Abstract
Endophytes are microbes and fungi that live inside plant tissues without damaging the host. Herein we examine the dynamic changes in the endophytic bacterial community in potato (Solanum tuberosum) tuber in response to pathogenic infection by Pectobacterium atrosepticum, which causes soft rot in numerous economically important crops. We quantified community changes using both cultivation and next-generation sequencing of the 16S rRNA gene and found that, despite observing significant variability in both the mass of macerated tissue and structure of the endophytic community between individual potato tubers, P. atrosepticum is always taken over by the endophytes during maceration. 16S rDNA sequencing revealed bacteria from the phyla Proteobacteria, Actinobacteria, Firmicutes, Bacteroidetes, Fusobacteria, Verrucomicrobia, Acidobacteria, TM7, and Deinococcus-Thermus. Prior to infection, Propionibacterium acnes is frequently among the dominant taxa, yet is out competed by relatively few dominant taxa as the infection proceeds. Two days post-infection, the most abundant sequences in macerated potato tissue are Gammaproteobacteria. The most dominant genera are Enterobacter and Pseudomonas. Eight days post-infection, the number of anaerobic pectolytic Clostridia increases, probably due to oxygen depletion. These results demonstrate that the pathogenesis is strictly initiated by the pathogen (sensu stricto) and proceeds with a major contribution from the endophytic community.
A wide diversity of bacteria interact with plants and form a broad spectrum of communities that often differ between plant species and contribute to the development and health of plants in various ways. Endophytes are often defined as microbes and fungi that live within plant tissues without either damaging the host or eliciting symptoms of plant disease1. However, some organisms that have been classified as endophytes seem to be latent pathogens and, under certain conditions, either induce or participate in host infections. Interestingly, both endophytic and pathogenic bacteria utilize similar molecular mechanisms to interact with their plant hosts2.
A considerable fraction of endophytes in plant roots originate from soil. It seems that a relatively small number of endophytes are recruited from the large pool of soil species and clones that exist within the bulk soil or the narrow region of soil that interacts with the plant root, termed the rhizosphere. The bacterial community that lives within root tissues, the root endosphere, is less diverse than the communities in the rhizosphere3.
In most plants, more endophytes are found in roots than in above-ground tissues4. The species composition of the bacterial communities that associate with plants fluctuates depending on the genotype and growth stage of the host plant, environmental conditions, and the type of soil5,6,7,8.
Endophytic communities are typically dominated by relatively few bacterial phyla. The most abundant bacterial species found within potato, rice, and Arabidopsis tissues are classified under the phyla Actinobacteria, Bacteroidetes, Proteobacteria, and Firmicutes5,9,10. There is a taxonomically narrow set of core root microbiota from the orders Actinomycetales, Burkholderiales, and Flavobacteriales found in three Arabidopsis species, and these remain stable under both natural and controlled environments11,12.
In potato (Solanum tuberosum), the species composition in the rhizosphere and endosphere depends on both the potato cultivar and the growth stage of the plant9,13,14.
Most plant microbiota studies address growing plants15,16,17,18, however, some have examined post-harvest endophyte communities. To the best of our knowledge, the most recent study of the microbial population dynamics within potato tubers during the storage was conducted by Perombelon and coworkers in 197919 and was limited to a few bacterial taxa.
Endophytes have historically been identified using cultivation-based methods. However, not all endophytes can be cultivated due to either unknown growth requirements or the presence of bacteria that are in a viable but not in a cultivatable state20. Various molecular techniques provide means to overcome these difficulties21,22.
We have previously shown that the resident bacterial community within potato tubers can survive in considerable numbers despite surface-sterilization with hypochlorite acid and prior to inoculation with the plant pathogen, Pectobacterium wasabiae23. Nevertheless, our previous study focused on pathogenesis and did not aim to describe changes within the endophytic bacterial community. Now we aim to cover this gap by describing how the endophytic microbial community within post-harvest potatoes changes in response to soft rot pathogen attack.
Results
To characterize the microbial communities in potato tuber, we used potatoes harvested in 2012 (Experiment 1) and 2013 (Experiment 2). Experiment 1 was performed using potato tubers that were stored until March 2013 while Experiment 2 was performed using tubers stored until December 2013. Because of this time difference we expected that the results of these experiments would differ while preserving some common features that could be interpreted as reflecting general aspects of changes in the potato microbiome in response to infection with Pectobacterium atrosepticum SCRI1043.
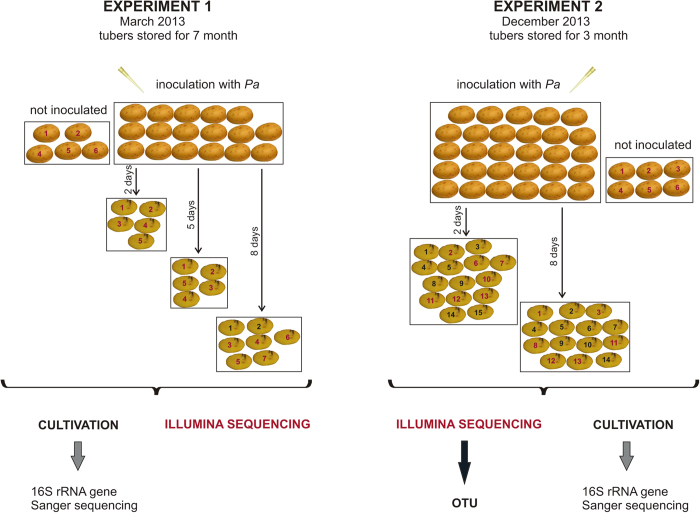
Every potato tuber was infected with the minimum number of P. atrosepticum cells (1.5 × 104 cells per ml) required to induce maceration (as determined by preliminary experiments). In Experiment 1, we sampled the macerated tissue from potato tubers incubated for either two, five, or eight days after infection with P. atrosepticum. The fifth day of sampling was not performed in Experiment 2 because the bacterial counts determined in Experiment 1 for the fifth and eighth day samples were quite similar. Figure 1 provides an overview of the experimental setup.
Figure 1. Schematic representation of the experimental setup.
Bacteria were cultivated from all the potatoes marked with numbers. Red numbers indicate that the Illumina sequencing was performed in addition to the cultivation. Pa designates inoculation with the pathogen, P. atrosepticum.
Cultivable bacteria in P. atrosepticum macerated potato tissue
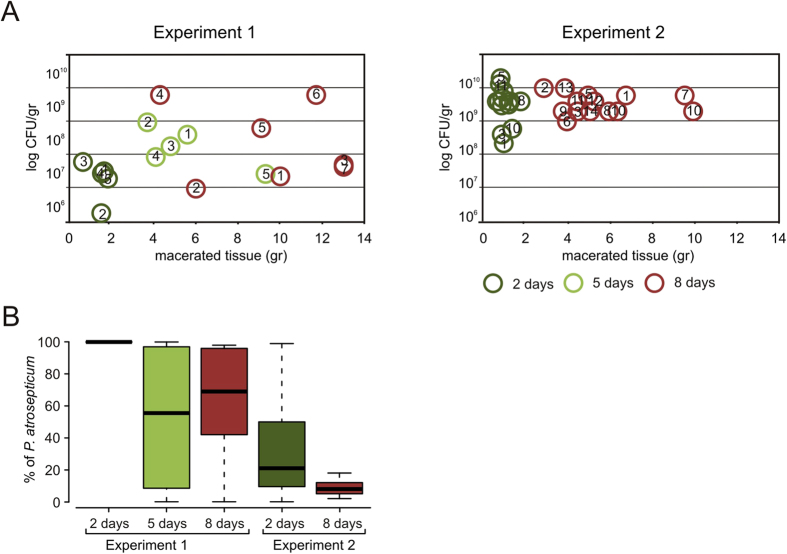
First, we examined the dynamics of cultivable bacteria in macerated potato tissue. Bacterial CFU counts of macerated potato tuber tissue started to rise quickly (Fig. 2). In both experiments, the CFU/g reached 108–1010 by the eighth day post-infection (Fig. 2A). Although the mass of macerated tissue was similar in the two experiments, fluctuations in the CFU/g for individual potatoes were significantly higher in the first. The CFU/g in uninfected potatoes ranged from 2 × 104–105 in both experiments.
Figure 2. Potato tuber maceration by P. atrosepticum and resident cultivable microbiome.
(A) The amount of macerated potato tissue and the content of cultivable bacteria in the macerated tissue. Numbering of potato samples, indicated inside the circle, is recurrent throughout the study. (B) The proportion of P. atrosepticum in the entire population of cultivated bacteria in the macerated potato tissue.
The quantity of P. atrosepticum among all cultivated bacteria was determined on the basis of 50 randomly picked colonies from each sample. The level of P. atrosepticum was quite different in the two experiments, although the mass of macerated tissue was similar (Fig. 2). In Experiment 1, no other bacteria, aside from P. atrosepticum, was present among the 50 colonies taken from the macerated tissue two days after infection. Other bacteria appeared on the fifth day post-infection, although the differences between individual potato tubers were remarkable. Similar infection levels occurred by the eighth day post-infection. By the second day after inoculation in Experiment 2, endophytic bacteria had gained ground in the macerated tissue within most tubers. P. atrosepticum continued to decrease and did not make up >20% of the total CFU by the eighth day, although the mass of macerated tissue varied between 3 to 11 g per tuber. There was no correlation between the total CFU, % of P. atrosepticum, and the amount of macerated tissue in either experimental series (Fig. 2).
