Abstract
Cancers mediated by viral etiology must exhibit deregulated cellular proliferation and evade immune recognition. The role of the retinoblastoma tumor suppressor (RB) pathway, which is lost at relatively high frequency in HCC, has recently been expanded to include the regulation of innate immune responsiveness. In this study, we investigated the coordinate impact of RB-loss on cell cycle control and immune function in the liver. We found that RB depletion in hepatoma cells resulted in a compromised immunological response to multiple stimuli and reduced the potential of these cells to recruit myeloid cells. Viral-mediated liver-specific RB deletion in vivo led to the induction of genes associated with proliferation and cell cycle entry as well as the significant attenuation of genes associated with immune function, as evidenced by decreases in cytokine and chemokine expression, leukocyte recruitment and hepatic inflammation. To determine if these changes in gene expression were instructive in human disease, we compared our liver-specific RB-loss gene signature to existing profiles of HCC and found that this signature was associated with disease progression and confers a worse prognosis.
Conclusion
Our data confirm that RB participates in the regulation of innate immunity in liver parenchymal cells both in vitro and in vivo and to our knowledge describes the first gene signature associated with HCC that includes both immunoregulatory and proliferative genes and that can also be attributed to the alteration of a single gene in vitro.
Keywords: Liver, Tumor suppressor, Immune Function, HCC, Gene Signature
INTRODUCTION
The tumor suppressive function of RB is largely the result of transcriptional repression of cell cycle regulation genes, particularly those controlled by E2F family transcription factors (1, 2), and functional inactivation of the tumor suppressor RB is a common event in nearly all human malignancy (3, 4). Multiple oncogenic viruses specifically inactivate RB (5) whereas others, such as hepatitis B and C, have been speculated to inactivate RB via indirect mechanisms such as sequestration or nuclear export (6, 7). Interestingly, expression of a large subset of immune response genes are attenuated in cancers resulting from these viruses (8).
The liver is constantly exposed to pathogens and must deftly distinguish tolerable agents from pathogens. Hepatocytes are capable of binding toll-like receptor (TLR) ligands (9, 10) as well as presenting exogenous antigens (11). Thus, hepatocytes play a vital role in the initiation of the immune response both directly and indirectly. Notably, cirrhosis and the transitional, early stages of HCC are characterized by chronic inflammation (12), but as the disease progresses local immune function is suppressed (13).
An increasing number of studies have demonstrated that RB is involved in immune function (14-17) and expression of a large subset of immune function-related genes is downregulated upon RB-loss in cell culture models (18). Further, RB-loss results in increased viral susceptibility, decreased TLR3 expression, reduced nuclear localization of the RELA (p65) subunit of NF-κB, and diminished production of cytokines and chemokines including IFNβ, interleukin 8, and Cxcl1 (19, 20). While these data suggest an important role for RB in regulating the immune response, particularly in response to viral infection, relatively few studies have investigated the impact of RB-loss on the response to specific immune stimuli and thus the exact mechanisms by which RB regulates these pathways as well as any potential relationships with the canonical role of RB in cell cycle regulation remain unclear.
Given that the regulation of innate immunity is a key role of the liver, in this report we will investigate the role RB-loss plays in the hepatic immune response and examine whether this has significant relevance in the development or progression of HCC.
MATERIALS AND METHODS
Transfection and stimulation of cell lines
HepG2 or Huh7 cells, transfected with either shRB or shNS as previously described (21), were stimulated with LPS (100ng/ml), PMA (10ng/ml), gardiquimod (1μg/ml), FITCODN 2395 (1μg/ml) (Invivogen, San Diego, CA), fMLP (0.2-2μg/ml, Sigma-Aldrich, St. Louis, MO) CpG DNA, or adenovirus (1×107 viral particles/ml) for up to 24 hours.
Quantitative real-time PCR (qRT-PCR)
For mice, primers were used at an annealing temperature of 60° C and 40 cycles. The ΔΔCT values were calculated after amplification using the Power SYBR Green reagent (Applied Biosystems) and the corresponding fold-change values were graphed. For humans, TaqMan primers for specified genes (Life Technologies) were used and multiplex qRT-PCR was performed on a BiomarkHD system (Fluidigm, San Francisco, CA). Prepared cDNA samples were preamplified for 14 cycles and resultant cDNA was then loaded on a 96x96 gene expression chip and run for 40 cycles.
Flow cytometry and in-cell western
For flow cytometry, harvested cells were stained for 30min with purified antibodies specific to CD16, CD74, TLR4, HLADR (Santa Cruz Biotechnology, Dallas, TX), or CD44 (BD Biosciences, San Jose, CA). Data were acquired on an LSR2 fluorescence activated cell sorter (BD Biosciences) and analyzed using FlowJo software (Treestar Inc., Ashland, OR). For in-cell western, cells were plated in 96-well plates and treated as indicated before fixation with methanol. Cells were probed with purified anti-CD74 antibody and donkey anti-goat DyLight 800-conjugated antibody (Thermo Fisher Scientific, Rockford, IL). Relative cell numbers were quantified using Sapphire700 (Li-Cor Biosciences, Lincoln, NE) and DRAQ5 (Thermo Fisher). Staining was visualized on an Odyssey CLx (Li-Cor).
Myeloid recruitment and differentiation
Hepatoma cells were plated in 24-well plates at a concentration of 50,000 cells/well. After 24 hours, 5,000 undifferentiated, serum-starved THP-1 cells were added to the top chamber of a 0.5-micron pore transwell. Membranes were washed after 4 hours and fixed in methanol. Cells were removed from the top of the membranes by gently scraping with a PBS-wetted cotton swab and membranes were visualized following DAPI staining.
Mice
Mice (Rb1f/f) containing LoxP sites flanking exon 19 of Rb1 with or without albumin-specific cre have been previously reported (22-24).
Ethics Statement
All mouse care, treatment and sacrifice were conducted using the highest standards for humane animal care in accordance with the NIH Guide for the Care and Use of Laboratory Animals.
