Abstract
The normal prostate as well as early stages and advanced prostate cancer (PCa) require a functional androgen receptor (AR) for growth and survival. The recent discovery of microRNAs (miRNAs) as novel effector molecules of AR disclosed the existence of an intricate network between AR, miRNAs and downstream target genes. In this study DUCaP cells, characterized by high content of wild-type AR and robust AR transcriptional activity, were chosen as the main experimental model. By integrative analysis of chromatin immunoprecipitation-sequencing (ChIP-seq) and microarray expression profiling data, miRNAs putatively bound and significantly regulated by AR were identified. A direct AR regulation of miR-22, miR-29a, and miR-17-92 cluster along with their host genes was confirmed. Interestingly, endogenous levels of miR-22 and miR-29a were found to be reduced in PCa cells expressing AR. In primary tumor samples, miR-22 and miR-29a were less abundant in the cancerous tissue compared with the benign counterpart. This specific expression pattern was associated with a differential DNA methylation of the genomic AR binding sites. The identification of laminin gamma 1 (LAMC1) and myeloid cell leukemia 1 (MCL1) as direct targets of miR-22 and miR-29a, respectively, suggested a tumor-suppressive role of these miRNAs. Indeed, transfection of miRNA mimics in PCa cells induced apoptosis and diminished cell migration and viability. Collectively, these data provide additional information regarding the complex regulatory machinery that guides miRNAs activity in PCa, highlighting an important contribution of miRNAs in the AR signaling.
Androgens and the androgen receptor (AR) are indispensable for the function of the prostate gland and drive growth and survival of prostate tumors. Androgens exert their biological effects through AR activation, inducing an AR-dependent gene expression program that promotes cell survival and cell cycle initiation while suppressing apoptosis (1). Because the AR is intimately linked to prostate cancer (PCa) biology, it is the primary therapeutic target in the treatment of PCa via androgen withdrawal strategies (2). Removal of testicular androgens by surgical or chemical castration aims to inhibit AR transcriptional activity. However, most tumors eventually relapse to an incurable phenotype termed castration resistant PCa, which is characterized by enhanced AR signaling in the absence of testicular androgens (3). Several mechanisms responsible for this event have been identified, such as AR mutations or overexpression, but they are still not entirely understood and most likely many more are involved (4). Thus, there is an urgent need for a thorough characterization of the AR downstream regulatory network in order to develop novel and more efficacious therapeutic approaches towards castration resistant PCa and also to identify promising biomarkers for the diagnosis, prognosis, and monitoring of PCa.
To date, much research has focused on identifying AR regulated protein-coding genes. Only recently, investigations have awakened attention to the interplay between AR and microRNAs (miRNAs or miRs) (5–8). Like many protein-coding genes, miRNAs are under control of transcription factors as well as epigenetic mechanisms whose deregulation leads to a loss or gain of miRNA function with possible implications in tumorigenesis (9, 10). miRNAs are short, noncoding, single-stranded RNAs that potentially regulate hundreds of mRNAs. They represent a family of powerful and evolutionarily conserved posttranscriptional regulators that lessen gene expression via binding preferentially but not exclusively to the 3′-untranslated region (UTR) of target mRNAs with a perfect or imperfect sequence match. Evidence of aberrant miRNA expression has been reported in numerous types of cancer, highlighting their ability to regulate important biological processes such as proliferation, differentiation, apoptosis, and angiogenesis (11, 12). In addition, an active involvement of miRNAs in metastasis promoting epithelial to mesenchymal transition has been shown (13, 14). As result of their up- or down-regulation, miRNAs can act as oncogenes or tumor suppressors (15), and the particular expression pattern of miRNAs may be used as a classifier to differentiate between normal and cancerous tissues and also for diagnosis and prognosis of various cancers (16–18). Specific miRNA signatures for AR positive and negative PCa cell lines as well as for malignant and nonmalignant tissues were previously determined via miRNA expression profiling studies (19–23).
Using a broader integrative approach, this work investigates the interaction between AR and miRNAs in vitro and in vivo, particularly in terms of miRNA regulation by AR, with the final goal to elucidate the activity and role of miRNAs in PCa. AR binding sites (ARBSs) in proximity of the host genes encoding miR-22, miR-29a, and miR-17-92 cluster were validated. In addition, androgen treatment and AR inhibition experiments confirmed the androgenic regulation of these miRNAs as well as their host genes. Furthermore, by screening various prostate cell lines, endogenous basal levels of miR-22 and miR-29a were found to be differentially modulated depending on the AR status of the cells. Identification of LAMC1 and MCL1 as direct targets of miR-22 and miR-29a, respectively, indicates their involvement in inhibiting pivotal oncogenic pathways, such as cell migration and survival. These findings suggest a tumor-suppressive role of these miRNAs consistent with their reduced expression in the cancerous tissue of primary tumor specimens.
Materials and Methods
Cell lines and reagents
Human PCa cell lines LNCaP, PC3, DU145, VCaP, and CWR22RV1 were obtained from ATCC. DUCaP and BPH-1 were a generous gift from Dr Schalken (Center for Molecular Life Science, Nijmegen, The Netherlands), PC3-AR and LAPC-4 from Dr Cato (Karlsruhe Institute of Technology, Karlsruhe, Germany), and RWPE-1 from Dr Watson (Conway Institute, Dublin, Ireland). EP156T were generated by immortalization of primary cells with human telomerase reverse transcriptase (24, 25). For routine culture, cells were maintained at 37°C in a humidified 5% CO2 atmosphere in RPMI 1640 (Lonza) supplemented with 10% fetal calf serum (FCS) (PAA), 2mM L-glutamine (Life Technologies), and antibiotics (100 U/mL of streptomycin and penicillin). LNCaP cells required, in addition, 2.5 g/L of D-glucose (Invitrogen), 10mM HEPES, and 1mM Na-pyruvate (Lonza). PC3-AR cells were cultured in the presence of geneticin (G418) (500 μg/mL; Life Technologies) to preserve the expression of AR, whereas 100nM dihydrotestosterone was added to the culture medium of LAPC-4 cells. VCaP cells were kept in DMEM low-glucose medium (Fisher), supplemented with 10% FCS, 2mM L-glutamine (Life Technologies), and 1.75 g/L of D-glucose (Invitrogen). EP156T and RWPE-1 cell lines were cultured as recommended (24, 25). Generally, for treatment with the synthetic androgen methyltrienolone (R1881) (1nM) or the antiandrogen enzalutamide (MDV3100) (10μM) cells were seeded in RPMI 1640 supplemented with 10% charcoal/dextran-treated FCS (Fischer) for 2 days before incubation with the indicated reagents for 24 and 48 hours, or as stated. Cell pellets were collected at the mentioned time point and frozen at −20°C for future use. Reagents and R1881 were purchased from Sigma-Aldrich unless otherwise specified. The antiandrogen MDV3100 was obtained from Eurasia.
Chromatin immunoprecipitation coupled with deep-sequencing (ChIP-seq) and analysis
ChIP was performed on androgen and vehicle treated DUCaP cells as previously described (26). ChIP AR-precipitated DNA fragments were then detected by deep sequencing. Peaks were identified by macs (v1.4) and annotated to the whole human genome sequence as described in more detail elsewhere (27).
ChIP-PCR
AR or control antibody ChIP-precipitated DNA samples, obtained according to the above mentioned protocol, were amplified by means of either PCR or real-time PCR using specific primers targeting ChIP-seq identified AR binding fragments in proximity of the miR-22, miR-29a, and miR-17-92 cluster host genes. PCR products were then separated and visualized with the FlashGel System (Lonza). Primer sequences used for amplification of ARBSs close to miRNA host genes were as follows: miR-22 ARBS_1 forward, 5′-AGCCCCATTGTCTGCCTTAG-3′ and reverse, 5′-CCAGACGCTTCCTCCTTACC-3′; miR-22 ARBS_2 forward, 5′-GAGGAGGGTGAGAGCAAGG-3′ and reverse, 5′-GTTGATGTTTGCCAGGTCATC-3′; miR-22 ARBs_3 forward, 5′-TATCTGTGATCGCGTGGGTA-3′ and reverse, 5′-ACCCCACCTTGACTTCAGC-3′; miR-29a ARBS_1 forward, 5′-TCTTTGGTGCCTGCCTACTT-3′ and reverse, 5′-CAGAAGGAAGAGCGAGTTCC-3′; miR-17-92-cluster ARBS_1 forward, 5′-CACCTCTTCTGACTGCTGGGCAT-3′ and reverse, 5′-CCCAAGGTAAACAGAAGAGCAGGG-3′; and miR-17-92 cluster ARBS_2 forward, 5′-AGGAGGTGCTCCTGATTGGGCT-3′ and reverse, 5′-TGAGCCTCCCCTCTCATGCCC-3′.
In silico detection of androgen responsive elements (AREs)
Detection of AREs within the ARBSs of miR-22, miR-29a, and the miR-17-92 cluster was performed using the MatInspector software. Briefly, ARBSs genomic sequences in FASTA format were retrieved from the University of California Santa Cruz Genome Browser (GRCh37/hg19) and analyzed for the presence of transcription factor binding motifs with MatInspector using the default parameters (28).
Gene microarray
Gene array expression profiling was performed at the Expression Profiling Unit of the Medical University Innsbruck. DUCaP and LNCaP cells were hormone deprived for 2 days and subsequently treated for 8 and 24 hours with 1nM R1881 or vehicle equivalent. Total RNA was amplified, labeled, and then hybridized to the HuGene-1.0 st v1 Affymetrix platform as previously described (29). Analysis of expression data has been carried out as previously indicated (29–31). Raw and preprocessed data have been submitted to Gene Expression Omnibus (GEO) (accession number GSE 50936).
RNA isolation, cDNA synthesis, and quantitative real-time PCR (qPCR)
Total RNA was extracted from collected cell pellets using Direct-sol MiniPrep kit (Zymo Research) according to the manufacturer's instructions. For each sample, RNA quantification and quality control were assessed by NanoDrop 2000c measurements (Thermo Scientific). A final amount of 5 ng of the template RNA was required for first-strand cDNA synthesis of miRNAs with the Universal cDNA Synthesis kit II (Exiqon). Otherwise, cDNA was generated from 1–0.5 μg of total RNA by means of iScript Select cDNA Synthesis kit (Bio-Rad Laboratories). qPCR with cDNA or ChIP-isolated DNA as template was carried out using SyBRgreen (SyBr) chemistry or Fam labeled TaqMan probe detection on an ABI 7500 Fast Sequence Detection System (Applied Biosystems). Real-Time PCRs were performed for 40 cycles with denaturation at 95°C for 15 seconds, followed by annealing and extension at 60°C for 1 minute. Primers and TaqMan probe for FKBP5 (Hs01561006_m1) as well as for MIR22HG (Hs02338582_m1), MIR17HG (Hs01382425_m1), SERPINF2 (Hs01018690_m1), SERPINF1 (Hs01106934_m1), WDR81 (Hs00912091_m1), LAMC1 (Hs00267056_m1), and MCL1 (Hs01050896_m1) were purchased from Applied Biosystems. Specific primers for miR-22-3p (204606), miR-29a-3p (204698), for the 3p-isoform of miR-17-92 cluster members except for miR-17-5p (204771, 204207, 204781, 204450, 204292, and 204258), and primers for SNORD38b (203901), SNORD44 (203902), and SNORD48 (203903), used for normalization along with miR-320b (205921) and miR-151a-3p (204566), were obtained from Exiqon. The other primers were custom-designed using the program primer-BLAST (32) and ordered from GenXpress. Respective sequences were as follows: AR probe, 5′-Fam-TGGTTTTCAATGAGTACCGCATGCACA-Tamra-3′; AR forward, 5′-AGGATGCTCTACTTCGCCCC-3′; and AR reverse, 5′-ACTGGCTGTACATCCGGGAC-3′; TBP probe, 5′-Fam-TCTTCACTCTTGGCTCCTGTGCACA-Tamra-3′; TBP forward, 5′-CACGAACCACGGCACTGATT-3′; and TBP reverse, 5′-TTTTCTTGCTGCCAGTCTGGAC-3′; and HPRT1 probe, 5′-Fam-TCAAGGTCGCAAGCTTGCTGGTGAAAAGGA-Tamra-3′; HPRT1 forward, 5′-GCTTTCCTTGGTCAGGCAGTA-3′; and HPRT1 reverse, 5′-GTCTGGCTTATATCCAACACTTCGT-3′.
