Abstract
Polycistronic microRNA clusters are a common feature of vertebrate genomes. The coordinated expression of miRNAs belonging to different seed families from a single transcription unit suggests functional cooperation, but this hypothesis has not been experimentally tested. Here we report the characterization of an allelic series of genetically engineered mice harboring selective targeted deletions of individual components of miR-17~92. Our results demonstrate the co-existence of functional cooperation and specialization among members of this cluster, identify a novel function for the miR-17 seed family in controlling axial patterning in vertebrates, and show that loss of miR-19 selectively impairs Myc-driven tumorigenesis in two models of human cancer. By integrating phenotypic analysis and gene expression profiling, we provide a genome-wide view of how components of a polycistronic miRNA-cluster affect gene expression in vivo. The reagents and datasets reported here will accelerate exploration of the complex biological functions of this important miRNA cluster.
INTRODUCTION
miRNAs modulate gene expression at the post-transcriptional level by repressing translation and by promoting destabilization of target mRNAs1–4. Approximately 60% of mammalian miRNAs are part of polycistronic clusters, and in many cases multiple seed families are represented within a single polycistron5. Coordinated expression of unrelated miRNAs from the same transcription unit could provide a simple mechanism for different miRNAs to act cooperatively, thus explaining why this gene structure has been so frequently favored by natural selection. To test this hypothesis we investigated the miR-17~92 cluster, a classic polycistronic miRNA gene with essential roles in development and disease6.
miR-17~92 consists of six highly conserved miRNAs belonging to four seed families (Fig. 1a). In mice, targeted deletion of the whole cluster causes a wide array of developmental defects affecting multiple organs and systems7, while in humans germline mono-allelic microdeletions involving miR-17~92 are responsible for the developmental defects observed in a subset of patients affected by Feingold Syndrome8 (FS2; OMIM 614326), an autosomal dominant disease characterized by short stature and variable combinations of craniofacial abnormalities, limb and digit malformations, gastro-intestinal atresia, and learning disabilities9.
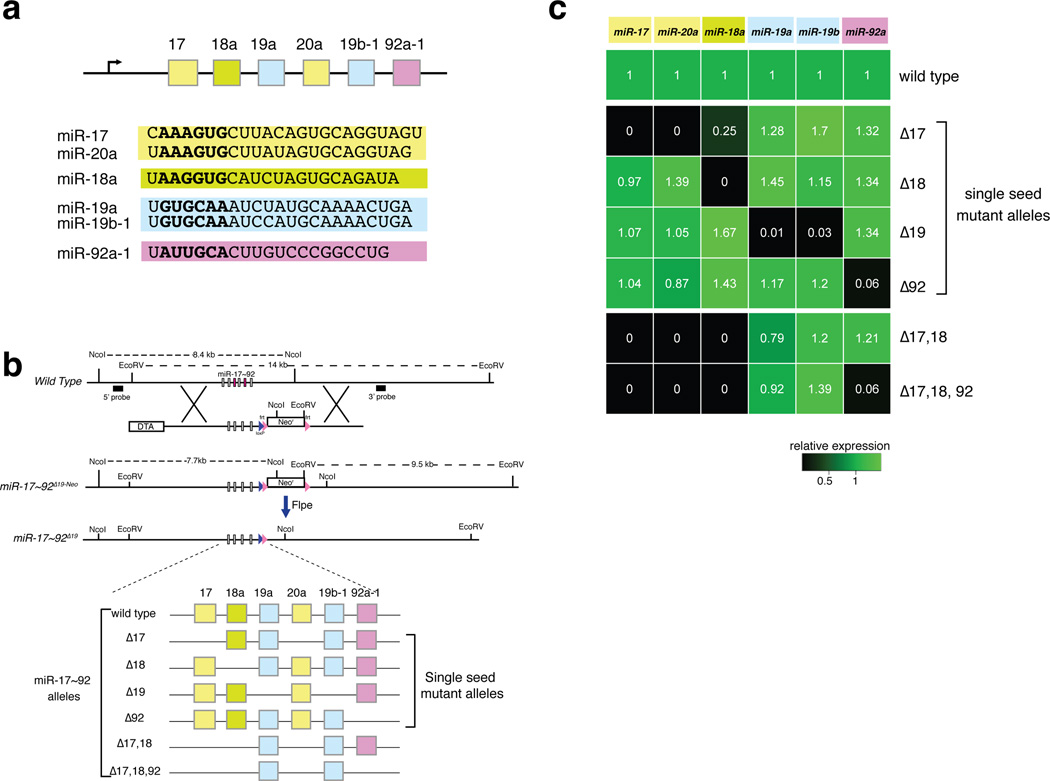
Figure 1. Generation of an allelic series of miR-17~92 mutant mice.
(a) Schematic of the miR-17~92 cluster. Sequence of the mature miRNAs is shown below, with the seed (nucleotides 2–7) in bold. Arrow: direction of transcription. (b) Top, Schematic of the targeting strategy, using the miR-17~92Δ19 allele as an example. DTA, diphtheria toxin gene. Bottom, Schematic of the miRNAs expressed by each allele. (c) Heat map showing miRNA expression (detected by RT-qPCR) in tails from homozygous mice relative to wild type controls (n = 3).
miR-17~92 is also a bona fide human oncogene. Focal amplifications of its locus are frequent in human diffuse large B-cell lymphomas10 and its components are overexpressed in many human cancers11–16 at least partially as result of direct transcriptional activation by members of the Myc family of oncogenic transcription factors17. In addition, ectopic expression of miR-17~92 accelerates or initiates tumor formation in several mouse models of human cancer12,18–24.
We reasoned that the multiple phenotypes caused by miR-17~92 deletion could be used to experimentally determine the extent of functional cooperation among the miRNAs encoded by a polycistronic cluster. Here we report the generation of an allelic series of miR-17~92 mutant mice and integrate their phenotypic characterization with RNA-seq analysis showing the consequences of selective miRNA inactivation on gene expression in the developing embryo. Our results provide novel insights into the biology of this important oncogenic miRNA cluster and pave the way for further mechanistic studies.
RESULTS
Generation of an allelic series of miR-17~92
We used homologous recombination in mouse embryonic stem cells to generate six miR-17~92 alleles (Fig. 1b, Supplementary Fig. 1a), each characterized by the deletion of genomic sequences corresponding to one or more of its pre-miRNAs. Four alleles harbor the deletion of the components of single “seed families”: miR-17~92Δ17, miR-17~92Δ18, miR-17~92Δ19, and miR-17~92Δ92. Two additional alleles, miR-17~92Δ17,18 and miR-17~92Δ17,18,92, are characterized by targeted deletion of more than one seed family. The miR-17~92Δ17,18 allele was generated because members of the miR-17 and miR-18 seed families have high sequence homology and their seeds differ only at position 4. Finally, the miR-17~92Δ17,18,92 allele retains only pre-miRNAs belonging to the miR-19 family and its generation was justified by reports indicating that this seed family is the key oncogenic determinant of the cluster21,25,26. After germline transmission of the targeted alleles we intercrossed heterozygous mice to generate homozygous mutant animals for each allele of the series (Table 1).
Table 1. Absolute numbers and frequency of genotypes obtained from heterozygous breedings.
p-Value, adjusted χ2 test. Genotypes were determined at postnatal day 10–12, except for the miR-17~92Δ17,18,92 allele that was also examined at birth (P0).
| +/+ (25%) |
+/Δ (50%) |
Δ/Δ (25%) |
Corrected p Value |
|
|---|---|---|---|---|
| Δ17 | 38/105 (36%) |
49/105 (46%) |
18/105 (18%) |
NS |
| Δ18 | 29/111 (26%) |
42/111 (38%) |
40/111 (36%) |
NS |
| Δ19 | 32/109 (29%) |
54/109 (50%) |
23/109 (21%) |
NS |
| Δ92 | 28/125 (22%) |
61/125 (49%) |
36/125 (29%) |
NS |
| Δ17,18 | 33/94 (35%) |
46/94 (49%) |
15/94 (16%) |
NS |
| Δ17,18,92 | 49/106 (46%) |
57/106 (54%) |
0/106 (0%) |
p<0.0005 |
|
Δ17,18,92 (P0) |
19/73 (26%) |
33/73 (45%) |
21/73 (28%) |
NS |
| Δ17~92 | 101/240 (42%) |
139/240 (58%) |
0/240 (0%) |
p<0.0005 |
We confirmed loss of expression of the deleted miRNAs in homozygous animals by both RT-qPCR and small RNA sequencing (Fig. 1c and Supplementary Fig. 1b,c). For each allele, we found that the expression of the remaining miRNAs was not substantially altered, with the notable exception of a significant reduction of miR-18a in miR-17~92Δ17/Δ17 mice. Expression profiling of wild type and mutant animals however, suggests that this reduction has very limited, if any, functional consequences (Supplementary Discussion). Finally, no changes in the sequences of the mature miRNAs encoded by the various miR-17~92 alleles were detected by small RNA sequencing (data not shown).