Adenovirus Delivery and Liver Damage Models
Adenoviral delivery was performed on male mice, anesthetized with isoflurane (2-chloro-2-(difluoromethoxy)-1,1,1-trifluoro-ethane) as previously described (22). Diethylnitrosamine (DEN) and 1,4-Bis-[2-(3,5-dichloropyridyloxy)] benzene (TCPOBOP) (3mg/kg) were administered as previously described (23, 25). Following sacrifice, livers were isolated and processed as previously described (24).
Gene Expression Array and Analysis
Total RNA was extracted and gene expression analysis was performed as previously described (24) using Affymetrix GeneChip for Mouse Genome 430 2.0 and an Affymetrix GeneChip Scanner 3000 with GeneChip Operating Software version 3.0. Following normalization (26), gene set enrichment analysis (GSEA) was performed (27). Significant differences were determined (28) in the TM4 MultiExperiment Viewer software package (29) with a 1.2-fold cut-off in expression change and FDR<25%. Ontology was done using DAVID (http://david.abcc.ncifcrf.gov/).
Immunoblotting
Membranes were incubated with: Santa Cruz Biotechnology: Lamin B (sc-6217), E2F1 (sc-193), p107 (sc-318), Mcm7 (sc-9966), PCNA (sc-56); Cell Signaling Technology: RB (9309).
Immunohistochemistry and immunofluorescence
Five-micron sections were cut from paraffin-embedded liver specimens. H&E staining was performed by standard methods. For immunohistochemistry, sections were heated at 55°C for 30 minutes, rehydrated, and blocked against endogenous peroxidases (Dako, Denmark) and biotin (Vector Labs, Burlingame, CA) as described by the manufacturers before overnight incubation with anti-Mac2 antibody (M3/38, Cedarlane Labs, Burlington, NC) or anti-CD74 (C-16, Santa Cruz Biotechnology). For immunofluorescence, sections were rehydrated and treated with citrate buffer before overnight incubation with anti-CD74 and visualization with Alexa fluor 546-conjugated donkey anti-goat secondary antibody and DAPI counterstain (Life Technologies).
Comparative gene expression analyses
Lesion data (GSE4108, GSE6764, GSE36376, GSE50579) were obtained from the Gene Expression Omnibus. A subset of genes herein identified as the liver specific RB-loss signature (Supplemental Table 1) was used to evaluate these datasets. A median-centered gene expression profile for all genes in the signature was used to order the samples by expression and observe phenotypic trends as a function of expression gradient. Additional survival information (GSE4108) was obtained from the authors and used to compare the survival curves based on liver specific RB-loss signature expression by Kaplan-Meier analysis. Significance was determined by the log rank-p value.
RESULTS
RB mediates hepatocyte immune responsiveness
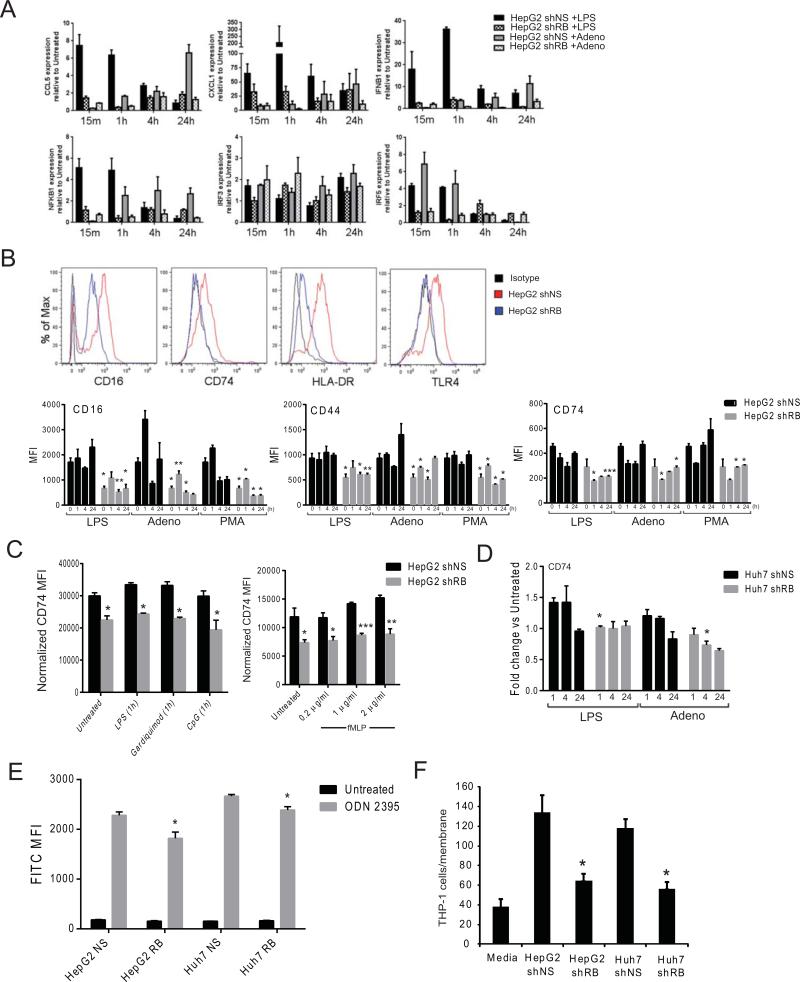
To investigate the role of RB in innate immunity, HepG2 cells were infected with a retroviral vector containing either a short-hairpin RNA specific for RB (HepG2shRB) or with a non-specific sequence (HepG2shNS) (21). Whereas HepG2shNS cells demonstrated a robust response to both LPS and adenovirus, this response was subdued in HepG2shRB cells as evidenced by decreased expression of cytokine and pro-inflammatory transcription factor genes (Figure 1A). One explanation for these data would be a decreased capacity for shRB cells to detect immune stimuli. To test this, expression of multiple immunoregulatory receptors was examined by flow cytometry. Expression of all examined receptors was reduced in HepG2shRB cells as compared to HepG2 shNS cells under basal conditions as well after stimulation with LPS (TLR4 agonist), adenovirus, or phorbol 12-myristate 13-acetate (PMA) for up to 24 hours (Figure 1B) or after acute stimulation with gardiquimod (TLR7 agonist), CpG DNA (TLR9 agonist), or N-formyl-Met-Leu-Phe (fMLP) (Figure 1C). Huh7 cells demonstrated a similar defect, as CD74 expression was less responsive to LPS and adenoviral stimulation in Huh7shRB cells as compared to Huh7shNS (Figure 1D). Functionally, RB-deficient hepatocytes displayed less uptake of CpG DNA (Figure 1E) and a diminished capacity for recruiting myeloid cells (Figure 1F). Taken together, these data indicate that RB regulates the ability of hepatoma cells to respond to diverse immune stimuli at least in part by regulating their ability to detect exogenous signals and contribute to a pro-inflammatory milieu.
