Abstract
Gastrointestinal stromal tumors (GIST) are the most frequently occurring digestive sarcomas. The prognosis of localized GIST is heterogeneous, notably for patients with an Armed Forces Institute of Pathology (AFIP) intermediate or high risk of relapse. Despite imatinib effectiveness, it is crucial to develop therapies able to overcome the resistance mechanisms. The immune system represents an attractive prognostic and therapeutic target. The Programmed cell Death 1 (PD1)/programmed cell death ligand 1 (PDL1) pathway is a key inhibitor of the immune response; recently, anti-PD1 and anti-PDL1 drugs showed very promising results in patients with solid tumors. However, PDL1 expression has never been studied in GIST. Our objective was to analyze PDL1 expression in a large series of clinical samples. We analyzed mRNA expression data of 139 operated imatinib-untreated localized GIST profiled using DNA microarrays and searched for correlations with histoclinical features including postoperative metastatic relapse. PDL1 expression was heterogeneous across tumors and was higher in AFIP low-risk than in high-risk samples, and in samples without than with metastatic relapse. PDL1 expression was associated with immunity-related parameters such as T–cell-specific and CD8+ T–cell-specific gene expression signatures and probabilities of activation of interferon α (IFNα), IFNγ, and tumor necrosis factor α (TNFα) pathways, suggesting positive correlation with a cytotoxic T-cell response. In multivariate analysis, the PDL1-low group was associated with a higher metastatic risk independently of the AFIP classification and the KIT mutational status. In conclusion, PDL1 expression refines the prediction of metastatic relapse in localized GIST and might improve our ability to better tailor adjuvant imatinib. In the metastatic setting, PDL1 expression might guide the use of PDL1 inhibitors, alone or associated with tyrosine kinase inhibitors.
Keywords: DNA microarray, gene expression, GIST, immune response, PDL1, prognosis
Abbreviations: AFIP, Armed Forces Institute of Pathology; FDR, false discovery rate; GEO, gene expression omnibus; GES, gene expression signatures; GIST, gastrointestinal stromal tumors; GO, gene ontology; IHC, immunohistochemistry; ISH, in situ hybridization; MFS, metastasis-free survival; MHC, major histocompatibility complex; NCBI, National Center for Biotechnology Information; NK cells, natural killer cells; PCA, principal component analysis; PD1, programmed cell death 1; PDL1, programmed cell death ligand 1; PDGFRA, platelet-derived growth factor receptor α; qRT-PCR, quantitative reverse transcription-polymerase chain reaction; REMARK, REcommendations for tumor MARKer; RMA, robust multichip average; ROC, receiver operating characteristic; TILs, tumor-infiltrating lymphocytes; Treg, regulatory T cells; WT, wild type
Introduction
Gastrointestinal stromal tumors (GIST) are the most frequently occurring digestive sarcomas.1,2 They are an exemplary model for molecular-based treatment within solid tumors because of the presence of activating KIT or PDGFRA oncogenic mutations in ∼85% of cases3 and the resulting high sensitivity to KIT and platelet-derived growth factor receptor α (PDGFRA) tyrosine kinase inhibitors (imatinib or sunitinib).4,5 In advanced stages, first-line imatinib increases both the response rate (70% vs. <10% with chemotherapy) and the median overall survival (76 months in the recent BFR14 clinical trial6 vs. <10 months with chemotherapy). In localized stages treated by complete surgical resection,2 adjuvant imatinib decreases the relapse rate7,8 and improves overall survival8 and is recommended for patients with intermediate or high metastatic risk according to the Armed Forces Institute of Pathology (AFIP) prognostic classification.9
Nevertheless, the current prognostic classifications remain imperfect with substantial heterogeneity within each class: for example, the 2-year relapse-free survival without adjuvant imatinib is ∼75% in the AFIP intermediate-risk patients and ∼50% in the high-risk patients.7 Clearly, these classifications, based on histoclinical features need to be refined.4,10-13 Another crucial objective is to develop new therapies able to overcome the molecular mechanisms of primary or secondary resistance to imatinib. Indeed, if not present initially, imatinib-resistant tumor clones emerge in most cases because of the mutagenic capacity of cancer cells, thus limiting the duration of tumor responses.
In this context, and even if our current understanding of the immune response in GIST remains limited when compared with other cancers, several data suggest that the exploration of the immune system is an interesting strategy. First, immune cells such as T cells, CD8+ T-cells, regulatory T cells (Treg), natural killer (NK) cells, and macrophages are present in clinical GIST samples.14-17 Second, their presence and/or activation have been associated with prognosis16 and/or response to imatinib.14,18,19 Third, and as observed with certain chemotherapy drugs,20 the antitumor action of imatinib is also due in part to indirect effects on immune cells, notably NK cells;21 and CD8+ T cells14; in the same line, the concurrent CTLA-4 blockade augments the efficacy of imatinib in mouse GIST by increasing IFNγ-producing CD8+ T cells.14 Finally, by contrast with targeted therapies, immunotherapy can adapt to the emergence of resistant clones thanks to the high adaptability of the immune response.
Like CTLA-4, the Programmed cell Death 1 (PD1) pathway is a key inhibitor of the immune response, regulating the balance between activation and inhibition signals. PD1 is expressed at the surface of various immune cells including T cells. PD1 activation by its ligand Programmed cell Death 1 ligand (PDL1), expressed by antigen-presenting cells (APCs) such as macrophages or B cells, regulates lymphocyte activation22-24 and promotes Treg cell development and function, allowing termination of the immune response. Cancer cells from different locations have acquired the capacity to express PDL1.25 The PD1-PDL1 pathway has, thus, been involved in cancer progression.26,27 Anti-PD1 and anti-PDL1 drugs28 are being tested in phase 3 clinical trials after very promising results in phase 2 trials.29-32 Durable responses have been observed, notably in melanoma and renal and lung carcinomas,33,34 and a relationship between PDL1 expression on cancer cells and objective response has been evidenced.34,35 PDL1 expression has never been studied in GIST.
Here, we analyzed PDL1 expression in 139 imatinib-untreated localized GIST and searched for correlations with histoclinical features including metastasis-free survival (MFS) after surgery. We show that PDL1 expression is heterogeneous and is an independent prognostic factor in multivariate analysis.
Results
Patients characteristics and PDL1 expression
Of the 159 available GIST samples, a total of 139 represented localized tumors from patients treated with primary complete surgery without adjuvant imatinib. Their characteristics are summarized in Table 1. Sixty-four percent of patients were male. The median patient age was 63 y The most frequent anatomical site was the stomach (78%), followed by the small intestine (15%). The mutational status was available for 138 samples: as expected, the most frequent mutations were KIT mutations, most frequently located in exon 11, followed by PDGFRA mutations, most frequently located in exon 18. No KIT and PDGFRA mutation was observed in 12% of samples (KIT and PDGFRA wild type, WT). Regarding the relapse risk defined according to the AFIP classification, 54% of samples were predicted as low risk, 18% as intermediate risk, and 28% as high risk. Twenty percent of patients experienced a metastatic relapse during follow-up. As shown in Figure S1A, PDL1 mRNA expression varied among these 139 tumors with a wide range of intensities over 3 decades in a log2 scale, suggesting a heterogeneous expression across clinical samples of GIST.
