Significance
Obligate plant-symbiotic, arbuscular mycorrhizal fungi (AMF) are major drivers of terrestrial ecosystems and host enigmatic Mollicutes-related endobacteria (MRE) in their cytoplasm. The genome analysis of a MRE living in the AMF Dentiscutata heterogama revealed it to represent a previously unidentified bacterial lineage of Mycoplasma-related species. DhMRE shows strongly reduced metabolic capacity and underwent trans-kingdom gene transfer: its genome codes for an arsenal of eukaryotic-like putative effector proteins, with nuclear encoded homologues in AMF and Mortierella. The MRE-fungus (-plant) association probably evolved in ancestors of Glomeromycota and Mucoromycotina. This calls for a targeted search for ancient effector proteins that play crucial roles in the MRE interaction with fungal hosts, and putatively also with plants.
Keywords: arbuscular mycorrhizal fungi, endobacteria, intracellular symbiont, Mycoplasma, horizontal gene transfer
Abstract
For more than 450 million years, arbuscular mycorrhizal fungi (AMF) have formed intimate, mutualistic symbioses with the vast majority of land plants and are major drivers in almost all terrestrial ecosystems. The obligate plant-symbiotic AMF host additional symbionts, so-called Mollicutes-related endobacteria (MRE). To uncover putative functional roles of these widespread but yet enigmatic MRE, we sequenced the genome of DhMRE living in the AMF Dentiscutata heterogama. Multilocus phylogenetic analyses showed that MRE form a previously unidentified lineage sister to the hominis group of Mycoplasma species. DhMRE possesses a strongly reduced metabolic capacity with 55% of the proteins having unknown function, which reflects unique adaptations to an intracellular lifestyle. We found evidence for transkingdom gene transfer between MRE and their AMF host. At least 27 annotated DhMRE proteins show similarities to nuclear-encoded proteins of the AMF Rhizophagus irregularis, which itself lacks MRE. Nuclear-encoded homologs could moreover be identified for another AMF, Gigaspora margarita, and surprisingly, also the non-AMF Mortierella verticillata. Our data indicate a possible origin of the MRE-fungus association in ancestors of the Glomeromycota and Mucoromycotina. The DhMRE genome encodes an arsenal of putative regulatory proteins with eukaryotic-like domains, some of them encoded in putative genomic islands. MRE are highly interesting candidates to study the evolution and interactions between an ancient, obligate endosymbiotic prokaryote with its obligate plant-symbiotic fungal host. Our data moreover may be used for further targeted searches for ancient effector-like proteins that may be key components in the regulation of the arbuscular mycorrhiza symbiosis.
Soil fungi from the Glomeromycota, which form arbuscular mycorrhiza (AM) symbioses with the vast majority of land plants, are major players in terrestrial ecosystems (1). This symbiosis originated more than 450 million years ago (2) and represents an impressive example of an ancient and stable mutualistic association: AM fungi (AMF) efficiently explore the soil with their fine mycelium and supply plants with inorganic nutrients and water, whereas the plants provide carbohydrates derived from photosynthesis in above-ground organs.
Interestingly, AMF, as biotrophic and obligate plant symbionts, themselves host additional endosymbionts in their cytoplasm, biotrophic endobacteria (3, 4). These endobacteria were electron-microscopically described in the 1970s as bacterium-like organisms (5). Two types were later defined, the first one being found only in members of the family Gigasporaceae. This Gram-negative bacterium was named Candidatus Glomeribacter gigasporarum (CaGg) and is related to Burkholderia (4). The second and much more widespread type represents the only known fungal endobacteria belonging to the Mollicutes (“Mollicutes-related endobacteria”; MRE), although related endobacteria (e.g., Mycoplasma and Phytoplasma species) are widespread as biotrophic parasites of humans, mammals, reptiles, fishes, arthropods, or plants. MRE were frequently detected in the intraradical and extraradical mycelium and in spores of AMF; however, they could never be detected free-living (3). Strikingly, MRE have recently been demonstrated to also occur in several non-AMF species from the genus Endogone (Mucoromycotina), where some members are also plant symbionts (6).
MRE are associated with all major phylogenetic lineages of AMF studied so far and, thus, indirectly also with more than 80% of all land plants. They are coccoid, located in the cytoplasm without a surrounding host-membrane, and appear to possess a Gram-positive cell wall, which is surprising because of the phylogenetic relationship with cell wall-lacking Mollicutes (3). During their long-lasting coevolution, MRE have formed distinct, monophyletic evolutionary lineages within their fungal hosts, with a 16S rRNA gene (16S) sequence divergence of up to 20% (3, 7).
We hypothesized that MRE play an important biological role in AM, consistent with the observation that they have been maintained as ancient endosymbionts in major evolutionary AMF lineages that separated hundreds of million years ago. To obtain hints for such roles and to shed light on MRE evolution, we analyzed the genome of a MRE colonizing the AMF Dentiscutata heterogama FL654 (syn.: Scutellospora heterogama; Gigasporaceae), termed DhMRE. Only three genomes of fungal endobacteria have been published so far: from CaGg, which colonize some AMF from the Gigasporaceae (8); from Burkholderia rhizoxinica, the endobacterium of Rhizopus microsporus, which participates in the production of rhizoxin (a potent antimitotic agent that acts as a virulence factor) (9); and from a betaproteobacterial endosymbiont associated with Mortierella elongata (10).
