Abstract
Associations between oral and systemic health are ancient. Oral opportunistic bacteria, particularly, Porphyromonas gingivalis and Fusobacterium nucleatum, have recently been deviated from their traditional roles and arguably ascended to central players based on their participations in complex co-dependent mechanisms of diverse systemic chronic diseases risk and pathogenesis, including cancers, rheumatoid-arthritis, and diabetes.
Keywords: Oral Microbes, P. gingivalis, F. nucleatum, Cancer, Chronic diseases, dysbiosis, small molecule danger signaling
1. Introduction
The oral microbial communities have evolved along with Homo sapiens and developed together with our dietary and hygienic habits over millions of years [1]. The first documented dental procedures date back to Neolithic times only about 8000 years BC [2] and dentistry and medicine have been linked in ancient records dating back to Egyptian times approximately 4000 years BC [3]. The importance of oral health for holistic wellbeing is also not a novel concept, as ancient civilizations of the Mediterranean, for example, had already noticed that teeth problems are associated with reproduction problems in women [4], and Hippocrates treated joint pain with tooth extractions (460-377 BC). With the discovery of microorganisms, and their causative link to diseases in the early 17th and throughout 18th century, the association between oral health and oral microbiota, became even more pronounced [3]. The concept of the single-pathogen causality became fashionable among the scientific community and Robert Koch postulated the criteria to establish a causative relationship between a microbe and disease. However many diseases and conditions remained unexplained by single pathogens. Moreover, with the development of the modern molecular tools the concept of the microbiome and the balance of the microbial communities that colonize the human body came to light [5, 6]. Currently the links between the oral microbial consortia and their interactions with the host in the maintenance of homeostasis and in the pathogenesis of many diseases have taken center stage [6]. This more novel concept attributes the observed systemic effects not to secondary dissemination and spread of specific microorganisms and/or their toxins, but to a dysbiotic change in the constitution and inter-microbial interactions of the healthy oral microbial community, leading to an immune response from the host, locally and systemically [3].
Yet, the truth seems to lie in between. Even within the concept of the oral microbiome as a dynamic society of over 700 species of inter-communicating microorganisms, many of which still not cultivable , certain key species, such as Porphyromonas gingivalis and Fusobacterium nucleatum have stolen the attention with their ability to modulate the balance of the microbiome and the subsequent interactions with the host mucosa and immunity [7-10]. Based on the recent archeological findings from calcified dental plaque, giving a snapshot information of the period when hunter-gatherer societies started converting to farming of domesticated animals and plants approximately 10000 years ago, the change in lifestyle and diet seem to have led to an explicit shift in the composition of the oral microbiota featuring increased presence of certain species, such as P. gingivalis [1]. This change in the microbiome also correlated with a more frequent occurrence of severe forms of periodontal disease in those populations and seems to suggest a correlation of the severity of chronic diseases of the oral cavity with the change in the environmental factors [1]. In today’s urbanized human society, aside from the diet there are many other contributing factors, such as environmental pollution, climate, lifestyle and distinct health habits, which may also simultaneously influence the oral communities and have been linked with a noted increase in the frequencies of a broader array of chronic diseases [11].
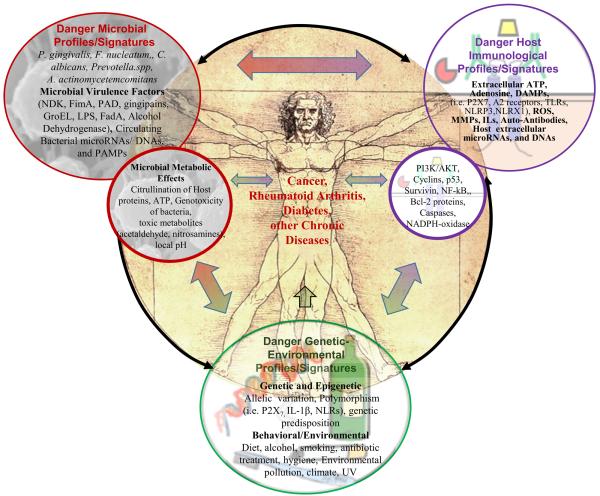
Apart from the now well established role in periodontal disease, a select group of oral microbes have increasingly become associated with chronic diseases such as orodigestive cancer, rheumatoid arthritis, diabetes, and some other severe chronic diseases. These new observations have critically transformed the traditionally accepted views especially on the opportunistic chronic pathogens of the oral cavity and how those microbes may have engaged with human health and disease [12-15]. Among those microorganisms, P. gingivalis has frequently been linked with the potential development and/or advancement of several distinct chronic diseases and has become a much highlighted topic of research. Thus, this review will critically examine the newly started to be characterized roles of a selective number of oral microbes in cancers, rheumatoid arthritis, diabetes, and chronic inflammatory conditions of the liver, kidneys and lungs. In addition, it outlines mechanistic connections among the highly complex cellular and metabolic molecular networks that appear to be predominantly co-shared by these seemingly unrelated chronic diseases and also modulated by the specific subset of oral opportunistic pathogens (The postulated interactions are illustrated in Diagram 1).
Diagram 1. Postulated Relationships of Oral Microbes to Diverse Chronic Diseases.
The oral microbiome and specific subset of pathogens, in particular, Porphyromonas gingivalis and a few others, have been suggested to play an important role in multiple anatomically and clinically unrelated chronic diseases such as periodontal disease, orodigestive and gastrointestinal cancers, rheumatoid arthritis, and diabetes. P. gingivalis, a key-stone pathogen, has been implicated in all these chronic diseases, whereas other oral microbes have been associated only with some of these conditions. The driving processes of these diseases appear to involve multifaceted complex interactions among the oral microbiome constituents, the host genetic, immunological behavioral factors, and environmental conditions working in concert to shape a local microenvironment that predisposes the host cells to inflammation, transformation and promotes the development and/or progression of these diseases. It is tempting to extrapolate the common pathways that seem to be shared among the etiologies of all these diseases, and which seem to be influenced by the microbiome. Thus the specific microbiome and host components can be disease inducers and may act as important risk modifiers and early diagnostic/prognostic factors for the management of these diseases. We accordingly call those potentially major disease determinants as ‘danger profiles or signatures’.
2. Cancer and oral microbes
Although late archeological evidence suggests that cancers have existed as far back as the earliest human civilizations, the fact that cancers are relatively rare in archaic mummified remains from old civilizations worldwide (4000 – 400BC) [11] highlights that the incidence of cancers may have increased with the development and industrialization of modern society (2012), when cancer mortality is second only to cardiovascular mortality [11, 16]. Indeed, a terracotta torso which was excavated in Anatolia (todays Turkey) dates from 200-100 BC, and resides in the collections of the Institute for Medical History in Jena, Germany, illustrates one of earliest accountable depiction of cancerous female breast in art, while the celebrated Renaissance sculpture “Notte” by Michelangelo is considered to be the first known portrayal of advanced breast cancer in modern history [17]. Similarly, the belief that cancers could be transmissible goes back as early as 16th century [17].
Currently, approximately a quarter of the worldwide malignancies are attributed to microbial contribution [18] and orodigestive cancers are among the top five leading causes of cancer mortalities [16]. One of the particularly devastating examples of orodigestive cancer mortality with escalating prevalence is oral squamous cell carcinoma (OSCC), which claims an estimated 7500 lives yearly in the United States [19]. Importantly, the increasing incidence of OSCC does not seem to correlate with the presence of commonly accepted risk factors, such as tobacco and alcohol consumption [20]. Further, Human Papilloma Virus, that has been attributed as major etiological agent for oropharyngeal cancers of the head and neck is only present in 2-4% of OSCCs [20]. Accordingly, there are no reliable diagnostic markers and/or risk factors present for early identification of OSCC cases along with the other cancers of the orodigestive tract, encompassing the oral cavity, upper intestinal tract, and pancreas. A number of recent epidemiological/clinical studies, including case-control and cohort studies, described strong associations of periodontal disease and/or tooth loss with cancers of the orodigestive system, and suggested the involvement of select oral microbes in the development of orodigestive malignancies [19]. Accordingly, a successful oral colonizer and well recognized periodontal pathogen, P. gingivalis was identified among the known microbial species of the oral cavity to have the highest correlation with OSCC [19, 21], followed by associations with pancreatic cancer, where P. gingivalis was found to be an important independent risk factor [19, 22, 23]. F. nucleatum, another well-known opportunistic microorganism which has been long considered as a commensal bacterium in the oral cavity, only recently was elevated to a pathogenic agent status [24] having a propensity for colonization also of the lower gastrointestinal tract, and proposed to take part in inflammatory bowel disease and exclusively in colorectal cancer [8, 12, 22, 24, 25].
