Abstract
Moonlighting proteins comprise a subset of multifunctional proteins that perform two or more biochemical functions that are not due to gene fusions, multiple splice variants, proteolytic fragments, or promiscuous enzyme activities. The project described herein focuses on a sub-set of moonlighting proteins that have a canonical biochemical function inside the cell and perform a second biochemical function on the cell surface in at least one species. The goal of this project is to consider the biophysical features of these moonlighting proteins to determine whether they have shared characteristics or defining features that might suggest why these particular proteins were adopted for a second function on the cell surface, or if these proteins resemble typical intracellular proteins. The latter might suggest that many other normally intracellular proteins found on the cell surface might also be moonlighting in this fashion. We have identified 30 types of proteins that have different functions inside the cell and on the cell surface. Some of these proteins are found to moonlight on the surface of multiple species, sometimes with different extracellular functions in different species, so there are a total of 98 proteins in the study set. Although a variety of intracellular proteins (enzymes, chaperones, etc.) are observed to be re-used on the cell surface, for the most part, these proteins were found to have physical characteristics typical of intracellular proteins. Many other intracellular proteins have also been found on the surface of bacterial pathogens and other organisms in proteomics experiments. It is quite possible that many of those proteins also have a moonlighting function on the cell surface. The increasing number and variety of known moonlighting proteins suggest that there may be more moonlighting proteins than previously thought, and moonlighting might be a common feature of many more proteins.
Introduction
Moonlighting proteins comprise a subset of multifunctional proteins that perform two or more biochemical functions that are not due to gene fusions, multiple splice variants, multiple proteolytic fragments with different functions, or promiscuous enzyme activities [1, 2]. Some of the first examples to be identified were some of the taxon specific crystallins, ubiquitous cytosolic enzymes that were adopted by a variety of species to help form the lens of the eye [3, 4]. Today, over 300 moonlighting proteins have been identified [5]. Many of the known moonlighting proteins are cytosolic enzymes, chaperones, or other proteins that exhibit a second function in other cellular locations, in other types of cells, as part of multi-protein complexes, when binding to DNA or RNA, or when the cellular concentration of a substrate, product, or other ligand changes. Data suggests, however, that there may be even more moonlighting proteins than previously thought, and increasingly more proteins with moonlighting capabilities are being uncovered.
The project described herein is focused on a sub-set of moonlighting proteins that are primarily intracellular, but perform a second biochemical function on the cell surface in at least one species. Intracellular proteins are frequently found on the cell surface during proteomics experiments. A few dozen of these intracellular/cell surface proteins have been demonstrated to have a distinct function in that location. In some pathogenic bacteria, protozoans and fungi, this extracellular function plays a key role in infection or virulence or, in the case of some nonpathogenic symbionts, in commensual interactions with a host species [6,7]. Colonization of the host requires adhesion of the bacterium or other cell type to the host, and many of these proteins have been shown to bind to proteins in the extracellular matrix or directly to host cells while some play other roles in invasion of host tissues.
The goal of this project is to consider the biophysical features of intracellular/cell surface moonlighting proteins to determine whether they have shared characteristics or defining features that might suggest why these particular proteins were adopted for a second function on the cell surface, or if these proteins are more likely to simply resemble typical intracellular proteins. The latter might suggest that many other normally intracellular proteins found on the cell surface during proteomics experiments might also have a moonlighting function in that location. We have identified 30 types of proteins that have different functions inside the cell and on the cell surface. Some of these proteins are found to moonlight on the surface of multiple species, sometimes with different extracellular functions in different species, so there are a total of 98 proteins in the study set.
Methods
Selection of proteins
The proteins in this study were selected with the criterion that the proteins have a biochemical function inside the cell and a second biochemical function on the cell surface. The proteins were identified by searching the literature for experimental evidence that each protein is performing a function on the surface, and is not just observed to be present on the surface.
From our analysis of the literature, we identified 30 different types of proteins, some which are found to moonlight with intracellular and cell surface functions in multiple organisms, for a total of 98 total proteins in the study. The FASTA sequence (the primary amino acid sequence) for each protein was obtained through the NCBI database (National Center for Bintechnology Information, http://www.ncbi.nlm.nih.gov), and the three-dimensional structures of each protein, when available, were obtained through the Protein Data Bank (www.rcsb.org, PDB) [8].
Programs used
Protparam (http://web.expasy.org /) [9] was used to calculate isoelectric point (pI), amino acid composition, aliphatic index, and GRAVY (grand average of hydropathy) score [10].
The CATH Protein Structure Classification Database [11] was used to identify the types of three-dimensional folds found in the proteins. For each of the 30 types of proteins in our study set, if there is a protein in our set with a X-ray crystal structure in the Protein Data bank, that protein was selected as the representative of that type of protein. If there was no structure for a type of protein in our list, the protein that has the highest sequence identity to a protein in the PDB was chosen as the representative for that type of protein. The CATH database was then searched using the FASTA sequence for each representative protein.
UIPred [12,13] was used to predict intrinsically disordered regions of each protein. The UIPred server (iupred.enzim.hu) was used to analyze the amino acid sequence of each protein. Amino acids that scored above 0.5 were noted as being in potentially disordered regions.
SignalP, (cbs.dtu.dk/services/SignalP/) [14] and Psort, (http://psort.hgc.jp/) [15–17] were used to identify potential signal sequences.
The UniProt database (http://www.uniprot.org/) [18] was searched using the amino acid sequence of each protein to identify Gene Ontology (GO) terms in the annotation in the categories of Process and Function for each protein (www.geneontology.org) [19]. The GO terms annotated in the UniProt database predominantly describe the intracellular functions of the proteins and were used to summarize the most common intracellular pathways or roles of the proteins.
Results
Selection of proteins for study (Table 1)
Table 1. Moonlighting Proteins Used In the Study.
| Intracellular Function | Surface Function | Species | UniProt | Reference |
|---|---|---|---|---|
| Alcohol acetaldenyde dehydrogenase | fibronectin, laminin, and type II collagen binding | Enteamoeba histolytica | Q24803 | [20] |
| Alcohol acetaldehyde dehydrogenase | Listeria adhesion protein (LAP) | Listeria monocytogenes | Q6Q3I2 | [21–24] |
| Aspartate ammonia lyase | plasminogen binding | Haemophilus influenzae | P44324 | [25] |
| Alcohol dehydrogenase (ADH1) | plasminogen binding | Candida albicans | P43067 | [26] |
| Bile salt hydrolase | plasminogen binding | Bifidobacterium lactis, B. bifidum, and B. longum | Q9KK62 | [27] |
| Peroxisomal catalase (CTA1) | plasminogen binidng | Candida albicans | O13289 | [26] |
| DnaK/Hsp70 | plasminogen binding | Bifidobacterium animalis | Q8G6W1 | [28] |
| DnaK/Hsp70 | binding to invertase | Lactococcus lactis | P0A3J0 | [29] |
| DnaK/Hsp70 | plasminogen binding | Mycobacterium tuberculosis | H8EVI1 | [30] |