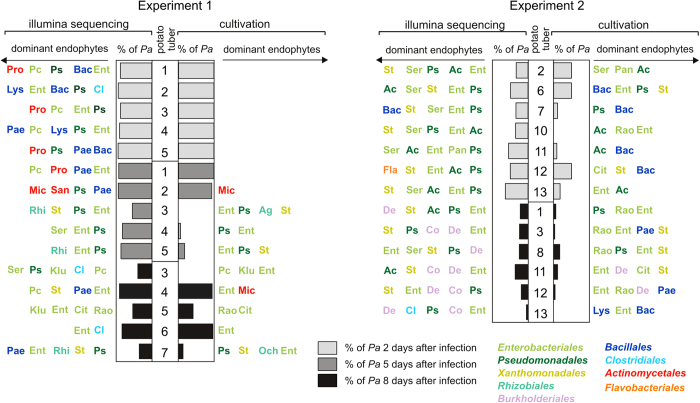
The endophytic bacteria were grouped by phenotypical and morphological characteristics and followed by 16S rDNA sequencing. Altogether, 82 different bacterial strains from four phyla: Proteobacteria, Actinobacteria, Bacteroidetes and Firmicutes - were isolated (Fig. 3). The Gammaproteabacteria were the most dominant cultivatable taxon and these were largely comprised of bacteria from two groups, Enterobacteriaceae and Enterobacter (Lelliottia). In second place was the Pseudomonaceae family with various species of Pseudomonas. These two families were those most prevalent in most samples from both experiments.
Figure 3. Comparison of cultivation and Illumina sequencing results.
The columns indicate the proportion of P. atrosepticum (Pa) in the entire population of cultivated and Illumina sequenced bacteria in the macerated potato tissue. Five most prevalent bacterial genera based on cultivation (right-hand side of the columns) and Illumina sequencing (left-hand side of the columns) that are present ≥1% in particular potato sample: Cit - Citrobacter; Ent - Enterobacter; Klu - Kluyvera; Pan - Pantoea; Pc - Pectobacterium carotovorum; Rao - Raoultella; Ser - Serratia; Ps - Pseudomonas; Ac - Acinetobacter; St - Stenotrophomonas; Ag - Agrobacterium; Och - Ochrobactrum; Rhi - Rhizobium; Co - Comamonas; De - Delftia; Bac - Bacillus; Lys - Lysinibacillus; Pae - Paenibacillus; Cl - Clostridium; Mi - Microbacterium; Pro - Propionibacterium; San - Sanguibacter; Fla - Flavobacterium. Strains are colored according to their respective order. The arrow indicates the direction of decreasing amount of bacteria.
A higher variability of prevalence in the samples occurred among the bacteria from the phylum Firmicutes, and more precisely Paenibacillus sp., Lysinibacillus fusiformis, and several Bacillus species. The number of Acinetobacter calcoaceticus detected was high, however, was exclusively found in the second day post-infection in Experiment 2.
Amplicon analyses by Illumina sequencing
Five to seven potato tubers were randomly chosen from each time-point in both experiments to follow bacterial community dynamics using rRNA gene mass-sequencing. Note that the designation of potato tubers (Fig. 2) for cultivated bacteria is kept the same for designation of the sequencing results. Approximately 300 bp of the 16S rRNA gene spanning the variable regions V1 and V2 was amplified and sequenced. In Experiment 1, this generated ~100 bp high quality reads that randomly cover the 300 bp V1–V2 region of 16S rRNA gene. In total, 2.6 × 106 reads were obtained with sequences per infected potato ranging from 9 × 103 to 5 × 105 high-quality tags. About 99.9% of the sequence reads obtained from samples of uninfected potato tubers were classified as chloroplasts and not further analyzed. The number of chloroplast reads in the infected potato samples was <1.5%. In Experiment 2, sequencing generated 250 bp high quality reads spanning V1 and V2. In total, 8.5 × 107 reads were obtained with sequences per infected potato ranging from 3.8 × 104 to 8.8 × 105 high-quality tags. As with Experiment 1, ~99.9% reads from uninfected potatoes were classified as chloroplasts and left unanalyzed. As a consequence, the number of reads of bacterial origin from uninfected potato samples was 2–3 magnitudes lower (61–1385 in Experiment 1 and 84–1914 in Experiment 2) than in the samples of macerated potato tissue.
Survival of P. atrosepticum during tuber maceration
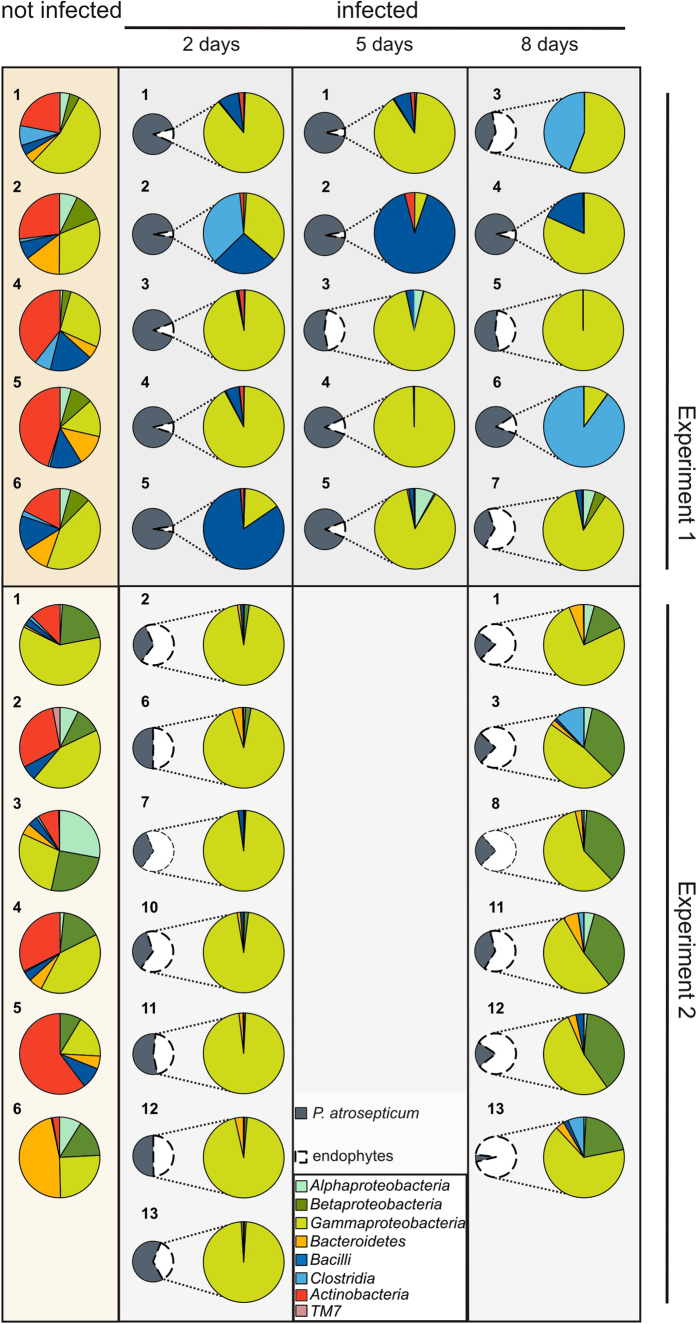
Survival of P. atrosepticum was assessed by counting all sequences that had >98% similarity with the P. atrosepticum 16S rRNA gene. A 98% cutoff was chosen to exclude reads derived from intrinsic Pectobacteria that inhabit the tuber, e.g., the identity of the region V1–V2 of P. atrosepticum and Pectobacterium carotovorum is 97%. Figure 4 provides the ratio of P. atrosepticum reads within the entire dataset of 16S rRNA gene reads. The survival results provided by Illumina sequencing agree with those obtained using bacterial cultivation (Fig. 3). Potatoes sampled in Experiment 1 displayed both higher absolute counts of P. atrosepticum and much higher variation in P. atrosepticum counts than the potatoes sampled in Experiment 2.
Figure 4. Phyla/class level distribution of all sequence reads from each potato sample.
Taxonomic assignments were made using the Ribosomal Database Project (50% confidence threshold). Small pie charts show the percentage of P. atrosepticum reads (dark grey 98% identity) among total sequence reads from individual potatoes. The larger pie charts show the composition of the dominant endophytic bacterial community, the colored segments represent bacterial phyla/classes that were most abundant in each sample. (The numbering of potato samples is recurrent throughout the study).
Diversity of the resident microflora in potato tubers
To assess the structure of the endophytic bacterial community in macerated potato tissue, we excluded P. atrosepticum sequences and analyzed the remaining sequences. All high quality reads from both uninfected and infected potatoes were classified using the RDP classifier24. Altogether, sequences from phyla Proteobacteria, Actinobacteria, Firmicutes, Bacteroidetes, Fusobacteria, Verrucomicrobia, Acidobacteria, TM7, and Deinococcus-Thermus, were detected. In both experiments, the dominating sequences all belong to the phyla Proteobacteria, Firmicutes, Actinobacteria, and Bacteroidetes (Fig. 4).
To characterize the community structures in more detail, sequences from the 19 samples of Experiment 2 were pooled and grouped into operational taxonomic units (OTU) based on >98% identity. The minimal permitted number of reads per OTU was five. Because the 100 bp sequences obtained from Experiment 1 randomly covered the 300 bp V1–V2 region, it was impossible to group these reads to generate OTUs. Therefore, the sequences of Experiment 1 were grouped into the OTUs using the results of Experiment 2 (Supplemental Data 1).
