Abstract
Motivation: The data gathered by the Pan-Cancer initiative has created an unprecedented opportunity for illuminating common features across different cancer types. However, separating tissue-specific features from across cancer signatures has proven to be challenging. One of the often-observed properties of the mutational landscape of cancer is the mutual exclusivity of cancer driving mutations. Even though studies based on individual cancer types suggested that mutually exclusive pairs often share the same functional pathway, the relationship between across cancer mutual exclusivity and functional connectivity has not been previously investigated.
Results: We introduce a classification of mutual exclusivity into three basic classes: within tissue type exclusivity, across tissue type exclusivity and between tissue type exclusivity. We then combined across-cancer mutual exclusivity with interactions data to uncover pan-cancer dysregulated pathways. Our new method, Mutual Exclusivity Module Cover (MEMCover) not only identified previously known Pan-Cancer dysregulated subnetworks but also novel subnetworks whose across cancer role has not been appreciated well before. In addition, we demonstrate the existence of mutual exclusivity hubs, putatively corresponding to cancer drivers with strong growth advantages. Finally, we show that while mutually exclusive pairs within or across cancer types are predominantly functionally interacting, the pairs in between cancer mutual exclusivity class are more often disconnected in functional networks.
Contact: przytyck@ncbi.nlm.nih.gov
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
The Pan-Cancer initiative surveyed genetic and epigenetic aberrations in cancer samples from thousands of cancer patients over 12 cancer types (Cancer Genome Atlas Research et al., 2013). This data opened the door to a systematic examination of the similarities and differences among multiple cancer types. However, integrative analysis for subtype classifications from five ‘omics’ platforms revealed that ‘cell-of-origin’ features dominate the molecular taxonomy of diverse tumor types rather than discovering common features across tissue types. Although such an outcome may be partly due to the tissue specificity of some cancer driver genes and pathways, it can also be attributed to the lack of power to untangle mixed signals of cell-type-specific and cancer-specific features (Cancer Genome Atlas Research et al., 2013).
Despite these strong tissue-specific signals, it has also become evident that many mutated genes, and even whole dysregulated subnetworks, are shared by multiple cancer types (Lawrence et al., 2014; Leiserson et al., 2013, 2014). Pathway-centric methods have emerged as a key approach to empower studies of complex diseases with heterogeneous signals by focusing on the genetic activities on the level of biological pathways (subnetworks) rather than individual genes. Several computational methods have been developed to utilize genotypic data for the identification of mutated subnetworks (Cho et al., 2012; Vandin et al., 2011, 2012a, b) and stratification of cancer subtypes (Hofree et al., 2013).
One of the often-observed properties of the mutational landscape of cancer is the mutual exclusivity of cancer driving mutations (Thomas et al., 2007). Mutually exclusive mutations observed among genes in the same functional pathway prompted the development of computational methods that utilize mutual exclusivity in the context of identifying pathways dysregulated in cancer as well as for the pathway independent identification of mutually exclusive cancer drivers (Ciriello et al., 2012, 2013, Leiserson et al., 2013; Szczurek and Beerenwinkel, 2014; Vandin et al., 2012a, b). However, the sets of mutually exclusive drivers previously obtained in Pan-Cancer analysis (Kandoth et al., 2013; Szczurek and Beerenwinkel, 2014) predominantly contain tissue type-specific genes and do not necessarily share common pathways, indicating that the previous definition of mutual exclusivity is not sufficient in Pan-Cancer analysis and it is necessary to recognize different mutual exclusivity classes.
To this end, we started by classifying mutual exclusivity patterns into three basic classes: within tissue type exclusivity (mutual exclusivity observed only in one cancer type), across tissue type exclusivity (mutual exclusivity common to several tissue types), and between tissue type exclusivity (mutual exclusivity between putative tissue-specific drivers). We developed statistical tests that allow us to identify the membership of gene sets to these mutual exclusivity classes. We then aimed to use the principle of mutual exclusivity for identification of subnetworks dysregulated across multiple cancer types. To identify Pan-Cancer dysregulated modules, we utilized the across cancer mutual exclusivity measure jointly with the interaction data from a network. Interestingly, we found that functionally interacting gene pairs are more likely to be involved in across tissue type mutual exclusivity while between type exclusivity is more often observed in gene pairs that are not functionally related.
Adapting the Module Cover approach (Kim et al., 2013), we uncovered subnetworks that are dysregualted in multiple cancer types. Module Cover is an extension of the classical Multi-Set Cover approach, utilized before to find disease marker genes for highly heterogeneous genomic data, to a network setting. This is an optimization technique that is particularly suitable in the context of heterogonous data. The approach essentially aims to identify a set of dysregulated subnetworks so that a desired subnetwork score is optimized, while at the same time, ensuring that altered genes from different cancer samples are present in the union of the selected subnetworks. Our proposed algorithm, Mutual Exclusivity Module Cover (MEMCover), uses the module cover strategy to combine mutual exclusivity feature with functional interactions and genomic aberration data from individual Pan-Cancer cases to uncover Pan-Cancer dysregulated subnetworks. Unlike previous approaches for finding a set of genetic aberrations exclusive between all pairs, MEMCover does not necessarily seek to identify subnetworks with complete exclusivity among the genes, (Leiserson et al., 2013; Szczurek and Beerenwinkel, 2014; Vandin et al., 2012a, b) but utilizes pairwise mutual exclusivity to complement interaction data for a better identification of subnetworks dysregulated in Pan-Cancer samples.
When applied to Pan-Cancer data, MEMCover identified a large number of subnetworks dysregulated across many cancer types. We found that many of the identified subnetworks are linked to cancer-related pathways, overlapping with all but one subnetwork with at least 10% coverage recently identified by HotNet2 in Pan-Caner analyses (Leiserson et al., 2014) despite the fact that MEMCover is based on completely different principle from HotNet2. We also identified subnetworks whose role across cancer types has not been appreciated previously.