| DnaK/Hsp70 | plasminogen binding | Neisseria meningitidis | A9M296 | [31] |
| Ef-Tu | attachment to human cells and mucins | Lactobacillus johnsonii | Q74JU6 | [32] |
| Ef-Tu | fibronectin binding | Mycoplasma pneumoniae | P23568 | [33] |
| Ef-Tu | factor H and plasminogen binding | Pseudonomas aeruginosa | B7V630 | [34] |
| Enolase | plasminogen binding | Aeromonas hydrophila | Q8GE63 | [35] |
| Enolase | plasminogen and laminin binding | Bacillus anthracis | D8H2L1 | [36] |
| Enolase | plasminogen binding | Bifidobacterium longum, B. bifidum, B. breve and B. lactisa | B7GTK2 | [27, 37] |
| Enolase | plasminogen binding | Borrelia burgdorferi | B7J1R2 | [38] |
| Enolase | plasminogen binding | Candida albicans | P30575 | [39] |
| Enolase | plasminogen binding | Homo sapiens | P06733 | [40, 41] |
| Enolase | plasminogen and laminin binding | Lactobacillus crispatus | Q5K117 | [42] |
| Enolase | plasminogen and laminin binding | Lactobacillus johnsonii | A3F8V9 | [42] |
| Enolase | fibronectin binding | Lactobacillus plantarum | Q5NJY7 | [43] |
| Enolase | plasminogen binding | Leishmania mexicana | Q3HL75 | [44] |
| Enolase | plasminogen binding | Neisseria meningitidis | E0N8L2 | [31] |
| Enolase | plasminogen binding | Onchocerca volvulus | Q7YZX3 | [45] |
| Enolase | fibronectin binding | Paracoccidioides brasiliensis | A5JQI1 | [46] |
| Enolase | plasminogen binding | Rattus norvegicus | Q5BJ93 | [47] |
| Enolase | plasminogen binding | Schistosoma bovis | B2LXU1 | [48] |
| Enolase | plasminogen and laminin binding | Staphylococcus aureus | E5R9G0 | [42, 49] |
| Enolase | plasminogen binding | Streptococcus anginosus and S. oralis | E7GW07 | [50] |
| Enolase | plasminogen binding | Streptococcus mutans | C6SQ43 | [51] |
| Enolase | plasminogen binding | Streptococcus pneumoniae | H8LG96 | [42, 52, 53] |
| Enolase | plasminogen binding | Streptococcus pyogenes | A3F8V6 | [42, 54] |
| Enolase | plasminogen and fibronectin binding | Streptococcus suis | C6GGT7 | [55] |
| Fructose 1,6-bisphosphate aldolase | plasminogen binding | Candida albicans | C4YHS0 | [26] |
| Fructose-1,6-bisphosphate aldolase | adhesin | Neisseria meningitidis | F0N9L0 | [56] |
| GAPDH | plasminogen binding | Bacillus anthracis | Q81X74 | [57] |
| GAPDH | plasminogen, fibronectin and laminin binidng | Candida albicans | Q5ADM7 | [26, 58] |
| GAPDH | NAD ribosylating activity | Escherichia coli | Q0TH49 | [59] |
| GAPDH | plasminogen binding | Lactobacillus crispatus | D5H2B9 | [60] |
| GAPDH | binds mucin and Caco-2 cells | Lactobacillus plantarum | F9UM10 | [61] |
| GAPDH | binds invertase | Lactococcus lactis | F2HK64 | [29] |
| GAPDH | binds mucin | Mycoplasma genitalium | J7HIM3 | [62] |
| GAPDH | adhesin | Neisseria meningitidis | C6S993 | [63] |
| GAPDH | fibronectin, laminin, and type I collagen binding | Paracoccidioides brasiliensis | Q8X1X3 | [64] |
| GAPDH | transferrin-binding protein and plasminogen binding | Staphylococcus aureus and S. epidermidis | D9RFF4 | [65] |
| GAPDH | plasminogen binding | Streptococcus anginosus and S. oralis | S6AWM2 | [50] |
| GAPDH | plasminogen binding | Streptococcus—group A | Q1J8I3 | [66] |
| GAPDH | plasminogen binding | Streptococcus agalactiae | Q9ALW2 | [67] |
| GAPDH | plasminogen binding | Streptococcus pneumoniae | Q97NL1 | [68] |
| GAPDH | fibronectin binding and binds uPAR/CD87 receptor on human cells | Streptococcus pyogenes | B5XJR2 | [69, 70] |
| GAPDH | plasminogen binding | Streptococcus suis | Q3Y454 | [71] |
| GAPDH | fibronectin, plasminogen, and collagen binding | Trichomonas vaginalis | Q27820 | [72] |
| Glucose 6-phosphate isomerase | laminin and collagen I binding | Lactobacillus crispatus | K1MPC4 | [73] |
| Glutamine synthetase | plasminogen binding | Bifidobacterium lactis, B. bifidum, and B. longum | C2GUH0 | [28] |
| Glutamine synthetase | fibronectin, laminin, collagen I and plasminogen binding | Lactobacillus crispatus | D5GYN9 | [73] |
| Glutamine synthetase | plasminogen and fibronectin binding | Mycobacterium tuberculosis | H8ESK0 | [30] |
| Histone H1 | thyroglobulin receptor | Mus musculus | P43274 | [74] |
| Hsp60/GroEL | cytotoxic activity | Aggregatibacter actinomycetemcomitans | C9R2H0 | [75] |
| Hsp60/GroEL | adhesin | Chlamydiae pneumoniae | P31681 | [76] |
| Hsp60/GroEL | adhesin | Clostridium difficile | Q9KKF0 | [77] |
| Hsp60/GroEL | adhesin to glycosphingolipids | Haemophilus ducreyi | P31294 | [78, 79] |
| Hsp60/GroEL | adhesin | Helicobacter pylori | Q8RNU2 | [80, 81] |
| Hsp60/GroEL | adhesin | Histoplasma capsulatum | P50142 | [82] |
| Hsp60/GroEL | receptor for HDL | Homo sapiens and Rattus rattus | P10809 | [83] |
| Hsp60/GroEL | adhesin, binds mucin | Lactobacillus johnsonii | F7SCR2 | [84] |
| Hsp60/GroEL | binds invertase | Lactococcus lactis | F2HIT2 | [29] |
| Hsp60/GroEL | adhesin | Legionella pneumophila | B2C318 | [85] |
| Hsp60/GroEL | adhesin | Listeria | Q8KP52 | [23] |
| Hsp60/GroEL | adhesin | Mycobacterium tuberculosis | H8EVS5 | [86] |
| Hsp60/GroEL | adhesin | Plesiomonas shigelloides | Q1EQW2 | [87] |
| Hsp60/GroEL | adhesin | Salmonella typhimurium | F5ZZ81 | [88] |
| Inosine 5'-monophosphate dehydrogenase | plasminogen binding | Staphylococcus aureus | D6SDD0 | [89] |
| Malate synthase | fibronectin and laminin binding | Mycobacterium tuberculosis | E2T9U8 | [90] |
| Ornithine carbamoyltransferase | fibronectin binding | Staphylococcus epidermidis | T0BSW2 | [91] |
| Pyruvate dehydrogenase (E1 beta subunit, PDH-B) | fibrinogen binding. | Mycoplasma pneumoniae | E1QCD9 | [33] |
| Peroxiredoxin | plasminogen binding | Neisseria meningitidis | J8V2K1 | [31] |
| 6-phosphofructokinase | binding to invertase | Lactococcus lactis | F2HMQ4 | [29] |
| 6-phosphofructokinase | plasminogen binding | Streptococcus oralis | E6KMA1 | [50] |
| Pyruvate-ferredoxin oxidoreductase (PFO) | adhesin | Trichomonas vaginalis | Q27089 | [92] |
| 6-phosphogluconate dehydrogenase | adhesin | Streptococcus pneumoniae | Q97SI6 | [93] |
| Phosphoglycerate kinase | plasminogen bindng | Candida albicans | P46273 | [26] |
| Phosphoglycerate kinase | plasminogen binding | Streptococcus agalactiae S8XU00 | S8XU00 | [94] |
| Phosphoglycerate kinase | plasminogen binding | Streptococcus anginosus and S. oralis | E7GZG8 | [50] |
| Phosphoglycerate kinase | plasminogen binding | Streptococcus pneumoniae | J1RST3 | [95] |
| Phosphoglycerate mutase | plasminogen binding | Bifidobacterium lactis, B. bifidum, and B. longum | S3DNJ2 | [27] |
| Phosphoglyceromutase | plasminogen binidng | Candida albicans | P82612 | [26] |
| Phosphoglycerate mutase | plasminogen binding | Streptococcus anginosus and S. oralis | E6IYJ0 | [50] |
| Pyruvate kinase | binds invertase | Lactococcus lactis | F2HMQ3 | [29] |
| Ribonucleotide reductase subunit 2 | plasminogen binding | Staphylococcus aureus | Q7A6T1 | [89] |
| Superoxide dismutase | adhesin | Mycobacterium avium | P47201 | [96, 97] |
| Superoxide dismutase | Adhesion | Mycobacterium tuberculosis | H8F202 | [97] |
| Transcription elongation factor (TEF1) | plasminogen binding | Candida albicans | C4YDJ3 | [26] |
| Triose phosphate isomerase | adhesin | Paracoccidioides brasiliensis | Q96VN5 | [98] |
| Triose phosphate isomerase | adhesin | Staphylococcus aureus | D9RMW0 | [99] |
| Triose phosphate isomerase | plasminogen binding | Streptococcus anginosus and S. oralis | E6J203 | [50] |
| Thiol-specific antioxidant protein (TSA1) | plasminogen binding | Candida albicans | C4YNZ5 | [26] |
The proteins in this study [20–99] were selected with the criteria that each protein has a biochemical function inside the cell and at least one different biochemical function on the cell surface. The proteins were identified by searching the literature for experimental evidence that each protein is performing a biochemical function on the cell surface. Proteins that were only observed to be present on the cell surface and have not had a function identified in that location were not included in the study. This latter requirement removes proteins that may have been observed on the cell surface as potential false-positives in proteomics experiments. In addition, for this study, proteins that are secreted but not attached to the cell surface were not included. From our analysis of the literature, we identified 30 different types of proteins, some which are found to moonlight with intracellular and cell surface functions in multiple organisms, for a total of 98 total proteins in the study.