Perinatal lethality, cardiac defects, and lung hypoplasia
Homozygous deletion of miR-17~92 results in fully penetrant perinatal lethality, a phenotype that has been attributed to severe lung hypoplasia and defective cardiac development7 (Table 1 and Supplementary Fig. 2). By contrast, heterozygous intercrosses of the single-seed mutants yielded viable homozygous adults at expected Mendelian ratios and even combined deletion of the miR-17 and miR-18 seed families was compatible with postnatal survival. For each of these strains, crosses between homozygous animals produced viable offspring (data not shown). Strikingly, we observed perinatal lethality only in miR-17~92Δ17,18,92/Δ17,18,92 mice (Table 1) and only these animals displayed lung hypoplasia (Supplementary Fig. 2). Defective heart development was only detected upon deletion of the entire cluster (Supplementary Fig. 2). Thus the perinatal lethality, cardiac defects, and lung hypoplasia observed in miR-17~92-null mice result from the concomitant loss of multiple components of the cluster, and provide a clear example of functional cooperation.
Although the characterization of miR-17~92-null animals7 and the results described above were performed on mice of mixed B6;129 background, during this study we observed that backcrossing the mutant alleles into a pure C57BL/6J genetic background results in generally more severe phenotypes. More specifically, we noticed that a smaller fraction of miR-17~92Δ/Δ embryos reaches full term and we observed significant perinatal lethality in mice harboring deletion of members of the miR-17 or miR-19 seed families (Supplementary Table 1).
Feingold Syndrome and axial patterning
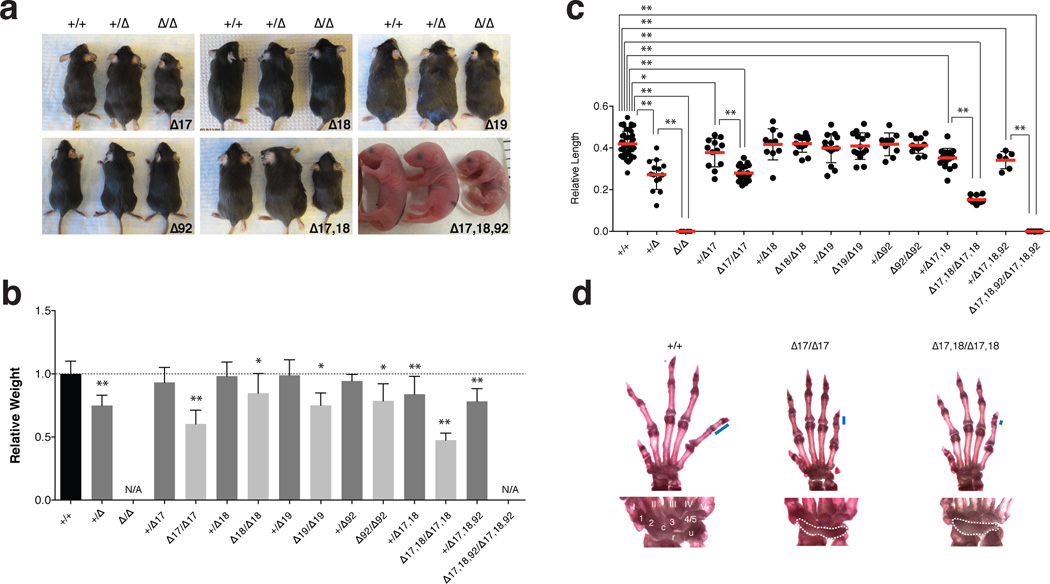
Hemizygosity at the miR-17~92 locus in mice phenocopies several of the key features of FS2, including size and digit abnormalities8. To define the relative contribution of each seed family to the pathogenesis of this syndrome, we examined mice from the miR-17~92 allelic series. Homozygous animals for each of the four single-seed mutant alleles were smaller than age- and sex-matched wild type controls (Fig. 2a, 2b). This phenotype was most severe in miR-17~92Δ17/Δ17 mice, which at 5 weeks of age weighed on average 40% less than wild type animals (p=7.47×10−7; two-tailed t-test). Co-deletion of the miR-17 and miR-18 families resulted in a more severe phenotype with a significant weight-reduction in both heterozygous (16%, p=4.05×10−4) and homozygous animals (52%, p=8.44×10−9). Finally, the weight of miR-17~92+/Δ17,18,92 animals was comparable to that of miR-17~92+/Δ mice (Fig. 2a, 2b).
Figure 2. Feingold Syndrome features in the miR-17~92 mutant mice.
(a) Macroscopic appearance of miR-17~92 allelic series’ animals. (b) Mean weight of 4–5 week-old +/+ (n = 77), +/Δ17~92 (n = 13), +/Δ17 (n = 21), Δ17/Δ17 (n = 11), +/Δ18 (n = 26), Δ18/Δ18 (n = 17), +/Δ19 (n = 19), Δ19/Δ19 (n = 6), +/Δ92 (n = 9), Δ92/Δ92 (n = 9), +/Δ17,18 (n = 22), Δ17,18/Δ17,18 (n = 7) and +/Δ17,18,92 (n = 12) animals,– normalized to sex- and age-matched controls. (c) Mean length of the 5th mesophalanx relative to the metacarpal bone. (d) Top, representative pictures of forelimb autopods of wild type, miR-17~92Δ17/Δ17 and miR-17~92Δ17,18/Δ17,18 adult animals. Blue bars represent length of mesophalanx; Bottom, detailed view of wrist bones. Digits are numbered I–V. Carpal bones, 1–3 and 4/5, c (central element), u (ulnare), and r (radiale). Fusion of carpal bones is highlighted with dashed line.
Error bars = s.d.,* p < 0.05, ** p < 0.001, 2-tailed t-test, N/A, not available.
A relative shortening of the 5th mesophalanx (brachymesophalangy) of the forelimb is another characteristic feature of FS2 recapitulated in miR-17~92+/Δ mice. In miR-17~92Δ/Δ animals, forelimb abnormalities are more severe and include absence of the 5th mesophalanx as well as fusion of the proximal carpal bones8 (Fig. 2c, Supplementary Figure 3a, and Supplementary Table 2). Among the single-seed mutants, only hemizygous or homozygous deletion of the miR-17 seed family resulted in a significant shortening of the 5th mesophalanx (Fig. 2c,d). A fraction of miR-17~92Δ17/Δ17 mice, but not mice homozygous for the other single-seed mutant alleles, also displayed fusion of the proximal carpal bones. miR-17~92Δ17,18/Δ17,18 mutant mice showed more severe phenotypes than those carrying a deletion of miR-17 alone, and further deletion of miR-92a (miR-17~92Δ17,18,92/Δ17,18,92) fully recapitulated the phenotypes seen in miR-17~92 loss of function mice (Fig. 2c and Supplementary Fig. 3a,b).