Transfection of antagomiRs and miRNA mimics
miRNA inhibitors, also known as antimiRs or antagomiRs, and miRNA mimics specifically designed for miR-22 and miR-29a were purchased from Exiqon. Semiconfluent PC3 and DU145 cells as well as LNCaP and DUCaP cells were transfected using Nanofectin siRNA kit (PAA) or Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions, the former ones with 50nM antagomiRs (4100870-011, 4101448-011) or the Negative Control A (199004-04), the latter ones with 50nM mimics (473552-001, 472650-001) or the Negative Control 4 (479903-001). miRNAs expression levels were evaluated by qPCR 72 or 48 hours after transfection of antagomiRs or mimics along with the quantification of target protein abundance via Western blot analysis.
Western blotting
Western blot analysis was performed with 40–50 μg of total protein extract and analyzed using the Odyssey IR Imaging System (LI-COR Biosciences) as previously described (33). The following primary antibodies were used: anti-GAPDH (1:100 000; Chemicon), anti-PARP p85 (1:1000; Promega), anti-LAMC1 (1:1000; Sigma-Aldrich), anti-Mcl-1 (1:500; Santa Cruz Biotechnology, Inc), and anti-AR (1:500; Santa Cruz Biotechnology, Inc). As secondary antibodies, IRDye 800-conjugate goat antimouse/rabbit (611-132-122; Rockland) and Alexa Fluor 680-conjugated goat antimouse/rabbit (a21058; Invitrogen) were employed.
Luciferase reporter gene assay
cDNA fragments of the LAMC1 (506 bp) or MCL1 (748 bp) 3′-UTRs predicted to encompass miR-22 and miR-29a target sites, respectively, were inserted into the SacI-XbaI restriction sites in the multiple cloning region located in the reporter gene 3′-UTR of the reporter vector pmiRNano-GLO (Promega). This bicistronic vector contains NanoLuc luciferase (NlucP) as the primary reporter gene and Firefly luciferase (Luc2) as control reporter for normalization. PC3 cells were transfected with the reporter vectors in combination with either miR-22 mimic, miR-29a mimic and the Negative Control 4 or antimiR-22, antimiR-29a, and the Negative Control A (all purchased from Exiqon), using jetPrime (Polyplus, VWR). The plasmid pCS2-Venus, kindly provided from Dr Ploner (Medical University of Innsbruck, Innsbruck, Austria), which expresses a bright GFP variant under the control of a CMV promoter, was added to visualize and control transfection efficiency. Plasmid vectors and miRNA mimics or antimiRs along with jetPrime transfection reagent were premixed in jetPrime transfection buffer and incubated for 15 minutes. In the meanwhile, PC3 cells were trypsinized and resuspended in RPMI 1640 medium supplemented with 3% charcoal-stripped FCS (Fischer). Cell suspension and transfection solution were mixed, incubated for additional 15 minutes and afterwards seeded in quadruplicates in 96-well plates in a final volume of 100 μL/well. As follows, the specific amounts used per well: 8000 PC3 cells, 40-ng reporter vector, 20-ng pCS2-Venus plasmid, 55nM (final concentration) miRNAs mimics or antimiRs, 0.6-μL transfection reagent, and 4-μL transfection buffer. After 36 hours, transfected PC3 cells underwent NanoLuc and Firefly luciferase activity measurements using the Chameleon 5025 instrument (HVD Life Sciences) and the Nano-Glo Dual-Luciferase Assay (Promega) according to the manufacturer's recommendations. The ratio NanoLuc reporter activity to Firefly control reporter activity was calculated for each well, and the measurement deviating the most from the mean value of the 4 replicates was excluded. Then, normalized mean reporter activities were further compared with the respective controls, either miRNA mimic or antimiR control.
Transwell cell migration assay
PC3 and LNCaP cells were cultured in 6-well plates and transfected with 50nM miRNA Negative Control number 1 (AM17110; Ambion) or miR-22 and miR-29a mimics (PM10203 and PM12499; Ambion). After 48 hours, cells were trypsinized and seeded onto transwell cell migration inserts (8-μm pore size; Becton Dickenson) in presence of 1% FCS medium at a density of 50 000 cells/well for PC3 and 100 000 cells/well for LNCaP. In the surrounding compartment, 10% FCS medium was added as a chemoattractant. On the next day, the cells that had not migrated were removed by cotton swabs, whereas the ones that migrated to the lower surface of the insert filters were fixed in ice-cold methanol, stained with DAPI, and counted as previously described (34).
Cell viability assay
Twenty-four hours before transfection with miR-22, miR-29a or control mimic (Exiqon), PC3 and LNCaP cells were seeded in 96-well plates in the appropriate growth medium at a density of 3000 and 9000 cells/well, respectively. Cell viability was measured 96 hours after transfection using the colorimetric reagent WST-1 (Roche). Absorbance values were evaluated with the Chameleon 5025 instrument (HVD Life Sciences) after incubation for 30 minutes or 1 hour, and the mean value of 3 replicates was used to calculate the percentage of viable cells relative to the control mimic.
miRNA expression levels according to The Cancer Genome Atlas (TCGA)
Illumina HiSeq miRNA expression data (level 3) of prostate adenocarcinoma were downloaded from TCGA (https://tcga-data.nci.nih.gov). A total of 421 tumor samples and 52 normal samples were available at the time of the query (August 2014). Only the expression data of 52 patients with miRNA expression levels available from both tumor and benign tissues and of 324 participants, whose complete clinical information was available, were considered and their normalized values were further evaluated for each miRNA under study.
Tissue microarray (TMA) and immunohistochemistry
A TMA of formalin-fixed paraffin-embedded tissue blocks of 41 untreated patients who underwent radical prostatectomy was constructed to investigate LAMC1 and Mcl-1 expression in malignant and benign prostate tissue. The use of the archive samples was approved by the Ethics Committee of the Medical University of Innsbruck (UN 3174 and AM3174). Paraffin-embedded tissue samples of tumor cases, for which total RNA extracted from the corresponding freshly frozen specimens was available, were chosen for the TMA. Three cancer tissue cores and 3 corresponding benign cores of each case were punched under the supervision of an uropathologist (G.S.). The TMA was assembled using a manual tissue arrayer (Beecher Instruments). In addition, a TMA of formalin-fixed paraffin-embedded cell line samples, including PC3, DU145, LNCaP, and DUCaP cells, was established. The following antibodies were used for immunostaining: anti-AR (1:200; Abcam), anti-Mcl-1 (1:200; Santa Cruz Biotechnology, Inc), and anti-LAMC1 (1:100; Santa Cruz Biotechnology, Inc). Immunostaining was performed on a Ventana Discovery instrument (Roche) using standard conditions. The sections were counterstained with hematoxylin, and images were taken as described in detail elsewhere (35). HistoQuest software version 3.5 (Tissue Gnostics) was used to quantify staining intensity and the percentage of stained cells within the cell line TMA cores. Multiplying these 2 values resulted in an immunostaining score. For tissue specimens, an uropathologist analyzed LAMC1 and Mcl-1 staining intensities by visual inspection in a blinded fashion adopting a semiquantitative scoring system (0 = negative, 1 = weak, 2 = moderate, 3 = strong) and AR immunostaining was evaluated using the HistoQuest software. For AR and Mcl-1, the final immunostaining score was calculated by multiplying the fraction of positive cells with the staining intensity. The LAMC1 antibody stained both the stromal and epithelial compartments of benign and tumor tissue and the staining intensity of benign and tumor epithelial cells was used as immunostaining score.
DNA methylation and gene expression analysis
Methylated DNA immunoprecipitates (MeDIPs) were obtained either from prostate tissues, including unmatched benign (n = 53) and tumor (n = 51) specimens, or from PCa cell lines (LNCaP, VCaP, DU145, PC3) using a specific antibody for 5-methyl cytosine and analyzed as previously described (36). In addition, total RNA extracted from the same tissue samples was amplified, labeled, hybridized to the Affymetrix 1.0 Human Exon ST array, and then evaluated according to Börno et al (36).
Statistical analysis
Statistical investigations were performed using the GraphPad Prism software version 5 (GraphPad Software, Inc). Real-time PCR data were normalized to the appropriate housekeeping gene and expressed as fold change of expression, 2−ΔCt. The 2-tailed Student's t test was chosen to assess differential expression of genes/miRNAs of interest between either treated-untreated cell lines (unpaired t test) or matched cancer-benign tissue specimens (paired t test).
In order to statistically test for differential expression of miRNAs in matched benign-tumor samples from the TCGA database, the raw miRNA-seq read count data was analyzed with 2 methods implemented as R packages: EBSeq, an empirical Bayesian differential expression analysis tool (37), and DESeq, a method based on negative binomial distribution (38). The miRNAs of interest were confirmed to have differential expression in tumor and benign samples by both methods. To visualize the miRNAs expression as boxplots, the count data was normalized to the estimated size factors, using DESeq. This is important to compare different samples to account for different sequencing depths. The ANOVA statistical test was used to analyze the miRNA-seq tumor data stratified according to the Gleason score, followed by multiple comparison with Tukey post hoc tests.
The AR immunohistochemistry scores were compared between tumor and benign tissues using the nonparametric Mann-Whitney test. The semiquantitative immunostaining scores for LAMC1 and Mcl-1 were categorized into 2 groups (none or low, medium or high) and the resulting frequencies were compared between tumor-benign tissues using the Fisher exact test to calculate the P value and the odds ratio.
Differences in DNA methylation between benign and tumor tissues were considered statistically significant for P < .05 according to the Mann-Whitney test with Benjamini and Hochberg correction.
Before using parametric tests to analyze data from tissue samples, the normal distribution assumption was tested with Shapiro-Wilk test. Statistical differences are indicated in graphs as *, P < .05; **, P < .01; ***, P < .001.
Results
miRNAs as potential members of the human androgenome
To elucidate the AR cistrome, ChIP-seq data were generated as previously described (27), using the DUCaP cell line model, which is characterized by an elevated level of wild-type AR protein and the presence of a TMPRSS2-ERG fusion gene (39). A total of 39 156 putative ARBSs were detected in DUCaP cells stimulated with the synthetic androgen R1881 when compared with untreated cells. Because androgen-regulated control elements can be located in a wide-range distance from the transcription start site (TSS) of a target gene (40–43), a gene was considered as a potential target of AR when its TSS was within a window of 25 kb either up- or downstream of the ARBS. A total of 14 635 genes annotated in the University of California Santa Cruz Genome Browser (NCBI36/hg18) were identified, including well-known androgen-responsive genes (eg, KLK3 [PSA], KLK2, FKBP5) as well as a total of 275 miRNA host genes.