Figure 1. RB-depleted HepG2 cells are less responsive to innate immune stimuli.
(A) qRT-PCR for gene expression of cytokines, chemokines, and transcription factors associated with the response to innate stimuli. Cells were cultured in triplicate and harvested after 15min - 24hr +/− LPS or adenovirus. (B) Histograms and MFI of HepG2shNS and HepG2shRB cells probed for the indicated cell surface receptor by flow cytometry. Cells were plated in triplicate and treated for 1-24hr in the presence or absence the indicated stimuli. Data are representative of at least three experiments. (C) MFI of CD74 expression as measured by in-cell western following 1hr exposure to the indicated stimuli. (D) Fold-change of CD74 MFI as measured by flow cytometry in Huh7shNS and Huh7shRB cells following treatment with the indicated stimuli. Cells were plated in triplicate and treated for 1-24hr in the presence or absence the indicated stimuli. (E) Quantitative measurement of TLR-agonist uptake by flow cytometry. HepG2shNS, HepG2shRB, Huh7shNS, and Huh7shRBcells were cultured in triplicate with ODN 2395 for 24hr. (F) Transwell THP-1 cell migration in response to chemotactic signals from HCC cell lines +/−RB for 4 hr. Following harvest, recruited cells were stained with DAPI and counted. Data represent the mean ± SEM and were compared by Welch's t-test. * P<0.05, ** P<0.001, *** P<0.005.
Acute RB deletion deregulates cell cycle-related gene expression in vivo
Adenoviral-mediated delivery of cre-recombinase in RBf/f mice was utilized to determine if RB-loss led to similar immune defects in vivo. Adenoviral transduction was specific to the liver and resulted in complete recombination (Supplemental Figure 1A) and subsequent RB protein ablation as previously reported (22). RB-loss was accompanied by a rapid induction in DNA synthesis, as measured by BrdU incorporation (Supplemental Figure 1A), and increased expression of proteins corresponding with DNA replication (Supplemental Figure 1B).
Liver-specific RB-loss revealed increased proliferative gene expression and decreased immune function gene expression
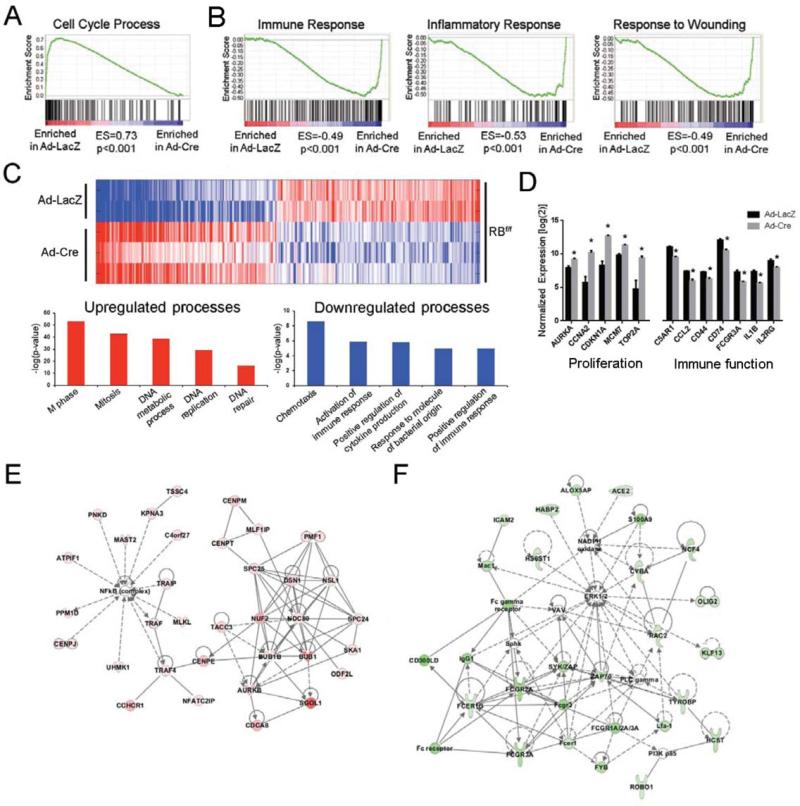
The majority of HCC cases result from chronic viral infection and thus this model of RB-loss was selected, in part, because it accounts for viral-mediated hepatitis concomitant with the loss of RB in an approximation of what occurs during HCC progression. To focus these studies on the acute response to viral challenge and RB deletion, RNA was isolated from livers 3 days following Adeno-Cre (Ad-Cre) or Adeno-LacZ (Ad-LacZ) infection for transcriptomic analyses. Analysis by GSEA revealed upregulation of overlapping gene ontologies related to cell cycle progression (Figure 2A) coupled with the downregulation of multiple ontologies related to different aspects of immune responsiveness and repair (Figure 2B). Further examination of the resultant liver specific RB-loss gene signature (Figure 2C, D) by gene ontology (Table 1) or Ingenuity pathway analysis (Figure 2E,F, Supplemental Figure 2) reinforced these observations, which are also consistent with previous studies (18, 30). Similar to our in vitro data (Figure 1B), these changes in gene expression are likely in part related to NF-κB and type-1 IFN dysfunction (Supplemental Figure 3A, B).
