Abstract
Voltage-gated sodium channels are essential for the initiation and propagation of the action potential in neurons and other excitable cells. Because of their critical roles in electrical signaling, sodium channels are targets of a variety of naturally occurring and synthetic neurotoxins, including several classes of insecticides. This review is intended to provide an update on the molecular biology of insect sodium channels and the molecular mechanism of pyrethroid resistance. Although mammalian and insect sodium channels share fundamental topological and functional properties, most insect species carry only one sodium channel gene, compared to multiple sodium channel genes found in each mammalian species. Recent studies showed that two posttranscriptional mechanisms, alternative splicing and RNA editing, are involved in generating functional diversity of sodium channels in insects. More than 50 sodium channel mutations have been identified to be responsible for or associated with knockdown resistance (kdr) to pyrethroids in various arthropod pests and disease vectors. Elucidation of molecular mechanism of kdr led to the identification of dual receptor sites of pyrethroids on insect sodium channels. Most of the kdr mutations appear to be located within or close to the two receptor sites. The accumulating knowledge of insect sodium channels and their interactions with insecticides provides a foundation for understanding the neurophysiology of sodium channels in vivo and the development of new and safer insecticides for effective control of arthropod pests and human disease vectors.
Keywords: sodium channel, pyrethroids, alternative splicing, RNA editing, knockdown resistance, pyrethroid receptor sites
1. Introduction
1.1. Voltage-gated sodium channels
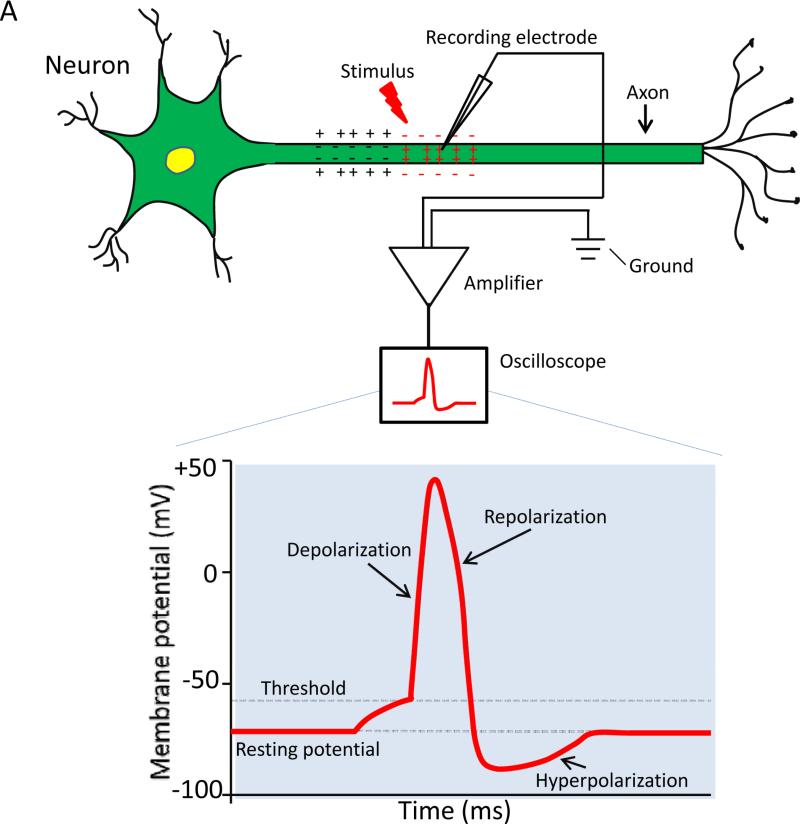
The nervous system enables animals to detect external and internal stimuli, integrate and process the information detected, and react with speed and coordination. It is hard to catch a cockroach and almost impossible to snatch a dragonfly out of the air because of the rapid electrical and chemical signaling within their nervous system. The electrical signals are comprised of action potentials (i.e., electrical impulses; Fig. 1A) propagating rapidly along axons and from one neuron to the next at synapses. The voltage-gated sodium channel is responsible for the initiation and propagation of action potentials along the axon (Fig. 1B).
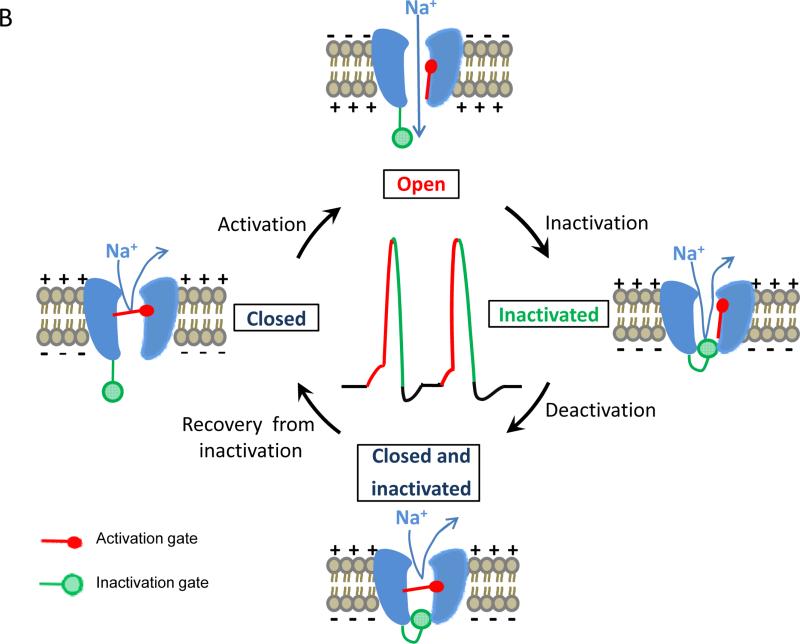
Fig. 1.
Voltage-gated sodium channels and the action potential. (A) Recording of an action potential. A recording microelectrode, which inserts inside the axon (intracellular recording), is connected to an amplifier. The amplifier compares the potential difference between the tip of the recording electrode and another electrode (called ground) placed in the solution bathing the neuron. The potential difference can be displayed using an oscilloscope. In response to membrane depolarization, such as a depolarizing stimulus indicated in A, sodium channels open (i.e., activated), resulting in further depolarization of the membrane as indicated by the rising phase of the action potential. Sodium channel inactivation together with potassium channel activation helps terminate the action potential (repolarization and hyperpolarization). (B) Gating (i.e., opening and closing) of voltage-gated sodium channels. Please see the text for explanation.
The sodium channel forms a pore in the membrane that is highly selective to sodium ions (Fig. 1B). The opening and closing of the sodium channel is regulated by two gating processes, activation and inactivation (Fig. 1B). When a neuron is at rest (i.e., not firing), sodium channels are closed. When the membrane of a neuron is depolarized, sodium channels are activated (open). Influx of sodium ions through activated sodium channels, which further depolarizes the membrane, is responsible for the rising phase of an action potential (Fig. 1B). Within a few milliseconds after sodium channel opening, sodium channels are rapidly inactivated. The inactivation process is partially responsible for the falling phase of an action potential (Fig. 1B) and plays an important role in the termination of an action potential. Upon repolarization, the sodium channel recovers from inactivation and deactivates (i.e., the activation gate closes). Deactivation and recovery from inactivation complete the transition from the inactivated state to the resting state of the sodium channel (Fig. 1B), which allows the cell membrane to regain its resting excitable properties and prepare to fire another action potential (Fig. 1B). As such, sodium channels play a critical role in controlling electrical signaling in the nervous system and regulating membrane excitability. In addition, in response to prolonged depolarization (seconds to minutes), sodium channels progressively enter into more stable, slow-inactivated states. This process is known as slow inactivation, which is important for regulating membrane excitability, action potential patterns and spike frequency adaptation.
1.2. Structure and function of sodium channels
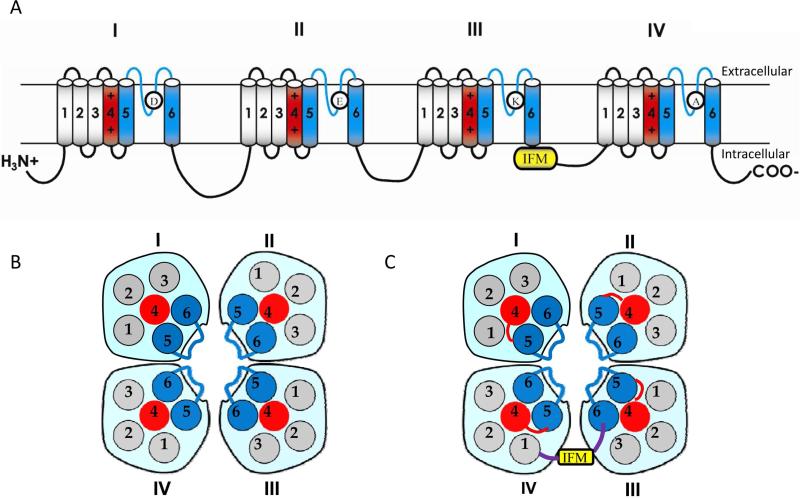
Mammalian sodium channels are composed of a pore-forming α-subunit and one or more β subunits. Multiple sodium channel α-subunits are found in mammals (Catterall, 2014; Goldin, 2002). Sodium channel α-subunits have four homologous domains (I-IV), each domain possessing six transmembrane segments (Fig. 2A). In each domain, segments 1-4 (S1-S4) constitute the voltage-sensing module, whereas S5, S6, and a membrane-reentrant loop connecting S5 and S6 segments (called the P-region) form the pore module (Fig. 2A-2C). β subunits (β1-β4) are small transmembrane proteins that possess an extracellular immunoglobulin domain, a single transmembrane segment, and a short intracellular C-terminal domain (Brackenbury and Isom, 2011; Catterall, 2000). Coexpression of β subunits modulates sodium channel expression and gating properties (Brackenbury and Isom, 2011; Catterall, 2000).
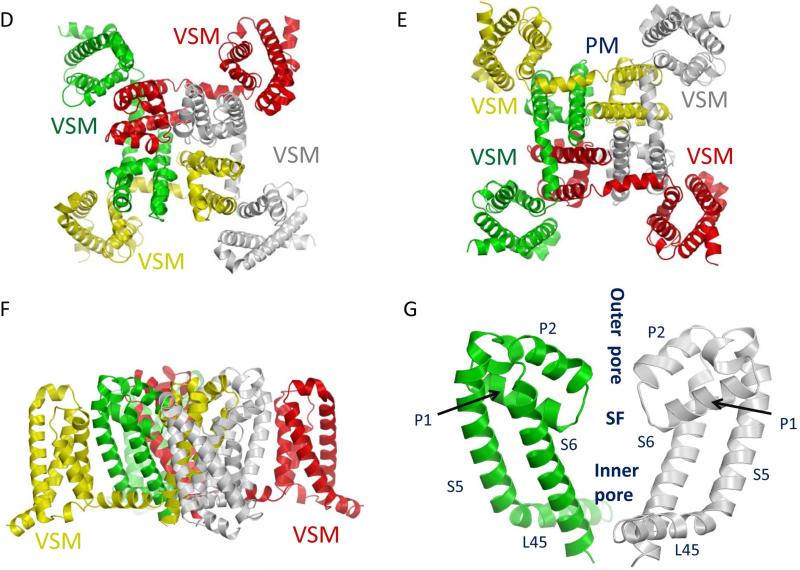
Fig. 2.
Structure of the voltage-gated sodium channel. (A) The topology of the sodium channel indicating the sequence features that are critical for channel function. The sodium channel protein contains four homologous repeats (I-IV), each having six transmembrane segments (1-6). The isoluecine in the IFM motif in mammalian sodium channels is substituted with a methionine in insect sodium channels. (B and C) Schematic representations of extracellular (B) and intracellular (C) views of sodium channels. (D-G) The X-ray structure of the closed NavAb sodium channel (Payandeh et al., 2011). Four subunits of NavAb in yellow, red, green and grey, respectively, correspond to domains I, II, III and IV in eukaryotic four-domain sodium channels. (D) Extracellular view. (E) Intracellular view indicating the four voltage-sensing modules (VSM), and the pore module (PM). (F) Side view. (G) Expanded side view of the pore module showing only two subunits for clarity. The inner and outer pores are separated by the selectivity-filter region (SF).
Significant progress has been made in the past two decades in the understanding of the domain structures and amino acid motifs/sequences required for specific gating properties of sodium channels, including channel activation and inactivation. Most information has been derived from studies of α-subunits of mammalian sodium channels, with which insect sodium channels share high levels of sequence and functional similarities. Below is a brief summary of the current understanding of the structure-function relationship of sodium channels. Readers are referred to comprehensive reviews (Catterall, 2000; Catterall, 2012; Catterall, 2014) on this topic.
The ion selectivity of sodium channels is determined by the amino acids D, E, K, and A (the selectivity-filter motif “DEKA”) in the analogous positions of domains I, II, III, and IV, respectively, of the α-subunit. Each S4 segment contains repeated motifs of a positively charged amino acid residue followed by two hydrophobic residues and serves as a voltage sensor of the sodium channel. In response to membrane depolarization, the S4 segments move outward, initiating conformational changes which lead to pore opening and inactivation of sodium channels. Short intracellular linkers between the S4 and S5 segments (L45) transmit the movements of the voltage sensing modules to the S6 segments during channel opening and closing. Fast-inactivation is achieved by the movement of an inactivation gate (formed mainly by the IFM motif in the short intracellular linker connecting domains III and IV), which physically occludes the open pore (Fig. 2A).
Since the publication of the first X-ray structure of a bacterial potassium channel KcsA (Doyle et al., 1998), homology models of sodium channels have been developed to predict binding sites of drugs, such as local anesthetics (Lipkind and Fozzard, 2005; Tikhonov and Zhorov, 2007) and toxins, such as tetrodotoxin (Lipkind and Fozzard, 2000; Tikhonov and Zhorov, 2005a) and batrachotoxin (Du et al., 2011; Tikhonov and Zhorov, 2005b). Currently, the mammalian voltage-gated potassium channel Kv1.2 crystallized in the open state (Long et al., 2005) and a bacterial sodium channel, NavAb, crystallized in the closed state (Payandeh et al., 2011) are used as reasonable templates to model eukaryotic four-domain sodium channels in the open and closed states, respectively. Four identical subunits of the bacterial sodium channel NavAb (i.e., a homotetramer) arrange around the pore axis in the way of four-fold rotational symmetry (Fig. 2D-2F) (Payandeh et al., 2011). Accordingly, the four voltage-sensing modules are symmetrically arranged around the outer rim of the pore module. The voltage sensing module of one subunit is closely associated with the pore-forming module of the adjacent subunit (Payandeh et al., 2011), as in the Kv1.2 channel (Long et al., 2005). This arrangement likely enforces concerted gating of the four subunits of homotetrameric sodium and potassium channels (Catterall, 2014). The Inner pore is lined by the lower two-thirds of the transmembrane S6 segments and the C-terminal parts of the P1 helices from the membrane-diving P-loops. The outer pore is lined by the P2 helices from the P-loops and the ascending limbs that connect helices P1 and P2 (Fig. 2G).
1.3. Sodium channels are targets of a variety of neurotoxins including insecticides
Because of their crucial roles in membrane excitability, sodium channels are targets of a broad range of naturally occurring neurotoxins, such as tetrodotoxin from the puffer fish and polypeptide toxins from scorpions and sea anemones (Catterall, 2000). They are also primary target sites of synthetic compounds including insecticides, such as dichlorodiphenyltrichloroethane (DDT) and pyrethroids, and therapeutic drugs, such as local anesthetics (Catterall et al., 2007; Narahashi, 1996, 2002). These sodium channel neurotoxins bind to their respective receptor sites on the sodium channel and alter various channel properties, including ion conductance, ion selectivity and/or channel gating (i.e., opening and closing). Because of the diverse pharmacological effects of these neurotoxins on the sodium channel, localization of the toxin-binding sites has been a powerful approach to probe the architecture of the sodium channel (Catterall et al., 2007; Cestele and Catterall, 2000; Gordon, 1997; Gurevitz, 2012; Wang and Wang, 2003; Zlotkin, 1999). For example, the outer pore of the sodium channel can be blocked by tetrodotoxin, saxitoxin and conotoxins; and the central cavity of the inner pore is targeted by local anesthetics, batrachotoxin, and sodium channel blocker insecticides (SCBIs). Furthermore, some polypeptide toxins from the venoms of scorpions, spiders and sea anemones specifically block or modify insect sodium channel gating, but have little effect on mammalian sodium channels (Bende et al., 2013; Bosmans and Tytgat, 2007; Gordon et al., 2007; Gur et al., 2011; Gurevitz et al., 2007; Klint et al., 2012; Moran et al., 2009; Moran et al., 2007; Skinner et al., 1992; Strugatsky et al., 2005). Such insect-specific activities suggest the potential use of these natural polypeptide toxins as bioinsecticides for insect pest control.