Species and types of organisms
Most of the proteins in the study are from bacteria. The bacterial species represented include typical Gram-positive and Gram-negative species, as well as mycobacteria, spirochetes, and mycoplasma. Most of the species represented are pathogenic, but some are considered “pro-biotic”, or members of the normal gut biota. A few of the proteins are found to moonlight in single-celled eukaryotic organisms, including protozoa and yeast fungi, or in multicellular eukaryotes, including mammals and worms.
Intracellular Functions
The majority of the proteins in the study are metabolic enzymes. Alll six Enzyme Commission (EC) groups are represented. Of the 23 types of enzymes, ten are in EC group 1 (oxidoreductases) and five are in group 2 (transferases). Most of the enzymes are ubiquitous or at least found in many species and function in central pathways in metabolism, including glycolysis, the citric acid cycle, the pentose phosphate pathway, or in nucleotide or amino acid metabolism. Other intracellular/cell surface moonlighting proteins include chaperones (heat shock protein 60 Hsp60/GroEL, heat shock protein 70 Hsp70/DnaK), as well as a protein synthesis elongation factor (Ef-Tu, elongation factor Tu), a transcription elongation factor (transcription elongation factor 1, TEF1), a thiol specific antioxidant protein (TSA1), and a histone (H1).
Extracellular functions
The majority of the intracellular/cell surface moonlighting proteins function on the cell surface in binding to extracellular matrix, as an adhesin to attach to host cells, or as a cell surface receptor for a soluble protein (Fig 1). For those proteins described as adhesins in Table 1, the specific host protein on the host cell surface has been identified in only a few cases. Streptococcus pyogenes glyceraldehyde 3-phosphate dehydrogenase (GAPDH) binds to the uPAR/CD87 receptor on human cells [70]. Haemophilus ducreyi Hsp60/GroEL binds to glycosphingolipids [78, 79]. On the surface of Listeria monocytogenes, alcohol acetaldehyde dehydrogenase is used to bind to a moonlighting Hsp60 on the surface of mammalian host cells, an interesting example of a moonlighting protein interacting with another moonlighting protein [22, 23].
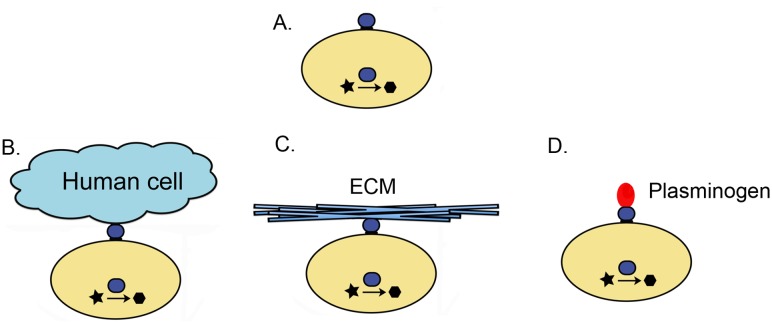
Fig 1. An increasing number of intracellular enzymes, chaperones, and other proteins are being found on the cell surface where they perform other functions.
A single protein can have a function inside the cell, for example and enzyme that converts a substrate (star) to a product (hexagon), and also be found on the cell surface (A). Some of these proteins moonlight as adhesins for binding to host cell surface proteins (B) or to extracellular matrix (C, ECM) and play a role in infection and virulence. Other proteins bind to the zymogen plasminogen and enable its conversion to plasmin, a broad specificity protease (D). The active protease is then used as an aide to degrade and invade host tissues.
Many of the bacterial moonlighting proteins bind to structural components of the host extracellular matrix, including fibronectin, laminin, and/or collagen, or to mucin, a component of the mucosal epithelial lining. These interactions enable a physical attachment to the host, whether it is for a pathogen invading host tissues or a commensual gut bacterium establishing a more symbiotic relationship with a mammalian host. When Lactococcus lactis Hsp60/GroEL, DnaK/Hsp70, GAPDH, pyruvate kinase and 6-phosphofructokinase bind yeast Invertase, a hyperglycosylated cell surface protein, the interaction may also assist in a symbiotic relationship between the bacterium and the yeast [29].
Several of the moonlighting proteins in the study are receptors for soluble proteins. Human and mouse Hsp60/GroEL are cell surface receptors for high density lipoprotein (HDL) [83], and mouse histone H1 serves as a thyroglobulin receptor to mediate thyroglobulin endocytosis [74]. Staphylococcal GAPDH serves as a transferrin binding protein to acquire iron from the host [65]. Binding to the soluble protein plasminogen aids in invasion of host tissues by many pathogens. Plasminogen is a precursor to plasmin, which is a serine protease present in blood that helps break down fibrin clots [100]. When an invading pathogen uses a surface moonlighting protein to bind plasminogen from the host, the plasminogen can be converted to plasmin, the active form of the protease, by tissue-type plasminogen activator (tPA) and urokinase. The plasmin that is now attached to the surface of the invading organism can be used as a general protease to degrade host extracellular matrix and basement membrane, thereby facilitating migration through tissues.
We note that in many of the published reports about the proteins in our study, the intention was to determine if the species being studied had any surface proteins that bind to a specific target protein (plasminogen, fibronectin, etc.) Each of these intracellular/cell surface proteins may also have other functions on the cell surface in addition to the ones that have been identified to date, for example some of the proteins that are known to bind to plasminogen might also bind to other host proteins.
Physical features
The primary amino acid sequence for each protein and the three-dimensional structures of each protein, when available, were studied using bioinformatics tools in order to ascertain general shared characteristics or defining features of these proteins as a set, be they structural features, sequence motifs, or biochemical properties. Any trends, patterns, or generalizations that we discover among these moonlighting proteins could aid in the identification of other proteins that may have a moonlighting function on the cell surface.
Signal sequences
In general, most proteins targeted to the cell surface contain an N-terminal signal peptide, but many of these intracellular/cell surface moonlighting proteins have been found to lack a signal sequence. SignalP [14] and Psort [15–17] were used to look for the presence of a signal sequence for targeting to the plasma membrane for the study set of 98 proteins. None of the proteins contain a signal peptide.
Molecular weight
The individual proteins of each specific type in our study varied very little in molecular weight regardless of species of origin (Fig 2a). Most of these moonlighting proteins are within the range of 200–600 amino acids. The longest were alcohol acetaldehyde dehydrogenase (866–870 amino acids) and pyruvate-ferredoxin oxidoreductase (1157 amino acids). Brocchieri and Karlin [101] found through a study of 5 eukaryotic genomes and 67 bacterial genomes that the median length of proteins in eukaryotes is 361 amino acids and the median length is only 267 amino acids in bacteria. Other studies of many genes from a genome or from whole genomes found similar averages for protein length [102]. Interestingly, most of the proteins in our study set are significantly longer than these median sizes. The eukaryotic proteins in our study contain from 196 to 1157 amino acids, with eight protein types containing over 400 amino acids, three types near the observed median length (between 330 and 360 amino acids) and 4 protein types containing fewer than 250 amino acids. The bacterial proteins ranged from 122 amino acids (peroxiredoxin) to 866 amino acids (acetaldehyde dehydrogenase) in length. Only three types of proteins contain fewer than the median of 267 amino acids observed by Brocchieri and Karlin (peroxiredoxin, superoxide dismutase, and phosphoglycerate mutase), and one protein was near the median length (triose phosphate isomerase (TPI), with 252 amino acids in Streptococcus TPI and 283 amino acids in Staphylococcus TPI), twenty of the bacterial protein types in the study contain over 300 amino acids.