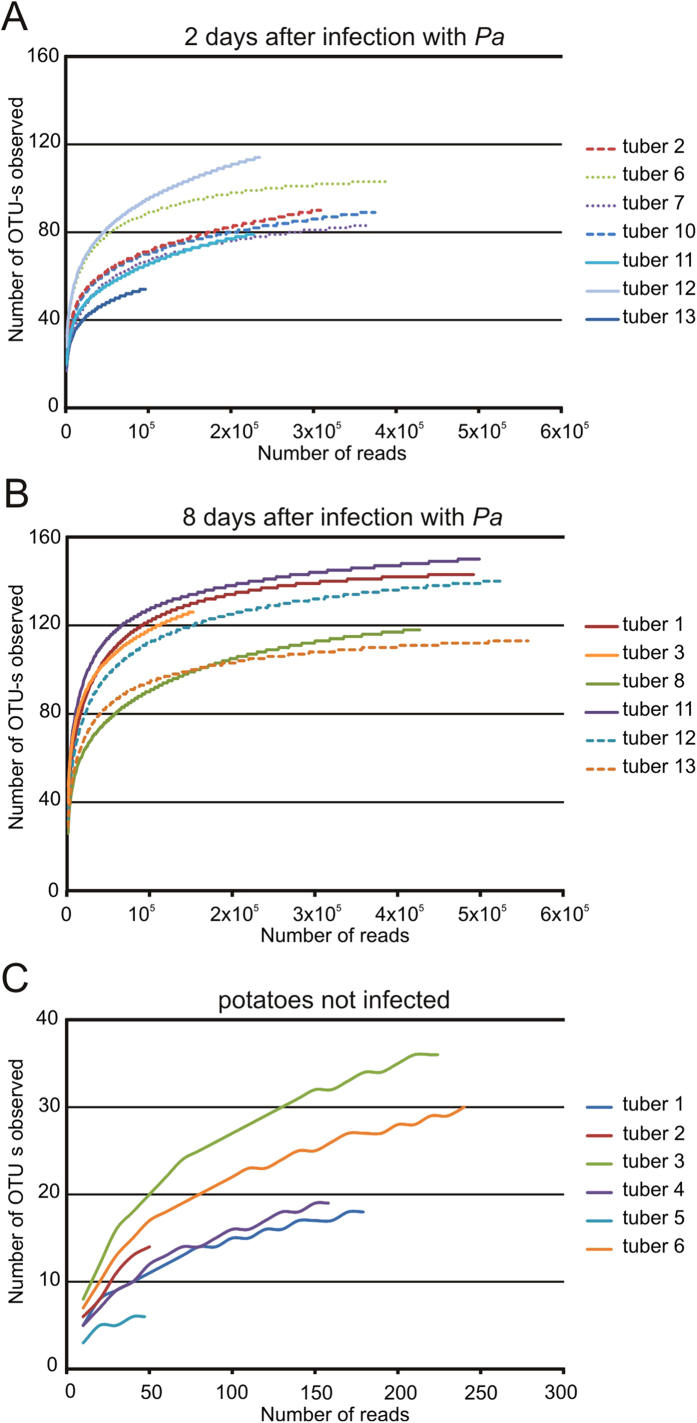
Although the sequencing was very deep (4,780,691 pooled sequences of Experiment 2), only 294 OTUs were detected in Experiment 2. Rarefaction curves show that bacterial diversity, as assessed by the number of OTUs, increased during maceration, starting from 50–114 OTUs on the second day post-infection and reached 113–150 OTUs by the eighth day (Fig. 5). The small amount of OTUs (6–36) detected within uninfected potato samples might be caused by the very low number of sequence reads from these samples, however, most rarefaction curves indicate saturation in the samples (Fig. 5).
Figure 5. Rarefaction analysis curves of endophytic bacterial 16S rDNA sequences of Experiment 2.
Sequences were clustered into OTU - s (≥5 sequences per OTU) using a sequence identity threshold of 98%. (A) Macerated potato tuber tissue samples after two days of incubation after infection with P. atrosepticum (Pa); (B) Macerated potato tuber tissue samples after eight days of incubation after infection with P. atrosepticum (Pa); (C) Uninfected potato tuber samples.
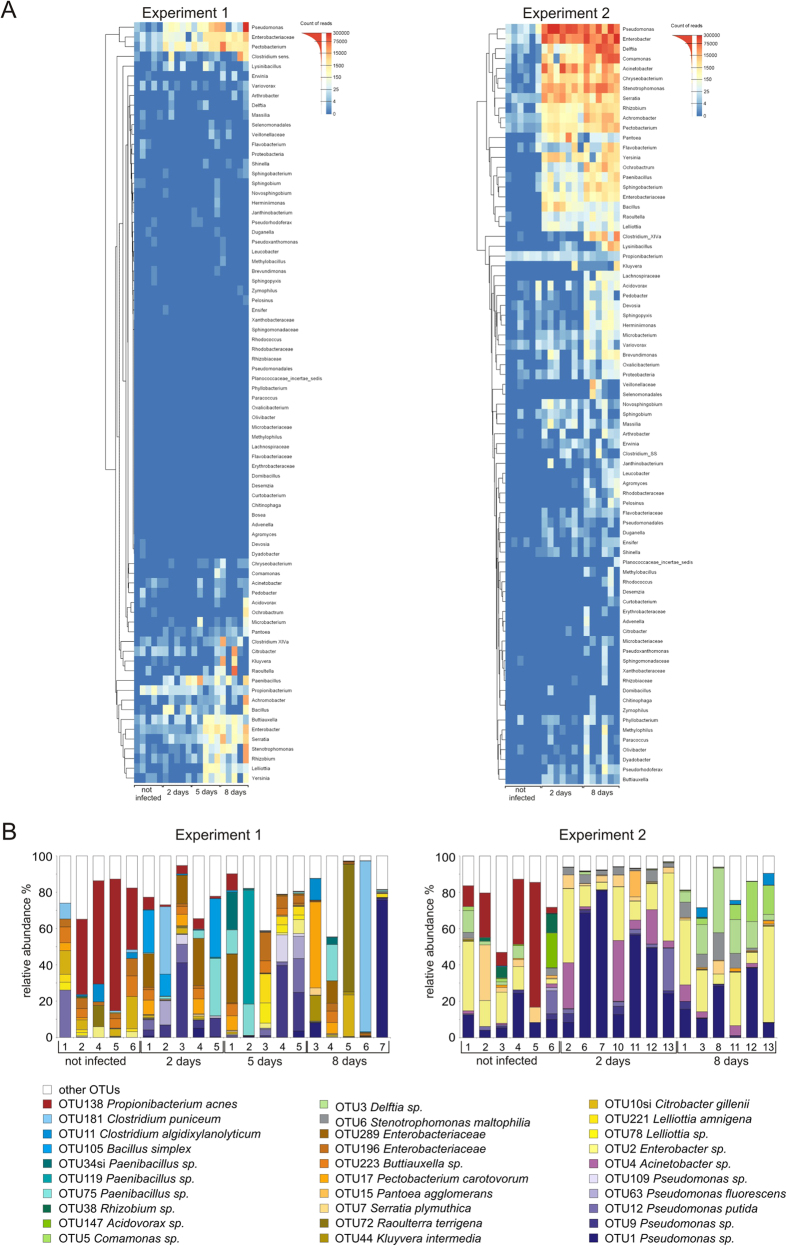
OTU sequences were classified at the genus level using the RDP classifier (Fig. 6). In cases where it was impossible to classify the OTUs at the genus level, the next higher taxonomical rank was used. OTUs generated based on the data from Experiment 2 were used to cluster the sequences in Experiment 1, which resulted in over 75% of the sequences being classified. The sequences were counted and used to make a heatmap as in Experiment 2 (Fig. 6A). To see the extent to which OTUs from Experiment 2 represent Experiment 1, the heatmap was based on the entire dataset of Experiment 1 as classified at the genus level by the RDP classifier. Only a few differences between the two heatmaps from Experiment 1 (Fig. S1) were found: the RDP classified total dataset contains more genera with very low sequence counts; a greater abundance of the family Lachnospiraceae (samples 3 and 6 from day eight), and of the enterobacterial genera Yersinia, Buttiauxella, and unclassified Enterobacteriaceae (sample 5 from day eight). We conclude that this classification method and analysis is robust.
Figure 6. Distribution of endophytic bacteria according to mass-sequencing results.
(A) Sequencing reads of Experiment 2 were clustered with AbundantOTU; all the chimeric clusters were removed56. Consensus sequences (OTUs) were classified using the RDP classifier 24 and clustered at the genus level. OTU sequences from Experiment 2 were used to cluster all similar sequences from Experiment 1. Data have been normalized using inverse hyperbolic sine transformation to make a heatmap. Heatmaps were made in R using pheatmap (parameters: clustering_method = average, clustering_distance = euclidean). (B) Dominating OTUs that are present ≥10% in any of the samples in Experiments 1 and 2.
There were a few very abundant OTUs in the samples from Experiment 2 (Fig. 6B). In the samples from the second day post-infection Pseudomonas sp. OTU1, Lelliottia amnigena, OTU2 and Acinetobacter calcoaceticus OTU4 covered 65–85% of all sequence reads. In the samples from the eighth day, four OTUs dominated, Pseudomonas sp. OTU1; Lelliottia amnigena OTU2; Delftia sp OTU3 and Comamonas sp. OTU5, and these covered between 53–80% of all sequence reads. OTU138 (Propionibacterium acnes) was highly abundant in uninfected potatoes.
The distribution of OTUs was highly variable in the potato tubers sampled during Experiment 1. For example, Pseudomonas sp. OTU1 covered 75% of all sequence reads in the sample 7 of eighth day, but <4% in other infected potato samples, and was not found in uninfected potatoes. The spectrum of dominant bacterial groups proved to be very different from Experiment 2, especially the dominance of Firmicutes (Bacillus, Paenibacillus, Clostridium). The only clear similarity between the two experiments was the abundance of Propionibacterium acnes OTU138 in uninfected potato samples.
Endophytic population dynamics in P. atrosepticum infected potato tubers
The succession of bacteria in P. atrosepticum infected potato tuber tissue developed uniformly in Experiment 2. During the first two days of maceration, the fast growing members of family Enterobacteriaceae (dominated by Enterobacter sp.) and Pseudomonaceae (dominated by various Pseudomonas strains and Acinetobacter sp.) began to flourish in the macerated tissues of all potatoes investigated. During the total progress of maceration (eight days), the Gammaproteobacteria were partially taken over by Betaproteobacteria, mainly from the closely related genera Comamonas and Delftia and to a lesser degree by other bacteria who are members of Alfaproteobacteria, Bacteroidetes, Clostridia, and Veillonellaceae.
In Experiment 1, the endophytic bacterial succession during maceration varied between the different potato tubers sampled: after two days post-infection with P. atrosepticum (samples 2 and 5), the members of phylum Firmicutes - Bacilli (Paenibacillus) and Clostridia (Clostridium puniceum) - dominate in some potato tubers, but not in others (samples 1, 3, and 4) Gammaproteobacteria (Enterobacter sp., Pseudomonas spp.). The same tendency continued after eight days of infection. In both experiments, the quantity of Clostridia increased as maceration progressed, probably as a consequence of oxygen depletion, although the amount remained much lower in Experiment 2 (Fig. 4). The number of representatives of bacteria that start to dominate over P. atrosepticum during maceration was already higher before infection, Betaproteobacteria in Experiment 2 and Firmicutes in Experiment 1 (Fig. 4).