Table 1.
Histoclinical characteristics of patients and tumors
| Characteristicsa | N (%) |
|---|---|
| Sex (139) | |
| Female | 50 (36%) |
| Male | 89 (64%) |
| Age, years (79) | |
| ≤60 | 31 (39%) |
| <60 | 48 (61%) |
| median (range) | 63 (26–85) |
| Site (139) | |
| Gastric | 109 (78%) |
| Small intestine | 21 (15%) |
| Other | 9 (6%) |
| Mutationa (138) | |
| KIT exon 11 | 88 (64%) |
| KIT exon 9 | 8 (6%) |
| PDGFRA exon 18 | 16 (12%) |
| Wild type (WT) | 17 (12%) |
| Otherb | 9 (7%) |
| AFIP risk (139) | |
| Low | 75 (54%) |
| Intermediate | 25 (18%) |
| High | 39 (28%) |
| Metastatic relapse (138) | |
| No | 111 (80%) |
| Yes | 27 (20%) |
aThe number of patients with available information is shown between brackets; bKIT exons 13 (n = 1) and 17 (n = 2), PDGFRA exons 12 (n = 5), and 14 (n = 1).
PDL1 expression and histoclinical and immune features
We searched for correlations between PDL1 expression (continuous value; Student's t-test) and histoclinical features (Table 2). PDL1 expression was not significantly associated with patients’ sex and age or with tumor site and mutational status. By contrast, PDL1 expression correlated with the AFIP classification and metastatic relapse. PDL1 expression was higher in low-risk samples than in high-risk samples (P = 0.0194), and in samples without metastatic relapse than in samples with metastatic relapse (P = 0.0029) , suggesting favorable prognostic value.
Table 2.
Correlations of PDL1 expression with histoclinical features
| Characteristics* | N | Average PDL1 expression (min–max) | P-value |
|---|---|---|---|
| Sex (139) | 0.07 | ||
| Female | 50 | 0.27 (−1.32–3.15) | |
| Male | 89 | −0.07 (−2.13–2.24) | |
| Age, years (79) | 0.649 | ||
| ≤60 | 31 | 0.16 (−1.5–2.24) | |
| <60 | 48 | 0.05 (−1.73–3.15) | |
| Site (139) | 0.37 | ||
| Gastric | 109 | 0.09 (−2.13–3.15) | |
| Small intestine | 21 | −0.21 (−1.61–1.21) | |
| Other | 9 | 0.23 (−0.41–1.19) | |
| Mutation (129) | 0.89 | ||
| KIT exon 11 | 88 | 0.08 (−2.13–2.36) | |
| KIT exon 9 | 8 | −0.18 (−0.67–0.48) | |
| PDGFRA exon 18 | 16 | −0.01 (−1.15–1.63) | |
| Wild type (WT) | 17 | −0.01 (−1.61–3.15) | |
| AFIP risk (139) | 0.0194 | ||
| Low | 75 | 0.2 (−1.75–3.15) | |
| Intermediate | 25 | 0.19 (−1.13–2.36) | |
| High | 39 | −0.32 (−2.13–1.67) | |
| Metastatic relapse (138) | 0.0029 | ||
| No | 111 | 0.18 (−1.75–3.15) | |
| Yes | 27 | −0.46 (−2.13–1.19) |
aThe number of patients with available information is shown within parentheses.
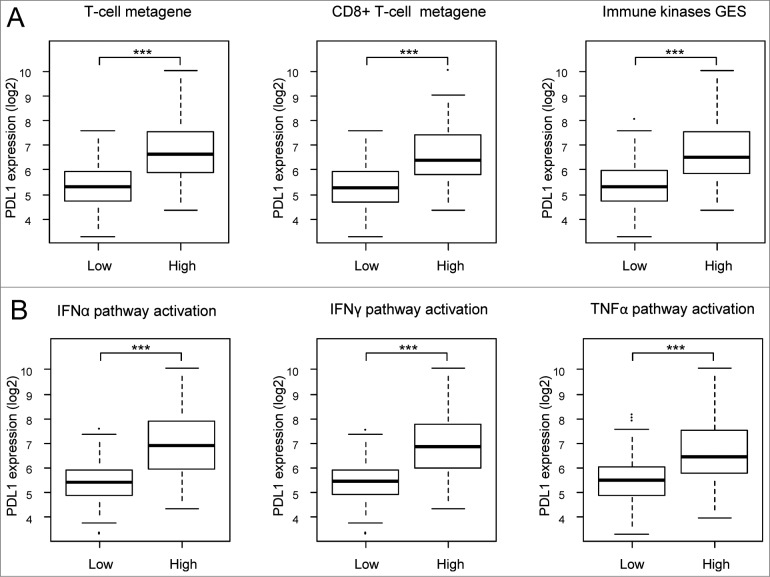
We then studied whether PDL1 expression was associated (Student's t-test) with immunity-related parameters in clinical GIST samples. First, we found a correlation with T–cell-specific, CD8+ T–cell-specific, and B–cell-specific gene expression signatures36; samples with higher expression of these signatures overexpressed PDL1 (P < 0.001; Fig. 1A). Second, PDL1 expression was higher (P < 0.001) in samples showing higher expression of an immune kinase signature reflecting the immune response and, particularly, cytotoxic T-cell response.37 Finally, we found that the probability of activation of IFNα, IFNγ, and tumor necrosis factor α (TNFα) pathways38 was associated (P < 0.001) with higher PDL1 expression (Fig. 1B). Altogether, these results suggested that PDL1 expression in samples of GIST is associated with antitumor T-cell response.
Figure 1.
Correlations of PDL1 expression with immune features. PDL1 expression levels reported as a box plot according to sample classifications based on gene expression signatures (A) including T cells (left) and CD8+ T cells (middle) metagenes and a prognostic immune kinase gene expression signature (right), and based on the probability of activation of immune pathways (B) including IFNα (left), IFNγ (middle), and TNFα (right). The P-values are indicated (Student's t-test) are indicated as follows: ***, P < 0.001.