We present the annotation of a MRE genome draft, the phylogenetic placement of MRE based on a multilocus analysis, the evolutionary implications about the origin of MRE-fungus (-plant) associations, and evidence for transkingdom horizontal gene transfer (HGT). Moreover, the genome presents genes coding for proteins with likely regulatory functions in the interaction between this obligate symbiotic prokaryote and its obligate plant-symbiotic fungal host.
Results
General Features of the DhMRE Genome.
To study the genomic organization and gene repertoire of an MRE living in AMF, we isolated the DNA of the endobacterial population from spores of D. heterogama FL654, which harbors MRE but not CaGg (SI Appendix, Fig. S1). Following Illumina sequencing and PCR based gap-closing approaches, three DhMRE genomic scaffolds could be defined. One is circular, with a size of 648,879 bp and 34.1% GC content (scaffold A), the second (scaffold B) is 60,375 bp in size with a GC content of 32.3%, and the third (scaffold C) is a putative plasmid of 3,779 bp and 34.1% GC content (SI Appendix, Fig. S2 and Tables S1 and S2). Tetranucleotide frequencies analyses validated the origin of scaffolds A and B (SI Appendix, Table S3). The total size of the DhMRE genome draft assembly is 713,033 bp, with an estimated sequence coverage of approximately 130-fold.
Most of the studied AMF possess two major phylotypes of MRE 16S rRNA genes co-occurring in the same spore (3, 7). The analysis of 16S rRNA gene PCR-amplicon derived clone libraries from D. heterogama also revealed these two major phylotypes (SI Appendix, Fig. S3). Phylotype I was more abundant (83%) than phylotype II (17%). The dominance of phylotype I in clone libraries rose to 96% after applying the genomic DNA extraction protocol developed for DhMRE (SI Appendix, Table S4), explaining why phylotype I was almost exclusively identified in the Illumina dataset (SI Appendix, Fig. S4). We speculate that the phylotype I hosting MRE might possess a less rigid cell envelope leading to digestion of its DNA in the DNase treatment applied. To ensure that reads from different phylotypes were not coassembled, we performed a QualitySNPng analysis (11) on the contig with the 16S rRNA gene (which is organized in a split rRNA operon). Only when relaxing the parameters (“minimal percentage of reads per allele” set to 15%), four haplotypes were discovered, but their very low read composition does not support the existence of a second phylotype in the assembled sequences.
Thus, the genome assembly presented here carries only one of the two major MRE 16S phylotypes and, therefore, putatively only one of the corresponding MRE genomes found in D. heterogama. This reduction in complexity enabled us to close most of the genome assembly.
Evolutionary Placement.
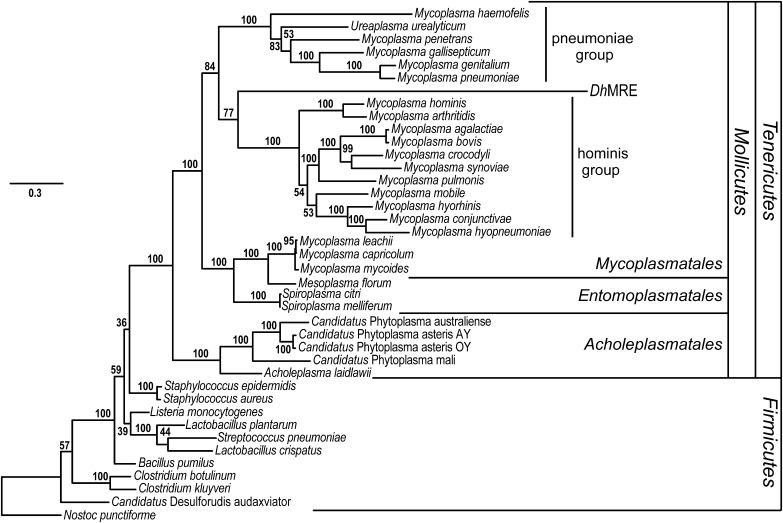
To determine the phylogenetic placement of DhMRE as a representative of the monophyletic MRE clade (3), a maximum likelihood phylogenetic analysis of amino acid sequences from 10 housekeeping genes from 38 bacteria was conducted (listed in SI Appendix, Table S5). The MRE lineage was firmly placed in the Mycoplasmatales (Fig. 1), most likely being closest related to the hominis group of Mycoplasma species.
Fig. 1.
Phylogenetic placement of DhMRE inferred by maximum likelihood phylogenetic analysis. For tree construction, 10 housekeeping genes from 28 Tenericutes and 10 Firmicutes bacteria species were used (listed in SI Appendix, Table S5). The phylogenetic analysis shows that DhMRE is placed in the order Mycoplasmatales. The pneumoniae and hominis mycoplasma groups are indicated in the tree. Nostoc punctiforme was used as outgroup. Bootstrap values are indicated at the branches of the tree (100 replicates).
Transkingdom Horizontal Gene Transfer Between DhMRE and the Fungal Host.
The analysis of the annotated genes has revealed that several DhMRE proteins share similarity with proteins coded by the nuclear genome of Rhizophagus irregularis, which is the only AMF with a sequenced genome (12, 13). All evaluations to date indicate that this fungus does not host endobacteria and separated evolutionary from D. heterogama more than 200 million years ago (3, 14). In total, we identified 17 proteins in scaffold A and 10 proteins in scaffold B showing best BLAST hits with an identity higher than 30% and query coverage above 35% against R. irregularis proteins. Among the 27 proteins, 21 (78%) have predicted functional domains, which could be related to bacteria-host association: eight are nonspecific tyrosine-kinases like proteins (TKLs; two of them located in putative genomic islands), seven contain leucine rich repeats (LRR) domains normally functioning in protein–protein interactions, and six contain an AIG1 domain, usually found in plant proteins thought to be involved in plant resistance to bacteria (15) (SI Appendix, Table S6).