It is possible that some taxa have positive or negative interactions between others. In Experiment 1 the complexity is high and considering the low number of potato tubers analyzed any association analysis is of limited value. In Experiment 2 there was a smaller number of dominating taxa and we performed an association analysis58. The results indicate that at day two the tubers have either a higher fraction of Endobacter (Lelliottia) and Acinetobacter or Pseudomonas (Fig. 7). These associations are not as significant in day eight, however, inspecting the corresponding OTUs presented in Fig. 6B suggests similar trend.
Figure 7.
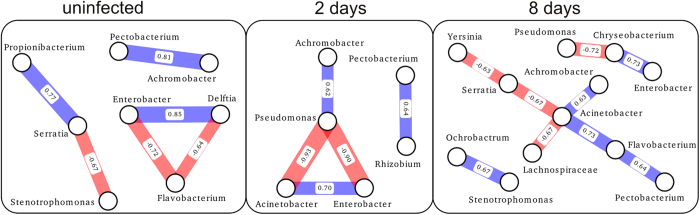
Genera level bacterial correlation networks during P. atrosepticum infection for Experiment 2. Each network was inferred by analyzing the fractional abundance of bacteria with one set of individual potatoes using SparCC58. Connections between nodes give the correlations between genera that form greater than 0.2% of the bacterial population in any given potato within one set. Only connections with correlation values >0.6 and pvalues < = 0.04 are included in the network. Red indicates a negative correlation and blue indicates a positive correlation and the width of each edge is proportional to the pvalue. The most significant relationship is the antagonistic triangle between Pseudomonas, Acinetobacter, and Enterobacter that emerges two days after infection.
Comparing bacterial isolates with Illumina sequencing data
Both methods clearly demonstrate that the fraction of P. atrosepticum decreases during infection (Fig. 3). As expected, not all of the taxa detected by sequencing are cultivatable under the conditions used. In general, the endophytic bacteria from potato tubers can be efficiently cultivated and the data are in agreement with sequencing results.
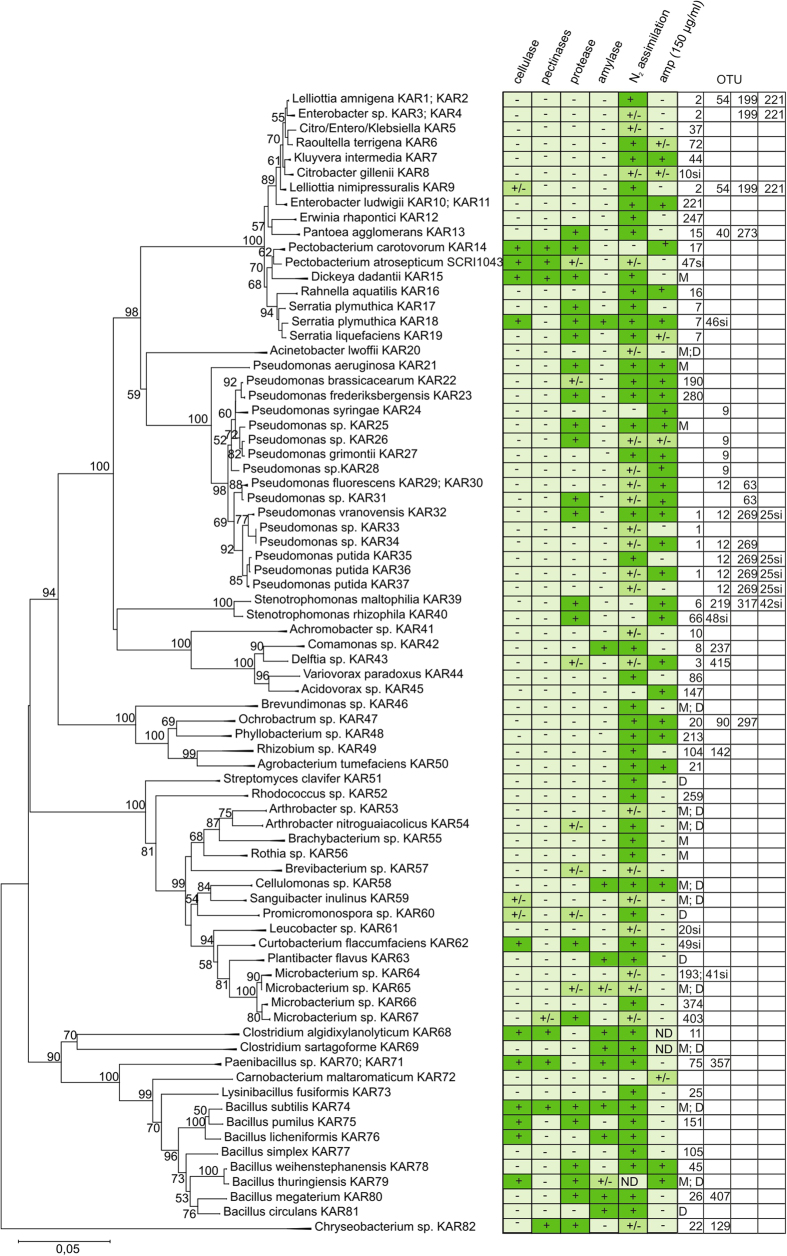
We investigated how isolated bacteria are represented in the pool of OTUs. The isolates are marked with the letters “KAR” and a corresponding number. To identify OTUs, a BLAST homology search was performed against the 16S rDNA sequences of cultivated strains using >98% sequence identity cutoff. A phylogenetic tree was constructed based on the nearly full-length 16S rRNA gene sequence and their closest evolutionary counterparts retrieved from the GenBank database (Fig. S2). Only bacteria isolated in the present work (KAR) are shown as tree branches in Fig. 8. The phenotypic descriptions and OTUs that display >99% identity to the 16S rDNA of isolated bacteria have also been added to the tree. A 99% identity cutoff was used because the two most abundant bacterial groups in the macerated potato tissue, Enterobacter (Lelliottia) and Pseudomonas cannot be discriminated with a lower cutoff. Raoultella terrigena KAR5 (OTU72), Kluyvera intermedia KAR7 (OTU44) and Citrobacter gillenii KAR8 (OTU10si) are well differentiated from each other at a 99% identity level, but fall into the same branch as Enterobacter isolates.
Figure 8. Phylogenetic tree of cultivable bacteria based on the 16S rRNA gene sequence comparison.
The tree was constructed from the isolated 16S rRNA bacterial sequences with their respective reference sequences from GenBank. Only bacteria isolated in this experiment are shown as tree branches (full tree is provided in additional Fig. 2). Numbers at the nodes indicate the percentages of occurrence in 1,000 bootstrapped trees; only values >50% are shown. The bar indicates 0.05 substitutions per nucleotide position. The degradation capacity of the main plant polymers CMC - carboxymethylcellulose, PGA-polygalacturonic acid, proteins (casein) and starch are provided. Fixation of N2 and sensitivity to ampicillin are also indicated. OTU-s matching to the 16S rDNA of isolated bacteria at >99% identity have been added to the tree. M - sequences in a single sequences pool of Experiment 1 matching with >98% identity to the corresponding cultivated 16S rDNA sequence; D - sequences in single sequences pool of Experiment 2 matching with >98% identity to the corresponding cultivated 16S rDNA.
A disturbance in the 16S rDNA phylogenetic tree is created by isolate KAR5 (OTU37) whose 16S rDNA sequence matches equally well with genera Citrobacter, Enterobacter, and Klebsiella. After cloning and sequencing 16S rDNA fragments from five clones, we found between one to three nucleotide differences in the sequences of this strain which do not change the phylogenetic affiliation of this strain. The quantity of the strain KAR5 (OTU37) found during the maceration process was distinctive; it increases during maceration while other Enterobacter strains decrease (Fig. 9).
Figure 9. Relative abundance of the main Enterobacter sp.
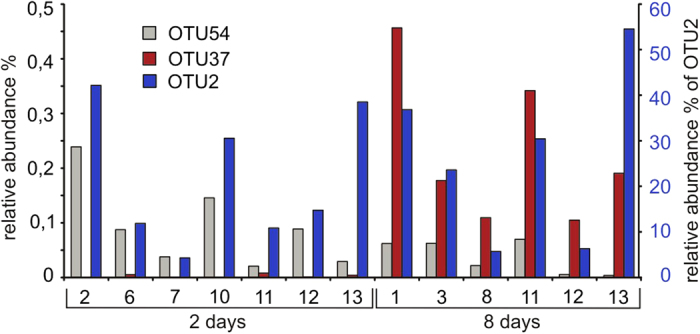
OTUs. Individual potatoes are indicated on the x-axis. Values for OTU37 and OTU54 are indicated on the left y-axis and values for OTU2 on the right y-axis.
As with the Enterobacter group, the most abundant Pseudomonas strains that belong to the Ps. putida group (OUT1; OTU12; OTU269, and OTU25si) and the Ps. fluorescence group (OTU9, OUT12 and OTU63) cannot be discriminated based on their 16S rDNA sequences, while other less abundant strains are clearly distinctive (Fig. 8).
Most of the isolates are represented by Illumina sequencing reads with minor exceptions: Brevibacterium sp. KAR46, Carnobacterium maltoaromaticum KAR72 and Bacillus licheniformis KAR76. The number of isolates that are represented with <5 16S rDNA sequences, i.e. not used to generate an OTU, are quite high. The absence of sequences that belong to Bacillus licheniformis is unexpected because this easily cultivatable and recognizable bacterium was present in a number of potato samples (data not shown). The proportion of Bacilli in rDNA sequencing samples was also clearly lower compared to cultivated samples (Fig. 3).