Finally, our analysis also suggests the existence of mutual exclusivity hubs—genes whose genetic aberrations are mutually exclusive with aberrations in a large number of other genes, both connected and not connected in the network, indicating that these mutual exclusivity hubs may correspond to cancer driver genes that have particularly strong growth advantages.
2 Classification of mutual exclusivity in pan-cancer
2.1 Mutual exclusivity classes
Although mutually exclusive mutations are often observed in cancer samples, the underlying cause of mutual exclusivity is not always clear and might vary depending on the context. Gene pairs with mutually exclusive aberrations might correspond to different drivers dysregulating the same pathway or might be tissue-specific cancer drivers dysregulating completely different pathways. A mutually exclusive pair can also be specific in only one tissue type or might be common to multiple cancer types. Observing different mutual exclusive patterns, we start by classifying mutual exclusivity into the following classes (Fig. 1):
Mutual Exclusivity Within a cancer type (WITTHIN_ME): mutual exclusivity is observed within only one individual tissue type. For example, the RB1 and EGFR pair has a significant WITHIN_ME relationship in GBM as shown in Figure 1A.
Mutual Exclusivity Between cancer types (BETWEEN_ME): mutual exclusivity is observed due to different tissue specificity of a given pair of genetic aberrations. Figure 1B shows an example of BETWEEN_ME, where von Hippel-Lindau (VHL) mutation is specific for KIRC, APC is enriched in CRC and EGFR in GBM.
Mutual Exclusivity Across multiple cancer types (ACROSS_ME): mutual exclusivity is observed in more than one tissue types. It may mean that mutual exclusivity is statistically significant in two or more cancer types or that the statistical significance is increased when considering multiple cancer types relative to within cancer type exclusivity. Figure 1C and D are two examples of ACROSS_ME class.
Fig. 1.
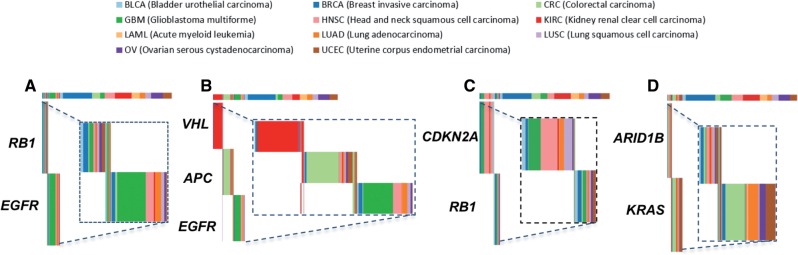
Examples of Mutual Exclusivity Classes. The top bar in each figure shows cancer samples in their cancer type color (see the legend above for the color coding of cancer types) and each row below the top bar indicates the presence or the absence of alteration of a given gene in the corresponding cancer samples. The insets show the zoom-in views for the set of patients with alterations in at least one gene. (A) RB1 and EGFR show WITHIN_ME pattern with respect to GBM. While, in the inset, we can see that the exclusivity may also be present in other cancer types it is not statistically significant (compare the width of the altered samples to the width for all samples in the top bar) (B) VHL, APC and EGFR show BETWEEN_ME pattern. VHL mutation is specific for KIRC, APC is enriched in CRC and EGFR in GBM. (C) and (D) show the examples of ACROSS_ME. The CDKN2A and RB1genes shown in panel C also satisfy WITHIN_ME with respect to GBM and LUSC while the ARID1B, KRAS pair shown in panel D is not significant in any TS_test of individual cancer type separately
It is important to keep in mind that this classification is data dependent, and expanding the dataset might change membership in some classes. In particular, ACROSS_ME pairs that are not detected as WITHIN_ME in the individual tissues (e.g. ARID1B, KRAS in Fig. 1D) are likely to be detected as such if a larger sample size was available.
2.2 Estimating mutual exclusivity classes
Permutation test is a commonly used technique to estimate mutual exclusivity in cancer mutation profiles (Ciriello et al., 2013; Vandin et al., 2012a, b). In the approach, the null model is created by permuting a given mutation profile while preserving the mutation rates of individual genes and samples. If the mutations of a pair of genes are mutually exclusive, they will collectively cover more samples than expected by chance. The significance of mutual exclusivity is measured by counting the number of random instances in which more samples are covered than in the original mutation profile. We extended the permutation technique to estimate each of the Pan-cancer ME classes defined in Section 2.1. We first describe the tests and then explain how they are used to assess the exclusivity classes of gene pairs.
Type Separate Permutation Test of an Individual Tissue Type (TS_test): Mutual Exclusivity within a cancer type is estimated separately by permuting mutation profiles independently for each cancer type. To preserve the mutation rates of each gene and each sample, in each iteration, two (gene, sample) pairs are randomly chosen and swapped. A permuted mutation profile is obtained after a certain number of iterations. For a gene g, let COVERC(g) be the set of samples in cancer type C with gene g mutated. Given a set of genes A, COVERC(A) = Ug in A COVERC(g). The significance of ME is estimated by counting the number of random instances in which |COVERc(A)| is bigger than the size in the original mutation profile. The size of COVERc(A) is likely to be bigger than expected if the mutations of the genes in A are mutually exclusive (Ciriello et al., 2013; Vandin et al., 2012a). In Figure 2A, gene 1 and gene 2 are mutually exclusive and the size of the cover in the original mutation is bigger than the permutated instances.
