kif2c, Spag5, Ckap5, Rcc2, Ccna2, Setd8, Prc1, Kif22, Ppm1a, Kntc1, Cdc25b, Uhrf1, Trip13, Tacc3, Hcfc1, Plk1, Cenpa, Tnfaip3, Rps27l, Kif20a, Aspm, Pbk, Miki67, Cdca2, Kif20b, Pbrm1, Aurkb, Pole, Birc5, Rbl1, Tfdp1, H2afx, Hdac1, Aurka, Cenpe, Ube2s, Nup37, Ube2c, E2f2, Ccnb2, Espl1, Ctbp1, Suv39h1, and Melk.
In summary, Table 2 presents the results of an analysis of the cell cycle mRNAs that are the most common constituents of cell cycle expression networks in multiple tissues that show species, sex and tissue variability. A total of between 1067 to 1295 cell cycle mRNAs that exist mice, rats and humans according to WebGestalt averaging 1128. Those involved in the cell cycle expression networks described in the nine studied tissue numbers 242. Table 2 emphasizes that of the 60 most common or core network constituents, Cdc20, Plk1, Birc5, Cenpe, Ccnb1, Mki67 and Aurka represent the top eight cell cycle expression network members. Therefore an average cell cycle expression network can be envisioned to be composed of approximately 65% core cell cycle network mRNAs and 35% species, sex or tissue specific mRNAs. It is of note that of those mRNAs listed in Table 2, 17 represent CIN signatures and 10 of these are CIN mitosis signatures: Cdc20, Ccnb2, Ube2c, Cdca8, Prc1, Espl1, Aurkb, Ect2, Nek2, and Cep55.
Table 2.
Core cell cycle expression network of Cdc20 covariates.
| 9 of 9 representations | Cdc20, Plk1 |
| 8 of 9 representations | Birc5, Cenpe, Ccnb2, Mki67, Aurka |
| 7 of 9 representations | Ube2c, Cdca8, Melk, Trip13, Kif20b, Uhrf1, Racgap1 |
| 6 of 9 representations | Sgol1, Sgol2, Cdca5, Mcm2, Ccna2, Nuf2, Prc1, Fignl1, Tacc3, Aspm, Espl1 |
| 5 of 9 representations | Mcm5, Psrc1, Aurkb, Spag5, Ect2, Kif11, Rad51, Ccdc99 |
| 4 of 9 representations | Fanci, Stmn1, Ckap2, Nusap1, Ccnb1, Cdca2, Clspn, D2Ertd750e, Tpx2 |
| 3 of 9 representations | Lig1, Casc5, Nek2, Ncapg, Cit, Zwilch, Mcm7, Cdk1, Cenpf, Plk4, Cep55, Cenph, Kif23, Cenpa, Exo1, Csk2, Cdca3, Ddf4, Mcm6 |
Data are based on an analysis of a set of nine databases that include: BXD female liver, BXD male liver, BHHBF2 male liver, BXD lung, BXD pituitary, BHHBF2 female adipose tissue, BHHBF2 male adipose tissue, HXBBXH rat liver, and human lymphoblasts.
IV. Are cell cycle expression networks evident in the adult central nervous system that does not have prominent proliferative potential?
Finally, it is important to state that the cell cycle expression networks are not evident in tissues that lack significant proliferative potential, i.e., the adult central nervous system. The adult human whole brain database (GSE5281 Human Brain Normal (Jul09) RMA) showed no evidence of a cell cycle expression network by analysis of the top 500 expression covariates with Cdc20. In addition, the adult BXD whole brain database (UCHSC RMA, Nov 06), the adult BXD cerebellum database (SJUT MAS5, Oct 03) and the adult BXD hippocampus database (Consortium RMA, Nov 06) also showed no evidence of cell cycle expression networks as determined by analysis of the top 100 and 500 expression covariates with either Cdc20 or Aurka. In addition, no cell cycle expression network was detected in evaluations of the microarray database of non-proliferative adult BXD retina (Normal HEI Retina (April 2010) RankInv) that was searched for Cdc20, Plk1 and Tpx2 covariates.
Discussion
The results establish that cell cycle expression networks exist in multiple proliferative tissues and that the characteristics of these networks show species, sex and tissue variability. These cell cycle expression networks are distinct from all previously described cell cycle pathways and complexes. The existence of these networks of genes in proliferative tissues argues strongly for a previously unrealized mechanism of transcription control. Without such a unique genetic-based transcription control system, how else might it be possible to explain the fact that the lung of BXD mice, for example, 94% of the top 65 mRNAs that are covariate with Cdc20 are other cell cycle mRNAs? To begin to evaluate how the coordinate control of cell cycle mRNA might be mediated, preliminary study are investigating the characteristics of quantitative trait loci (QTL) for cell cycle networks. Initial QTL mapping studies suggest the existence of comparable species, sex and tissue variability. If these genetic regulators can be identified, they hold the potential to represent not only a new paradigm for the genetic control of transcription but also personalized targets for drug develop for cancer prevention and therapy.