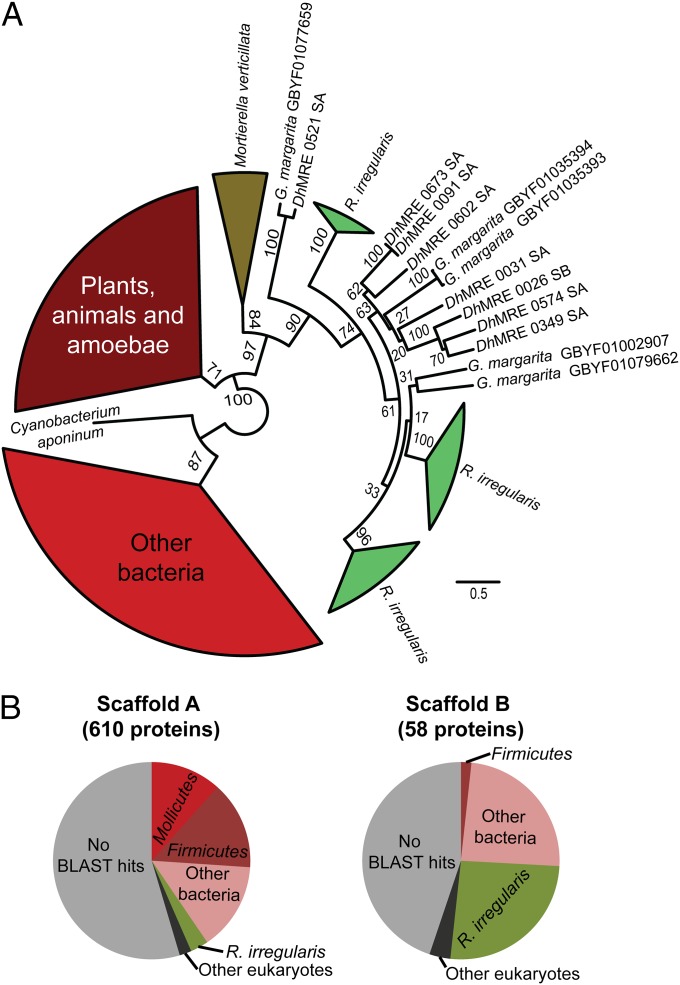
We performed phylogenetic analyses to identify the putative origin of the DhMRE proteins sharing similarity with nuclear-encoded R. irregularis proteins, including homologous nuclear-encoded proteins from Gigaspora margarita BEG34, an AMF not hosting MRE (Fig. 2 and SI Appendix, Fig. S5). The phylogenetic analysis of the DhMRE TKLs show them to robustly cluster together with protein kinases from the AMF R. irregularis, G. margarita, and also from the non-AMF Mortierella verticillata (Mortierellales). This clade is clearly separated from other prokaryotic and eukaryotic protein kinases (Fig. 2 and SI Appendix, Fig. S5).
Fig. 2.
Maximum likelihood phylogenetic reconstruction of the protein kinases candidates for horizontal gene transfer (A) and taxonomic composition of the DhMRE scaffolds according to sequence similarity (B). BLASTP-based affiliation was based on the best-hit with a cutoff value of e−03. For each protein, BLASTP hits in the nonredundant database were recovered by using the program Blast2GO, and the classification of the corresponding organism was extracted according to NCBI taxonomy. SA, scaffold A; SB, scaffold B.
The other main group of DhMRE proteins similar to nuclear-encoded proteins of R. irregularis possesses an AIG1 domain. DhMRE, Rhizophagus and Gigaspora AIG1 domains also form a monophyletic clade. The only other AIG1 domains that also clustered in this clade are from the soil amoeba Dictyostelium fasciculatum. According to the phylogenetic analysis, the ancestor of these domains is eukaryotic, but not fungal (SI Appendix, Fig. S6).
Proteins with Eukaryotic Domains.
Analysis of the DhMRE genome revealed an exceptionally large arsenal of proteins (including some with homologs to nuclear-encoded R. irregularis proteins) with eukaryotic-like domains that are likely to interfere with eukaryotic cellular functions. Several of the coding genes are located in putative genomic islands and/or near transposases. At least 29 proteins carry domains predominantly found in eukaryotic proteins (4% of the total proteome). The most abundant eukaryotic domains are LRR (SI Appendix, Fig. S7), which are versatile binding motifs involved in protein–protein interactions and in the interaction of microbes with host cells (16). One gene in scaffold B codes for a protein with a heterokaryon incompatibility domain (HET domain), which is part of the nonself recognition systems in ascomycetes (17).
The DhMRE proteome also contains proteins related to the eukaryotic ubiquitination system that controls the degradation of proteins by the proteasome and may interfere with host transcriptional gene activation or repression, nuclear transport and enzyme activity (SI Appendix, Fig. S7). We found three proteases with a small ubiquitin-like modifier (SUMO)/sentrin domain, reversibly regulating the SUMO modification pathway in eukaryotes (18), one of them predicted to be secreted. Another protein predicted to be secreted carries an ovarian tumor (OTU)-like cysteine protease domain, which is also putatively involved in the ubiquitination system.
