Abstract
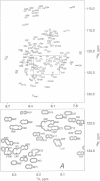
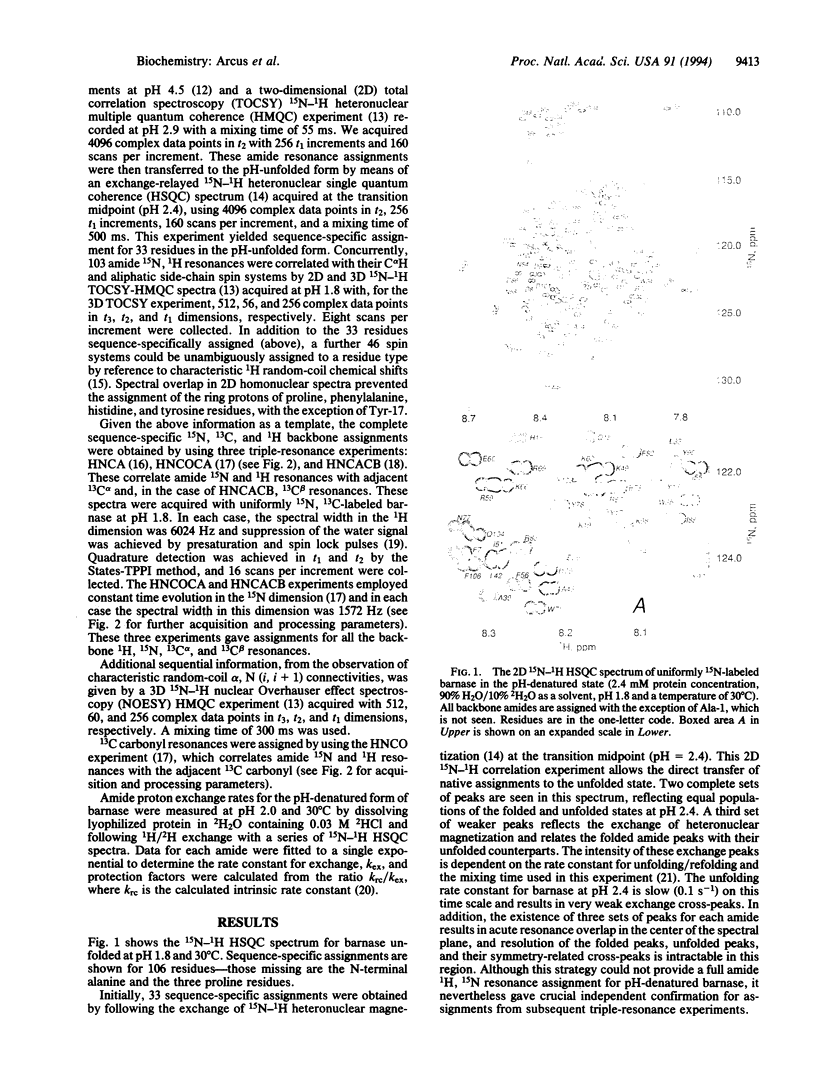
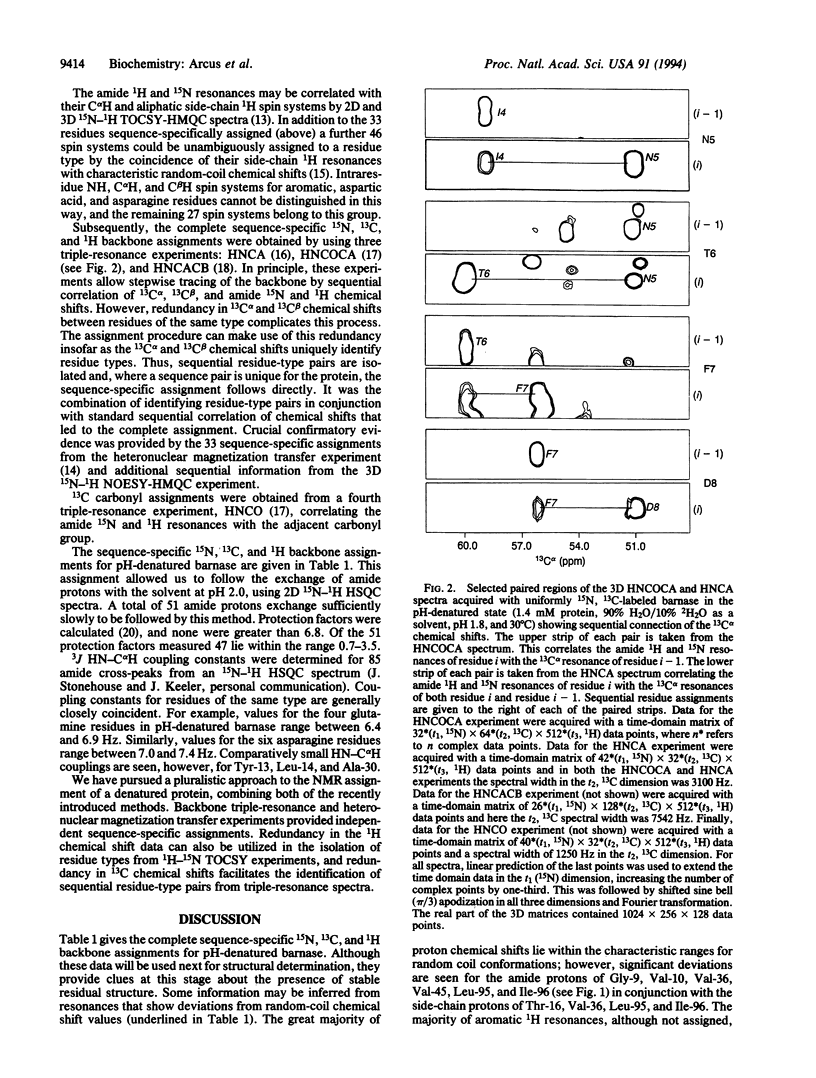
The structures of the major folding intermediate, the transition state for folding, and the folded state of barnase have been previously characterized. We now add a further step toward a complete picture of the folding of barnase by reporting the backbone 15N, 13C, and 1H NMR assignments for barnase unfolded at pH 1.8 and 30 degrees C. These assignments, which were obtained from a combination of heteronuclear magnetization transfer and backbone triple-resonance NMR experiments, constitute the first stage in the structural characterization of this denatured state by NMR. Interresidue nuclear Overhauser effect contacts and deviations from 1H random-coil chemical shifts provide evidence for stable residual structure. The structured regions span residues in the native protein that contain its major alpha-helix and central strands of the beta-sheet. Earlier experiments have shown that these regions are predominantly intact in the major folding intermediate and that their docking is partly rate determining in folding.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bai Y., Milne J. S., Mayne L., Englander S. W. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993 Sep;17(1):75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bycroft M., Sheppard R. N., Lau F. T., Fersht A. R. Sequential assignment of the 1H nuclear magnetic resonance spectrum of barnase. Biochemistry. 1990 Aug 14;29(32):7425–7432. doi: 10.1021/bi00484a011. [DOI] [PubMed] [Google Scholar]
- Farmer B. T., 2nd, Venters R. A., Spicer L. D., Wittekind M. G., Müller L. A refocused and optimized HNCA: increased sensitivity and resolution in large macromolecules. J Biomol NMR. 1992 Mar;2(2):195–202. doi: 10.1007/BF01875530. [DOI] [PubMed] [Google Scholar]
- Fersht A. R. The sixth Datta Lecture. Protein folding and stability: the pathway of folding of barnase. FEBS Lett. 1993 Jun 28;325(1-2):5–16. doi: 10.1016/0014-5793(93)81405-o. [DOI] [PubMed] [Google Scholar]