Figure 2. Liver specific RB-loss gene signature is defined by increased proliferation and decreased immune responsiveness.
(A-B) GSEA of RB-flox mice following Ad-LacZ or Ad-Cre infection. (C) Heat map representing altered gene expression following Ad-LacZ or Ad-Cre infection of RB-flox mice. Red indicates increased expression, blue indicates decreased expression. (D) Expression of selected genes from the liver specific RB-loss gene signature representing proliferation and immune response genes. * represents P<0.05. (E) Molecular network of upregulated genes most significantly affected by RB-loss as indicated by IPA. Functional associations included “Cell Cycle, Cellular Assembly and Organization, DNA Replication, Recombination, and Repair”. (F) Molecular network of downregulated genes most significantly affected by RB-loss as indicated by IPA. Functional associations included “Cellular Movement, Immune Cell Trafficking, Cell-To-Cell Signaling and Interaction”.
Table 1.
Selected genes from top liver-specific RB-loss gene signature GO term clusters
| Representative GO Term (ID) | Selected Genes | Enrichment Score | P-value |
|---|---|---|---|
| Upregulated | |||
| Cluster 1: Cell cycle (0007049) | HAUS8, DSN1, E2F1, E2F2, H2AFX, RAD21, AURKA, AURKB, ANAPC4, BRCA2, CDC5, CENPE, CCNA2, CDK2, INCENP, MCM7, MDM2, MYB, PIN1, PLK1 | 51.77 | 1.4E-74 |
| Cluster 2: DNA metabolic process (0006259) | ASF1A, BLM, DNMT1, APAF1, BRCA1, BRCA2, CHEK1, CCNE1, POLA1, RBBP7, PARP1, TOP2A | 22.37 | 1.5E-40 |
| Cluster 3: M phase of meiotic cell cycle (0051327) | FBXO43, FBXO5, NEK2, EXO1, PTTG1, SMC3, TRIP13, TOPBP1 | 9.72 | 1.7E-11 |
| Cluster 4: Microtubule cytoskeleton organization (0000226) | HAUS1, HAUS4, BIRC5, KIF11, RANBP1, CENPE, CENPJ, CKAP5 | 9.23 | 1.7E-13 |
| Cluster 5: Chromosome organization (0051276) | ASF1A, DNMT1, H1FX, CBX6, CDCA5, HAT1, HELLS, NUSAP1 | 5.76 | 1.1E-14 |
| Downregulated | |||
| Cluster 1: Immune response (0006955) | BCL3, CD14, CD74, CCL2, CCL5, CXCL1, CFP, IGJ, IL1B, NLRP3, TLR2, TLR4, TLR7, TGFB1 | 10.06 | 2.1E-12 |
| Cluster 2: Taxis (0042330) | FCER1G, RAC2, S100A9, CCL2, CCL5, CXCL10, C5AR1, FPR1, ITGB2, | 5.78 | 4.5E-9 |
| Cluster 3: Activation of immune response (0002253) | CD3E, FCER1G, C1QA, C1QB, MASP1, NFKBIA, PLCG2, PTPRC | 3.94 | 1.8E-6 |
| Cluster 4: Response to molecule of bacterial origin (0002237) | CD14, C5AR1, IL1B, PLCG2, SLC11A1, TLR2, TLR4 | 3.75 | 1.4E-5 |
| Cluster 5: Enzyme linked receptor protein signaling pathway (0007167) | ARAP1, AXL, SMAD7, CSF1R, CTGF, FOXC2, ID1, PDGFRA, TGFBR3 | 3.35 | 2.6E-4 |
Genes selected from the top liver-specific RB-loss gene signature annotation clusters represent both increased (cellular proliferation) and decreased (immune function) processes. Expression of all selected genes was significantly different between groups as defined in the Materials and Methods. GO terms/IDs and selected genes represent the top ontology from each cluster. Enrichment scores and P-values represent the entire cluster.
Liver-specific RB-loss gene signature accurately predicts immune-related gene deregulation in vivo
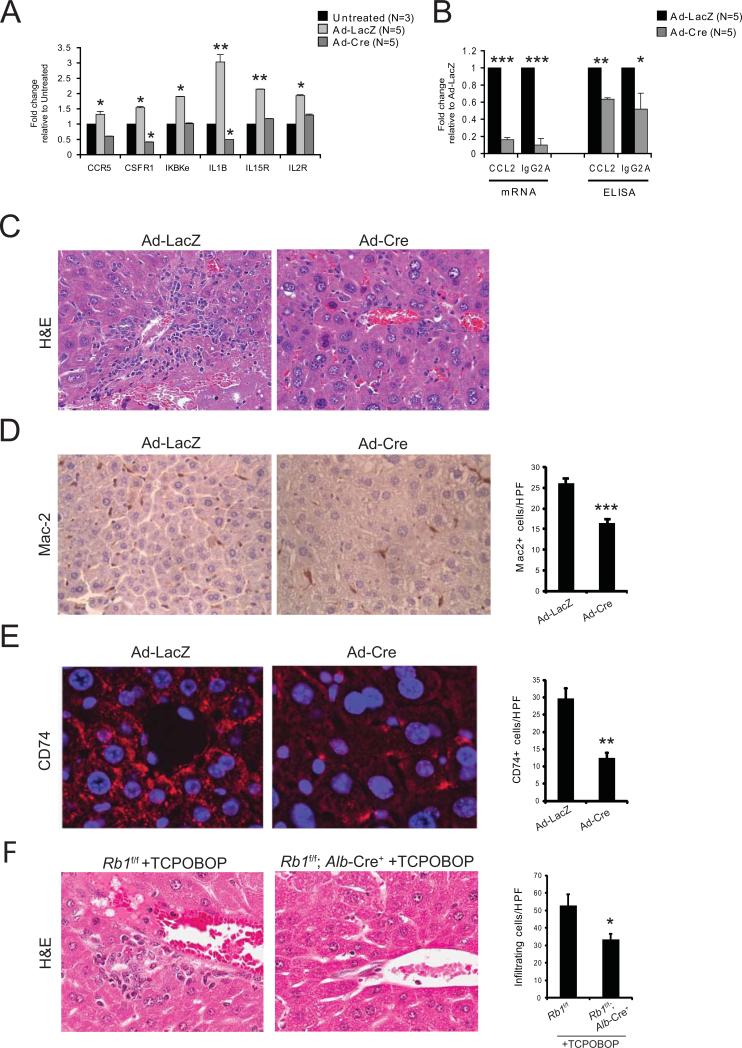