Although specific data about the role of other species of the oral ecosystems are very sparse, the oral microbial profiles as “danger signatures” with potential to drive the host to a dysbiotic state are newly postulated for some other chronic disease progressions. Along the same lines, a recent clinical cohort study, showed the shifts in the abundance of operational taxonomic units (OTUs) in OSCC cancer and pre-cancer patients, when compared to healthy subjects [20]. The study described increased amounts of phylum Bacteroidetes, and specifically genera Porphyromonas and Prevotella, and decreased Firmicutes and Actinobacterium phyla, and particularly Streptotococcus and Rothia species in swabs of cancer subjects compared to clinically normal patients using 16S qPCR detection and metagenomics analysis [20]. More critically, in the same study a significant increase of P. gingivalis was detected in both pre-malignant and malignant samples of patients, when compared to healthy controls. Whereas Prevotella species were increased only in oral cancer patients, but not in pre-malignant patients, suggesting a potentially secondary colonization of cancer tissues by Prevotella species, perhaps due to favorable microenvironmental changes occurring during oral carcinogenesis processes [20]. Further, the same study displayed a reduction of F. nucleatum in oral cancer and oral pre-cancer patients contrasting the microorganism’s newly demonstrated role in colorectal cancer, while total abundance of Fusobacteria species was increased in those oral cancer patients [20]. Overall, despite the moderately small sample size, these data pointed to the presence of conserved oral microbial profiles in the samples of premalignant versus malignant tissues, as well as healthy individuals. The pathogenesis of OSCC is considered as a multi-step process and the premalignant lesions of the oral mucosa, are strongly indicated as early precursors of OSCC. It is therefore tempting to hypothesize that distinct oral microbial-profiles associated with different clinical outcomes and/or stages of disease could serve as ’danger signatures’ and help to identify the cancer susceptible populations.
The pathogeneses of cancers are in general conceptualized to be far complex, while the accumulated evidence collectively emphasizes the occurrence of highly complex inter-microbial interactions within the oral microbiome that can concurrently modulate and also be swayed by the host and environmental factors. All these interactions in return could lead to a local environment favorable to dysplastic transformation. Indeed, the concept that persistent microbes that cannot be cleared from the host, continue to participate in an ongoing combat that damages host tissues and promotes malignancy is not new. In late 1800s, French surgeon Jean Nicholas Marjolin originally noted the formation of neoplastic growth in chronic wounds which was subsequently named as ‘Marjolin ulcers’ to indicate any malignant transformation mainly originating from chronic persistent infections with chronic inflammatory features [18]. For example, sinus tract squamous cell carcinomas are described to be the long-lasting inflammatory results of chronic osteomyelitis in the sinus tract, with polymicrobial etiology. In the following sections, we will systematically feature the specific oral microorganisms that have emerged to be strongly connected with risk of cancer and provide insights into the mechanisms by these microbes promote the development of cancer.
Porphyromonas gingivalis
P. gingivalis, is a gram-negative opportunistic anaerobe that has long been known for its role as a major pathogen, strongly contributing in development of severe chronic inflammations of oral tissues, and can also successfully replicate, survive, and spread from cell-to-cell within the oral epithelium while evading the host immune response for prolonged periods [19, 26]. The microorganism can also colonize in various parts of the oral mucosa, including dorsal/ventral sides of the tongue, floor of the mouth, and buccal mucosa, which are described to be primary lesion sites during OSCC initial presentation [26]. From the currently known cultivable constituents of the oral microbial community P. gingivalis is the most highly associated organism with cancers of the orodigestive tract, including OSCC. To date multiple lines and levels of evidence from case-control and cohort-epidemiological/clinical survey studies have demonstrated several degrees of association of P. gingivalis with cancers of the oral cavity [19, 22] and as a important risk modifier for pancreatic cancer [19, 23]. Using 16S rRNA pyrosequencing, a 2013 case-control study has also described increased presence of Porphyromonadaceae family together with Fusobacteriaceae and four other OTUs, including Eubacteriaceae, Clostridiales, Staphylococcaceae and Campylobacteraceae, in fecal samples of colorectal cancer patients versus healthy controls [27] suggesting a possible synergistic effect among these bacterial phyla, that contain both P. gingivalis and F. nucleatum, in colorectal cancer. Another study analyzing OSCC and healthy control tissues from a relatively small sample size, was the first to describe a P. gingivalis detection in OSCC tissues, by showing significant increase of P. gingivalis strain ATCC 33277 in the cancer samples compared to healthy samples [28]. The study used antibody staining against the commensal microbe Streptococcus gordonii as a control, which showed no significant distribution difference [28]. In 2012, a large European prospective cohort study demonstrated an association of anti-P. gingivalis antibodies in pre-diagnostic blood samples of pancreatic cancer patients, and specifically against two type strains (ATCC 53978 and 33277), with an significant increased risk of pancreatic cancer [23]. This finding implies that the microbial danger is perhaps determined at the subspecies level and the outcome could be contributing to the degrees of predispositions.
P. gingivalis strain 33277 in vitro has been shown to activate pro-survival phosphatidylinositol-3-kinase (PI3K)/protein kinase B (AKT) pathway in the primary oral epithelial cells for effective colonization and the infected epithelial cells harboring high number of intracellular P. gingivalis can undergo successful cell division and display enhanced proliferation [19, 29-31]. Induction of the PI3K/AKT signaling along with the epithelial mesenchymal transition (EMT) have been suggested to be among the first steps to carcinogenesis. P. gingivalis has been found to carry plentiful virulence factors to promote oral chronic inflammation, which seem to also target complex network of molecular pathways culminating in neoplastic transformation and cancerogenesis. Such P. gingivalis virulence molecules and their known pathways include: distinctive fimbriae that have been shown to attenuate the host p53-mediated tumor-suppression pathways and cell-cycle progression in primary oral epithelial cells [19, 26]; cysteine proteases (gingipains) that have been suggested to be majorly involved in the activation of matrix metalloproteinases (MMPs), particularly MMP9, which are recognized to be associated with increased metastatic dissemination of carcinoma cells [19, 26]; and a nucleoside diphosphate kinase homologue (NDK), which has been demonstrated to modulate purinergic danger signaling, induce imbalance in oxidative stress metabolism, and inhibit pro-apoptotic and pro-inflammatory mechanisms in primary oral epithelial cells [32, 33]. A recent 2014 study in mice implicated another potential virulence factor from P. gingivalis – GroEL, which caused in tumor volume and weight acceleration, and in increased mortality rate of BALB/c mice implanted with a mouse-colon-carcinoma cell line (C26) [34]. GroEL is a bacterial homologue of eukaryotic heat shock protein 60 (HSP60), which is known to play a role in proper folding of mitochondrial proteins, and recently proposed to be a target for anti-cancer therapy, as well as a prognostic marker for prostate cancer [34, 35]. This study examined the effect of GroEL using a recombinant protein approach and mice carrying an already established tumor cell line (C26). Thus, the design of the study was not maximized to demonstrate direct pro-cancer effect of this putative factor in a clinically relevant cell culture model such as primary oral epithelial cells. Established oral cancer cell lines have been proven to have very limited correlation to the genetic makeup and other key attributes of human oral cancers [36].