Type Restricted Permutation Test with All Pan Cancer Samples (TR_test): In this test, we run permutation for each cancer separately as in the TS_test and compute the normalized sum of cover size, NCOVER(A)=∑C NCOVERC(A) where NCOVERC(A) = |COVERc(A)|/|C| and |C| is the number of samples in cancer type C. The cover size is divided by |C| to remove the impact of different sample sizes of cancer types. The empirical P-value is computed based on the rank of NCOVER(A). Figure 2B gives an illustration of TR test, where the significance of ME is increased by combining the samples from cancer type A and B. Note that because the mutations are permuted separately for each type, the mutations from different cancer types will never be swapped with each other, which prevents a pair of genes whose mutations are specific to two different cancer types (i.e. BETWEEN_ME class) from being assigned a low P-value (the second panel in Fig. 2C)
Type Oblivious Permutation Test with All Pan Samples (TO_test): We applied the permutation test described in TS_test to all Pan-Cancer samples and estimate the significance of the mutual exclusivity. That is, we considered all cancer samples and repeatedly performed swapping of two randomly selected (gene, sample) pairs regardless of their cancer types. The bottom panel in Figure 2C shows an illustration of TO test. The mutations are specific to two different cancer types and the mutual exclusivity may be significant in TO test but not in TR_test.
Fig. 2.
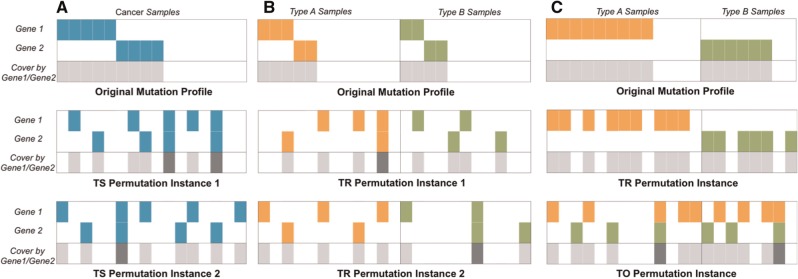
Illustrations of Different Permutation Tests. (A) TS Test: the first two rows of each panel show the mutation profiles of gene 1 and gene 2 and the bottom row shows the samples covered by either gene 1 or gene 2. The darker gray color means that the samples are covered by both of the genes. The permuted instances cover less samples than the original mutation profile, indicating the significance of mutual exclusivity (B) TR test: when two cancer types are considered together, gene 1 and gene 2 are mutually exclusive in both cancer types but they may not be statistically significant when tested separately in each cancer type as only small subset of samples have mutations. The mutual exclusivity of rare mutations can be picked up when the samples are combined and tested together. (C) TO versus TR test: When the mutations of a pair of genes have BETWEEN_ME relationship, they will not be found statistically significant in TR test as permutation is performed separately, but may be significant in TO test
The gene pairs passing TS_test for exactly one tissue type with a more significant P-value than in TR_test are naturally assigned to the WIHTIN_ME class. The ACROSS_ME class contains the gene pairs that pass TS_test for more than one tissue or pass TR_test (with a more significant P-value than in TS_test). Note that by summing up the size of covers across all cancer types, the statistically power to identify ME patterns existing in multiple cancer types can be increased, allowing for detecting across-cancer mutual exclusivity that might not be detected when each tissue type is considered individually.
Finally, we assign a pair to the BETWEEN_ME class if the pair pass TO_test but none of the other tests. Indeed, TO_test can potentially capture mutually exclusive pairs of all three types. However, in the case where the overall mutation rates of a given set of genes are low and the mutations are associated with only a few cancer types, permuting samples across cancer types may wipe out existing ME patterns especially for WITHIN_ME and ACROSS ME classes. Thus, TS_test and TR_test are more accurate for the purpose of detecting these two classes, whereas TO_test is used to capture gene sets in the BETWEEN_ME class provided that other classes of ME are not present. We confirmed the correctness of this assignment in the discussion below.
2.3 ME classes of interacting gene pairs
Dataset: We obtained Pan-Cancer mutation profiles by collecting somatic mutation and Copy Number Variation (CNV) data for a set of 11 different types (COAD and READ are combined into one type) of cancers from TCGA with total of 3182 cancer samples. We then created 10 000 randomly permuted profiles using both type-separate and type-oblivious permutations. Note that TS and TR tests use the same permutation method by permuting samples independently in each cancer type but differ in the way to compute P-values. For each permutation instance, we performed edge swapping 1000*|E| times (|E| is the number of edges in the network) as suggested in the previous study (Milo et al., 2003).
As we focus on finding cancer drivers belonging to the same pathways and their mutual exclusivity patterns, we utilized a functional interaction network, HumanNet downloaded from http://www.functionalnet.org/humannet/ and performed the ME test to classify the interacting pairs into different ME classes. HumanNet is a functional gene network, including 18 714 validated protein-encoding genes of Homo Sapiens and 474 913 interactions. The network is significantly dense compared with other available human interaction networks as the network is constructed by a Bayesian integration of 21 different types of ‘omics’ data including expression profiles, protein interactions, genetic interactions etc. We tested with another interaction network and found similar results (Section 3.3 and Supplementary Material). We also tested non-interacting pairs and discuss the results in Section 3.3.
Among more than 450 K pairs we tested, there are over 3000 pairs of genes that are significant in the TO_test (P < 0.01) but none in other tests. The gene pairs in the category are most likely to have BETWEEN_ME relationships and each gene has its distinct tissue specificity. To confirm that these genes indeed have tissue specificity, we performed a hypergeometric test for each cancer type and examined if the gene is significantly tissue specific, i.e., has significantly more (or less) mutated samples in a given cancer type (P-value cutoff < 0.01). For example, VHL mutation is significantly overrepresented in KIRC and underrepresented in all other cancer types. Mutually exclusive tissue specificity for a pair of genes is subsequently tested by checking if there is at least one cancer type in which the mutation of one gene is overrepresented and the other is underrepresented (or vice versa). We found that for the pairs that were significant only in the TO_TEST, 73% of the interactions (2405 out of 3292 pairs) have mutually exclusive tissue specificity and thus confirmed that they can be properly assigned to the BETWEEN_ME class while for the interactions in other categories, only 29% of them (1753 out of 5986 pairs) have mutually exclusive tissue specificity.