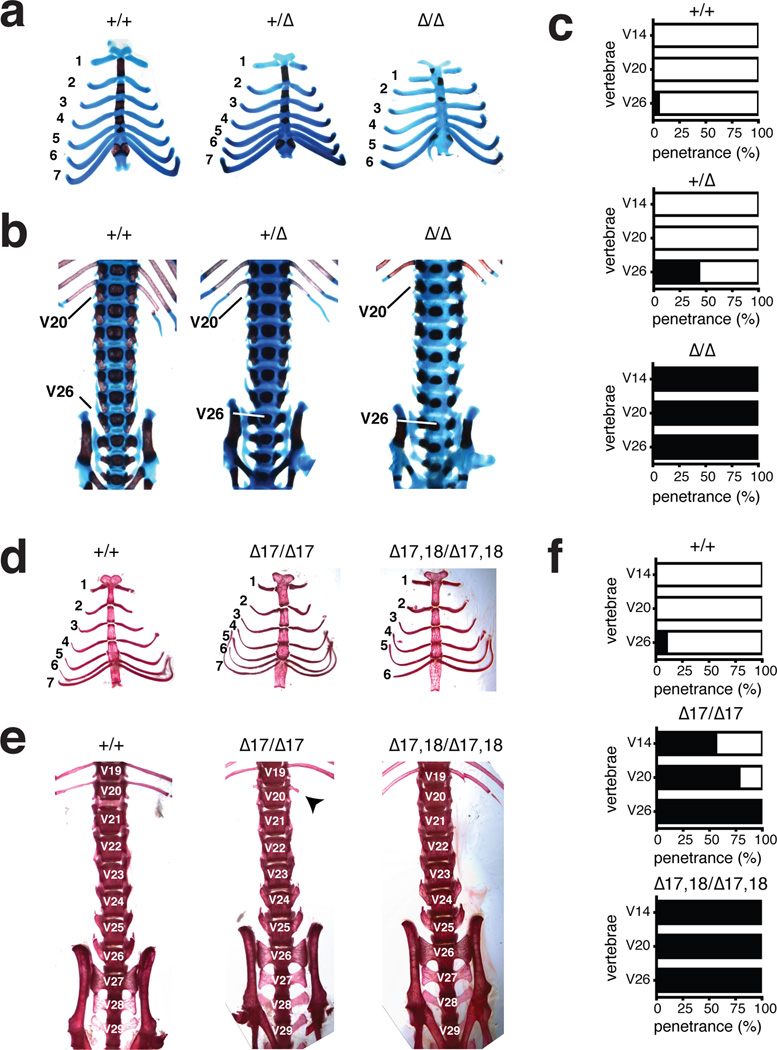
The role of miR-17~92 in FS2 compelled us to further analyze skeletal development in the absence of the entire cluster or of its individual components (see Supplementary Tables 2–4). During this analysis we found that loss of miR-17~92 affected a key event during the formation of the mammalian skeleton: its patterning along the antero-posterior axis, which leads to the differentiation of morphologically distinct skeletal segments27,28 (Supplementary Fig. 3c). Homeotic transformations in miR-17~92Δ/Δ mice were fully penetrant and affected multiple skeletal segments (Fig. 3a–c and Supplemental Fig. 3c,d). Specifically, V14–which in wild type mice is the last vertebra of the rib cage–was transformed into the first vertebra with floating ribs (i.e., ribs that do not attach to the sternum; n=10/10) (Fig.3a). We observed a unilateral (n=3/10) or bilateral (n=7/10) loss of ribs on V20, and a transformation of V26 from the last lumbar into the first sacral vertebra (n=10/10) (Fig. 3b, 3c Supplementary Figure 3d and Supplementary Table 3). Transformations at the lumbar/sacral level were also observed in 46% of heterozygous mice, indicating a dose-dependent role of this cluster in axial patterning.
Figure 3. MiR-17~92 regulates axial patterning.
(a) Alcian blue/alizarin red stained rib cages from miR-17~92+/+, miR-17~92+/Δ, and miR-17~92Δ/Δ E18.5 embryos. (b) Ventral view of the axial skeleton. The positions of V20 and V26 are indicated (c) Percentage of animals showing vertebral transformations (black) or normal vertebral identity (white) in miR-17~92+/+ (n = 20), miR-17~92+/Δ (n = 28), and miR-17~92Δ/Δ (n = 10) animals. (d) Alizarin-red stained rib cages from wild type, miR-17~92Δ17/Δ17 and miR-17~92Δ17,18/Δ17,18 adult mice. (e) Ventral view of axial skeleton. Arrow indicates a rudimental rib (f) Percentage of mutant animals showing vertebral transformations (black) or normal vertebral identity (white) in miR-17~92+/+ (n = 97), miR-17~92Δ17/Δ17 (n = 9), and miR-17~92Δ17,18/Δ17,18 (n = 7) animals.
We next examined axial patterning in animals from the allelic series (Fig. 3d–f and Supplementary Table 4). Among the single-seed mutants, only those harboring deletion of the miR-17 seed family displayed transformations similar to those seen in the absence of the full cluster. Simultaneous deletion of miR-17 and miR-18 families was enough to fully recapitulate the phenotypes seen in miR-17~92-null mice. These results reveal a previously unappreciated role for miR-17~92 in patterning of the axial skeleton, identify the miR-17 seed family as the main effector of this function, and provide a direct link between mutation of a miRNA gene and homeotic transformations.
The miR-17 and miR-18 families regulate B-cell development
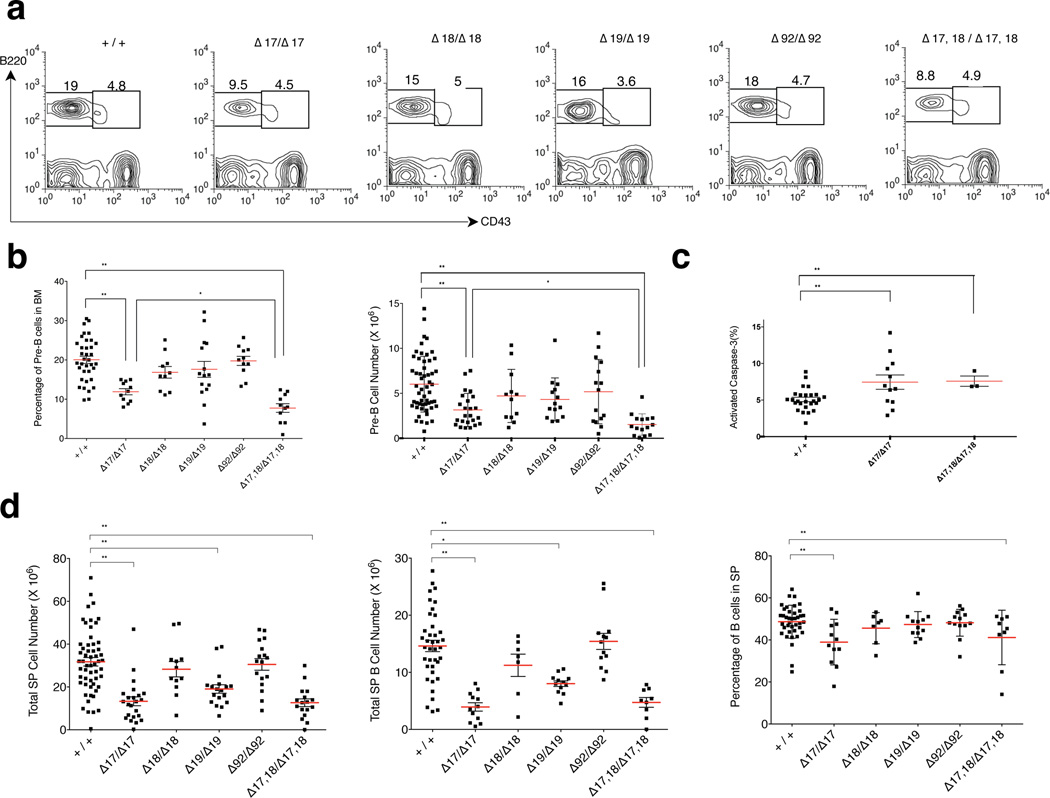
In miR-17~92-deficient mice, early B-cell development is characterized by a partial block at the pro-B to pre-B transition due to increased apoptosis7. Among the single seed mutant mice, only miR-17~92Δ17/Δ17 animals displayed a statistically significant reduction of pre-B-cells (Fig. 4a, 4b). This phenotype was exacerbated in miR-17~92Δ17,18/Δ17,18 mice, once more demonstrating cooperation among these two highly related miRNA families. Consistent with a predominant role for the miR-17 seed family in regulating the pro- to pre-B-cell transition, we observed a significant increase in the fraction of apoptotic pre-B-cells in miR-17~92Δ17/Δ17 and miR-17~92Δ17,18/Δ17,18 mice (Fig. 4c), specifically at the pre-BII stage (Supplementary Fig.4a, 4b). These mice also displayed a decrease in the ratio of splenic B-cells to T-cells and myeloid cells. However, further maturation of B-cells in the spleen was unaffected and the spleens appeared histologically normal (Fig. 4d and Supplementary Fig. 4d, 4e).
Figure 4. Analysis of B-cell development in the miR-17~92 allelic series.