PDL1 expression and metastatic relapse
The follow-up was available for 138 patients: 27 experienced a metastatic relapse and 111 did not. As shown in Table 1, lower PDL1 expression was associated with more frequent metastatic relapse. We repeated the analysis using PDL1 expression as a binary variable. We first defined the optimal expression cut-off associated with the occurrence of metastatic relapse in a learning set of 92 samples: measured at 5.08, it resulted into an odds ratio (OR) for relapse of 4.74 ([1.30–17.68], P = 0.0083; Fisher's exact test) in the PDL1-low group versus the PDL1-high group. We confirmed its discriminatory prognostic value in the validation set of 46 samples with an OR for relapse of 8.80 ([1.04–115.54], P = 0.023; Fisher's exact test) in the PDL1-low vs. PDL1-high groups (Table S1).
Of the 138 samples, 27 were in the PDL1-low group (20%) and 111 in the PDL1-high group (80%). In the univariate analysis (Table 3), PDL1-low group (P = 0.0002; logit test) , AFIP intermediate-risk classification (P = 0.0065), and AFIP high-risk classification (P < 0.0001) were associated with a higher risk of relapse, whereas patient age, anatomical site, and PDGFRA exon 18 mutations were not. A trend (P < 0.10) toward worse prognosis was observed for KIT exon 11 and KIT exon 9 mutations, and male gender. In the multivariate analysis, PDL1-low group (P = 0.0056; logit test), AFIP intermediate-risk classification (P = 0.0226), and AFIP high-risk classification (P < 0.0001) remained significant, whereas gender and mutations did not, even if a trend existed for KIT exon 11 mutation versus WT status (Table 3). Of note, and to verify the absence of overfitting with regard to the independent prognostic value of PDL1, we repeated the same multivariate analysis in the validation set only: 2 variables – the PDL1 group (P = 0.0097; logit test) and the AFIP high-risk classification (P = 0.0004) remained significant (data not shown).Stratification of patients according to the PDL1 group and, respectively, the AFIP risk and the mutational status identified subgroups with different relapse rates (Table 2). For example, PDL1 expression affected the clinical outcome of AFIP high-risk patients: the PDL1-low group showed a higher rate of relapse (79%) than the PDL1-high group (46%). Similarly, PDL1 expression affected the clinical outcome of patients with a KIT exon 11 mutation (56% relapse rate in the PDL1-low group vs. 16% in the PDL1-high group) and patients with wild-type (WT) GIST (20% relapse rate in the PDL1-low group versus 0% in the PDL1-high group), leading to similar relapse rate between KIT exon 11-PDL1-high GIST (16%) and WT-PDL1-low GIST (20%).
Table 3.
Univariate and multivariate prognostic analyses for metastasis-free survival
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| N | Odds ratio [95%CI] | P-value | N | Odds ratio [95%CI] | P-value | |
| Sex, | ||||||
| Male vs. Female | 138 | 1.13 [1.00–1.26] | 0.09 | 128 | 1.08 [0.98–1.19] | 0.20 |
| Age, | ||||||
| ≤60 vs. >60 years | 78 | 0.94 [0.81–1.08] | 0.44 | |||
| Site, | ||||||
| Other vs. Gastric | 138 | 1.18 [0.94–1.48] | 0.23 | |||
| Small intestine vs. gastric | 138 | 1.13 [0.96–1.32] | 0.21 | |||
| Mutation, | ||||||
| KIT exon 11 vs. WT | 128 | 1.20 [1.01–1.43] | 0.09 | 128 | 1.15 [1.00–1.32] | 0.10 |
| KIT exon 9 vs. WT | 128 | 1.37 [1.03–1.82] | 0.07 | 128 | 1.30 [1.04–1.63] | 0.06 |
| PDGFRA exon 18 vs. WT | 128 | 1.00 [0.80–1.26] | 0.98 | 128 | 1.08 [0.90–1.30] | 0.48 |
| AFIP risk, | ||||||
| Intermediate vs. low | 138 | 1.22 [1.08–1.38] | 0.0065 | 128 | 1.19 [1.05–1.35] | 0.0226 |
| High vs. low | 138 | 1.78 [1.61–1.98] | < 0.0001 | 128 | 1.66 [1.49–1.86] | < 0.0001 |
| PDL1 expression, High vs. low | 138 | 0.74 [0.64–0.84] | 0.0002 | 128 | 0.81 [0.72–0.92] | 0.0056 |
PDL1 expression and associated biological processes
Supervised analysis comparing the whole-genome expression profiles of the PDL1-low (n = 27) and PDL1-high (n = 111) samples identified 303 differentially expressed genes, which were all overexpressed in the PDL1-high samples (Table S3). Ontological analysis (Table S4) showed that these genes were particularly involved in immune response regulation, and more specifically in T-cell activation. Indeed, many top genes (correlated with PDL1) were major histocompatibility complex (MHC)-related molecules, that is, involved in the processing of endogenous antigens and presentation to cytotoxic and helper T cells. Among them, we found not only numerous human leukocyte antigen (HLA)-I or HLA–I-related molecules (HLA-A-C, HLA-E, HLA-G, HLA-F, all members of the BTN3A family, KLRG1, etc.), but also HLA-II molecules (HLA-DR, HLA-DP, HLA-DQ, HLA-DM, CD74, etc.) and molecules involved in the degradation of cytosolic peptides across the endoplasmic reticulum into the membrane-bound compartment where class I molecules assemble (TAP1, TAPBP, PSMB8–10, CTSS, etc). This “antigen presentation” signature was associated with a strong antitumor response, as suggested by the numerous genes testifying of TH1 activation (many IFN-related genes, IL12RB1, IL18BP, IL18, IL2RB, IL2RG, IL15, IL7 and IL7R, STAT1, ITK, LCK, JAK2, LAG3, CD69, etc.) and cytotoxic effector molecules (GZMA, GZMK, GZMH, PRF1, C1-complement members, CD52, FASLG, etc). This signature suggested that cytotoxic T cells are major mediators of such antitumor response, which was further confirmed with the increased expression of transcripts related to CD8+ T-cells (CD160, CD2, CD53, CD8A, CD3D, CD247, PTPRC, etc). Other actors of the immune system, notably APCs, certainly play an important role as well (especially, macrophages or related APCs expressing CD163, CD209, CD14, TLR3, or TLR8, etc). Both protagonists were probably recruited within the tumor through the following cytokines/cytokine receptors whose genes were found included in the signature: CCR5, CXCR6, CCL4 and CCL5, CXCL9–11, CD97, LTB, etc Altogether, this PDL1-high signature indicated a strong ability of PDL1-high GIST microenvironment to trigger an efficient antitumor response, principally conducted by cytotoxic CD8+ cells.
Discussion
We analyzed PDL1 expression in 139 imatinib-untreated, operated, localized GIST and showed that high expression was an independent favorable prognostic factor for metastatic relapse. To our knowledge, this is the first study analyzing PDL1 expression in GIST.