In parallel, a microarray expression analysis was performed by means of the Affymetrix HuGene-1.0 st v1 microarray platform to identify androgen-regulated miRNA host genes, defined as genes harboring miRNA sequences within one of their exons or introns. The androgen-sensitive cell line, DUCaP, was used to investigate changes in the AR transcriptional program induced by 8 and 24 hours of exposure to androgen. Using a Benjamini-Hochberg corrected P < .05 as cut-off, 71 miRNA host genes were found to be significantly androgen regulated at both time points in DUCaP cells (either up or down). The integration of expression profiling data with ChIP-seq information disclosed that 32 host genes were significantly androgen-regulated and bound by AR in DUCaP cells (Figure 1A). In addition, MIR22HG was included in the list as a highly AR regulated gene harboring ARBSs up to 36 kb upstream of the TSS. On the basis of the aforementioned criteria as well as of their reported deregulated expression in tumors and their predicted downstream targets belonging to cancer hallmark pathways, miR-22, miR-29a, and miR-17-92 cluster were selected for further studies (Table 1).
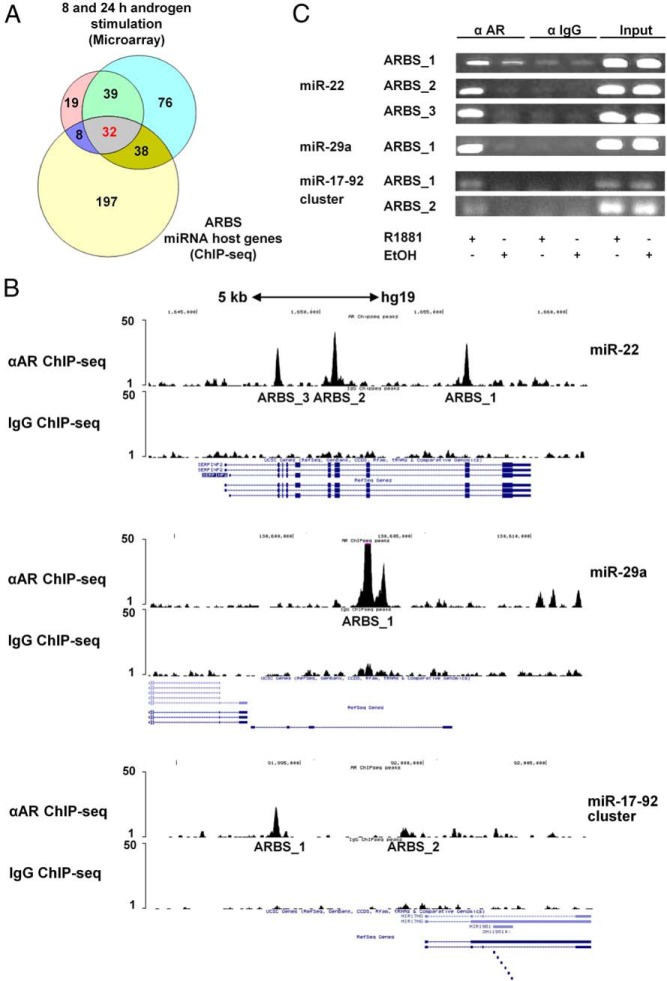
Figure 1.
Identification of miR-22, miR-29a, and miR-17-92 cluster as members of the human androgenome. A, Venn diagram of potential androgen-regulated and AR-target miRNA host genes according to gene expression microarray and ChIP-seq analyses in androgen-stimulated DUCaP cells. B, ChIP-seq tracks of miR-22, miR-29a, and miR-17-92 cluster ARBSs genomic regions in DUCaP cells after androgen treatment. DNA reads of anti-AR and control IgG ChIP samples were aligned to the human consensus sequence (GRCh37/hg19) to identify ARBSs. Three ARBSs for miR-22, a single one for miR-29a, and 2 for miR-17-92 cluster were detected. Peaks height reflects the relative AR affinity for each binding site. A schematic depiction of the 3 host genes and the identified ARBSs is presented in Supplemental Figure 1. C, Validation of the ARBSs in the selected miRNAs via PCR amplification of independent anti-AR (α-AR) ChIP-samples obtained from DUCaP cells stimulated for 1 hour with 1nM R1881. Ethanol-treated (EtOH) and control IgG-immunoprecipitated (α-IgG) cells together with the inputs were used as controls. The binding sites present in miR-17-92 cluster host gene, which showed the weakest enrichment, were further confirmed by Sanger DNA sequencing.
Table 1.
Putative AR Target miRNAs According to ChIP-seq and Microarray Analysis
| HG Name | miRNA | Microarray |
ChIP-Seq |
Target | ||
|---|---|---|---|---|---|---|
| BHp | M | Score | Fold | |||
| ALDH4A1 | miR-1290 | 2.007E-06 | 1.019 | 129.11 | 30.70 | KIF13B |
| EU154352 | miR-29c | 9.158E-03 | 0.716 | 97.38 | 11.69 | MCL1 |
| NVL | miR-320b-2 | 1.470E-02 | −0.325 | 107.87 | 16.37 | TFRC |
| LYST | miR-1537 | 1.447E-02 | −0.330 | 187.71 | 24.56 | MYCN |
| SHOC2 | miR-548e | 3.571E-08 | 0.914 | 52.97 | 9.98 | FOXJ3 |
| C10orf118 | miR-2110 | 2.843E-08 | 1.085 | 275.26 | 28.76 | PIK3R3 |
| RPS6KA4 | miR-1237 | 3.971E-07 | −0.800 | 568.80 | 19.47 | ADAM19 |
| PDE2A | miR-139 | 5.366E-09 | −1.156 | 55.37 | 8.82 | TCF4 |
| ARRB1 | miR-326 | 2.723E-06 | −0.933 | 228.18 | 16.37 | NOB1 |
| HOXC4 | miR-615 | 3.552E-03 | −0.605 | 584.54 | 31.78 | IGF2R |
| MIR17HG | miR-17–92-cluster | 5.286E-05 | −1.347 | 245.48 | 28.07 | PTEN |
| EVL | miR-342 | 8.209E-03 | −0.367 | 1125.46 | 38.41 | SREBP-2 |
| TLE3 | miR-629 | 5.193E-04 | −0.442 | 1103.09 | 31.69 | AKT3 |
| WWP2 | miR-140 | 1.502E-03 | 0.380 | 278.43 | 49.11 | ADAM10 |
| MAP2K4 | miR-744 | 1.028E-04 | 0.871 | 594.48 | 54.03 | TGFβ1 |
| WIPI1 | miR-635 | 1.490E-13 | 4.112 | 1407.54 | 46.66 | P53AIP1 |
| DNM2 | miR-638 | 3.220E-08 | 0.933 | 1058.57 | 94.14 | SOX2 |
| ATF2 | miR-933 | 2.663E-02 | −0.250 | 174.48 | 16.37 | MEF2A |
| ZNF385B | miR-1258 | 3.971E-10 | −2.341 | 268.43 | 47.75 | HPSE |
| C21orf34 | miR-99a | 3.232E-06 | −1.826 | 1335.18 | 66.74 | MTOR |
| CTDSPL | miR-26a-1 | 3.094E-05 | −0.610 | 113.88 | 16.37 | PTEN |
| FOXP1 | miR-1284 | 1.570E-06 | −0.634 | 267.97 | 38.20 | RAB10 |
| TNIK | miR-569 | 6.275E-11 | −1.667 | 664.44 | 73.67 | RAB8b |
| FAM114A1 | miR-574 | 6.254E-06 | 0.885 | 90.38 | 23.76 | Cullin-2 |
| ARL15 | miR-581 | 1.358E-06 | −0.863 | 330.29 | 34.79 | EEF1A1 |
| KLHL3 | miR-874 | 3.602E-10 | −1.487 | 818.45 | 37.91 | HDAC1 |
| GALNT10 | miR-1294 | 1.863E-02 | 0.442 | 340.31 | 40.02 | PARM1 |
| MGAT4B | miR-1229 | 1.526E-02 | −0.280 | 97.40 | 10.91 | BHLHE41 |
| RNF130 | miR-340 | 5.275E-14 | −1.499 | 76.21 | 13.10 | ROCK1 |
| SND1 | miR-593 | 7.202E-06 | 0.696 | 1240.59 | 46.66 | RAB21 |
| MIR29HG | miR-29a | 1.067E-02 | 0.801 | 1366.96 | 31.72 | MCL1 |
| CHPF2 | miR-671 | 6.446E-09 | 0.938 | 86.86 | 10.00 | NFYA |
| MIR22HG | miR-22 | 1.702E-10 | 1.682 | 336.79 | 37.61 | LAMC1 |
Thirty-three miRNA host genes, which are androgen regulated after 8 and 24 hours and harbor a single or more ARBSs or are located close to ARBSs were identified. The table includes host gene (HG) names, their resident miRNA(s), and androgen regulation properties along with either already validated or predicted major downstream targets of the corresponding miRNA. The miRNAs selected for further studies are underlined. BHp, Benjamini-Horchberg corrected P value, M, extent of androgen regulation (log2); Score, quality value of specific ARBS; Fold, enrichment in AR binding to that specific ARBS.
ARBSs upstream of miRNA host genes
According to ChIP-seq results obtained from androgen stimulated DUCaP cells, AR binds close to MIR22HG at 3 different sites, approximately 28, 30, and 36 kb upstream of the TSS. Notably, all 3 sites are located within another protein-coding gene named SERPINF2. Conversely, the hypothetical miR-29a host gene (MIR29HG) harbors one single ARBS located circa 6 kb upstream. For the miR-17-92 cluster, 2 ARBSs were identified located in the promoter region of the host gene (MIR17HG), about 0.6 and 5.4 kb upstream of the TSS, respectively (Figure 1B and Supplemental Figure 1). ChIP-seq revealed ARBSs with diverse affinity for AR as depicted by different peak heights of DNA enrichment (Figure 1B). The analysis of these genomic loci with MatInspector predicted the presence of at least 1 and up to 5 AREs within each of the identified ARBS, with the exception of the miR-17-92 cluster ARBS_1 (Supplemental Table 1).
The interaction between AR and the candidate miRNA host genes was further validated in DUCaP cells treated with 1nM R1881 for 1 hour by AR-ChIP and subsequent qPCR or PCR amplification of the ARBS DNA fragments. All ARBSs exhibited direct binding of AR to miR-22, miR-29a, and miR-17-92 cluster host genes at these genomic loci (Figure 1C).
Androgen regulation of miRNAs and their host genes in PCa cells
As described above, microarray expression profiling of AR-positive cell line DUCaP suggested an important role of the AR in regulating miR-22, miR-29a, and miR-17-92 cluster expression. Therefore, the same experimental model together with an additional androgen-dependent cell line, LNCaP, was adopted to evaluate the response of each miRNA to R1881 treatment, choosing as time points 24 and 48 hours. Both LNCaP and DUCaP cells showed a time-dependent increase in miR-22 as well as miR-29a levels after incubation with R1881. In particular, miR-22 displayed the most significant and highest induction of approximately 11-fold in the DUCaP cell line upon 48 hours, whereas miR-29a expression was up-regulated almost 4-fold at the same time point. In LNCaP cells, miR-22 was significantly regulated up to 3-fold after 48 hours, whereas miR-29a showed increased levels, corresponding to almost 6-fold regulation, already after 24 hours. In both models, the most consistent changes in miR-17-92 cluster expression were detected after 24 hours of stimulation albeit in opposing directions. Under these conditions, in DUCaP cells, all cluster members, with the exception of miR-92a, were found to be negatively regulated with a decrease of approximately 2-fold for miR-19a and a significant down-regulation of miR-19b and miR-17-5p. In LNCaP cells, however, apart from miR-19a, androgen treatment stimulated miR-17-92 cluster expression with an increase of 1.5-fold for miR-20a and an upward trend without statistical significance for the rest of the cluster members (Figure 2A).