(a) Flow cytometry plots showing the percentage of bone marrow pro- (IgM−, B220+, CD43+) and pre-B cells (IgM−, B220+, CD43−) in 5–6-week-old wild type and homozygous mutant mice. (b) Quantification of the pre-B-cell percentage and absolute pre-B cell number (n > 10). (c) Percentage of cleaved caspase-3 positive pre-B-cells in 5–6-week-old wild type, miR-17~92Δ17/Δ17 and miR-17~92Δ17,18/Δ17,18 animals (n > 3). (d) Total number of splenocytes and B-cells as well as percentage of B-cells in the spleen. Red bars indicate average. * p < 0.05, ** p < 0.001, 2-tailed t-test.
Based on these results we conclude that members of the miR-17 and miR-18 seed families play a central role in modulating pre-B cell survival and early B cell development.
Loss of miR-19 impairs Myc-driven tumorigenesis
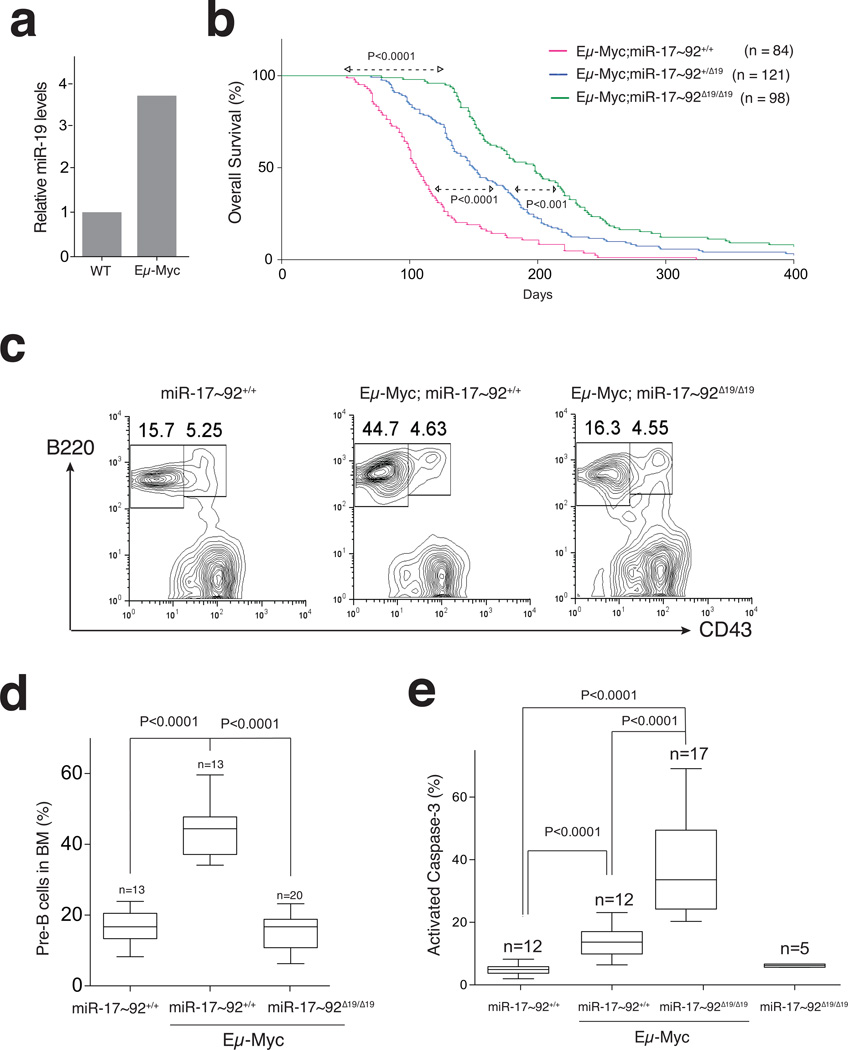
Ectopic expression studies previously demonstrated that the tumor-promoting activity of miR-17~92 is largely determined by miR-19a and miR-19b-1 (hereafter referred to as miR-19). miR-19 overexpression can recapitulate the ability of miR-17~92 to cooperate with the Myc oncogene in driving B-cell lymphomagenesis21 and can rescue the increased apoptosis observed in Eμ-Myc lymphoma cell lines upon acute deletion of miR-17~9226. Based on these results, and on the observation that miR-17~92 is a direct transcriptional target of Myc17 (Fig. 5a), we hypothesized that upregulation of endogenous miR-19 by Myc might play a role in tumor initiation and progression even in the absence of ectopic expression of these miRNAs.
Figure 5. Impaired Myc-driven lymphomagenesis in miR-19-deficient mice.
(a) Expression of miR-19b in pre-B-cells from 5 week-old wild type and Eμ-Myc mice (RT-qPCR; p < 0.01; 2-tailed t-test). (b) Kaplan-Meier plots of Eμ-Myc carrying the indicated miR-17~92 alelles. (p-values: Mantel-Cox test). (c) Representative FACS and (d) Box and whiskers plots showing the fraction of pre-B-cells in the bone marrow of 5 week-old mice. (e) Box and whiskers plots showing the percentage of cleaved caspase-3 positive pre-B-cells (median, 25th and 75th percentiles, and min and max values are plotted; p-values: 2-tailed t-test; Error bars: s.d.).
To test this hypothesis, cohorts of Eμ-Myc;miR-17~92+/+, Eμ-Myc;miR-17~92+/Δ19, and Eμ-Myc;miR-17~92Δ19/Δ19 animals were monitored for the appearance of palpable lymphomas and overall survival. While control Eμ-Myc animals developed lymphomas with the expected latency, deletion of miR-19 resulted in a dosage-dependent increase in tumor-free and overall survival (Fig. 5b and Supplementary Fig. 5a). miR-19-deficient mice that eventually succumbed to the disease developed tumors markedly biased towards a more mature immunophenotype (Supplementary Fig.5b). Deletion of miR-18a or miR-92a-1 had minimal or no impact on Eμ-Myc lymphomagenesis (Supplementary Fig. 5d), while targeted deletion of the miR-17 seed family resulted in a significant delay in tumor latency, presumably a consequence of the reduced number of pre-B cells (Supplementary Fig. 5d). The increased latency and the shift in the immunophenotype of lymphomas in Eμ-Myc;miR-17~92Δ19/Δ19 mice suggest that the miR-19 seed family plays a permissive role in the transformation of early B-cell progenitors by Myc.
To determine whether absence of miR-19 impaired the early stages of Myc-driven lymphomagenesis, we examined the bone marrow of young (5 week-old) Eμ-Myc mice, which is typically characterized by a pre-neoplastic accumulation of pre-B-cells29,30 (Fig. 5c, 5d). Strikingly, this accumulation was entirely absent in age-matched Eμ-Myc;miR-17~92Δ19/Δ19 animals. This was not due to an inability of the Myc oncogene to promote hyperproliferation in the absence of miR-19 (Supplementary Fig. 5c, 5e). Rather, the increase in pre-B-cell proliferation was counterbalanced by a concomitant marked increase in apoptosis (Fig. 5e).
It is worth noting that this role of miR-19 in suppressing apoptosis becomes evident only in the context of oncogenic levels of Myc, while loss of miR-19 expression doesn’t impair normal pre-B cell development or survival (Fig.4a and 5e).
We next sought to determine whether miR-19 plays a more general role in Myc-driven tumorigenesis. Up to one-third of prostate cancers harbor genomic amplification of the MYC locus31,32, and mice expressing a Myc transgene controlled by a prostate-specific promoter (Hi-Myc mice) develop invasive prostatic adenocarcinomas within 6–12 months of age33.
To examine the role of miR-19 in this context, we aged cohorts of Hi-Myc;miR-17~92+/+ and Hi-Myc;miR-17~92Δ19/Δ19 mice. At 8 weeks of age, Hi-Myc;miR-17~92+/+ male mice displayed extensive signs of prostatic intraepithelial neoplasia (PIN) (Supplementary Fig. 6a), as previously described33. By contrast, the prostate of age-matched Hi-Myc;miR-17~92Δ19/Δ19 mice appeared largely normal with only occasional foci of PIN. Immunohistochemistry for cleaved caspase-3 showed that also in this context deletion of miR-19 caused a significant increase in apoptosis in response to the Myc oncogene (Supplementary Fig. 6b, 6c).