An intriguing possibility is that the three dimensional organization of regulatory machinery in nuclear matrix-associated microenvironments may account for such transcriptional regulatory plasticity and complexity. The temporal and spatial organization of nuclear matrix microenvironments make it possible for threshold concentrations of genes and cognate factors to exist and facilitate functional interactions. This type architectural organization of regulatory machinery for gene expression suggests that matrix configurations may account for different aspects of biological control. More specifically, localized scaffolding of regulatory macromolecules at strategic promoter sites and focal compartmentalization of genes, transcripts, and regulatory factors within intra-nuclear microenvironments provides an infrastructure for combinatorial control of transcription that reflects the complex three-dimensional structure of nuclear architecture (Stein et al., 2004; Zaidi et al., 2007, 2005). Regulatory complexes are retained at target gene loci during mitosis and are distributed symmetrically to progeny cells at the completion of cell division providing additional dimensions to epigenetic control (Pande et al., 2009; Stein et al., 2009, 2006; Young et al., 2007, 2004; Zaidi et al., 2010). It has also been proposed that abnormalities in nuclear matrix organization may account for changes in gene expression patterns in cancer (Stein, 2007).
The >2fold enrichment of mitosis mRNAs with cell cycle expression networks is also important. This finding links the cell cycle expression networks to the findings of Carter et al, on the existence of a chromosomal instability (CIN) signature of mRNAs that is recognized to be a characteristic of most cancers that commonly develops early in the process of carcinogenesis (Carter et al., 2006). Of the 70 gene CIN signatures Carter et al described, those most mechanistically linked to CIN were 29 genes functionally linked to mitosis. Of these, 69% are represented as components of the cell cycle expression networks in the tissues evaluated in this study. This strongly suggest that when additional tissues are analyzed, even more functional CIN mitosis signatures will be found to be components of cell cycle expression networks. Thereby the possibility can be envisioned wherein aberrations in the control of cell cycle networks would lead to aberrations in mitosis as a step in the early stages of carcinogenesis that are linked to the development of CIN.
It will be informative to evaluate from the perspective of the cell cycle expression networks herein described, the expression of mitotic genes that epigenetically exhibit mitotic bookmarking where transcription factors remain associated with target genes during cell division (Stein et al., 2011; Zaidi et al., 2014, 2012, 2011). It will also be important to evaluate the importance of cell cycle expression networks in human embryonic stem cells that exhibit unrestricted proliferation, while retaining stringent cell cycle control. These cells have a dramatically reduced G1 period establishing competency for initiation of DNA replication, rapidly following emergence from mitosis (Ghule et al., 2008; Kapinas et al., 2013; Medina et al., 2012). Equally important could be an evaluation of cell cycle expression networks with gene regulatory characteristics that are operative in cancer stem cells that exhibit a prolonged G1 period (Cabrera et al., 2015; D’Angelo and Wicha, 2010).
The cell cycle is of seminal importance in the process of carcinogenesis (Hanahan and Weinberg, 2011; Scott and Florine, 1982; Scott and Maercklein, 1985; Scott et al., 1988; Wier and Scott, 1985; Wille and Scott, 1986; Wille et al., 1982). The central mechanism of the early stages of carcinogenesis involves the sequential accumulation of mutations that activate oncogenes and disrupt suppressor genes combined with multiple rounds of clonal selection and clonal evolution. More specifically, the early stages of carcinogenesis are associated with the progressive generation and selection of aberrant cell clones that express the following biological characteristics: First, dysregulated cell proliferation resulting from cell cycle and mitosis defects and the production of auto-regulatory growth stimulation to expand the proliferative pool of aberrant cells to support further clonal selection; Second, decreased cell cycle–linked cellular differentiation and cellular senescence that normally cause the loss of cells from the proliferative pool. This allows the proliferative pool of aberrant cells to expand and support further clonal selection. Third, increased cell survival and decreased apoptosis to expand and maintain the proliferative pool of aberrant cells to support further clonal selection.
Cell-to-cell and cell-to-environment communication pathways that are linked to the control of the cell cycle are the most important biological targets of genetic mutations and epigenetic changes that initially lead to aberrant cell proliferation, differentiation, and survival characteristics that are the hallmarks of the early stages of carcinogenesis. These include acquisition of the ability of emerging cancer cells to auto-stimulate their own proliferation.
Another dimension to control of cell proliferation that is emerging is the relationships between sequential and simultaneously integrated regulatory networks that contribute to cell cycle and growth control. Here there are lessons to be learned about the role and scope of regulatory factors that are being identified by genomic and proteomic strategies. The extent that genetic predispositions and epigenetic mechanisms are operative can be highly informative. The static and dynamic components to cell cycle machinery from architectural as well as regulatory perspectives are becoming increasingly evident. Cyclic remodeling of chromatin structure and nucleosome organization in relation to DNA replication and the coupling of histone biosynthesis with DNA synthesis are linked relationships that require further understanding. The sequence of regulatory processes that are obligatory for the fidelity of mitotic apparatus assembly and activity, engagement of a mitotic cycle that supports chromosome organization and distribution to progeny cells without compromising chromosome/chromatin integrity or genome stability are being actively pursued. Insight is accruing into conditions when regulatory events that occur during the G1/S, G2 and mitotic phase in the cell cycle are modified in a physiologically responsive manner and as a consequence of pathology. The cyclic organization and reconfiguration of chromosomal territories during the proliferation cycle (both interphase and mitosis) to support cell cycle progression and tissue specific gene expression during interphase is being actively investigated. The mitotic distribution of regulatory information to progeny cells during mitosis is emerging as a novel and powerful dimension to epigenetic control. The boundaries that define regulatory pathways and focally organized regulatory machinery within the cell nucleus is providing a basis for relating the temporal and spatial parameters of cell cycle control with dynamic modifications required to sustain the structural and functional properties of cells and tissues in an in vivo context.