Among the well characterized features of P. gingivalis as a persistent colonizer is the ability to alter the apoptotic and immune responses elicited by oral epithelial cells upon infection. P. gingivalis inhibits multiple apoptotic cell-death pathways in primary oral epithelial cells induced by potent chemical agents as well as extracellular adenosine-5'-triphosphate (ATP) [26]. As a biologically relevant pro-apoptotic molecule and a key danger signal (DS) ATP has become best recognized for its major participation in modulation of inflammation and cell-death [30, 33, 37]. ATP released from inflamed tissues acts through ionotropic purinergic receptor P2X7, to activate specific pro-inflammatory signaling cascades, apoptosis and to eliminate intracellular pathogens [33, 38]. The latest clinical findings also highlight the significance of purinergic signaling in the human oral cavity. Both saliva and plasma-derived gingival crevicular fluid, have been found to contain markedly high levels of purines during periodontal disease using unbiased metabolomics profiling [38, 39]. Similarly, our studies on human oral epithelium revealed novel roles for ATP and its metabolite ‘adenosine’ (another host derived DS molecule) in the context of P. gingivalis and host interaction [37, 40]. We showed that P. gingivalis modulates ATP-induced reactive oxygen species (ROS) formed through P2X7/NADPH-oxidase andmitochondria interactome, and effectively inhibits ATP/P2X7 mediated host cell-death in human primary oral epithelial cells via its secreted effector, NDK.[37, 38, 40]. Our recent studies also indicate that P. gingivalis can impact on the ATP-mediated inflammasome pathway and subsequent interleukin-1β secretion via P2X7 signaling [30, 33, 40]. Further, activation of adenosine 2a (A2a) receptor results in elevated intracellular P. gingivalis replication, which also correlates with significantly higher levels of anti-inflammatory cyclic AMP production during the infection of oral epithelial cells. Pharmacological A2a receptor antagonisms and knockdown via RNA interference significantly reduce metabolically active intracellular P. gingivalis [37].
Although the afore mentioned anti-apoptotic and immuno-modulatory actions of P. gingivalis appear to serve the microorganism’s primary goal which is to efficaciously colonize and survive in the oral epithelium, the targeted complex networks of molecules are also closely engaged and shared by inflammation and cancer molecular pathways [19, 38]. For example, overexpression of P2X7 receptors and the significance of P2X7, as well as A2a receptors’ signaling, have been critically underlined in several distinct types of cancers [19, 41-43]. Those include lung, breast, prostate, neuroblastoma, and leukemia cancers, suggesting the danger receptors concertedly could be potential drug targets for effective therapy options in cancer. Conjointly, human homologues of P. gingivalis’ effector NDK, that is shown to act as an inhibitor of ATP/P2X7 receptor coupling, commonly known as “non-metastatic cell expressed proteins” (NMEs), have been studied for their participations in various forms of human cancers, such as lung, breast, thyroid and neuroblastoma cancers, as well as for a role in metastasis and regulation of p53-mediated gene transcription [19]. Intriguingly, a 2006 study showed that human NDK (NME23) was among the few significantly overexpressed proteins in surgically excised OSCC tissue, compared to control tissue of same individuals, pointing to a potential role of the bacterial NDK not only as a putative virulence element in the OSCC process, but also as a microbial signature molecule for early detection of orodigestive cancers in high-risk individuals [44]. Moreover, P. gingivalis, and other oral pathogens such as Aggregatibacter actinomycetemcomitans are recently described for their modulation of interleukin-1β (IL-1β) secretion through NRLP3 inflammasome complex activated by ATP [26, 33, 38, 40]. IL-1β has been implicated in the development of diverse chronic diseases and lately in various types of cancers including gastric and colon cancers, as well as oral and esophageal squamous cell carcinomas [38], further underscoring the ability of P. gingivalis to jointly modulate specific inflammation and cancer pathways that appear to be also co-dependent in nature.
Currently, among the known virulence molecules of P. gingivalis, there is not a significantly attributed molecular determinant that could be strongly linked to the illustrated association of P. gingivalis with orodigestive cancers. One can presume that the answer involves the concerted play of multiple virulence factors and molecular mechanisms to drive such change, and very likely the potential synergistic ability of P. gingivalis with other oral microbial species are contributory to the postulated malignant transformation and progression in the oral cavity and upper digestive tract (see Diagram 1). Accordingly, we will continue to underline other pertaining bacterial species and their putative mechanisms of tumorigenic actions below.
Fusobacterium nucleatum
As mentioned briefly above, F. nucleatum is an anaerobic bacterium of the oral cavity that until recently was considered to be a commensal colonizer, and an important bridging organism between early and late colonizers of dental biofilm associated with periodontal disease. However, in the last decade it has been slowly accepted to have a pathogenic potential especially in the lower intestinal tract, where it has been associated with gastrointestinal disorders and specifically colorectal cancers [24]. Currently there are few studies showing mainly clinical association of F. nucleatum with colorectal cancer [8, 25]. A 2014 European cohort survey involving 122 colorectal cancer patients showed that F. nucleatum is over-represented in colorectal tumor tissues compared to normal tissues in colorectal cancer patients, and moreover F. nucleatum bacterial load seemed to increase with disease progression from pre-malignant (adenoma) to colorectal cancer (adenocarcinoma), thus possibly being related to cancer mortality as well [8]. However, this study represented a relatively limited sample size when compared to major gastric cancer epidemiological cohort studies [45], and other oral bacterial species were not investigated. An earlier study also pointed to colorectal carcinoma over-representation of Fusobacterium species [25]. This study also observed significant co-occurrence within individual colorectal tumors of the anaerobic microbial genera Fusobacterium, Leptotrichia and Campylobacter, which besides being part of the oral microbial community are also found in the lower gastrointestinal tract [25]. This co-occurrence was also associated with over-expression of numerous host genes, including the pro-inflammatory chemokine interleukin-8 (IL-8) gene [25]. The same study also described the isolation of a novel, significantly different and potentially more virulent strain of Campylobacter showae from a colorectal tumor specimen that aggregated with a previously isolated tumor strain of F. nucleatum [25]. Unlike P. gingivalis, which has been shown to possess specific virulence factors likely involved in stages of carcinogenesis, F. nucleatum’s proposed carcinogenic potential has been attributed mostly to its pro-inflammatory effect through the induction of various cytokines, such as tumor necrosis factor alpha, IL-6, IL-8, IL-10, IL-12 and production of ROS within the colon lining epithelial cells that can ultimately lead to dysplasia and development of cancer [24, 46]. It is intriguing to mention that despite its belonging to the oral microbial community F. nucleatum seems to cause most severe problems in extraoral sites such as the lower gastrointestinal tract, placenta and abscesses [47], which suggests that the microorganism’s higher pathogenicity may result from the difference in the host cell types that it targets and the extreme environment conditions in the lower intestinal tract, as well as possible synergistic effects with lower intestinal microbiota such as proposed with members of the phylum Enterobacteriaceae, mostly represented by E. coli [24]. A 2013 complex experimental study has implicated F. nucleatum adhesin A molecule (FadA) with direct binding to E-cadherin leading to triggering of local inflammation and β-catenin signaling, which are well known pathways in the EMT process [48]. EMT signaling is one of the early events in malignant transformation. This study however utilized ex vivo adenocarcinoma tissues and in vitro colorectal carcinoma cell line, both of which represent already transformed cancer phenotypic cells that are very difficult to interpret for their mesenchymal transformation capacity. Additionally, the study did not present any microbial and/or host mechanism for the proposed pathways.. In order to be able to validate and distinguish the individual roles of F. nucleatum and the other partnering microorganisms in the proposed etiology of cancer, large epidemiological studies encompassing greater sample size, resembling previous studies on the role of Helicobacter pylori in gastric cancers, and substantiated with relevant in vitro and in vivo mechanistic approaches are prudent. Similarly, in the following section we will relay present information about other potential oral microbes that are suggested to be involved in cancers of the orodigestive and lower gastrointestinal tract.