Reversing the analyses to examine how well the sequence reads are covered by cultivatable taxa showed that most of the dominant bacteria can be cultivated, including Clostridia which was isolated anaerobically from frozen samples after its presence was detected by mass sequencing. The anticipated number of different OTUs (294) proved to be more numerous than the number of cultivated bacterial strains (82). One dominant bacterial strain from the macerated tissue from Experiment 2, Acinetobacter calcoaceticus (OTU4), was not found in our library of cultivated bacteria which we isolated and identified by MALDI phenotyping, but lost during the purification process. This also occurred with some less dominant Actinobacteria spp.
Phenotypic characterization of isolated bacterial strains
Microbial endophytes that colonize potato tissue are widespread and some of the non-pathogenic endophytes could turn into phytopathogens that are able to induce infection symptoms. The virulence potential of potato endophytes was assessed by their ability to produce extracellular enzymes and their ability to attack plant cell wall components (the most important virulence factors of soft rot bacteria). Degradation of pectic substances (PGA), cellulose (CMC), starch and proteins (casein), was tested on indicator plates. Bacteria that belong to the phylum Firmicutes, Paenibacillus sp, Bacillus subtilis, and the anaerobic species Clostridium algidixylanolyticum, can all degrade the main polymers in a potato tuber, including pectin, cellulose, and starch (Fig. 8). The Enterobacterial plant pathogens, Pectobacterium carotovorum and Dickeya dadantii, degrade plant cell wall components, but not starch. Some Bacilli, Chryseobacterium sp., Curtobacterium flaccumfaciens, and Sanguibacter inulinus, can hydrolase either pectin or cellulose. Starch is degraded by only a few bacteria and their number tended to increase during the maceration process when the slow growing and oxygen-sensitive Clostridia begin to dominate over other bacteria.
To characterize potential symbionts, bacterial assimilation of N2 under conditions of low oxygen was followed by the growth of bacteria on nitrogen deficient agar medium modified from a previously published method25. This modified medium allows one to grow a wider spectrum of bacterial taxa than those variants previously reported. We found that N2 assimilation is intrinsic to the majority of endophytic bacteria in potato. This feature was less common among pseudomonads and representatives of the phylum Actinobacteria, however, we might not have found a suitable medium to detect N2 assimilation for these bacteria.
While isolating pseudomonads on LB agar supplemented with ampicillin to eliminate other endophytic ampicillin-sensitive bacteria, some Pseudomonas strains also seemed to be sensitive to ampicillin. Ampicillin sensitivity of all isolated bacteria was therefore explored. Unexpectedly, the sensitivity of bacteria was not predictable by their systematic affiliation. Several enterobacteria and bacilli are not sensitive, while pseudomonads can be sensitive to ampicillin at 150 μg/ml, a technique generally used to distinguish these bacteria.
Discussion
The recent development of second generation sequencing techniques has enabled considerable advances in describing plant endophyte communities; however, much less attention has been given to study changes in the composition of the microbial community during plant decay. Here, we infected surface-sterilized potatoes with the pectolytic plant pathogen P. atrosepticum using a method adopted in the laboratory to study plant-pathogen interactions. The experiment was carried out twice with two main differences between them, (i) the year of harvesting (and thereby conditions of plant cultivation) and (ii) the length of storage. Although the results of the two experiments are variable, several common features were observed.
Sequencing analysis shows the presence of bacteria in tubers from the phyla Proteobacteria, Actinobacteria, Firmicutes, Bacteroidetes, Fusobacteria, Verrucomicrobia, Acidobacteria, TM7, and Deinococcus-Thermus. This agrees with other reports that have demonstrated that the members of phyla Proteobacteria, Actinobacteria, Firmicutes, and Bacteroidetes dominate in several plants, including potato9,26,27. The most abundant species in macerated potato tissues belong to the class, Gammaproteobacteria - Pseudomonas sp. (putida) and Enterobacter sp. (Leliottia). Strains of the genus Enterobacter frequently are described in studies on different plant endophytes8,28,29,30,31. These strains are commensals, or non-pathogenic microbes, that invade the intercellular space of the host, but are usually dormant. They often have a close relationship with the host plants, as suggested by their widespread ability to assimilate N2. We found that, despite their mutualistic nature, these strains begin to proliferate exponentially when pectolytic pathogens initiate an infection process. The same is true for pseudomonads, which are well known plant growth-promoting bacteria8,29,32,33, yet begin to flourish in Pa-infected potato tubers. Pseudomonas and Enterobacter strains probably do not help P. atrosepticum to decay tuber tissue directly, but utilize sugars and sugar acids liberated during the decay process. Our data also suggest that the pathogenesis process can proceed with the domination of either Pseudomonas or Enterobacter. This interaction might be considered when analyzing other datasets on plant microbiomes.
Like many genera within the family Enterobacteriaceae, Enterobacter is polyphyletic based on the 16S rRNA gene34,35,36. A possible reclassification of E. amnigenus and E. nimipressuralis into the novel genus Lelliottia as Lelliottia amnigena comb. nov. and Lelliottia nimipressuralis comb. nov., respectively, was introduced36. The dominant enterobacterial isolates in our infected potato tubers belong to this genus, but cannot be discriminated at the species level by their 16S rDNA sequences. The high number of diverse Enterobacter/Lelliottia strains in the macerated potato tissue can form a platform for specialization into diverse micro niches during infection, thereby increasing the fitness of this taxonomic group.
In both experiments, bacteria that are rare at the beginning of the infection begin to multiply during maceration. In Experiment 2, Comamonas, Delftia, and other closely related betaproteobacteria, started to out-compete the sugar-consuming Enterobacter and Pseudomonas strains. Bacteria that belong to the family Comamonadaceae prefer organic acids and amino acids rather than carbohydrates, and can degrade a wide variety of complex aromatic compounds (according to Bergey’s Manual of Systematic Bacteriology). The Comamonas sp. KAR42 that we isolated also possess amylase activity that may provide a growth advantage in potato tubers. Another large group of bacteria that can dominate over gammaproteobacteria during the maceration process is comprised of different members from the phylum Firmicutes (mainly in Experiment 1). These are the most versatile decomposers of plant polymers, namely Paenibacillus sp. KAR70/KAR71, Bacillus subtilis KAR74 and Clostridium algidixylanolyticum KAR68, which can all degrade the main polymers found in potato tubers - pectin, cellulose, and starch. With respect to plant pathogenicity, the possession of pectolytic and to some extent cellulolytic activity is of greater importance to bacteria than amylolytic activity. This expected distribution of enzymatic activities was indeed observed in the endophytes of potato tubers (Fig. 8). The number of anaerobic Clostridia begins to increase during maceration, and in some potatoes, Clostridia, Clostridium puniceum (OTU181) and Clostridium algidixylanolyticum KAR68 (OTU11), overgrew both P. atrosepticum and other bacteria. Clostridium is one of the major cellulolytic/pectinolytic and fermentative bacterial groups in anaerobic environments, however, few reports discuss bacteria that are able to degrade pectic substances within the genus Clostridium. Between 1970–1980, it was found that pectolytic, i.e. pathogenic Clostridia, can frequently be isolated from decaying plant material. Clostridia with high pectolytic and tuber-rotting capabilities were described both in the United Kingdom37 and the United States of America38. Clostridium puniceum may cause potato soft rot39 and pectolytic Clostridia causes cavity spots in carrots40. Clostridium puniceum and C. saccharobutylicumstrain P262 have been found to cause post-harvest disease in sweetpotato41. We observed that Paenibacillus sp., Clostridium algidixylanolyticum, and Clostridium puniceum, not only support P. atrosepticum during plant decay, but in some cases dominate P. atrosepticum within the infection site. This is probably because these organisms possess amylase activity29 and while P. atrosepticum cannot degrade starch directly, it does provide access to an underutilized nutrient niche. Overall, the succession of bacteria in P. atrosepticum infected potato tuber could proceed as follows: P. atrosepticum breaks down the plant cell wall thereby generating large amounts of free sugars and sugar acids. Bacteria (Enterobacteriaceae and Pseudomonadaceae) that consume these energetically favorable sugars have an advantage during this phase of infection. Later, the bacteria that specialize in consuming energetically less favorable compounds (Comamonadaceae) begin to take over the infection site. During tuber tissue consumption, oxygen is depleted and the diffusion of atmospheric oxygen into the macerated tissue is reduced because most of the bacteria produce slime during this growth phase. Thus, strictly anaerobic Clostridia, which can degrade cell wall components, begin to expand and thereby extend tuber decay together with P. atrosepticum, or even supersede it.
N2 fixation is widespread among bacterial plant endophytes. Diazotrophy is well described among Rhizobiaceae, but also among plant-associated Enterobacteriaceae, Bacilli and other species42,43,44. Relying on genome data, the chromosome of plant pathogen P. atrosepticum strain SCRI1043 that was used in our potato infection experiments contains genes for molybdenum-dependent nitrogenase synthesis, an enzyme that catalyzes the fixation of nitrogen. However, the ability of P. atrosepticum to fix N2 was found to be very weak under our experimental conditions. Another enterobacterial plant pathogen, Pectobacterium carotovorum KAR14, did not fix N2 under these experimental conditions, but Dickeya dadantii KAR15 did so quite efficiently (Fig. 8). Bacillus subtilis KAR74, Paenibacillus sp. KAR70/71 and Clostridium algidixylanolyticum KAR68, which can efficiently degrade all plant polymers, are also efficient in N2 fixation. Consequently, bacteria that behave as pathogens during some stages of their life can also be mutualistic (i.e. beneficial to the host) at other stages45.
An interesting finding is the abundance of actinobacterial Propionibacterium acnes in uninfected potato samples, which might have been caused by human contamination when handling the tubers during harvest time and storage. Another explanation is based on recent finding that the human opportunistic pathogen, Propionibacterium acnes, could have been horizontally transferred to the grapevine, Vitis vinifera, possibly during the Neolithic period when the grapevine was domesticated46. We speculate that this phenomenon could be more general, and Propionibacterium acnes may have extended the host circle to include domesticated potato.
In conclusion, our data show that bacterial plant pathogenesis is not a process where the role of a particular pathogen can be singled out but involves complex contributions from the endophytic community.