The current findings provide the foundation for new perspectives on how it might be possible to develop personalized cancer prevention drugs. If the genetic regulators of various cell cycle expression networks can successfully be identified in a large sex and tissue specific human populations via GWAS SNP analysis, the genes linked to such GWAS SNPs have the potential to represent a new class of targets for personalized cancer prevention drugs. It is envisioned that such drugs could be used to perhaps stabilize the integrity of the cell cycle expression networks and thereby antagonize the development of the early stages of cancer development.
Acknowledgments
Contract Grant Sponsor: National Institutes of Health; Contract Grant Number: R01 CA139322
Footnotes
No Conflict of Interest
References
- Aguilar R, Grandy R, Meza D, Sepulveda H, Pihan P, van Wijnen AJ, Lian JB, Stein GS, Stein JL, Montecino M. A functional N-terminal domain in C/EBPbeta-LAP* is required for interacting with SWI/SNF and to repress Ric-8B gene transcription in osteoblasts. J. Cell. Physiol. 2014;229:1521–1528. doi: 10.1002/jcp.24595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asghar U, Witkiewicz AK, Turner NC, Knudsen ES. The history and future of targeting cyclin-dependent kinases in cancer therapy. Nat. Rev. Drug Discov. 2015;14:130–146. doi: 10.1038/nrd4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz F, van Wijnen aJ, Stein JL, Stein GS. HiNF-D (CDP-cut/CDC2/cyclin A/pRB-complex) influences the timing of IRF-2-dependent cell cycle activation of human histone H4 gene transcription at the G1/S phase transition. J. Cell. Physiol. 1998;177:453–464. doi: 10.1002/(SICI)1097-4652(199812)177:3<453::AID-JCP8>3.0.CO;2-F. doi:10.1002/(SICI)1097-4652(199812)177:3<453::AID-JCP8>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Bertoli C, Skotheim JM, de Bruin RAM. Control of cell cycle transcription during G1 and S phases. Nat. Rev. Mol. Cell Biol. 2013;14:518–528. doi: 10.1038/nrm3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagosklonny MV, Pardee AB. The restriction point of the cell cycle. cc. 2002;1:103–110. [PubMed] [Google Scholar]
- Cabrera MC, Hollingsworth RE, Hurt EM. Cancer stem cell plasticity and tumor hierarchy. World. J. Stem Cells. 2015;7:27–36. doi: 10.4252/wjsc.v7.i1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet. 2006;38:1043–1048. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- Curtis C. Genomic profiling of breast cancers. Curr. Opin. Obstet. {&} Gynecol. 2015;27:34–39. doi: 10.1097/GCO.0000000000000145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo RC, Wicha MS. Stem cells in normal development and cancer. Prog. Mol. Biol. Transl. Sci. 2010;95:113–158. doi: 10.1016/B978-0-12-385071-3.00006-X. [DOI] [PubMed] [Google Scholar]
- Datla US, Scovill NC, Brokamp AJ, Kim E, Asch AS, Lee M-H. Role of PUF-8/PUF protein in stem cell control, sperm-oocyte decision and cell fate reprogramming. J. Cell. Physiol. 2014;229:1306–1311. doi: 10.1002/jcp.24618. [DOI] [PubMed] [Google Scholar]
- De Lichtenberg U, Jensen LJ, Brunak S, Bork P. Dynamic complex formation during the yeast cell cycle. Science (80-.) 2005;307:724–727. doi: 10.1126/science.1105103. [DOI] [PubMed] [Google Scholar]
- De Oliveira Dal’Molin C, Quek L-E, Saa PA, Nielsen LK. A multi-tissue genome-scale metabolic modeling framework for the analysis of whole plant systems. Front. Plant Sci. 2015;6:4. doi: 10.3389/fpls.2015.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desautels D, Harlos C, Czaykowski P. The advent of precision therapy in gastrointestinal malignancies: Targeting the human epidermal growth factor receptor family in colorectal and esophagogastric cancer. J. Carcinog. 2014;13:13. doi: 10.4103/1477-3163.145609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudakovic A, Camilleri E, Riester SM, Lewallen EA, Kvasha S, Chen X, Radel DJ, Anderson JM, Nair AA, Evans JM, Krych AJ, Smith J, Deyle DR, Stein JL, Stein GS, Im H-J, Cool SM, Westendorf JJ, Kakar S, Dietz AB, van Wijnen AJ. High-resolution molecular validation of self-renewal and spontaneous differentiation in clinical-grade adipose-tissue derived human mesenchymal stem cells. J. Cell. Biochem. 2014;115:1816–1828. doi: 10.1002/jcb.24852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulknor RA, Olekson MA, Nativ NI, Ghodbane M, Gray AJ, Berthiaume F. Mesenchymal stromal cells reverse hypoxia-mediated suppression of alpha-smooth muscle actin expression in human dermal fibroblasts. Biochem. Biophys. Res. Commun. 2015 doi: 10.1016/j.bbrc.2015.01.013. [DOI] [PubMed] [Google Scholar]