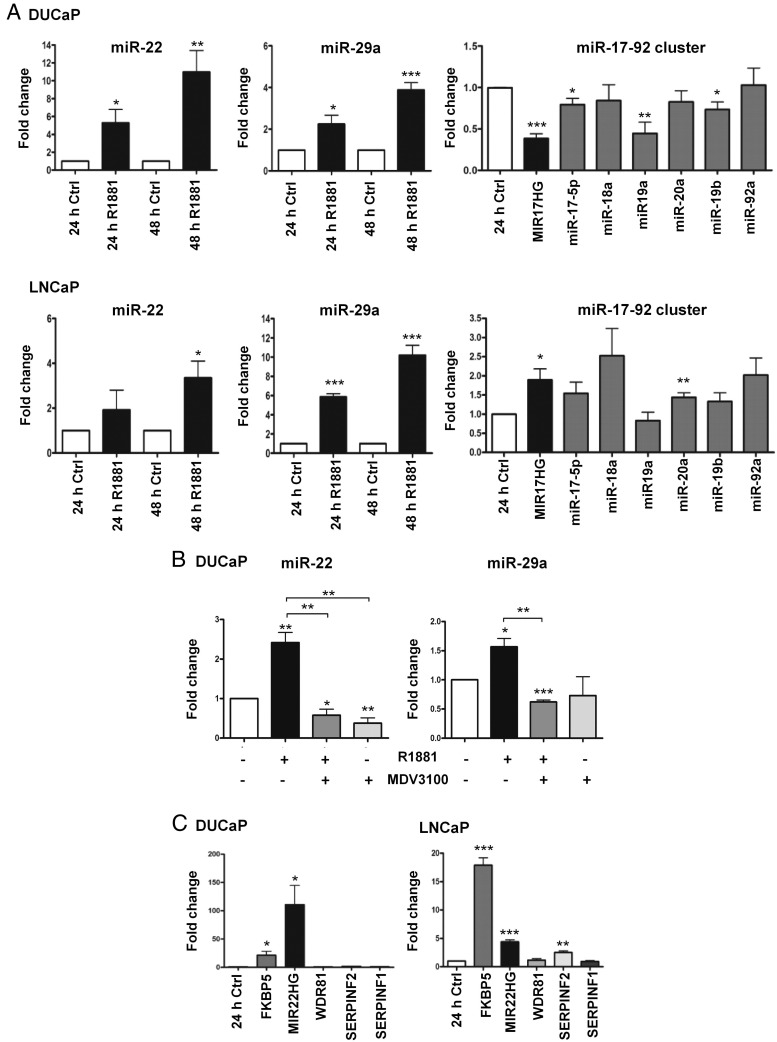
Figure 2.
Androgen regulation of the selected miRNAs and their host genes. A, Androgen regulation of miR-22, miR-29a, and miR-17-92 cluster components along with MIR17HG in DUCaP and LNCaP cell lines was confirmed by qPCR. LNCaP and DUCaP cells were treated either with 1nM R1881 or equivalent vehicle (Ctrl) before RNA isolation. For miR-22 and miR-29a, the mature isoform -3p was analyzed. B, Attenuation of androgen regulation of miR-22 and miR-29a by antiandrogen treatment. DUCaP cells were treated with 10μM MDV3100 either alone or in combination with 1nM R1881 for 24 hours, and, afterwards, miRNAs levels were analyzed by real-time PCR. C, Robust increase of MIR22HG expression but not of other genes adjacent to the ARBSs upon androgen stimulation, as shown by qPCR in DUCaP and LNCaP cell lines. TBP or HPRT1 were used to normalize gene expression. The direct AR target gene FKBP5 was included as positive control. A–C, miRNAs expression was normalized to the small nucleolar RNAs SNORD44 or SNORD38b. Each bar represents mean values ± SEM of at least 3 independent experiments (unpaired Student's t test; *, P < .05; **, P < .01; ***, P < .001).
AR inhibition by MDV3100, a new generation antagonist, which prevents ligand binding as well as nuclear translocation of AR and its association with DNA (44), efficiently attenuated androgen-mediated induction of miR-22 and miR-29a in DUCaP cells. Expression of both miRNAs was almost 3 times lower in cells treated for 24 hours with MDV3100 (10 μM) either alone or in combination with R1881 (1nM) (Figure 2B).
To better understand whether members of a miRNA cluster or resident miRNA are coregulated with the corresponding host gene, MIR17HG as well as MIR22HG expression was assessed in response to 24 hours of androgen stimulation. MIR29HG resides within a long-intergenic nonprotein coding gene, located inside a common fragile site (FRA7H) whose sequence is continuously updated. Thus, it was not feasible in the current study to investigate its modulation. The similar regulation of MIR17HG and the miR-17-92 cluster elements (down in DUCaP and up in LNCaP cells) confirmed that miRNAs and their host genes are potentially coregulated by AR as unique transcription units (Figure 2A). Indeed, also MIR22HG expression was androgen induced in both LNCaP and DUCaP by 4.4- and 110-fold, respectively. The well-characterized androgen-sensitive gene FKBP5 served as control (Figure 2C). Due to the fact that the ARBSs for MIR22HG/miR-22 are within the gene SERPINF2 and the nearby genomic region includes 2 additional genes called SERPINF1 and WDR81, located upstream and downstream of the ARBSs, it was additionally studied if androgen treatment has an impact on the expression of these adjacent genes. In the DUCaP cell line, the expression of these 3 genes was not affected by 24 hours incubation with R1881 but in the LNCaP cells SERPINF2 expression increased 2.5-fold upon R1881 treatment. Nevertheless, this regulation was not as prominent as the up-regulation of MIR22HG. Thus, miR-22 together with its host gene was considered to be the main transcriptional target of AR at that genomic locus (Figure 2C).
miR-22 and miR-29a abundance in various prostate cell lines
The strong sensitivity of miR-22 and miR-29a to treatment with androgen and antiandrogen suggested an association of their expression with the AR status of prostate cells. Consequently, a panel of benign and malignant AR positive and negative prostate cell lines were screened for the abundance of these 2 miRNAs under basal cell culture conditions. miR-22 and miR-29a were typically expressed at higher levels in AR-negative compared with AR-positive lines (Figure 3A). Moreover, the comparison between AR-negative PC3 cells and their clonal derivative cell line PC3-AR, obtained by stable transfection with a plasmid harboring the coding sequence of the human AR (Supplemental Figure 2A), clearly revealed that the presence of AR modulates miR-22 and miR-29a expression (Figure 3A). Also, in the cellular context of PC3-AR, androgen stimulation for 48 hours resulted in a significant increase of miR-22 expression together with its corresponding host gene and the control AR target gene FKBP5. In contrast, no change was observed for miR-29a levels (Supplemental Figure 2B). This corroborated the idea of AR coregulation of host genes and resident miRNAs and suggested miR-22, whose expression was strongly dependent on AR, as a consistent mediator of androgenic and AR effects.
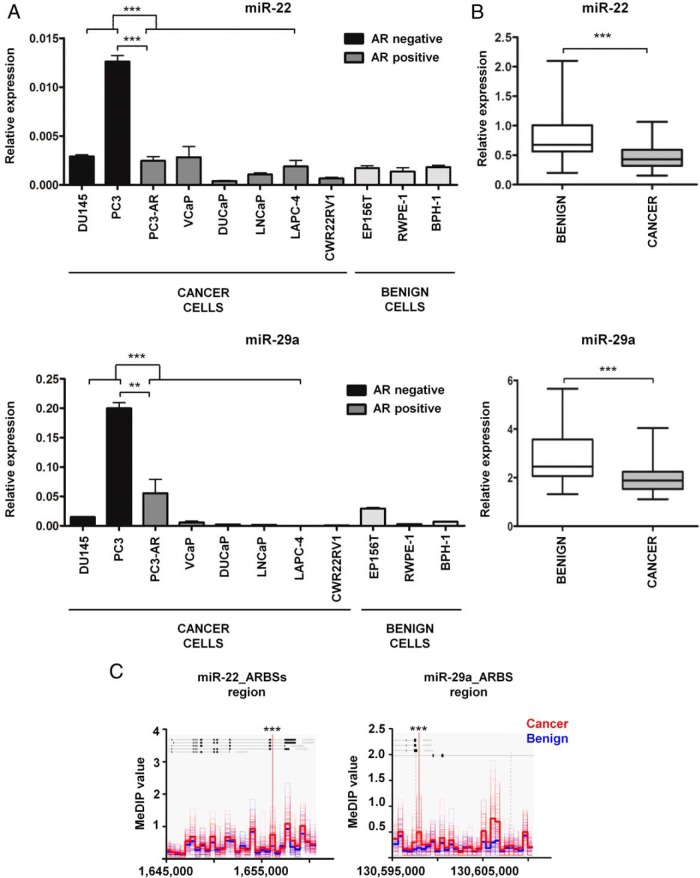
Figure 3.
Basal levels of miR-22 and miR-29a are decreased in AR-positive PCa cell lines and in primary prostate tumors. A, miR-22 and miR-29a basal levels were measured by qPCR in various prostate cell lines. miRNA expression was normalized to the small nucleolar RNA, SNORD44. Each bar represents mean value ± SEM of 3 independent experiments (unpaired Student's t test; *, P < .05; **, P < .01; ***, P < .001). Cell lines were grouped according to their AR and malignant/benign status, as specified below the graph. B, miR-22 and miR-29a expression determined by qPCR in a cohort of 41 matched nonmalignant and malignant tissue sections isolated from radical prostatectomy specimens. miRNA expression was normalized using a panel of normalizers: SNORD44, SNORD48, miR-151a–3p, and miR-320b, (paired Student's t test; *, P < .05; **, P < .01; ***, P < .001). C, Differential DNA methylation of miR-22 and miR-29a ARBSs genomic regions. DNA methylation data were obtained from benign (n = 53) and cancer (n = 51) DNA samples using MeDIP-seq and further analyzed for the presence of differences in the genomic region encompassing miR-22 and miR-29a ARBSs. Depicted are the MeDIP-seq values. Significance was tested with the Mann-Whitney test and further corrected using the Benjamini-Hochberg approach (Mann-Whitney test; *, Benjamini-Horchberg corrected P value (BHp) < .05; **, BHp < .01; ***, BHp < .001).
miR-22 and miR-29a expression in primary prostate tumors
In order to determine miR-22 and miR29a expression patterns in benign and malignant prostate tissue, total RNA was isolated from a cohort of 41 matched cancer-benign tissue samples, obtained from patients who underwent radical prostatectomy after tumor diagnosis, and analyzed by means of qPCR. The comparison between normal and tumor specimens disclosed a higher relative expression of both miRNAs in benign tissue (Figure 3B). When the patients were further classified into subgroups according either to Gleason score (low ≤ 6, medium = 7, high > 7) or tumor stage (low = T2a–T2c, high = T3a–T4) no significant difference in miR-22 and miR-29a levels was found (data not shown).
For independent validation of our results, an in silico analysis was carried out using miRNA-sequencing expression data from a larger panel of PCa tissue specimens accessible from TCGA (45). Within a group of 52 patients for which paired benign-tumor information was available, both miRNAs showed a slight but significantly reduced expression in the cancerous tissue (Supplemental Figure 3A). Notably, the stratification of 324 patients according to Gleason score displayed a significant reduction of miR-29a in the high Gleason class (Gleason score > 7) and no difference for miR-22 (Supplemental Figure 3B).