Materials and Methods
Plant material and sampling
The commercial potato (Solanum tuberosum) cultivar “Ants” was grown under normal field conditions at the Jõgeva Plant Breeding Institute in two consecutive years (2012 and 2013). The soil characteristics of the experimental fields and crop protection measures were similar in both experiments (Supplemental material). The tubers were stored at 7 °C and 92% humidity post-harvest prior to randomly sampling healthy tubers with equal size within the weight range of 80–90 grams. Potatoes harvested in 2012 were stored until March 2013 and used to conduct Experiment 1, while those harvested in 2013 were stored until December 2013 and used to conduct Experiment 2.
Surface sterilization and virulence assays
Potato tubers were first washed with tap water followed by surface sterilization in 10% sodium hypochlorite (NaOCl) for 10 min, followed by three rinsing cycles with sterile water and an additional rinse with 96% ethanol prior to air-drying. Each tuber was placed into a plastic seedling box on top of a moist sterile paper towel. Pectobacterium atrosepticum SCRI1043 (herein referred to as Pa)47 was grown overnight in M9 medium48 supplemented with 0.4% polygalacturonic acid (PGA) as the sole carbon source. The cells were spun down at 5000 g for 5 min. The supernatant was passed through a filter with pore size 0.2 μm. The cells were washed once with 10 mM MgSO4 prior to centrifugation and then suspended in the sterile supernatant from the overnight culture. The tubers were stabbed with a pipette tip to create a cavity to inoculate 30 μl of diluted bacterial suspension (5 × 105 cells per ml). The tubers were then incubated in the dark at room temperature and 100% humidity for either two, five, or eight days. At the end of the incubation period, each potato was cut in half and the softened tissue was scraped out, weighed, and used to identify and quantify the composition of its bacterial community. The macerated tissue was frozen and stored at −80 °C prior to DNA extraction.
For the isolation of endophytic bacteria from uninfected potatoes, 15 g from each surface-sterilized tuber, including peel, were cut into pieces and homogenized with a garlic smasher. About 15 ml sterile 0.9% NaCl was added to the mixture, vortexed vigorously, and filtered through cotton fabric (instruments and materials for this procedure were sterile). The filtrate was centrifuged for 30 min at 5000 g and the resulting pellet was used for the extraction of genomic DNA.
Counting, isolation and identification of cultivable bacteria
The macerated and crude uninfected potato tissue was homogenized together with an equal volume of sterile 0.9% NaCl. Dilutions for CFU counting were plated on R2A agar (LabM), and the plates were incubated for one week at room temperature. We then determined the CFU/g of either macerated or crude tissue for each potato tuber. To distinguish between different bacterial strains, including P. atrosepticum, 50 colonies were successively picked from R2A agar and streaked on six different agar plates containing: carboxymethylcellulose (CMC), polygalacturonic acid (PGA), skimmed milk for semi-quantitative agar plate assays for the production of extracellular cellulases; hydrolyses of pectic substances; and proteases49. After 2–5 days of incubation, the exoenzyme activity was measured according to the halo on the agar plates, where the size is proportional to the amount of secreted enzyme. Agar plates containing 0.2% starch were used for the detection of amylase activity50. The presence of clearance halo after Lugol staining was observed after 5–10 days of incubation. Luria–Bertani (LB) agar medium was supplemented with ampicillin (150 μg/ml) or polymyxin B (5 μg/ml)51. Assimilation of N2 was studied on the agar plates containing (gL−1): MgSO4 0.2; K2HPO4 0.8; KH2PO4 0.2; CaSO4 0.13; FeCl3 0.00145; Na2MoO4 0.000253; sucrose 10; mannitol 10; agar 15 (modified protocol of Wang et al.25). The plates were kept under microaerophilic conditions for between one to two weeks. All incubations were performed at room temperature.
Bacteria were grouped on the basis of their enzymatic activities and morphological structure. In Experiment 1, representatives of the groups were identified by 16S rRNA gene sequencing. In Experiment 2, the first round of identification was made by MALDI-TOF phenotyping (Bruker Daltonik MALDI Biotyper), which was later confirmed by 16S rRNA gene sequencing. All isolates were identified by Sanger sequencing ~1500 bases of a near complete section of the 16S rRNA gene, using the primer pair PCRI 5′AGAGTTTGATCATGGCTCAG and PCRII 5′TACGGTTACCTTGTTACGACTT adapted from the primers fD2 and rP152. Phylogenetic affiliations of the sequences obtained from this method were assessed using the Ribosome Database Project Classifier24. The 16S rRNA gene nucleotide sequences of isolated bacterial strains were deposited in the GenBank database under accession numbers KAR1 - KR054963; KAR2 - KR054964; KAR3 - KR054965; KAR4 - KR054966; KAR5 - KR054967; KAR6 - KR054968; KAR7 - KR054969; KAR8 - KR054970; KAR9 - KR054971; KAR10 - KR054972; KAR11 - KR054973; KAR12 - KR054974; KAR13 - KR054975; KAR14 - KR054976; KAR15 - KR054977; KAR16 - KR054978; KAR17 - KR054979; KAR18 - KR054980; KAR19 - KR054981; KAR20 - KR054982; KAR21 - KR054983; KAR22 - KR054984; KAR23 - KR054985; KAR24 - KR054986; KAR25 - KR054987; KAR26 - KR054988; KAR27 - KR054989; KAR28 - KR054990; KAR29 - KR054991; KAR30 - KR054992; KAR31 - KR054993; KAR32 - KR054994; KAR33 - KR054995; KAR34 - KR054996; KAR35 - KR054997; KAR36 - KR054998; KAR37 - KR054999; KAR39 - KR055000; KAR40 - KR055001; KAR41 - KR055002; KAR42 - KR055003; KAR43 - KR055004; KAR44 - KR055005; KAR45 - KR055006; KAR46 - KR055007; KAR47 - KR055008; KAR48 - KR055009; KAR49 - KR055010; KAR50 - KR055011; KAR51 - KR055012; KAR52 - KR055013; KAR53 - KR055014; KAR54 - KR055015; KAR55 - KR055016; KAR56 - KR055017; KAR57 - KR055018; KAR58 - KR055019; KAR59 - KR055020; KAR60 - KR055021; KAR61 - KR055022; KAR62 - KR055023; KAR63 - KR055024; KAR64 - KR055025; KAR65 - KR055026; KAR67 - KR055027; KAR68 - KR055028; KAR69 - KR055029; KAR70 - KR055030; KAR71 - KR055031; KAR72 - KR055032; KAR73 - KR055033; KAR74 - KR055034; KAR75 - KR055035; KAR76 - KR055036; KAR77 - KR055037; KAR78 - KR055038; KAR79 - KR055039; KAR80 - KR055040; KAR81 - KR055041; KAR82 - KR055042.
The 16S rDNA sequences of isolates (marked as “KAR”) were combined with the closest matching sequences from the Ribosome Database Project38. A phylogenetic tree (additional Fig. 2) was constructed in MEGA653 using the neighbor-joining algorithm with “maximum composite likelihood” as the substitution model. Bootstrapping with 1000 repetitions was used to test the correctness of the tree. A smaller tree (Fig. 8) was derived by condensing the reference sequences with those of the isolates. The resulting branches were named after the isolated “KAR” sequences and all branches that had <50% bootstrap support were removed.
Isolation and cultivation of Clostridia
Macerated potato tuber tissue was incubated in 50% ethanol for 1h and streaked on Fastidious Anaerobic Agar plate (FAA) and 10% potato extract containing agar plates, which were kept under anaerobic conditions for one week either at 37 °C or room temperature.
Total-community DNA isolation
DNA was isolated from 250 mg of bacterial cells obtained from either macerated or crude potato tuber tissue. The cells were suspended in the lysis buffer with beads from a PowerSoil DNA kit (MoBio, USA). The cells were lysed by bead beating with Precellys® 24 (Bertin Technologies, France) at 5000 rpm for 3 × 60 s followed by purification of the extract with phenol, phenol:chloroform:isoamylalcohol (25:24:1) and chloroform:isoamylalcohol (24:1), and subsequently isopropanol precipitation. The quantity and quality of DNA extracts were determined spectrophotometrically (NanoDrop 2000c).
Amplification of the bacterial 16S rRNA genes
The taxonomic composition of the endophytic bacterial community was also analyzed using deep-sequencing by amplifying the hypervariable V1–V2 region of the 16S rRNA gene using an 8R/357R primer set54. PCR amplifications were performed in triplicate and then pooled. The PCR reaction was carried out at 95 °C for 5 min followed by 15 cycles each of 95 °C for 45 s, 50 °C for 45 s, and 72 °C for 30 s, followed by 10 min at 72 °C. The pooled PCR products were cleaned using an UltraClean PCR Clean-Up Kit (MoBio, USA), and both the quantity and quality was determined spectrophotometrically (NanoDrop 2000c). Sequencing used an Illumina MiSeq (San Diego, USA).
Sequencing of 16S rDNA
In Experiment 1 (March 2013), 20 bacterial 16S rRNA gene amplicons were prepared for sequencing using Nextera XT sample preparation kits (Illumina, San Diego, USA). The purified DNA of the amplicons was first quantified with a Qubit 2.0 Fluorometer (Invitrogen, Grand Island, USA) and 2200 TapeStation (Agilent Technologies, Santa Clara, USA). A sample containing 1 ng of DNA was used to prepare the sequencing libraries using the NexteraXT protocol. Libraries were validated by qPCR with Kapa Library Quantification Kit (KapaBiosystems, Woburn, USA) to optimize cluster generation. Twenty ssDNA Nextera XT libraries were pooled and sequenced on a MiSeq v2 flowcell with paired-end (PE), 250-bp reads yielding in 20 M reads (Illumina CSPro lab, Estonian Genome Centre, University of Tartu, Estonia).