- Ferrell JEJ. Feedback loops and reciprocal regulation: recurring motifs in the systems biology of the cell cycle. Curr Opin Cell Biol. 2013;25:676–686. doi: 10.1016/j.ceb.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford HL, Pardee AB. Cancer and the cell cycle. J. Cell. Biochem. 1999;(Suppl 32-3):166–172. doi: 10.1002/(sici)1097-4644(1999)75:32+<166::aid-jcb20>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Fu J, Hagan IM, Glover DM. The Centrosome and Its Duplication Cycle. Cold Spring Harb. Perspect. Biol. 2015;7 doi: 10.1101/cshperspect.a015800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghule PN, Dominski Z, Yang X-C, Marzluff WF, Becker KA, Harper JW, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Staged assembly of histone gene expression machinery at subnuclear foci in the abbreviated cell cycle of human embryonic stem cells. Proc. Natl. Acad. Sci. U. S. A. 2008;105:16964–16969. doi: 10.1073/pnas.0809273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghule PN, Xie R-L, Medina R, Colby JL, Jones SN, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Fidelity of histone gene regulation is obligatory for genome replication and stability. Mol. Cell. Biol. 2014;34:2650–2659. doi: 10.1128/MCB.01567-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glade Bender J, Verma A, Schiffman JD. Translating genomic discoveries to the clinic in pediatric oncology. Curr. Opin. Pediatr. 2015;27:34–43. doi: 10.1097/MOP.0000000000000172. [DOI] [PubMed] [Google Scholar]
- Gordon JAR, Lisle JW, Alman BA, Lian JB. Disruption of crosstalk between mesenchymal stromal and tumor cells in bone marrow as a therapeutic target to prevent metastatic bone disease. J. Cell. Physiol. 2014;229:1884–1886. doi: 10.1002/jcp.24692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hartmann A, Schreiber F. Integrative analysis of metabolic models - from structure to dynamics. Front. Bioeng. Biotechnol. 2014;2:91. doi: 10.3389/fbioe.2014.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrup K. Post-mitotic role of the cell cycle machinery. Curr Opin Cell Biol. 2013;25:711–716. doi: 10.1016/j.ceb.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongsma MLM, Berlin I, Neefjes J. On the move: organelle dynamics during mitosis. Trends Cell Biol. 2014 doi: 10.1016/j.tcb.2014.10.005. [DOI] [PubMed] [Google Scholar]
- Jorgensen JT. Companion diagnostics: the key to personalized medicine. Expert Rev. Mol. Diagn. 2015;15:153–156. doi: 10.1586/14737159.2015.1002470. [DOI] [PubMed] [Google Scholar]
- Kapinas K, Grandy R, Ghule P, Medina R, Becker K, Pardee A, Zaidi SK, Lian J, Stein J, van Wijnen A, Stein G. The abbreviated pluripotent cell cycle. J. Cell. Physiol. 2013;228:9–20. doi: 10.1002/jcp.24104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapinas K, Kim H, Mandeville M, Martin-Buley LA, Croce CM, Lian JB, van Wijnen AJ, Stein JL, Altieri DC, Stein GS. microRNA-mediated survivin control of pluripotency. J. Cell. Physiol. 2015;230:63–70. doi: 10.1002/jcp.24681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasperbauer JL, Neel HB, Scott RE., 3rd Proliferation and differentiation characteristics of normal human squamous mucosal cells of the upper aerodigestive tract. Ann. Otol. Rhinol. Laryngol. 1990;99:29–37. doi: 10.1177/000348949009900105. [DOI] [PubMed] [Google Scholar]
- Krawisz BR, Scott RE. Coupling of proadipocyte growth arrest and differentiation. I. Induction by heparinized medium containing human plasma. J Cell Biol. 1982;94:394–399. doi: 10.1083/jcb.94.2.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kummar S, Williams PM, Lih C-J, Polley EC, Chen AP, Rubinstein LV, Zhao Y, Simon RM, Conley BA, Doroshow JH. Application of molecular profiling in clinical trials for advanced metastatic cancers. J. Natl. Cancer Inst. 2015;107 doi: 10.1093/jnci/djv003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamerton LF. The mitotic cycle and cell population control. J. Clin. Pathol. Suppl. (R. Coll. Pathol) 1974;7:19–25. [PMC free article] [PubMed] [Google Scholar]