DDT and pyrethroid insecticides are among the earliest synthetic compounds that were identified to act on sodium channels (Narahashi, 2000). Pyrethroids enhance activation and inhibit deactivation and inactivation, resulting in prolonged channel opening. At the cellular level, pyrethroids disrupt nerve function, causing repetitive discharges, membrane depolarization, and synaptic disturbances (Narahashi, 1996, 2000; Silver et al., 2014; Soderlund, 2012). Another group of insecticides, including indoxacarb and metaflumizone, represents a new class of sodium channel-targeting insecticides with a mode of action distinct from that of DDT and pyrethroids. They inhibit sodium current and are known as SCBIs (Silver et al., 2014; Silver et al., 2010; Wing et al., 2005) or sodium channel inhibitors, SCIs; (von Stein and Soderlund, 2012). In insects, indoxacarb is metabolically converted to N-decarbomethoxyllated JW062 (DCJW), a more active metabolite, whereas mammals convert indoxacarb into nontoxic metabolites. This difference in metabolism contributes to the selective toxicity of indoxacarb to insect pests (Silver et al., 2009; von Stein and Soderlund, 2012; Wing et al., 2005).
2. Sodium Channels in Insect Species
2.1. Identification of Sodium Channel Genes
The first insect sodium channel gene (the para gene, later named DmNav) was cloned from Drosophila melanogaster based on temperature-sensitive paralytic mutations (Loughney et al., 1989). The overall structure and amino acid sequence of the DmNav protein shares a high similarity with those of mammalian sodium channel α-subunits. The structural features that are critical for mammalian sodium channel function, including ion selectivity and channel gating (Fig. 2A), are conserved in the DmNav sodium channel (Dong, 2007; Loughney et al., 1989; Soderlund, 2005).
In the early 1990s, pyrethroid resistance was found to be genetically linked to DmNav-like sequences in house flies and cockroaches (Soderlund, 2005). These exciting findings triggered a wave of studies that resulted in the isolation of partial or full-length cDNAs of DmNav orthologs from many arthropod pests and human disease vectors (Table S1). More recently, genome sequencing is providing sequences of DmNav-family sodium channel genes from additional insect species (Table S1).
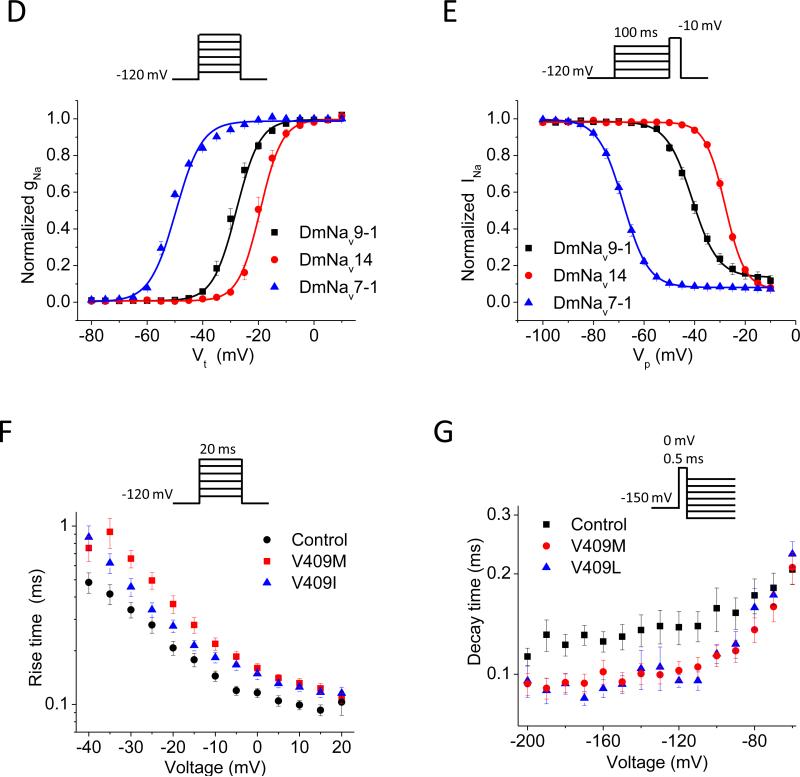
Functional characterization in the Xenopus oocyte expression system confirmed that DmNav encode functional sodium channels (Feng et al., 1995; Warmke et al., 1997). These significant advances set the stage for molecular, functional and pharmacological characterization of insect sodium channels from other species. To date, four DmNav orthologs have been functionally expressed in Xenopus oocytes. These include, Vssc1 from the house fly Musca domestica (Smith and Soderlund, 1998), paraCSMA (later named BgNav) from the German cockroach Blattella germanica (Liu et al., 2000; Tan et al., 2002a), VmNav from the varroa mite Varroa destructor (Du et al., 2009b), and AaNav from the yellow-fever mosquito Aedes aegypti (Du et al., 2013). Sodium channel activation controls the rate and voltage dependence of opening of the sodium channel after depolarization, whereas inactivation controls the rate and voltage-dependence of the subsequent inactivation gate-mediated closing of the sodium channel during a sustained depolarization. Fig. 3D-3G illustrates these major gating properties of DmNav and BgNav channels expressed in oocytes.
Fig. 3.
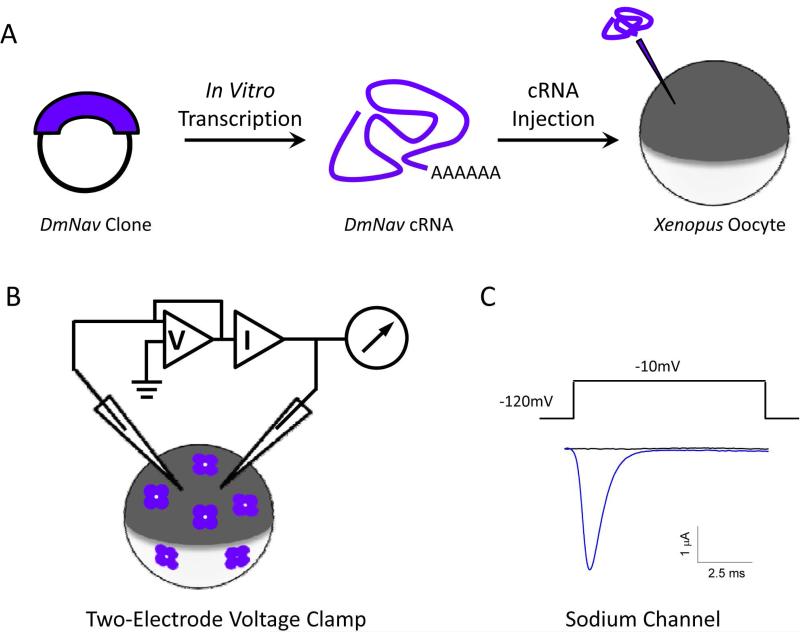
Functional characterization of insect sodium channels expressed in Xenopus oocytes using the voltage-clamp technique. (A) Expression of insect sodium channels in Xenopus oocytes. Full-length cDNA clones encoding insect sodium channels are used as templates to synthesize capped-RNA (cRNA) in vitro. The cRNA is then injected into oocytes using a microinjector, and is often co-injected with cRNA of either TipE or a TipE-ortholog to enhance sodium channel expression. Insect sodium channels are expressed on the surface of the injected oocytes several days post-injection. (B) Two-electrode voltage clamp. The membrane of the oocyte is penetrated by two microelectrodes, one for measuring the voltage across the membrane while the other for injecting current into the cell to keep the voltage constant (i.e., voltage clamp). By measuring the amount of current injected, the system can determine the amplitude and time course of the ionic current flowing across the membrane at a given depolarization step, as shown in C. (C) A trace of an inward sodium current showing the rapid activation and inactivation of DmNav sodium channels. The current was elicited by a step depolarization to −10 mV from a holding potential of −120 mV. (D and E) Voltage dependence of activation (D) and inactivation (E) of three DmNav variants with different gating properties. (F and G) Activation kinetics (F) and deactivation kinetics (G) of BgNav1-1 (control) and mutants carrying V409M or V409L mutation (i.e., V410M/L in Vssc1). The kinetics of activation and deactivation (i.e., the rate of opening and closing of the activation gate were determined by the cell-attached macropatch (Oliveira et al., 2013; Warmke et al., 1997). Both mutations slow activation kinetics and accelerate deactivation kinetics (Oliveira et al., 2013).
In the American cockroach (Periplaneta americana), besides a DmNav ortholog, a DmNav-like gene called PaFPC (Periplaneta americana four P-domain channel) was reported (Moignot et al., 2009). In PaFPC, the DEKA motif that determines ion selectivity is conserved, but the MFM motif that is critical for fast inactivation is not. PaFPC transcripts are expressed in both neuronal and non-neuronal tissues (Moignot et al., 2009). The functionality of PaFPC remains unknown. Analysis of the currently sequenced insect genomes suggests that most insects have only one DmNav-family sodium channel gene with two exceptions (Table S1). The red flour beetle (Tribolium casteneum) has a second DmNav-like gene lacking the MFM motif, like PaFPC (GenBank accession number: NM_001165909 in Table S1). In addition, in Acyrthosiphon pisum, domains I and II, and domains III and IV, appear to be encoded by two different genes (Table S1). Whether these two-domain proteins co-assemble into a functional sodium channel remains to be determined.
2.2. DSC1 and its Orthologs in Insects
In addition to DmNav, Drosophila also has a sodium channel-like gene named Drosophila Sodium Channel 1 (DSC1). DSC1 was cloned by probing Drosophila genomic DNA libraries with an eel sodium channel cDNA (Okamoto et al., 1987; Ramaswami and Tanouye, 1989; Salkoff et al., 1987). DSC1 orthologs were subsequently isolated from B. germanica and Heliothis virescens (Liu et al., 2001; Park et al., 1999) and have now been found in all insect species with a complete genome sequence (Cui et al., 2012; Liu et al., 2001; Park et al., 1999). For a long time, DSC1 had been considered a sodium channel gene because its overall topology and sequence are similar to those of sodium channels, consisting of four homologous repeats each having six transmembrane segments (Kulkarni et al., 2002; Zhang et al., 2011). However, the DEKA motif, which determines the ion selectivity for voltage-gated sodium channels, including insect sodium channels, is not conserved in the DSC1 family of genes (Zhou et al., 2004). Instead, DSC1 and its orthologs all contain a DEEA motif (Cui et al., 2012). Functional analysis of DSC1-family channels in Xenopus oocytes revealed that they represent a novel type of voltage-gated cation channels with high permeability to Ca2+ (Zhang et al., 2011; Zhou et al., 2004).
2.3. Auxiliary Subunits of Insect Sodium Channels
As mentioned above, β subunits of mammalian sodium channels play important roles in modulating the expression and gating of mammalian sodium channels (Brackenbury and Isom, 2011; Cestele and Catterall, 2000). Surprisingly, no orthologs of mammalian β subunits are found in insects. Instead, an unrelated protein, TipE in D. melanogaster (Feng et al., 1995; Warmke et al., 1997) and its orthologs in other insects (Bourdin et al., 2013; Du et al., 2013; Lee et al., 2000) facilitate robust expression of insect sodium channels in Xenopus oocytes and are considered as auxiliary subunits of insect sodium channels. Besides TipE, there are also three to four TipE-homologous genes (TEH1-4) in D. melanogaster and other insect species (Derst et al., 2006). TipE and TEH proteins contain two transmembrane segments, an extracellular linker connecting the two transmembrane segments and intracellular C- and N-termini (Derst et al., 2006; Feng et al., 1995). TEH1 is expressed exclusively in the nervous system, whereas the transcripts of other TEHs and TipE are detected in both neuronal and non-neuronal tissues, such as fat body and gut (Derst et al., 2006).
TipE, TEH1, TEH2 and TEH3 have been shown to not only enhance the peak current of DmNav channels expressed in Xenopus oocytes, but also modulate various gating properties of DmNav channels (Derst et al., 2006; Wang et al., 2013; Warmke et al., 1997). Like β subunits of mammalian sodium channels, TipE and TEH proteins have both shared as well as distinct modulating effects on various sodium channel variants (Table S2), indicating that TipE and TEH proteins are functionally analogous to β subunits. TipE and TEH1 orthologs from other species also have similar modulating effects (Bourdin et al., 2013; Du et al., 2013; Lee et al., 2000).
3. Insects achieve functional diversity of sodium channels via alternative splicing and RNA editing
Mammals such as humans and mice express nine α-subunit isoforms with different gating properties (Goldin et al., 2000). The nine α-subunit genes are differentially expressed in various cell types, tissues, and developmental stages, presumably to fulfill unique physiological roles in specific cells (Catterall, 2000; Frank and Catterall, 2003; Goldin et al., 2000). Because most insects have only a single sodium channel gene, a question arises as to how insects achieve functional diversity of sodium channels from a single gene. An initial clue came from molecular characterization of the DmNav gene in Drosophila species, which revealed extensive alternative splicing of the DmNav transcript (Loughney et al., 1989; Thackeray and Ganetzky, 1994, 1995). Subsequent analyses of DmNav-orthologs from other insect species, showed that many of the alternative splicing sites identified in DmNav are conserved in DmNav orthologs (Chang et al., 2009; Davies et al., 2007; Lee et al., 2002; Park et al., 1999; Shao et al., 2009; Song et al., 2004; Sonoda et al., 2006; Sonoda et al., 2008; Tan et al., 2002a). In addition, extensive RNA editing, another post-transcriptional mechanism, is found in sodium channel transcripts of D. melanogaster (Hanrahan et al., 2000; Olson et al., 2008; Reenan et al., 2000) and B. germanica (Liu et al., 2004; Song et al., 2004). Functional analyses in Xenopus oocytes of DmNav and BgNav splicing and/or RNA-editing variants have revealed an impressive spectrum of both drastic and subtle differences in channel expression and channel gating properties (Du et al., 2009c; Lin et al., 2009; Liu et al., 2004; Olson et al., 2008; Song et al., 2004; Tan et al., 2002a). These findings suggest the importance of alternative splicing and RNA editing in diversifying sodium channel activities in vivo. This line of study represents a major effort in the study of insect sodium channels in the past decade.
3.1. Alternative splicing of sodium channel transcripts
3.1.1. In Drosophila
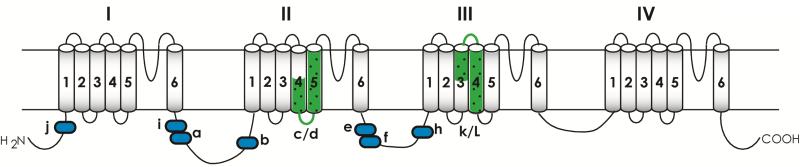
A total of 11 alternatively spliced exons, seven optional exons (a, b, e, f, h, i, j) in intracellular linkers and four mutually exclusive exons (c/d and k/l) in transmembrane domains (Fig. 4), have been identified in DmNav transcripts from adults of D. melanogaster (Lee et al., 2002; Lin et al., 2009; Loughney et al., 1989; O'Dowd et al. 1995; Thackeray and Ganetzky, 1994, 1995). These alternative splice sites are conserved in D. virilis (Thackeray and Ganetzky, 1995). Although thousands of splicing types are theoretically possible from various combinations of mutually exclusive and optional exons in D. melanogaster, the actual number of splice types seems far more limited. From 64 full-length cDNA clones isolated from D. melanogaster adults, 29 splicing types were uncovered (Olson et al., 2008). Fifty DmNav cDNA clones from late-stage embryos could be grouped into 27 splice types (Lin et al., 2009). Interestingly, only three adult splice types (including the most abundant type in adults) and four additional optional exons are found in the embryo (Lin et al., 2009). The major difference in splicing between adults and embryos appears to be the usage of exons j and f. The usage of exon j increases from 10% in the embryo to 89% in the adult, whereas inclusion of exon f decreases from 78% to 10%. However, the physiological importance of these differences remains to be investigated.
Fig. 4.