In Experiment 2 (December 2013), 19 bacterial 16S rRNA gene libraries were constructed by two-step PCR. Two 5′overhang Illumina adapters,
Forward primer 5′TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG and
Reverse primer 5′GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG,
were added to the 8F /357R primer set and used in the first round of PCR, with purification carried out as described above. Purified amplicons were quantified using Qubit 2.0 Fluorometer (Invitrogen, Grand Island, USA) and 2200 TapeStation (Agilent Technologies, Santa Clara, USA). A sample containing 1 ng of DNA was used in the second round of PCR (12 cycles) with the primers from the Nextera XT sample preparation kit. PCR products were purified and normalized using the Nextera XT protocol provided by Illumina. Libraries were validated by qPCR with Kapa Library Quantification Kits (Kapa Biosystems, Woburn, USA) to optimize cluster generation. In total, 19 ssDNA libraries were pooled and sequenced on MiSeq v2 flowcell, with paired-end (PE), 250 bp reads yielding 15.8 M reads (Illumina CSPro lab, Estonian Genome Centre, University of Tartu, Estonia). PE reads were stitched together using MiSeq Reporter software (Illumina, San Diego, USA).
Raw sequence data processing
Based on our quality assessment, only first set of sequences from the paired group was used in Experiment 1. The raw reads were trimmed from the 3′ end using the program fastq_quality_trimmer (parameters: -t 35 -l 200), followed by quality filtering with fastq_quality_filter (parameters: -q 30 -p 98) from the FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/). PCR primers and sequencing adapters were removed using the program cutadapt 1.2.1 (parameters: -e 0.2 -n 2 -O 10 -m 100) (https://pypi.python.org/pypi/cutadapt/1.2.1). After trimming, all sequences <100 bp were discarded. In Experiment 2, the first 20 nucleotides from the 5′ end of the stitched raw reads were trimmed using the program fastx_trimmer to remove all 8F PCR primer sequences. The reads were converted to their reverse complement sequences and the first 17 nucleotides from the 5′ end were trimmed to remove all 357R primer sequences. Subsequently, the reads were filtered using fastq_quality_filter (parameters: -q 30 -p 98), sorted, and written to the file “Pectobacterium“ if they provided a BLASTN hit against the 16S rRNA gene of P. atrosepticum SCRI1043 with an identity cutoff of 98% and a match length cutoff of either 100 bp (Experiment 1) or 250 bp (Experiment 2). Chloroplast sequences were removed after comparison with the 16S rRNA gene of S. tuberosum isolate DM1-3-516-R44 chloroplast (JF772170) at BLASTN cutoff values of 98% and either 100 bp (Experiment 1) or 250 bp (Experiment 2). All remaining reads with an identity cutoff 98% and match length cutoff of either 100 bp (Experiment 1) or 250 bp (Experiment 2) were classified with RDP multiclassifier, an application derived from the RDP Classifier24, against the Ribosomal Database Project (RDP) taxonomy training set 9 (trainset9_032012.rdp.fasta) with the default confidence threshold set at 80%. These reads were sorted and written to the file “Other 16S“.
Sequence processing and assignment of OTUs
All high-quality sequences of the file “Other 16S“ of Experiment 2 were clustered into OTUs and consensus sequences that represented all sequences in that group were derived using AbundantOTU55 program (parameters: -abundantonly –d 0.02). Minimum cluster size allowed was five sequences and the lowest intracluster identity allowed was 98%. Single reads that did not belong to any clusters of at least five sequences were clustered again using AbundantOTU with the same parameters as above, however, most of these sequences still did not cluster. All consensus sequences were classified using the RDP classifier24 Version 2.8, June 2014. The training set was RDP 16S rRNA training set 9 with a confidence threshold of 80%. In further analysis, a 50% confidence threshold was used to determine the taxonomic position of each read, however, when the confidence of the lowest taxonomic assignment was <50%, the next higher taxon which had at least 50% confidence was used. To avoid chloroplast sequences, all consensus sequences classified as Streptophyta were removed from further analysis.
Chimeras were removed using UCHIME56 from the mothur57 package (chimera.uchime function with parameters: mindiv = 2.0, reference database “rdp gold“ from UCHIME homepage). First, all consensus sequences were analyzed and then all chimeric sequences were removed. All single reads which would not cluster were analyzed and then all chimeras were removed. This resulted in a final set of 294 OTUs and a total of 4,777,229 reads and 3,462 single reads for Experiment 2.
Rarefaction analysis was performed in mothur (rarefaction.single with parameters freq = 10 for samples K1–K6 and freq = 1,000 for other samples).
The final 294 OTUs were identified further by blasting them against a cultivated bacteria dataset using MegaBLAST (parameters: -p 98 -D 3 -v 5 -b 5 -W 52). A top-scoring hit for each consensus with at least 98% of the consensus sequence aligned was chosen as the closest identifiable bacterial strain for each OTU. Because the reads from Experiment 1 are not aligned and correspond to different 100 bp sequences within the V1–V2 hypervariable region, we could not cluster them as in Experiment 2. Instead, we took all 294 OTU consensus sequences from Experiment 2 and then clustered the sequences from Experiment 1 to these using MegaBLAST as above. All reads that had <6 unaligned positions were assigned to the OTU with the highest-scoring match. In total, 539,068 reads were assigned, and of the remaining 180,050 reads, 16,106 were chimeric (using UCHIME as above). The 163,944 unclustered reads were then compared to the cultivated bacteria dataset with MegaBLAST using the above parameters.
Correlation network analysis
Correlation network analysis was performed to identify the most significant antagonistic and synergistic relationships between bacteria from uninfected potatoes and sets of potatoes infected for a given length of time. We utilized the SparCC method and software developed by Friedman and Alm58. To avoid spurious correlations that can arise using data with very small or zero values, we excluded genera that form less than 0.2% of the bacterial population in any given potato or were not detected in more than two potatoes within a given set. After computing the correlations with the main SparCC script (-i 20, -x 10, -t 0.1) we generated bootstrap files using the SparCC script MakeBootstraps (-n 100) and computed correlations for each of these bootstrap replacement files using the main SparCC script with the same parameters. The significance of each correlation was then computed from the bootstrapped results (N = 100) using the SparCC script PseudoPvals. The resulting genera level correlation networks were analyzed using NetworkX (https://networkx.github.io). One network visualization was created for each of the three sets of potatoes from Experiment 2 (uninfected, two days post-infection, and eight days post-infection) using matplotlib (http://matplotlib.org) and combined to create Fig. 7.
Additional Information
How to cite this article: Kõiv, V. et al. Microbial population dynamics in response to Pectobacterium atrosepticum infection in potato tubers. Sci. Rep. 5, 11606; doi: 10.1038/srep11606 (2015).
Supplementary Material
Acknowledgments
This work was funded by grants SF0180088s08, SF0180026s09, IUT2-22 and RLOMRCELM from the Estonian Ministry of Education and Research, EU ERDF through the Estonian Centres of Excellence in Genomics and Chemical Biology and grant SLOMRARENG1. We thank Tiiu Rööp for her valuable technical assistance during cultivation of anaerobic bacteria. We also thank Mati Koppel (Jõgeva PI) for providing potato tubers and Triinu Kõressaar for help with preliminary Illumina sequence analysis.
Footnotes
Author Contributions V.K. conceived the project, coordinated the study, performed experiments, and wrote the paper; M.Ro. analysed Illumina sequences and wrote the paper; E.V. analysed preliminary Illumina sequences and wrote the paper; K.T. performed experiments; P.-A.K. performed experiments and wrote the paper; D.S. performed the network analysis and wrote the paper; M.R. coordinated the study; T.T. coordinated the study, contributed materials and reagents and wrote the paper; A.M. coordinated the study, contributed materials and reagents, and wrote the paper.