We also note that 135 interactions were found to be significant in more than one cancer types in TS-test (P < 0.01) and all but 15 of those interactions were also captured by TR_test, suggesting TR_test captures most of ACROSS_ME pairs. In addition, there are almost 4000 pairs that are significant in the TR_test but none in the TS_test, demonstrating the increased power of the TR_test by combining samples from multiple tissue types.
2.4 ACROSS_ME pairs are clustered
We conjectured that if the existence of ACROSS_ME pairs is due to the fact that the corresponding genes belong to the same pathways, the ACROSS_ME pairs should be close to each other in an interaction network. To examine if ME edges are more clustered than expected, we selected 4722 edges with empirical P-value < 0.01 (TR_TEST) and constructed a subnetwork consisting of those selected edges. The network included 217 connected components with only 160 edges (∼3%) not adjacent to other ME edges. We also created 100 random subnetworks using a random sampling of the same number of edges (a degree preserving permutation test via edge shuffling is not applicable in this case because ME scores are tied with gene pairs and original ME scores cannot be preserved after shuffling edges) and found that the random networks have an average of over 853 connected components with more than 603 single edges on average. The ME subnetwork also has a clustering coefficient of 0.015, confirming it is more clustered than expected (empirical P-value < 0.01). We note that nodes with high degree in HumanNet are more likely to have ME edges (Spearman Correlation Coefficient: 0.41) and the clustering of ME edges can be due to the existence of those hubs. However, we found that random subnetworks of the same size have even higher correlation in degree distributions (the Average Spearman Correlation Coefficient = 0.59) i.e., hubs are more likely to be selected in random sampling but the random networks are not as clustered as the ME network. This suggests that ME edges are clearly not randomly placed in the network but there may be some nodes incident to many ME edges, namely ME hubs. For example, KRAS and TP53 have as many as 236 and 141 ACROSS_ME edges, respectively. ME hubs may not necessarily be the nodes with the highest degree in the original network. We discuss ME hubs in more detail below (Section 3.3). The ME degree of a node (the number of ME edges incident to the node) also has a slight correlation with the number samples for which the gene is altered (Spearman Correlation Coefficient = 0.19).
3 Finding pan-cancer dysregulaed modules using functional network and mutual exclusivity
3.1 The MEMCover algorithm
Motivated by the fact that ME edges are more clustered than expected, in the next step we aim to find a set of gene modules where genes in the same module are mutually exclusive and/or functionally related. We adapted the Module Cover algorithm, which was developed to identify a set of disease signature modules and successfully uncovered several important cancer related subnetworks in GBM and Ovarian Cancer samples (Kim et al., 2013). In the Module Cover algorithm, genes are greedily selected to cover disease samples while simultaneously creating modules of ‘closely related’ genes. In MEMCover, we improved the method by utilizing ACROSS_ME levels obtained in the previous section when we compute the weights of edges.
Like Module Cover, MEMCover finds a set of modules with the minimum total cost while covering all disease samples at least k times. We say that a gene covers a sample [cover(c, g) = 1] if the gene is altered in the sample. Because we assume that each cancer case has more than one driver gene, we require that each sample has to be covered at least k times where k is the input parameter of algorithm (see parameter selection in Supplementary Material). Formally, the goal is to find a minimum cost set of modules (subnetworks) that covers all disease cases at least k times. In other words, we search for a module set that minimizes while for each disease sample c, . Module cost Cost(M) is computed based on the edge weights inside module M as follows:
| (1) |
where . The module cost decreases as the average weight inside module increases, which ensures a selection of subnetworks with heavy weight edges. We define edge weight (−1 ≤ w(x, y) ≤ 1) based on the confidence score in HumanNet and its ACROSS_ME score. We describe the choice of edge weights in Section 3.2 and the parameter selection section in the Supplementary Material in more detail.
Because the optimization problem defined above is NP-hard, MEMCover greedily selects genes and creates modules: in each iteration, we greedily choose a gene with minimum IC(g)/Benefit(g). Benefit(g) is defined to be the number of samples covered by a gene g which are not yet covered k times. The increase in the cost when adding a gene g depends on whether the gene creates a separate module by itself or is added to an existing module. The cost is 1 if it creates a separate module and for the latter case (added to an existing module), the cost is defined as IC(g) = minΜi ε P(g) (Cost(−Cost(Mi))), where P(g) is the subset of existing modules connected to g. The gene is added in a way that it incurs the minimum increased cost. If the minimum cost of gene g is obtained when gene g is added to an existing module M, the module is updated accordingly otherwise a new module is created. We repeat this until we cover all samples by the required number of times.
3.1.1 Post-processing
In the post-processing in MEMCover, we refined the modules by (i) merging two modules if the average edge weight between them is positive, (ii) allowing a gene to be added to more than one module if the average edge weight between the gene and a module is positive.
Merging modules: we consider all possible pairs of selected modules and greedily merge two modules if the average weight between module pairs is positive. More specifically, in each iteration, we take the module pair with the heaviest average weight between the two modules and repeatedly merge them until there is no such pair.
Overlapping modules: so far a gene can belong to only one module. In the overlapping phase, we consider every pair of a selected gene and a module (which does not already include the gene) and check if the average weight between the gene and the module is positive. As in the merging step, we start overlapping with the highest weight pairs first until there is no pair satisfying the condition.
3.2 ME scores help finding cancer signature modules
We utilize the CNV and mutation data for the same 3182 Pan-Cancer samples described in Section 2 and performed MEMCover. A simple extension of the MEMCover method allows us to assign a different coverage rate for different type of alterations. Because somatic mutations are relatively rare and accurate compared to CNV data (Hofree et al., 2013), we weight the coverage rate higher for somatic mutations than CNV. Among different parameters, we chose k = 15 and the coverage rate of mutations (mr) = 3 [i.e. cover(c, g) = 3 if g has mutation and cover(c, g) = 1 if g has CNV]. The parameters result in selecting 5–15 covering alterations for each cancer sample, a number roughly consistent with the expected number of cancer drivers. See Supplementary Materials B for further discussion on the parameter selection.