Other oral microbes
A few clinical studies have also described increased presence of Streptococcus anginosus within head and neck squamous cell carcinomas, including the OSCC, although the associations were based only on the uncharacteristic detection of the bacteria within the tumor tissue [21, 49]. In one of these studies other oral species such as Clavibacter michiganensis, Fusobacterium naviforme and Ralstonia insidiosa were found to be more prevalent in tumorous than in non-tumorous samples [21]. Candida albicans on the other hand, which is a known opportunistic colonizer of mucosal tissues in hosts with diminished immune capacity, has been proposed to play a role in carcinogenesis by metabolizing consumed products, and especially alcohol, into locally carcinogenic products like acetaldehyde and nitrosamines [50].
Since P. gingivalis has recently been ascribed to a role of being key modulator of the oral microbiome [47] and considerable amount of studies have highlighted the bacterium’s synergy with other oral microbes, it is conceivable to assume that these associations may be the result of a cumulative effect of the interaction of P. gingivalis with other members of the polymicrobial consortia, in concert with host and environment factors. In support of this view, several studies have shown that co-infections of P. gingivalis and F. nucleatum are mutually beneficial for the survival and proliferation of both species, although oral microbial co-infection studies are generally lacking [38]. The ability of P. gingivalis to evade innate immune responses in oral epithelial cells by depleting host-derived danger signal molecules like extracellular ATP can possibly also be beneficial for the survival and proliferation of other opportunistic species that otherwise elicit rigorous inflammatory response or rely on compromised immunity of the host to elicit their pathogenic potential, such as C. albicans [50]. C. albicans as being another potential microbe which has recently been associated with tumors, and specifically adds to the molecular pool of microbial factors that have direct pro-cancerogenic potential, such as acetaldehyde and nitrosamines [50]. Additionally, pretreatment of Ca9-22, a human oral epithelial cancer cell line, and gingival fibroblasts with heat killed C. albicans has been shown to increase P. gingivalis invasion in these cells [51], further complicating the disease-associated inter-microbial relationships within the oral microbiome and its potential link with cancer.
The current limited knowledge of the role of the oral microbes as communities in orodigestive and lowerintestinal (e.g. colorectal) cancers points to a cumulative role of a disordered inter-microbial communications and their relationship to the host biology. In addition to those major elements local mucosal micro-environmental factors, derived from the host, could all culminate in genomic, proteomic, and metabolomic molecular level alterations towards oncogenesis. Despite the all complexity, there is strong indication that specific persistent bacteria, such as P. gingivalis and F. nucleatum, may have a significant role driving these potentially dysbiotic processes, as well as molecularly step-wise contributing to the pre-cancer and cancer-transformation events in the host tissues. Therefore the increased presence of the specific microbial profiles and virulence molecules may be used as early warning ‘danger’ signals for timely diagnosis and management of cancers respectively (Diagram 1). Additionally, the high complexity of the cancer-driving events related to the microbiome, the host, and the environment, may explain the differences in the susceptibility of different individuals to malignant transformation, and highlights the need for individual-targeted approach for possible prevention and management of those cancers, and likely other chronic diseases associated with alterations in oral microbial communities (Diagram 1).
3. Rheumatoid arthritis and oral microbes
Rheumatoid arthritis (RA) is a debilitating autoimmune chronic disease that is characterized by systemic inflammation, often progressive irreversible joint and bone damage, with peri-articular osteoporosis, bone erosions, narrowing of joint spaces, thickening of synovial membranes, and net loss of cartilage in affected patients [52]. Joint problems have been correlated with oral diseases to our knowledge since the time of Hippocrates, but the association of periodontal disease, and specifically Porphyromonas gingivalis, with rheumatoid arthritis is much more recent [53]. This association is also to date perhaps one of the most studied oral versus systemic chronic disease links [53, 54]. Multiple epidemiologic and clinical studies have found varying relations between periodontal disease and RA [10, 53]. Still, the majority of the studies that have shaped our perspective on the connection between these two chronic diseases are based on clinical co-occurrence of periodontitis and RA, common pathobiologic inflammatory and tissue/bone destructive processes in the two-diseases, and most importantly disease-specific circulating autoantibodies found in sera of RA patients and periodontitis patients [10, 53]. The fact that there are patients exhibiting only one of the diseases, and that RA and periodontal disease seem to fuel each other, while often it is not clear which one was developed first, highlights a controversy and gaps in the current understanding of the character of the purported association. Additionally, the fact that the disease-specific autoantibodies precede the onset of RA for long periods also sets many currently unanswered questions on the actual mechanisms of RA development and progression [55]. As players in the etiology of periodontal disease, several oral bacteria have been studied for association with RA etiology. Clinical studies have detected P. gingivalis, Prevotella intermedia, and Prevotella nigrescens DNA in sera and synovial fluids of RA patients [56], and antibodies against P. gingivalis, A. actinomycetemcomitans and few other oral microbiota constituents have also been found in sera of patients suffering from RA, compared to healthy controls [53, 54].
In the recent years, P. gingivalis has emerged as a major mechanistic link between the observed connections of RA and periodontitis [10, 53, 56]. To date, P. gingivalis has been found to be the primary microorganism whose special molecular actions have been systemically tied with RA development and progression. P. gingivalis is also the only described prokaryote to secrete a peptidyl-arginine deiminase (PAD) enzyme which specifically converts arginine amino acids within a peptide into citrulline residues (peptidylcitrulline) [10, 53]. This amino acid transformation may lead to the induction of protein conformational changes and exposing those proteins to host antigen recognition mechanisms for the production of anti-citrullinated peptide antibodies, hence creating opportunity for triggering of autoimmune inflammation in the host [53]. Indeed, this molecular action seems to be supported by the clinical detection of autoantibodies against citrullinated host peptides in patients suffering from rheumatoid arthritis [53, 56]. Additionally, the MMPs that have been shown to be induced by P. gingivalis’ gingipains are considered to contribute to the destruction of synovial cartilages and oral connective tissues during RA progression and periodontal disease [56], further creating opportunity for exposure of normally immune-isolated host peptides to the citrullination activity of P. gingivalis enzymes and immune system recognition. There are currently few mechanistic experimental studies, attempting to elucidate the microbial participation in RA etiology, and the majority of them rely on murine models of RA and/or periodontal disease [53, 57, 58]. For example, mouse models of experimental RA and periodontitis have been used to confirm experimental exacerbation of collagen induced RA in mice upon P. gingivalis infection and to verify the effect of P. gingivalis PAD in the development of anti-citrullinated-protein antibodies, using PAD mutant strain of P. gingivalis [53, 57]. Mice, however, do not seem to be a relevant model for human RA development, as it has been shown that mouse rarely develops anti-citrullinated protein antibodies, and may exhibit independent citrulline reactivity in control sera [57, 59].
Despite all the specific attributes, there is currently a controversial debate on the indicated role of auto-citrullination of the P. gingivalis’ PAD enzyme as source of antigen for anti-P. gingivalis antibody production detected in RA patients [53], which has been recently disputed by in vitro P. gingivalis studies with bacterial culture and recombinant protein technique, showing that outside the host, P. gingivalis’ PAD does not auto-citrullinate itself [55]. This finding also emphasizes that even with the proposed specific mechanisms of P. gingivalis involvement in RA etiology, the potential influence of the other constituents of the oral microbiome cannot be ruled out. Apart from the specific citrullination processes, the proposed pro-inflammatory mechanisms and cytokine inductions are shared between RA, periodontal disease and cancer. For example, the MMPs are involved mainly in extracellular matrix destruction and bone resorption processes in RA, and in cancer these host enzymes have been modulated to possibly aid metastasis [19]. Intriguingly, active recent research has emphasized the potential central role of P2X7 signaling and inflammasome activation as aspects of the etiologic mechanisms and risk factors for RA [19, 60], hence adding another plausible participation of P. gingivalis and co-employed bacterial virulence factors in RA etiology. Further, P2X7 receptor allelic variations have been proposed to be potential risk modifiers in RA and cancer [19, 38]. This, once again further supports the potential perpetual cross-talk and possible co-utilization of analogous cellular/molecular networks in these chronic diseases.