References
- Hallmann J., Quadt-Hallmann A., Mahaffee W. F. & Kloepper J. W. Bacterial endophytes in agricultural crops. Canadian Journal of Microbiology. 43, 895–914 (1997). [Google Scholar]
- Hentschel U., Steinert M. & Hacker J. Common molecular mechanisms of symbiosis and pathogenesis. Trends Microbiol. 8, 226–31 (2000). [DOI] [PubMed] [Google Scholar]
- Knief C. et al. Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice. ISME J 6, 1378–90 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblueth M. & Martínez-Romero E. Rhizobium etli maize populations and their competitiveness for root colonization. Arch. Microbiol. 181, 337–44 (2004). [DOI] [PubMed] [Google Scholar]
- Bulgarelli D. et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488, 91–5 (2012). [DOI] [PubMed] [Google Scholar]
- Donn S., Kirkegaard J. A., Perera G., Richardson A. E. & Watt M. Evolution of bacterial communities in the wheat crop rhizosphere. Environ. Microbiol. (2014). 10.1111/1462-2920.12452. [DOI] [PubMed] [Google Scholar]
- Schreiter S. et al. Effect of the soil type on the microbiome in the rhizosphere of field-grown lettuce. Front Microbiol 5, 144 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A. et al. Whole genome sequencing and analysis of plant growth promoting bacteria isolated from the rhizosphere of plantation crops coconut, cocoa and arecanut. PLoS ONE 9, e104259 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manter D. K., Delgado J. A., Holm D. G. & Stong R. A. Pyrosequencing reveals a highly diverse and cultivar-specific bacterial endophyte community in potato roots. Microb. Ecol. 60, 157–66 (2010). [DOI] [PubMed] [Google Scholar]
- Hardoim P. R. et al. Rice root-associated bacteria: insights into community structures across 10 cultivars. FEMS Microbiol. Ecol. 77, 154–64 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg D. S. et al. Defining the core Arabidopsis thaliana root microbiome. Nature 488, 86–90 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaeppi K., Dombrowski N., Oter R. G., Ver Loren van Themaat E. & Schulze-Lefert P. Quantitative divergence of the bacterial root microbiota in Arabidopsis thaliana relatives. Proc. Natl. Acad. Sci. USA. 111, 585–92 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- İnceoğlu Ö., Falcão Salles J. & van Elsas J. D. Soil and cultivar type shape the bacterial community in the potato rhizosphere. Microb. Ecol. 63, 460–70 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert N. et al. PhyloChip hybridization uncovered an enormous bacterial diversity in the rhizosphere of different potato cultivars: many common and few cultivar-dependent taxa. FEMS Microbiol. Ecol. 75, 497–506 (2011). [DOI] [PubMed] [Google Scholar]
- Andreote F. D. et al. Endophytic colonization of potato (Solanum tuberosum L.) by a novel competent bacterial endophyte, Pseudomonas putida strain P9, and its effect on associated bacterial communities. Appl. Environ. Microbiol. 75, 3396–406 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreote F. D., Rocha U. N., Araújo W. L., Azevedo J. L. & van Overbeek L. S. Effect of bacterial inoculation, plant genotype and developmental stage on root-associated and endophytic bacterial communities in potato (Solanum tuberosum). Antonie Van Leeuwenhoek 97, 389–99 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter B., Pfeifer U., Schwab H. & Sessitsch A. Response of endophytic bacterial communities in potato plants to infection with Erwinia carotovora subsp. atroseptica. Appl. Environ. Microbiol. 68, 2261–8 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbeva P., Overbeek L. S. S., Vuurde J. W. L. W. & Elsas J. D. D. Analysis of Endophytic Bacterial Communities of Potato by Plating and Denaturing Gradient Gel Electrophoresis (DGGE) of 16S rDNA Based PCR Fragments. Microb. Ecol. 41, 369–383 (2001). [DOI] [PubMed] [Google Scholar]
- Pérombelon M. C. M., Gullings-Handley J. & Kelman A. Population Dynamics of Erwinia carotovora and Pectolytic Clostridium spp. in Relation to Decay of Potatoes. Phytopathology 69, 167–173 (1979). [Google Scholar]
- Tholozan J. L., Cappelier J. M., Tissier J. P., Delattre G. & Federighi M. Physiological characterization of viable-but-nonculturable Campylobacter jejuni cells. Appl. Environ. Microbiol. 65, 1110–6 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston-Monje D. & Raizada M. N. Conservation and diversity of seed associated endophytes in Zea across boundaries of evolution, ethnography and ecology. PLoS ONE 6, e20396 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulgarelli D., Schlaeppi K., Spaepen S., Ver Loren van Themaat E. & Schulze-Lefert P. Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol 64, 807–38 (2013). [DOI] [PubMed] [Google Scholar]
- Kõiv V. et al. Lack of RsmA-mediated control results in constant hypervirulence, cell elongation, and hyperflagellation in Pectobacterium wasabiae. PLoS ONE 8, e54248 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Garrity G. M., Tiedje J. M. & Cole J. R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–7 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. et al. A minimal nitrogen fixation gene cluster from Paenibacillus sp. WLY78 enables expression of active nitrogenase in Escherichia coli. PLoS Genet. 9, e1003865 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez-Santacruz H. A., Hernandez-Leon R., Orozco-Mosqueda M. C., Velazquez-Sepulveda I. & Santoyo G. Diversity of bacterial endophytes in roots of Mexican husk tomato plants (Physalis ixocarpa) and their detection in the rhizosphere. Genet. Mol. Res. 9, 2372–80 (2010). [DOI] [PubMed] [Google Scholar]
- Bodenhausen N., Horton M. W. & Bergelson J. Bacterial communities associated with the leaves and the roots of Arabidopsis thaliana. PLoS ONE 8, e56329 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghavi S. et al. Genome sequence of the plant growth promoting endophytic bacterium Enterobacter sp. 638. PLoS Genet. 6, e1000943 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudeja S. S., Giri R., Saini R., Suneja-Madan P. & Kothe E. Interaction of endophytic microbes with legumes. J. Basic Microbiol. 52, 248–60 (2012). [DOI] [PubMed] [Google Scholar]
- George P., Gupta A., Gopal M., Thomas L. & Thomas G. V. Multifarious beneficial traits and plant growth promoting potential of Serratia marcescens KiSII and Enterobacter sp. RNF 267 isolated from the rhizosphere of coconut palms (Cocos nucifera L.). World J. Microbiol. Biotechnol. 29, 109–17 (2013). [DOI] [PubMed] [Google Scholar]
- Liu W.-Y. Y., Wong C.-F. F., Chung K. M., Jiang J.-W. W. & Leung F. C. Comparative genome analysis of Enterobacter cloacae. PLoS ONE 8, e74487 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J., Jiang W., Cheng Z., Heikkila J. J. & Glick B. R. The complete genome sequence of the plant growth-promoting bacterium Pseudomonas sp. UW4. PLoS ONE 8, e58640 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hol W. H., Bezemer T. M. & Biere A. Getting the ecology into interactions between plants and the plant growth-promoting bacterium Pseudomonas fluorescens. Front Plant Sci 4, 81 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan R. et al. Enterobacter turicensis sp. nov. and Enterobacter helveticus sp. nov., isolated from fruit powder. Int. J. Syst. Evol. Microbiol. 57, 820–6 (2007). [DOI] [PubMed] [Google Scholar]
- Madhaiyan M. et al. Enterobacter arachidis sp. nov., a plant-growth-promoting diazotrophic bacterium isolated from rhizosphere soil of groundnut. Int. J. Syst. Evol. Microbiol. 60, 1559–64 (2010). [DOI] [PubMed] [Google Scholar]
- Brady C., Cleenwerck I., Venter S., Coutinho T. & De Vos P. Taxonomic evaluation of the genus Enterobacter based on multilocus sequence analysis (MLSA): proposal to reclassify E. nimipressuralis and E. amnigenus into Lelliottia gen. nov. as Lelliottia nimipressuralis comb. nov. and Lelliottia amnigena comb. nov., respectively, E. gergoviae and E. pyrinus into Pluralibacter gen. nov. as Pluralibacter gergoviae comb. nov. and Pluralibacter pyrinus comb. nov., respectively, E. cowanii, E. radicincitans, E. oryzae and E. arachidis into Kosakonia gen. nov. as Kosakonia cowanii comb. nov., Kosakonia radicincitans comb. nov., Kosakonia oryzae comb. nov. and Kosakonia arachidis comb. nov., respectively, and E. turicensis, E. helveticus and E. pulveris into Cronobacter as Cronobacter zurichensis nom. nov., Cronobacter helveticus comb. nov. and Cronobacter pulveris comb. nov., respectively, and emended description of the genera Enterobacter and Cronobacter. Syst. Appl. Microbiol. 36, 309–19 (2013). [DOI] [PubMed] [Google Scholar]
- Lund B. M. Isolation of pectolytic clostridia from potatoes. J. Appl. Bacteriol. 35, 609–14 (1972). [DOI] [PubMed] [Google Scholar]
- Lund B. M. & Kelman A. Determination of the potential for developement of bacterial soft rot of potatoes. Am. Potato J. 54, 211–225 (1977). [Google Scholar]
- Lund B. M., Brocklehurst T. F. & Wyatt G. M. Characterization of strains of Clostridium puniceum sp. nov., a pink-pigmented, pectolytic bacterium. Journal of General Microbiology 122, 17–26 (1981). [Google Scholar]
- Perry D. A. & Harrison J. G. Pectolytic anaerobic bacteria cause symptoms of cavity spot in carrots. Nature 269, 509–5 10 (1977). [Google Scholar]
- Da Silva W. L. Sweetpotato Storage Root Rots: Flooding-Associated Bacterial Soft Rot Caused by Clostridium spp. and Infection by Fungal End Rot Pathogens Prior to Harvest. Lousiana State University Master’s Thesis (2013).
- Minamisawa K. et al. Anaerobic nitrogen-fixing consortia consisting of clostridia isolated from gramineous plants. Appl. Environ. Microbiol. 70, 3096–102 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Peng M. & Li Y. Phylogenetic diversity of nitrogen-fixing bacteria and the nifH gene from mangrove rhizosphere soil. Can. J. Microbiol. 58, 531–9 (2012). [DOI] [PubMed] [Google Scholar]
- Dos Santos P. C., Fang Z., Mason S. W., Setubal J. C. & Dixon R. Distribution of nitrogen fixation and nitrogenase-like sequences amongst microbial genomes. BMC Genomics 13, 162 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton A. C., Fitt B. D., Atkins S. D., Walters D. R. & Daniell T. J. Pathogenesis, parasitism and mutualism in the trophic space of microbe-plant interactions. Trends Microbiol. 18, 365–73 (2010). [DOI] [PubMed] [Google Scholar]
- Campisano A. et al. Interkingdom transfer of the acne-causing agent, Propionibacterium acnes, from human to grapevine. Mol. Biol. Evol. 31, 1059–65 (2014). [DOI] [PubMed] [Google Scholar]
- Hinton J. C., Perombelon M. C. & Salmond G. P. Efficient transformation of Erwinia carotovora subsp. carotovora and E. carotovora subsp. atroseptica. J. Bacteriol. 161, 786–8 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J. & Russell D. W. Molecular Cloning: a Laboratory Manual 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory (2001). [Google Scholar]
- Chatterjee A., Cui Y., Liu Y., Dumenyo C. K. & Chatterjee A. K. Inactivation of rsmA leads to overproduction of extracellular pectinases, cellulases, and proteases in Erwinia carotovora subsp. carotovora in the absence of the starvation/cell density-sensing signal, N-(3-oxohexanoyl)-L-homoserine lactone. Appl. Environ. Microbiol. 61, 1959–67 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedder E. B. Starch agar, a useful culture medium. J. Infect. Dis. 16, 385 (1915). [Google Scholar]
- Miller J. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory. (1972). [Google Scholar]
- Weisburg W. G., Barns S. M., Pelletier D. A. & Lane D. J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A. & Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30, 2725–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armougom F. & Raoult D. Exploring Microbial Diversity Using 16S rRNA High-Throughput Methods. Journal of Computer Science & Systems Biology 02, 7492 (2009). [Google Scholar]
- Ye Y. Identification and Quantification of Abundant Species from Pyrosequences of 16S rRNA by Consensus Alignment. Proceedings (IEEE Int Conf Bioinformatics Biomed) 2010, 153–157 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C., Haas B. J., Clemente J. C., Quince C. & Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–200 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss P. D. et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–41 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J. & Alm E. Inferring Correlation Networks from Genomic Survey Data. PLoS Comput Biol (2012). 10.1371/journal.pcbi.1002687. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.