Overall, DhMRE encodes a large repertoire of proteins with eukaryotic domains, some of them already characterized in prokaryotic effector proteins. Moreover, a subset of them has probably gone through HGT with the host (SI Appendix, Table S6).
Functional Annotation of Predicted Protein-Coding Genes.
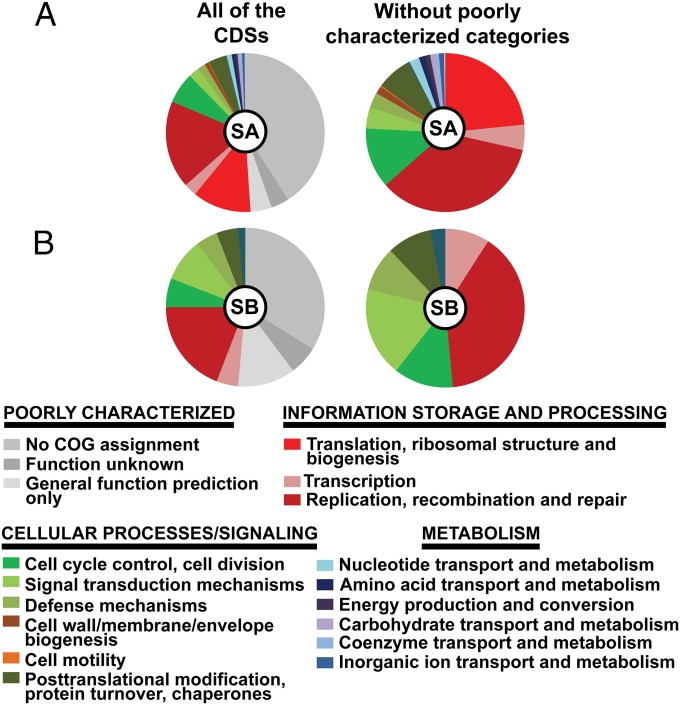
The gene repertoire of DhMRE comprises 90 of the 100 essential cluster of orthologous groups (COG) conserved in 99% of bacteria (SI Appendix, Table S7) (19). The COG functional categories with the most representatives in scaffold A are category L, involved in DNA replication, recombination, and repair and category J, involved in translation and ribosomal structure biogenesis, mostly represented by the large number of ribosomal proteins and tRNA synthetases (Fig. 3). Fifty-five percent of the DhMRE proteins are annotated as proteins of unknown function (PUF), with no significant similarity (<30%) in the SwissProt and TrEMBL databases. Thus, the reduced DhMRE genome encodes for a large number of unique proteins with unknown function.
Fig. 3.
Functional classification of annotated protein-coding genes for the scaffolds A (A) and B (B). The functional categorization of each protein-coding gene was classified according to the cluster of orthologs groups (COG assignments). SA, scaffold A; SB, scaffold B.
We could not identify a canonical origin of replication (oriC), the gene dnaA coding for the DNA replication initiator protein DnaA, or proteins related to cell wall biosynthesis or bacterial cell division (e.g., FtsZ). To validate these findings, we performed a narrowed protein sequence similarity search with HMMER (20) against a full D. heterogama spore-derived metagenome (including all contaminant sequences) peptides database. There was no match to any Mycoplasma-related sequences, indicating that these genes are absent in the DhMRE genome draft.
In summary, DhMRE possesses some general features of endosymbionts, like a reduced genome, strict dependence of the host, and the absence of the proteins DnaA and FtsZ. Surprisingly, we could not identify any protein related to the cell wall biosynthesis pathway, although a Gram-positive cell wall was indicated by electron microscopy.
DhMRE Has Strongly Reduced Metabolic Capacities.
The number of DhMRE proteins assigned to the COG of metabolic categories is only 29 (5.1%; Fig. 3) and, thus, the metabolic capacity of DhMRE is extremely reduced.
DhMRE is not enclosed by a host membrane and has direct access to nutrients from the host cytoplasm. However, we found only a low number of transporter genes. Two code for proteins related to ATP-binding cassette transporters and three for a xenobiotics-transporting type of ATPase protein that often mediate multidrug resistance (21). We annotated a putative arginine-ornithine antiporter, which could play a key role in DhMRE nutrition. This antiporter may import arginine, which is a prominent storage and transport amino acid in AMF (22). However, we could not identify any of the known genes involved in the catabolism of arginine.
To compare the metabolic capacity of DhMRE with other obligate symbionts, we performed a pathway completion analysis (number of reactions present in a given organism for a pathway in relation to the total number of reactions in the same pathway defined in the KEGG databases) and a hierarchical clustering analysis. DhMRE obtained low pathway completion values. As a consequence of convergent evolution, the hierarchical clustering groups DhMRE with obligate insect endosymbionts with highly reduced metabolic capacities, such as Candidatus Carsonella ruddii, Candidatus Sulcia muelleri, and Candidatus Zinderia insecticola (SI Appendix, Fig. S8).
In conclusion, DhMRE has strongly reduced metabolic capacities and appears to fully depend on its host, but surprisingly only a small number of putative nutrient transporters could be annotated.
Discussion
The genome analysis of a member of the widespread obligate symbiotic MRE living within obligate plant-symbiotic AMF provide first insights, to our knowledge, into the evolution of MRE and interactions with their fungal host. The DhMRE genome revealed HGT events between the fungal host and MRE, an arsenal of proteins with eukaryotic domains that are likely involved in host–cell interaction and, despite the much-reduced genome, a large repertoire of unknown, unique proteins. Furthermore, DhMRE has extremely reduced metabolic capacities. This set of biotrophic adaptations raises new questions about the role of these endobacteria in the evolution of the fungal host and about transkingdom interactions of symbionts.