Next we examined gene expression patterns in mice injected with Ad-Cre or Ad-LacZ, as compared to untreated mice. Three days following administration of Ad-LacZ, expression of Ccr5, Csfr1, Ikbke, Il1b, Il15r, and Il2r was increased, while Ad-Cre treated mice displayed reduced expression of these genes (Figure 3A). Similar results were found in the expression of Ccl2 and IgG2a at both the mRNA and protein level (Figure 3B). These data collectively suggest a decreased inflammatory state in Cre-positive livers as compared to Cre-negative livers, associated with the loss of RB and adenoviral-mediated hepatitis. Histological examination of livers isolated 6 days following infection confirmed that livers from Ad-LacZ-treated mice displayed a more robust inflammatory response, measured by immune cell infiltration, as compared to livers from Ad-Cre-treated mice (Figure 3C). This was evidenced by a significant decrease in macrophage recruitment, as determined by Mac-2 staining (Figure 3D). Furthermore, while RB-sufficient hepatocytes, particularly those surrounding central venules, expressed high levels of CD74, this expression was largely suppressed in RB-deficient livers (Figure 3E). The decreased infiltration of immune cells was also present in Rb1f/f; Alb-cre+ mice following treatment with TCPOBOP (Figure 3F). Thus, as suggested by our in vitro data, the deregulation of hepatic immunity that results from liver-specific RB-loss extends beyond viral-mediated hepatitis in vivo.
Figure 3. Ablation of Rb results in decreased hepatic inflammation and leukocyte recruitment.
(A) qRT-PCR for selected RB/E2F-target and immune response genes in each condition. (B) qRT-PCR and ELISA analyses for Ccl2 and Igg2a transcripts and protein secretion from mouse livers in each condition. (C) Representative H&E staining of liver sections from mice following RB ablation (n=5) as compared to mice with RB (n=5). (D) Representative IHC and related quantitation of hepatic macrophages (Mac-2+) following infection with Ad-Cre (n=5) or Ad-LacZ (n=5). Five high-power fields (63x objective) were selected randomly and counted for each sample. (E) Representative immunofluorescence staining (63×) and related quantitation of CD74 expression in Ad-Cre mice (n=5) as compared to Ad-LacZ mice (n=5). (F) Representative H&E staining and related quantitation of RB-sufficient or RB-deficient liver sections from mice (n=3 per group) 3 days after TCPOBOP injection. Data represent the mean ± SEM and were compared by Welch's t-test. * P<0.05, ** P<0.001, *** P<0.005.
Murine liver-specific RB-loss gene signature correlates with progression to HCC and poor clinical outcome
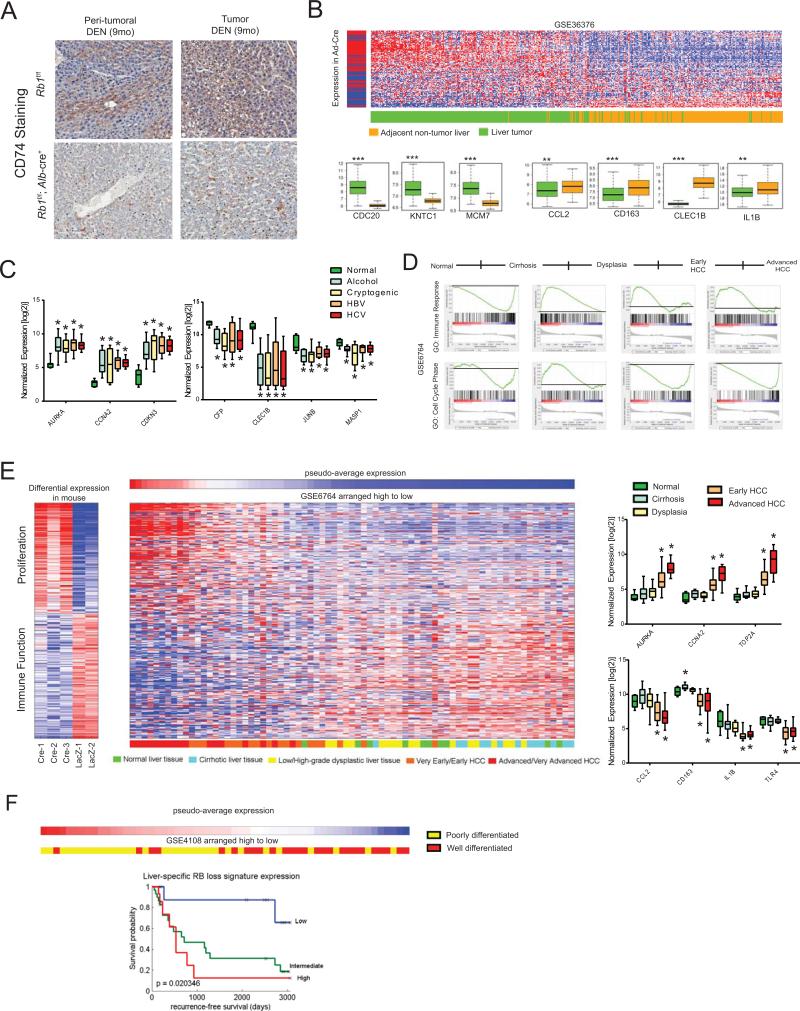
Following DEN injection, mice with liver-specific RB loss developed more tumors than RB-sufficient controls, as expected (data not shown); however, tumoral and peri-tumoral areas displayed decreased expression of CD74 (Figure 4A). Given our previous findings and this possible association between CD74-expression and hepatocarcinogenesis, we analyzed existing human HCC gene expression datasets retrieved from the Gene Expression Omnibus (31) for evidence of the liver specific RB-loss gene signature (Supplemental Table 1). This signature was able to differentiate liver tumors from adjacent non-tumor tissue (Figure 4B) and was independent of disease etiology (Figure 4C). Subsequent GSEA revealed that while there was positive enrichment of pro-inflammatory genes as normal liver developed into cirrhosis, gene ontologies similar to the liver-specific RB-loss gene signature (Table 1) began to be negatively enriched at the onset of dysplasia and this trend continued as HCC progressed (Figure 4D). These findings were further supported by examination of individual disease state gene signatures, wherein the liver-specific RB-loss gene signature was not overtly associated with cirrhosis or liver dysplasia, but demonstrated a reciprocal relationship with the progression of HCC (Figure 4E).