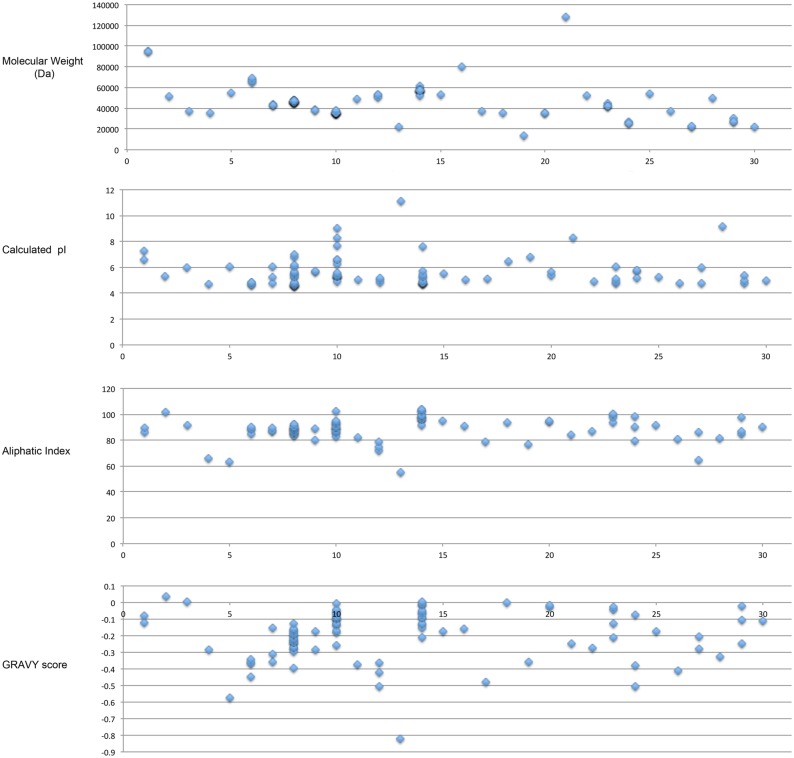
Fig 2. Physical features of intracellular proteins that moonlight on the cell surface.
Protein molecular weight (A), calculated pI (B), aliphatic index (C), and GRAVY score (D) are shown for each type of protein. For some types of proteins, more than one score is shown because the protein is found to moonlight in more than one organism, and the score was determined for each protein in each organism in which it moonlights. The type of enzyme is indicated by number on the x-axis: 1. Alcohol acetaldehyde dehydrogenase. 2. Aspartase. 3. Alcohol dehydrogenase. 4. Bile salt hydrolase. 5. Peroxisomal catalase. 6. DnaK. 7. Ef-Tu. 8. Enolase. 9. Fructose 1,6-bisphosphate aldolase. 10. GAPDH. 11. Glucose 6-phosphate isomerase. 12. Glutamine synthetase. 13. Histone H1. 14. Hsp60/GroEL. 15. IMPDH. 16. Malate synthase. 17. Ornithine carbamoyl transferase. 18. Pyruvate dehydrogenase. 19. Peroxiredoxin. 20. 6-phosphofructokinase. 21. Pyruvate-ferredoxin oxidoreductase. 22. 6-phosphogluconate dehydrogenase. 23. Phosphoglycerate kinase. 24. Phosphoglyceromutase. 25. Pyruvate kinase. 26. Ribonucleotide reductase. 27. Superoxide dismutase. 28. Transcription elongation factor. 29. Triose phosphate isomerase. 30. Thiol specific antioxidant protein TSA1. Calculations were performed with ProtParam [9].
Theoretical pI
The calculated isoelectric points (pI) have been found to differ among proteins that localize to different sub-cellular locations [103]. Proteins are generally least soluble near their isoelectric points, which means that a protein’s pI needs to be different from its environmental pH for the protein to be adequately soluble. Cytoplasmic pH is usually around 7, so proteins with a pI greater than or less than this pH are favored. The calculated pIs for known cytosolic proteins have been found to center around 5.5 [103]. The pI’s of most of the proteins in our study lie between 4.5–6.5 (Fig 2b), which is typical of cytosolic proteins.
Aliphatic index
The aliphatic index is a measure of the relative volume occupied by aliphatic side chains—alanine, valine, isoleucine, and leucine. A higher aliphatic index is an indicator of higher thermostability, and also an indicator of solubility in a cell when the protein is overexpressed [104]. Most cytosolic enzymes have aliphatic indexes around 80–100, as did most of the proteins in this study set (Fig 2C) [104]. However, Histone H1 (55.3), which is a nuclear protein, CTA1 peroxisomal catalase (64), bile salt hydrolase (66), glutamine synthase (72–79), peroxiredoxin (77), and one of the superoxide dismutases (65) had aliphatic indexes below 80.
GRAVY score
The Grand Average of Hydropathy (GRAVY) for a protein is calculated as the sum of hydropathy values of all the amino acids, divided by the number of residues in the sequence [10]. More hydrophobic/non-polar residues are given more positive values, whereas more polar/ionic residues are given more negative values. The overall GRAVY scores for the proteins in this study were negative or zero for all except aspartase, which was slightly above zero at 0.036 (Fig 2d). These scores are typical for soluble proteins. None of the proteins had sequences of hydrophobic amino acids sufficiently long enough to indicate the presence of a transmembrane domain.
Intrinsically disordered regions of proteins allow many different conformational states, and in many proteins enable highly specific interactions with multiple binding partners [105]. Intrinsically disordered regions also may be more tolerant of mutations than more structured domains, and thereby might provide the material for evolution of an additional function. We studied the amino acid sequences of the intracellular/cell surface moonlighting proteins using UIPred to identify potential regions of disorder [12, 13]. Analysis using UIPred identified only very short regions of most of the proteins that scored above 0.5 and are most likely surface loops. Only Histone H1 contained longer intrinsically disordered regions. These results suggest that most of the intracellular/cell surface moonlighting proteins in this study set are not likely to be the type of moonlighting proteins that interact with multiple proteins by folding into different conformations with different binding partners.
Type of three-dimensional fold
Different protein folds vary in their stability and their tolerance of amino acid sequence substitutions, insertions and deletions. In addition, some folds might make better “scaffolds” for evolution of new functions, because of additional stability, for example. The CATH protein structure classification system is a hierarchical classification of protein domains [11]. Of the 30 different types of proteins in our list, there were 58 CATH domain classifications represented (some proteins contained multiple domains). Thirty-nine domains were in Class 3 (containing a significant amount of both alpha-helical and beta sheet secondary structure elements), 13 were in class 1 (mostly alpha-helical), and only 6 domains were in Class 2 (mostly beta sheet). Consistent with the IUPred results, no domains were classified as having very little secondary structure (Class 4). The CATH Architectures (arrangements of secondary structures) that were represented most often include 1.10 or mainly alpha/orthogonal bundle, 1.20 mainly alpha/up-down bundle, 3.20 alpha-beta/alpha-beta barrel, 3.30 alpha-beta/2-layer sandwich, 3.40 alpha beta/3-layer (aba) sandwich. Two topology or fold groups were represented at least six different proteins: 3.20.20 TIM barrel and 3.40.50 Rossmann fold. TIM barrels and alpha/beta sandwiches (which includes the Rossman fold) are two of the most common architectures for proteins in general.
Discussion
A goal of this bioinformatics analyses was to determine if there are any characteristics or trends that could define and help identify intracellular/cell surface moonlighting proteins. Although a variety of intracellular proteins (enzymes, chaperones, etc.) are observed to be re-used on the cell surface, for the most part they seem to be typical intracellular proteins. Other than the few exceptions mentioned above, they do not exhibit extremes in calculated pI, disorder, stability, or other characteristics. The types of three-dimensional folds are diverse and common among many proteins.
The question remains as to why and how these proteins obtained a second function. Like many of the other known moonlighting proteins, most of the proteins in this study are ubiquitous enzymes in central metabolism or ubiquitous chaperone proteins. They are likely to have been adopted for a second function because organisms evolve by utilizing and building upon components they already possess, and these proteins are available in many organisms. Many of the moonlighting proteins in this study, as well as many described elsewhere [1–7, 106, 107], are essential housekeeping proteins. These proteins first arose billions of years ago and are expressed in many species and cell types, making them available targets for organisms to modify and use to develop a new function.