Even more dramatic were the consequences on tumor progression; In Hi-Myc control mice, invasive adenocarcinomas were detected by 8 months of age and were present in all animals by 10 months (Supplementary Fig. 6d, 6e). In contrast miR-19-deficient mice showed a largely unaffected prostate even at 12 months of age; by 14 months only one-third of miR-19-null animals displayed histological evidence of invasive cancer. Delayed tumor progression was confirmed by non-invasive imaging studies (Supplementary Fig. 6f, 6g).
By demonstrating a critical function in two distinct Myc-driven cancer models our results establish miR-19 as a central component of the Myc oncogenic network.
Genome-wide analysis of miR-17~92 targets in vivo
To gain insights into the physiologic targets of miR-17~92, we next sought to explore how targeted deletion of its individual components affects gene expression in vivo. Although miRNA-mediated regulation ultimately controls protein output, previous studies comparing proteomic and RNA-expression profiling have demonstrated that most, if not all, of the changes in protein levels can be explained by changes in mRNA levels34–36.
We performed RNA-seq experiments on RNAs from hearts and tail buds micro-dissected from the various miR-17~92 mutant embryos. These tissues were chosen because loss of miR-17~92 in these tissues results in some of the most prominent developmental defects described here (Fig.1–3). We used only embryos at 20–22 somites stage, to minimize variability and because this stage immediately precedes the appearance of the first detectable developmental abnormalities in miR-17~92-null mice.
Cumulative distribution fraction plots of log2 fold-changes revealed that in each mutant strain the predicted targets of the deleted miRNA(s) were preferentially upregulated compared to wild type embryos (Supplementary Fig. 7 and Supplementary Table 5), demonstrating that our analysis captures changes in gene expression caused by targeted deletion of miR-17~92 and of its various components.
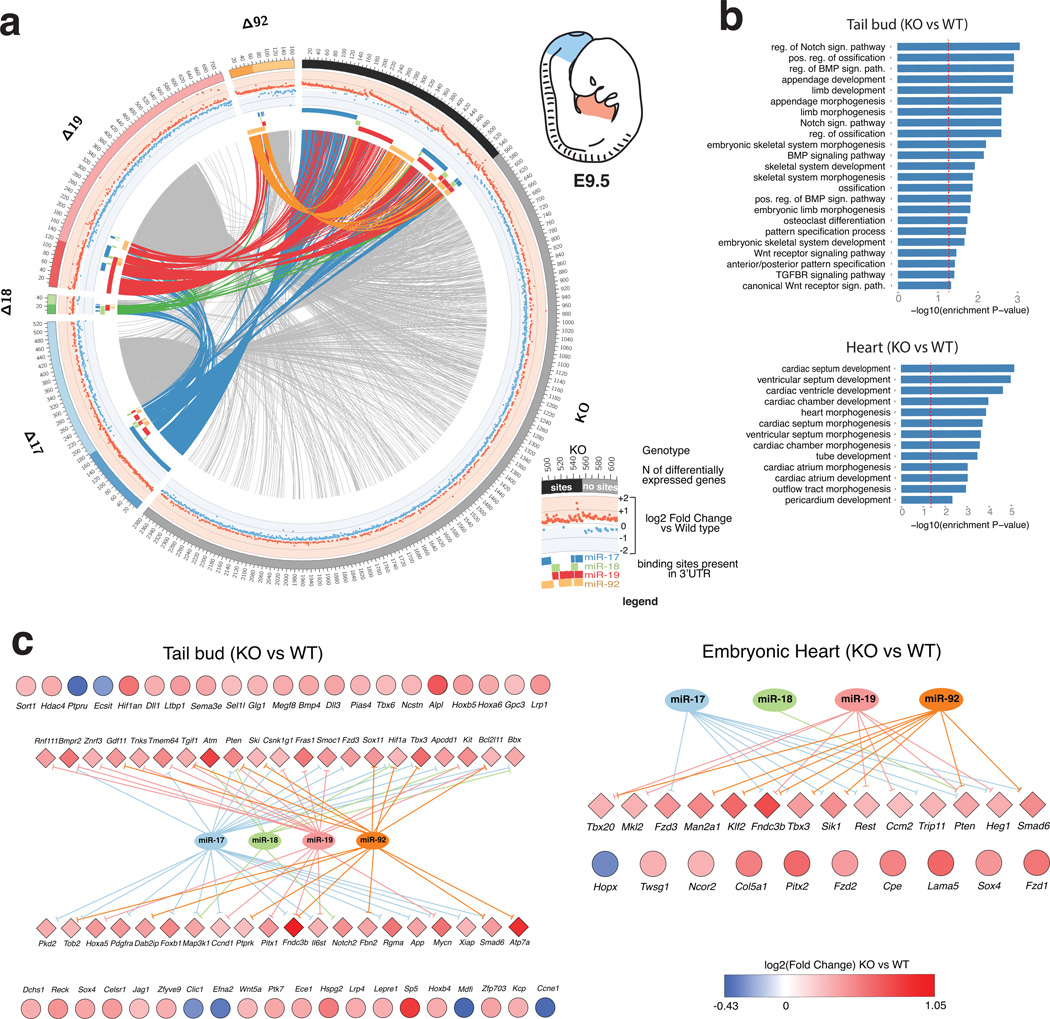
To examine the relative contribution of each seed family and their functional interactions in the modulation of targets, we next visualized the genes significantly deregulated in each genotype using the Circos package37 (Fig. 6a and Supplementary Fig. 8–11). In these plots genes deregulated in two or more mutant mice are joined by links. For each gene, the plots also provide information regarding the direction and amplitude of deregulation (in the middle track), and the presence or absence of predicted binding sites for the various members of miR-17~92 (in the inner track) (see Fig. 6a inset and legend for additional details).
Figure 6. Genome-wide identification of genes regulated by members of the miR-17~92 cluster.
(a) Circos plot of differentially expressed genes (FDR <= 0.1) in the tail bud of miR-17~92 mutants embryos. Each sector of the plot correspond to a different miR-17~92 mutant allele. To reduce complexity, only the complete knock-out (KO) and the four single-seed mutant alleles are shown (for additional Circos plots—including ones generated from the heart dataset—see Supplementary Figure 8–11). Genes with predicted binding sites for one or more of the deleted miRNAs are grouped under the darker portion of each sector. A scatter plot of the log2 fold-change is presented in the intermediate circle, with up- and down-regulated genes labeled in red and blue, respectively. Presence of predicted binding sites for the various components of miR-17~92 is indicated by a colored bar in the inner circle. Genes differentially expressed in more than one genotype are joined by links. Links are colored if the gene has predicted binding sites for miR-17~92, otherwise they are shown in light gray. (b) Bar plot of GO terms enriched among the most significant differentially expressed genes (FDR <= 0.01 and |og2 FC >= 0.26) in miR-17~92Δ/Δ tail bud (top) or heart (bottom). Only terms relevant to cardiac or skeletal development are shown. A red dashed vertical line indicates marks a nominal enrichment p-Value of 0.05. (c) Genes contributing to the enriched GO terms shown in (b). Diamonds indicate predicted miR-17~92 targets, circles indicate presumed indirect targets. Color reflects the log2 FC value.
These plots provide a high-level view of the regulatory activity of miR-17~92 in vivo and illustrate, at the molecular level, the complex interactions existing among the various components of miR-17~92. Several conclusions can be drawn from their analysis. First, it is immediately evident that the four seed-families encoded by miR-17~92 differ greatly in the number of genes they regulate in vivo. For example, only a handful of genes are affected by loss of miR-18a, while the miR-17 and the miR-19 seed families modulate the expression of the largest number of genes. It is also worth noting that the extent of gene regulation by the cluster is context dependent: nearly four times as many genes are affected by loss of miR-17~92 in the tail bud compared to the embryonic heart.
Another conclusion stems from the observation that the number of genes deregulated upon deletion of the entire miR-17~92 cluster is substantially greater than the sum of the genes deregulated in the single-seed mutants (Fig. 6a). This finding supports the idea that the various members of the miR-17~92 cluster cooperatively regulate, directly or indirectly, a large number of genes. Interestingly, the number of genes differentially expressed in the miR-17~92Δ17,18 double-seed mutants is much greater than the sum of the genes deregulated in the miR-17~92Δ17 and miR-17~92Δ18 single seed mutants (Supplementary Fig. 8–11), indicating a strong cooperation between these two closely related miRNA families.