Alternative splicing of DmNav transcripts. Optional exons are illustrated in blue blocks; and mutually exclusive exons (c/d and l/k) are highlighted in green. Four optional exons detected in larvae of D. melanogaster (Lin et al., 2009) are not indicated.
While transcripts lacking the optional exons j, i, a, b, e, f, and h still produce functional channels in Xenopus oocytes, inclusion or exclusion of optional exons f, j or h alters the voltage-dependence of channel activation or inactivation (Lin et al., 2009). These findings indicate that optional exons serve a modulatory function regulating neuronal excitability. Furthermore, exclusion of optional exon b increases the current expression of DmNav, BgNav or VmNav channels in oocytes (Du et al., 2009b; Song et al., 2004). Exons a and b contain consensus sequences for phosphorylation. Down-regulation of sodium current expression by protein kinases has been documented for mammalian sodium channels (Numann et al., 1991; Smith and Goldin, 1996, 1997, 2000; West et al., 1991). It is therefore possible that exon b may serve as a regulatory on-or-off switch to regulate sodium channel function and neuronal excitability in response to second messengers in specific tissues or cell types.
Persistent sodium currents, which do not inactivate completely even with prolonged depolarization, contributes to membrane depolarization (Crill, 1996; Kiss, 2008; Taylor, 1993). Widespread expression of of persistent current have been detected in various neurons in the vertebrate and invertebrate species and could be important in regulating neuronal activities, such as bursts of neuroactivity frequency and generating pacemaker behavior (Crill, 1996; Kiss, 2008; Stafstrom, 2007; Taylor, 1993). Interestingly, mutually exclusive exons k/l, encoding IIIS3-IIIS4 (i.e., part of IIIS3 and III4 and the extracellular linker connecting IIIS3 and IIIS4), modulate the generation of persistent current in D. melanogaster (Lin et al., 2009). Persistent current ranges from 1.5 to 2.4% of transient current in exon k-containing sodium channels expressed in oocytes, but ranges from 4.1 to 9.5% in sodium channels with exon l (Lin et al., 2009). Pasilla, an RNA-binding protein, is required to regulate the relative proportions of DmNav transcripts that contain exons k/l in vivo (Park et al., 2004). Apparently, splicing to include exon l at the expense of exon k is associated with increased neuroactivity (Lin et al., 2012). Increased persistent sodium current in D. melanogaster slamdance mutants is the physiological basis for developing this insect system as a seizure model (Marley and Baines, 2011).
3.1.2. In other species
Many of the alternative splice sites identified in D. melanogaster are conserved in other insect species, including M. domestica (Lee et al., 2002), B. germanica (Song et al., 2004; Tan et al., 2002a), H. virescens (Park et al., 1999), Plutella xylostella (Sonoda et al., 2006; Sonoda et al., 2008), Bombyx mori (Shao et al., 2009), Ae. aegypti (Chang et al., 2009), Anopheles gambiae, Culex pipiens, Ae. aegypti, Ae. albopictus, Apis mellifera, Nasonia vitripennis, Pediculus humanis, Tribolium castaneum (Davies et al., 2007). Two mutually exclusive exons corresponding to Drosophila exons l/k were conserved in all insect species mentioned above, except for T. castaneum which has only exon l (Davies et al., 2007; Shao et al., 2009). Conservation of these two alternative exons and functional modulation of sodium channel function by exons l/k described in 3.1.1 suggest an important role of the two alternative exons in insect neurophysiology. However, less conservation was found for the other pair of mutually exclusive exons corresponding to Drosophila exons c/d, both exons c/d were conserved in M. domestica, H. virescens, Ae. aegypti, An. gambiae, P. xylostella, C. pipiens and B. mori (Shao et al., 2009), but only exon c in B. germanica, A. mellifera and N. vitripennis (Davies et al., 2007; Song et al., 2004); and only exon d in T. castaneum (Davies et al., 2007). Furthermore, there are also variations at the intron donor and acceptor splice sites that extend or shorten exons. For example, exons equivalent to exon f in D. melanogaster can be 24 or 30 nucleotides long (Thackeray and Ganetzky, 1994). Exon c is extended by an additional 16 nucleotides in N. vitripennis (Davies et al., 2007). Intron donor splice site variation excludes G1111 in the BgNav transcripts.
Sequence analysis of cDNA clones show that most optional exons identified in D. melanogaster are conserved in M. domestica (Lee et al., 2002), B. germanica (Tan et al., 2002a), Ae. Aegypti (Chang et al., 2009), An. gambiae (Davies et al., 2007) and P. xylostella (Sonoda et al., 2008). Although most of exons corresponsing to optional exons in Drosophila were found in the genome of B. mori, high-throughput Solexa sequencing of cDNAs from B. mori revealed exons corresponding to exons j and f are optional, exons corresponsding to exons a, b and l are not optional and a new optional exon unique to B. mori was identified (Shao et al., 2009). Therefore, detailed analysis of sodium channel cDNAs is necessary to confirm optional exons in other species. Furthermore, the frequence of alternative exon usage can be different among these species. For example, more than 60% of the DmNav transcripts from D. melanogaster and D. virilis adults (Thackeray and Ganetzky, 1995) and about 50% of AaNav (Chang et al., 2009) contain exon b, but less than 20% of BgNav (Song et al., 2004) and Vssc1 (Lee et al., 2002) transcripts contain exon b. Only 3-6% of DmNav and BgNav transcripts contain exon k (Song et al., 2004). In contrast, most of Vssc1 transcripts contain exon k (Lee et al., 2002). Developmental regulation of alternative exon usage has also been documented in several species (Lee et al., 2002; Shao et al., 2009; Sonoda et al., 2006).
Conservation of one particular alternatively splicing site appears to extend beyond insect sodium channels. An exon G3 in BgNav, one of three mutually exclusive exons, called G1 G2 and G3, in BgNav (Tan et al., 2002a), has no sequence homology with either G1 or G2, and possesses a stop codon, whereas exons G1 and G2 correspond to exons l and k in DmNav, respectively. One of 69 BgNav variants contains exon G3 producing a truncated protein containing only the first two domains and are not functional when expressed in oocytes (Tan et al., 2002a). Remarkably, at this splicing site, VmNav contains an optional exon (named exon 3) (Wang et al., 2003). Like BgNav varaints carrying exon G3, VmNav variants that exclude exon 3 would also produce a truncated protein. Furthermore, the exon/intron arrangement of this region in insects and mites bears a striking similarity with that of the vertebrate sodium channel protein Nav1.6 (Scn8a) in mice, humans, and fish (Plummer et al., 1997). Like exon G3 in BgNav, the mammalian counterparts also possess a mutually exclusive exon containing a stop codon that would generate a truncated protein containing only the first two domains. Therefore, alternative splicing in this region (encoding IIIS3-S4) appears to be of a very ancient origin and is preserved in organisms predating the evolutionary divergence of arthropods and vertebrates. This level of conservation suggests an unknown physiological role of the two-domain sodium channel proteins in the nervous system in diverse animals.
3.2. RNA editing of sodium channel transcripts
RNA editing is a well-known post-transcriptional modification that could significantly change protein function by introducing site-specific alterations in gene transcripts. RNA editing results in conversion of one base to another or insertion and deletion of nucleotides. As such, RNA editing has the potential to cause amino acid substitutions, splice site variations or alteration in the level of transcripts (Bass, 2001; Nishikura, 2010). The most prevalent type of RNA editing is adenosine-to-inosine (A-to-I) editing, catalyzed by the Adenosine Deaminase Acting on RNA (ADAR) family of enzymes. ADARs bind double-stranded RNAs and deaminate adenosine to inosine, which is recognized as guanosine by the translational machinery. In Drosophila, a wide range of coding mRNAs associated with signaling in the nervous system undergo A-to-I editing, and loss of editing results in extreme behavioral defects (Jepson and Reenan, 2009; Jepson et al., 2011; Jin et al., 2007; Jones et al., 2009; Maldonado et al., 2013; Palladino et al., 2000). Eleven A-to-I editing sites have been identified in the DmNav transcript (Hanrahan et al., 2000; Palladino et al., 2000; Reenan et al., 2000; Rieder et al., 2013). The actual number of RNA editing sites may be much greater, because the transcript region examined in these studies represents only a portion of the complete DmNav open reading frame. Nine of the eleven editing events result in amino acid changes. Some of these RNA editing sites are conserved in different species of Drosophila (Hanrahan et al., 2000); however, whether these editing events cause functional alterations remain to be determined. Surprisingly, none of these A-to-I editing sites in DmNav are found in BgNav. Instead, two unique A-to-I RNA editing sites and three U-to-C editing sites are present in BgNav, each resulting in an amino acid change in the transmembrane segments (Liu et al., 2004; Song et al., 2004). One of the U-to-C editing sites, F1919S, is also found in DmNav (Liu et al., 2004). RNA editing of DmNav provides an excellent opportunity to study the mechanism of RNA editing by ADAR (Reenan et al., 2000; Rieder et al., 2013).
Two RNA editing events of BgNav transcripts cause sutble shifts in the voltage dependence of activation and/or inactivation (Song et al., 2004), supporting the hypothesis that RNA editing in vivo may be involved in the fine-tuning of the neuronal activity (Seeburg, 2000). RNA editing events have also been shown to contribute to the generation of persistent current (Liu et al., 2004). A U-to-C RNA editing event resulting in a F1919S substitution at the C-terminal domain of BgNav4 is responsible for this persistent current. Editing at the same site in DmNav1 is also detected and produces a similar increase in persistent current (Liu et al., 2004).
In conclusion, accumulative experimental evidence shows the activity of sodium channels are modulated by alternative splicing and RNA editing; and also suggests the importance of these two posttranscriptional mechanisms in fine-tuning neuronal excitability in vivo. Even small changes in sodium channel expression and/or function could be biologically relevant, as underscored by sodium channel mutations that cause various behavioral defects in D. melanogaster (Garber et al., 2012; Lilly et al., 1994; Lindsay et al., 2008; Parker et al., 2011; Sun et al., 2012; Wang et al., 1997). In addition, identification of alternative splicing and RNA-editing variants that have unique gating properties provided a valuable resource to gain insights into specific amino acid sequences/residues that are critical for channel function and interactions of sodium channels with insecticides and other toxins (Du et al., 2006; Du et al., 2009c; Du et al., 2010; Silver et al., 2014; Song et al., 2011; Song et al., 2006; Tan et al., 2002a).
4. Naturally occurring mutations in sodium channels that confer knockdown resistance (kdr) to DDT and pyrethroids
Pyrethroids are extensively used to control agricultural arthropod pests and vectors of human diseases because of their relatively low mammalian toxicity and potent insecticidal properties. However, in the past several decades intensive use of DDT and pyrethroids has led to the development of resistance in many pest populations. One major form of pyrethroid resistance is kdr, which involves reduced target-site sensitivity to DDT and pyrethroids and confers cross-resistance to DDT (Soderlund and Bloomquist, 1990). Since its initial report in the house fly (Milani, 1956), kdr or kdr-like resistance has been documented globally in almost all agriculturally important arthropod pests and disease vectors (Rinkevich et al., 2013; Soderlund, 2005, 2012).
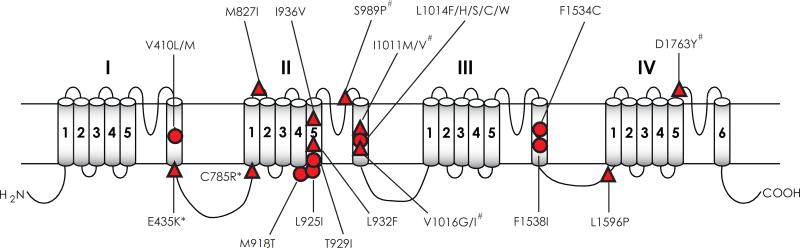
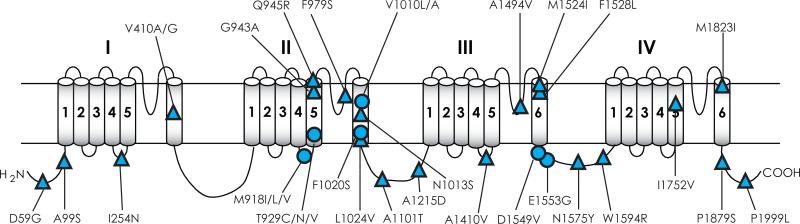
Study of the mechanism of kdr in the past two decades led to the identification of more than 50 sodium channel mutations or combinations of mutations that are associated with pyrethroid resistance in arthropod species (Table 1 and Fig. 5). Some of these mutations are detected in more than one species, whereas others are found only in a single species. Many mutations (Fig. 5A) reduce the pyrethroid sensitivity of Vssc1, BgNav, DmNav or AaNav sodium channels expressed in Xenopus oocytes, confirming their role in pyrethroid resistance. The involvement of many other mutations (Fig. 5B) in kdr remains to be investigated.
Table 1.
Sodium channel mutations that have been associated with kdr in arthropod species.