Regulation of miR-22 and miR-29a expression in PCa by DNA methylation
Despite an increment of AR and enhanced AR signaling in PCa (46), both androgen-up-regulated miR-22 and miR-29a showed decreased expression in tumor tissue (Figure 3B). Consequently, an additional regulation level of their expression, which overrules the androgen-AR axis, was hypothesized to be involved. As a potential mechanism, epigenetic silencing by DNA methylation was investigated, particularly because partial demethylation of the miR-29a promoter in LNCaP cells by isoflavone or 5′-aza-2′-deoxycytidine treatment has been shown to induce an up-regulation of miR-29a levels (47). Therefore, a DNA methylation dataset generated by MeDIP-seq of PCa specimens was examined for the methylation profile of the genomic regions encompassing miR-22 and miR-29a ARBSs. As visible from the methylation density plots in Figure 3C, a significant hypermethylation of both genomic loci was uncovered in cancer tissue compared with the benign one. Similarly, the MeDIP analysis of the same genomic regions performed on LNCaP, VCaP, DU145, and PC3 cell line DNA samples showed the highest methylation level in the AR-positive LNCaP and VCaP cells (Supplemental Figure 4). In contrast, in the AR-negative PC3 and DU145 cells, characterized by elevated basal levels of miR-22 and miR-29a (Figure 3A), the degree of methylation was lower (Supplemental Figure 4).
Identification of miR-22 and miR-29a target genes
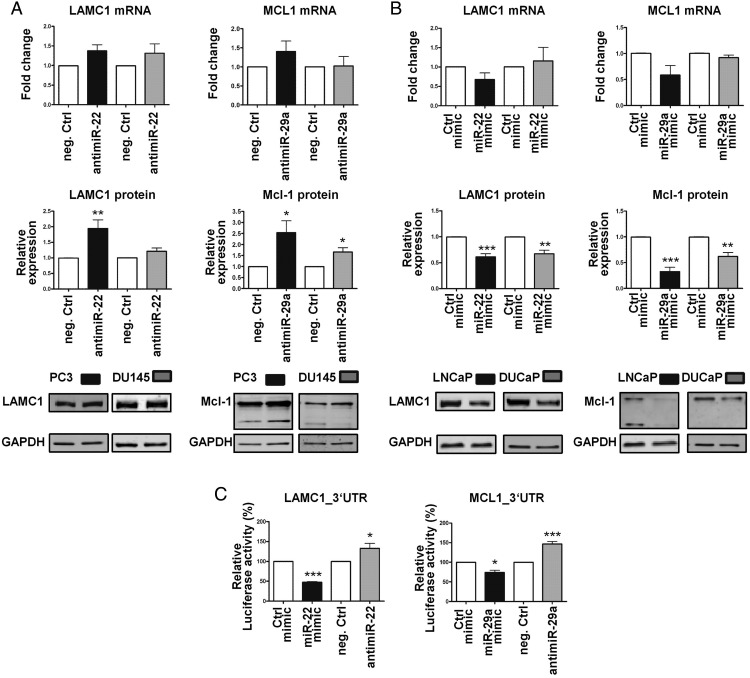
To determine potential downstream targets of miR-22 and miR-29a, different target prediction tools were employed, including TargetScan (http://www.targetscan.org/) and miRWalk (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/index.html). PTEN, a well-known tumor suppressor gene, appeared in the list of both miRNAs as an already validated target. Among other predicted targets of miR-22, LAMC1 was of particular interest given that it encodes a noncollagenous component of the basement membrane involved in a wide variety of cancer-relevant biological processes, including cell adhesion, differentiation, and migration (48). The most promising target of miR-29a was MCL1, an antiapoptotic gene member of the Bcl-2 family, which enhances cell survival. A direct interaction of the MCL1 transcript with miR-29 family is supported by eleven public argonaute crosslinking immunoprecipitation (AGO-CLIP) datasets (49). To evaluate whether LAMC1 and MCL1 transcripts are bona fide targets of miR-22 and miR-29a, respectively, specific antimiRs or mimics together with the appropriate negative controls were employed for the functional inhibition or overexpression of both miRNAs. For this purpose, PC3 and DU145 cells were chosen as experimental models for antimiR treatments due to their higher basal levels of both miRNAs. Conversely, induction of miR-22 and miR-29a levels via mimics usage was performed in the AR-positive cell lines LNCaP and DUCaP based on their reduced expression of both miRNAs. Transfection with antimiR-22 significantly augmented LAMC1 protein levels compared with the negative control but did not significantly alter mRNA levels after 72 hours. Similarly, inhibition of miR-29a via antimiR treatment increased Mcl-1 protein levels consistently in both cell lines without affecting mRNA levels (Figure 4A). LAMC1 and Mcl-1 proteins were clearly reduced 48 hours after miR-22 and miR-29a mimic transfection, respectively (Figure 4B). However, despite a huge increment of miR-22 and miR-29a levels in LNCaP and DUCaP cells (Supplemental Figure 5A), mimics transfection had no significant consequences on LAMC1 and MCL1 transcript levels. As expected by the strong modulation of protein levels, a visible impairment of cellular growth and attachment was observed (Supplemental Figure 5B) The miR-29a mimic also slightly reduced LAMC1 protein but this effect was milder and not significant in comparison with the one generated by miR-22 mimic (Supplemental Figure 5C).
Figure 4.
miR-22 and miR-29a target LAMC1 and Mcl-1 in PCa cells. A, Increased LAMC1 or Mcl-1 protein expression in the PC3 and DU145 cells 72 hours after transfection with 50nM antimiR-22 or antimiR-29a, respectively, but no change in their mRNA levels. B, Reduced LAMC1 or Mcl-1 protein content in LNCaP and DUCaP cells 48 hours after transfection with 50nM miR-22 mimic or miR-29a mimic, respectively, without any significant effects on their transcripts. A and B, Protein values were quantified using the Odyssey IR Imaging System and normalized to GAPDH, qPCR data to TBP, and levels of both were further normalized to the appropriate controls (neg. Ctrl, Ctrl mimic). Each bar represents mean value ± SEM of at least 3 independent experiments (unpaired Student's t test; *, P < .05; **, P < .01; ***, P < .001). C, Validation of miR-22 and miR-29a target sites in the LAMC1 and MCL1 3′-UTRs by reporter gene assay. A luciferase reporter vector containing partial sequences of the LAMC1 or the MCL1 3′-UTRs, harboring the predicted miRNA target sites, in the 3′-UTR of a NanoLuc luciferase gene was used to confirm the regulation by miR-22 and miR-29a, respectively. PC3 cells were transfected for 36 hours with a combination of reporter and control vectors along with miR-22 mimic/antimiR-22 or miR-29a mimic/antimiR-29a (55nM). Afterwards, NanoLuc luciferase reporter gene and Firefly luciferase control reporter activities were measured using a Nano-Glo Dual-Luciferase assay. Reporter gene activity was normalized to the control reporter activity, and the values were expressed relative to the appropriate miRNA mimic or antimiR control treatment. Each bar represents mean value ± SEM of at least 3 independent experiments (unpaired Student's t test; *, P < .05; **, P < .01; ***, P < .001).
Regulation of LAMC1 and MCL1 expression by miR-22 and miR-29a, respectively, was further investigated by reporter gene assays. 3′-UTR cDNA fragments containing the predicted miRNA target sites of the 2 genes were generated by RT-PCR and cloned into the 3′-UTR of a luciferase reporter gene. Thirty-six hours after cotransfection of PC3 cells with reporter gene and either antimiRs or miRNA mimics, a significant up- or down-regulation of the reporter gene activity was measured confirming the predicted modulation by miR-22 and miR-29a (Figure 4C).
Regulation of cancer pathways by miR-22 and miR-29a
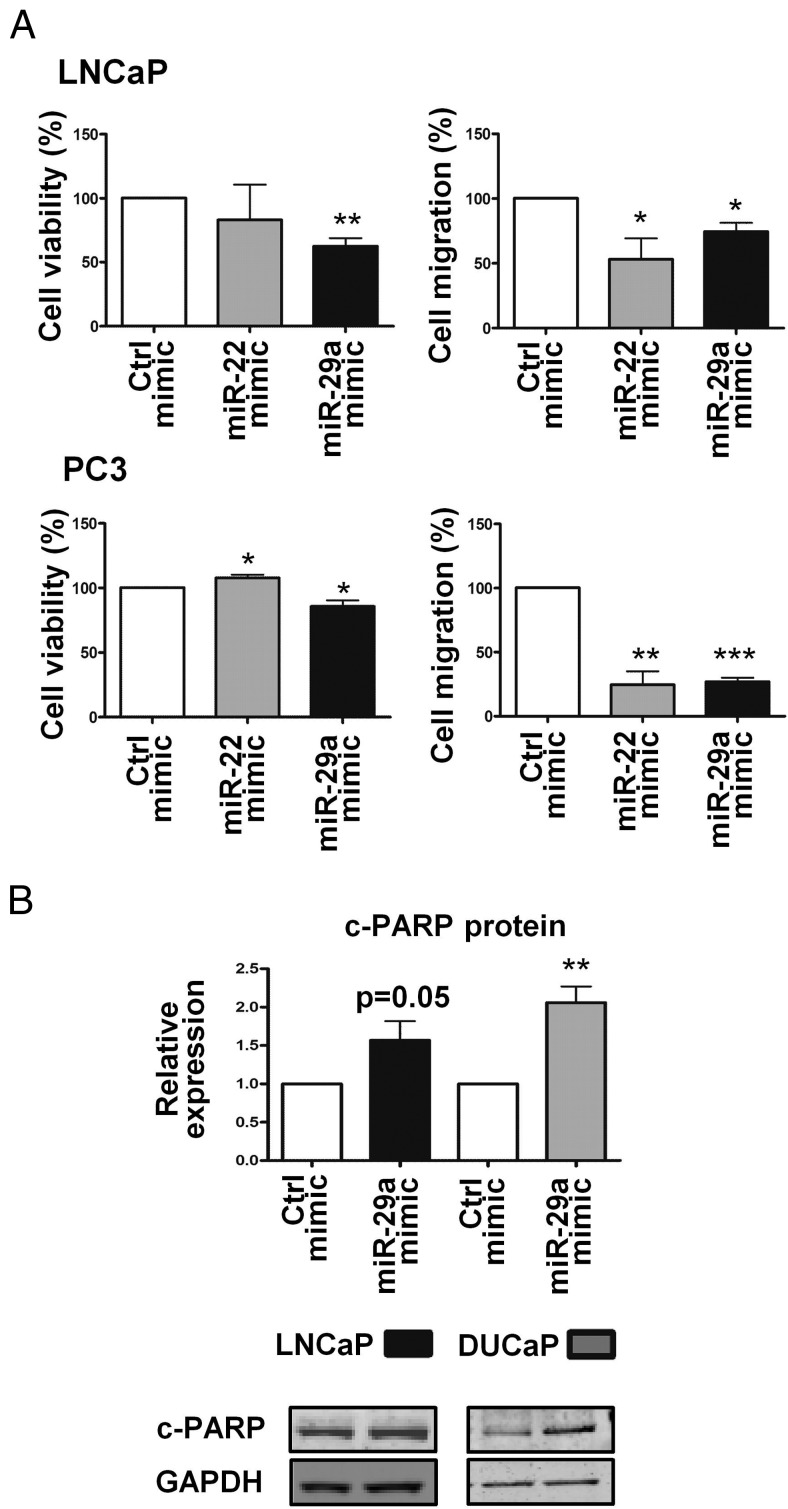
The down-regulation of LAMC1 protein, a component of the basement membrane, has been associated with reduced neuronal cell migration (50), whereas deficiency of Mcl-1, a strong survival factor in tumorigenic cells, including PCa cells (51–54), has been demonstrated to increase apoptosis (55). Our data suggested a link of these functional predictions with miR-22 and miR-29a effects. This was tested in PCa cells, which were transfected with the corresponding miRNA mimics (50nM). As shown in Figure 5A, both miRNAs significantly impaired cell migration, and in addition, miR-29a reduced cell viability of LNCaP and PC3 cells by 35% and 15%, respectively.