Figure 4. Liver-specific Rb-loss signature correlates to HCC progression and poor outcome in human disease.
(A) Representative CD74 staining of RB-deficient (n=5) and RB-sufficient (n=5) livers isolated from mice 9 months after DEN treatment. (B) Clustering of patient liver samples and adjacent non-tumor liver tissue (n=433) based on liver-specific RB-loss signature expression. Gene expression box plots represent selected RB-loss signature genes. (C) Gene expression box plots for representative genes selected from the liver-specific RB-loss signature in normal liver tissue (n=7) or HCC associated with alcohol abuse (n=8), cryptogenic cause (n=11), HBV (n=8), or HCV (n=9). (D) Patient liver sample GSEA grouped by disease state (n=75). (E) Clustering of patient liver samples (n=75) from varying disease states, based on liver-specific RB-loss signature expression. Gene expression box plots represent selected RB-loss signature genes. (F) Kaplan-Meier plots indicating survival probability in human patients (n=58) with High, Intermediate, or Low expression of the liver-specific RB-loss signature. * P<0.05
These observations suggest that RB pathway disruption plays a transformative role for RB in disease progression. In sum, our findings suggest that RB-loss, at least as it relates to human disease, is likely a later stage event that promotes lesion de-differentiation and aggressiveness. In accordance with this assertion, the liver-specific RB-loss signature was predictive of poor outcome in patients, with even intermediate signature expression correlating to decreased survival (Figure 4F). Taken together, these findings indicate that RB pathway disruption, which results in both increased cellular proliferation as well as decreased immune function, is a defining event for HCC progression in human disease, and could play a critical role in determining the aggressiveness and outcome of individual disease cases.
DISCUSSION
Multiple reports have identified gene signatures in HCC associated with increased proliferation and decreased chromosomal stability (32). Meanwhile, in viral-mediated oncogenesis there is an inherent evolutionary benefit to immune evasion and a gene signature composed solely of immune-mediated genes that correlates with improved patient prognosis in early-stage HCC has recently been reported (33). RB-loss has been associated with the progression of HCC (34) and in the present study we have markedly diminished RB expression both in vitro and in vivo, to examine the role of RB in hepatic immunity.
RB-loss led to decreased inflammatory responses in vitro (Figure 1) and in vivo (Figures 2-3), coinciding with previous reports that have indicated a role for RB in immune signaling and pathogen clearance (14, 15, 19, 20). Gene expression analysis of RB-deficient mouse livers allowed for the identification of a liver-specific RB-loss gene signature that we have demonstrated aligns with HCC disease progression and decreased patient survival (Figure 4). Although our signature shares some characteristics with subtype 2 HCC, originally described by Hoshida, et al. (35), to our knowledge this is the first report of a gene signature associated with RB-loss that aligns with cancer progression based on both increased proliferation and decreased immune function.
Interestingly, several of the receptors and chemokines we have found to be decreased upon loss of RB have previously been associated with HCC progression. Ccl5 contributes to antitumor activity and polymorphisms in either the Ccl5 or Ccr5 gene increases HCC susceptibility (36, 37). Defects in expression and allelic variations of various HLA genes, including HLA-DR, are associated with the disease (38). CD74-deficient animals experience enhanced fibrosis in response to CCl4 treatment (39) and primary biliary cirrhosis patients display enhanced levels of circulating CD74 which neutralizes circulating macrophage migration inhibitory factor (40). Additionally, the pro-inflammatory senescence associated secretory phenotype (SASP) promotes obesity-induced hepatic cancer (41). Paradoxically, TLR4 sensing of LPS from gut microbiota is required for initiation of HCC (42) and increased cytokine production has been correlated with improved prognosis in early stage HCC (33), while we report here that loss of RB results in decreased expression of many immune function genes (Figure 1, Table 1). Several of these genes overlap with the previously reported immune gene signature, yet we have demonstrated that this is associated instead with progression of HCC and decreased survival. In each case, the previously reported findings were associated with early stage events in disease development or progression while our data suggest that the decrease in immune responsiveness associated with RB-loss becomes more prevalent as HCC progresses (Figure 4D-E). These findings are congruent with reports suggesting that immune function is greatly diminished in advanced HCC (13).
Here, we report a potential role for RB-loss as a molecular switch separating the cytokine/chemokine producing signature of early HCC from the non-responsive immune profile of advanced HCC. This effect would occur in concert with the cell cycle effects of RB-loss and may result in a situation where cells are proliferating rapidly but can no longer be recognized by the immune system. In summary, our findings suggest that RB plays a critical role in promoting the hepatic immune response and that RB loss may be a key checkpoint in the cancer-immunity cycle in HCC by increasing the ability of tumors to escape immune surveillance as well as by promoting cellular proliferation.