| Mutation1 | Species | Original Numbering2 | Reference3,4 |
|---|---|---|---|
| A99S | Culex quinquefasciatus | A109S | (Xu et al., 2012) |
| I254N | Drosophila melanogaster | I286N | (Pittendrigh et al., 1997) |
| V410M | Helicoverpa zea | V421M | (Hopkins and Pietrantonio, 2010) |
| Heliothis virescens | V421M | (Park et al., 1997) | |
| V410L | Cimex luctularis | V419L | (Yoon et al., 2008) |
| V410A | Helicoverpa zea | V421A | (Hopkins and Pietrantonio, 2010) |
| V410G | Helicoverpa zea | V421G | (Hopkins and Pietrantonio, 2010) |
| D59G+E435K+C785R+L1014F+P1999L | Blattella germanica | D58G+E434K+C764R+L993F+P1888L | (Liu et al., 2000) |
| M827I | Pediculus humanus capitis | M815I | (Hodgdon et al., 2010) |
| M827I+T929I | Pediculus humanus capitis | M815I+T917I | (Hodgdon et al., 2010) |
| M827I+T929I+L932F | Pediculus humanus capitis | M815I+T917I+L920F | (Lee et al., 2003) |
| Pediculus humanus corporis | M815I+T917I+L920F | (Drali et al., 2012) | |
| M827I+L932F | Pediculus humanus capitis | M815I+L920F | (Hodgdon et al., 2010) |
| M918T | Aphis gossypii | (Marshall et al., 2012) | |
| Tetranychus evansi | (Nyoni et al., 2011) | ||
| M918T+L1014F | Haematobia irritans irritans | (Guerrero et al., 1997) | |
| Liriomyza huidobrensis | (Davies et al., 2007) | ||
| Musca domestica | (Williamson et al., 1996) | ||
| Myzus persicae | (Eleftherianos et al., 2008) | ||
| Thrips tabaci | (Toda and Morishita, 2009) | ||
| Tuta absoluta | (Haddi et al., 2012) | ||
| M918L | Aphis gossypii | (Carletto et al., 2009) | |
| Hyelella azteca | (Weston et al., 2013) | ||
| M918L+L925I | Trialeurodes vaporariorum | (Karatolos et al., 2012) | |
| M918L+V1010A | Thrips tabaci | (Wu et al., 2013) | |
| M918V | Bemisia tabaci | (Morin et al., 2002) | |
| M918I+L1014F | Plutella xylostella | (Sonoda et al., 2008) | |
| L925I | Bemisia tabaci | (Morin et al., 2002) | |
| Cimex luctularis | (Yoon et al., 2008) | ||
| Hyelella azteca | (Weston et al., 2013) | ||
| Trialeurodes vaporariorum | (Karatolos et al., 2012) | ||
| Rhipicephalus microplus | (Morgan et al., 2009) | ||
| L925V | Varroa destructor | (Gonzalez-Cabrera et al., 2013) | |
| T929I | Thrips tabaci | (Toda and Morishita, 2009) | |
| Thrips palmi | (Bao and Sonoda, 2012) | ||
| Trialeurodes vaporariorum | (Karatolos et al., 2012) | ||
| Leptinotarsa decemlineata | (Rinkevich et al., 2012) | ||
| Sitophilus zeamais | (Araujo et al., 2011) | ||
| T929I+L932F | Pediculus humanus capitis | (Lee et al., 2000) | |
| T929I+L1014F | Frankliniella occidentalis | (Forcioli et al., 2002) | |
| Leptinotarsa decemlineata | (Rinkevich et al., 2012) | ||
| Plutella xylostella | (Schuler et al., 1998) | ||
| Tuta absoluta | (Haddi et al., 2012) | ||
| T929I+L1014F+A1101T+P1879S | Plutella xylostella | (Sonoda, 2010) | |
| T929C | Frankliniella occidentalis | (Forcioli et al., 2002) | |
| T929C+L1014F | Frankliniella occidentalis | (Forcioli et al., 2002) | |
| T929V | Bemisia tabaci | (Roditakis et al., 2006) | |
| Ctenocephalides felis | (Bass et al., 2004) | ||
| Frankliniella occidentalis | (Forcioli et al., 2002) | ||
| T929V+L1014F | Ctenocephalides felis | (Bass et al., 2004) | |
| T929N+L1014F | Leptinotarsa decemlineata | (Rinkevich et al., 2012) | |
| G933V5 | Rhipicephalus microplus | G72V | (Jonsson et al., 2010) |
| I936V | Helicoverpa zea | I951V | (Hopkins and Pietrantonio, 2010) |
| G943A | Pediculus humanus capitis | (Kristensen, 2005) | |
| Q945R | Lepeophtheirus salmonis | (Fallang et al., 2005) | |
| I1011M | Aedes aegypti | I104M | (Brengues et al., 2003) |
| I1011V | Aedes aegypti | (Saavedra-Rodriguez et al., 2007) | |
| L1014F | Anopheles gambiae | (Martinez-Torres et al., 1998) | |
| Anopheles stephensi | L31F | (Enayati et al., 2003) | |
| Anopheles subpictus | (Karunaratne et al., 2007) | ||
| Aphis gossypii | (Marshall et al., 2012) | ||
| Blattella germanica | L993F | (Dong, 2007; Miyazaki et al., 1996) | |
| Ctenocephalides felis | (Bass et al., 2004) | ||
| Culex pipiens pipiens | (Martinez-Torres et al., 1999a) | ||
| Culex pipiens pallens | (Chen et al., 2010) | ||
| Culex pipiens quinquefasciatus | (Xu et al., 2005) | ||
| Cydia pomonella | (Brun-Barale et al., 2005) | ||
| Frankliniella occidentalis | (Forcioli et al., 2002) | ||
| Haematobia irritans irritans | (Guerrero et al., 1997) | ||
| Haematobia irritans exigua | (Rothwell et al., 2011) | ||
| Leptinotarsa decemlineata | (Lee et al., 1999) | ||
| Liriomyza huidobrensis | (Davies et al., 2007) | ||
| Liriomyza sativae | (Davies et al., 2007) | ||
| Meligethes aeneus | (Nauen et al., 2012) | ||
| Musca domestica | (Ingles et al., 1996; Miyazaki et al., 1996; Williamson et al., 1996) | ||
| Myzus persicae | (Martinez-Torres et al., 1999b) | ||
| Sitobion avenae | (Foster et al., 2013) | ||
| Triatoma infestans | (Fabro et al., 2012) | ||
| L1014F+F979S | Myzus persicae | (Cassanelli et al., 2005) | |
| L1014F+N1575Y | Anopheles gambiae | (Jones et al., 2012) | |
| L1014S | Anopheles arabiensis | (Stump et al., 2004) | |
| Anopheles culicifacies | (Singh et al., 2010) | ||
| Anopheles gambiae | (Ranson et al., 2000) | ||
| Anopheles parilae | (Verhaeghen et al., 2010) | ||
| Anopheles peditaeniatus | (Verhaeghen et al., 2010) | ||
| Anopheles sacharovi | (Luleyap et al., 2002) | ||
| Anopheles sinensis | (Verhaeghen et al., 2010) | ||
| Anopheles vagus | (Verhaeghen et al., 2010) | ||
| Culex pipiens pallens | (Chen et al., 2010) | ||
| Culex pipiens pipiens | (Martinez-Torres et al., 1999b) | ||
| L1014S+V1010L | Anopheles culicifacies | (Singh et al., 2010) | |
| L1014H | Helicoverpa zea | L1029H | (Hopkins and Pietrantonio, 2010) |
| Heliothis virescens | L1029H | (Park et al., 1997) | |
| Liriomyza trifolii | (Davies et al., 2007) | ||
| Musca domestica | (Liu and Pridgeon, 2002; Rinkevich et al., 2006) | ||
| Stomoxys calcitrans | (Olafson et al., 2011) | ||
| L1014C | Anopheles sinensis | (Kim et al., 2007) | |
| Anopheles albimanus | (Lol et al., 2013) | ||
| Culex pipiens pipiens | (Wang et al., 2012) | ||
| L1014W | Anopheles sinensis | (Tan et al., 2012) | |
| V1016G | Aedes aegypti | V109G | (Brengues et al., 2003) |
| V1016G+S989P | Aedes aegypti | (Srisawat et al., 2010) | |
| V1016G+D1763Y | Aedes aegypti | D1794Y | (Chang et al., 2009) |
| V1016I | Aedes aegypti | (Saavedra-Rodriguez et al., 2007) | |
| N1013S | Anopheles sinensis | (Tan et al., 2012) | |
| F1020S | Blattella germanica | F999S | (Pridgeon et al., 2002) |
| Plutella xylostella | (Endersby et al., 2011) | ||
| L1024V | Tetranychus urticae | L1022V | (Kwon et al., 2010) |
| A1101T+P1879S | Plutella xylostella | A1060T+P1836S | (Sonoda et al., 2008) |
| A1410V | Drosophila melanogaster | A1549V | (Pittendrigh et al., 1997) |
| A1494V | Drosophila melanogaster | A1648V | (Pittendrigh et al., 1997) |
| F1534C | Aedes aegypti | F1269C | (Kawada et al., 2009) |
| Aedes albopictus | (Kasai et al., 2011) | ||
| F1538I | Rhipicephalus microplus | F1550I | (He et al., 1999) |
| Tetranychus cinnabarinus | (Feng et al., 2011) | ||
| F1538I+A1215D6 | Tetranychus urticae | (Tsagkarakou et al., 2009) | |
| M1524I | Drosophila melanogaster | (Pittendrigh et al., 1997) | |
| F1528L+M1823I7 | Varroa destructor | F758L+M1055I | (Wang et al., 2002) |
| F1528L+L1596P8+I1752V+M1823I7 | Varroa destructor | F758L+L826P+I982V+M1055I | (Wang et al., 2002) |
| D1549V+E1553G | Helicoverpa armigera | D1561V+E1565G | (Head et al., 1998) |
| Heliothis virescens | D1561V+E1565G | (Head et al., 1998) | |
| W1594R | Culex quinquefasciatus | W1573R | (Xu et al., 2012) |
Mutations are numbered according to the amino acid sequence of Vssc1 deposited in GenBank (Accession no: AAB47604). See Fig. 5 for the locations of the mutations.
Refers to the numbering of mutations in the original paper.
The first report of the mutation in the literature.
Full citations are available in Supplementary References.
C993V is resistant to deltamethrin (Usherwood et al., 2007).
D in Vssc1 at this position.
M in VdNav1, but V in Vssc1 at this position.
P is in Vssc1 and other insect sodium channels, and P1596L increases sensitivity of BgNav1-1a to fluvalinate (Liu et al., 2006).
Fig. 5.
Mutations in sodium channels associated with pyrethroid resistance in arthropod species. (A) Mutations that have been examined in Xenopus oocytes. (B) Mutations that have not been examined in oocytes. Circles denote those detected in more than one species and triangles indicate those found in a single species. See Table 1 for details on these mutations. The four mutations marked with # have been examined in oocytes, but do not reduce the sensitivity of AaNav1-1 channels to permethrin or deltamethrin. The two mutations marked with * alone do not confer pyrethroid resistance, but enhanced pyrethroid resistance mediated by L1014F or V410M. References on functional characterization of mutations in Xenopus oocytes are listed below: V410L/M (Oliveira et al., 2013; (Lee et al., 1999; Lee and Soderlund, 2001; Liu et al., 2002; Oliveira et al., 2013; Zhao et al., 2000); E435K + C785R + L1014F (Tan et al., 2002b); M827I, T929I and L932F (Usherwood et al., 2007; Vais et al., 2001; Yoon et al., 2008); M918T (Lee et al., 1999; Usherwood et al., 2005; Vais et al., 2001); L925I and I936V (Usherwood et al., 2007); L1014F/S/H (Burton et al., 2011; Du et al., 2013; Smith et al., 1997; Tan et al., 2002b; Tan et al., 2005; Usherwood et al., 2005; Vais et al., 2000); V1016G/I, I1011M/V, S989P , and D1763Y (Du et al., 2013); F1534C (Du et al., 2013; Hu et al., 2011); F1538I (Tan et al., 2005).
The L1014F mutation in IIS6 was the first resistance-associated mutation that was detected and confirmed as a cause of kdr (Rinkevich et al., 2013). Since initial reports of the L1014F mutation in M. domestica and B. germanica, various substitutions (C, H, S, or W) at this position have been found in pyrethroid-resistant populations across evolutionarily divergent insect groups (Table 1; Fig. 5A). Divergent substitutions have been shown at other sites, including V410 (M/A/G/L) in IS6, M918 (T/L/V) in the linker connecting S4 and S5 in domain II, T929 (I/C/N/V) in IIS5, I1011M/V and V1016G/I in IIS6 (Table 1). Different substitutions could confer different levels of resistance when examined in Xenopus oocytes. For example, sodium channels with the L1014F, L1014H and L1014S mutations provide variable levels of protection to Type I or Type II pyrethroids or DDT (Burton et al., 2011). Furthermore, mutations, I1011M and V1016G in IIS6, reduce the sensitivity of the AaNav1-1 channel to two pyrethroids permethrin and deltamethrin, but different substiutions, I1011V and V1016I, have no effect (Du et al., 2013). Since I1011 and V1016 are located in the pyrethroid receptor sites and are critical for pyrethroid binding (see below), I1011V and V1016I mutations could affect the action of other pyrethroids and/or DDT. Such cases have been documented for two other kdr mutations, M918T in the linker connecting S4 and S5 in domain II and F1534C in IIIS6, both of which are also located in the pyrethroid receptor sites (see below). M918T provides extremely high levels of protection against permethrin and deltamethrin (Vais et al., 2000), but does not provide protection from DDT (Usherwood et al., 2005), and F1534C confers sodium channel resistance to type I, but not type II pyrethroids (Hu et al., 2011).
Co-occurrence of more than one kdr mutation often leads to greater reduction of sodium channel sensitivity to pyrethroids than individual mutations. For example, the L1014F or M918T mutation alone caused about a 5-10 fold reduction in the sensitivity of the DmNav channel to deltamethrin, but the double mutations L1014F+M918T almost abolished the sensitivity of the DmNav channel to deltamethrin (Lee et al., 1999; Vais et al., 2000). Similarly, the T929I mutation in combination with M827I and L932F completely eliminated the permethrin sensitivity of Vssc1 channels (Yoon et al., 2008). Two mutations, E435K and C785R, in the linker sequence connecting domains I and II are found to co-exist with L1014F in the cockroach. Each mutation alone did not reduce the sensitivity of the BgNav to deltamethrin (Tan et al., 2002b). However, when either the E435K or C785R mutation was combined with the L1014F mutation, the channel sensitivity is reduced by 100-fold. Concomitant presence of all three mutations reduced channel sensitivity to deltamethrin by 500-fold (Tan et al., 2002b). Similarly, E435K and C785R mutations also further reduced the sensitivity of the V410M channel to pyrethroids in the cockroach (Liu et al., 2002). These two mutations can therefore be considered as enhancers of the V410M and L1014F mutations (Liu et al., 2002; Tan et al., 2002b). More recently, a N1575Y mutation in the linker sequence connecting domains III and IV was reported along with L1014 in pyrethroid-resistant populations of the African malaria mosquito A. gambiae (Jones et al., 2012). Mosquitoes carrying the double mutations (N1575Y + L1014F) are more resistant to permethrin than mosquitoes carrying only L1014F (Jones et al., 2012). Multiple nonsynonymous and synonymous mutations and mutation combinations were identified in permethrin-resistant Culex quinquefasciatus populations (Li et al., 2012; Xu et al., 2012). However, the involvement of synonymous mutations in pyrethroid resistance remains to be examined. In addition, mutations, S989P and D1763Y, are often found to co-exist with V1016G in pyrethroid-resistant field populations of Ae. Aegypti (Rinkevich et al., 2013). Unexpectedly, neither V1016G/S989P nor V1016G/D1763Y is more resistant to pyrethroids than the single mutants (Du et al., 2013). These results suggest that S989P and D1763Y are not enhancers of the V1016G kdr mutation. However, it remains possible that these mutations could compensate for potential fitness disadvantage caused by the V1016G mutation.
5. kdr mutations define the receptor sites of pyrethroids
Identification of kdr mutations has proven to be very valuable for elucidating the mechanisms of pyrethroid binding and action on sodium channels. Because kdr mutations reduce the activity of pyrethroids on insect sodium channels, a logical hypothesis is that some of the kdr mutations occur at the pyrethroid-binding site and confer resistance by reducing pyrethroid binding. Unfortunately, direct radioligand binding to determine the effect of kdr mutations on pyrethroid binding is not feasible because of extremely high levels of nonspecific binding of pyrethroids to membranes (Dong and Scott, 1994; Pauron et al., 1989; Rossignol, 1988). As alternative approaches, computational modeling of pyrethroid receptor sites on insect sodium channels and electrophysiological and pharmacological analyses of kdr mutant sodium channels expressed in Xenopus oocytes have been successfully employed to shed light on the mechanism of action of and resistance to pyrethroids.
5.1. Evidence for dual pyrethroid receptor sites
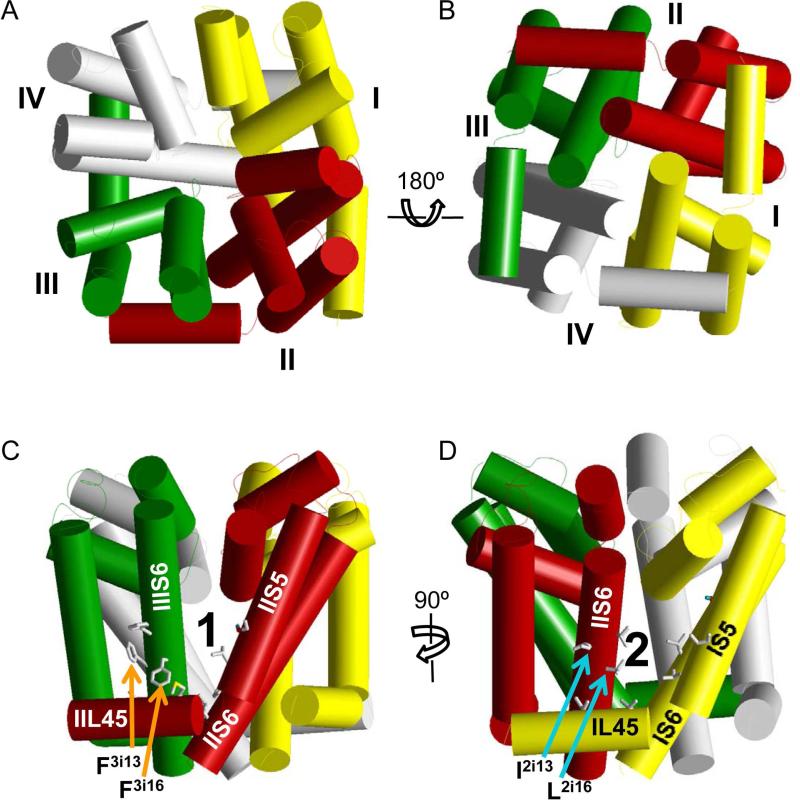
In the absence of X-ray structures of eukaryotic sodium channels, homology models of insect sodium channels using Kv1.2 crystallized in the open state (Long et al., 2005) and NavAb crystallized in the closed state (Payandeh et al., 2011) as templates have been used in the analysis of pyrethroid-sodium channel interactions. Although sequences of individual repeat domains in a eukaryotic sodium channel are different, the four-fold rotational pseudo-symmetry is believed to characterize folding of repeat domains around the pore axis (Fig. 6A and 6B). From the extracellular view (Fig. 6A), repeat domains I, II, III, and IV are arranged clockwise (Dudley et al., 2000). To better visualize structurally analogous positions of amino acid residues in the four homologous domains within the same channel and also make comparisons with sodium channels from other species, we label sodium channel residues using nomenclature that is universal for P-loop ion channels (Du et al., 2010; Zhorov and Tikhonov, 2004). A residue is labeled by the domain number (1 to 4), segment type (k, the L45 linker; i, the inner helix; o, the outer helix), and the relative number of the residue in the segment. For example, the kdr mutation, L1014F, in IIS6 becomes L2i16F under the new nomenclature (Fig. 7).