The fact that the association between oral health and joint problems has long been documented and yet RA remains a world-wide burden of society and the exact mechanisms have still not been pinpointed, highlights the urge for furthering this scientific field. The dissecting of the RA etiological mechanisms will similarly require a highly integrated approach supported by novel methodologies and relevant animal and in vitro experimental models that reflect better the complexity of human oral and systemic changes occurring in RA patients. A current understanding on the potential microbial and multifactorial etiology of the development and progression of RA is presented in Diagram 1 along with proposed co-dependent pathways potentially linking oral microbiome with diabetes and cancer etiology.
4. Diabetes and oral microbes
Diabetes mellitus is a major metabolic chronic disease that poses a severe medical, social and economic problem globally. The two-way relationship between diabetes and periodontal disease has been long debated and currently it is widely accepted that there is a reciprocal feedback between these two chronic diseases [61]. This does not come as a surprise since oral health is dependent on the metabolism and diet of the host, which in turn are dependent on the microbiome constitution and the health of the oral cavity. A recent study, examined the available metadata on the association of periodontal disease and diabetes and concluded that while there is substantial information on potential mechanistic pathways which support a close association between diabetes and periodontitis, there is still a real need for well controlled longitudinal clinical studies using larger patient groups, integrated with studies of in appropriate vivo models and cells/tissues in vitro [62]. From a clinical perspective, an example of the observed controversy is demonstrated by a few studies that have suggested a direct link between P. gingivalis’ persistence in subgingival microbial biofilm and glycemic control in type 2 diabetes patients [62]. In contrast a recent in vivo mice study has shown no alteration of onset or severity of type 2 diabetes by P. gingivalis-induced periodontitis [63]. A 2014 large serological study involving 534 diabetes and metabolic syndrome affected patients and 820 matched controls examined serum antibody levels against A. actinomycetemcomitans and P. gingivalis and detected significant association between metabolic syndrome and anti-A. actinomycetemcomitans antibodies [64]. An earlier clinical cohort study aiming to correlate bacterial counts of key periodontal pathogens, such as P. gingivalis, Treponema denticola, Tannerela forsythia and A. actinomycetemcomitans, in gingival sulci with severity of glycemic control in diabetes mellitus patients versus non-diabetic controls, also showed high positive association of the specific bacterial counts with poor glycemic control [65]. On a more mechanistic level however, diabetes has been shown to prolong inflammatory responses to bacterial stimuli through cytokine dysregulation and particularly tumor necrosis factor-α (TNFα) and the chemokines MCP-1 and MCP-2 [62]. All these molecular actions can be involved in inflammation and are likely to exacerbate the burdens on the immune system, that are associated with type 2 diabetes progression (Diagram 1). A more novel possible mechanistic link that may be at this point speculatively drawn between P. gingivalis and diabetes is the already proposed role of the bacterium in modulation of purinergic P2X7 receptor signaling. Interestingly, P2X7 receptor expression was recently also strongly correlated with reduced metabolic control in type 2 diabetes patients [66]. NLRP3 inflammasome activation and subsequent IL-1β secretion have been gradually recognized to partake in the prolonged hyperglycemic toxicity in pancreatic islets, contributing to the destruction of β-cells in the pancreas, and dysregulation of glucose-induced insulin secretion associated with type 2 diabetes [38]. This role of NLRP3 inflammasome is also emphasized by the recently detected sustained beneficial effect of an IL-1 receptor antagonist in type 2 diabetes clinical trials [38]. A very recent mouse model study also proposed a possible role of P. gingivalis PAD enzyme in citrullination of host 78kDa glucose regulated protein (GRP78), which is a member of the heat shock protein family, and was shown to be an autoantigen in type 1 diabetes etiology [67]. This interesting preliminary finding further mechanistically links the distinct chronic diseases’ pathophysiologies that P. gingivalis has been shown to contribute to.
In spite of all recently investigated connections, the molecular basis of the etiology of diabetes is still largely unresolved. The bi-directional relationship of diabetes with periodontal disease, and the occasional study describing association of systemic exposure to other oral pathogens (e.g. A. actinomycetemcomitans) with diabetes severity and progression, suggest that the observed etiologic features of diabetes may be again the result of concerted metabolic interactions of the microbiome components along with host genetic predisposition factors and immune responses (Diagram 1). Notably, as with cancer and RA, a repeating pattern of a likely association with purinergic signaling, along with inflammasome activation and cytokine secretion is observed in diabetes as well, prompting intriguing questions on the conceivable etiologic links and differences among these diseases, that may determine each disease’s specific manifestations.
5. Other chronic diseases and oral microbes
In the past decade the oral microbiome has become a popular research target for a number of other chronic diseases that have not previously been associated with oral health. Among these chronic diseases newly discovered connections with the oral microbiome and/or its specific constituent species have been found with lung inflammatory diseases [68], non-alcoholic fatty liver disease [69], and chronic kidney disease [70].
Currently only few studies exist, elucidating associations between oral microbiome and chronic lung, kidney or liver diseases and the molecular mechanisms are generally unsolved. A 2014 prospective outpatient study examining a total of 94 bronchoscopy patients showed a statistically significant correlation between the presences of P. gingivalis, A. actinomycetemcomitans, T. forsythia, and T. denticola in bronchial samples and in saliva samples [68]. The same study also showed increased concentration of MMP8 in the bronchial fluid of patients with periodontal pathogens detectable in the lung compared to patients who didn’t harbor periodontal pathogens (p = 0.09) [68]. Similar association was observed between P. gingivalis prevalence in oral swabs and non-alcoholic fatty liver disease (NAFLD) in a 2012 cohort study involving 102 patients with non-alcoholic steatohepatitis, 48 with non-alcoholic fatty liver and 60 non-NAFLD control subjects [69]. P. gingivalis was found at significantly higher frequency in NAFLD patients than in the control subjects (46.7% vs. 21.7% respectively; odds ratio of 3.16). In addition, P. gingivalis in non-alcoholic steatohepatitis patients was found at markedly higher frequency than in the non-NAFLD controls (52.0%, odds ratio of 3.91) [69]. Likewise, in chronic kidney disease, a Japanese study involving 215 elderly participants observed that individuals with elevated serum antibody levels to P. gingivalis were ~3 times more likely to have decreased kidney function, compared to people with lower antibody levels [70].
All these studies have established clinical associations between the oral microbiome and selective pathogens like P. gingivalis, with these chronic inflammatory diseases. At present the observed phenomena, however remain unexplained in a mechanistic manner and further studies are very much needed to elucidate the pathway dependencies of these new associations and the common pathway nodes and specific effects that they may share.
6. Current caveats and outlook on future oral microbiome and chronic disease challenges
The past and present of the scientific knowledge on oral and systemic chronic diseases have been gradually shaped by multidimensional lines of clinical/ epidemiological associations, to perturbed microbial and host phenotypes, as well as molecular mechanistic and micro-environmental etiological connections. The appreciation of the oral microbes for the well-being of the entire body, including even embryonic development is evolving rapidly with the newly forming knowledge on the human microbiome. Yet there is still much unknown about the inter-microbial dynamics as well as their complex interface within either healthy individuals or diseased mucosal sites. The accumulated evidence, although it is perhaps preliminary and not specific enough in some circumstances, consistently points that specific oral microbes expressively involve in increased risk of a number of chronic diseases, although the causal direction of these relations have not yet been entirely ascertained. The identification of opportunistic persistent pathogens such as, P. gingivalis and F. nucleatum that are able to alter the microbiome structure/function and host cellular responses, and can also colonize and allocate their virulence factors in anatomically distant locations/organs, is an essential initial step for unraveling the causation and pathogenesis behind these increasingly plaguing chronic diseases of modern man. There is a nascent view that restoration of our ancestral microbial ecology, which may be entirely lost in some populations, could be therapeutic or prophylactic strategy in these chronic diseases with presumed disordered microbiome etiology. The difficulty of reproducing the close human counterpart of oral microbial consortia in in vitro and/or in vivo systems, encoding the microbe-microbe communications and the associated host factors are presently significant step-limiting factors. In parallel, the co-dependent host pathways that may be involved in the different chronic diseases are an equally intriguing future research objective for the development of highly targeted approach to the management of more than one of these chronic diseases.