Our analysis was based on mRNA expression measured using DNA microarrays rather than on immunohistochemistry (IHC). In recent years, PDL1 expression in cancer has most often been studied at the protein level using IHC, but discordant results have been reported across studies, notably in prognostic studies.39 The main reason for these divergences is the absence of standardization of PDL1 IHC, an issue that remains unsolved today. Many antibodies are available but lack specificity and reproducibility,40,41 the optimal positivity cut-off is not defined, and staining interpretation suffers from subjectivity. These limitations led to the use of alternative methods, such as mRNA analysis based on in situ hybridization (ISH),42 DNA microarrays,43 or quantitative real-time polymerase chain reaction (qRT-PCR).44 A positive relationship between protein and mRNA expression has been reported.42 One limitation of DNA microarray- or qRT–PCR-based measurements is that they quantify expression level of both tumor and non-tumor cells present in the sample. This is particularly critical for carcinomas, although results are consistent with those reported using ISH.42 Our mRNA analysis based on DNA microarrays allowed us to avoid the limitations of IHC and to work on a relatively large pooled series of samples.
We showed that PDL1 expression was heterogeneous in GIST samples with a range of values over 3 decades on logarithmic scale, providing the opportunity to search for correlations with histoclinical features. Significant association was found with the AFIP classification and metastatic relapse. PDL1 expression was higher in AFIP low-risk samples than in high-risk samples. In literature, other immune tumor features have been found associated with the AFIP classification: in a series of 57 localized GIST, NK tumor-infiltrating lymphocytes (TILs) were more frequent and Tregs were less frequent in low/intermediate-risk samples than in high-risk samples,16 whereas no correlation was evidenced with CD3+ cells. Such association and ours with PDL1 expression suggest that the immune microenvironment may be, in part, driven by GIST cell-intrinsic features. In our series, PDL1 expression was also higher in samples without metastatic relapse than in samples with metastatic relapse, suggesting favorable prognostic value. This was confirmed in univariate and multivariate analyses, which identified not only the AFIP classification but also the PDL1-based tumor classification as independent prognostic variables. Patients of the PDL1-low group experienced more metastatic relapses than patients in the PDL1-high group. PDL1-based classification added prognostic information to the AFIP classification: for example, whereas AFIP high-risk patients displayed a 58% relapse rate, those included in the PDL1-low group displayed a 79% rate vs. 46% in the PDL1-high group. Such favorable prognostic value of high PDL1 expression has already been reported in other cancers, such as breast cancer,42 lung cancer,39 colorectal cancer,45 and Merkel cell carcinoma,46 but has never been studied in GIST.
The prognostic classifications, currently based on features such as anatomical site, pathological tumor size, mitotic count, tumor rupture, and mutational status, are imperfect and efforts are ongoing to improve them.4,10-13 Several tumor cell-intrinsic molecular prognosticators have been suggested, mainly based on proteins47-51 or gene signatures52,53 related to cell proliferation. Potential immune prognostic/predictive parameters have been more recently reported. In metastatic patients treated with imatinib, the NK-cell IFNγ production after 2 months of treatment18 and an alternative transcript of NKp30 gene19 were predictors for survival. In localized GIST, a low blood neutrophil-to-lymphocyte ratio was an independent favorable prognostic parameter,54 as were strong tumor infiltrates of CD3+ T cells and of NK cells.16 The favorable prognostic value of high PDL1 expression that we observed is in agreement with these data. Our observation, a priori counterintuitive given the immunosuppressive function of PDL1, may be explained by the fact that PDL1 expression is a consequence of a strong cytotoxic immune response (associated with lower rate of metastatic relapse16), and is induced in response to a homeostatic negative feedback loop, such as the one associated with IDO overexpression. For example, in breast cancer, high PDL1 mRNA expression is induced in tumor microenvironment by activated TILs42,55 through the massive release of IFNγ.43,56 Our results seem to corroborate this hypothesis because PDL1 expression positively correlated with expression signatures reflecting the immune response, and particularly cytotoxic T-cell response36,37, and the probability of activation of IFNα, IFNγ, and TNFα pathways.38 Furthermore, we identified a robust cytotoxic immune response signature in the PDL1-up GIST group, which involved differentiated CD8+ T cells, but also other actors of antitumor immunity such as γδ-T cells, NK cells, macrophages and related APCs, B cells, etc. These cells were differentiated TH1-biased cells (eomesodermin, IL-12 and IFN-induced pathways), clearly endowed with cytotoxic effector functions (CD160, granzymes, perforin, complement-related molecules, etc).
Cancer immunosurveillance relies on effector/memory tumor-infiltrating CD8+ T cells with a TH1 profile. Our results suggest a major role of the immune microenvironment, notably cytotoxic cells, in influencing the clinical outcome of GIST patients independently from other tumor cell-intrinsic features, such as mitotic index or mutational status. This natural immunosurveillance system, which essentially involves activated cytotoxic cells, most probably due to an efficient antigen presentation, might control the PDL1-high group of GIST. The composition of the immune environment, and particular cytotoxic T cells and related APCs, might shape the tumor microenvironment in the early stages. Further studies linking PDL1 transcript and PDL1 protein expression should now be launched to establish whether PDL1 could be a new biomarker able to refine the current methods of risk stratification in GIST. Finally, the use of anti-PDL1 or anti-PD1 antibodies might help reactivate an inhibited antitumor response or enhance the efficiency of the TH1 response already initiated. This might be particularly interesting in combination with imatinib administration, as shown in murine models with CTLA-4 blockade.14
In conclusion, we showed that PDL1 mRNA expression is heterogeneous in GIST samples and is associated with AFIP classification and metastatic relapse. Samples with low expression are associated with a higher risk of metastatic relapse independently from the AFIP classification and the mutational status, suggesting that PDL1 expression cooperates with tumor cell-intrinsic features to influence survival. The strength of our results lies in its originality (the first one describing PDL1 expression in GIST), the analysis of a homogeneously treated population (surgery without adjuvant imatinib), the biological and clinical relevance of PDL1 expression and its independent prognostic value in multivariate analysis. Limitations include its retrospective nature, the size of our series, although relatively important for a rare tumor and when compared to other biological prognostic studies published to date, and the analysis at the mRNA level on whole tissue samples rather than protein level. Analysis of larger imatinib-untreated patients series, retrospective, then prospective, is warranted to confirm our observation, as well as protein analyses when reliable antibodies are commercially available. IHC analyses will help determine whether PDL1 is predominantly expressed in tumor or immune cells in clinical GIST samples. If confirmed, PDL1 expression might refine the prediction of metastatic relapse and improve our ability to better tailor adjuvant imatinib. In the metastatic setting, PDL1 expression might guide the use of PDL1-inhibitors alone or in association with tyrosine kinase inhibitors.