Figure 5.
Regulation of cell viability, migration, and apoptosis by miR-22 and miR-29a. A, Impairment of cell viability and migration. LNCaP or PC3 cells were transfected with 50nM miR-22 or miR-29a mimic. Cell viability was determined after 96 hours using the WST-1 colorimetric assay whereas the number of migrated cells was assessed 72 hours after transfection via cell counting of DAPI-stained migrated cells. Values were normalized to the mimic control. Each bar represents the mean value ± SEM of 3 independent experiments (unpaired Student's t test; *, P < .05; **, P < .01; ***, P < .001). B, Apoptosis induction. The effect of miR-29a targeting the prosurvival factor Mcl-1 was evaluated in LNCaP and DUCaP cells. Transient transfection with miR-29a mimic enhanced PARP cleavage. Cleaved PARP (c-PARP) protein levels, determined by Western blot analysis, were quantified using the Odyssey IR Imaging System and normalized to the housekeeping protein GAPDH. Each bar represents the mean value ± SEM of at least 3 independent experiments (unpaired Student's t test; *, P < .05; **, P < .01; ***, P < .001).
To evaluate miR-29a impact on apoptosis, cleavage of PARP was adopted as apoptosis marker. Up-regulation of miR-29a led to an increase of cleaved PARP protein levels in LNCaP and especially in the DUCaP cells (Figure 5B).
Expression of LAMC1 and Mcl-1 in prostate cell lines and PCa tissue samples
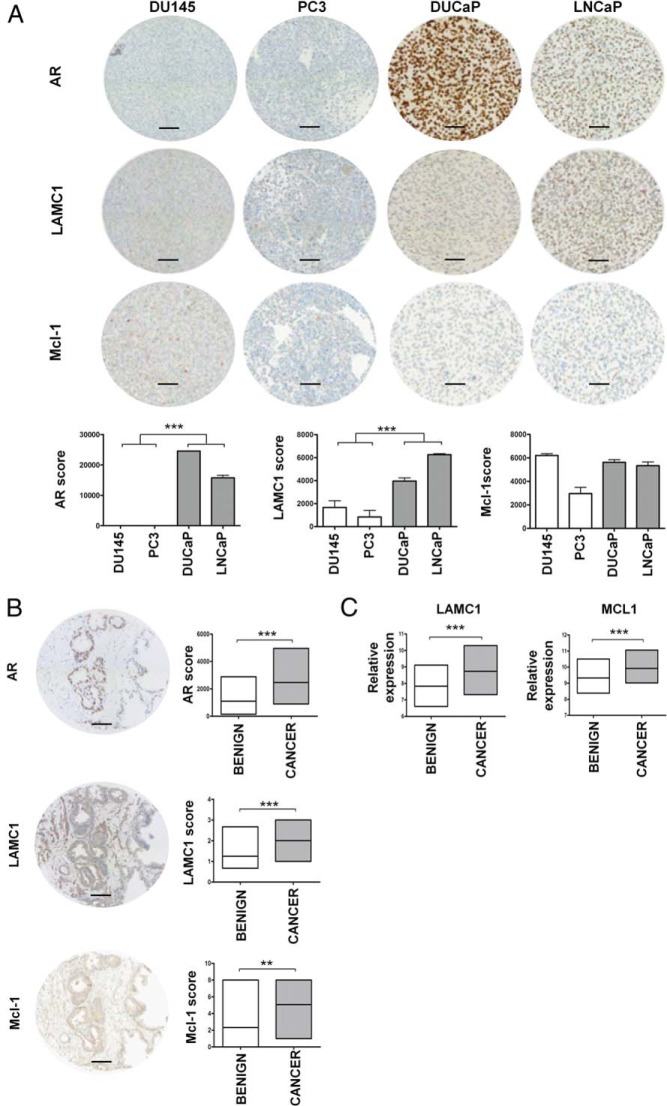
Because screening of PCa cell lines had revealed that both miRNAs were differentially expressed depending on AR presence/absence, a TMA containing 2 AR-positive and 2 AR-negative cell lines was stained using antibodies against AR, LAMC1, and Mcl-1. As hypothesized, in the AR-negative PC3 and DU145 cells, which have a high copy number of miR-22 and miR-29a (Figure 3A), the miRNA target proteins were expressed at lower levels than in AR-positive DUCaP and LNCaP cells, which express only moderate levels of both miRNAs. Grouping the cell lines according to their AR status showed stronger LAMC1 and Mcl-1 staining in AR-positive cells, although the differences were not significant for Mcl-1 (Figure 6A).
Figure 6.
LAMC1 and Mcl-1 expression in PCa cell lines and in benign/cancer prostate tissues. A, Representative TMA cores of paraffin-embedded PCa cell lines stained with anti-AR, anti-LAMC1, and anti-Mcl-1 antibodies along with the immunostaining score values calculated using the HistoQuest software version 3.5. Each bar represents mean value ± SEM of 3 independent experiments (unpaired Student's t test; *, P < .05; **, P < .01; ***, P < .001). B, Anti-AR, anti-LAMC1, and anti-Mcl-1 immunostaining of representative cores of a primary tumor. Original magnification 20/0.5 digital camera. Scale bar, 100 μm. The immunostaining score values summarized in the graphs for tumor and benign tissue cores were obtained by visual inspection in a blinded fashion for anti-LAMC1 and anti-Mcl-1 (Fisher exact test; *, P < .05; **, P < .01; ***, P < .001) while for anti-AR were calculated with the HistoQuest software version 3.5 (Mann-Whitney test; *, P < .05; **, P < .01; ***, P < .001). C, LAMC1 and MCL1 mRNA expression according to gene expression analysis of an independent cohort of unmatched benign (n = 48) and cancer (n = 47) RNA samples isolated from primary tumors. Log 2 values of the normalized array signal are indicated. (Mann-Whitney test; *, P < .05; **, P < .01; ***, P < .001).
As a consequence of miR-22 and miR-29a down-regulation in cancer tissue, an up-regulation of LAMC1 and Mcl-1 proteins was expected and investigated by immunohistochemistry. For this purpose, a tissue array containing cancer and benign tissue samples was immunostained with antibodies against AR, LAMC1, and Mcl-1, respectively. As shown in Figure 6B, all 3 proteins were up-regulated in cancer tissue compared with their benign counterparts. As hypothesized, higher miR-22 and miR-29a levels in the benign tissue were associated with a decreased amount of their target proteins, LAMC1 and Mcl-1. Consistent with this finding, gene expression array analysis of an independent cohort of benign (n = 48) and cancer (n = 47) tissue samples obtained from primary tumors revealed an up-regulation of LAMC1 and MCL1 transcripts in the malignant tissue (Figure 6C).
Discussion
Identification of AR relevant downstream targets is essential to develop a broader and combined therapeutic approach that might improve the responsiveness of PCa to anticancer treatment and also to define new biomarkers useful for the tumor diagnosis, prognosis, and monitoring. Recently, it has been observed that altered expression of miRNAs is associated with progression of androgen-sensitive prostate cells to antiandrogen therapy resistance (56). Moreover, some studies described the involvement of AR signaling in the regulation of specific miRNAs in PCa (6, 57). Hence, relying on an integrative approach, the present work tried to elucidate the impact of the AR signaling axis on miRNAs expression in PCa to better understand miRNA regulation, their role, and their contribution in mediating AR-induced effects.
The combination of ChIP-seq and microarray expression profiling data, both generated from androgen stimulated DUCaP cells, identified 32 significantly androgen-regulated and putatively direct AR-target miRNAs. In view of reported aberrant expression in other types of cancer and the involvement of their predicted targets in oncogenic pathways, miR-22, miR-29a, and the miR-17-92 cluster were chosen for further validation.
Indeed, miR-17-92 cluster is a well-known oncomiR composed of 6 miRNAs, which can potentially affect multiple oncogenic processes. Expression of these miRNAs promotes cell proliferation, suppresses apoptosis of cancer cells, and induces tumor angiogenesis (58). miR-22 can act as a tumor suppressor on one hand, reducing the levels of metastasis-specific signaling proteins in breast cancer cells (59). However, on the other hand, it can behave as oncomiR, promoting cell survival via PTEN repression (60). Also, miR-29a has a double role, inhibiting cell proliferation in human gastric cancer (61) and contributing to metastasis in breast cancer patients (62). Involvement of these miRNAs in tumor biology was further supported by pathway analyses, which showed enrichment for breast cancer and other cancer-associated terms.
In the present study, we demonstrated that miR-22, miR-29a, and the miR-17-92 cluster harbor ARBSs and that their expression is sensitive to androgen stimulation in DUCaP and LNCaP cells. miR-22 and miR-29a were robustly up-regulated by AR activation in a time-dependent manner, whereas more moderate changes in the expression of the miR-17-92 cluster members were observed, which is in line with the moderate AR enrichment at the corresponding binding sites. In DUCaP cells, most cluster miRNAs were down-regulated, with miR-19a as the most significantly repressed miRNA. miR-19a, as well as miR-19b, has been reported to be responsible for the oncogenic activity of the cluster (63). Conversely, in the LNCaP cell line, the miR-17-92 cluster expression was overall enhanced upon androgen treatment. This finding supports the hypothesis that miRNA regulation and maturation is sensitive to the cellular milieu and conditions, such as cell-cell contact and tumor microenvironment (64, 65).
The moderate fold change of the miR-17-92 cluster components upon androgen stimulation compared with the extent of AR regulation of miR-22 and miR-29a, and the influence of the cellular context suggested that AR does not represent the main modulator of this cluster, although it may have an accessory role and interact with other transcription factors. Therefore, the subsequent part of this work focused on the characterization of miR-22 and miR-29a function.
Comparison of basal expression levels in various prostate cell lines and in the AR-positive derivative PC3-AR revealed that miR-22 and miR-29a are more abundant in AR null cells despite the positive AR regulation. A possible mechanism underlying this specific basal expression pattern is epigenetic modulation of expression. Indeed, MeDIP analysis of AR positive and negative PCa cell lines showed hypermethylation of miR-22 and miR-29a genomic regions harboring the ARBSs in the AR-positive cells, whereas in the AR-negative cells, with the highest basal expression of miR-22 and miR-29a, the identical genomic positions were hypomethylated. In line with this mechanism, partial demethylation of the miR-29a promoter in LNCaP cells by isoflavone or 5′-aza-2′-deoxycytidine treatment led to up-regulation of miR-29a levels and, in turns, inhibition of PCa cell growth and invasion (47).
Epigenetic regulation by differential DNA methylation was also identified as a likely reason for higher basal levels of miR-22 and miR-29a in benign prostate tissue compared with the malignant counterpart. Indeed, a differential methylation of the DNA regions comprising the AR regulatory elements of miR-22 and miR-29a was confirmed. Numerous miRNA host genes as well as miRNAs promoters are epigenetically regulated in PCa (36, 66). For instance, miR-193b and miR-34a are tightly controlled by promoter methylation in PCa (9).