Supplementary Material
ACKNOWLEDGMENTS
The authors of this manuscript recognize all who participated in scientific discussion and feedback related to this manuscript, including Dr. Linda Greenbaum of Thomas Jefferson University, and especially those from the Erik Knudsen laboratory. E.S. Knudsen is supported by grants from the National Cancer Institute.
Financial support: ESK is supported by grants from the National Cancer Institute
List of abbreviations
- HCC
hepatocellular carcinoma
- NF-κB
nuclear factor kappa-light-chain-enhancer of activated B cells
- RB
retinoblastoma protein
- TLR
toll-like receptor
Footnotes
Disclosures: The authors have nothing to disclose.
References
- 1.Blais A, Dynlacht BD. Hitting their targets: an emerging picture of E2F and cell cycle control. Curr Opin Genet Dev. 2004;14:527–532. doi: 10.1016/j.gde.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 2.Harbour JW, Dean DC. Rb function in cell-cycle regulation and apoptosis. Nat Cell Biol. 2000;2:E65–67. doi: 10.1038/35008695. [DOI] [PubMed] [Google Scholar]
- 3.Burkhart DL, Sage J. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat Rev Cancer. 2008;8:671–682. doi: 10.1038/nrc2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knudsen ES, Knudsen KE. Tailoring to RB: tumour suppressor status and therapeutic response. Nat Rev Cancer. 2008;8:714–724. doi: 10.1038/nrc2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ludlow JW, Skuse GR. Viral oncoprotein binding to pRB, p107, p130, and p300. Virus Res. 1995;35:113–121. doi: 10.1016/0168-1702(94)00094-s. [DOI] [PubMed] [Google Scholar]
- 6.Choi BH, Choi M, Jeon HY, Rho HM. Hepatitis B viral X protein overcomes inhibition of E2F1 activity by pRb on the human Rb gene promoter. DNA Cell Biol. 2001;20:75–80. doi: 10.1089/104454901750070274. [DOI] [PubMed] [Google Scholar]
- 7.Munakata T, Liang Y, Kim S, McGivern DR, Huibregtse J, Nomoto A, Lemon SM. Hepatitis C virus induces E6AP-dependent degradation of the retinoblastoma protein. PLoS Pathog. 2007;3:1335–1347. doi: 10.1371/journal.ppat.0030139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Le Buanec H, Lachgar A, D'Anna R, Zagury JF, Bizzini B, Bernard J, Ittele D, et al. Induction of cellular immunosuppression by the human papillomavirus type 16 E7 oncogenic protein. Biomed Pharmacother. 1999;53:323–328. doi: 10.1016/s0753-3322(00)88505-4. [DOI] [PubMed] [Google Scholar]
- 9.Vodovotz Y, Liu S, McCloskey C, Shapiro R, Green A, Billiar TR. The hepatocyte as a microbial product-responsive cell. J Endotoxin Res. 2001;7:365–373. [PubMed] [Google Scholar]
- 10.Xia C, Lu M, Zhang Z, Meng Z, Shi C. TLRs antiviral effect on hepatitis B virus in HepG2 cells. J Appl Microbiol. 2008;105:1720–1727. doi: 10.1111/j.1365-2672.2008.03896.x. [DOI] [PubMed] [Google Scholar]
- 11.Herkel J, Jagemann B, Wiegard C, Lazaro JF, Lueth S, Kanzler S, Blessing M, et al. MHC class II-expressing hepatocytes function as antigen-presenting cells and activate specific CD4 T lymphocyutes. Hepatology. 2003;37:1079–1085. doi: 10.1053/jhep.2003.50191. [DOI] [PubMed] [Google Scholar]
- 12.Stauffer JK, Scarzello AJ, Jiang Q, Wiltrout RH. Chronic inflammation, immune escape, and oncogenesis in the liver: a unique neighborhood for novel intersections. Hepatology. 2012;56:1567–1574. doi: 10.1002/hep.25674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greten TF, Duffy AG, Korangy F. Hepatocellular carcinoma from an immunologic perspective. Clin Cancer Res. 2013;19:6678–6685. doi: 10.1158/1078-0432.CCR-13-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu Y, Ussery GD, Jacim M, Tschickardt M, Boss JM, Blanck G. Retinoblastoma protein regulation of surface CD74 (invariant chain) expression in breast carcinoma cells. Mol Immunol. 1994;31:1365–1368. doi: 10.1016/0161-5890(94)90055-8. [DOI] [PubMed] [Google Scholar]
- 15.Lu Y, Ussery GD, Muncaster MM, Gallie BL, Blanck G. Evidence for retinoblastoma protein (RB) dependent and independent IFN-gamma responses: RB coordinately rescues IFN-gamma induction of MHC class II gene transcription in noninducible breast carcinoma cells. Oncogene. 1994;9:1015–1019. [PubMed] [Google Scholar]
- 16.Tschickardt ME, Lu Y, Jacim M, Ussery GD, Steimle V, Mach B, Blanck G. RB and a novel E2F-1 binding protein in MHC class II deficient B-cell lines and normal IFN-gamma induction of the class IL transactivator CIITA in class II non-inducible RB-defective tumor lines. Int J Cancer. 1995;62:461–465. doi: 10.1002/ijc.2910620417. [DOI] [PubMed] [Google Scholar]
- 17.Osborne A, Tschickardt M, Blanck G. Retinoblastoma protein expression facilitates chromatin remodeling at the HLA-DRA promoter. Nucleic Acids Res. 1997;25:5095–5102. doi: 10.1093/nar/25.24.5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Markey MP, Bergseid J, Bosco EE, Stengel K, Xu H, Mayhew CN, Schwemberger SJ, et al. Loss of the retinoblastoma tumor suppressor: differential action on transcriptional programs related to cell cycle control and immune function. Oncogene. 2007;26:6307–6318. doi: 10.1038/sj.onc.1210450. [DOI] [PubMed] [Google Scholar]