Patients and Methods
Tumor samples
We collected histoclinical and gene expression data of clinical GIST samples from our 2 databases57,58 and 3 public data sets53,59,60 from the National Center for Biotechnology Information (NCBI)/Genbank GEO and ArrayExpress databases. The 5 datasets are described in Table S5. Samples were profiled using whole-genome DNA microarrays: Affymetrix U133 Plus 2.057–59, and Agilent 44K.53,60 The pooled data set contained 159 samples, including 139 with localized GIST treated with primary surgery without adjuvant imatinib. The study was approved by our institutional board.
Gene expression data analysis
Data analysis required pre-analytic processing. The first step was to normalize each data set separately: we used quantile normalization for the available processed Agilent data and Robust Multichip Average (RMA)61 with the non-parametric quantile algorithm for the raw Affymetrix data. Normalization was done in R using Bioconductor and associated packages. Then, hybridization probes were mapped across the 2 technological platforms represented as previously reported.62 When multiple probes mapped to the same GeneID, we retained the 1 with the highest variance in a particular dataset. We then merged the 5 data sets by using COMBAT (empirical Bayes)63 as batch effects removal method, included in the inSilicoMerging R/Bioconductor package.64 The final merged set included 16 759 genes in log2-transformed data. The accuracy of normalization was controlled by principal component analysis (PCA) (Fig. S1B).
PDL1 (CD274) tumor expression was measured by analyzing different probe sets whose identity and specificity were verified using the NCBI program BLASTN 2.2.29+ (Table S6). Analysis was done by using both continuous and binary values. Because of the involvement of PDL1 in immunity, we searched for correlations of its expression with other immune features of tumors. We thus applied different immune multigene classifiers to each tumor in each dataset separately, including metagenes associated with different immune populations such as T cells, CD8+ T cells, and B cells 36, gene expression signatures (GES) of immune pathway activity such as IFNα, IFNγ, and TNFα pathways38 and a prognostic immune kinase GES.37 To define the optimal cut-off of PDL1 expression associated with the occurrence of metastatic relapse, we divided our population into 2 randomly selected sets: a learning set to define the cut-off by using a ROC curve (n = 92) and a validation set to validate it in independent samples (n = 46). Once validated, the cut-off was applied to all samples to define the PDL1-low group (expression inferior to the cut-off) and the PDL1-high group (expression superior or equal to the cut-off). Finally, to explore the biological pathways linked to PDL1 expression in GIST, we applied a supervised analysis to the 139 samples to compare the whole-genome expression profiles of 14 546 filtered (expression level above background) genes between the PDL1-high versus PDL1-low groups. We used Significant Analysis of Microarrays (SAM)65 algorithm and P-values, corrected for multiple comparisons, were considered significant only if the false discovery rate (FDR) was smaller than 0.001. Ontological analysis of the resulting gene list was based on GO biological processes of the Database for Annotation, Vizualization and Integrated Discovery (DAVID; http://david.abcc.ncifcrf.gov/) and on the BioCarta database (http://www.biocarta.com/support/howto/path.asp).
Statistical analyses
Correlations between PDL1 expression and histoclinical factors were calculated with the Student's t-test or One-way ANOVA when appropriate for expression assessed as continuous variable and the Fisher's exact test for expression assessed as binary variable (PDL1-low and PDL1-high). The primary endpoint was the occurrence of metastatic relapse during follow-up, the delay of relapse and follow-up being not available in 2 data sets.53,60 Univariate and multivariate analyses were done using a logistic regression analysis using the lm function (R's statistical package) (significance estimated by specifying a binomial family for model with a logit link). The variables tested in univariate analysis included the sample groups based on PDL1 expression (PDL1-low and PDL1-high), patient age and gender, tumor site and mutational status, and the AFIP classification (high vs. intermediate vs. low-risk). Multivariate analysis incorporated all variables with a P-value inferior to 10% in univariate analysis. All statistical tests were 2-sided at the 5% level of significance. Statistical analysis was done using the survival package (version 2.30) in the R software (version 2.9.1). We wrote the article in accordance with the criteria specified in the reporting recommendations for tumor marker prognostic studies (REMARK).66
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the Institut Paoli-Calmettes, Inserm, Institut National du Cancer, and Comité de Corse du Sud de la Ligue Nationale contre le Cancer.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1. Corless CL, Barnett CM, Heinrich MC. Gastrointestinal stromal tumours: origin and molecular oncology. Nat Rev Cancer 2011; 11:865-78; PMID: [DOI] [PubMed] [Google Scholar]
- 2. ESMO/European Sarcoma Network Working Group Gastrointestinal stromal tumours: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2014; Suppl 3:iii21-6. [DOI] [PubMed] [Google Scholar]
- 3. Kitamura Y, Hirota S, Nishida T. Gastrointestinal stromal tumors (GIST): a model for molecule; PU1 -based diagnosis and treatment of solid tumors. Cancer Sci 2003; 94:315-20; PMID:; http://dx.doi.org/ 10.1111/j.1349-7006.2003.tb01439.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Joensuu H, Dematteo RP. The management of gastrointestinal stromal tumors: a model for targeted and multidisciplinary therapy of malignancy. Annu Rev Med 2011; 63:247-58; PMID:; http://dx.doi.org/ 10.1146/annurev-med-043010-091813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Antonescu CR. The GIST paradigm: lessons for other kinase-driven cancers. J Pathol 2011; 223:251-61; PMID:; http://dx.doi.org/ 10.1002/path.2798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Le Cesne A, Ray-Coquard I, Bui BN, Adenis A, Rios M, Bertucci F, Duffaud F, Chevreau C, Cupissol D, Cioffi A, et al. Discontinuation of imatinib in patients with advanced gastrointestinal stromal tumours after 3 years of treatment: an open-label multicentre randomised phase 3 trial. Lancet Oncol 2010; 11:942-9; PMID:; http://dx.doi.org/ 10.1016/S1470-2045(10)70222-9 [DOI] [PubMed] [Google Scholar]