The edge weight w(e) is defined for each interaction e in HumanNet to be (hn(e) + me(e))/2−f(θ), where hn(e) (0 < hn(e) ≤ 1) is the normalized HumanNet confidence score and me(e) is the normalized −log(P-value) in TR_test (both values are normalized by dividing by the maximum of each). f(θ) is a parameter that we can use to control the trade-off between the number of modules and the average weights within each module; that is to determine how low the two scores (hn(e) and me(e)) are allowed to include a gene in a module instead of creating a separate module. We define θ to be the percentage of edges whose weights remain positive and f(θ) is the corresponding weight threshold (when edges are sorted by their unadjusted weights). Recall that by the definition of the module cost in (1), positive edge weights reduce the module cost and therefore, the bigger θ is [and the less f(θ)], the more edges have positive scores and MEMCover will select bigger modules in general. To compare the impact of combining mutual exclusivity scores in the edge weights, we also ran the algorithm with HumanNet-only weight scheme where w(e) = hn(e)−f(θ).
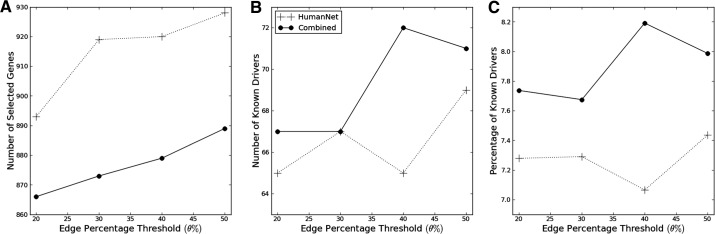
To examine the quality of modules, we utilized a set of 138 known cancer driver genes in (Vogelstein et al., 2013), and checked how many known cancer drivers were identified by MEMCover. Although varying parameters gives different results in terms of the number of selected genes, the number of selected drivers, etc., we found that combining mutual exclusivity scores into edge weights consistently improves the quality of modules as measured by identifying more known cancer drivers while selecting a less number of candidate genes, thus having a high percentage of known cancer drivers (Fig. 3). In particular, we ran MEMCover with varying θ’s in the range of 20, 30, 40 and 50% of the top weighted edges and found that θ = 40% finds the highest number and percentage of known cancer drivers (72 out of 138). In the following subsections, we presented a set of modules obtained with θ = 40%.
Fig. 3.
Genes Selected by MEMCover: more cancer driver genes were selected when ME data is combined. The results are shown for k = 15, mw = 3 and varying θ of 20–50% for top edges (x-axis) (A) Number of total genes selected (B) number of known cancer driver genes selected (C) percentage of known driver genes in the selected gene set
3.3 Properties of pan-cancer dysregulated subnetworks identified by MEMCover
3.3.1 MEMCover identifies subnetworks covering a high percentage of pan-cancer cases
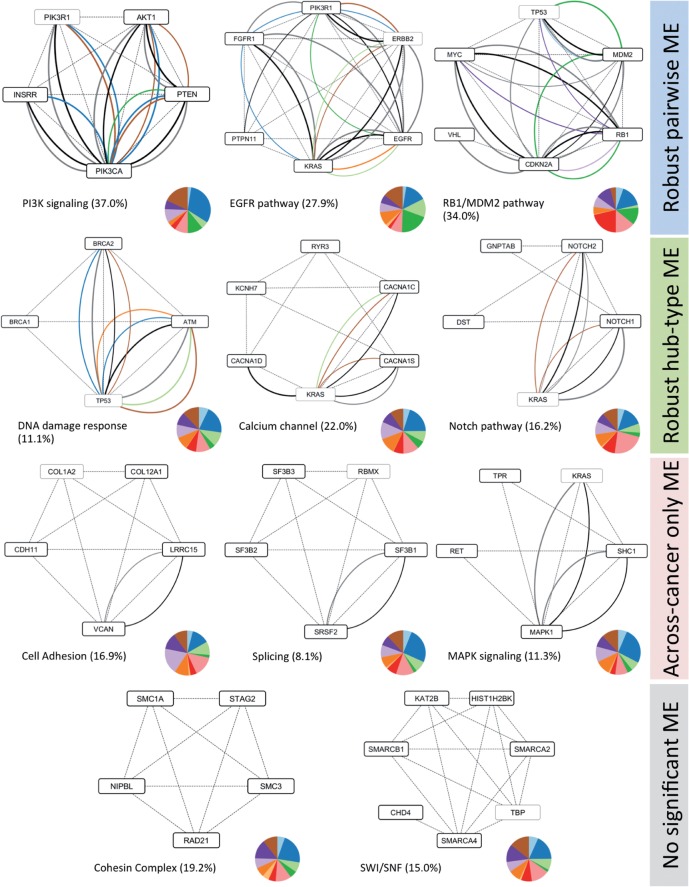
MEMCover algorithm selected 536 modules of size 1–8 (including 325 non-singletons) with 879 genes in total. Nearly half of the modules covered at least 5% of samples and 62 modules provide coverage ∼10%. These statistics are based on the modules before the post-processing extension to allow overlaps. After the overlap allowing module extension, ∼10% of genes (86) belong to more than one module while KRAS, TP53, EGFR, ERBB2, CTNNB1, PPM1L, PRKACA belong to more than five modules. A representative subset of modules is shown in Figure 6 and the full list of modules is provided in Supplementary Materials.
Fig. 6.