Evolution of DhMRE.
DhMRE belongs to a bacterial lineage closely related to the pneumoniae and hominis mycoplasma groups, which mainly occur as biotrophic extracellular parasites of animals (23). The DhMRE gene homologs we detected in the nuclear genome of M. verticillata (Mortierellales), together with the recent findings of MRE in Endogone (Endogonales) species (24), indicate that the MRE association with fungi was already present in a common ancestor of the Glomeromycota and Mucoromycotina (25).
The phylogenetic position of DhMRE in-between mycoplasma groups indicates that its ancestor had shifted from a eukaryotic, probably animal, host to a common ancestor of the Glomeromycota and Mycoromycotina, resulting in the ancient MRE-fungus association. Host shifts are not uncommon for Mycoplasma species, e.g., causing the Mycoplasma gallisepticum outbreak in wild house finches (26).
Although MRE are phylogenetically embedded in the Mycoplasmatales and neither bacteria from Mycoplasmatales nor from lineages branching earlier (Spiroplasma, Phytoplasma, Acholeplasma) possess a cell wall, MRE surprisingly present a structure that was interpreted as being a Gram-positive cell wall (3). However, the nature of the MRE cell wall was never experimentally confirmed and the genome data presented here indicate that this structure does not represent a prokaryotic murein sacculus.
Mechanisms Promoting DhMRE Evolution and Diversification.
The DhMRE draft genome lacks some genes for DNA repair, for example ung, coding for uracil-DNA glycosylase, which is in line with the suggestion that species of the Mycoplasmatales are constitutive mutators because of the lack of some components of the SOS response and DNA repair machinery (27, 28). High mutation rates also play an important role in adaptation processes and may have helped the last common ancestor of the MRE in its shift to a fungal host.
Another important mechanism for rapid evolution and adaptation in Mycoplasmatales is HGT (29, 30). It appears to be also a major driving force in MRE evolution. Transkingdom HGT events between MRE and AMF could have helped MRE to use acquired eukaryotic genes to exploit fungal metabolism or other functions, but it could also have been a key element enabling the fungi to control the plant host, as indicated by the fact of such genes being kept in the MRE-free R. irregularis. Horizontal gene transfer and evolution of eukaryotic-like proteins to effector proteins have been proposed to enable the intracellular life of L. pneumophila (31). A similar scenario may have happened in the ancestral MRE-AMF association, or in even more ancestral fungi, like such from the Mucoromycotina.
A major group of genes that underwent HGT between AMF and DhMRE encode proteins related to signal transduction, in particular TKLs. The origin of these TKLs is uncertain, but the sequencing of genomes from more AMF and Endogone- and Mortierella-related fungi and their MRE will clarify this point. The nuclear genome of R. irregularis showed TKL gene family expansions, and several TKL-containing proteins are strongly expressed in germinating spores and intraradical mycelia, indicating their functional relevance (12). The roles that the protein tyrosine kinases that underwent HGT, and which have homologs in the nuclear genomes of species from the Glomeromycota and Mortierellales, might play in signal transduction in early diverging fungal lineages becomes a highly interesting question.
The other major group of genes likely to have undergone HGT between AMF and DhMRE encode proteins with an AIG1 domain, which appears to be involved in plant resistance to bacteria. The surprising finding of AIG1 homologs existing in the soil-living amoeba D. fasciculatum makes it tempting to speculate about an early association between the ancestors of MRE and amoeba. In this context, it may be noted that it was suggested that L. pneumophila evolved some of its host-interaction systems in associations with amoeba (32).
Phylogenetic analyses indicated different possible scenarios for MRE HGT and evolution. Perhaps the most parsimonious scenario that we can envisage is an ancestral HGT from animals to biotrophic MRE ancestors, with subsequent HGT to the AMF host in the AM symbiosis, followed by a loss of the MRE symbionts in some AMF, such as certain members of the genera Rhizophagus and Gigaspora. The alternative of an ancestral HGT from animals or amoeba to the fungi followed by HGT from the fungi to the bacteria appears less likely. The sequencing of more MRE and AMF genomes will shed more light on this open question.
DhMRE also possess several genes encoding eukaryotic-like proteins carrying domains known from effector proteins. For example, SUMO and OTU proteases that could interfere with the host ubiquitination system. Different microbial effector proteins targeting the host ubiquitination system have been characterized, e.g., the effectors XopD, AvrXv4, and AvrBsT from Xanthomonas, and YopJ from Yersinia mimic SUMO protease activity, disrupting the regulation of the SUMO pathway and altering the activity or stability of the host proteins (33). DhMRE, or their fungal host, might use such proteins with eukaryotic domains to communicate with or regulate host processes.
In addition to the proteins described above, the proteome of DhMRE also contains a high proportion (1.5%) of proteins with LRR domains, similar to intracellular bacteria associated with amoebae (1.3%) (34), but higher than in Wolbachia or Rickettsia endobacteria (usually below 0.25%). These proteins are also candidates for playing a role in the interaction between DhMRE and the fungal host.
Taking all together, DhMRE genome sequencing has highlighted its plasticity, with both localized (mutations) and broad (genome reduction, HGT) processes altering its genetic content, in close coevolution with its hosts.
Overall Traits of the Small, Mycoplasma-Like DhMRE Genome.