- Lim S, Kaldis P. Cdks, cyclins and CKIs: roles beyond cell cycle regulation. Development. 2013;140:3079–3093. doi: 10.1242/dev.091744. [DOI] [PubMed] [Google Scholar]
- Liu L-J, Xie R, Hussain S, Lian JB, Rivera-Perez J, Jones SN, Stein JL, Stein GS, van Wijnen AJ. Functional coupling of transcription factor HiNF-P and histone H4 gene expression during pre- and post-natal mouse development. Gene. 2011;483:1–10. doi: 10.1016/j.gene.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- London N, Biggins S. Signalling dynamics in the spindle checkpoint response. Nat. Rev. Mol. Cell Biol. 2014;15:736–747. doi: 10.1038/nrm3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Camacho C, van Wijnen AJ, Lian JB, Stein JL, Stein GS. Core binding factor beta (CBFbeta) is retained in the midbody during cytokinesis. J. Cell. Physiol. 2014;229:1466–1474. doi: 10.1002/jcp.24588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Mejia IC, Fajas L. Cell cycle regulation of mitochondrial function. Curr Opin Cell Biol. 2014;33C:19–25. doi: 10.1016/j.ceb.2014.10.006. [DOI] [PubMed] [Google Scholar]
- Mao R, Zielke CL, Zielke HR, Pevsner J. Global up-regulation of chromosome 21 gene expression in the developing Down syndrome brain. Genomics. 2003;81:457–467. doi: 10.1016/s0888-7543(03)00035-1. [DOI] [PubMed] [Google Scholar]
- Marquet P, Longeray P-H, Barlesi F, Ameye V, Auge P, Cazeneuve B, Chatelut E, Diaz I, Divine M, Froguel P, Goni S, Gueyffier F, Hoog-Labouret N, Mourah S, Morin-Surroca M, Perche O, Perin-Dureau F, Pigeon M, Tisseau A, Verstuyft C. Translational Research: Precision Medicine, Personalized Medicine, Targeted Therapies: Marketing or Science? Therapie. 2015 doi: 10.2515/therapie/2014231. [DOI] [PubMed] [Google Scholar]
- Mason RR, Watt MJ. Unraveling the roles of PLIN5: linking cell biology to physiology. Trends Endocrinol. Metab. 2015 doi: 10.1016/j.tem.2015.01.005. [DOI] [PubMed] [Google Scholar]
- Meadows JC, Millar JBA. Sharpening the anaphase switch. Biochem. Soc. Trans. 2015;43:19–22. doi: 10.1042/BST20140250. [DOI] [PubMed] [Google Scholar]
- Medina R, Ghule PN, Cruzat F, Barutcu AR, Montecino M, Stein JL, van Wijnen AJ, Stein GS. Epigenetic control of cell cycle-dependent histone gene expression is a principal component of the abbreviated pluripotent cell cycle. Mol. Cell. Biol. 2012;32:3860–3871. doi: 10.1128/MCB.00736-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miele A, Braastad CD, Holmes WF, Mitra P, Medina R, Xie R, Zaidi SK, Ye X, Wei Y, Harper JW, van Wijnen AJ, Stein JL, Stein GS. HiNF-P directly links the cyclin E/CDK2/p220NPAT pathway to histone H4 gene regulation at the G1/S phase cell cycle transition. Mol. Cell. Biol. 2005;25:6140–6153. doi: 10.1128/MCB.25.14.6140-6153.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra P, Xie R, Harper JW, Stein JL, Stein GS, van Wijnen AJ. HiNF-P is a bifunctional regulator of cell cycle controlled histone H4 gene transcription. J. Cell. Biochem. 2007;101:181–191. doi: 10.1002/jcb.21157. [DOI] [PubMed] [Google Scholar]
- Mundade R, Ozer HG, Wei H, Prabhu L, Lu T. Role of ChIP-seq in the discovery of transcription factor binding sites, differential gene regulation mechanism, epigenetic marks and beyond. cc. 2014;13:2847–2852. doi: 10.4161/15384101.2014.949201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberhardt MA, Gianchandani EP. Genome-scale modeling and human disease: an overview. Front. Physiol. 2014;5:527. doi: 10.3389/fphys.2014.00527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pande S, Ali SA, Dowdy C, Zaidi SK, Ito K, Ito Y, Montecino MA, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Subnuclear targeting of the Runx3 tumor suppressor and its epigenetic association with mitotic chromosomes. J. Cell. Physiol. 2009;218:473–479. doi: 10.1002/jcp.21630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee AB. Regulation, restriction, and reminiscences. J. Biol. Chem. 2002;277:26709–26716. doi: 10.1074/jbc.R200013200. [DOI] [PubMed] [Google Scholar]
- Pardee AB, Li CJ, Reddy GPV. Regulation in S phase by E2F. cc. 2004;3:1091–1094. [PubMed] [Google Scholar]
- Pardee AB, Qiao M. A devious cube of cancer. J. Cell. Physiol. 2008;216:1–2. doi: 10.1002/jcp.21425. [DOI] [PubMed] [Google Scholar]