A new binding function can result if a protein’s structure is modified to create a new binding site on the protein surface, and binding to another protein is the key characteristic of the second function of most of the proteins in this study. Modification of a short amino acid sequence could be sufficient to form a new protein-protein interaction site. In general, proteins appear to contain many more amino acids than are required to form their active site, leaving a lot of surface amino acids that are not involved in the first function and are therefore not under as much selective pressure. For example, the active site amino acid residues of the glycolytic enzyme phosphoglucose isomerase have shown to be highly conserved in over 126 species, however the solvent-exposed areas are not as carefully conserved and contain a multitude of loops, pockets, clefts and other structural features that have been modified during billions of years of evolution and could have easily developed a new binding site, yielding a new moonlighting function for this protein [108]. It is interesting to note that one trend of the proteins in this study is that they tend to be somewhat longer than the median length of a cytosolic protein. Perhaps having more amino acids than the average protein increases the probability that a small surface region can be modified by evolution to form a new protein binding site without affecting the original function of the protein.
A large number of the proteins in the study bind plasminogen on the cell surface. The binding site for plasminogen has been found in several cases to be a short lysine-containing amino acid sequence usually at the C-terminus of the protein, although found internally in some enolases [109, 110]. Addition of a plasminogen binding site by replacement of a few surface or C-terminal residues with lysines is just the kind of modification that could add a second function without significant changes to the overall protein structure or original function. It is also possible that proteins that already contain lysines in appropriate places could be adopted for a second function if they were to be expressed in a new location, for example on the cell surface. This adoption of proteins for a new function by changes in expression without significant modification of the protein structure is how several ubiquitous enzymes became taxon specific crystallins [111].
Another possible reason for the increased sequence length is suggested by an observation by Ghosh and Dill that longer proteins tend to be more stable than shorter proteins [112]. Because the moonlighting function of each of the proteins in our study involves being displayed on the cell surface, in a harsher environment than in the cell cytoplasm, proteins with an above average level of protein stability may have been selected for this function. Another correlation with protein length is the degree of amino acid sequence conservation, with conserved proteins tending to be longer [113]. This is consistent with many of the moonlighting proteins in our list being conserved proteins with original functions in central metabolism.
These moonlighting intracellular/cell surface proteins not only need a method to interact with another protein, but also they need a mechanism to be transported across the cell membrane and a mechanism to become attached to the cell surface. None of the proteins have been found to possess a signal peptide for targeting to the cell membrane or a sequence motif for attachment to the cell surface, for example the LPXTG motif that is involved in attachment to the cell surface in Gram-positive bacteria [114]. How these intracellular proteins end up located outside of the cell and attached to the cell surface is an active area of inquiry in this field.
Many other intracellular proteins have also been found on the surface of bacterial pathogens and other organisms in proteomics experiments. It is quite possible that they also have a moonlighting function on the cell surface. The increasing number and variety of known moonlighting proteins suggest that there may be more moonlighting proteins than previously thought, and moonlighting might be a common feature of many more proteins.
Funding Statement
The research reported herein was supported by a Sarah Madonna Kabbes Award for Undergraduate Research to V.A. and a University of Illinois Honors College Research Grant (www.uic.edu/honors/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jeffery CJ.Moonlighting proteins. Trends Biochem Sci 1999;24: 8–11. [DOI] [PubMed] [Google Scholar]
- 2. Jeffery CJ. Multifunctional proteins: examples of gene sharing. Ann Med 2003;35: 28–3. [DOI] [PubMed] [Google Scholar]
- 3. Wistow G, Piatigorsky J. Recruitment of enzymes as lens structural proteins. Science 1987;236: 1554–6. [DOI] [PubMed] [Google Scholar]
- 4. Piatigorsky J, Wistow GJ. Enzyme/crystallins: gene sharing as an evolutionary strategy. Cell 1989;57: 197–9. [DOI] [PubMed] [Google Scholar]
- 5. Mani M, Chen C, Amblee V, Liu H, Mathur T, Zwicke G, et al. Moonlighting Proteins Database (MoonProt): A database of proteins that are known to moonlight. Nucleic Acids Research.2015;42, D277–D282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Henderson B, Martin A. Bacterial virulence in the moonlight: multitasking bacterial moonlighting proteins are virulence determinants in infectious disease. Infect Immun 2011;79: 3476–91. 10.1128/IAI.00179-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Henderson B, Martin A. Bacterial moonlighting proteins and bacterial virulence. Curr Top Microbiol Immunol. 2013;358:155–213. 10.1007/82_2011_188 [DOI] [PubMed] [Google Scholar]
- 8. Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. The Protein Data Bank. Nucleic Acids Research 2000;28: 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M.R., Appel R.D., et al. Protein Identification and Analysis Tools on the ExPASy Server In:Walker JM, editor. The Proteomics Protocols Handbook, New York: Humana Press; 2005. pp. 571–607. [Google Scholar]
- 10. Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157: 105–32. [DOI] [PubMed] [Google Scholar]
- 11. Sillitoe I, Cuff AL, Dessailly BH, Dawson NL, Furnham N, Lee D, et al. New functional families (FunFams) in CATH to improve the mapping of conserved functional sites to 3D structures. Nucleic Acids Res. 2013;41: D490–8. 10.1093/nar/gks1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dosztányi Z, Csizmók V, Tompa P, Simon I. The Pairwise Energy Content Estimated from Amino Acid Composition Discriminates between Folded and Intrinsically Unstructured Proteins J Mol Biol. 2005;347: 827–839. [DOI] [PubMed] [Google Scholar]
- 13. Dosztányi Z, Csizmók V, Tompa P, Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21: 3433–3434. [DOI] [PubMed] [Google Scholar]
- 14. Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions Nature Methods. 2011;8: 785–786. 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- 15. Nakai K, Horton P. PSORT: a program for detecting the sorting signals of proteins and predicting their subcellular localization. Trends Biochem Sci.1999;24: 34–35. [DOI] [PubMed] [Google Scholar]
- 16. Nakai K, Kanehisa M. Expert system for predicting protein localization sites in Gram-negative bacteria. PROTEINS: Structure, Function, and Genetics. 1991;11: 95–110. [DOI] [PubMed] [Google Scholar]
- 17. Nakai K, Kanehisa M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics. 1992;14: 897–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. The UniProt Consortium. Update on activities at the Universal Protein Resource (UniProt) in 2013. Nucleic Acids Res. 2013;41: D43–D47. 10.1093/nar/gks1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25: 25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yang W, Li E, Kairong T, Stanley SL Jr., Entamoeba histolytica has an alcohol dehydrogenase homologous to the multifuctional adhE gene product of Escherichia coli . Mol Biochem Parasitol. 1994;64: 253–260. [DOI] [PubMed] [Google Scholar]