The DhMRE genome shares a set of characteristics with small genomes of endobacteria, reflecting a high level of adaptation to an obligate intracellular lifestyle. For example, the absence of glycolysis, the tricarboxylic acid cycle, and a functional respiratory chain together indicate that DhMRE could generate energy only by substrate-level phosphorylation through the degradation of substrates obtained from its host, as the amoeba symbiont Candidatus Amoebophilus asiaticus (34). Another trait occasionally associated to intracellular symbionts (35) is the absence of the dnaA gene, which has been considered as an indication of a more direct control of DNA replication and regulation of endosymbionts propagation by the host.
In conclusion, the DhMRE genome possesses typical features of members of the Mollicutes, such as low GC content, a reduced genome size, high gene density, and 55% of its genes coding for PUF, which is even more than normally found in other species from the Mollicutes (36, 37). In our opinion, the occurrence of MRE in evolutionary lineages of AMF that separated more than 400 million years ago points to a mutualistic association, but it remains unknown how the MRE impact the AMF or, for that matter, the plant host. The large number of PUF as yet may hide important functions of the MRE in the symbiosis. We found transkingdom HGT of putatively regulatory genes of which homologs are retained in MRE-free AMF and, in general, a large arsenal of proteins with eukaryotic domains. Thus, our study may open the way for a more targeted search for effectors playing roles in AM. Taking the ancestral nature of the MRE–fungus association into account, the analysis of putative effector proteins with eukaryotic like domains and derived from HGT might lead to the identification of crucial effectors in bacteria-fungus-plant associations.
Material and Methods
Biological Material.
D. heterogama FL654 spores hosting MRE but not CaGg were used for genome sequencing. FL654 was produced in vitro in monoxenic culture on root organs by The Energy and Resources Institute. The spores were handled in a sterile manner under a laminar flow hood.
Endobacteria DNA Extraction for Sequencing.
For DhMRE genomic DNA preparation, a new protocol was established, attempting to enrich the DhMRE fraction and to remove AMF nuclear DNA (protocol details in SI Appendix, SI Materials and Methods). To obtain 50 ng of DNA, ∼300,000 spores from D. heterogama were used. The noted DNA isolation procedure was specifically designed with the aim to isolate intact small and rigid appearing endobacteria. Shearing and osmotic forces were used to rupture nuclei and mitochondria, the DNA of which was then removed by DNase digestion. In this process, we seem to have selectively enriched only one of the two MRE phylotypes known from previous Sanger sequencing approaches. In a semiquantitative approach to study this aspect, we constructed and analyzed clone libraries (details in SI Appendix, SI Materials and Methods).
DNA Sequencing and Sequence Assembly.
Illumina sequencing was performed by using the Illumina MiSeq platform at the Genomics Service Unit of the Ludwig-Maximilian-University Munich Biocenter, generating 41 × 106 paired end 150-bp raw reads. The paired-end reads were quality trimmed by using CLC workbench v5 (CLC Bio), under the default parameters. Trimmed reads were mapped against the three main bacterial contaminant (Delftia acidovorans, Stenotrophomonas maltophilia, Achromobacter xylosoxidans) genomes to remove contaminant reads. The remaining, cleaned reads were assembled by using the CLC de novo assembly algorithm, identifying 24 DhMRE contigs (details in SI Appendix, SI Materials and Methods). After assembly, the 24 remaining gaps were closed by using conventional directed PCR and primer-walking approaches followed by Sanger sequencing.
Annotation and Metabolic Reconstruction.
The DhMRE genome was integrated into the MicroScope platform to perform automatic and manual annotation and comparative analysis with other species from Mycoplasmatales and Firmicutes. AMIGene software was used to predict coding DNA sequences (CDSs). More information on the syntaxic and functional annotation process is given in ref. 38.
Phylogenetic Analyses.
For the multilocus phylogenetic reconstruction, 10 housekeeping genes (single loci genes coding for eight ribosomal proteins, the translation initiation factor IF-2, and the DNA-directed RNA polymerase subunit beta) often used for multilocus analyses (8) were selected from 38 bacteria (listed in SI Appendix, Table S5). The sequences were manually concatenated. The final length of the concatenated sequences was 2,900 aa after removing ambiguously aligned positions.
To study DhMRE proteins candidate for HGT by phylogenetic analyses, homologs were selected from the NCBI database (details in SI Appendix, SI Materials and Methods). For all phylogenetic analyses, protein sequences were aligned by using MAFFT under default parameters (39). JalView (40) was used to manually remove ambiguously aligned positions and regions. Highly divergent sequences were also removed from the alignment. Only the conserved parts of the alignment were selected for the maximum likelihood analysis. Best-fitting evolutionary models for the alignment were determined by its Bayesian information criterion values with ProtTest version 3.4. Maximum likelihood phylogenetic analyses were computed through the CIPRES web portal with RAxML-HPC2 on XSEDE (41) with the best-fit model by using 100 bootstraps.
Accession Numbers and Alignments.
Alignments and phylogenetic trees were deposited at TreeBase under submission ID 17164 for the multilocus analysis (Fig. 1), 17266 for the protein kinase analysis (Fig. 2), and 17268 for the AIG domain protein analysis (SI Appendix, Fig. S6).
The DhMRE genome assembly and annotation were deposited at the European Molecular Biology Laboratory (EMBL)-European Bioinformatics (EBI) European Nucleotide Archive under accession no. PRJEB8356.