The last and perhaps most important conclusion is that although loss of miR-17~92 or of its individual components results in the reproducible deregulation of hundreds of genes, the amplitude of this effect is invariably very modest, with the vast majority of genes changing by less than one log compared to wild type (Fig. 6, Supplementary Fig. 8–11 and Supplementary Table 5). Interestingly, this is not only true for differentially expressed genes that have predicted binding sites for miR-17~92 in their 3’UTR—the “direct targets”—but also for genes that are regulated indirectly by miR-17~92. This observation indicates that the modest effects on the direct targets are not amplified downstream and has important implications because it suggests that, rather than acting as genetic switches for specific signaling pathways or transcription factors, the miRNAs of miR-17~92 act as fine-tuners, ensuring that the expression of a large number of genes stays within a very narrow range at critical developmental stages. As a consequence, it is unlikely that the complexity of phenotypes observed in the various mutant mice described here can be assigned to loss of repression of one or a few key targets.
To gain further insights into the genes regulated by miR-17~92 we searched for enrichment of Gene Ontology (GO) terms within the subset of genes deregulated in miR-17~92Δ/Δ embryos (Fig. 6b and Supplementary Table 6). Consistent with the phenotypes observed in miR-17~92-null mice, we found significant enrichment of GO terms associated with cardiac development, and in particular with the development of the ventricular septum in the heart data set. Similarly, GO terms related to ossification, skeletal development and patterning were significantly over-represented in the tail bud dataset. The differentially expressed genes contributing to these terms, their fold change compared to wild type embryos, and their predicted interactions with members of the miR-17~92 cluster, are shown in Figure 6c.
Many of the genes contributing to the GO terms in the heart, such as Tbx3, Tbx20, Smad6, Heg1, Klf2, and Trip11 are direct targets of the cluster for which there is experimental evidence that perturbation of their expression is sufficient to induce cardiac defects in vertebrates38–44. Analogously, GO terms from the tail bud dataset include direct and indirect targets of miR-17~92 involved in mesoderm formation (Bmp4 and Bmpr2)45,46, specification (Tbx6)47, segmentation (Notch2, Jag1, Dll3, Dll1)48,49, axial patterning (Gdf11, Hoxa5, Hoxa6, Hoxb4)50,51, and limb development (Tbx3, Bmp4)52,53.
Gene Ontology assignments are imperfect and limited by our current knowledge of embryonic development, hence it is likely that this analysis under-estimates the number of genes whose deregulation in miR-17~92 deficient animals contributes to the developmental defects. Nevertheless, the picture emerging from our work is consistent with a model in which members of the miR-17~92 cluster have evolved to coordinately fine-tune the expression of a wide network of genes involved in multiple aspects of mammalian development. We propose that it is the concomitant loss of fine-tuning of these genes, rather than the deregulation of a single or a few key targets, that causes the complex phenotype observed in miR-17~92 mutant mice.
DISCUSSION
The allelic series of miR-17~92 mutant mice and the associated RNA-seq dataset reported here offer an unprecedented view of the functional organization of a multifamily miRNA cluster. There are at least five aspects of this work that we believe are of general relevance.
First, we provide direct genetic evidence that strong functional specialization and cooperation can coexist among members of the same polycistronic cluster. A clear example of functional specialization is represented by the unique role played by the miR-17 seed family in modulating axial patterning. By contrast, the perinatal lethality, cardiac defects, and lung hypoplasia are clear examples of functional cooperation. We also identify cases in which one seed family plays a dominant, but not unique role, as exemplified by the role of the miR-17 family in pre-B cell survival and in digit development.
Second, we uncover an unexpected role for miR-17~92 in modulating skeletal patterning, as shown by the occurrence of fully penetrant homeotic transformations in miR-17~92Δ/Δ and miR-17~92Δ17/Δ17 mice. Despite multiple reports linking miRNAs to embryonic patterning54–56, loss of function mutations of a miRNA gene have not previously been linked to disrupted patterning of embryonic axes in mice. This discovery also lends support to the idea posited by Tanzer and colleagues57 that the expansion of the miR-17 family is temporally and functionally connected to the origin of vertebrates and with the concomitant genome-wide duplications that resulted in the formation of the four Hox clusters.
Third, we define members of the miR-19 seed family as key components of the oncogenic network orchestrated by Myc. The observation that miR-19-deficient mice are remarkably resistant to Myc-driven tumorigenesis in two distinct tumor models demonstrates that this miRNA family is a critical downstream effector of Myc and plays a crucial role in Myc-driven tumor initiation and progression. In this context, a surprising finding is that in the absence of oncogenic levels of Myc loss of miR-19 expression has no obvious detrimental effects on pre-B cell differentiation or on prostate homeostasis. This has obvious therapeutic implications because it suggests that pharmacologic inhibition of miR-19 in patients affected by tumors expressing oncogenic levels of MYC could have anticancer effects with little toxicity. It is tempting to speculate that miR-19 induction by Myc might be part of the physiologic response to conditions requiring the rapid and transient expansion of specific cell populations in response to acute stresses.
Fourth, the RNA-seq dataset derived from this allelic series of mutant mice provides a unique, and to some extent surprising, view of the physiologic targets of a polycistronic miRNA cluster in vivo. We have already discussed some of its specific implications with respect to miR-17~92 functions. Here we would like to emphasize that our findings strongly support the notion that in animals many miRNAs have evolved not to act as genetic switches of specific pathways or individual targets, but to modulate the expression of large gene networks. If our interpretation is correct, the search for individual “key targets” that can explain the biological properties of a given miRNA might prove futile, if not misleading.
Fifth, the six novel miR-17~92 mutant strains reported here represent a valuable resource for the non-coding RNA field, as they will accelerate and facilitate further mechanistic investigations into the complex developmental and oncogenic functions of this cluster. Likewise, the RNA-seq datasets we have generated will provide the scientific community unprecedented opportunities to explore the molecular determinants of miRNA targeting in a physiologic in vivo setting.
METHODS
Mouse husbandry and generation of miR-17~92 mutant mice
miR-17~92Δ, Eμ-Myc, and Hi-Myc mice have been previously described7,29,33. Targeting constructs for the miR-17~92 mutant alleles were generated by modifying the original miR-17~92 conditional targeting construct. Briefly, a 2.2kb fragment containing one loxP site and the sequence for the six pre-miRNAs was retrieved through PmlI/XhoI digestion and cloned in the multiple cloning site of pBS KS+. Pre-miRNA sequences were deleted through PCR-mediated mutagenesis after which the mutated PmlI/XhoI fragment was re-cloned into the targeting vector. V6.5 ES cells (obtained from Rudolf Jaenisch and tested negative for mycoplasma contamination) were electroporated with the various linearized targeting constructs and selected in standard ES medium containing G418 (Sigma) for 10 days. Recombinant clones were identified by Southern blot using probes that hybridize outside the 5’ and the 3’ homology regions as previously described7. Two clones for each allele were injected into C57BL/6 blastocysts to generate chimeric mice. To delete the Neo cassette, mice heterozygous for each allele were crossed to beta-Actin-Flpe mice58. miR-17~92Δ mice were genotyped as previously described7. Animals carrying the remaining miR-17~92 mutant alleles were genotyped using a three-primer PCR (p1: CCTCAAGGGAAAGATGGCAAAC, final concentration 0.4µM; p2: ATAGCCTGAAACCAACTGTGC, final concentration 0.2µM; p3: TAAAGCGCATGCTCCAGAC, final concentration 0.2µM), which amplifies a 155bp band from a wild type allele (p1/p2), a 237bp band from all targeted alleles (p1/p3), and a 368 bp band following deletion of the Neo cassette (p1/p2). Mice were maintained in both mixed 129SvJae × C57BL/6J (B6;129) and pure C57BL/6J backgrounds. All data related to skeletal defects were collected in mice backcrossed to C57BL/6 for at least 7 generations. Remaining data were collected in animals from B6;129 colonies. No specific randomization procedure was required. For the lymphoma and prostate cancer experiments, the technician monitoring mouse health status was blind to genotype. All studies and procedures were approved by the Memorial Sloan-Kettering Cancer Center Institutional Animal Care and Use Committee.
