Abstract
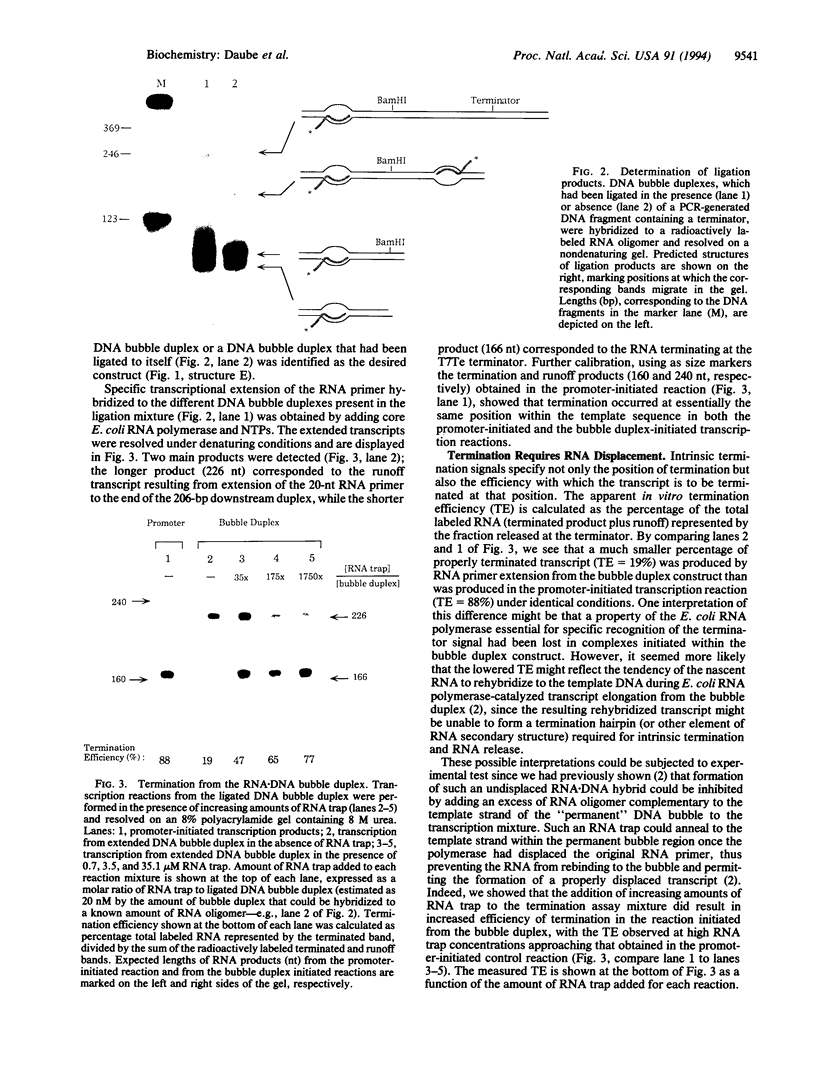
Functional transcription elongation complexes can be formed by adding RNA polymerase in trans to a preformed nucleic acid construct. This construct consists of a double-stranded DNA fragment that contains a noncomplementary (permanent DNA bubble) region into which an RNA primer oligonucleotide has been hybridized. By ligating a DNA fragment containing the strong intrinsic terminator T7Te to the RNA.DNA bubble duplex, we show here that Escherichia coli core RNA polymerase-catalyzed transcription, initiated from such a construct, terminates at the predicted position. Furthermore, we show that the termination efficiency obtained is comparable to that observed in a control reaction initiated with the E. coli holopolymerase from the T7A1 promoter if an RNA oligomer trap is used to permit proper displacement of the nascent RNA from the DNA template strand. The trap oligomer is complementary to the template strand of the permanent DNA bubble and prevents rehybridization of the nascent RNA at this site. Varying the amount of RNA trap that is added permits us to modulate the extent of total RNA displacement. Our results show that RNA displacement and termination efficiency are directly correlated, suggesting that intrinsic termination requires that the nascent RNA be free to assume its solution conformation. Several models of intrinsic termination are presented and discussed in light of these data.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adhya S., Gottesman M. Control of transcription termination. Annu Rev Biochem. 1978;47:967–996. doi: 10.1146/annurev.bi.47.070178.004535. [DOI] [PubMed] [Google Scholar]
- Campbell F. E., Jr, Setzer D. R. Transcription termination by RNA polymerase III: uncoupling of polymerase release from termination signal recognition. Mol Cell Biol. 1992 May;12(5):2260–2272. doi: 10.1128/mcb.12.5.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A. Control of transcription termination by RNA-binding proteins. Annu Rev Biochem. 1993;62:893–930. doi: 10.1146/annurev.bi.62.070193.004333. [DOI] [PubMed] [Google Scholar]
- Daube S. S., von Hippel P. H. Functional transcription elongation complexes from synthetic RNA-DNA bubble duplexes. Science. 1992 Nov 20;258(5086):1320–1324. doi: 10.1126/science.1280856. [DOI] [PubMed] [Google Scholar]
- Daube S. S., von Hippel P. H. RNA displacement pathways during transcription from synthetic RNA-DNA bubble duplexes. Biochemistry. 1994 Jan 11;33(1):340–347. doi: 10.1021/bi00167a044. [DOI] [PubMed] [Google Scholar]
- Farnham P. J., Platt T. A model for transcription termination suggested by studies on the trp attenuator in vitro using base analogs. Cell. 1980 Jul;20(3):739–748. doi: 10.1016/0092-8674(80)90320-7. [DOI] [PubMed] [Google Scholar]
- Goliger J. A., Yang X. J., Guo H. C., Roberts J. W. Early transcribed sequences affect termination efficiency of Escherichia coli RNA polymerase. J Mol Biol. 1989 Jan 20;205(2):331–341. doi: 10.1016/0022-2836(89)90344-6. [DOI] [PubMed] [Google Scholar]