G. margarita proteins used for phylogenetic analyses are part of the G. margarita BEG34 Transcriptome Shotgun Assembly (TSA) project (Bioproject PRJNA267628; Biosamples SAMN03216569-SAMN03216586), which was deposited at DNA Data Bank of Japan/EMBL/GenBank under the accession GBYF00000000. The sequences used in the analyses are GBYF01079662, GBYF01002907, GBYF01035393, GBYF01077659, GBYF01035394, GBYF01010756, GBYF01025162, GBYF01016789 and belong to the first TSA version, GBYF01000000.
Supplementary Material
Acknowledgments
We thank German Research Foundation (Deutsche Forschungsgemeinschaft) for funding Grant SCHU1203/14.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The DhMRE genome assembly and annotation were deposited at the EMBL-EBI European Nucleotide Archive (accession no. PRJEB8356). Alignments and phylogenetic trees were deposited at TreeBase under submission ID 17164 for the multilocus analysis (Fig. 1), 17266 for the protein kinase analysis (Fig. 2), and 17268 for the AIG domain protein analysis (SI Appendix, Fig. S6). Gigaspora margarita proteins used for phylogenetic analyses are part of the G. margarita BEG34 Transcriptome Shotgun Assembly project (Bioproject PRJNA267628; Biosamples SAMN03216569-SAMN03216586), which was deposited at DDBJ/EMBL/GenBank (accession no. GBYF00000000). The sequences used in the analyses are GBYF01079662, GBYF01002907, GBYF01035393, GBYF01077659, GBYF01035394, GBYF01010756, GBYF01025162, and GBYF01016789 and belong to the first TSA version, GBYF01000000.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1501540112/-/DCSupplemental.
See Commentary on page 7622.
References
- 1.van der Heijden MGA, Bardgett RD, van Straalen NM. The unseen majority: Soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol Lett. 2008;11(3):296–310. doi: 10.1111/j.1461-0248.2007.01139.x. [DOI] [PubMed] [Google Scholar]
- 2.Schüßler A, Walker C. In: Evolution of the ‘Plant-Symbiotic’ Fungal Phylum, Glomeromycota. Evolution of Fungi and Fungal-Like Organisms, The Mycota. Pöggeler S, Wöstemeyer J, editors. Springer; Berlin: 2011. pp. 163–185. [Google Scholar]
- 3.Naumann M, Schüßler A, Bonfante P. The obligate endobacteria of arbuscular mycorrhizal fungi are ancient heritable components related to the Mollicutes. ISME J. 2010;4(7):862–871. doi: 10.1038/ismej.2010.21. [DOI] [PubMed] [Google Scholar]
- 4.Bianciotto V, Lumini E, Bonfante P, Vandamme P. ‘Candidatus glomeribacter gigasporarum’ gen. nov., sp. nov., an endosymbiont of arbuscular mycorrhizal fungi. Int J Syst Evol Microbiol. 2003;53(Pt 1):121–124. doi: 10.1099/ijs.0.02382-0. [DOI] [PubMed] [Google Scholar]
- 5.Mosse B. Honey-coloured, sessile Endogone spores: II. Changes in fine structure during spore development. Arch Microbiol. 1970;74(2):129–145. [Google Scholar]
- 6.Desirò A, Faccio A, Kaech A, Bidartondo MI, Bonfante P. Endogone, one of the oldest plant-associated fungi, host unique Mollicutes-related endobacteria. New Phytol. 2015;205(4):1464–1472. doi: 10.1111/nph.13136. [DOI] [PubMed] [Google Scholar]
- 7.Desirò A, et al. Detection of a novel intracellular microbiome hosted in arbuscular mycorrhizal fungi. ISME J. 2014;8(2):257–270. doi: 10.1038/ismej.2013.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghignone S, et al. The genome of the obligate endobacterium of an AM fungus reveals an interphylum network of nutritional interactions. ISME J. 2012;6(1):136–145. doi: 10.1038/ismej.2011.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Partida-Martinez LP, Hertweck C. Pathogenic fungus harbours endosymbiotic bacteria for toxin production. Nature. 2005;437(7060):884–888. doi: 10.1038/nature03997. [DOI] [PubMed] [Google Scholar]
- 10.Fujimura R, et al. Draft genome sequence of the betaproteobacterial endosymbiont associated with the fungus Mortierella elongata FMR23-6. Genome Announc. 2014;2(6):e01272-14. doi: 10.1128/genomeA.01272-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nijveen H, van Kaauwen M, Esselink DG, Hoegen B, Vosman B. QualitySNPng: A user-friendly SNP detection and visualization tool. Nucleic Acids Res. 2013;41(web server issue) W1:W587–590. doi: 10.1093/nar/gkt333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tisserant E, et al. Genome of an arbuscular mycorrhizal fungus provides insight into the oldest plant symbiosis. Proc Natl Acad Sci USA. 2013;110(50):20117–20122. doi: 10.1073/pnas.1313452110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin K, et al. Single nucleus genome sequencing reveals high similarity among nuclei of an endomycorrhizal fungus. PLoS Genet. 2014;10(1):e1004078. doi: 10.1371/journal.pgen.1004078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Redecker D, Kodner R, Graham LE. Glomalean fungi from the Ordovician. Science. 2000;289(5486):1920–1921. doi: 10.1126/science.289.5486.1920. [DOI] [PubMed] [Google Scholar]