- 21. Jagadeesan B, Koo OK, Kim KP, Burkholder KM, Mishra KK, Aroonnual A, et al. LAP, an alcohol acetaldehyde dehydrogenase enzyme in Listeria, promotes bacterial adhesion to enterocyte-like Caco-2 cells only in pathogenic species. Microbiology. 2010;156: 2782–2795. 10.1099/mic.0.036509-0 [DOI] [PubMed] [Google Scholar]
- 22. Kim KP, Jagadeesan B, Burkholder KM, Jaradat ZW, Wampler JL, Lathrop AA, et al. Adhesion characteristics of Listeria adhesion protein (LAP)-expressing Escherichia coli to Caco-2 cells and of recombinant LAP to eukaryotic receptor Hsp60 as examined in a surface plasmon resonance sensor. FEMS Microbiol Lett. 2006;256: 324–32. [DOI] [PubMed] [Google Scholar]
- 23. Wampler JL, Kim KP, Jaradat Z, Bhunia AK. Heat shock protein 60 acts as a receptor for the Listeria adhesion protein in Caco-2 cells. Infect Immun.2004;72: 931–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Burkholder KM, Bhunia AK. Listeria monocytogenes uses Listeria adhesion protein (LAP) to promote bacterial transepithelial translocation and induces expression of LAP receptor Hsp60. Infect Immun. 2010;78: 5062–73. 10.1128/IAI.00516-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sjöström I, Gröndahl H, Falk G, Kronvall G, Ullberg M. Purification and characterisation of a plasminogen-binding protein from Haemophilus influenzae. Sequence determination reveals identity with aspartase. Biochim Biophys Acta. 1997;1324: 182–90. [DOI] [PubMed] [Google Scholar]
- 26. Crowe JD, Sievwright IK, Auld GC, Moore NR, Gow NA, Booth NA. Candida albicans binds human plasminogen: identification of eight plasminogen-binding proteins. Mol Microbiol. 2003;47: 1637–1651. [DOI] [PubMed] [Google Scholar]
- 27. Candela M, Bergmann S, Vici M, Vitali B, Turroni S, Eikmanns BJ, et al. (2007) Binding of human plasminogen to Bifidobacterium . J Bacteriol. 2007;189: 5929–5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Candela M, Centanni M, Fiori J, Biagi E, Turroni S, Orrico C, et al. DnaK from Bifidobacterium animalis subsp. lactis is a surface-exposed human plasminogen receptor upregulated in response to bile salts. Microbiology. 2010;156: 1609–1618. 10.1099/mic.0.038307-0 [DOI] [PubMed] [Google Scholar]
- 29. Katakura Y, Sano R, Hashimoto T, Ninomiya K, Shioya S. Lactic acid bacteria display on the cell surface cytosolic proteins that recognize yeast mannan. Appl Microbiol Biotechnol. 2010;86: 319–326. 10.1007/s00253-009-2295-y [DOI] [PubMed] [Google Scholar]
- 30. Xolalpa W, Vallecillo AJ, Lara M, Mendoza-Hernandez G, Comini M, Spallek R, et al. Identification of novel bacterial plasminogen-binding proteins in the human pathogen Mycobacterium tuberculosis . Proteomics. 2007;7: 3332–41. [DOI] [PubMed] [Google Scholar]
- 31. Knaust A, Weber MV, Hammerschmidt S, Bergmann S, Frosch M, Kurzai O. Cytosolic proteins contribute to surface plasminogen recruitment of Neisseria meningitidis . J Bacteriol. 2007;189: 3246–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Granato D, Bergonzelli GE, Pridmore RD, Marvin L, Rouvet M, Corthésy-Theulaz IE. Cell surface-associated elongation factor Tu mediates the attachment of Lactobacillus johnsonii NCC533 (La1) to human intestinal cells and mucins. Infect Immun. 2004;72: 2160–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dallo SF, Kannan TR, Blaylock MW, Baseman JB. Elongation factor Tu and E1 beta subunit of pyruvate dehydrogenase complex act as fibronectin binding proteins in Mycoplasma pneumoniae . Mol Microbiol. 2002;46: 1041–51. [DOI] [PubMed] [Google Scholar]
- 34. Kunert A, Losse J, Gruszin C, Hühn M, Kaendler K, Mikkat S, et al. Immune evasion of the human pathogen Pseudomonas aeruginosa: elongation factor Tuf is a factor H and plasminogen binding protein. J Immunol. 2007;179: 2979–88. [DOI] [PubMed] [Google Scholar]
- 35. Sha J, Erova TE, Alyea RA, Wang S, Olano JP, Pancholi V, et al. Surface-expressed enolase contributes to the pathogenesis of clinical isolate SSU of Aeromonas hydrophila . J Bacteriol. 2009;191: 3095–107. 10.1128/JB.00005-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Agarwal S, Kulshreshtha P, Bambah Mukku D, Bhatnagar R. Alpha-enolase binds to human plasminogen on the surface of Bacillus anthracis . Biochim Biophys Acta. 2008;1784: 986–994. 10.1016/j.bbapap.2008.03.017 [DOI] [PubMed] [Google Scholar]
- 37. Candela M, Biagi E, Centanni M, Turroni S, Vici M, Musiani F, et al. Bifidobacterial enolase, a cell surface receptor for human plasminogen involved in the interaction with the host. Microbiology. 2009;155: 3294–303. 10.1099/mic.0.028795-0 [DOI] [PubMed] [Google Scholar]
- 38. Floden AM, Watt JA, Brissette CA. Borrelia burgdorferi Enolase Is a Surface-Exposed Plasminogen Binding Protein. PLoS One. 2001;6: e27502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jong AY, Chen SH, Stins MF, Kim KS, Tuan TL, Huang SH. Binding of Candida albicans enolase to plasmin(ogen) results in enhanced invasion of human brain microvascular endothelial cells. J Med Microbiol. 2003;52: 615–22. [DOI] [PubMed] [Google Scholar]
- 40. Miles LA, Dahlberg CM, Plescia J, Felez J, Kato K, Plow EF. Role of cell-surface lysines in plasmin(ogen)-binding to cells: identification of alpha-enolase as a candidate plasminogen receptor. Biochemistry. 1991;30: 1682–1691. [DOI] [PubMed] [Google Scholar]
- 41. Redlitz A, Fowler BJ, Plow EF, Miles LA. The role of an enolase-related molecule in plasminogen binding to cells. Eur J Biochem. 1985;227: 407–15. [DOI] [PubMed] [Google Scholar]
- 42. Antikainen J, Kuparinen V, Lähteenmäki K, Korhonen TK. Enolases from Gram-positive bacterial pathogens and commensal lactobacilli share functional similarity in virulence-associated traits. FEMS Immunol Med Microbiol. 2007;51: 526–34. [DOI] [PubMed] [Google Scholar]
- 43. Castaldo C, Vastano V, Siciliano RA, Candela M, Vici M, Muscariello L, et al. Surface displaced alfa-enolase of Lactobacillus plantarum is a fibronectin binding protein. Microb Cell Fact. 2009;8: 14 10.1186/1475-2859-8-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vanegas G, Quiñones W, Carrasco-López C, Concepción JL, Albericio F, Avilán L. Enolase as a plasminogen binding protein in Leishmania mexicana . Parasitol Res. 2007;101: 1511–6. [DOI] [PubMed] [Google Scholar]
- 45. Jolodar A, Fischer P, Bergmann S, Buttner DW, Hammerschmidt S, Brattig NW. Molecular cloning of an a-enolase from the human filarial parasite Onchocerca volvulus that binds human plasminogen. Biochim Biophys Acta. 2003;1627: 111–120. [DOI] [PubMed] [Google Scholar]
- 46. Donofrio FC, Calil AC, Miranda ET, Almeida AM, Benard G, Soares CP. Enolase from Paracoccidioides brasiliensis: isolation and identification as a fibronectin-binding protein. J Med Microbiol. 2009;58: 706–13. 10.1099/jmm.0.003830-0 [DOI] [PubMed] [Google Scholar]
- 47. Nakajima K, Hamanoue M, Takemoto N, Hattori T, Kato K, Kohsaka S. Plasminogen binds specifically to a alpha-enolase on rat neuronal plasma membrane. J Neurochem. 1994;63: 2048–2057. [DOI] [PubMed] [Google Scholar]
- 48. de la Torre-Escudero E, Manzano-Román R, Pérez-Sánchez R, Siles-Lucas M, Oleaga A. Cloning and characterization of a plasminogen-binding surface associated enolase from Schistosoma bovis . Vet Parasitol. 2010;173: 76–84. 10.1016/j.vetpar.2010.06.011 [DOI] [PubMed] [Google Scholar]
- 49. Carneiro CR, Postol E, Nomizo R, Reis LF, Brentani RR. Identification of enolase as a laminin-binding protein on the surface of Staphylococcus aureus . Microbes Infect. 2004;6: 604–608. [DOI] [PubMed] [Google Scholar]