Fig. 6.
Modeling the pyrethroid receptor sites in the AaNav1-1 channel. (A and B) An NavAb-based model of the pore-forming module of the mosquito sodium channel AaNav1-1. Helices are shown as cylinders and repeat domains I, II, III, and IV are yellow, red, green and gray, respectively. The four repeat domains are arranged clockwise in the extracellular view (A) and counterclockwise at the cytoplasmic view (B). Side views of the sodium channel, where side chains of pyrethroid-sensing residues in Site 1 (C) and Site 2 (D) are shown by sticks. Note that Site 1 is located at the interface between domains II and III (O'Reilly et al., 2006; Usherwood et al., 2007); whereas Site 2, is located at the interface between domains I and II domain (Du et al., 2013).
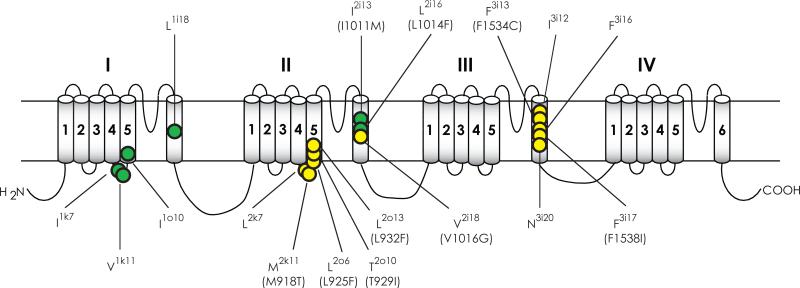
Fig. 7.
Pyrethroid-sensing residues in Site 1 or Site 2 of an insect sodium channel. Pyrethroid-sensing residues are indicated in solid yellow circles for Site 1 residues and solid green circles for Site 2 residues. Mutations that are detected in pyrethroid-resistant field populations at the corresponding positions are indicated in brackets. See Table 2 for the amino acid sequence alignment of Kv1.2, NavAb, and AaNav1-1 channels in these regions that contain pyrethroid-sensing residues.
The first homology model of the house fly sodium channel (O'Reilly et al., 2006), which is based on the X-ray structure of the open Kv1.2 channel, predicts that pyrethroids bind to the lipid-exposed interface formed by the linker helix connecting S4 and S5 in domain II (IIL45) and helices IIS5 and IIIS6 (the IIL45-IIS5-IIIS6 triangle). Intrigingly, residues Mk211 (M918), L2o6 (L925), T2o10 (T929), F3i13 (F1534), and F3i16 (F1537) are exposed into the interface between the linker-helix IIL45 and transmembrane helices IIS5 and IIIS6 (Fig. 6C). Henceforth we refer to this interface as pyrethroid-binding site 1 (Site 1). DDT and several pyrethroids (fenvalerate, acrinathrin, bifenthrin, deltamethrin, permethrin) have been docked into Site 1 to predict atomistic details of pyrethroid-channel interactions (O'Reilly et al., 2006; Usherwood et al., 2007). In the obtained structural models, pyrethroids make multiple contacts with helices IIL45, IIS5, and IIIS6 that would stabilize the channel in the open state. The smaller DDT molecule is docked between helices IIS5 and IIIS6 and does not reach the IIL45 helix, thus making fewer contacts with the sodium channel. The existence of this binding site is further supported by experimental evidence from systematic site-directed mutagenesis studies (Du et al., 2009a; Usherwood et al., 2007), which identified additional pyrethroid-sensing residues at this site.
Although several kdr mutations are located in Site 1 (O'Reilly et al., 2006), other kdr mutations, such as V409M and L1014F (i.e., V1i19M and L2i16F), are located far from Site 1. These mutations were initially suggested to affect pyrethroid action allosterically by impeding the bending motions of the S6 helices, which are necessary for channel opening (O'Reilly et al., 2006). Intriguingly, however, the positions of these kdr mutations are analogous to those in Site 1, but in a different domain. For example, the pyrethroid-sensing residue F1537 (F3i16) in Site 1 is at the analogous position in domain III as L1014 (L2i16) in domain II (Fig. 6C and 6D; Table 2). This symmetry suggested the possibility that a second pyrethroid binding site may be formed by helices IL45, IS5, and IIS6 (i.e., the IL45-IS5-IIS6 triangle, which would be analogous to the Site 1 IIL45-IIS5-IIIS6 triangle). To explore this possibility, homology models of a mosquito sodium channel (AaNav1-1) in both the open and closed states were constructed based on the X-ray structures of NavAb (Payandeh et al., 2011) and Kv1.2 (Long et al., 2005) as templates. The models predicted specific residues in the second binding site (Site 2 hereinafter). Indeed, mutations I1k7A and V1k11A in IL45, I1o10C in IS5 and L1i18G in IS6 reduced the sensitivity of the AaNav1–1 channel to pyrethroids, thus supporting the existence of Site 2 (Du et al., 2013). As predicted, the kdr mutations I2i13M and L2i16F/S (i.e., I1011M and L1014F/S in Vssc1) that are detected in mosquito and other insect pest species are located in Site 2. Furthermore, mutational analysis of additional residues, which are identified as molecular determinants of Site 2 in the mosquito channel model, supports the existence of this site (Du et al., 2013). This dual receptor-site model for pyrethroids (Fig. 6C and 6D) is consistent with a previous study that reveal cooperative binding of two or more deltamethrin molecules to each DmNav channel expressed in Xenopus oocytes (Vais et al., 2000; Vais et al., 2003). Most of the pyrethroid-sensing residues in Site 1 have analogues in Site 2 indicating that the two sites are rather symmetric (Table 2 and Fig. 7). However, unlike the IIS5 mutations in Site 1, some of the corresponding IS5 mutations in Site 2 do not have a strong effect on pyrethroid action, suggesting that the precise architecture of Sites 1 and 2 are not identical (Du et al., 2013). This is not surprising given the essential sequential asymmetry of individual repeat domains.
Table 2.
Sequence alignment of segments L45/S5 and S6 of Kv1.2, NavAb, and AaNav1-1 channels.
| Channel | Segment | # | k1 | |
k11 | |
o11 | |
o11 | |
o21 | |
|
|---|---|---|---|---|---|---|---|---|
| Kv1.2 | L45/S5 | 323 | SKGLQILGQT | LK | ASMRELGLLI | FFLFIGVILF | SSAVYFAE | |
| NavAb | L45/S5 | 113 | VPQMRKIVSA | LI | SVIPGMLSVI | ALMTLFFYIF | AIMATQLF | |
| AaNav1-1 | I | L45/S5 | 281 | VPGLKTIVGA | VI | ESVKNLRDVI | ILTMFSLSVF | ALMGLQIY |
| II | L45/S5 | 917 | WPTLNLLISI | MG | RTMGALGNLT | FVLCIIIFIF | AVMGMQLF | |
| III | L45/S5 | 1419 | MQGMRVVVNA | LV | QAIPSIFNVL | LVCLIFWLIF | AIMGVQLF | |
| IV | L45/S5 | 1737 | AKGIRTLLFA | LA | MSLPALFNIC | LLLFLVMFIF | AIFGMSFF | |
| i1 | |
i11 | |
i21 | |
||||
|---|---|---|---|---|---|---|
| Kv1.2 | S6 | 385 | IGGKIVGSLC | AIAGVLTIAL | PVPVIVSNFN | |
| NavAb | S6 | 192 | PYAWVFFIPF | IFVVTFVMIN | LVVAICVDAM | |
| AaNav1-1 | I | S6 | 429 | PWHMLFFIVI | IFLGSFYLVN | LILAIVAMSY |
| II | S6 | 1006 | VSCIPFFLAT | VVIGNLVVLN | LFLALLLSNF | |
| III | S6 | 1540 | IYMYLYFVFF | IIFGSFFTLN | LFIGVIIDNF | |
| IV | S6 | 1840 | TIGITYLLAY | LVISFLIVIN | MYIAVILENY |
a Position of a residue is designated by a symbol, which identifies a segment, and a relative position of the residue in the segment. Symbols “k”, “o”, and “i” represent, respectively, the S4-S5 linker, the outer helix, and the inner helix.
b Pyrethroid-sensing residues are highlighted in yellow for Site 1 and in green for Site 2. These residues are identified either by functional analysis of kdr mutations or by systematic mutational studies guided by pyrethroid receptor site models (Du et al., 2009; Du et al., 2013; Usherwod et al., 2007).
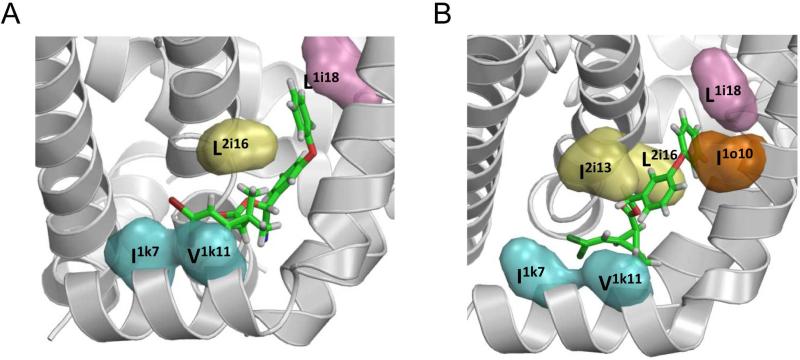
In the predicted binding modes of deltamethrin and 1R-cis-permethrin in Site 2 of the open AaNav1-1 channel, the CCl2 or CBr2 group binds between helices IL45 and IIS6, while the phenyl group binds between helices IS6 and IIS6 (Du et al., 2013). The bulky dimethylcyclopropane moiety fits between helices IL45, IS5, IS6, and IIS6 into a space below the gating-hinge position i14. In the completely extended conformation of deltamethrin, the most distant heavy atoms (a bromine and para-carbon in the terminal phenyl ring) are ~14 Å apart. The Cβ atoms of I1k7 and L1i18, which are the most distant pyrethroid-sensing residues in both the Kv1.2- and NavAb-based homology models of the AaNav1–1 channel, are 18.8 and 21.4 Å apart, respectively. Therefore, simultaneous interaction of deltamethrin with both I1k7 and L1i18 in the closed channel seems less likely than in the open channel. Thus, molecular modeling supports the notion that pyrethroids clamp the interface between repeat domains I and II in its open state and thus increase the open-state probability (Du et al., 2013). In the Kv1.2-based homology models of open housefly sodium channels with pyrethroids bound in Site 1 (Usherwood et al., 2007), the dimethylcyclopropane moiety of deltamethrin and permethrin binds “above” the gating hinge, in a region between the two inner helices and the pore helix, while the pyrethroid-sensing residues V2i18 (Du et al., 2013) and L2k7 (Usherwood et al., 2007), which are analogous to Site 2 pyrethroid-sensing residues L1i18 and I1k7, are seemingly not involved in ligand binding. Further experimental and theoretical studies are necessary to elaborate detailed models of binding of different pyrethroids and DDT in Sites 1 and 2 of insect sodium channels.
More recently, O'Reilly and associates used the pyrethroid receptor Site 1 model to explain selective toxicity of pyrethroids as acaricides towards mites/ticks vs. insects (O'Reilly et al., 2014). Mite/tick sodium channel contains a glycine, alanine or valine in the middle of helix IIS5 (G933 based on the amino acid residue numbering of the Vssc1 protein), whereas a cysteine is found in insect sodium channels at the corresponding position. Acaricidal pyrethroids, such as flumethrin and fluvalinate, have larger moieties in place of the 2,2-dibromoethenyl moiety of deltamethrin. The authors propose that the large moieties bind in the void space created by the small side chain of the glycine, alanine or valine residue in mite/tick sodium channels, but clash with the cysteine residue in insect sodium channels.
5.2. kdr mutations reduce pyrethroid binding and/or alter channel gating
Experimental evidence has been obtained showing that kdr mutations reduce pyrethroid effects either by reducing pyrethroid binding and/or by altering the gating properties (kinetics or voltage dependence) of sodium channels. For example, introducing M918T (Mk211T) and T929I (T2o10I) mutations into wild type DmNav channels reduce pyrethroid binding from two deltamethrins per channel to one molecule of deltamethrin for each sodium channel based on Hill plot analysis (Vais et al., 2000; Vais et al., 2003). Similarly, Schild plot analysis, which uses competitive binding of active and inactive isomers to determine the binding affinity of the inactive isomer (Lund and Narahashi, 1982; Tan et al., 2005), showed that the F1519I (F3i17I) mutation in IIIS6 and the L993F(L2i16F) mutation in IIS6 of a cockroach sodium channel, equivalent to F1538I (F3i17I) in Site 1 and L1014F (L2i16F) in Site 2 in the numbering of the Vssc1, respectively, reduces the binding of the inactive 1S-cis permethrin isomer (Tan et al., 2005). Thus, mutations in both Sites 1 and 2, as predicted by computer modeling, have been confirmed to reduce pyrethroid binding to sodium channels. Future analysis of other kdr mutations and pyrethroid-sensing residues predicted in Sites 1 and 2 will be valuable to further examine the dual-pyrethroid binding site model.
Besides altering pyrethroid binding, kdr mutations have also been shown to alter sodium channel gating kinetics (i.e., the rate of closing and opening), which indirectly affects the pyrethroid sensitivity of sodium channels. For example, the L1014F (L2i16F) mutation and the super-kdr mutation, M918T (M2k11T) enhance closed-state inactivation (Vais et al., 2000). This altered gating could contribute to channel resistance to pyrethroids because pyrethroids preferably bind to sodium channels in the open state. Similarly, two kdr mutations, V409M/L (V1i19M/L; equivalent to V410 in Vssc1), in IS6 slowed activation and accelerated deactivation kinetics (Oliveira et al., 2013), which could in principle counteract the action of pyrethroids (pyrethroids enhance activation and inhibit deactivation). Interestingly, however, another substitution at this amino acid position, V409I (V1i19I), which reduced channel sensitivity to pyrethroids, did not alter sodium channel gating kinetics (Oliveira et al., 2013). Furthermore, the reduction in pyrethroid sensitivity caused by V409M/L (V1i19M/L) mutations is greater than that by the V409I (V1i19I) mutation. Thus, the higher level of resistance to pyrethroids provided by the V409M/L (V1i19M/L) mutations, as opposed to the V409I (V1i19I) mutation, may result from the altered gating kinetics caused by V409M/L (V1i19M/L). A similar scenario has been documented for substitutions of an aspartic acid residue D802 in IIS1 of a pyrethroid-resistant BgNav variant from the German cockroach. Substitution at this position with glycine or lysine each reduced channel sensitivity to pyrethroids (Du et al., 2010), but the reduction in sensitivity was much greater for D802K. Like V409M/L, the D802K substitution slowed channel activation and accelerated deactivation kinetics, potentially antagonizing the effects of pyrethroids (Du et al., 2010). These findings indicate that kdr mutations could contribute to resistance of sodium channels to pyrethroids indirectly by counteracting the effects of pyrethroids on sodium channel gating.
Several kdr mutations have been shown to shift the voltage-dependence of activation in the positive direction, making sodium channels less likely to open, which could again indirectly antagonize the actions of pyrethroids. For example, the L1014F/S (L2i16F/S) mutation in IIS6 shifts the voltage dependence of activation and inactivation of DmNav and Vssc1 channels to more positive potentials (Burton et al., 2011; Lee et al., 1999; Smith et al., 1997; Vais et al., 2001). Similarly, the V409M (V1i19M) mutation in IS6 causes a positive shift in the voltage dependence of activation of BgNav1-1a channels (Oliveira et al., 2013), which is also consistent with measurements made from neurons of pyrethroid-resistant Heliothis virescens adults carrying the V409M (V1i19M) mutation (Lee et al., 1999). However, the V409L/I (V1i19L/I) mutations do not alter the voltage-dependence of activation of BgNav1-1a channels, yet V409L mutation confers a similar level of resistance to pyrethroids as V409M in Xenopus oocytes (Oliveira et al., 2013). These results suggest that a positive shift in the voltage dependence of activation per se may not be the major cause of pyrethroid resistance.