- 7. Dematteo RP, Ballman KV, Antonescu CR, Maki RG, Pisters PW, Demetri GD, Blackstein ME, Blanke CD, von Mehren M, Brennan MF, et al. Adjuvant imatinib mesylate after resection of localised, primary gastrointestinal stromal tumour: a randomised, double-blind, placebo-controlled trial. Lancet 2009; 373:1097-104; PMID:; http://dx.doi.org/ 10.1016/S0140-6736(09)60500-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Joensuu H, Eriksson M, Sundby Hall K, Hartmann JT, Pink D, Schütte J, Ramadori G, Hohenberger P, Duyster J, Al-Batran SE, et al. One vs three years of adjuvant imatinib for operable gastrointestinal stromal tumor: a randomized trial. JAMA 2012; 307:1265–1272; PMID:22453568 [DOI] [PubMed] [Google Scholar]
- 9. Miettinen M, Lasota J. Gastrointestinal stromal tumors: pathology and prognosis at different sites. Semin Diagn Pathol 2006; 23:70-83; PMID:; http://dx.doi.org/ 10.1053/j.semdp.2006.09.001 [DOI] [PubMed] [Google Scholar]
- 10. Gold JS, Gonen M, Gutierrez A, Broto JM, Garcia-del-Muro X, Smyrk TC, Maki RG, Singer S, Brennan MF, Antonescu CR, et al. Development and validation of a prognostic nomogram for recurrence-free survival after complete surgical resection of localised primary gastrointestinal stromal tumour: a retrospective analysis. Lancet Oncol 2009; 10:1045-52; PMID:; http://dx.doi.org/ 10.1016/S1470-2045(09)70242-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rossi S, Miceli R, Messerini L, Bearzi I, Mazzoleni G, Capella C, Arrigoni G, Sonzogni A, Sidoni A, Toffolatti L, et al. Natural history of imatinib-naive GISTs: a retrospective analysis of 929 cases with long-term follow-up and development of a survival nomogram based on mitotic index and size as continuous variables. Am J Surg Pathol 2011; 35:1646-56; PMID:; http://dx.doi.org/ 10.1097/PAS.0b013e31822d63a7 [DOI] [PubMed] [Google Scholar]
- 12. Patel S. Navigating risk stratification systems for the management of patients with GIST. Ann Surg Oncol 2011; 18:1698-704; PMID:; http://dx.doi.org/ 10.1245/s10434-010-1496-z [DOI] [PubMed] [Google Scholar]
- 13. Wozniak A, Rutkowski P, Schoffski P, Ray-Coquard I, Hostein I, Schildhaus HU, Le Cesne A, Bylina E, Limon J, Blay JY, et al. Tumor genotype is an independent prognostic factor in primary gastrointestinal stromal tumors of gastric origin: a European multicenter analysis based on ConticaGIST. Clin Cancer Res 2014; 20(23):6105-16; PMID:; http://dx.doi.org/ 10.1158/1078-0432.CCR-14-1677 [DOI] [PubMed] [Google Scholar]
- 14. Balachandran VP, Cavnar MJ, Zeng S, Bamboat ZM, Ocuin LM, Obaid H, Sorenson EC, Popow R, Ariyan C, Rossi F, et al. Imatinib potentiates antitumor T cell responses in gastrointestinal stromal tumor through the inhibition of Ido. Nat Med 2011; 17:1094-100; PMID:; http://dx.doi.org/ 10.1038/nm.2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cameron S, Haller F, Dudas J, Moriconi F, Gunawan B, Armbrust T, Langer C, Fuzesi L, Ramadori G. Immune cells in primary gastrointestinal stromal tumors. Eur J Gastroenterol Hepatol 2008; 20:327-34; PMID:; http://dx.doi.org/ 10.1097/MEG.0b013e3282f3a403 [DOI] [PubMed] [Google Scholar]
- 16. Rusakiewicz S, Semeraro M, Sarabi M, Desbois M, Locher C, Mendez R, Vimond N, Concha A, Garrido F, Isambert N, et al. Immune infiltrates are prognostic factors in localized gastrointestinal stromal tumors. Cancer Res 2013; 73:3499-510; PMID:; http://dx.doi.org/ 10.1158/0008-5472.CAN-13-0371 [DOI] [PubMed] [Google Scholar]
- 17. van Dongen M, Savage ND, Jordanova ES, Briaire-de Bruijn IH, Walburg KV, Ottenhoff TH, Hogendoorn PC, van der Burg SH, Gelderblom H, van Hall T. Anti-inflammatory M2 type macrophages characterize metastasized and tyrosine kinase inhibitor-treated gastrointestinal stromal tumors. Int J Cancer 2010; 127:899-909; PMID: [DOI] [PubMed] [Google Scholar]
- 18. Menard C, Blay JY, Borg C, Michiels S, Ghiringhelli F, Robert C, Nonn C, Chaput N, Taieb J, Delahaye NF, et al. Natural killer cell IFN-gamma levels predict long-term survival with imatinib mesylate therapy in gastrointestinal stromal tumor-bearing patients. Cancer Res 2009; 69:3563-9; PMID:; http://dx.doi.org/ 10.1158/0008-5472.CAN-08-3807 [DOI] [PubMed] [Google Scholar]
- 19. Delahaye NF, Rusakiewicz S, Martins I, Menard C, Roux S, Lyonnet L, Paul P, Sarabi M, Chaput N, Semeraro M, et al. Alternatively spliced NKp30 isoforms affect the prognosis of gastrointestinal stromal tumors. Nat Med 2011; 17:700-7; PMID:; http://dx.doi.org/ 10.1038/nm.2366 [DOI] [PubMed] [Google Scholar]
- 20. Zitvogel L, Kepp O, Kroemer G. Immune parameters affecting the efficacy of chemotherapeutic regimens. Nat Rev Clin Oncol 2011; 8:151-60; PMID:; http://dx.doi.org/ 10.1038/nrclinonc.2010.223 [DOI] [PubMed] [Google Scholar]
- 21. Borg C, Terme M, Taieb J, Menard C, Flament C, Robert C, Maruyama K, Wakasugi H, Angevin E, Thielemans K, et al. Novel mode of action of c-kit tyrosine kinase inhibitors leading to NK cell-dependent antitumor effects. J Clin Invest 2004; 114:379-88; PMID:; http://dx.doi.org/ 10.1172/JCI21102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Amarnath S, Mangus CW, Wang JC, Wei F, He A, Kapoor V, Foley JE, Massey PR, Felizardo TC, Riley JL, et al. The PDL1-PD1 axis converts human TH1 cells into regulatory T cells. Sci Translational Med 2011; 3:111ra20; http://dx.doi.org/ 10.1126/scitranslmed.3003130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Blank C, Mackensen A. Contribution of the PD-L1/PD-1 pathway to T-cell exhaustion: an update on implications for chronic infections and tumor evasion. Cancer Immunol Immunother: CII 2007; 56:739-45; http://dx.doi.org/ 10.1007/s00262-006-0272-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Francisco LM, Salinas VH, Brown KE, Vanguri VK, Freeman GJ, Kuchroo VK, Sharpe AH. PD-L1 regulates the development, maintenance, and function of induced regulatory T cells. J Exp Med 2009; 206:3015-29; PMID:; http://dx.doi.org/ 10.1084/jem.20090847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gatalica Z, Snyder C, Maney T, Ghazalpour A, Holterman DA, Xiao N, Overberg P, Rose I, Basu GD, Vranic S, et al. Programmed cell death 1 (PD-1) and its ligand (PD-L1) in common cancers and their correlation with molecular cancer type. Cancer Epidemiol Biomarkers Prev 2014; 23(12):2965-70; PMID:; http://dx.doi.org/ 10.1158/1055-9965.EPI-14-0654 [DOI] [PubMed] [Google Scholar]