The current systems biology approaches to study causation and pathogeneses of human diseases heavily employ murine/rodent models and the recent introduction of genomically “humanized mice” approach is receiving plenty of attention. While these in vivo models seem to provide biologically helpful information depending on the questions asked, they lack the complexity of human local and systemic levels which renders them mostly not very suitable as models of chronic diseases, as shown in the RA pathogenesis studies. Nevertheless, well integrated multidisciplinary research involving large-sample clinical/epidemiological studies accompanied by systems biology approach using pertinent mechanistic studies in biologically relevant human cell cultures and animal models combined with bioinformatics analyses can harness some important solutions.
Acknowledgements
We would like to acknowledge the support of the NIDCR grant R01DE016593. The authors regret that a number of studies could only be cross-referenced indirectly through comprehensive reviews, due to reference number restriction.
Abbreviations
- NDK
nucleoside diphosphate kinase
- FimA
P. gingivalis major fimbriae protein
- PAD
peptidyl-arginine-deiminase enzyme
- GroEL
bacterial counterpart of human heat shock protein 60
- LPS
lipopolysaccharide
- FadA
F. nucleatum’s adhesin A
- PAMPs
pathogen-associated molecular patterns
- DAMPs
danger-associated molecular patterns
- TLRs
toll-like receptors
- NLRs
NOD-like receptors
- ROS
reactive oxygen species
- MMPs
matrix metalloproteinases
- ILs
interleukins
- PI3K/AKT
phosphatidylinositol-3-kinase/protein kinase B pathway
- NF-kB
nuclear factor kappa beta
- Bcl2
“B-cell lymphoma 2” regulator pro-apoptotic protein
- NADPH-oxidase
nicotinamide adenine dinucleotide phosphate-oxidase
- UV
ultraviolet light
References
- [1].Adler CJ, Dobney K, Weyrich LS, Kaidonis J, Walker AW, Haak W, Bradshaw CJA, Townsend G, Soltysiak A, Alt KW, Parkhill J, Cooper A. Sequencing ancient calcified dental plaque shows changes in oral microbiota with dietary shifts of the Neolithic and Industrial revolutions. Nat Genet. 2013;45:450–455. doi: 10.1038/ng.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Coppa A, Bondioli L, Cucina A, Frayer DW, Jarrige C, Jarrige JF, Quivron G, Rossi M, Vidale M, Macchiarelli R. Palaeontology: early Neolithic tradition of dentistry. Nature. 2006;440:755–756. doi: 10.1038/440755a. [DOI] [PubMed] [Google Scholar]
- [3].Vieira CLZ, Caramelli B. The history of dentistry and medicine relationship: could the mouth finally return to the body? Oral Dis. 2009;15:538–546. doi: 10.1111/j.1601-0825.2009.01589.x. [DOI] [PubMed] [Google Scholar]
- [4].Gold SI. Periodontics. The past. Part (I). Early sources. Journal of clinical periodontology. 1985;12:79–97. doi: 10.1111/j.1600-051x.1985.tb01367.x. [DOI] [PubMed] [Google Scholar]
- [5].Integrative HMPRNC The Integrative Human Microbiome Project: dynamic analysis of microbiome-host omics profiles during periods of human health and disease. Cell host & microbe. 2014;16:276–289. doi: 10.1016/j.chom.2014.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Lampe JW. The Human Microbiome Project: getting to the guts of the matter in cancer epidemiology. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2008;17:2523–2524. doi: 10.1158/1055-9965.EPI-08-0792. [DOI] [PubMed] [Google Scholar]
- [7].Jenkinson HF. Beyond the oral microbiome. Environ Microbiol. 2011;13:3077–3087. doi: 10.1111/j.1462-2920.2011.02573.x. [DOI] [PubMed] [Google Scholar]
- [8].Flanagan L, Schmid J, Ebert M, Soucek P, Kunicka T, Liska V, Bruha J, Neary P, Dezeeuw N, Tommasino M, Jenab M, Prehn JH, Hughes DJ. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur J Clin Microbiol Infect Dis. 2014;33:1381–1390. doi: 10.1007/s10096-014-2081-3. [DOI] [PubMed] [Google Scholar]
- [9].Castrillon CA, Hincapie JP, Yepes FL, Roldan N, Moreno SM, Contreras A, Botero JE. Occurrence of red complex microorganisms and Aggregatibacter actinomycetemcomitans in patients with diabetes. Journal of investigative and clinical dentistry. 2013 doi: 10.1111/jicd.12051. [DOI] [PubMed] [Google Scholar]
- [10].Ogrendik M. Rheumatoid arthritis is an autoimmune disease caused by periodontal pathogens. International journal of general medicine. 2013;6:383–386. doi: 10.2147/IJGM.S45929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].David AR, Zimmerman MR. Cancer: an old disease, a new disease or something in between? Nat Rev Cancer. 2010;10:728–733. doi: 10.1038/nrc2914. [DOI] [PubMed] [Google Scholar]
- [12].Ahn J, Chen CY, Hayes RB. Oral microbiome and oral and gastrointestinal cancer risk. Cancer Causes Control. 2012;23:399–404. doi: 10.1007/s10552-011-9892-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Gollwitzer ES, Marsland BJ. Microbiota abnormalities in inflammatory airway diseases - potential for therapy. Pharmacol Ther. 2013 doi: 10.1016/j.pharmthera.2013.08.002. [DOI] [PubMed] [Google Scholar]
- [14].Bingham CO, 3rd, Moni M. Periodontal disease and rheumatoid arthritis: the evidence accumulates for complex pathobiologic interactions. Curr Opin Rheumatol. 2013;25:345–353. doi: 10.1097/BOR.0b013e32835fb8ec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Bastos JA, Diniz CG, Bastos MG, Vilela EM, Silva VL, Chaoubah A, Souza-Costa DC, Andrade LC. Identification of periodontal pathogens and severity of periodontitis in patients with and without chronic kidney disease. Arch Oral Biol. 2011;56:804–811. doi: 10.1016/j.archoralbio.2010.12.006. [DOI] [PubMed] [Google Scholar]
- [16].Organization WH. GLOBOCAN 2012: All Cancers (excluding non-melanoma skin cancer) Estimated Incidence. Mortality and Prevalence Worldwide in 2012. 2012 doi: 10.1093/jnci/djx205. [DOI] [PubMed] [Google Scholar]
- [17].Dahlgren AL. Michelangelo and medicine. Journal of the Royal Society of Medicine. 2003;96:256. doi: 10.1258/jrsm.96.5.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Samaras V, Rafailidis PI, Mourtzoukou EG, Peppas G, Falagas ME. Chronic bacterial and parasitic infections and cancer: a review. Journal of infection in developing countries. 2010;4:267–281. doi: 10.3855/jidc.819. [DOI] [PubMed] [Google Scholar]
- [19].Atanasova KR, Yilmaz O. Looking in the Porphyromonas gingivalis cabinet of curiosities: the microbium, the host and cancer association. Mol Oral Microbiol. 2014;29:55–66. doi: 10.1111/omi.12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Schmidt BL, Kuczynski J, Bhattacharya A, Huey B, Corby PM, Queiroz EL, Nightingale K, Kerr AR, DeLacure MD, Veeramachaneni R, Olshen AB, Albertson DG, Muy-Teck T. Changes in abundance of oral microbiota associated with oral cancer. PLoS One. 2014;9:e98741. doi: 10.1371/journal.pone.0098741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hooper SJ, Crean SJ, Fardy MJ, Lewis MA, Spratt DA, Wade WG, Wilson MJ. A molecular analysis of the bacteria present within oral squamous cell carcinoma. J Med Microbiol. 2007;56:1651–1659. doi: 10.1099/jmm.0.46918-0. [DOI] [PubMed] [Google Scholar]