- Jones D. N., Bycroft M., Lubienski M. J., Fersht A. R. Identification of the barstar binding site of barnase by NMR spectroscopy and hydrogen-deuterium exchange. FEBS Lett. 1993 Sep 27;331(1-2):165–172. doi: 10.1016/0014-5793(93)80319-p. [DOI] [PubMed] [Google Scholar]
- Logan T. M., Thériault Y., Fesik S. W. Structural characterization of the FK506 binding protein unfolded in urea and guanidine hydrochloride. J Mol Biol. 1994 Feb 18;236(2):637–648. doi: 10.1006/jmbi.1994.1173. [DOI] [PubMed] [Google Scholar]
- Marion D., Driscoll P. C., Kay L. E., Wingfield P. T., Bax A., Gronenborn A. M., Clore G. M. Overcoming the overlap problem in the assignment of 1H NMR spectra of larger proteins by use of three-dimensional heteronuclear 1H-15N Hartmann-Hahn-multiple quantum coherence and nuclear Overhauser-multiple quantum coherence spectroscopy: application to interleukin 1 beta. Biochemistry. 1989 Jul 25;28(15):6150–6156. doi: 10.1021/bi00441a004. [DOI] [PubMed] [Google Scholar]
- Mossakowska D. E., Nyberg K., Fersht A. R. Kinetic characterization of the recombinant ribonuclease from Bacillus amyloliquefaciens (barnase) and investigation of key residues in catalysis by site-directed mutagenesis. Biochemistry. 1989 May 2;28(9):3843–3850. doi: 10.1021/bi00435a033. [DOI] [PubMed] [Google Scholar]
- Neri D., Billeter M., Wider G., Wüthrich K. NMR determination of residual structure in a urea-denatured protein, the 434-repressor. Science. 1992 Sep 11;257(5076):1559–1563. doi: 10.1126/science.1523410. [DOI] [PubMed] [Google Scholar]
- Neri D., Wider G., Wüthrich K. Complete 15N and 1H NMR assignments for the amino-terminal domain of the phage 434 repressor in the urea-unfolded form. Proc Natl Acad Sci U S A. 1992 May 15;89(10):4397–4401. doi: 10.1073/pnas.89.10.4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddon C. J., Hartley R. W. Expression of Bacillus amyloliquefaciens extracellular ribonuclease (barnase) in Escherichia coli following an inactivating mutation. Gene. 1987;53(1):11–19. doi: 10.1016/0378-1119(87)90088-6. [DOI] [PubMed] [Google Scholar]