Consistent with the pattern of DNA methylation, miR-22 and miR-29a expression profiling of clinical samples revealed reduced expression of both miRNAs in the cancerous tissue relative to the benign counterpart, suggesting a potential tumor-suppressive role of these miRNAs. Recently, it has been observed that tumor cells, characterized by loss of cell contact inhibition, escape the tight control of miRNA biogenesis mediated by Hippo signaling in a cell density-dependent manner (67). Indeed, a significant down-regulation of miR-22 and miR-29a also in hormone-refractory carcinomas was previously detected by 2 independent microarray studies performed on benign prostatic hyperplasia (BPH) and castration resistant PCa specimens (19, 57). Additionally, the miRNA-seq information available in TCGA database comprising a larger cohort of PCa patients confirmed the reduced levels of miR-22 and miR-29a when comparing the cancerous and the benign areas of the examined tissues, even though not so pronounced as in our qPCR analysis. A significant decrease in miR-29a expression with increasing Gleason score was also recognized in the TCGA expression dataset.
The different extent of regulation observed between miRNA-seq data and qPCR results is probably related to various factors, such as the examination of diverse patient cohorts and the use of 2 different techniques, which also implies different normalization methods. Indeed, normalization represents an important aspect of miRNA expression profiling, especially regarding tissue specimens, which requires minimizing the variability between cancer and benign counterparts within the same patient (68). Due to the fact that a specific miRNA normalizer has not yet been established, a panel of housekeeping RNAs, including specific miRNAs and small nucleolar RNAs, was chosen in this study for qPCR evaluation of the clinical samples and assessment of miR-22 and miR-29a expression.
The combination of prediction tools, antagomiRs, and mimics transfection, together with luciferase reporter gene assay and immunohistochemistry, confirmed that LAMC1 and Mcl-1 proteins are downstream targets of miR-22 and miR-29a, respectively. Mcl-1 has already been shown to be a target of another component of the miR-29 family, namely miR-29b, in the KMCH cell line (69). More recently, it has been also demonstrated that the miR-29 family members can modulate PCa cell migration and invasion via targeting LAMC1 (70). Indeed, a mild effect on LAMC1 protein was recognized in DUCaP cells upon miR-29a mimic transfection. However, in our experimental settings the extent of LAMC1 down-regulation by miR-29a was weaker and minor compared with the strong effect of miR-22.
Both LAMC1 and MCL1 act as oncogenes. The LAMC1 gene encodes for the laminin γ1 subunit of laminin, which is one of the earliest and the most ubiquitously expressed subunits (71). LAMC1 is involved in cell adhesion, proliferation and migration. The interaction of cancer cells with laminin was identified as a key event in tumor invasion and metastasis. Indeed, invading tumor cells attach to laminin and this interaction increases their metastatic potential (48). Thus, up-regulation of LAMC1 expression and its secretion can be considered as a kind of auto-stimulation of cancer cells. Deregulation of LAMC1 has been observed in human hepatocellular carcinoma, breast cancer, glioblastoma, and its direct knock-down decreases the ability of MDA-MB-231 cells to form colonies (72). As to PCa, senescence-induced alterations of laminin chain expression have been reported to increase in vitro migration and in vivo tumorigenicity of PCa cell (73).
MCL1 is a member of the antiapoptotic Bcl-2 family and its overexpression is associated with cancer progression and clinical relapse (74). Mcl-1 promotes survival of cancer cells and its up-regulation is a major mechanism of resistance to anticancer agents, such as the Bcl-2 antagonists (75). In the context of PCa, it has been reported that the proinflammatory cytokine IL-6, whose serum levels are increased in patients with metastatic PCa (76), is responsible for resistance to apoptosis and increased Mcl-1 protein expression in LNCaP-IL6+ cells, a model system for advanced PCa (77). Recently, Santer et al (78) demonstrated an up-regulation of Mcl-1 in response to androgen deprivation and an enhanced antitumor effect of androgen deprivation in combination with Mcl-1 inhibition. Knockdown of Mcl-1 and the other Bcl-2 family components has been demonstrated to impair both migration and invasion of colorectal cancer cells, indicating their potential contribution to metastasis in addition to the well-known antiapoptotic effects (79).
AR-stimulated miR-22 and miR-29a, which trigger decreased expression of LAMC1 and Mcl-1 proteins reducing cell migration and viability and enhancing apoptosis, are an example of AR-regulated miRNAs with tumor-suppressive function. Similarly, miR-135a and miR-200b, which attenuate PCa cell migration, invasion and growth, are up-regulated by AR (80, 81).
In summary, data herein demonstrate that miR-22 and miR-29a are novel members of the AR cistrome, since they are significantly regulated upon androgen stimulation. Moreover, their residual gene loci harbor ARBSs and their expression is negatively affected by antiandrogen treatment. Regulation of 2 predicted cancer-associated targets, LAMC1 and Mcl-1, was confirmed. In addition, further information regarding the complex regulatory machinery that controls miRNA activities in PCa cells was provided, disclosing a simultaneous modulation of host genes and the corresponding miRNAs in response to AR activation as well as an active involvement of epigenetic mechanisms in addition to AR regulation.
Acknowledgments
We thank Dr Ummuhan Demir for helpful discussions, Dr Natalie Sampson for editing the manuscript, and Irma Sottsas Mag. Eberhard Steiner, Dr Andrea Eigentler, and Dr Elisabeth Tafatsch for technical assistance.
This work was supported by the Molecular Cell Biology and Oncology (MCBO) doctoral college funded by the Austrian Research Fund (FWF, Project W1101). N.N. was supported by the Intramural program of National Human Genome Research Institute (NHGRI), NIH; and M.R.S. by the Lichtenberg program (VolkswagenStiftung).
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- AR
- androgen receptor
- ARBS
- AR binding site
- ARE
- androgen responsive element
- ChIP
- chromatin immunoprecipitation
- ChIP-seq
- chromatin immunoprecipitation coupled with deep-sequencing
- FCS
- fetal calf serum
- LAMC1
- laminin gamma 1 protein
- Mcl-1
- myeloid cell leukemia 1 protein
- MDV3100
- enzalutamide
- MeDIP
- methylated DNA immunoprecipitate
- miRNA
- microRNA
- PCa
- prostate cancer
- qPCR
- quantitative real-time PCR
- R1881
- methyltrienolone
- TCGA
- The Cancer Genome Atlas
- TMA
- tissue microarray
- TSS
- transcription start site
- UTR
- untranslated region.
References
- 1. Knudsen KE, Scher HI. Starving the addiction: new opportunities for durable suppression of AR signaling in prostate cancer. Clin Cancer Res. 2009;15(15):4792–4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hodgson MC, Bowden WA, Agoulnik IU. Androgen receptor footprint on the way to prostate cancer progression. World J Urol. 2012;30(3):279–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shen MM, Abate-Shen C. Molecular genetics of prostate cancer: new prospects for old challenges. Genes Dev. 2010;24(18):1967–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Feldman BJ, Feldman D. The development of androgen-independent prostate cancer. Nat Rev Cancer. 2001;1(1):34–45. [DOI] [PubMed] [Google Scholar]
- 5. Waltering KK, Porkka KP, Jalava SE, et al. Androgen regulation of micro-RNAs in prostate cancer. Prostate. 2011;71(6):604–614. [DOI] [PubMed] [Google Scholar]
- 6. Ribas J, Ni X, Haffner M, et al. miR-21: an androgen receptor-regulated microRNA that promotes hormone-dependent and hormone-independent prostate cancer growth. Cancer Res. 2009;69(18):7165–7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. ChunJiao S, Huan C, ChaoYang X, GuoMei R. Uncovering the roles of miRNAs and their relationship with androgen receptor in prostate cancer. IUBMB Life. 2014;66(6):379–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Coppola V, De Maria R, Bonci D. MicroRNAs and prostate cancer. Endocr Relat Cancer. 2010;17(1):F1–F17. [DOI] [PubMed] [Google Scholar]
- 9. Paone A, Galli R, Fabbri M. MicroRNAs as new characters in the plot between epigenetics and prostate cancer. Front Genet. 2011;2:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jerónimo C, Bastian PJ, Bjartell A, et al. Epigenetics in prostate cancer: biologic and clinical relevance. Eur Urol. 2011;60(4):753–766. [DOI] [PubMed] [Google Scholar]
- 11. van Rooij E. The art of microRNA research. Circ Res. 2011;108(2):219–234. [DOI] [PubMed] [Google Scholar]
- 12. Chira P, Vareli K, Sainis I, Papandreou C, Briasoulis E. Alterations of MicroRNAs in solid cancers and their prognostic value. Cancers. 2010;2(2):1328–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gururajan M, Josson S, Chu GC, et al. miR-154* and miR-379 in the DLK1-DIO3 microRNA mega-cluster regulate epithelial to mesenchymal transition and bone metastasis of prostate cancer. Clin Cancer Res. 2014;20(24):6559–6569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Josson S, Gururajan M, Hu P, et al. miR-409-3p/-5p promotes tumorigenesis, epithelial-to-mesenchymal transition, and bone metastasis of human prostate cancer. Clin Cancer Res. 2014;20(17):4636–4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Di Leva G, Croce CM. Roles of small RNAs in tumor formation. Trends Mol Med. 2010;16(6):257–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Selth LA, Townley SL, Bert AG, et al. Circulating microRNAs predict biochemical recurrence in prostate cancer patients. Br J Cancer. 2013;109(3):641–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schrauder MG, Strick R, Schulz-Wendtland R, et al. Circulating micro-RNAs as potential blood-based markers for early stage breast cancer detection. PLoS One. 2012;7(1):e29770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim WT, Kim WJ. MicroRNAs in prostate cancer. Prostate Int. 2013;1(1):3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TL, Visakorpi T. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007;67(13):6130–6135. [DOI] [PubMed] [Google Scholar]
- 20. Ozen M, Creighton CJ, Ozdemir M, Ittmann M. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene. 2008;27(12):1788–1793. [DOI] [PubMed] [Google Scholar]
- 21. Ambs S, Prueitt RL, Yi M, et al. Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Res. 2008;68(15):6162–6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. DeVere White RW, Vinall RL, Tepper CG, Shi XB. MicroRNAs and their potential for translation in prostate cancer. Urol Oncol. 2009;27(3):307–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tong AW, Fulgham P, Jay C, et al. MicroRNA profile analysis of human prostate cancers. Cancer Gene Ther. 2009;16(3):206–216. [DOI] [PubMed] [Google Scholar]
- 24. Kogan I, Goldfinger N, Milyavsky M, et al. hTERT-immortalized prostate epithelial and stromal-derived cells: an authentic in vitro model for differentiation and carcinogenesis. Cancer Res. 2006;66(7):3531–3540. [DOI] [PubMed] [Google Scholar]
- 25. Madar S, Brosh R, Buganim Y, et al. Modulated expression of WFDC1 during carcinogenesis and cellular senescence. Carcinogenesis. 2009;30(1):20–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang Q, Carroll JS, Brown M. Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol Cell. 2005;19(5):631–642. [DOI] [PubMed] [Google Scholar]