- 50. Kinnby B, Booth NA, Svensater G. Plasminogen binding by oral streptococci from dental plaque and inflammatory lesions. Microbiology. 2008;154: 924–931. 10.1099/mic.0.2007/013235-0 [DOI] [PubMed] [Google Scholar]
- 51. Jones MN, Holt RG. Cloning and characterization of an alpha-enolase of the oral pathogen Streptococcus mutans that binds human plasminogen. Biochem Biophys Res Communication. 2007;364: 924–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kolberg J, Aase A, Bergmann S, Herstad TK, Rødal G, Frank R. Streptococcus pneumoniae enolase is important for plasminogen binding despite low abundance of enolase protein on the bacterial cell surface. Microbiology. 2006;152: 1307–17. [DOI] [PubMed] [Google Scholar]
- 53. Bergmann S, Rohde M, Chhatwal GS, Hammerschmidt S. alpha-Enolase of Streptococcus pneumoniae is a plasmin(ogen)- binding protein displayed on the bacterial cell surface. Mol Microbiol. 2001;40: 1273–1287. [DOI] [PubMed] [Google Scholar]
- 54. Pancholi V, Fischetti VA. alpha-enolase, a novel strong plasmin(ogen) binding protein on the surface of pathogenic streptococci. J Biol Chem. 1998;273: 14503–14515. [DOI] [PubMed] [Google Scholar]
- 55. Esgleas M, Li Y, Hancock MA, Harel J, Dubreuil JD, Gottschalk M.Isolation and characterization of alpha-enolase, a novel fibronectin-binding protein from Streptococcus suis . Microbiology. 2008;154: 2668–2679. 10.1099/mic.0.2008/017145-0 [DOI] [PubMed] [Google Scholar]
- 56. Tunio SA, Oldfield NJ, Berry A, Ala'Aldeen DA, Wooldridge KG, Turner DP. The moonlighting protein fructose-1, 6-bisphosphate aldolase of Neisseria meningitidis: surface localization and role in host cell adhesion. Mol Microbiol. 2010;76: 605–15. 10.1111/j.1365-2958.2010.07098.x [DOI] [PubMed] [Google Scholar]
- 57. Matta SK, Agarwal S, Bhatnagar R. Surface localized and extracellular Glyceraldehyde-3-phosphate dehydrogenase of Bacillus anthracis is a plasminogen binding protein. Biochim Biophys Acta. 2010;1804: 2111–20. 10.1016/j.bbapap.2010.08.004 [DOI] [PubMed] [Google Scholar]
- 58. Gozalbo D, Gil-Navarro I, Azorin I, Renau-Piqueras J, Martinez JP, Gil ML. The cell wall-associated glyceraldehyde-3-phosphate dehydrogenase of Candida albicans is also a fibronectin and laminin binding protein. Infect Immun. 1998;66: 2052–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Aguilera L, Giménez R, Badia J, Aguilar J, Baldoma L. NAD+-dependent post-translational modification of Escherichia coli glyceraldehyde-3-phosphate dehydrogenase. Int Microbiol. 2009;12: 187–192. [PubMed] [Google Scholar]
- 60. Hurmalainen V, Edelman S, Antikainen J, Baumann M, Lähteenmäki K, Korhonen TK. Extracellular proteins of Lactobacillus crispatus enhance activation of human plasminogen. Microbiology. 2007;153: 1112–1122. [DOI] [PubMed] [Google Scholar]
- 61. Kinoshita H, Uchida H, Kawai Y, Kawasaki T, Wakahara N, Matsuo H, et al. Cell surface Lactobacillus plantarum LA 318 glyceraldehyde-3-phosphate dehydrogenase (GAPDH) adheres to human colonic mucin. J Appl Microbiol. 2008;104: 1667–1674. 10.1111/j.1365-2672.2007.03679.x [DOI] [PubMed] [Google Scholar]
- 62. Alvarez RA, Blaylock MW, Baseman JB. Surface localized glyceraldehyde-3-phosphate dehydrogenase of Mycoplasma genitalium binds mucin. Mol Microbiol. 2003;48:1417–1425. [DOI] [PubMed] [Google Scholar]
- 63. Tunio SA, Oldfield NJ, Ala'Aldeen DA, Wooldridge KG, Turner DP. The role of glyceraldehyde 3-phosphate dehydrogenase (GapA-1) in Neisseria meningitidis adherence to human cells. BMC Microbiol. 2010;10: 280 10.1186/1471-2180-10-280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Barbosa MS, Bao SN, Andreotti PF, de Faria FP, Felipe MS, dos Santos Feitosa L, et al. (2006) Glyceraldehyde 3-phosphate dehydrogenase of Paracoccidioides brasiliensis is a cell surface protein, involved in fungal adhesion to extracellular matrix proteins and interaction with cells. Infect Immun. 2006;74: 382–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Modun B, Williams P. The staphylococcal transferrin-binding protein is a cell wall glyceraldehyde-3-phosphate dehydrogenase. Infect Immun. 1999;67: 1086–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Winram SB, Lottenberg R. The plasmin-binding protein Plr of group A streptococci is identified as glyceraldehyde-3-phosphate dehydrogenase. Microbiology. 1996;142: 2311–2320. [DOI] [PubMed] [Google Scholar]
- 67. Seifert KN, McArthur WP, Bleiweis AS, Brady LJ. Characterization of group B streptococcal glyceraldehyde-3-phosphate dehydrogenase: surface localization, enzymatic activity, and protein-protein interactions. J Microbiol. 2003;49: 350–6. [DOI] [PubMed] [Google Scholar]
- 68. Bergmann S, Rohde M, Hammerschmidt S. Glyceraldehyde-3-phosphate dehydrogenase of Streptococcus pneumoniae is a surface-displayed plasminogen-binding protein. Infect Immun. 2004;72: 2416–2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Pancholi V, Fischetti VA. A major surface protein on group A streptococci is a glyceraldehyde-3-phosphate-dehydrogenase with multiple binding activity. J Exp Med. 1992;176: 415–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Jin H, Song YP, Boel G, Kochar J, Pancholi V. Group A streptococcal surface GAPDH, SDH, recognizes uPAR/CD87 as its receptor on the human pharyngeal cell and mediates bacterial adherence to host cells. J Mol Biol. 2005;350: 27–41. [DOI] [PubMed] [Google Scholar]
- 71. Jobin MC, Brassard J, Quessy S, Gottschalk M, Grenier D. Acquisition of host plasmin activity by the Swine pathogen Streptococcus suis serotype 2. Infect Immun. 2004;72: 606–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Lama A, Kucknoor A, Mundodi V, Alderete JF. Glyceraldehyde-3-phosphate dehydrogenase is a surface-associated, fibronectin-binding protein of Trichomonas vaginalis . Infect Immun. 2009;77: 2703–2711. 10.1128/IAI.00157-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kainulainen V, Loimaranta V, Pekkala A, Edelman S, Antikainen J, Kylväjä R. (2012) Glutamine synthetase and glucose-6-phosphate isomerase are adhesive moonlighting proteins of Lactobacillus crispatus released by epithelial cathelicidin LL-37. J Bacteriol. 2012;194: 2509–19. 10.1128/JB.06704-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Brix K, Summa W, Lottspeich F, Herzog V. Extracellularly occurring histone H1 mediates the binding of thyroglobulin to the cell surface of mouse macrophages. J Clin Invest. 1998;102: 283–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Goulhen F, Hafezi A, Uitto VJ, Hinode D, Nakamura R, Grenier D, et al. Subcellular localization and cytotoxic activity of the GroEL-like protein isolated from Actinobacillus actinomycetemcomitans . Infect Immun. 1998;66: 5307–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wuppermann FN, Mölleken K, Julien M, Jantos CA, Hegemann JH. Chlamydia pneumoniae GroEL1 protein is cell surface associated and required for infection of HEp-2 cells. J Bacteriol. 2008;190: 3757–67. 10.1128/JB.01638-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Hennequin C, Porcheray F, Waligora-Dupriet A, Collignon A, Barc M, Bourlioux P, et al. GroEL (Hsp60) of Clostridium difficile is involved in cell adherence. Microbiology. 2001;147: 87–96. [DOI] [PubMed] [Google Scholar]
- 78. Frisk A, Ison CA, Lagergård T. GroEL heat shock protein of Haemophilus ducreyi: association with cell surface and capacity to bind to eukaryotic cells. Infect Immun. 1998;66: 1252–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Pantzar M, Teneberg S, Lagergård T. Binding of Haemophilus ducreyi to carbohydrate receptors is mediated by the 58.5-kDa GroEL heat shock protein. Microbes Infect. 2006;8: 2452–8. [DOI] [PubMed] [Google Scholar]
- 80. Kamiya S, Yamaguchi H, Osaki T, Taguchi H. A virulence factor of Helicobacter pylori: role of heat shock protein in mucosal inflammation after H. pylori infection. J Clin Gastroenterol. 1998;27 Suppl 1: S35–9. [DOI] [PubMed] [Google Scholar]