- 26. Iwai Y, Ishida M, Tanaka Y, Okazaki T, Honjo T, Minato N. Involvement of PD-L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc Natl Acad Sci U S A 2002; 99:12293-7; PMID:; http://dx.doi.org/ 10.1073/pnas.192461099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Reiss KA, Forde PM, Brahmer JR. Harnessing the power of the immune system via blockade of PD-1 and PD-L1: a promising new anticancer strategy. Immunotherapy 2014; 6:459-75; PMID:; http://dx.doi.org/ 10.2217/imt.14.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer 2012; 12:252-64; PMID:; http://dx.doi.org/ 10.1038/nrc3239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Aranda F, Vacchelli E, Eggermont A, Galon J, Sautes-Fridman C, Tartour E, Zitvogel L, Kroemer G, Galluzzi L. Trial Watch: peptide vaccines in cancer therapy. Oncoimmunology 2013; 2:e26621; PMID:; http://dx.doi.org/ 10.4161/onci.26621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Eggermont AM, Kroemer G, Zitvogel L. Immunotherapy and the concept of a clinical cure. Eur J Cancer 2013; 49:2965-7; PMID:; http://dx.doi.org/ 10.1016/j.ejca.2013.06.019 [DOI] [PubMed] [Google Scholar]
- 31. Robert C, Soria JC, Eggermont AM. Drug of the year: programmed death-1 receptor/programmed death-1 ligand-1 receptor monoclonal antibodies. Eur J Cancer 2013; 49:2968-71; PMID:; http://dx.doi.org/ 10.1016/j.ejca.2013.07.001 [DOI] [PubMed] [Google Scholar]
- 32. Zitvogel L, Kroemer G. Targeting PD-1/PD-L1 interactions for cancer immunotherapy. Oncoimmunology 2012; 1:1223-5; PMID:; http://dx.doi.org/ 10.4161/onci.21335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ, Topalian SL, Hwu P, Drake CG, Camacho LH, Kauh J, Odunsi K, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med 2012; 366:2455-65; PMID:; http://dx.doi.org/ 10.1056/NEJMoa1200694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med 2012; 366:2443-54; PMID:; http://dx.doi.org/ 10.1056/NEJMoa1200690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Taube JM, Klein A, Brahmer JR, Xu H, Pan X, Kim JH, Chen L, Pardoll DM, Topalian SL, Anders RA. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin Cancer Res 2014; 20:5064-74; PMID:; http://dx.doi.org/ 10.1158/1078-0432.CCR-13-3271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Palmer C, Diehn M, Alizadeh AA, Brown PO. Cell-type specific gene expression profiles of leukocytes in human peripheral blood. BMC Genomics 2006; 7:115; PMID:; http://dx.doi.org/ 10.1186/1471-2164-7-115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sabatier R, Finetti P, Mamessier E, Raynaud S, Cervera N, Lambaudie E, Jacquemier J, Viens P, Birnbaum D, Bertucci F. Kinome expression profiling and prognosis of basal breast cancers. Mol Cancer 2011; 10:86; PMID:; http://dx.doi.org/ 10.1186/1476-4598-10-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gatza ML, Lucas JE, Barry WT, Kim JW, Wang Q, Crawford MD, Datto MB, Kelley M, Mathey-Prevot B, Potti A, et al. A pathway-based classification of human breast cancer. Proc Natl Acad Sci U S A 2010; 107:6994-9; PMID:; http://dx.doi.org/ 10.1073/pnas.0912708107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Velcheti V, Schalper KA, Carvajal DE, Anagnostou VK, Syrigos KN, Sznol M, Herbst RS, Gettinger SN, Chen L, Rimm DL. Programmed death ligand-1 expression in non-small cell lung cancer. Lab Invest 2014; 94:107-16; PMID:; http://dx.doi.org/ 10.1038/labinvest.2013.130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gadiot J, Hooijkaas AI, Kaiser AD, van Tinteren H, van Boven H, Blank C. Overall survival and PD-L1 expression in metastasized malignant melanoma. Cancer 2011; 117:2192-201; PMID:; http://dx.doi.org/ 10.1002/cncr.25747 [DOI] [PubMed] [Google Scholar]
- 41. Rimm D, Schalper K, Pusztai L. Unvalidated antibodies and misleading results. Breast Cancer Res Treat 2014; 147:457-8; PMID:; http://dx.doi.org/ 10.1007/s10549-014-3061-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schalper KA, Velcheti V, Carvajal D, Wimberly H, Brown J, Pusztai L, Rimm DL. In situ tumor PD-L1 mRNA expression is associated with increased TILs and better outcome in breast carcinomas. Clin Cancer Res 2014; 20:2773-82; PMID:; http://dx.doi.org/ 10.1158/1078-0432.CCR-13-2702 [DOI] [PubMed] [Google Scholar]
- 43. Soliman H, Khalil F, Antonia S. PD-L1 expression is increased in a subset of basal type breast cancer cells. PLoS One 2014; 9:e88557; PMID:; http://dx.doi.org/ 10.1371/journal.pone.0088557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shen JK, Cote GM, Choy E, Yang P, Harmon D, Schwab J, Nielsen GP, Chebib I, Ferrone S, Wang X, et al. Programmed cell death ligand 1 expression in osteosarcoma. Cancer immunology research 2014; 2:690-8; PMID:; http://dx.doi.org/ 10.1158/2326-6066.CIR-13-0224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Droeser RA, Hirt C, Viehl CT, Frey DM, Nebiker C, Huber X, Zlobec I, Eppenberger-Castori S, Tzankov A, Rosso R, et al. Clinical impact of programmed cell death ligand 1 expression in colorectal cancer. Eur J Cancer 2013; 49:2233-42; PMID:; http://dx.doi.org/ 10.1016/j.ejca.2013.02.015 [DOI] [PubMed] [Google Scholar]