- Johnston H. M., Roth J. R. DNA sequence changes of mutations altering attenuation control of the histidine operon of Salmonella typhimurium. J Mol Biol. 1981 Feb 5;145(4):735–756. doi: 10.1016/0022-2836(81)90312-0. [DOI] [PubMed] [Google Scholar]
- Lee F., Yanofsky C. Transcription termination at the trp operon attenuators of Escherichia coli and Salmonella typhimurium: RNA secondary structure and regulation of termination. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4365–4369. doi: 10.1073/pnas.74.10.4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F. H., Tinoco I., Jr DNA-RNA hybrid duplexes containing oligo(dA:rU) sequences are exceptionally unstable and may facilitate termination of transcription. Nucleic Acids Res. 1980 May 24;8(10):2295–2299. doi: 10.1093/nar/8.10.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt T. Transcription termination and the regulation of gene expression. Annu Rev Biochem. 1986;55:339–372. doi: 10.1146/annurev.bi.55.070186.002011. [DOI] [PubMed] [Google Scholar]
- Reynolds R., Bermúdez-Cruz R. M., Chamberlin M. J. Parameters affecting transcription termination by Escherichia coli RNA polymerase. I. Analysis of 13 rho-independent terminators. J Mol Biol. 1992 Mar 5;224(1):31–51. doi: 10.1016/0022-2836(92)90574-4. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H. Genetic and physical mapping of the late region of bacteriophage T7 DNA by use of cloned fragments of T7 DNA. J Mol Biol. 1981 Dec 15;153(3):503–525. doi: 10.1016/0022-2836(81)90405-8. [DOI] [PubMed] [Google Scholar]
- Telesnitsky A. P., Chamberlin M. J. Sequences linked to prokaryotic promoters can affect the efficiency of downstream termination sites. J Mol Biol. 1989 Jan 20;205(2):315–330. doi: 10.1016/0022-2836(89)90343-4. [DOI] [PubMed] [Google Scholar]
- Tomizawa J., Masukata H. Factor-independent termination of transcription in a stretch of deoxyadenosine residues in the template DNA. Cell. 1987 Nov 20;51(4):623–630. doi: 10.1016/0092-8674(87)90131-0. [DOI] [PubMed] [Google Scholar]
- Travers A. A., Burgessrr Cyclic re-use of the RNA polymerase sigma factor. Nature. 1969 May 10;222(5193):537–540. doi: 10.1038/222537a0. [DOI] [PubMed] [Google Scholar]
- Yager T. D., von Hippel P. H. A thermodynamic analysis of RNA transcript elongation and termination in Escherichia coli. Biochemistry. 1991 Jan 29;30(4):1097–1118. doi: 10.1021/bi00218a032. [DOI] [PubMed] [Google Scholar]
- d'Aubenton Carafa Y., Brody E., Thermes C. Prediction of rho-independent Escherichia coli transcription terminators. A statistical analysis of their RNA stem-loop structures. J Mol Biol. 1990 Dec 20;216(4):835–858. doi: 10.1016/s0022-2836(99)80005-9. [DOI] [PubMed] [Google Scholar]
- von Hippel P. H., Yager T. D. Transcript elongation and termination are competitive kinetic processes. Proc Natl Acad Sci U S A. 1991 Mar 15;88(6):2307–2311. doi: 10.1073/pnas.88.6.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]