Skeletal preparations
E18.5 fetuses were eviscerated, soaked in water for 2–4 h, and placed in a 65°C water bath for 1 min before skinning. Adult mice were euthanized by CO2 asphyxiation. Eviscerated and skinned animals were fixed in 100% ethanol, stained for cartilage with alcian blue (150 mg/l alcian blue 8GX, 80% ethanol, 20% acetic acid), followed by bone staining with alizarin red (50 mg/l alizarin red S in 2% KOH). Skeletons were stored in clearing solution (40% glycerol, 20% benzyl alcohol, 30% ethanol). Images were captured with a Zeiss Stereo Discovery V8 microscope and processed in Photoshop. Measurements were performed in ImageJ on digitalized images. The investigator scoring for skeletal defects was blind to the genotype.
miRNA detection by qPCR
Total RNA from cells or homogenized tissues was extracted using Trizol (Invitrogen) following manufacturer’s instructions. Expression of miRNAs was determined with TaqMan miRNA expression assays (Applied Biosystems) according to the manufacturer's instructions. Assays were performed on a minimum of three biological replicates and normalized to sno135.
Flow cytometry analysis
Single-cell suspensions prepared from bone marrow, spleen, and tumor samples were stained in FACS buffer (0.5%FBS and 2mM EDTA in PBS) for B-, T-, and myeloid cells markers using the following antibodies: B220, IgD, CD8, CD11b, CD25, CD43, CD44, CD117, BP-1, CD24/HSA (BD PharMingen); IgM, TCRβ, and CD4 (eBioscience). Flow cytometry was performed on FACSCalibur or LSRII (BD Biosciences) flow cytometers. FACS data were analyzed with FlowJo (TreeStar). Apoptosis was measured using the Caspase Detection Kit (Red-VAD-FMK or FITC-VAD-FMK, Calbiochem). Cell cycle analysis of pre-B cell was measured using Click-iT® EdU Cell Proliferation Assays (Life technology) and FITC BrdU Flow Kit (BD Pharmingen).
Histology
Eosin/Haematoxylin and immunohistochemistry stainings were performed on 5μm sections of formalin-fixed, paraffin-embedded tissues according to standard protocols. Cleaved caspase-3 was detected with an antibody against Asp175 (Cell Signaling) diluted 1:200 in PBS; 2% BSA. Images were acquired using a Zeiss Apotome microscope equipped with a Zeiss AxioCam MRM camera.
Magnetic resonance imaging (MRI)
Mouse prostate MR images were acquired on a Bruker 4.7T Biospec scanner by the Animal Imaging Core Facility at Memorial Sloan-Kettering Cancer Center. A total of 35 images were acquired every 0.9mm per animal and used for volumetric calculations.
Small RNA sequencing
Small RNA libraries were prepared by previously published protocol59 and sequenced as single read 50bp on a single HiSeq2500 lane that generated more than 180M reads. The sequencing adaptor (TCGTATGCCGTCTTCTGCTTG) was trimmed from the 3’ end of the reads using FLEXBar tool and libraries demuxed using a custom python script. Reads were mapped to the reference mouse genome (mm9) using STAR aligner60 with parameter --outFilterMismatchNmax 1 to minimize mismatches to the genome. The resulting sam files were converted to bam and indexed using samtools61. MiRNA read counts were computed using featureCounts62 using miRNA mm9 coordinates downloaded from mirBase (version 18). Multi-mapping reads were counted according to the number of locations they were mapped to and reads mapping to more than one mature miRNA were counted equally. Finally, miRNA read counts were normalized by geometric mean implemented in DESeq R package.
RNA-seq experiment and computational analysis
Total RNAs was extracted from microdissected tail buds and embryonic hearts (20–22 somite stage; E9.5) of three embryos/genotype (48 samples in total). Libraries were prepared using the TruSeq SBS Kit v3 kit according to Illumina protocol and sequenced as single read 50 on 12 lanes generating an average of 34M reads per sample. The adapter sequence GATCGGAAGAGC was trimmed using FLEXBar (version 2.4) and reads were mapped to the mm9 mouse genome using STAR aligners (version 2.3.0e_r291) with parameter –outFilterMismatchNmax 563.
RNA-seq reads aligned to the genome were counted at each gene locus, and count values were transformed to Reads Per Kilobase per Million (RPKM) by using the total number of uniquely mapping reads to all the genes and the length of each gene. For every tissue, a gene was called expressed if it had 5 RPKM or more in at least 2/3 samples. These genes were further analyzed. Differential gene expression analysis was performed with edgeR64, contrasting each of the targeted deletions of members of the miR-17~92 cluster (Δ17; Δ18; Δ19; Δ92; Δ17,18; and Δ17,18,92) against the wild type condition.
Circos plots were generated using the Circos software37 differentially expressed genes were selected using a 10% false discovery rate cutoff. Predicted target sites were downloaded from TargetScan v6.2.
Enrichment of Gene Ontologies65 in the Biological Processes category was calculated for genes differentially expressed (abs(log2-FC) >= 0.26 and FDR <= 1%) in each of the knockouts and in each of the tissues using the Bioconductor GOstats package66, using as background all the genes expressed in each condition; Reported are GO terms filtered by size (between 15 and 250 genes) to focus on terms that are not too general or too specific.
Supplementary Material
Acknowledgements
We thank Jennifer Hollenstein for editing the manuscript and members of the Ventura lab for helpful discussions. The authors greatly acknowledge Weill Cornell Epigenomics Core contribution. This work was funded by grants from NIH/NCI (R01CA149707 to A.V. and Core Grant P30 CA 008748), the STARR Consortium (to A.V. and D.B.), the Geoffrey Beene Cancer Foundation (to A.V.), the Gabrielle’s Angel Foundation (to A.V.), the Leukemia Lymphoma Society (to Y.C.H.), the American Italian Cancer Foundation (to C.B.) and an NIH training grant (F31CA168356 to C.P.C.).
Footnotes
URLs
Targetscan: http://www.targetscan.org/
Author contributions
AV, JAV, YCH and PM conceived the project and designed the experiments. AV, JAV and YCH wrote the manuscript. AV, EY, PO, and PM generated the mouse strains. YCH characterized overall viability. The hematopoietic phenotypes were characterized by YCH and PM; JAV, EY, and LS, the skeletal phenotypes; and PM, BC, and CB characterized the oncogenic phenotypes. YCH, JAV and CPC generated the small RNA libraries. YCH and JAV generated the RNA-seq libraries. AV, DB, CL, IS, and AG did the computational analysis of the RNA-seq data.
Statement on competing financial interests
The authors declare no competing financial interests.