- 46. Lipson EJ, Vincent JG, Loyo M, Kagohara LT, Luber BS, Wang H, Xu H, Nayar SK, Wang TS, Sidransky D, et al. PD-L1 expression in the Merkel cell carcinoma microenvironment: association with inflammation, Merkel cell polyomavirus and overall survival. Cancer Immunol Res 2013; 1:54-63; PMID:; http://dx.doi.org/ 10.1158/2326-6066.CIR-13-0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ricci R, Arena V, Castri F, Martini M, Maggiano N, Murazio M, Pacelli F, Potenza AE, Vecchio FM, Larocca LM. Role of p16/INK4a in gastrointestinal stromal tumor progression. Am J Clin Pathol 2004; 122:35-43; PMID:; http://dx.doi.org/ 10.1309/MJ4XN2M57HNC8X5H [DOI] [PubMed] [Google Scholar]
- 48. Nakamura N, Yamamoto H, Yao T, Oda Y, Nishiyama K, Imamura M, Yamada T, Nawata H, Tsuneyoshi M. Prognostic significance of expressions of cell-cycle regulatory proteins in gastrointestinal stromal tumor and the relevance of the risk grade. Hum Pathol 2005; 36:828-37; PMID:; http://dx.doi.org/ 10.1016/j.humpath.2005.03.012 [DOI] [PubMed] [Google Scholar]
- 49. Romeo S, Debiec-Rychter M, Van Glabbeke M, Van Paassen H, Comite P, Van Eijk R, Oosting J, Verweij J, Terrier P, Schneider U, et al. Cell cycle/apoptosis molecule expression correlates with imatinib response in patients with advanced gastrointestinal stromal tumors. Clin Cancer Res 2009; 15:4191-8; PMID:; http://dx.doi.org/ 10.1158/1078-0432.CCR-08-3297 [DOI] [PubMed] [Google Scholar]
- 50. Sabah M, Cummins R, Leader M, Kay E. Altered expression of cell cycle regulatory proteins in gastrointestinal stromal tumors: markers with potential prognostic implications. Hum Pathol 2006; 37:648-55; PMID:; http://dx.doi.org/ 10.1016/j.humpath.2006.01.023 [DOI] [PubMed] [Google Scholar]
- 51. Schneider-Stock R, Boltze C, Lasota J, Peters B, Corless CL, Ruemmele P, Terracciano L, Pross M, Insabato L, Di Vizio D, et al. Loss of p16 protein defines high-risk patients with gastrointestinal stromal tumors: a tissue microarray study. Clin Cancer Res 2005; 11:638-45; PMID: [PubMed] [Google Scholar]
- 52. Bertucci F, Finetti P, Ostrowski J, Kim WK, Kim H, Pantaleo MA, Astolfi A, Polkowski M, Birnbaum D. Genomic Grade Index predicts postoperative clinical outcome of GIST. Br J Cancer 2012; 107:1433-41; PMID:; http://dx.doi.org/ 10.1038/bjc.2012.390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lagarde P, Perot G, Kauffmann A, Brulard C, Dapremont V, Hostein I, Neuville A, Wozniak A, Sciot R, Schoffski P, et al. Mitotic checkpoints and chromosome instability are strong predictors of clinical outcome in gastrointestinal stromal tumors. Clin Cancer Res 2011; 18(3):826-38; PMID: [DOI] [PubMed] [Google Scholar]
- 54. Perez DR, Baser RE, Cavnar MJ, Balachandran VP, Antonescu CR, Tap WD, Strong VE, Brennan MF, Coit DG, Singer S, et al. Blood neutrophil-to-lymphocyte ratio is prognostic in gastrointestinal stromal tumor. Ann Surg Oncol 2013; 20:593-9; PMID:; http://dx.doi.org/ 10.1245/s10434-012-2682-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ghebeh H, Barhoush E, Tulbah A, Elkum N, Al-Tweigeri T, Dermime S. FOXP3+ Tregs and B7-H1+/PD-1+ T lymphocytes co-infiltrate the tumor tissues of high-risk breast cancer patients: Implication for immunotherapy. BMC Cancer 2008; 8:57; PMID:; http://dx.doi.org/ 10.1186/1471-2407-8-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Spranger S, Spaapen RM, Zha Y, Williams J, Meng Y, Ha TT, Gajewski TF. Up-regulation of PD-L1, IDO, and T(regs) in the melanoma tumor microenvironment is driven by CD8(+) T cells. Sci Translational Med 2013; 5:200ra116; http://dx.doi.org/ 10.1126/scitranslmed.3006504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Astolfi A, Nannini M, Pantaleo MA, Di Battista M, Heinrich MC, Santini D, Catena F, Corless CL, Maleddu A, Saponara M, et al. A molecular portrait of gastrointestinal stromal tumors: an integrative analysis of gene expression profiling and high-resolution genomic copy number. Lab Invest 2010; 90:1285-94; PMID:; http://dx.doi.org/ 10.1038/labinvest.2010.110 [DOI] [PubMed] [Google Scholar]
- 58. Ostrowski J, Polkowski M, Paziewska A, Skrzypczak M, Goryca K, Rubel T, Kokoszynska K, Rutkowski P, Nowecki ZI, Vel Dobosz AJ, et al. Functional features of gene expression profiles differentiating gastrointestinal stromal tumours according to KIT mutations and expression. BMC Cancer 2009; 9:413; PMID:; http://dx.doi.org/ 10.1186/1471-2407-9-413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yamaguchi U, Nakayama R, Honda K, Ichikawa H, Hasegawa T, Shitashige M, Ono M, Shoji A, Sakuma T, Kuwabara H, et al. Distinct gene expression-defined classes of gastrointestinal stromal tumor. J Clin Oncol 2008; 26:4100-8; PMID:; http://dx.doi.org/ 10.1200/JCO.2007.14.2331 [DOI] [PubMed] [Google Scholar]
- 60. Lee EJ, Kang G, Kang SW, Jang KT, Lee J, Park JO, Park CK, Sohn TS, Kim S, Kim KM. GSTT1 copy number gain and ZNF overexpression are predictors of poor response to imatinib in gastrointestinal stromal tumors. PLoS One 2013; 8:e77219; PMID:; http://dx.doi.org/ 10.1371/journal.pone.0077219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003; 4:249-64; PMID:; http://dx.doi.org/ 10.1093/biostatistics/4.2.249 [DOI] [PubMed] [Google Scholar]
- 62. Bertucci F, Finetti P, Viens P, Birnbaum D. EndoPredict predicts for the response to neoadjuvant chemotherapy in ER-positive, HER2-negative breast cancer. Cancer letters 2014; 355:70-5; PMID:; http://dx.doi.org/ 10.1016/j.canlet.2014.09.014 [DOI] [PubMed] [Google Scholar]
- 63. Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007; 8:118-27; PMID:; http://dx.doi.org/ 10.1093/biostatistics/kxj037 [DOI] [PubMed] [Google Scholar]
- 64. Taminau J, Steenhoff D, Coletta A, Meganck S, Lazar C, de Schaetzen V, Duque R, Molter C, Bersini H, Nowe A, et al. inSilicoDb: an R/Bioconductor package for accessing human Affymetrix expert-curated datasets from GEO. Bioinformatics 2011; 27:3204-5; PMID:; http://dx.doi.org/ 10.1093/bioinformatics/btr529 [DOI] [PubMed] [Google Scholar]
- 65. Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 2001; 98:5116-21; PMID:; http://dx.doi.org/ 10.1073/pnas.091062498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. Reporting recommendations for tumor marker prognostic studies. J Clin Oncol 2005; 23:9067-72; PMID:; http://dx.doi.org/ 10.1200/JCO.2004.01.0454 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.