- 27. Bu H, Schweiger MR, Manke T, et al. Anterior gradient 2 and 3–two prototype androgen-responsive genes transcriptionally upregulated by androgens and by oestrogens in prostate cancer cells. FEBS J. 2013;280(5):1249–1266. [DOI] [PubMed] [Google Scholar]
- 28. Cartharius K, Frech K, Grote K, et al. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21(13):2933–2942. [DOI] [PubMed] [Google Scholar]
- 29. Martowicz A, Rainer J, Lelong J, Spizzo G, Gastl G, Untergasser G. EpCAM overexpression prolongs proliferative capacity of primary human breast epithelial cells and supports hyperplastic growth. Mol Cancer. 2013;12:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hoefer J, Kern J, Ofer P, et al. SOCS2 correlates with malignancy and exerts growth-promoting effects in prostate cancer. Endocr Relat Cancer. 2014;21(2):175–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rainer J, Lelong J, Bindreither D, et al. Research resource: transcriptional response to glucocorticoids in childhood acute lymphoblastic leukemia. Mol Endocrinol. 2012;26(1):178–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jian Ye, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13(134). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Puhr M, Santer FR, Neuwirt H, et al. Down-regulation of suppressor of cytokine signaling-3 causes prostate cancer cell death through activation of the extrinsic and intrinsic apoptosis pathways. Cancer Res. 2009;69(18):7375–7384. [DOI] [PubMed] [Google Scholar]
- 34. Bu H, Bormann S, Schäfer G, et al. The anterior gradient 2 (AGR2) gene is overexpressed in prostate cancer and may be useful as a urine sediment marker for prostate cancer detection. Prostate. 2011;71(6):575–587. [DOI] [PubMed] [Google Scholar]
- 35. Puhr M, Hoefer J, Schäfer G, et al. Epithelial-to-mesenchymal transition leads to docetaxel resistance in prostate cancer and is mediated by reduced expression of miR-200c and miR-205. Am J Pathol. 2012;181(6):2188–2201. [DOI] [PubMed] [Google Scholar]
- 36. Börno ST, Fischer A, Kerick M, et al. Genome-wide DNA methylation events in TMPRSS2-ERG fusion-negative prostate cancers implicate an EZH2-dependent mechanism with miR-26a hypermethylation. Cancer Discov. 2012;2(11):1024–1035. [DOI] [PubMed] [Google Scholar]
- 37. Leng N, Dawson JA, Thomson JA, et al. EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics. 2013;29(8):1035–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sun C, Dobi A, Mohamed A, Li H, et al. TMPRSS2-ERG fusion, a common genomic alteration in prostate cancer activates C-MYC and abrogates prostate epithelial differentiation. Oncogene. 2008;27(40):5348–5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cleutjens KB, van der Korput HA, van Eekelen CC, van Rooij HC, Faber PW, Trapman J. An androgen response element in a far upstream enhancer region is essential for high, androgen-regulated activity of the prostate-specific antigen promoter. Mol Endocrinol. 1997;11(2):148–161. [DOI] [PubMed] [Google Scholar]
- 41. Makkonen H, Kauhanen M, Paakinaho V, Jääskeläinen T, Palvimo JJ. Long-range activation of FKBP51 transcription by the androgen receptor via distal intronic enhancers. Nucleic Acids Res. 2009;37(12):4135–4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Prescott J, Jariwala U, Jia L, et al. Androgen receptor-mediated repression of novel target genes. Prostate. 2007;67(13):1371–1383. [DOI] [PubMed] [Google Scholar]
- 43. Massie CE, Lynch A, Ramos-Montoya A, et al. The androgen receptor fuels prostate cancer by regulating central metabolism and biosynthesis. EMBO J. 2011;30(13):2719–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen Y, Clegg NJ, Scher HI. Antiandrogens and androgen depleting therapies in prostate cancer: new agents for an established target. Lancet Oncol. 2009;10(10):981–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cancer Genome Atlas Research Network, Weinstein JN, Collisson EA, Mills GB, et al. The cancer genome atlas pan-cancer analysis project. Nat Genet. 2013;45(10):1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhou Y, Bolton EC, Jones JO. Androgens and androgen receptor signaling in prostate tumorigenesis. J Mol Endocrinol. 2015;54(1):R15–R29. [DOI] [PubMed] [Google Scholar]
- 47. Li Y, Kong D, Ahmad A, Bao B, Dyson G, Sarkar FH. Epigenetic deregulation of miR-29a and miR-1256 by isoflavone contributes to the inhibition of prostate cancer cell growth and invasion. Epigenetics. 2012;7(8):940–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Givant-Horwitz V, Davidson B, Reich R. Laminin-induced signaling in tumor cells. Cancer Lett. 2005;223(1):1–10. [DOI] [PubMed] [Google Scholar]
- 49. Hamilton MP, Rajapakshe K, Hartig SM, et al. Identification of a pan-cancer oncogenic microRNA superfamily anchored by a central core seed motif. Nat Commun. 2013;4:2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chen ZL, Haegeli V, Yu H, Strickland S. Cortical deficiency of laminin γ1 impairs the AKT/GSK-3β signaling pathway and leads to defects in neurite outgrowth and neuronal migration. Dev Biol. 2009;327(1):158–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Song L, Coppola D, Livingston S, Cress D, Haura EB. Mcl-1 regulates survival and sensitivity to diverse apoptotic stimuli in human non-small cell lung cancer cells. Cancer Biol Ther. 2005;4(3):267–276. [DOI] [PubMed] [Google Scholar]
- 52. Quinn BA, Dash R, Azab B, et al. Targeting Mcl-1 for the therapy of cancer. Expert Opin Investig Drugs. 2011;20(10):1397–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jackson RS, 2nd, Placzek W, Fernandez A, et al. Sabutoclax, a Mcl-1 antagonist, inhibits tumorigenesis in transgenic mouse and human xenograft models of prostate cancer. Neoplasia. 2012;14(7):656–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Son JK, Varadarajan S, Bratton SB. TRAIL-activated stress kinases suppress apoptosis through transcriptional upregulation of MCL-1. Cell Death Differ. 2010;17(8):1288–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yancey D, Nelson KC, Baiz D, et al. BAD dephosphorylation and decreased expression of MCL-1 induce rapid apoptosis in prostate cancer cells. PLoS One. 2013;8(9):e74561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ottman R, Nguyen C, Lorch R, Chakrabarti R. MicroRNA expressions associated with progression of prostate cancer cells to antiandrogen therapy resistance. Mol Cancer. 2014;13(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Jalava SE, Urbanucci A, Latonen L, et al. Androgen-regulated miR-32 targets BTG2 and is overexpressed in castration-resistant prostate cancer. Oncogene. 2012;31(41):4460–4471. [DOI] [PubMed] [Google Scholar]
- 58. Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133(2):217–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Patel JB, Appaiah HN, Burnett RM, et al. Control of EVI-1 oncogene expression in metastatic breast cancer cells through microRNA miR-22. Oncogene. 2011;30(11):1290–1301. [DOI] [PubMed] [Google Scholar]
- 60. Tan G, Shi Y, Wu ZH. MicroRNA-22 promotes cell survival upon UV radiation by repressing PTEN. Biochem Biophys Res Commun. 2012;417(1):546–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cui Y, Su WY, Xing J, et al. MiR-29a inhibits cell proliferation and induces cell cycle arrest through the downregulation of p42.3 in human gastric cancer. PLoS One. 2011;6(10):e25872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gebeshuber CA, Zatloukal K, Martinez J. miR-29a suppresses tristetraprolin, which is a regulator of epithelial polarity and metastasis. EMBO Rep. 2009;10(4):400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Olive V, Bennett MJ, Walker JC, et al. miR-19 is a key oncogenic component of mir-17-92. Genes Dev. 2009;23(24):2839–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hwang HW, Wentzel EA, Mendell JT. Cell-cell contact globally activates microRNA biogenesis. Proc Natl Acad Sci USA. 2009;106(17):7016–7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Musumeci M, Coppola V, Addario A, et al. Control of tumor and microenvironment cross-talk by miR-15a and miR-16 in prostate cancer. Oncogene. 2011;30(41):4231–4242. [DOI] [PubMed] [Google Scholar]
- 66. Gu L, Frommel SC, Oakes CC, et al. BAZ2A (TIP5) is involved in epigenetic alterations in prostate cancer and its overexpression predicts disease recurrence. Nat Genet. 2015;47(1):22–30. [DOI] [PubMed] [Google Scholar]
- 67. Mori M, Triboulet R, Mohseni M, et al. Hippo signaling regulates microprocessor and links cell-density-dependent miRNA biogenesis to cancer. Cell. 2014;156(5):893–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Schaefer A, Jung M, Miller K, et al. Suitable reference genes for relative quantification of miRNA expression in prostate cancer. Exp Mol Med. 2010;42(11):749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Mott JL, Kobayashi S, Bronk SF, Gores GJ. mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene. 2007;26(42):6133–6140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Nishikawa R, Goto Y, Kojima S, et al. Tumor-suppressive microRNA-29s inhibit cancer cell migration and invasion via targeting LAMC1 in prostate cancer. Int J Oncol. 2014;45(1):401–410. [DOI] [PubMed] [Google Scholar]
- 71. Smyth N, Vatansever HS, Murray P, et al. Absence of basement membranes after targeting the LAMC1 gene results in embryonic lethality due to failure of endoderm differentiation. J Cell Biol. 1999;144(1):151–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Piovan C, Palmieri D, Di Leva G, et al. Oncosuppressive role of p53-induced miR-205 in triple negative breast cancer. Mol Oncol. 2012;6(4):458–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sprenger CC, Drivdahl RH, Woodke LB, et al. Senescence-induced alterations of laminin chain expression modulate tumorigenicity of prostate cancer cells. Neoplasia. 2008;10(12):1350–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wuillème-Toumi S, Robillard N, Gomez P, et al. Mcl-1 is overexpressed in multiple myeloma and associated with relapse and shorter survival. Leukemia. 2005;19(7):1248–1252. [DOI] [PubMed] [Google Scholar]
- 75. Boiani M, Daniel C, Liu X, Hogarty MD, Marnett LJ. The stress protein BAG3 stabilizes Mcl-1 protein and promotes survival of cancer cells and resistance to antagonist ABT-737. J Biol Chem. 2013;288(10):6980–6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Adler HL, McCurdy MA, Kattan MW, Timme TL, Scardino PT, Thompson TC. Elevated levels of circulating interleukin-6 and transforming growth factor-β 1 in patients with metastatic prostatic carcinoma. J Urol. 1999;161(1):182–187. [PubMed] [Google Scholar]
- 77. Cavarretta IT, Neuwirt H, Untergasser G, et al. The antiapoptotic effect of IL-6 autocrine loop in a cellular model of advanced prostate cancer is mediated by Mcl-1. Oncogene. 2007;26(20):2822–2832. [DOI] [PubMed] [Google Scholar]
- 78. Santer FR, Erb HH, Oh SJ, et al. Mechanistic rationale for MCL1 inhibition during androgen deprivation therapy. Oncotarget. 2015;6(8):6105–6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Koehler BC, Scherr AL, Lorenz S, et al. Beyond cell death - antiapoptotic Bcl-2 proteins regulate migration and invasion of colorectal cancer cells in vitro. PLoS One. 2013;8(10):e76446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kroiss A, Vincent S, Decaussin-Petrucci M, et al. Androgen-regulated microRNA-135a decreases prostate cancer cell migration and invasion through downregulating ROCK1 and ROCK2. Oncogene. 2015;34(22):2846–2855. [DOI] [PubMed] [Google Scholar]
- 81. Williams LV, Veliceasa D, Vinokour E, Volpert OV. miR-200b inhibits prostate cancer EMT, growth and metastasis. PLoS One. 2013;8(12):e83991. [DOI] [PMC free article] [PubMed] [Google Scholar]