Representative modules obtained by MEMCover algorithm. Dashed lines correspond to HumanNet edges, solid lines are for WITHIN_ME colored by its tissue type using Pan-Cancer color coding (see the legend in Fig. 1). Black and gray edges represent ACROSS_ME and BETWEEN_ME, respectively. For each module, we show the percentage of samples with alterations in the module and a pie chart with the distribution of the number of samples with at least one mutation in each tissue type. For genes belonging to more than modules, they were counted according to module membership before extending them to allow overlap (therefore counted only once). Genes are not counted for the coverage and for the pie chart in a given subnetwork are marked as light gray boxes. Not pictured modules include, among others, an additional subnetwork related to SWI/SNF [PBRM1, TOX2, SMARCA4], MHC class one members [HLA, HLB], exocytosis related group [TRPC4, EXOC4, EXOC3, EXOC7, RALA]. The full list of subnetworks is provided in Supplementary Materials. The networks are created using Cytoscape (Shannon et al., 2003)
3.3.2 Mutual exclusivity patterns in selected modules
The selection of modules in MEMCover is guided by not mutual exclusivity only but also HumanNet connectivity and sample coverage. Thus modules uncovered by MEMCover are not necessarily required to contain mutually exclusive pairs. Indeed, as illustrated in Figure 6, we observed modules with robust pairwise exclusivity present between many different pairs, modules with mutual exclusivity converging to only one ‘hub’ gene, modules with mutual exclusivity significant in ACROSS_ME (but not WITHIN_ME), and modules with no significant mutual exclusivity.
3.3.3 Mutual exclusivity hubs
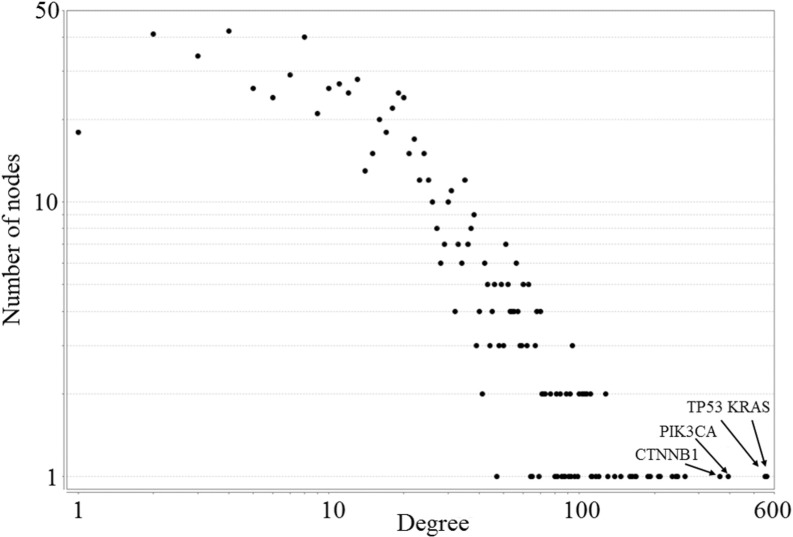
Modules with mutual exclusivity involving one particular gene strongly suggest the existence of ACROSS_ME/WITHIN_ME hubs. To confirm this, we examined the distribution of the number of ME partners among all the genes selected by MEMCover (Fig. 4). We checked all pairs for their mutual exclusivity regardless of their connectivity in HumanNet and chose the partner for which P-value < 0.01 in TR or TS test and where the P-value in TR/TS_test is more significant than in TO_test). Indeed, such Mutual Exclusivity hubs (ME hubs) can be clearly identified (with KRAS, TP53, PIK3CA, CTNNB1, DVL3, GRB7, CTCF at the top of the list, see the Supplementary Table for the extended list). Note that Mutual Exclusivity hub partners are not necessarily HumanNet neighbors. This suggests that ME hubs are likely to have more significant growth advantages than other cancer driver genes. We found that the ME degrees in known driver genes are significantly higher compared with the ME degrees in other genes (P-value < 10−3, Mann-Whitney U test).
Fig. 4.
The distribution of the number of mutually exclusive partners among all the genes selected by MEMCover (regardless of the connectivity in HumanNet)
3.3.4 Non-interacting pairs are enriched with BETWEEN_ME class
The existence of ME hubs calls for re-examining the assumption that mutual exclusivity occurs predominantly between drivers from the same pathway. Towards this end we divided all possible pairs of genes selected by MEMCover into two groups: those that are connected in HumanNet and those that are not. We then considered the distribution of mutual exclusivity classes within each of these two groups. We found that despite the existence of ME hubs whose influence might extend beyond one pathway, the ACROSS_ME class is enriched for HumanNet neighbors while non-HumanNet pairs show a higher percentage of BETWEEN_ME pairs (Fig. 5A). Interestingly, there is only one BETWEEN_ME edge in our top modules (modules with 10% or better coverage)—the edge between VHL and CDKN2A in the module related to the RB1/MDM2 pathway. Although these two genes are connected in HumanNet, the functional relation between them is not obvious. VHL gene (VHL tumor suppressor, E3 ubiquitin protein ligase) is the primary gene for the common, non-hereditary form of clear cell kidney cancer (Cancer Genome Atlas Research, 2013) while CDKN2A and RB1 are hallmarks of lung squamous cell carcinoma (Cancer Genome Atlas Research, 2012) and glioblastoma (Cancer Genome Atlas Research, 2008). Thus, while CDKN2A and RB1 are mutually exclusive within both cancer types, VHL and CDKN2A show between type exclusivity only. We have also tested the hypothesis with another network. The result with a PPI network (HINT+) (Das and Yu, 2012; Leiserson et al., 2014; Yu et al., 2011) was consistent with the one with HumanNet (Fig. 5A) that across-cancer and within cancer mutually exclusive pairs are enriched among pairs of interacting genes while between-cancer exclusivities are enriched for non-interacting gene pairs (Supplementary Material C for more discussion). Interestingly, we found that the physical network includes a bigger proportion of within cancer exclusivity than the functional interaction network does, which suggests that the mutations within the same protein complex are likely to be cancer drivers for the same tissue.
Fig. 5.