6. The molecular basis of differential pyrethroid sensitivities between insect and mammalian sodium channels
Insect sodium channels are much more sensitive to pyrethroids than mammalian sodium channels (Du et al., 2013; Vais et al., 2000; Warmke et al., 1997), which contributes to the selective toxicity of pyrethroids between mammals and insects. However, differential pyrethroid sensitivities among mammalian sodium channels have been documented. Rat dorsal root ganglion (DRG) neurons have two types of sodium current, tetrodotoxin-sensitive (TTX-S) and tetrodotoxin-resistant (TTX-R). TTX-S channels were less sensitive to pyrethroids than TTX-R sodium channels in the same neurons (Ginsburg and Narahashi, 1993; Song and Narahashi, 1996; Tatebayashi and Narahashi, 1994). More recent comparison of pyrethroid sensitivity among mammalian sodium channels expressed in Xenopus oocytes confirmed and extended these pharmacological observations. Rat Nav1.2, Nav1.4, and Nav1.7 channels are almost completely insensitive to pyrethroids, whereas rat Nav1.3; Nav1.6 and Nav1.8 sodium channels are more sensitive to pyrethroids (Choi and Soderlund, 2006; Du et al., 2013; Peng et al., 2009; Tan and Soderlund, 2009, 2010, 2011; Vais et al., 1997; Wang et al., 2001; Warmke et al., 1997). Identification of pyrethroid-sensing residues in insect sodium channels has been valuable for elucidating the molecular basis of selective toxicity of pyrethroids between mammals and insects. For example, at the position corresponding to the M918T (M2k11T) kdr mutation in IIL45, mammalian sodium channels possess an isoleucine. Substitution of the isoleucine in the rNav1.2 channel with a methionine (insect version) increased channel sensitivity to pyrethroids (Vais et al., 2000). Substitutions of leucine in IL45 and methionine in IS5 of rNav1.2 and rNav1.4, respectively, with valine and isoleucine, which occur in insect sodium channels, also enhanced the sensitivity of rat sodium channels to pyrethroids (Du et al., 2013). As mentioned above, V409I/L (V1i19I/L) kdr mutations reduced cockroach sodium channel sensitivity to pyrethroids (Oliveira et al., 2013). A valine is present in Nav1.3, Nav1.5, Nav1.6 and Nav1.8 channels, but an isoleucine is found in Nav1.1, Nav1.2, and Nav1.4, which are more resistant to pyrethroids than Nav1.3, Nav1.5, Nav1.6 and Nav1.8 channels (Soderlund, 2012). As expected, a valine substitution of isoleucine (I433V, I1i19V) enhanced the sensitivity of rNav1.4 channels to deltamethrin (Oliveira et al., 2013). Importantly, many of the “pyrethoid-insensitive” residues are conserved in all known mammalian sodium channels. These results suggest that isoleucine in IIL45 (Site 1), leucine in IL45 (Site 2) and methionine in IS5 (Site 2) contribute to the lower sensitivity of mammalian sodium channels to pyrethroids.
7. Conclusions
Since the initial cloning of the first insect sodium channel gene more than two decades ago, significant progress has been made in our understanding of the electrophysiology, pharmacology, and molecular biology of insect sodium channels. Successful functional expression of insect sodium channels in Xenopus oocytes had a major impact on insect sodium channel research. As highlighted in the current review, insects generate a remarkable array of structurally and functionally diverse sodium channel variants from a single sodium channel gene via alternative splicing and RNA editing. While it has been exciting to observe diverse functional properties of splice and RNA editing variants in Xenopus oocytes, the next challenge is to determine the role of various splicing and RNA editing variants in shaping up the dynamic electrical activity of the insect nervous system in vivo and how they ultimately modulate specific sensory, motor, and integrative functions at the whole organism level.
The great interest in studying insect sodium channels in the past two decades has been in large part motivated by the exciting connection between pyrethroid resistance and sodium channel mutations. The intensive research efforts in this area have greatly improved our understanding of the molecular basis of pyrethroid resistance and pyrethroid action on the sodium channel. In particular, it has been rewarding to note that identification of kdr mutations has led to the development of methods for early detection of kdr resistance in field populations as a novel means of resistance management. Nevertheless, it seems inevitable that pyrethroid resistance, already detected globally, will eventually diminish the effectiveness of existing pyrethroids, which would represent a major loss of an effective and safe measure against insect pests and disease vectors that threaten agriculture and human health globally. Further advances in fundamental knowledge of insect sodium channels are needed for deepening our understanding of the role of sodium channels in basic insect biology and interactions with various neurotoxins and for preserving the use of current sodium channel insecticides (e.g., pyrethroids) and the development of a new generation of safe and highly effective sodium channel-targeting insecticides (exemplified by SCBIs).
Supplementary Material
Highlights.
Insect sodium channel genes undergo extensive alternative splicing and RNA editing
Alternative splicing and RNA editing generate an array of functionally diverse sodium channels in insects
Mutations in sodium channels confer knockdown resistance (kdr) to pyrethroids
Sodium channels have two receptor sites for pyrethroids
Fig. 8.
A Kv1.2-based model of the open AaNav1–1 channel with deltamethrin (A) and 1R-cis-permethrin (B) bound to Site 2. Cyan, orange, pink, and yellow surfaces are side chains of pyrethroid-sensing residues in segments IL45, IS5, IS6, and IIS6, respectively. The halogen atoms of pyrethroids bind between IL45 and IIS6. The terminal phenyl rings of deltamethrin and 1R-cis-permethrin bind between IS6 and IIS6. Reproduced with permission from PNAS.
Acknowledgement
The authors would like to thank Dong lab members for their contributions to the research on insect sodium channels and the mechanism of action and resistance of sodium channel-targeting insecticides. Funding on sodium channel research in Dong lab is provided by the National Science Foundation (IBN 9808156 and IBN 9808156), the National Institutions of Health (GM057440), the United State Department of Agriculture Cooperative State Research, Education and Extension Service Grant (35607) and the Binational Agricultual Research and Development Fund (IS-3480-03).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bass BL. RNA editing. Oxford University Press; 2001. [Google Scholar]
- Bende NS, Kang E, Herzig V, Bosmans F, Nicholson GM, Mobli M, King GF. The insecticidal neurotoxin Aps III is an atypical knottin peptide that potently blocks insect voltage-gated sodium channels. Biochem Pharmacol. 2013;85:1542–1554. doi: 10.1016/j.bcp.2013.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosmans F, Tytgat J. Sea anemone venom as a source of insecticidal peptides acting on voltage-gated Na+ channels. Toxicon. 2007;49:550–560. doi: 10.1016/j.toxicon.2006.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdin CM, Moignot B, Wang L, Murillo L, Juchaux M, Quinchard S, Lapied B, Guerineau NC, Dong K, Legros C. Intron retention in mRNA encoding ancillary subunit of insect voltage-gated sodium channel modulates channel expression, gating regulation and drug sensitivity. PLoS One. 2013;8:e67290. doi: 10.1371/journal.pone.0067290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brackenbury WJ, Isom LL. Na Channel β Subunits: Overachievers of the Ion Channel Family. Front Pharmacol. 2011;2:53. doi: 10.3389/fphar.2011.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton MJ, Mellor IR, Duce IR, Davies TG, Field LM, Williamson MS. Differential resistance of insect sodium channels with kdr mutations to deltamethrin, permethrin and DDT. Insect Biochem Mol Biol. 2011;41:723–732. doi: 10.1016/j.ibmb.2011.05.004. [DOI] [PubMed] [Google Scholar]
- Catterall WA. From ionic currents to molecular mechanisms: the structure and function of voltage-gated sodium channels. Neuron. 2000;26:13–25. doi: 10.1016/s0896-6273(00)81133-2. [DOI] [PubMed] [Google Scholar]
- Catterall WA. Voltage-gated sodium channels at 60: structure, function and pathophysiology. J Physiol. 2012;590:2577–2589. doi: 10.1113/jphysiol.2011.224204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catterall WA. Sodium channels, inherited epilepsy, and antiepileptic drugs. Annu Rev Pharmacol Toxicol. 2014;54:317–338. doi: 10.1146/annurev-pharmtox-011112-140232. [DOI] [PubMed] [Google Scholar]
- Catterall WA, Cestele S, Yarov-Yarovoy V, Yu FH, Konoki K, Scheuer T. Voltage-gated ion channels and gating modifier toxins. Toxicon. 2007;49:124–141. doi: 10.1016/j.toxicon.2006.09.022. [DOI] [PubMed] [Google Scholar]
- Cestele S, Catterall WA. Molecular mechanisms of neurotoxin action on voltage-gated sodium channels. Biochimie. 2000;82:883–892. doi: 10.1016/s0300-9084(00)01174-3. [DOI] [PubMed] [Google Scholar]
- Chang C, Shen WK, Wang TT, Lin YH, Hsu EL, Dai SM. A novel amino acid substitution in a voltage-gated sodium channel is associated with knockdown resistance to permethrin in Aedes aegypti. Insect Biochem Mol Biol. 2009;39:272–278. doi: 10.1016/j.ibmb.2009.01.001. [DOI] [PubMed] [Google Scholar]
- Choi JS, Soderlund DM. Structure-activity relationships for the action of 11 pyrethroid insecticides on rat Na v 1.8 sodium channels expressed in Xenopus oocytes. Toxicol Appl Pharmacol. 2006;211:233–244. doi: 10.1016/j.taap.2005.06.022. [DOI] [PubMed] [Google Scholar]
- Crill WE. Persistent sodium current in mammalian central neurons. Annu Rev Physiol. 1996;58:349–362. doi: 10.1146/annurev.ph.58.030196.002025. [DOI] [PubMed] [Google Scholar]
- Cui YJ, Yu LL, Xu HJ, Dong K, Zhang CX. Molecular characterization of DSC1 orthologs in invertebrate species. Insect Biochem Mol Biol. 2012;42:353–359. doi: 10.1016/j.ibmb.2012.01.005. [DOI] [PubMed] [Google Scholar]
- Davies TG, Field LM, Usherwood PN, Williamson MS. A comparative study of voltage-gated sodium channels in the Insecta: implications for pyrethroid resistance in Anopheline and other Neopteran species. Insect Mol Biol. 2007;16:361–375. doi: 10.1111/j.1365-2583.2007.00733.x. [DOI] [PubMed] [Google Scholar]
- Derst C, Walther C, Veh RW, Wicher D, Heinemann SH. Four novel sequences in Drosophila melanogaster homologous to the auxiliary Para sodium channel subunit TipE. Biochem Biophys Res Commun. 2006;339:939–948. doi: 10.1016/j.bbrc.2005.11.096. [DOI] [PubMed] [Google Scholar]
- Dong K. A single amino acid change in the para sodium channel protein is associated with knockdown-resistance (kdr) to pyrethroid insecticides in German cockroach. Insect Biochem Mol Biol. 1997;27:93–100. doi: 10.1016/s0965-1748(96)00082-3. [DOI] [PubMed] [Google Scholar]
- Dong K. Insect sodium channels and insecticide resistance. Invert Neurosci. 2007;7:17–30. doi: 10.1007/s10158-006-0036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong K, Scott JG. Linkage of kdr-type resistance and the para-homologous sodium channel gene in German cockroaches (Blattella germanica). Insect Biochem Mol Biol. 1994;24:647–654. doi: 10.1016/0965-1748(94)90051-5. [DOI] [PubMed] [Google Scholar]
- Doyle DA, Morais Cabral J, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- Du Y, Garden DP, Wang L, Zhorov BS, Dong K. Identification of new batrachotoxin-sensing residues in segment IIIS6 of the sodium channel. J Biol Chem. 2011;286:13151–13160. doi: 10.1074/jbc.M110.208496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Lee JE, Nomura Y, Zhang T, Zhorov BS, Dong K. Identification of a cluster of residues in transmembrane segment 6 of domain III of the cockroach sodium channel essential for the action of pyrethroid insecticides. Biochem. J. 2009a;419:377–385. doi: 10.1042/BJ20082082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Liu Z, Nomura Y, Khambay B, Dong K. An alanine in segment 3 of domain III (IIIS3) of the cockroach sodium channel contributes to the low pyrethroid sensitivity of an alternative splice variant. Insect Biochem Mol Biol. 2006;36:161–168. doi: 10.1016/j.ibmb.2005.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Nomura Y, Liu Z, Huang ZY, Dong K. Functional expression of an arachnid sodium channel reveals residues responsible for tetrodotoxin resistance in invertebrate sodium channels. J Biol Chem. 2009b;284:33869–33875. doi: 10.1074/jbc.M109.045690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Nomura Y, Luo N, Liu Z, Lee JE, Khambay B, Dong K. Molecular determinants on the insect sodium channel for the specific action of type II pyrethroid insecticides. Toxicol Appl Pharmacol. 2009c;234:266–272. doi: 10.1016/j.taap.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Nomura Y, Satar G, Hu Z, Nauen R, He SY, Zhorov BS, Dong K. Molecular evidence for dual pyrethroid-receptor sites on a mosquito sodium channel. Proc Nat Acad Sci. 2013;110:11785–11790. doi: 10.1073/pnas.1305118110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Song W, Groome JR, Nomura Y, Luo N, Dong K. A negative charge in transmembrane segment 1 of domain II of the cockroach sodium channel is critical for channel gating and action of pyrethroid insecticides. Toxicol Appl Pharmacol. 2010;247:53–59. doi: 10.1016/j.taap.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley SC, Jr., Chang N, Hall J, Lipkind G, Fozzard HA, French RJ. mu-conotoxin GIIIA interactions with the voltage-gated Na+ channel predict a clockwise arrangement of the domains. J Gen Physiol. 2000;116:679–690. doi: 10.1085/jgp.116.5.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G, Deak P.t., Chopra M, Hall LM. Cloning and functional analysis of tipE, a novel membrane protein that enhances drosophila para sodium channel function. Cell. 1995;82:1001–1011. doi: 10.1016/0092-8674(95)90279-1. [DOI] [PubMed] [Google Scholar]
- Frank HY, Catterall WA. Overview of the voltage-gated sodium channel family. Genome Bio. 2003;4:207. doi: 10.1186/gb-2003-4-3-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber G, Smith LA, Reenan RA, Rogina B. Effect of sodium channel abundance on Drosophila development, reproductive capacity and aging. Fly (Austin) 2012;6:57–67. doi: 10.4161/fly.18570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsburg KS, Narahashi T. Differential sensitivity of tetrodotoxin-sensitive and tetrodotoxin-resistant sodium channels to the insecticide allethrin in rat dorsal root ganglion neurons. Brain Res. 1993;627:239–248. doi: 10.1016/0006-8993(93)90326-i. [DOI] [PubMed] [Google Scholar]
- Goldin AL. Evolution of voltage-gated Na+ channels. J Exp Biol. 2002;205:575–584. doi: 10.1242/jeb.205.5.575. [DOI] [PubMed] [Google Scholar]
- Goldin AL, Barchi RL, Caldwell JH, Hofmann F, Howe JR, Hunter JC, Kallen RG, Mandel G, Meisler MH, Netter YB. Nomenclature of voltage-gated sodium channels. Neuron. 2000;28:365. doi: 10.1016/s0896-6273(00)00116-1. [DOI] [PubMed] [Google Scholar]
- Gordon D. A new approach to insect-pest control--combination of neurotoxins interacting with voltage sensitive sodium channels to increase selectivity and specificity. Invert Neurosci. 1997;3:103–116. doi: 10.1007/BF02480365. [DOI] [PubMed] [Google Scholar]
- Gordon D, Karbat I, Ilan N, Cohen L, Kahn R, Gilles N, Dong K, Stuhmer W, Tytgat J, Gurevitz M. The differential preference of scorpion alpha-toxins for insect or mammalian sodium channels: implications for improved insect control. Toxicon. 2007;49:452–472. doi: 10.1016/j.toxicon.2006.11.016. [DOI] [PubMed] [Google Scholar]