- 19.Garcia MA, Gallego P, Campagna M, Gonzalez-Santamaria J, Martinez G, Marcos-Villar L, Vidal A, et al. Activation of NF-kB pathway by virus infection requires Rb expression. PLoS One. 2009;4:e6422. doi: 10.1371/journal.pone.0006422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taura M, Suico MA, Koyama K, Komatsu K, Miyakita R, Matsumoto C, Kudo E, et al. Rb/E2F1 regulates the innate immune receptor Toll-like receptor 3 in epithelial cells. Mol Cell Biol. 2012;32:1581–1590. doi: 10.1128/MCB.06454-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rivadeneira DB, Mayhew CN, Thangavel C, Sotillo E, Reed CA, Grana X, Knudsen ES. Proliferative suppression by CDK4/6 inhibition: complex function of the retinoblastoma pathway in liver tissue and hepatoma cells. Gastroenterology. 2010;138:1920–1930. doi: 10.1053/j.gastro.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mayhew CN, Bosco EE, Fox SR, Okaya T, Tarapore P, Schwemberger SJ, Babcock GF, et al. Liver-specific pRB loss results in ectopic cell cycle entry and aberrant ploidy. Cancer Res. 2005;65:4568–4577. doi: 10.1158/0008-5472.CAN-04-4221. [DOI] [PubMed] [Google Scholar]
- 23.Reed CA, Mayhew CN, McClendon AK, Knudsen ES. Unique impact of RB loss on hepatic proliferation: tumorigenic stresses uncover distinct pathways of cell cycle control. J Biol Chem. 2010;285:1089–1096. doi: 10.1074/jbc.M109.043380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bourgo RJ, Ehmer U, Sage J, Knudsen ES. RB deletion disrupts coordination between DNA replication licensing and mitotic entry in vivo. Mol Biol Cell. 2011;22:931–939. doi: 10.1091/mbc.E10-11-0895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reed C, Hutcheson J, Mayhew CN, Witkiewicz AK, Knudsen ES. RB Tumor Suppressive Function in Response to Xenobiotic Hepatocarcinogens. Am J Pathol. 2014 doi: 10.1016/j.ajpath.2014.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bartkova J, Hamerlik P, Stockhausen MT, Ehrmann J, Hlobilkova A, Laursen H, Kalita O, et al. Replication stress and oxidative damage contribute to aberrant constitutive activation of DNA damage signalling in human gliomas. Oncogene. doi: 10.1038/onc.2010.249. [DOI] [PubMed] [Google Scholar]
- 27.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 30.Ertel A, Dean JL, Rui H, Liu C, Witkiewicz AK, Knudsen KE, Knudsen ES. RB-pathway disruption in breast cancer: differential association with disease subtypes, disease-specific prognosis and therapeutic response. Cell Cycle. 2010;9:4153–4163. doi: 10.4161/cc.9.20.13454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wurmbach E, Chen YB, Khitrov G, Zhang W, Roayaie S, Schwartz M, Fiel I, et al. Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology. 2007;45:938–947. doi: 10.1002/hep.21622. [DOI] [PubMed] [Google Scholar]
- 32.Andrisani OM, Studach L, Merle P. Gene signatures in hepatocellular carcinoma (HCC). Semin Cancer Biol. 2011;21:4–9. doi: 10.1016/j.semcancer.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chew V, Chen J, Lee D, Loh E, Lee J, Lim KH, Weber A, et al. Chemokine-driven lymphocyte infiltration: an early intratumoural event determining long-term survival in resectable hepatocellular carcinoma. Gut. 2012;61:427–438. doi: 10.1136/gutjnl-2011-300509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mayhew CN, Carter SL, Fox SR, Sexton CR, Reed CA, Srinivasan SV, Liu X, et al. RB loss abrogates cell cycle control and genome integrity to promote liver tumorigenesis. Gastroenterology. 2007;133:976–984. doi: 10.1053/j.gastro.2007.06.025. [DOI] [PubMed] [Google Scholar]
- 35.Hoshida Y, Nijman SM, Kobayashi M, Chan JA, Brunet JP, Chiang DY, Villanueva A, et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 2009;69:7385–7392. doi: 10.1158/0008-5472.CAN-09-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J, Liu H, Li L, Wu H, Wang C, Yan Z, Wang Y, et al. The combination of an oxygen-dependent degradation domain-regulated adenovirus expressing the chemokine RANTES/CCL5 and NK-92 cells exerts enhanced antitumor activity in hepatocellular carcinoma. Oncol Rep. 2013;29:895–902. doi: 10.3892/or.2012.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsai HT, Yang SF, Chen DR, Chan SE. CCL5-28, CCL5-403, and CCR5 genetic polymorphisms and their synergic effect with alcohol and tobacco consumptions increase susceptibility to hepatocellular carcinoma. Med Oncol. 2012;29:2771–2779. doi: 10.1007/s12032-012-0189-9. [DOI] [PubMed] [Google Scholar]
- 38.Singh R, Kaul R, Kaul A, Khan K. A comparative review of HLA associations with hepatitis B and C viral infections across global populations. World J Gastroenterol. 2007;13:1770–1787. doi: 10.3748/wjg.v13.i12.1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heinrichs D, Knauel M, Offermanns C, Berres ML, Nellen A, Leng L, Schmitz P, et al. Macrophage migration inhibitory factor (MIF) exerts antifibrotic effects in experimental liver fibrosis via CD74. Proc Natl Acad Sci U S A. 2011;108:17444–17449. doi: 10.1073/pnas.1107023108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Assis DN, Leng L, Du X, Zhang CK, Grieb G, Merk M, Garcia AB, et al. The role of macrophage migration inhibitory factor in autoimmune liver disease. Hepatology. 2014;59:580–591. doi: 10.1002/hep.26664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoshimoto S, Loo TM, Atarashi K, Kanda H, Sato S, Oyadomari S, Iwakura Y, et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature. 2013;499:97–101. doi: 10.1038/nature12347. [DOI] [PubMed] [Google Scholar]
- 42.Dapito DH, Mencin A, Gwak GY, Pradere JP, Jang MK, Mederacke I, Caviglia JM, et al. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell. 2012;21:504–516. doi: 10.1016/j.ccr.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.