- 81. Yamaguchi H, Osaki T, Kurihara N, Taguchi H, Hanawa T, Yamamoto T, et al. Heat-shock protein 60 homologue of Helicobacter pylori is associated with adhesion of H. pylori to human gastric epithelial cells. J Med Microbiol. 1997;46: 825–31. [DOI] [PubMed] [Google Scholar]
- 82. Long KH, Gomez FJ, Morris RE, Newman SL. Identification of heat shock protein 60 as the ligand on Histoplasma capsulatum that mediates binding to CD18 receptors on human macrophages. J Immunol. 2001;170: 487–94. [DOI] [PubMed] [Google Scholar]
- 83. Bocharov AV, Vishnyakova TG, Baranova IN, Remaley AT, Patterson AP, Eggerman TL. Heat shock protein 60 is a high-affinity high-density lipoprotein binding protein. Biochem Biophys Res Commun. 2000;277: 228–235. [DOI] [PubMed] [Google Scholar]
- 84. Bergonzelli GE, Granato D, Pridmore RD, Marvin-Guy LF, Donnicola D, Corthésy-Theulaz IE. GroEL of Lactobacillus johnsonii La1 (NCC 533) is cell surface associated: potential role in interactions with the host and the gastric pathogen Helicobacter pylori . Infect Immun. 2006;74: 425–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Garduño RA, Garduño E, Hoffman PS. Surface-associated hsp60 chaperonin of Legionella pneumophila mediates invasion in a HeLa cell model. Infect Immun. 1998;66: 4602–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Hickey TB, Thorson LM, Speert DP, Daffé M, Stokes RW. Mycobacterium tuberculosis Cpn60.2 and DnaK are located on the bacterial surface, where Cpn60.2 facilitates efficient bacterial association with macrophages. Infect Immun. 2009;77: 3389e401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Tsugawa H, Ito H, Ohshima M, Okawa Y. Cell adherence-promoted activity of Plesiomonas shigelloides groEL. J Med Microbiol. 2007;56: 23–9. [DOI] [PubMed] [Google Scholar]
- 88. Ensgraber M, Loos M. A 66-kilodalton heat shock protein of Salmonella typhimurium is responsible for binding of the bacterium to intestinal mucus. Infect Immun. 1992;60: 3072–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Mölkänen T, Tyynelä J, Helin J, Kalkkinen N, Kuusela P. Enhanced activation of bound plasminogen on Staphylococcus aureus by staphylokinase. FEBS Lett. 2002;517: 72–78. [DOI] [PubMed] [Google Scholar]
- 90. Kinhikar AG, Vargas D, Li H, Mahaffey SB, Hinds L, Belisle JT, et al. Mycobacterium tuberculosis malate synthase is a laminin-binding adhesin. Mol Microbiol. 2006;60: 999–13. [DOI] [PubMed] [Google Scholar]
- 91. Hussain M, Peters G, Chhatwal GS, Herrmann M. A lithium chloride-extracted, broad-spectrum-adhesive 42-kilodalton protein of Staphylococcus epidermidis is ornithine carbamoyltransferase. Infect Immun. 1999;67: 6688–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Meza-Cervantez P, González-Robles A, Cárdenas-Guerra RE, Ortega-López J, Saavedra E, Pineda E, Arroyo R. Pyruvate:ferredoxin oxidoreductase (PFO) is a surface-associated cell-binding protein in Trichomonas vaginalis and is involved in trichomonal adherence to host cells. Microbiology. 2011;157: 3469–82. 10.1099/mic.0.053033-0 [DOI] [PubMed] [Google Scholar]
- 93. Daniely D, Portnoi M, Shagan M, Porgador A, Givon-Lavi N, Ling E, et al. Pneumococcal 6-phosphogluconate-dehydrogenase, a putative adhesin, induces protective immune response in mice. Clin Exp Immunol. 2006;144: 254–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Boone TJ, Burnham CA, Tyrrell GJ. Binding of group B streptococcal phosphoglycerate kinase to plasminogen and actin. Microb Pathog. 2011;51: 255–261. 10.1016/j.micpath.2011.06.005 [DOI] [PubMed] [Google Scholar]
- 95. Fulde M, Bernardo-García N, Rohde M, Nachtigall N, Frank R, Preissner KT, et al. Pneumococcal phosphoglycerate kinase interacts with plasminogen and its tissue activator. Thromb Haemost. 2013;111(3). [DOI] [PubMed] [Google Scholar]
- 96. Reddy VM, Suleman FG. Mycobacterium avium superoxide dismutase binds to epithelial cell aldolase, glyceraldehyde-3-phosphate dehydrogenase and cyclophilin A. Microb Pathog. 2004;36: 67–74. [DOI] [PubMed] [Google Scholar]
- 97. Reddy VM, Kumar B. Interaction of Mycobacterium avium complex with human respiratory epithelial cells. J Infect Dis. 2000;181: 1189–93. [DOI] [PubMed] [Google Scholar]
- 98. Pereira LA, Báo SN, Barbosa MS, da Silva JL, Felipe MS, de Santana JM, et al. Analysis of the Paracoccidioides brasiliensis triosephosphate isomerase suggests the potential for adhesin function. FEMS Yeast Res. 2007;7: 1381–8. [DOI] [PubMed] [Google Scholar]
- 99. Yamaguchi M, Ikeda R, Nishimura M, Kawamoto S. Localization by scanning immunoelectron microscopy of triosephosphate isomerase, the molecules responsible for contact-mediated killing of Cryptococcus, on the surface of Staphylococcus. Microbiol Immunol. 2010;54: 368–370. 10.1111/j.1348-0421.2010.00225.x [DOI] [PubMed] [Google Scholar]
- 100. Collen D, Verstraete M. Molecular biology of human plasminogen. II. Metabolism in physiological and some pathological conditions in man. Thromb Diath Haemorrh. 1975;34: 403–408. [PubMed] [Google Scholar]
- 101. Brocchieri L, Karlin S. Protein length in eukaryotic and prokaryotic proteomes. Nucleic Acids Res. 2005;33: 3390–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Skovgaard M, Jensen LJ, Brunak S, Ussery D, Krogh A. On the total number of genes and their length distribution in complete microbial genomes. Trends Genet. 2001;17:425–8. [DOI] [PubMed] [Google Scholar]
- 103. Schwartz R, Ting CS, King J. Whole proteome pI values correlate with subcellular localizations of proteins for organisms within the three domains of life. Genome Res. 2001;11: 703–9. [DOI] [PubMed] [Google Scholar]
- 104. Ikai A. Thermostability and aliphatic index of globular proteins. J Biochem. 1980;88: 1895–8. [PubMed] [Google Scholar]
- 105. Tompa P, Szász C, Buday L. Structural disorder throws new light on moonlighting. Trends Biochem Sci. 2005;30: 484–9. [DOI] [PubMed] [Google Scholar]
- 106. Jeffery CJ. Moonlighting proteins--an update. Mol Biosyst. 2009;5: 345–50. 10.1039/b900658n [DOI] [PubMed] [Google Scholar]
- 107. Gancedo C, Flores CL. Moonlighting proteins in yeasts. Microbiol Mol Biol Rev. 2008;72: 197–210. 10.1128/MMBR.00036-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Jeffery CJ, Bahnson BJ, Chien W, Ringe D, Petsko GA. Crystal structure of rabbit phosphoglucose isomerase, a glycolytic enzyme that moonlights as neuroleukin, autocrine motility factor, and differentiation mediator. Biochemistry. 2000;39: 955–64. [DOI] [PubMed] [Google Scholar]
- 109. Bergmann S, Wild D, Diekmann O, Frank R, Bracht D, Chhatwal GS. Identification of a novel plasmin(ogen)-binding motif in surface displayed alpha-enolase of Streptococcus pneumoniae . Mol Microbiol. 2003;49: 411–23. [DOI] [PubMed] [Google Scholar]
- 110. Ehinger S, Schubert WD, Bergmann S, Hammerschmidt S, Heinz DW. Plasmin(ogen)-binding alpha-enolase from Streptococcus pneumoniae: crystal structure and evaluation of plasmin(ogen)-binding sites. J Mol Biol. 2004;343: 997–1005. [DOI] [PubMed] [Google Scholar]
- 111. Piatigorsky J. Lens crystallins and their genes: diversity and tissue-specific expression. FASEB J. 1989;3: 1933–40. [DOI] [PubMed] [Google Scholar]
- 112. Ghosh K, Dill KA. Computing protein stabilities from their chain lengths. Proc Natl Acad Sci U S A. 2009;106:10649–54. 10.1073/pnas.0903995106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Lipman DJ, Souvorov A, Koonin EV, Panchenko AR, Tatusova TA. The relationship of protein conservation and sequence length. BMC Evol Biol. 2002;2:20–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Schneewind O, Mihaylova-Petkov D, Model P. Cell wall sorting signals in surface proteins of Gram-positive bacteria. EMBO J. 2993;12: 4803–11. [DOI] [PMC free article] [PubMed] [Google Scholar]