- Gur M, Kahn R, Karbat I, Regev N, Wang J, Catterall WA, Gordon D, Gurevitz M. Elucidation of the molecular basis of selective recognition uncovers the interaction site for the core domain of scorpion alpha-toxins on sodium channels. J Biol Chem. 2011;286:35209–35217. doi: 10.1074/jbc.M111.259507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevitz M. Mapping of scorpion toxin receptor sites at voltage-gated sodium channels. Toxicon. 2012;60:502–511. doi: 10.1016/j.toxicon.2012.03.022. [DOI] [PubMed] [Google Scholar]
- Gurevitz M, Karbat I, Cohen L, Ilan N, Kahn R, Turkov M, Stankiewicz M, Stuhmer W, Dong K, Gordon D. The insecticidal potential of scorpion beta-toxins. Toxicon. 2007;49:473–489. doi: 10.1016/j.toxicon.2006.11.015. [DOI] [PubMed] [Google Scholar]
- Hanrahan CJ, Palladino MJ, Ganetzky B, Reenan RA. RNA editing of the Drosophila para Na+ channel transcript. Evolutionary conservation and developmental regulation. Genetics. 2000;155:1149–1160. doi: 10.1093/genetics/155.3.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Du Y, Nomura Y, Dong K. A sodium channel mutation identified in Aedes aegypti selectively reduces cockroach sodium channel sensitivity to type I, but not type II pyrethroids. Insect Biochem. Molec. Biol. 2011;41:9–13. doi: 10.1016/j.ibmb.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingles PJ, Adams PM, Knipple DC, Soderlund DM. Characterization of voltage-sensitive sodium channel gene coding sequences from insecticide-susceptible and knockdown-resistant house fly strains. Insect Biochem Mol Biol. 1996;26:319–326. doi: 10.1016/0965-1748(95)00093-3. [DOI] [PubMed] [Google Scholar]
- Jepson JE, Reenan RA. Adenosine-to-inosine genetic recoding is required in the adult stage nervous system for coordinated behavior in Drosophila. J Biol Chem. 2009;284:31391–31400. doi: 10.1074/jbc.M109.035048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jepson JE, Savva YA, Jay KA, Reenan RA. Visualizing adenosine-to-inosine RNA editing in the Drosophila nervous system. Nat Methods. 2011;9:189–194. doi: 10.1038/nmeth.1827. [DOI] [PubMed] [Google Scholar]
- Jin Y, Tian N, Cao J, Liang J, Yang Z, Lv J. RNA editing and alternative splicing of the insect nAChR subunit alpha6 transcript: evolutionary conservation, divergence and regulation. BMC Evol Biol. 2007;7:98. doi: 10.1186/1471-2148-7-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AK, Buckingham SD, Papadaki M, Yokota M, Sattelle BM, Matsuda K, Sattelle DB. Splice-variant- and stage-specific RNA editing of the Drosophila GABA receptor modulates agonist potency. J Neurosci. 2009;29:4287–4292. doi: 10.1523/JNEUROSCI.5251-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CM, Liyanapathirana M, Agossa FR, Weetman D, Ranson H, Donnelly MJ, Wilding CS. Footprints of positive selection associated with a mutation (N1575Y) in the voltage-gated sodium channel of Anopheles gambiae. Proc Natl Acad Sci. 2012;109:6614–6619. doi: 10.1073/pnas.1201475109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T. Persistent Na-channels: origin and function. A review. Acta Biol Hung. 2008;59(Suppl):1–12. doi: 10.1556/ABiol.59.2008.Suppl.1. [DOI] [PubMed] [Google Scholar]
- Klint JK, Senff S, Rupasinghe DB, Er SY, Herzig V, Nicholson GM, King GF. Spider-venom peptides that target voltage-gated sodium channels: pharmacological tools and potential therapeutic leads. Toxicon. 2012;60:478–491. doi: 10.1016/j.toxicon.2012.04.337. [DOI] [PubMed] [Google Scholar]
- Kulkarni NH, Yamamoto AH, Robinson KO, Mackay TFC, Anholt RRH. The DSC1 channel, encoded by the smi60E locus, contributes to odor-guided behavior in Drosophila melanogaster. Genetics. 2002;161:1507–1516. doi: 10.1093/genetics/161.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Ingles PJ, Knipple DC, Soderlund DM. Developmental regulation of alternative exon usage in the house fly Vssc1 sodium channel gene. Invert Neurosci. 2002;4:125–133. doi: 10.1007/s10158-001-0014-1. [DOI] [PubMed] [Google Scholar]
- Lee SH, Smith TJ, Ingles PJ, Soderlund DM. Cloning and functional characterization of a putative sodium channel auxiliary subunit gene from the house fly (Musca domestica). Insect Biochem Mol Biol. 2000;30:479–487. doi: 10.1016/s0965-1748(00)00019-9. [DOI] [PubMed] [Google Scholar]
- Lee SH, Smith TJ, Knipple DC, Soderlund DM. Mutations in the house fly Vssc1 sodium channel gene associated with super-kdr resistance abolish the pyrethroid sensitivity of Vssc1/tipE sodium channels expressed in Xenopus oocytes. Insect Biochem Mol Biol. 1999;29:185–194. doi: 10.1016/s0965-1748(98)00122-2. [DOI] [PubMed] [Google Scholar]
- Lee SH, Soderlund DM. The V410M mutation associated with pyrethroid resistance in Heliothis virescens reduces the pyrethroid sensitivity of house fly sodium channels expressed in Xenopus oocytes. Insect Biochem Mol Biol. 2001;31:19–29. doi: 10.1016/s0965-1748(00)00089-8. [DOI] [PubMed] [Google Scholar]
- Li T, Zhang L, Reid WR, Xu Q, Dong K, Liu N. Multiple mutations and mutation combinations in the sodium channel of permethrin resistant mosquitoes, Culex quinquefasciatus. Sci Rep. 2012;2:781. doi: 10.1038/srep00781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilly B, Galewsky S, Firulli AB, Schulz RA, Olson EN. D-MEF2: a MADS box transcription factor expressed in differentiating mesoderm and muscle cell lineages during Drosophila embryogenesis. Proc Natl Acad Sci U S A. 1994;91:5662–5666. doi: 10.1073/pnas.91.12.5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WH, Gunay C, Marley R, Prinz AA, Baines RA. Activity-dependent alternative splicing increases persistent sodium current and promotes seizure. J Neurosci. 2012;32:7267–7277. doi: 10.1523/JNEUROSCI.6042-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WH, Wright DE, Muraro NI, Baines RA. Alternative splicing in the voltage-gated sodium channel DmNav regulates activation, inactivation, and persistent current. J Neurophysiol. 2009;102:1994–2006. doi: 10.1152/jn.00613.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay HA, Baines R, ffrench-Constant R, Lilley K, Jacobs HT, O'Dell KM. The dominant cold-sensitive out-cold mutants of Drosophila melanogaster have novel missense mutations in the voltage-gated sodium channel gene paralytic. Genetics. 2008;180:873–884. doi: 10.1534/genetics.108.090951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipkind GM, Fozzard HA. KcsA crystal structure as framework for a molecular model of the Na(+) channel pore. Biochemistry. 2000;39:8161–8170. doi: 10.1021/bi000486w. [DOI] [PubMed] [Google Scholar]
- Lipkind GM, Fozzard HA. Molecular modeling of local anesthetic drug binding by voltage-gated sodium channels. Mol Pharmacol. 2005;68:1611–1622. doi: 10.1124/mol.105.014803. [DOI] [PubMed] [Google Scholar]
- Liu Z, Chung I, Dong K. Alternative splicing of the BSC1 gene generates tissue-specific isoforms in the German cockroach. Insect Biochem Mol Biol. 2001;31:703–713. doi: 10.1016/s0965-1748(00)00178-8. [DOI] [PubMed] [Google Scholar]
- Liu Z, Song W, Dong K. Persistent tetrodotoxin-sensitive sodium current resulting from U-to-C RNA editing of an insect sodium channel. Proc Natl Acad Sci U S A. 2004;101:11862–11867. doi: 10.1073/pnas.0307695101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Tan J, Valles SM, Dong K. Synergistic interaction between two cockroach sodium channel mutations and a tobacco budworm sodium channel mutation in reducing channel sensitivity to a pyrethroid insecticide. Insect Biochem Mol Biol. 2002;32:397–404. doi: 10.1016/s0965-1748(01)00116-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Valles SM, Dong K. Novel point mutations in the German cockroach para sodium channel gene are associated with knockdown resistance (kdr) to pyrethroid insecticides. Insect Biochem Mol Biol. 2000;30:991–997. doi: 10.1016/s0965-1748(00)00074-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long SB, Campbell EB, Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- Loughney K, Kreber R, Ganetzky B. Molecular analysis of the para locus, a sodium channel gene in Drosophila. Cell. 1989;58:1143–1154. doi: 10.1016/0092-8674(89)90512-6. [DOI] [PubMed] [Google Scholar]
- Lund AE, Narahashi T. Dose-dependent interaction of the pyrethroid isomers with sodium channels of squid axon membranes. Neurotoxicology. 1982;3:11–24. [PubMed] [Google Scholar]
- Maldonado C, Alicea D, Gonzalez M, Bykhovskaia M, Marie B. Adar is essential for optimal presynaptic function. Mol Cell Neurosci. 2013;52:173–180. doi: 10.1016/j.mcn.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marley R, Baines RA. Increased persistent Na+ current contributes to seizure in the slamdance bang-sensitive Drosophila mutant. J Neurophysiol. 2011;106:18–29. doi: 10.1152/jn.00808.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani R. Mendelian behavior of resistance to the knock-down action of DDT and correlation between knock-down and mortality in Musca domestica L. Rend Ist Sup Sanit. 1956;19:1107–1143. [PubMed] [Google Scholar]
- Miyazaki M, Ohyama K, Dunlap DY, Matsumura F. Cloning and sequencing of the para-type sodium channel gene from susceptible and kdr-resistant German cockroaches (Blattella germanica) and house fly (Musca domestica). Mol Gen Genet. 1996;252:61–68. [PubMed] [Google Scholar]
- Moignot B, Lemaire C, Quinchard S, Lapied B, Legros C. The discovery of a novel sodium channel in the cockroach Periplaneta americana: Evidence for an early duplication of the para-like gene. Insect Biochem Molec Biol. 2009;39:814–823. doi: 10.1016/j.ibmb.2009.09.006. n.d. [DOI] [PubMed] [Google Scholar]
- Moran Y, Gordon D, Gurevitz M. Sea anemone toxins affecting voltage-gated sodium channels--molecular and evolutionary features. Toxicon. 2009;54:1089–1101. doi: 10.1016/j.toxicon.2009.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran Y, Kahn R, Cohen L, Gur M, Karbat I, Gordon D, Gurevitz M. Molecular analysis of the sea anemone toxin Av3 reveals selectivity to insects and demonstrates the heterogeneity of receptor site-3 on voltage-gated Na+ channels. Biochem. J. 2007;406:41–48. doi: 10.1042/BJ20070233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narahashi T. Neuronal ion channels as the target sites of insecticides. Pharmacol Toxicol. 1996;78:1–14. doi: 10.1111/j.1600-0773.1996.tb00234.x. [DOI] [PubMed] [Google Scholar]
- Narahashi T. Neuroreceptors and ion channels as the basis for drug action: past, present, and future. J Pharmacol Exp Ther. 2000;294:1–26. [PubMed] [Google Scholar]
- Narahashi T. Nerve membrane ion channels as the target site of insecticides. Mini Rev Medic Chem. 2002;2:419–432. doi: 10.2174/1389557023405927. [DOI] [PubMed] [Google Scholar]
- Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu Rev Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numann R, Catterall WA, Scheuer T. Functional modulation of brain sodium channels by protein kinase C phosphorylation. science. 1991;254:115–118. doi: 10.1126/science.1656525. [DOI] [PubMed] [Google Scholar]
- O'Dowd DK. Voltage-gated currents and firing properties of embryonic Drosophila neurons grown in a chemically defined medium. J Neurobiol. 1995;27:113–126. doi: 10.1002/neu.480270111. [DOI] [PubMed] [Google Scholar]
- O'Reilly AO, Khambay BP, Williamson MS, Field LM, Wallace BA, Davies TG. Modelling insecticide-binding sites in the voltage-gated sodium channel. Biochem J. 2006;396:255–263. doi: 10.1042/BJ20051925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Reilly AO, Williamson MS, Gonzalez-Cabrera J, Turberg A, Field LM, Wallace BA, Davies TG. Predictive 3D modelling of the interactions of pyrethroids with the voltage-gated sodium channels of ticks and mites. Pest Manag Sci. 2014;70:369–377. doi: 10.1002/ps.3561. [DOI] [PubMed] [Google Scholar]
- Okamoto H, Sakai K, Goto S, Takasu-Ishikawa E, Hotta Y. Isolation of Drosophila genomic clones homologous to the eel sodium channel gene. Proc Jpn Acad, B. 1987;63:284–288. [Google Scholar]
- Oliveira EE, Du Y, Nomura Y, Dong K. A residue in the transmembrane segment 6 of domain I in insect and mammalian sodium channels regulate differential sensitivities to pyrethroid insecticides. Neurotoxicology. 2013;38C:42–50. doi: 10.1016/j.neuro.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson RO, Liu ZQ, Nomura Y, Song WZ, Dong K. Molecular and functional characterization of voltage-gated sodium channel variants from Drosophila melanogaster. Insect Biochem Mol Biol. 2008;38:604–610. doi: 10.1016/j.ibmb.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palladino MJ, Keegan LP, O'Connell MA, Reenan RA. A-to-I pre-mRNA editing in Drosophila is primarily involved in adult nervous system function and integrity. Cell. 2000;102:437–449. doi: 10.1016/s0092-8674(00)00049-0. [DOI] [PubMed] [Google Scholar]
- Park JW, Parisky K, Celotto AM, Reenan RA, Graveley BR. Identification of alternative splicing regulators by RNA interference in Drosophila. Proc Natl Acad Sci U S A. 2004;101:15974–15979. doi: 10.1073/pnas.0407004101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y, Taylor MF, Feyereisen R. Voltage-gated sodium channel genes hscp and hDSC1 of Heliothis virescens F. genomic organization. Insect Mol Biol. 1999;8:161–170. doi: 10.1046/j.1365-2583.1999.820161.x. [DOI] [PubMed] [Google Scholar]
- Parker L, Howlett IC, Rusan ZM, Tanouye MA. Seizure and epilepsy: studies of seizure disorders in Drosophila. Int Rev Neurobiol. 2011;99:1–21. doi: 10.1016/B978-0-12-387003-2.00001-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauron D, Barhanin J, Amichot M, Pralavorio M, Berge JB, Lazdunski M. Pyrethroid receptor in the insect sodium channel: alteration of its properties in pyrethroid-resistant flies. Biochemistry. 1989;28:1673–1677. [Google Scholar]
- Payandeh J, Scheuer T, Zheng N, Catterall WA. The crystal structure of a voltage-gated sodium channel. Nature. 2011;475:353–358. doi: 10.1038/nature10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng F, Mellor IR, Williamson MS, Davies TG, Field LM, Usherwood PN. Single channel study of deltamethrin interactions with wild-type and mutated rat NaV1.2 sodium channels expressed in Xenopus oocytes. Neurotoxicology. 2009;30:358–367. doi: 10.1016/j.neuro.2009.02.016. [DOI] [PubMed] [Google Scholar]
- Plummer NW, McBurney MW, Meisler MH. Alternative splicing of the sodium channel SCN8A predicts a truncated two-domain protein in fetal brain and non-neuronal cells. J Biol Chem. 1997;272:24008–24015. doi: 10.1074/jbc.272.38.24008. [DOI] [PubMed] [Google Scholar]
- Ramaswami M, Tanouye MA. Two sodium-channel genes in Drosophila: implications for channel diversity. Proc Nat Acad Sci. 1989;86:2079–2082. doi: 10.1073/pnas.86.6.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reenan RA, Hanrahan CJ, Ganetzky B. The mle(napts) RNA helicase mutation in drosophila results in a splicing catastrophe of the para Na+ channel transcript in a region of RNA editing. Neuron. 2000;25:139–149. doi: 10.1016/s0896-6273(00)80878-8. [DOI] [PubMed] [Google Scholar]
- Rieder LE, Staber CJ, Hoopengardner B, Reenan RA. Tertiary structural elements determine the extent and specificity of messenger RNA editing. Nat Commun. 2013;4:2232. doi: 10.1038/ncomms3232. [DOI] [PubMed] [Google Scholar]