- [22].Ahn J, Segers S, Hayes RB. Periodontal disease, Porphyromonas gingivalis serum antibody levels and orodigestive cancer mortality. Carcinogenesis. 2012;33:1055–1058. doi: 10.1093/carcin/bgs112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Michaud DS, Izard J, Wilhelm-Benartzi CS, You DH, Grote VA, Tjonneland A, Dahm CC, Overvad K, Jenab M, Fedirko V, Boutron-Ruault MC, Clavel-Chapelon F, Racine A, Kaaks R, Boeing H, Foerster J, Trichopoulou A, Lagiou P, Trichopoulos D, Sacerdote C, Sieri S, Palli D, Tumino R, Panico S, Siersema PD, Peeters PH, Lund E, Barricarte A, Huerta JM, Molina-Montes E, Dorronsoro M, Quiros JR, Duell EJ, Ye W, Sund M, Lindkvist B, Johansen D, Khaw KT, Wareham N, Travis RC, Vineis P, Bueno-de-Mesquita HB, Riboli E. Plasma antibodies to oral bacteria and risk of pancreatic cancer in a large European prospective cohort study. Gut. 2012 doi: 10.1136/gutjnl-2012-303006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Han YW. Fusobacterium nucleatum: a commensal-turned pathogen. Current opinion in microbiology. 2015;23C:141–147. doi: 10.1016/j.mib.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Warren RL, Freeman DJ, Pleasance S, Watson P, Moore RA, Cochrane K, Allen-Vercoe E, Holt RA. Co-occurrence of anaerobic bacteria in colorectal carcinomas. Microbiome. 2013;1:16. doi: 10.1186/2049-2618-1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Yilmaz O. The chronicles of Porphyromonas gingivalis: the microbium, the human oral epithelium and their interplay. Microbiology. 2008;154:2897–2903. doi: 10.1099/mic.0.2008/021220-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wu N, Yang X, Zhang RF, Li J, Xiao X, Hu YF, Chen YF, Yang FL, Lu N, Wang ZY, Luan CG, Liu YL, Wang BH, Xiang C, Wang YZ, Zhao FQ, Gao GF, Wang SY, Li LJ, Zhang HZ, Zhu BL. Dysbiosis Signature of Fecal Microbiota in Colorectal Cancer Patients. Microb Ecol. 2013;66:462–470. doi: 10.1007/s00248-013-0245-9. [DOI] [PubMed] [Google Scholar]
- [28].Katz J, Onate MD, Pauley KM, Bhattacharyya I, Cha S. Presence of Porphyromonas gingivalis in gingival squamous cell carcinoma. International journal of oral science. 2011;3:209–215. doi: 10.4248/IJOS11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Yilmaz O, Jungas T, Verbeke P, Ojcius DM. Activation of the phosphatidylinositol 3-kinase/Akt pathway contributes to survival of primary epithelial cells infected with the periodontal pathogen Porphyromonas gingivalis. Infect Immun. 2004;72:3743–3751. doi: 10.1128/IAI.72.7.3743-3751.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Yilmaz O, Yao L, Maeda K, Rose TM, Lewis EL, Duman M, Lamont RJ, Ojcius DM. ATP scavenging by the intracellular pathogen Porphyromonas gingivalis inhibits P2X7-mediated host-cell apoptosis. Cell Microbiol. 2008;10:863–875. doi: 10.1111/j.1462-5822.2007.01089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Yao L, Jermanus C, Barbetta B, Choi C, Verbeke P, Ojcius DM, Yilmaz O. Porphyromonas gingivalis infection sequesters pro-apoptotic Bad through Akt in primary gingival epithelial cells. Mol Oral Microbiol. 2010;25:89–101. doi: 10.1111/j.2041-1014.2010.00569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Spooner R, Yilmaz O. Nucleoside-diphosphate-kinase: a pleiotropic effector in microbial colonization under interdisciplinary characterization. Microbes Infect. 2012;14:228–237. doi: 10.1016/j.micinf.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Yilmaz O, Sater AA, Yao LY, Koutouzis T, Pettengill M, Ojcius DM. ATP-dependent activation of an inflammasome in primary gingival epithelial cells infected by Porphyromonas gingivalis. Cell Microbiol. 2010;12:188–198. doi: 10.1111/j.1462-5822.2009.01390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Lin FY, Huang CY, Lu HY, Shih CM, Tsao NW, Shyue SK, Lin CY, Chang YJ, Tsai CS, Lin YW, Lin SJ. The GroEL protein of Porphyromonas gingivalis accelerates tumor growth by enhancing endothelial progenitor cell function and neovascularization. Mol Oral Microbiol. 2014 doi: 10.1111/omi.12083. [DOI] [PubMed] [Google Scholar]
- [35].Pace A, Barone G, Lauria A, Martorana A, Piccionello AP, Pierro P, Terenzi A, Almerico AM, Buscemi S, Campanella C, Angileri F, Carini F, Zummo G, de Macario EC, Cappello F, Macario AJ. Hsp60, a novel target for antitumor therapy: structure-function features and prospective drugs design. Curr Pharm Des. 2013;19:2757–2764. doi: 10.2174/1381612811319150011. [DOI] [PubMed] [Google Scholar]
- [36].Zhao M, Sano D, Pickering CR, Jasser SA, Henderson YC, Clayman GL, Sturgis EM, Ow TJ, Lotan R, Carey TE, Sacks PG, Grandis JR, Sidransky D, Heldin NE, Myers JN. Assembly and initial characterization of a panel of 85 genomically validated cell lines from diverse head and neck tumor sites. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:7248–7264. doi: 10.1158/1078-0432.CCR-11-0690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Spooner R, DeGuzman J, Lee KL, Yilmaz O. Danger signal adenosine via adenosine 2a receptor stimulates growth of Porphyromonas gingivalis in primary gingival epithelial cells. Mol Oral Microbiol. 2014;29:67–78. doi: 10.1111/omi.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Yilmaz O, Lee K. The inflammasome and danger molecule signaling: at the crossroads of inflammation and pathogen persistence in the oral cavity. Periodontol 2000. 2015 doi: 10.1111/prd.12084. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Barnes VM, Teles R, Trivedi HM, Devizio W, Xu T, Mitchell MW, Milburn MV, Guo L. Acceleration of Purine Degradation by Periodontal Diseases. J Dent Res. 2009;88:851–855. doi: 10.1177/0022034509341967. [DOI] [PubMed] [Google Scholar]
- [40].Choi CH, Spooner R, DeGuzman J, Koutouzis T, Ojcius DM, Yilmaz O. Porphyromonas gingivalis-nucleoside-diphosphate-kinase inhibits ATP-induced reactive-oxygen-species via P2X7 receptor/NADPH-oxidase signalling and contributes to persistence. Cell Microbiol. 2013;15:961–976. doi: 10.1111/cmi.12089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Roger S, Pelegrin P. P2X7 receptor antagonism in the treatment of cancers. Expert Opin Investig Drugs. 2011;20:875–880. doi: 10.1517/13543784.2011.583918. [DOI] [PubMed] [Google Scholar]
- [42].Huang S, Chen Y, Wu W, Ouyang N, Chen J, Li H, Liu X, Su F, Lin L, Yao Y. miR-150 promotes human breast cancer growth and malignant behavior by targeting the pro-apoptotic purinergic P2X7 receptor. PLoS One. 2013;8:e80707. doi: 10.1371/journal.pone.0080707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Sitkovsky MV, Hatfield S, Abbott R, Belikoff B, Lukashev D, Ohta A. Hostile, hypoxia-A2-adenosinergic tumor biology as the next barrier to overcome for tumor immunologists. Cancer immunology research. 2014;2:598–605. doi: 10.1158/2326-6066.CIR-14-0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Turhani D, Krapfenbauer K, Thurnher D, Langen H, Fountoulakis M. Identification of differentially expressed, tumor-associated proteins in oral squamous cell carcinoma by proteomic analysis. Electrophoresis. 2006;27:1417–1423. doi: 10.1002/elps.200500510. [DOI] [PubMed] [Google Scholar]