- Petrey D, Chen TS, Deng L, Garzon JI, Hwang H, Lasso G, Lee H, Silkov A, Honig B. Template-based prediction of protein function. Curr. Opin. Struct. Biol. 2015;32C:33–38. doi: 10.1016/j.sbi.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack JR, Sorlie T, Perou CM, Rees CA, Jeffrey SS, Lonning PE, Tibshirani R, Botstein D, Borresen-Dale A-L, Brown PO. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc. Natl. Acad. Sci. U. S. A. 2002;99:12963–12968. doi: 10.1073/pnas.162471999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricke RM, van Deursen JM. Aneuploidy in health, disease, and aging. J Cell Biol. 2013;201:11–21. doi: 10.1083/jcb.201301061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy AL, Singer DS. Core promoters in transcription: old problem, new insights. Trends Biochem. Sci. 2015 doi: 10.1016/j.tibs.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin MA. Toward a prostate cancer precision medicine. Urol. Oncol. 2014 doi: 10.1016/j.urolonc.2014.10.011. [DOI] [PubMed] [Google Scholar]
- Schwanbeck R. The role of epigenetic mechanisms in notch signaling during development. J. Cell. Physiol. 2015;230:969–981. doi: 10.1002/jcp.24851. [DOI] [PubMed] [Google Scholar]
- Scott RE, Florine DL. Cell cycle models for the aberrant coupling of growth arrest and differentiation in hyperplasia, metaplasia, and neoplasia. Am. J. Pathol. 1982;107:342–348. [PMC free article] [PubMed] [Google Scholar]
- Scott RE, Florine DL, Wille JJJ, Yun K. Coupling of growth arrest and differentiation at a distinct state in the G1 phase of the cell cycle: GD. Proc. Natl. Acad. Sci. U. S. A. 1982a;79:845–849. doi: 10.1073/pnas.79.3.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RE, Hoerl BJ, Wille JJJ, Florine DL, Krawisz BR, Yun K. Coupling of proadipocyte growth arrest and differentiation. II. A cell cycle model for the physiological control of cell proliferation. J Cell Biol. 1982b;94:400–405. doi: 10.1083/jcb.94.2.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RE, Maercklein PB. An initiator of carcinogenesis selectively and stably inhibits stem cell differentiation: a concept that initiation of carcinogenesis involves multiple phases. Proc. Natl. Acad. Sci. U. S. A. 1985;82:2995–2999. doi: 10.1073/pnas.82.9.2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RE, Wilke MS, Wille JJJ, Pittelkow MR, Hsu BM, Kasperbauer JL. Human squamous carcinoma cells express complex defects in the control of proliferation and differentiation. Am. J. Pathol. 1988;133:374–380. [PMC free article] [PubMed] [Google Scholar]
- Sharma BK, Patil M, Satyanarayana A. Negative regulators of brown adipose tissue (BAT)-mediated thermogenesis. J. Cell. Physiol. 2014;229:1901–1907. doi: 10.1002/jcp.24664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks RL, Seibel-Ross EI, Wier ML, Scott RE. Differentiation, dedifferentiation, and transdifferentiation of BALB/c 3T3 T mesenchymal stem cells: potential significance in metaplasia and neoplasia. Cancer Res. 1986;46:5312–5319. [PubMed] [Google Scholar]
- Stachowiak MK, Birkaya B, Aletta JM, Narla ST, Benson CA, Decker B, Stachowiak EK. Nuclear FGF Receptor-1 and CREB Binding Protein: An Integrative Signaling Module. J. Cell. Physiol. 2015;230:989–1002. doi: 10.1002/jcp.24879. [DOI] [PubMed] [Google Scholar]
- Stein GS. Gene expression in nuclear microenvironments for biological control and cancer. Cancer Biol. {&} Ther. 2007;6:1817–1821. doi: 10.4161/cbt.6.11.5294. [DOI] [PubMed] [Google Scholar]
- Stein GS, Lian JB, van Wijnen AJ, Stein JL, Javed A, Montecino M, Zaidi SK, Young D, Choi J-Y, Gutierrez S, Pockwinse S. Nuclear microenvironments support assembly and organization of the transcriptional regulatory machinery for cell proliferation and differentiation. J. Cell. Biochem. 2004;91:287–302. doi: 10.1002/jcb.10777. [DOI] [PubMed] [Google Scholar]
- Stein GS, Stein JL, van Wijnen AJ, Lian JB, Zaidi SK, Nickerson JA, Montecino MA, Young DW. An architectural genetic and epigenetic perspective. Integr. Biol. (Camb) 2011;3:297–303. doi: 10.1039/c0ib00103a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein GS, van Wijnen AJ, Stein JL, Lian JB, Montecino M, Zaidi SK, Braastad C. An architectural perspective of cell-cycle control at the G1/S phase cell-cycle transition. J. Cell. Physiol. 2006;209:706–710. doi: 10.1002/jcp.20843. [DOI] [PubMed] [Google Scholar]