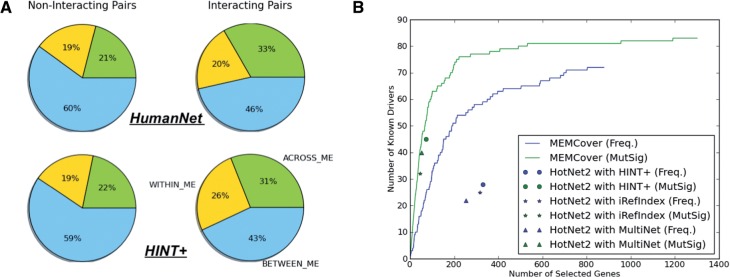
(A) The difference in the distribution in mutual exclusivity classes for interacting and non-interaacting pairs. The first row is for HumanNet and the second row for HINT+ (B) Comparison on the number of known drivers selected between MEMCover and HotNet2. MEMCover finds more driver genes than HotNet2 for the same number of selected genes and the same gene scoring scheme
3.4 MEMCover identifies new pan-cancer dysregulated subnetworks as well as known subnetworks
Due to their connectivity in HumanNet, modules uncovered by MEMCover include genes from specific molecular pathways/complexes and their regulators (Fig. 6). Despite very different algorithmic approach, differences in underlying interaction network, and different search criteria, all but one (ASCOM complex) of the six subnetworks with 10% or more coverage and most of subnetworks with coverage 2–10% reported with HotNet2 (Leiserson et al., 2014) overlap with our pathways. Because MEMCover is optimized, among others things, to find subnetworks with high coverage it found nearly all subnetworks with 10% or better coverage that were reported in (Leiserson et al., 2014) and a large number of non-reported subnetworks. Not surprisingly, our modules are not as related to the sets of mutually exclusive Pan-Cancer genetic aberrations identified in a recent study (Szczurek and Beerenwinkel, 2014), in which they focused on the identification of exclusive cancer drivers and zoomed on tissue type specific genes (for example VHL, APC, EGFR analyzed in Fig 1B) rather than genes which share common pathways (See also Supplementary Table A3).
In addition to finding expected subnetworks, an interesting example we found is the subnetwork containing splicing factor 3B protein complex (SF3B) together with the RBMX gene—a gene implicated in tissue-specific regulation of gene transcription and alternative splicing of several pre-mRNAs and the SRSF2 gene—an (SR)-rich pre-mRNA splicing factors, which constitute part of the spliceosome. Indeed, SF3B1 and SRSF2 have been recently found to be significantly mutated in chronic lymphocytic leukemia (CMML) (Je et al., 2013; Wang et al., 2011). Our analysis shows that spliceosomal machinery is dysregulated across many cancer types and displays ACROSS_ME exclusivity between SF3B and SRSF2 genes.
As for the cohesin complex, this multi-subunit protein complex plays an integral role in sister chromatid cohesion, DNA repair, and meiosis. Proteins from this complex are known to positively regulate the transcription of genes known to be dysregulated in cancer, such as Runx1, Runx3 and Myc as reviewed in (Rhodes et al., 2011). In addition, a recent study reported recurrent mutations and deletions involving multiple components of the cohesin complex, including STAG2, RAD21, SMC1A and SMC3, in different myeloid neoplasms (Kon et al., 2013).
Finally, in the case of calcium channel, there is evidence that the ryanodine receptor 3 gene (RYR3), which encodes a large protein that forms a calcium channel, is important for the growth, morphology, and migration of breast cancer cells (Zhang et al., 2011).
3.5 Comparison with alternative approaches
We compared our results with the pan-cancer subnetworks obtained using HotNet2 (Leiserson et al., 2014). Different from our approach, the method is based on a heat diffusion method to identify significantly mutated subnetworks and mutual exclusivity information was not used in identifying the subnetworks. Leiserson et al. reported that the results with two gene scoring schemes—frequency and MutSig scores. Our results presented in Section 3.4 uses frequency scores for cover rate. For comparison, we also ran MEMCover using MutSig scores as gene cover rate (Fig. 5B). Note that the number of genes selected is different depending on the methods and different parameters and that different networks have been used. Here, we considered the genes selected by MEMCover in the order in which they are added and counted the number of known drivers in the selected subset of genes in each iteration. Our algorithm consistently finds more known driver genes for the same number of selected genes compared with the set obtained by HotNet2. We note that known drivers tend to have much higher MutSig scores, and thus in the context of this comparison, the improvement from using MutSig is expected. But using MutSig scores may reduce the likelihood to find rare de novo cancer drivers as mentioned in the article (Leiserson et al., 2014).
We also compared our results with the work related to mutual exclusivity for Pan-Cancer (Kandoth et al., 2013; Szczurek and Beerenwinkel, 2014). Their approaches differ from our work in that the goal is to find a set of mutually exclusive genes regardless of their interaction relationships. The outcomes of the algorithm include a small number of modules (2–3 modules), resulting in 10–20 selected genes in total, most of which are known drivers. As briefly mentioned in Section 3.4, while it is true that the identified genes are mutually exclusive in Pan-Cancer samples, we found that they predominantly have BETWEEN_ME relationships (Supplementary Fig. S3). In particular, the first Pan-Cancer ME set identified by Dendrix (Kandoth et al., 2013) is mostly in BETWEEN_ME class, consistent with their claim (Supplementary Fig. S3A). The second module identified after removing tissue specificity contained more ACROSS_ME pairs, which confirms that our classification of ME types is valid. MEMCover identifies all 10 genes in both modules. Three modules of size five are identified by another study of mutual exclusivity in Pan-Cancer (Szczurek and Beerenwinkel, 2014). The modules include three meta-genes (a meta gene is a group of genes which have the same mutation profiles) and the modules are highly overlapped. We classified all pairs (with one representative from each meta-gene) in the modules into different ME types (Supplementary Fig. S3C–E), and found most of genes have BETWEEN_ME relationships and very few ACROSS_ME or functional interactions.