- [45].Yan S, Li B, Bai ZZ, Wu JQ, Xie DW, Ma YC, Ma XX, Zhao JH, Guo XJ. Clinical epidemiology of gastric cancer in Hehuang valley of China: a 10-year epidemiological study of gastric cancer. World journal of gastroenterology : WJG. 2014;20:10486–10494. doi: 10.3748/wjg.v20.i30.10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Allen-Vercoe E, Jobin C. Fusobacterium and Enterobacteriaceae: Important players for CRC? Immunol Lett. 2014 doi: 10.1016/j.imlet.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Hajishengallis G. Periodontitis: from microbial immune subversion to systemic inflammation. Nature reviews. Immunology. 2014;15:30–44. doi: 10.1038/nri3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell host & microbe. 2013;14:195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Morita E, Narikiyo M, Yano A, Nishimura E, Igaki H, Sasaki H, Terada M, Hanada N, Kawabe R. Different frequencies of Streptococcus anginosus infection in oral cancer and esophageal cancer. Cancer science. 2003;94:492–496. doi: 10.1111/j.1349-7006.2003.tb01471.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Ramirez-Garcia A, Rementeria A, Aguirre-Urizar JM, Moragues MD, Antoran A, Pellon A, Abad-Diaz-de-Cerio A, Hernando FL. Candida albicans and cancer: Can this yeast induce cancer development or progression? Critical reviews in microbiology. 2014:1–13. doi: 10.3109/1040841X.2014.913004. [DOI] [PubMed] [Google Scholar]
- [51].Tamai R, Sugamata M, Kiyoura Y. Candida albicans enhances invasion of human gingival epithelial cells and gingival fibroblasts by Porphyromonas gingivalis. Microb Pathog. 2011;51:250–254. doi: 10.1016/j.micpath.2011.06.009. [DOI] [PubMed] [Google Scholar]
- [52].Schett G, Coates LC, Ash ZR, Finzel S, Conaghan PG. Structural damage in rheumatoid arthritis, psoriatic arthritis, and ankylosing spondylitis: traditional views, novel insights gained from TNF blockade, and concepts for the future. Arthritis Res Ther. 2011;13(Suppl 1):S4. doi: 10.1186/1478-6354-13-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Koziel J, Mydel P, Potempa J. The link between periodontal disease and rheumatoid arthritis: an updated review. Current rheumatology reports. 2014;16:408. doi: 10.1007/s11926-014-0408-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Totaro MC, Cattani P, Ria F, Tolusso B, Gremese E, Fedele AL, D'Onghia S, Marchetti S, Sante GD, Canestri S, Ferraccioli G. Porphyromonas gingivalis and the pathogenesis of rheumatoid arthritis: analysis of various compartments including the synovial tissue. Arthritis Res Ther. 2013;15:R66. doi: 10.1186/ar4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Konig MF, Paracha AS, Moni M, Bingham CO, 3rd, Andrade F. Defining the role of Porphyromonas gingivalis peptidylarginine deiminase (PPAD) in rheumatoid arthritis through the study of PPAD biology. Ann Rheum Dis. 2014 doi: 10.1136/annrheumdis-2014-205385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Kaur S, White S, Bartold PM. Periodontal disease and rheumatoid arthritis: a systematic review. J Dent Res. 2013;92:399–408. doi: 10.1177/0022034513483142. [DOI] [PubMed] [Google Scholar]
- [57].Gully N, Bright R, Marino V, Marchant C, Cantley M, Haynes D, Butler C, Dashper S, Reynolds E, Bartold M. Porphyromonas gingivalis peptidylarginine deiminase, a key contributor in the pathogenesis of experimental periodontal disease and experimental arthritis. PLoS One. 2014;9:e100838. doi: 10.1371/journal.pone.0100838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Lin FY, Hsiao FP, Huang CY, Shih CM, Tsao NW, Tsai CS, Yang SF, Chang NC, Hung SL, Lin YW. Porphyromonas gingivalis GroEL induces osteoclastogenesis of periodontal ligament cells and enhances alveolar bone resorption in rats. PLoS One. 2014;9:e102450. doi: 10.1371/journal.pone.0102450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Cantaert T, Teitsma C, Tak PP, Baeten D. Presence and role of anti-citrullinated protein antibodies in experimental arthritis models. Arthritis Rheum. 2013;65:939–948. doi: 10.1002/art.37839. [DOI] [PubMed] [Google Scholar]
- [60].Shaw PJ, McDermott MF, Kanneganti TD. Inflammasomes and autoimmunity. Trends in molecular medicine. 2011;17:57–64. doi: 10.1016/j.molmed.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Stanko P, Izakovicova Holla L. Bidirectional association between diabetes mellitus and inflammatory periodontal disease. A review, Biomedical papers of the Medical Faculty of the University Palacky, Olomouc. Czechoslovakia. 2014;158:35–38. doi: 10.5507/bp.2014.005. [DOI] [PubMed] [Google Scholar]
- [62].Taylor JJ, Preshaw PM, Lalla E. A review of the evidence for pathogenic mechanisms that may link periodontitis and diabetes. J Clin Periodontol. 2013;40(Suppl 14):S113–134. doi: 10.1111/jcpe.12059. [DOI] [PubMed] [Google Scholar]
- [63].Li H, Yang H, Ding Y, Aprecio R, Zhang W, Wang Q, Li Y. Experimental periodontitis induced by Porphyromonas gingivalis does not alter the onset or severity of diabetes in mice. J Periodontal Res. 2013;48:582–590. doi: 10.1111/jre.12041. [DOI] [PubMed] [Google Scholar]
- [64].Hyvarinen K, Salminen A, Salomaa V, Pussinen PJ. Systemic exposure to a common periodontal pathogen and missing teeth are associated with metabolic syndrome. Acta diabetologica. 2014 doi: 10.1007/s00592-014-0586-y. [DOI] [PubMed] [Google Scholar]
- [65].Aemaimanan P, Amimanan P, Taweechaisupapong S. Quantification of key periodontal pathogens in insulin-dependent type 2 diabetic and non-diabetic patients with generalized chronic periodontitis. Anaerobe. 2013;22:64–68. doi: 10.1016/j.anaerobe.2013.06.010. [DOI] [PubMed] [Google Scholar]
- [66].Garcia-Hernandez MH, Portales-Cervantes L, Cortez-Espinosa N, Vargas-Morales JM, Fritche Salazar JF, Rivera-Lopez E, Rodriguez-Rivera JG, Quezada-Calvillo R, Portales-Perez DP. Expression and function of P2X(7) receptor and CD39/Entpd1 in patients with type 2 diabetes and their association with biochemical parameters. Cell Immunol. 2011;269:135–143. doi: 10.1016/j.cellimm.2011.03.022. [DOI] [PubMed] [Google Scholar]
- [67].Rondas D, Crevecoeur I, D'Hertog W, Ferreira GB, Staes A, Garg AD, Eizirik DL, Agostinis P, Gevaert K, Overbergh L, Mathieu C. Citrullinated glucose-regulated protein 78 is an autoantigen in type 1 diabetes. Diabetes. 2014 doi: 10.2337/db14-0621. [DOI] [PubMed] [Google Scholar]
- [68].Schmidlin PR, Fachinger P, Tini G, Graber S, Seifert B, Dombrowa S, Irani S. Shared microbiome in gums and the lung in an outpatient population. The Journal of infection. 2014 doi: 10.1016/j.jinf.2014.10.005. [DOI] [PubMed] [Google Scholar]
- [69].Yoneda M, Naka S, Nakano K, Wada K, Endo H, Mawatari H, Imajo K, Nomura R, Hokamura K, Ono M, Murata S, Tohnai I, Sumida Y, Shima T, Kuboniwa M, Umemura K, Kamisaki Y, Amano A, Okanoue T, Ooshima T, Nakajima A. Involvement of a periodontal pathogen, Porphyromonas gingivalis on the pathogenesis of non-alcoholic fatty liver disease. BMC gastroenterology. 2012;12:16. doi: 10.1186/1471-230X-12-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Iwasaki M, Taylor GW, Manz MC, Kaneko N, Imai S, Yoshihara A, Miyazaki H. Serum antibody to Porphyromonas gingivalis in chronic kidney disease. J Dent Res. 2012;91:828–833. doi: 10.1177/0022034512455063. [DOI] [PubMed] [Google Scholar]