- Stein GS, Zaidi SK, Stein JL, Lian JB, van Wijnen AJ, Montecino M, Young DW, Javed A, Pratap J, Choi J-Y, Ali SA, Pande S, Hassan MQ. Transcription-factor-mediated epigenetic control of cell fate and lineage commitment. Biochem. Cell Biol. 2009;87:1–6. doi: 10.1139/o08-094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J, Ghule PN, Shimamura A, Stein JL, Greenblatt M. Spindle microtubule dysfunction and cancer predisposition. J. Cell. Physiol. 2014;229:1881–1883. doi: 10.1002/jcp.24691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T-J, Tao R, Han Y-Q, Xu G, Liu J, Han Y-F. Wnt3a promotes human umbilical cord mesenchymal stem cells to differentiate into epidermal-like cells. Eur. Rev. Med. Pharmacol. Sci. 2015;19:86–91. [PubMed] [Google Scholar]
- Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science (80-.) 2007;317:916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- Torres EM, Williams BR, Amon A. Aneuploidy: cells losing their balance. Genetics. 2008;179:737–746. doi: 10.1534/genetics.108.090878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortolina L, Duffy DJ, Maffei M, Castagnino N, Carmody AM, Kolch W, Kholodenko BN, De Ambrosi C, Barla A, Biganzoli EM, Nencioni A, Patrone F, Ballestrero A, Zoppoli G, Verri A, Parodi S. Advances in dynamic modeling of colorectal cancer signaling-network regions, a path toward targeted therapies. Oncotarget. 2014 doi: 10.18632/oncotarget.3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsafrir D, Bacolod M, Selvanayagam Z, Tsafrir I, Shia J, Zeng Z, Liu H, Krier C, Stengel RF, Barany F, Gerald WL, Paty PB, Domany E, Notterman DA. Relationship of gene expression and chromosomal abnormalities in colorectal cancer. Cancer Res. 2006;66:2129–2137. doi: 10.1158/0008-5472.CAN-05-2569. [DOI] [PubMed] [Google Scholar]
- Vadia S, Levin PA. Growth rate and cell size: a re-examination of the growth law. Curr. Opin. Microbiol. 2015;24C:96–103. doi: 10.1016/j.mib.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan PS, Aziz F, van Wijnen aJ, Wu S, Harada H, Taniguchi T, Soprano KJ, Stein JL, Stein GS. Activation of a cell-cycle-regulated histone gene by the oncogenic transcription factor IRF-2. Nature. 1995;377:362–365. doi: 10.1038/377362a0. [DOI] [PubMed] [Google Scholar]
- Vaughan PS, van der Meijden CM, Aziz F, Harada H, Taniguchi T, van Wijnen aJ, Stein JL, Stein GS. Cell cycle regulation of histone H4 gene transcription requires the oncogenic factor IRF-2. J. Biol. Chem. 1998;273:194–199. doi: 10.1074/jbc.273.1.194. [DOI] [PubMed] [Google Scholar]
- Voskas D, Ling LS, Woodgett JR. Signals controlling un-differentiated states in embryonic stem and cancer cells: role of the phosphatidylinositol 3’ kinase pathway. J. Cell. Physiol. 2014;229:1312–1322. doi: 10.1002/jcp.24603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber AM, Ryan AJ. ATM and ATR as therapeutic targets in cancer. Pharmacol. {&} Ther. 2014 doi: 10.1016/j.pharmthera.2014.12.001. [DOI] [PubMed] [Google Scholar]
- Weinstein IB. Disorders in cell circuitry during multistage carcinogenesis: the role of homeostasis. Carcinogenesis. 2000;21:857–864. doi: 10.1093/carcin/21.5.857. [DOI] [PubMed] [Google Scholar]
- Wier ML, Scott RE. Defective control of terminal differentiation and its role in carcinogenesis in the 3T3 T proadipocyte stem cell line. Cancer Res. 1985;45:3339–3346. [PubMed] [Google Scholar]
- Wilke MS, Hsu BM, Wille JJJ, Pittelkow MR, Scott RE. Biologic mechanisms for the regulation of normal human keratinocyte proliferation and differentiation. Am. J. Pathol. 1988;131:171–181. [PMC free article] [PubMed] [Google Scholar]
- Wille JJJ, Maercklein PB, Scott RE. Neoplastic transformation and defective control of cell proliferation and differentiation. Cancer Res. 1982;42:5139–5146. [PubMed] [Google Scholar]
- Wille JJJ, Pittelkow MR, Scott RE. Normal and transformed human prokeratinocytes express divergent effects of a tumor promoter on cell cycle-mediated control of proliferation and differentiation. Carcinogenesis. 1985;6:1181–1187. doi: 10.1093/carcin/6.8.1181. [DOI] [PubMed] [Google Scholar]
- Wille JJJ, Pittelkow MR, Shipley GD, Scott RE. Integrated control of growth and differentiation of normal human prokeratinocytes cultured in serum-free medium: clonal analyses, growth kinetics, and cell cycle studies. J. Cell. Physiol. 1984;121:31–44. doi: 10.1002/jcp.1041210106. [DOI] [PubMed] [Google Scholar]