- 15.Reuber TL, Ausubel FM. Isolation of Arabidopsis genes that differentiate between resistance responses mediated by the RPS2 and RPM1 disease resistance genes. Plant Cell. 1996;8(2):241–249. doi: 10.1105/tpc.8.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marino M, Braun L, Cossart P, Ghosh P. A framework for interpreting the leucine-rich repeats of the Listeria internalins. Proc Natl Acad Sci USA. 2000;97(16):8784–8788. doi: 10.1073/pnas.97.16.8784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glass NL, Kaneko I. Fatal attraction: Nonself recognition and heterokaryon incompatibility in filamentous fungi. Eukaryot Cell. 2003;2(1):1–8. doi: 10.1128/EC.2.1.1-8.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hickey CM, Wilson NR, Hochstrasser M. Function and regulation of SUMO proteases. Nat Rev Mol Cell Biol. 2012;13(12):755–766. doi: 10.1038/nrm3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merhej V, Royer-Carenzi M, Pontarotti P, Raoult D. Massive comparative genomic analysis reveals convergent evolution of specialized bacteria. Biol Direct. 2009;4(1):13. doi: 10.1186/1745-6150-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finn RD, Clements J, Eddy SR. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011;39(web server issue) suppl 2:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishino K, Yamaguchi A. Role of xenobiotic transporters in bacterial drug resistance and virulence. IUBMB Life. 2008;60(9):569–574. doi: 10.1002/iub.90. [DOI] [PubMed] [Google Scholar]
- 22.Tian C, et al. Regulation of the nitrogen transfer pathway in the arbuscular mycorrhizal symbiosis: Gene characterization and the coordination of expression with nitrogen flux. Plant Physiol. 2010;153(3):1175–1187. doi: 10.1104/pp.110.156430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosengarten R, et al. Host-pathogen interactions in mycoplasma pathogenesis: Virulence and survival strategies of minimalist prokaryotes. Int J Med Microbiol. 2000;290(1):15–25. doi: 10.1016/S1438-4221(00)80099-5. [DOI] [PubMed] [Google Scholar]
- 24.Nadimi M, Beaudet D, Forget L, Hijri M, Lang BF. Group I intron-mediated trans-splicing in mitochondria of Gigaspora rosea and a robust phylogenetic affiliation of arbuscular mycorrhizal fungi with Mortierellales. Mol Biol Evol. 2012;29(9):2199–2210. doi: 10.1093/molbev/mss088. [DOI] [PubMed] [Google Scholar]
- 25.Hibbett DS, et al. A higher-level phylogenetic classification of the Fungi. Mycol Res. 2007;111(Pt 5):509–547. doi: 10.1016/j.mycres.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 26.Delaney NF, et al. Ultrafast evolution and loss of CRISPRs following a host shift in a novel wildlife pathogen, Mycoplasma gallisepticum. PLoS Genet. 2012;8(2):e1002511. doi: 10.1371/journal.pgen.1002511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carvalho FM, et al. DNA repair in reduced genome: The Mycoplasma model. Gene. 2005;360(2):111–119. doi: 10.1016/j.gene.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 28.Citti C, Blanchard A. Mycoplasmas and their host: Emerging and re-emerging minimal pathogens. Trends Microbiol. 2013;21(4):196–203. doi: 10.1016/j.tim.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 29.Sirand-Pugnet P, et al. Being pathogenic, plastic, and sexual while living with a nearly minimal bacterial genome. PLoS Genet. 2007;3(5):e75. doi: 10.1371/journal.pgen.0030075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sirand-Pugnet P, Citti C, Barré A, Blanchard A. Evolution of mollicutes: Down a bumpy road with twists and turns. Res Microbiol. 2007;158(10):754–766. doi: 10.1016/j.resmic.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 31.Al-Quadan T, Price CT, Abu Kwaik Y. Exploitation of evolutionarily conserved amoeba and mammalian processes by Legionella. Trends Microbiol. 2012;20(6):299–306. doi: 10.1016/j.tim.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Molmeret M, Horn M, Wagner M, Santic M, Abu Kwaik Y. Amoebae as training grounds for intracellular bacterial pathogens. Appl Environ Microbiol. 2005;71(1):20–28. doi: 10.1128/AEM.71.1.20-28.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hotson A, Mudgett MB. Cysteine proteases in phytopathogenic bacteria: Identification of plant targets and activation of innate immunity. Curr Opin Plant Biol. 2004;7(4):384–390. doi: 10.1016/j.pbi.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 34.Schmitz-Esser S, et al. The genome of the amoeba symbiont “Candidatus Amoebophilus asiaticus” reveals common mechanisms for host cell interaction among amoeba-associated bacteria. J Bacteriol. 2010;192(4):1045–1057. doi: 10.1128/JB.01379-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakabachi A, et al. The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science. 2006;314(5797):267. doi: 10.1126/science.1134196. [DOI] [PubMed] [Google Scholar]
- 36.Chambaud I, et al. The complete genome sequence of the murine respiratory pathogen Mycoplasma pulmonis. Nucleic Acids Res. 2001;29(10):2145–2153. doi: 10.1093/nar/29.10.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tran-Nguyen LTT, Kube M, Schneider B, Reinhardt R, Gibb KS. Comparative genome analysis of “Candidatus Phytoplasma australiense” (subgroup tuf-Australia I; rp-A) and “Ca. Phytoplasma asteris” Strains OY-M and AY-WB. J Bacteriol. 2008;190(11):3979–3991. doi: 10.1128/JB.01301-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vallenet D, et al. MicroScope—an integrated microbial resource for the curation and comparative analysis of genomic and metabolic data. Nucleic Acids Res. 2013;41(Database issue):D636–D647. doi: 10.1093/nar/gks1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33(2):511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25(9):1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.