MeMo took an approach similar to ours, combining permutation based mutual exclusivity scores with interaction data (Ciriello et al., 2013) and identified cancer related modules in GBM and ovarian cancer. Pan-cancer analysis with MeMo is not available and was reported as challenging to scale the algorithm for Pan-cancer samples (Ciriello et al., 2013; Vandin et al., 2012a, b).
4 Discussion
In this study, we focused on systematic analysis of mutual exclusivity in the context of Pan-Cancer data and on the application of this principle for guiding discovery of Pan-Cancer dysregulated subnetworks. It has been previously proposed that mutually exclusive genomic events are suggestive of a functional linkage of the altered genes. Indeed, classifying mutual exclusivity into three types, BETWEEN_ME (mutual exclusivity between putative tissue-specific drivers), WITHIN_ME (mutual exclusivity observable in one cancer type), and ACROSS_ME (common mutual exclusivity across multiple cancer types based on type restricted permutation test), we found that BETWEEN_ME is more frequent among functionally unrelated genes. Thus, our analysis demonstrates the importance of distinguishing mutual exclusivity classes in Pan-Cancer analyses.
Interestingly, the distribution of the number of partners in ACROSS_ME or WIHIN_ME relationships has a scale-free property and revealed the existence of ME hubs. We conjecture that these hubs have growth advantages that exceed the growth advantage imposed by other drivers. If so, ranking according to the number of non-BETWEEN_ME partners could be used to prioritize cancer drivers.
Our results indicate that mutual exclusivity analysis provides valuable information which, when utilized together with interaction data, can guide a discovery of Pan-Cancer dysregulated subnetworks. Indeed, we found that our new algorithm, MEMCover that uses the module cover optimization strategy to combine functional interactions, mutual exclusivity and genomic aberration frequency, identified many Pan-Cancer dysregulated subnetworks including previously known subnetworks as well as several new subnetworks whose across-cancer role has not been well appreciated previously
Supplementary Material
Acknowledgements
This work was supported in part by the Intramural Research Program of the National Institutes of Health, National Library of Medicine.
Conflict of Interest: none declared.
References
- Cancer Genome Atlas Research, N. (2008) Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature, 455, 1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research, N. (2012) Comprehensive genomic characterization of squamous cell lung cancers. Nature, 489, 519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research, N. (2013) Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature, 499, 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research, N. et al.. (2013) The cancer genome atlas pan-cancer analysis project. Nat. Genet., 45, 1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho D.Y., et al. (2012) Chapter 5: network biology approach to complex diseases. PLoS Comput. Biol., 8, e1002820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciriello G., et al. (2012) Mutual exclusivity analysis identifies oncogenic network modules. Genome Res., 22, 398–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciriello G., et al. (2013) Using MEMo to discover mutual exclusivity modules in cancer. Curr. Protoc. Bioinform., Chapter 8:Unit 8 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das J., Yu H. (2012) HINT: high-quality protein interactomes and their applications in understanding human disease. BMC Syst. Biol., 6, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofree M., et al. (2013) Network-based stratification of tumor mutations. Nat. Methods, 10, 1108–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Je E.M., et al. (2013) Mutational analysis of splicing machinery genes SF3B1, U2AF1 and SRSF2 in myelodysplasia and other common tumors. Int. J. Cancer, 133, 260–265. [DOI] [PubMed] [Google Scholar]
- Kandoth C., et al. (2013) Mutational landscape and significance across 12 major cancer types. Nature, 502, 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.A., et al. (2013) Module cover—a new approach to genotype-phenotype studies. In: Proceedings of Pacific Symposium on Biocomputing, Vol. 18, pp. 135–146. [PMC free article] [PubMed] [Google Scholar]
- Kon A., et al. (2013) Recurrent mutations in multiple components of the cohesin complex in myeloid neoplasms. Nat. Genet., 45, 1232–1237. [DOI] [PubMed] [Google Scholar]
- Lawrence M.S., et al. (2014) Discovery and saturation analysis of cancer genes across 21 tumour types. Nature, 505, 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiserson M.D., et al. (2013) Simultaneous identification of multiple driver pathways in cancer. PLoS Comput. Biol., 9, e1003054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiserson M.D., et al. (2014) Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nat. Genet., 47, 106–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milo R., et al. (2003) On the Uniform Generation of Random Graphs with Prescribed Degree Sequences. arXiv:cond-mat/0312028. [Google Scholar]
- Rhodes J.M., et al. (2011) Gene regulation by cohesin in cancer: is the ring an unexpected party to proliferation? Mol. Cancer Res., 9, 1587–1607. [DOI] [PubMed] [Google Scholar]
- Shannon P., et al. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res., 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczurek E., Beerenwinkel N. (2014) Modeling mutual exclusivity of cancer mutations. PLoS Comput. Biol., 10, e1003503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R.K., et al. (2007) High-throughput oncogene mutation profiling in human cancer. Nat. Genet., 39, 347–351. [DOI] [PubMed] [Google Scholar]
- Vandin F., et al. (2011) Algorithms for detecting significantly mutated pathways in cancer. J. Comput. Biol., 18, 507–522. [DOI] [PubMed] [Google Scholar]
- Vandin F., et al. (2012a) De novo discovery of mutated driver pathways in cancer. Genome Res., 22, 375–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandin F., et al. (2012b) Discovery of mutated subnetworks associated with clinical data in cancer. In: Proceedings of Pacific Symposium on Biocomputing, Vol. 17, pp. 55–66. [PubMed] [Google Scholar]
- Vogelstein B., et al. (2013) Cancer genome landscapes. Science, 339, 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., et al. (2011) SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N. Engl. J. Med., 365, 2497–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., et al. (2011) Next-generation sequencing to generate interactome datasets. Nat. Methods, 8, 478–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., et al. (2011) Functional SNP in the microRNA-367 binding site in the 3'UTR of the calcium channel ryanodine receptor gene 3 (RYR3) affects breast cancer risk and calcification. Proc. Natl. Acad. Sci. U.S.A., 108, 13653–13658. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.