REFERENCES
- 1.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lai EC. Micro RNAs are complementary to 3' UTR sequence motifs that mediate negative post-transcriptional regulation. Nature genetics. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- 3.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 4.Lim LP, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 5.Chiang HR, et al. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes & development. 2010;24:992–1009. doi: 10.1101/gad.1884710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Concepcion CP, Bonetti C, Ventura A. The microRNA-17-92 family of microRNA clusters in development and disease. Cancer journal. 2012;18:262–267. doi: 10.1097/PPO.0b013e318258b60a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ventura a, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 similar to 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Pontual L, et al. Germline deletion of the miR-17 approximately 92 cluster causes skeletal and growth defects in humans. Nature genetics. 2011;43:1026–1030. doi: 10.1038/ng.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feingold M, Hall BD, Lacassie Y, Martinez-Frias ML. Syndrome of microcephaly, facial and hand abnormalities, tracheoesophageal fistula, duodenal atresia, and developmental delay. Am J Med Genet. 1997;69:245–249. [PubMed] [Google Scholar]
- 10.Ota A, et al. Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer research. 2004;64:3087–3095. doi: 10.1158/0008-5472.can-03-3773. [DOI] [PubMed] [Google Scholar]
- 11.Hayashita Y, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer research. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 12.He L, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumps C, et al. Focal DNA copy number changes in neuroblastoma target MYCN regulated genes. PloS one. 2013;8:e52321. doi: 10.1371/journal.pone.0052321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mi S, et al. Aberrant overexpression and function of the miR-17-92 cluster in MLL-rearranged acute leukemia. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:3710–3715. doi: 10.1073/pnas.0914900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schetter AJ, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thayanithy V, et al. Perturbation of 14q32 miRNAs-cMYC gene network in osteosarcoma. Bone. 2012;50:171–181. doi: 10.1016/j.bone.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 18.Conkrite K, et al. miR-17~92 cooperates with RB pathway mutations to promote retinoblastoma. Genes & development. 2011;25:1734–1745. doi: 10.1101/gad.17027411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dews M, et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nature genetics. 2006;38:1060–1065. doi: 10.1038/ng1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin HY, et al. MicroRNA-17~92 plays a causative role in lymphomagenesis by coordinating multiple oncogenic pathways. The EMBO journal. 2013;32:2377–2391. doi: 10.1038/emboj.2013.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olive V, et al. miR-19 is a key oncogenic component of mir-17-92. Genes & development. 2009;23:2839–2849. doi: 10.1101/gad.1861409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olive V, et al. A component of the mir-17-92 polycistronic oncomir promotes oncogene-dependent apoptosis. eLife. 2013;2:e00822. doi: 10.7554/eLife.00822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sandhu SK, et al. B-cell malignancies in microRNA Emu-miR-17~92 transgenic mice. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:18208–18213. doi: 10.1073/pnas.1315365110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uziel T, et al. The miR-17~92 cluster collaborates with the Sonic Hedgehog pathway in medulloblastoma. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:2812–2817. doi: 10.1073/pnas.0809579106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mavrakis KJ, et al. Genome-wide RNA-mediated interference screen identifies miR-19 targets in Notch-induced T-cell acute lymphoblastic leukaemia. Nature cell biology. 2010;12:372–379. doi: 10.1038/ncb2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mu P, et al. Genetic dissection of the miR-17~92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes & development. 2009;23:2806–2811. doi: 10.1101/gad.1872909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saga Y, Takeda H. The making of the somite: molecular events in vertebrate segmentation. Nature reviews. Genetics. 2001;2:835–845. doi: 10.1038/35098552. [DOI] [PubMed] [Google Scholar]
- 28.Wellik DM. Hox patterning of the vertebrate axial skeleton. Developmental dynamics : an official publication of the American Association of Anatomists. 2007;236:2454–2463. doi: 10.1002/dvdy.21286. [DOI] [PubMed] [Google Scholar]
- 29.Adams JM, et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- 30.Langdon WY, Harris AW, Cory S, Adams JM. The c-myc oncogene perturbs B lymphocyte development in E-mu-myc transgenic mice. Cell. 1986;47:11–18. doi: 10.1016/0092-8674(86)90361-2. [DOI] [PubMed] [Google Scholar]
- 31.Jenkins RB, Qian J, Lieber MM, Bostwick DG. Detection of c-myc oncogene amplification and chromosomal anomalies in metastatic prostatic carcinoma by fluorescence in situ hybridization. Cancer research. 1997;57:524–531. [PubMed] [Google Scholar]
- 32.Qian J, Jenkins RB, Bostwick DG. Detection of chromosomal anomalies and c-myc gene amplification in the cribriform pattern of prostatic intraepithelial neoplasia and carcinoma by fluorescence in situ hybridization. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 1997;10:1113–1119. [PubMed] [Google Scholar]
- 33.Ellwood-Yen K, et al. Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer cell. 2003;4:223–238. doi: 10.1016/s1535-6108(03)00197-1. [DOI] [PubMed] [Google Scholar]
- 34.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Selbach M, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 36.Eichhorn SW, et al. mRNA destabilization is the dominant effect of mammalian microRNAs by the time substantial repression ensues. Molecular cell. 2014;56:104–115. doi: 10.1016/j.molcel.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krzywinski M, et al. Circos: an information aesthetic for comparative genomics. Genome research. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Galvin KM, et al. A role for smad6 in development and homeostasis of the cardiovascular system. Nature genetics. 2000;24:171–174. doi: 10.1038/72835. [DOI] [PubMed] [Google Scholar]
- 39.Meneghini V, Odent S, Platonova N, Egeo A, Merlo GR. Novel TBX3 mutation data in families with ulnar-mammary syndrome indicate a genotype-phenotype relationship: mutations that do not disrupt the T-domain are associated with less severe limb defects. European journal of medical genetics. 2006;49:151–158. doi: 10.1016/j.ejmg.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 40.Stennard FA, et al. Murine T-box transcription factor Tbx20 acts as a repressor during heart development, and is essential for adult heart integrity, function and adaptation. Development. 2005;132:2451–2462. doi: 10.1242/dev.01799. [DOI] [PubMed] [Google Scholar]
- 41.Kirk EP, et al. Mutations in cardiac T-box factor gene TBX20 are associated with diverse cardiac pathologies, including defects of septation and valvulogenesis and cardiomyopathy. American journal of human genetics. 2007;81:280–291. doi: 10.1086/519530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kleaveland B, et al. Regulation of cardiovascular development and integrity by the heart of glass-cerebral cavernous malformation protein pathway. Nature medicine. 2009;15:169–176. doi: 10.1038/nm.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee JS, et al. Klf2 is an essential regulator of vascular hemodynamic forces in vivo. Developmental cell. 2006;11:845–857. doi: 10.1016/j.devcel.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 44.Follit JA, et al. The Golgin GMAP210/TRIP11 anchors IFT20 to the Golgi complex. PLoS genetics. 2008;4 doi: 10.1371/journal.pgen.1000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beppu H, et al. BMP type II receptor is required for gastrulation and early development of mouse embryos. Developmental biology. 2000;221:249–258. doi: 10.1006/dbio.2000.9670. [DOI] [PubMed] [Google Scholar]
- 46.Winnier G, Blessing M, Labosky PA, Hogan BL. Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes & development. 1995;9:2105–2116. doi: 10.1101/gad.9.17.2105. [DOI] [PubMed] [Google Scholar]
- 47.Chapman DL, Papaioannou VE. Three neural tubes in mouse embryos with mutations in the T-box gene Tbx6. Nature. 1998;391:695–697. doi: 10.1038/35624. [DOI] [PubMed] [Google Scholar]
- 48.Jiang YJ, et al. Notch signalling and the synchronization of the somite segmentation clock. Nature. 2000;408:475–479. doi: 10.1038/35044091. [DOI] [PubMed] [Google Scholar]
- 49.Pourquie O. The segmentation clock: converting embryonic time into spatial pattern. Science. 2003;301 doi: 10.1126/science.1085887. [DOI] [PubMed] [Google Scholar]
- 50.Deschamps J, van Nes J. Developmental regulation of the Hox genes during axial morphogenesis in the mouse. Development. 2005;132:2931–2942. doi: 10.1242/dev.01897. [DOI] [PubMed] [Google Scholar]
- 51.McPherron AC, Lawler AM, Lee SJ. Regulation of anterior/posterior patterning of the axial skeleton by growth/differentiation factor 11. Nature genetics. 1999;22:260–264. doi: 10.1038/10320. [DOI] [PubMed] [Google Scholar]
- 52.Selever J, Liu W, Lu MF, Behringer RR, Martin JF. Bmp4 in limb bud mesoderm regulates digit pattern by controlling AER development. Developmental biology. 2004;276:268–279. doi: 10.1016/j.ydbio.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 53.Davenport TG, Jerome-Majewska LA, Papaioannou VE. Mammary gland, limb and yolk sac defects in mice lacking Tbx3, the gene mutated in human ulnar mammary syndrome. Development. 2003;130:2263–2273. doi: 10.1242/dev.00431. [DOI] [PubMed] [Google Scholar]
- 54.Yekta S, Tabin CJ, Bartel DP. MicroRNAs in the Hox network: an apparent link to posterior prevalence. Nature reviews. Genetics. 2008;9:789–796. doi: 10.1038/nrg2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hornstein E, et al. The microRNA miR-196 acts upstream of Hoxb8 and Shh in limb development. Nature. 2005;438:671–674. doi: 10.1038/nature04138. [DOI] [PubMed] [Google Scholar]
- 56.Ronshaugen M, Biemar F, Piel J, Levine M, Lai EC. The Drosophila microRNA iab-4 causes a dominant homeotic transformation of halteres to wings. Genes & development. 2005;19:2947–2952. doi: 10.1101/gad.1372505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tanzer A, Stadler PF. Molecular evolution of a microRNA cluster. Journal of molecular biology. 2004;339:327–335. doi: 10.1016/j.jmb.2004.03.065. [DOI] [PubMed] [Google Scholar]
- 58.Rodriguez CI, et al. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nature genetics. 2000;25:139–140. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- 59.Hafner M, et al. Barcoded cDNA library preparation for small RNA profiling by next-generation sequencing. Methods. 2012;58:164–170. doi: 10.1016/j.ymeth.2012.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liao Y, Smyth GK, Shi W. feature Counts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2013 doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 63.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature genetics. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23:257–258. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.