- Wille JJJ, Scott RE. Suppression of tumorigenicity by the cell-cycle-dependent control of cellular differentiation and proliferation. Int. J. Cancer. 1986;37:875–881. doi: 10.1002/ijc.2910370613. [DOI] [PubMed] [Google Scholar]
- Williams SE, Fuchs E. Oriented divisions, fate decisions. Curr Opin Cell Biol. 2013;25:749–758. doi: 10.1016/j.ceb.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie R, Medina R, Zhang Y, Hussain S, Colby J, Ghule P, Sundararajan S, Keeler M, Liu L-J, van der Deen M, Mitra P, Lian JB, Rivera-Perez JA, Jones SN, Stein JL, van Wijnen AJ, Stein GS. The histone gene activator HINFP is a nonredundant cyclin E/CDK2 effector during early embryonic cell cycles. Proc. Natl. Acad. Sci. U. S. A. 2009;106:12359–12364. doi: 10.1073/pnas.0905651106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie R, van Wijnen aJ, van Der Meijden C, Luong MX, Stein JL, Stein GS. The cell cycle control element of histone H4 gene transcription is maximally responsive to interferon regulatory factor pairs IRF-1/IRF-3 and IRF-1/IRF-7. J. Biol. Chem. 2001;276:18624–18632. doi: 10.1074/jbc.M010391200. [DOI] [PubMed] [Google Scholar]
- Yang H-J, Ratnapriya R, Cogliati T, Kim J-W, Swaroop A. Vision from next generation sequencing: Multi-dimensional genome-wide analysis for producing gene regulatory networks underlying retinal development, aging and disease. Prog. Retin. Eye Res. 2015 doi: 10.1016/j.preteyeres.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Ferrell JEJ. The Cdk1-APC/C cell cycle oscillator circuit functions as a time-delayed, ultrasensitive switch. Nat. Cell Biol. 2013;15:519–525. doi: 10.1038/ncb2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young DW, Hassan MQ, Pratap J, Galindo M, Zaidi SK, Lee S, Yang X, Xie R, Javed A, Underwood JM, Furcinitti P, Imbalzano AN, Penman S, Nickerson Ja, Montecino Ma, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Mitotic occupancy and lineage-specific transcriptional control of rRNA genes by Runx2. Nature. 2007;445:442–6. doi: 10.1038/nature05473. [DOI] [PubMed] [Google Scholar]
- Young DW, Zaidi SK, Furcinitti PS, Javed A, van Wijnen AJ, Stein JL, Lian JB, Stein GS. Quantitative signature for architectural organization of regulatory factors using intranuclear informatics. J. Cell Sci. 2004;117:4889–4896. doi: 10.1242/jcs.01229. [DOI] [PubMed] [Google Scholar]
- Zaidi SK, Grandy RA, Lopez-Camacho C, Montecino M, van Wijnen AJ, Lian JB, Stein JL, Stein GS. Bookmarking target genes in mitosis: a shared epigenetic trait of phenotypic transcription factors and oncogenes? Cancer Res. 2014;74:420–425. doi: 10.1158/0008-5472.CAN-13-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SK, Trombly DJ, Dowdy CR, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Epigenetic mechanisms in leukemia. Adv. Biol. Regul. 2012;52:369–76. doi: 10.1016/j.jbior.2012.05.001. [DOI] [PubMed] [Google Scholar]
- Zaidi SK, van Wijnen AJ, Lian JB, Stein JL, Stein GS. Targeting deregulated epigenetic control in cancer. J. Cell. Physiol. 2013;228:2103–2108. doi: 10.1002/jcp.24387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SK, Young DW, Choi J-Y, Pratap J, Javed A, Montecino M, Stein JL, van Wijnen AJ, Lian JB, Stein GS. The dynamic organization of gene-regulatory machinery in nuclear microenvironments. EMBO Rep. 2005;6:128–133. doi: 10.1038/sj.embor.7400337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SK, Young DW, Javed A, Pratap J, Montecino M, van Wijnen A, Lian JB, Stein JL, Stein GS. Nuclear microenvironments in biological control and cancer. Nat. Rev. Cancer. 2007;7:454–463. doi: 10.1038/nrc2149. [DOI] [PubMed] [Google Scholar]
- Zaidi SK, Young DW, Montecino Ma, Lian JB, van Wijnen AJ, Stein JL, Stein GS. Mitotic bookmarking of genes: a novel dimension to epigenetic control. Nat. Rev. Genet. 2010;11:583–9. doi: 10.1038/nrg2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SK, Young DW, Montecino M, van Wijnen AJ, Stein JL, Lian JB, Stein GS. Bookmarking the genome: maintenance of epigenetic information. J. Biol. Chem. 2011;286:18355–18361. doi: 10.1074/jbc.R110.197061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Yuan R, Hood L, Ao P. Endogenous molecular-cellular hierarchical modeling of prostate carcinogenesis uncovers robust structure. Prog. Biophys. Mol. Biol. 2015 doi: 10.1016/j.pbiomolbio.2015.01.004. [DOI] [PubMed] [Google Scholar]