- Rinkevich FD, Du Y, Dong K. Diversity and convergence of sodium channel mutations involved in tesistance to pyrethroids. Pestic Biochem Physiol. 2013;106:93–100. doi: 10.1016/j.pestbp.2013.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossignol DP. Reduction in number of nerve membrane sodium channels in pyrethroid resistant house flies. Pestic Biochem Physiol. 1988;32:146–152. [Google Scholar]
- Salkoff L, Butler A, Wei A, Scavarda N, Giffen K, Ifune C, Goodman R, Mandel G. Genomic organization and deduced amino acid sequence of a putative sodium channel gene in Drosophila. Science. 1987;237:744–749. doi: 10.1126/science.2441469. [DOI] [PubMed] [Google Scholar]
- Seeburg PH. RNA helicase participates in the editing game. Neuron. 2000;25:261–263. doi: 10.1016/s0896-6273(00)80891-0. [DOI] [PubMed] [Google Scholar]
- Shao YM, Dong K, Tang ZH, Zhang CX. Molecular characterization of a sodium channel gene from the Silkworm Bombyx mori. Insect Biochem Mol Biol. 2009;39:145–151. doi: 10.1016/j.ibmb.2008.10.005. [DOI] [PubMed] [Google Scholar]
- Silver KS, Du Y, Nomura Y, Oliveira EE, Salgado VL, Zhorov BS, Dong K. Voltage-gated sodium channels as insecticide target sites. Pestic Biochem Physiol. 2014 doi: 10.1016/B978-0-12-417010-0.00005-7. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver KS, Nomura Y, Salgado VL, Dong K. Role of the sixth transmembrane segment of domain IV of the cockroach sodium channel in the action of sodium channel blocker insecticides. Neurotoxicology. 2009;30:613–621. doi: 10.1016/j.neuro.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver KS, Song W, Nomura Y, Salgado VL, Dong K. Mechanism of action of sodium channel blocker insecticides (SCBIs) on insect sodium channels. Pestic Biochem Physiol. 2010;97:87–92. doi: 10.1016/j.pestbp.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner WS, Dennis PA, Li JP, Quistad GB. Identification of insecticidal peptides from venom of the trap-door spider, Aptostichus schlingeri (Ctenizidae). Toxicon. 1992;30:1043–1050. doi: 10.1016/0041-0101(92)90049-b. [DOI] [PubMed] [Google Scholar]
- Smith RD, Goldin AL. Phosphorylation of brain sodium channels in the I--II linker modulates channel function in Xenopus oocytes. J Neurosci. 1996;16:1965–1974. doi: 10.1523/JNEUROSCI.16-06-01965.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RD, Goldin AL. Phosphorylation at a single site in the rat brain sodium channel is necessary and sufficient for current reduction by protein kinase A. J Neurosci. 1997;17:6086–6093. doi: 10.1523/JNEUROSCI.17-16-06086.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RD, Goldin AL. Potentiation of rat brain sodium channel currents by PKA in Xenopus oocytes involves the I-II linker. Am J Physiol Cell Physiol. 2000;278:C638–645. doi: 10.1152/ajpcell.2000.278.4.C638. [DOI] [PubMed] [Google Scholar]
- Smith TJ, Lee SH, Ingles PJ, Knipple DC, Soderlund DM. The L1014F point mutation in the house fly Vssc1 sodium channel confers knockdown resistance to pyrethroids. Insect Biochem Mol Biol. 1997;27:807–812. doi: 10.1016/s0965-1748(97)00065-9. [DOI] [PubMed] [Google Scholar]
- Smith TJ, Soderlund DM. Action of the pyrethroid insecticide cypermethrin on rat brain IIa sodium channels expressed in xenopus oocytes. Neurotoxicology. 1998;19:823–832. [PubMed] [Google Scholar]
- Soderlund DM. Sodium channels. In: Gilbert LI, Iatrou K, Gill SS, editors. Comprehensive molecular insect science. Elsevier; Amsterdam: 2005. pp. 1–24. [Google Scholar]
- Soderlund DM. Molecular mechanisms of pyrethroid insecticide neurotoxicity: recent advances. Arch Toxicol. 2012;86:165–181. doi: 10.1007/s00204-011-0726-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderlund DM, Bloomquist JR. Molecular Mechanisms of Insecticide Resistance. In: Roush RT, Tabashnik BE, editors. Pesticide Resistance in Arthropods. Chapman and Hall; New York and London: 1990. p. 58. [Google Scholar]
- Song JH, Narahashi T. Differential effects of the pyrethroid tetramethrin on tetrodotoxin-sensitive and tetrodotoxin-resistant single sodium channels. Brain Res. 1996;712:258–264. doi: 10.1016/0006-8993(95)01449-7. [DOI] [PubMed] [Google Scholar]
- Song W, Du Y, Liu Z, Luo N, Turkov M, Gordon D, Gurevitz M, Goldin AL, Dong K. Substitutions in the domain III voltage-sensing module enhance the sensitivity of an insect sodium channel to a scorpion beta-toxin. J Biol Chem. 2011;286:15781–15788. doi: 10.1074/jbc.M110.217000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Liu Z, Dong K. Molecular basis of differential sensitivity of insect sodium channels to DCJW, a bioactive metabolite of the oxadiazine insecticide indoxacarb. Neurotoxicology. 2006;27:237–244. doi: 10.1016/j.neuro.2005.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Liu Z, Tan J, Nomura Y, Dong K. RNA editing generates tissue-specific sodium channels with distinct gating properties. J Biol Chem. 2004;279:32554–32561. doi: 10.1074/jbc.M402392200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda S, Igaki C, Ashfaq M, Tsumuki H. Pyrethroid-resistant diamondback moth expresses alternatively spliced sodium channel transcripts with and without T929I mutation. Insect Biochem Mol Biol. 2006;36:904–910. doi: 10.1016/j.ibmb.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Sonoda S, Tsukahara Y, Ashfaq M, Tsumuki H. Genomic organization of the para-sodium channel alpha-subunit genes from the pyrethroid-resistant and -susceptible strains of the diamondback moth. Arch Insect Biochem Physiol. 2008;69:1–12. doi: 10.1002/arch.20246. [DOI] [PubMed] [Google Scholar]
- Stafstrom CE. Persistent sodium current and its role in epilepsy. Epilepsy Curr. 2007;7:15–22. doi: 10.1111/j.1535-7511.2007.00156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strugatsky D, Zilberberg N, Stankiewicz M, Ilan N, Turkov M, Cohen L, Pelhate M, Gilles N, Gordon D, Gurevitz M. Genetic polymorphism and expression of a highly potent scorpion depressant toxin enable refinement of the effects on insect Na channels and illuminate the key role of Asn-58. Biochemistry. 2005;44:9179–9187. doi: 10.1021/bi050235t. [DOI] [PubMed] [Google Scholar]
- Sun L, Gilligan J, Staber C, Schutte RJ, Nguyen V, O'Dowd DK, Reenan R. A knock-in model of human epilepsy in Drosophila reveals a novel cellular mechanism associated with heat-induced seizure. J Neurosci. 2012;32:14145–14155. doi: 10.1523/JNEUROSCI.2932-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, Liu Z, Nomura Y, Goldin AL, Dong K. Alternative splicing of an insect sodium channel gene generates pharmacologically distinct sodium channels. J Neurosci. 2002a;22:5300–5309. doi: 10.1523/JNEUROSCI.22-13-05300.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, Liu Z, Tsai TD, Valles SM, Goldin AL, Dong K. Novel sodium channel gene mutations in Blattella germanica reduce the sensitivity of expressed channels to deltamethrin. Insect Biochem Mol Biol. 2002b;32:445–454. doi: 10.1016/s0965-1748(01)00122-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, Liu Z, Wang R, Huang ZY, Chen AC, Gurevitz M, Dong K. Identification of amino acid residues in the insect sodium channel critical for pyrethroid binding. Mol Pharmacol. 2005;67:513–522. doi: 10.1124/mol.104.006205. [DOI] [PubMed] [Google Scholar]
- Tan J, Soderlund DM. Human and rat Nav1.3 voltage-gated sodium channels differ in inactivation properties and sensitivity to the pyrethroid insecticide tefluthrin. Neurotoxicology. 2009;30:81–89. doi: 10.1016/j.neuro.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, Soderlund DM. Divergent actions of the pyrethroid insecticides S-bioallethrin, tefluthrin, and deltamethrin on rat Nav1.6 sodium channels. Toxicol Appl Pharmacol. 2010;247:229–237. doi: 10.1016/j.taap.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, Soderlund DM. Actions of Tefluthrin on Rat Nav1.7 Voltage-Gated Sodium Channels Expressed in Xenopus Oocytes. Pestic Biochem Physiol. 2011;101:21–26. doi: 10.1016/j.pestbp.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatebayashi H, Narahashi T. Differential mechanism of action of the pyrethroid tetramethrin on tetrodotoxin-sensitive and tetrodotoxin-resistant sodium channels. J Pharmacol Exp Ther. 1994;270:595–603. [PubMed] [Google Scholar]
- Taylor CP. Na+ currents that fail to inactivate. Trends Neurosci. 1993;16:455–460. doi: 10.1016/0166-2236(93)90077-y. [DOI] [PubMed] [Google Scholar]
- Thackeray JR, Ganetzky B. Developmentally regulated alternative splicing generates a complex array of Drosophila para sodium channel isoforms. J Neurosci. 1994;14:2569–2578. doi: 10.1523/JNEUROSCI.14-05-02569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thackeray JR, Ganetzky B. Conserved alternative splicing patterns and splicing signals in the Drosophila sodium channel gene para. Genetics. 1995;141:203–214. doi: 10.1093/genetics/141.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonov DB, Zhorov BS. Modeling P-loops domain of sodium channel: homology with potassium channels and interaction with ligands. Biophys J. 2005a;88:184–197. doi: 10.1529/biophysj.104.048173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonov DB, Zhorov BS. Sodium channel activators: model of binding inside the pore and a possible mechanism of action. FEBS letters. 2005b;579:4207–4212. doi: 10.1016/j.febslet.2005.07.017. [DOI] [PubMed] [Google Scholar]
- Tikhonov DB, Zhorov BS. Sodium channels: ionic model of slow inactivation and state-dependent drug binding. Biophys J. 2007;93:1557–1570. doi: 10.1529/biophysj.106.100248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usherwood PN, Davies TG, Mellor IR, O'Reilly AO, Peng F, Vais H, Khambay BP, Field LM, Williamson MS. Mutations in DIIS5 and the DIIS4-S5 linker of Drosophila melanogaster sodium channel define binding domains for pyrethroids and DDT. FEBS Lett. 2007;581:5485–5492. doi: 10.1016/j.febslet.2007.10.057. [DOI] [PubMed] [Google Scholar]
- Usherwood PNR, Vais H, Khambay BPS, Davies TGE, Williamson MS. Sensitivity of the Drosophila para sodium channel to DDT is not lowered by the super-kdr mutation M918T on the IIS4-S5 linker that profoundly reduces sensitivity to permethrin and deltamethrin. FEBS Lett. 2005;579:6317–6325. doi: 10.1016/j.febslet.2005.09.096. [DOI] [PubMed] [Google Scholar]
- Vais H, Atkinson S, Eldursi N, Devonshire AL, Williamson MS, Usherwood PN. A single amino acid change makes a rat neuronal sodium channel highly sensitive to pyrethroid insecticides. FEBS Lett. 2000;470:135–138. doi: 10.1016/s0014-5793(00)01305-3. [DOI] [PubMed] [Google Scholar]
- Vais H, Atkinson S, Pluteanu F, Goodson SJ, Devonshire AL, Williamson MS, Usherwood PN. Mutations of the para sodium channel of Drosophila melanogaster identify putative binding sites for pyrethroids. Mol Pharmacol. 2003;64:914–922. doi: 10.1124/mol.64.4.914. [DOI] [PubMed] [Google Scholar]
- Vais H, Williamson MS, Devonshire AL, Usherwood PN. The molecular interactions of pyrethroid insecticides with insect and mammalian sodium channels. Pest Manag Sci. 2001;57:877–888. doi: 10.1002/ps.392. [DOI] [PubMed] [Google Scholar]
- Vais H, Williamson MS, Hick CA, Eldursi N, Devonshire AL, Usherwood PN. Functional analysis of a rat sodium channel carrying a mutation for insect knock-down resistance (kdr) to pyrethroids. FEBS Lett. 1997;413:327–332. doi: 10.1016/s0014-5793(97)00931-9. [DOI] [PubMed] [Google Scholar]
- von Stein RT, Soderlund DM. Role of the local anesthetic receptor in the state-dependent inhibition of voltage-gated sodium channels by the insecticide metaflumizone. Mol Pharmacol. 2012;81:366–374. doi: 10.1124/mol.111.075283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Nomura Y, Du Y, Dong K. Differential effects of TipE and a TipE-homologous protein on modulation of gating properties of sodium channels from Drosophila melanogaster. PLoS One. 2013;8:e67551. doi: 10.1371/journal.pone.0067551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Huang ZY, Dong K. Molecular characterization of an arachnid sodium channel gene from the varroa mite (Varroa destructor). Insect Biochem Mol Biol. 2003;33:733–739. doi: 10.1016/s0965-1748(03)00068-7. [DOI] [PubMed] [Google Scholar]
- Wang SY, Barile M, Wang GK. A phenylalanine residue at segment D3-S6 in Nav1.4 voltage-gated Na+ channels is critical for pyrethroid action. Mol Pharmacol. 2001;60:620–628. [PubMed] [Google Scholar]
- Wang SY, Wang GK. Voltage-gated sodium channels as primary targets of diverse lipid-soluble neurotoxins. Cellular signalling. 2003;15:151–159. doi: 10.1016/s0898-6568(02)00085-2. [DOI] [PubMed] [Google Scholar]
- Wang XJ, Reynolds ER, Deak P, Hall LM. The seizure locus encodes the Drosophila homolog of the HERG potassium channel. J Neurosci. 1997;17:882–890. doi: 10.1523/JNEUROSCI.17-03-00882.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warmke JW, Reenan RAG, Wang PY, Qian S, Arena JP, Wang JX, Wunderler D, Liu K, Kaczorowski GJ, VanderPloeg LHT, Ganetzky B, Cohen CJ. Functional expression of Drosophila para sodium channels - Modulation by the membrane protein TipE and toxin pharmacology. J Gen Physiol. 1997;110:119–133. doi: 10.1085/jgp.110.2.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West JW, Numann R, Murphy BJ, Scheuer T, Catterall WA. A phosphorylation site in the Na+ channel required for modulation by protein kinase C. Science. 1991;254:866–868. doi: 10.1126/science.1658937. [DOI] [PubMed] [Google Scholar]
- Williamson MS, Martinez-Torres D, Hick CA, Devonshire AL. Identification of mutations in the housefly para-type sodium channel gene associated with knockdown resistance (kdr) to pyrethroid insecticides. Mol Gen Genet. 1996;252:51–60. doi: 10.1007/BF02173204. [DOI] [PubMed] [Google Scholar]
- Wing KD, Andaloro JT, McCann SF, Salgado VL. Indoxacarb and the sodium channel blocker insecticides: chemistry, physiology and biology in insects. Elsevier; New York: 2005. [Google Scholar]
- Xu Q, Zhang L, Li T, Zhang L, He L, Dong K, Liu N. Evolutionary adaptation of the amino acid and codon usage of the mosquito sodium channel following insecticide selection in the field mosquitoes. PloS one. 2012;7:e47609. doi: 10.1371/journal.pone.0047609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon KS, Kwon DH, Strycharz JP, Hollingsworth CS, Lee SH, Clark JM. Biochemical and molecular analysis of deltamethrin resistance in the common bed bug (Hemiptera: Cimicidae). J Med Entomol. 2008;45:1092–1101. doi: 10.1603/0022-2585(2008)45[1092:bamaod]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Zhang T, Liu Z, Song W, Du Y, Dong K. Molecular characterization and functional expression of the DSC1 channel. Insect Biochem Mol Biol. 2011;41:451–458. doi: 10.1016/j.ibmb.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Park Y, Adams ME. Functional and evolutionary consequences of pyrethroid resistance mutations in S6 transmembrane segments of a voltage-gated sodium channel. Biochem Biophys Res Commun. 2000;278:516–521. doi: 10.1006/bbrc.2000.3832. [DOI] [PubMed] [Google Scholar]
- Zhorov BS, Tikhonov DB. Potassium, sodium, calcium and glutamate-gated channels: pore architecture and ligand action. J. Neurochem. 2004;88:782–799. doi: 10.1111/j.1471-4159.2004.02261.x. [DOI] [PubMed] [Google Scholar]
- Zhou W, Chung I, Liu Z, Goldin AL, Dong K. A voltage-gated calcium-selective channel encoded by a sodium channel-like gene. Neuron. 2004;42:101–112. doi: 10.1016/s0896-6273(04)00148-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlotkin E. The insect voltage-gated sodium channel as target of insecticides. Annu Rev Entomol. 1999;44:429–455. doi: 10.1146/annurev.ento.44.1.429. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.