Abstract
ADP-ribosylation is a post-translational modification where single units (mono-ADP-ribosylation) or polymeric chains (poly-ADP-ribosylation) of ADP-ribose are conjugated to proteins by ADP-ribosyltransferases. This post-translational modification and the ADP-ribosyltransferases (also known as PARPs) responsible for its synthesis have been found to play a role in nearly all major cellular processes, including DNA repair, transcription, translation, cell signaling, and cell death. Furthermore, dysregulation of ADP-ribosylation has been linked to diseases including cancers, diabetes, neurodegenerative disorders, and heart failure, leading to the development of therapeutic PARP inhibitors, many of which are currently in clinical trials. The study of this therapeutically important modification has recently been bolstered by the application of mass spectrometry-based proteomics, arguably the most powerful tool for the unbiased analysis of protein modifications. Unfortunately, progress has been hampered by the inherent challenges that stem from the physicochemical properties of ADP-ribose, which as a post-translational modification is highly charged, heterogeneous (linear or branched polymers, as well as monomers), labile, and found on a wide range of amino acid acceptors. In this Perspective, we discuss the progress that has been made in addressing these challenges, including the recent breakthroughs in proteomics techniques to identify ADP-ribosylation sites, and future developments to provide a proteome-wide view of the many cellular processes regulated by ADP-ribosylation.
Daniels et al. discuss the recent breakthroughs in proteomics techniques to identify ADP-ribosylation sites and future developments to provide a proteome-wide view of the many cellular processes regulated by ADP-ribosylation.
Main Text
Introduction
ADP-ribosylation refers to the transfer of the ADP-ribose group from NAD+ to target proteins post-translationally. This post-translational modification (PTM) can be added onto amino acids of diverse chemistry, including aspartate, glutamate, lysine, arginine, and cysteine. ADP-ribose groups can be attached singly as mono(ADP-ribose) (MAR) or in polymeric chains as poly(ADP-ribose) (PAR) by the enzymatically active members of the family of 17 human ADP-ribosyltransferases (ARTs), commonly known as poly(ADP-ribose) polymerases (PARPs) (Hottiger et al., 2010, Vyas et al., 2014), as well as a subset of NAD+-dependent sirtuins (Houtkooper et al., 2012). Together, MAR and PAR regulate fundamental cellular processes through their roles as signaling molecules (Aredia and Scovassi, 2014, Perraud et al., 2001) and post-translational modifications (Feijs et al., 2013b, Gibson and Kraus, 2012). In addition, ADP-ribosylation has been shown to be a therapeutically important modification in cancers, neurodegenerative diseases, ischemia, and inflammatory disorders (Curtin and Szabo, 2013), where PARPs are hotly pursued drug targets by pharmaceutical companies (Steffen et al., 2013). Over a hundred clinical trials for the treatment of cancers have been carried out for PARP-1 inhibitors and many ongoing trials are in late stages (Garber, 2013, Lord et al., 2015). Notably, these anti-cancer drugs can also cross-react with other PARPs (Wahlberg et al., 2012), which are increasingly appreciated for their multifaceted roles in the cell (Figure 1 ; Table S1) (Gibson and Kraus, 2012, Vyas et al., 2013). Identifying the substrate specificities of these PARPs will help elucidate distinct functions of this 17-member family and may have therapeutic implications in designing PARP inhibitor-based therapies. Recent advances in mass spectrometry (MS)-based methods for characterizing ADP-ribosylated proteins have opened up unprecedented possibilities to explore the functions of this family of enzymes and provide insights into the clinical relevance of this under-studied protein modification.
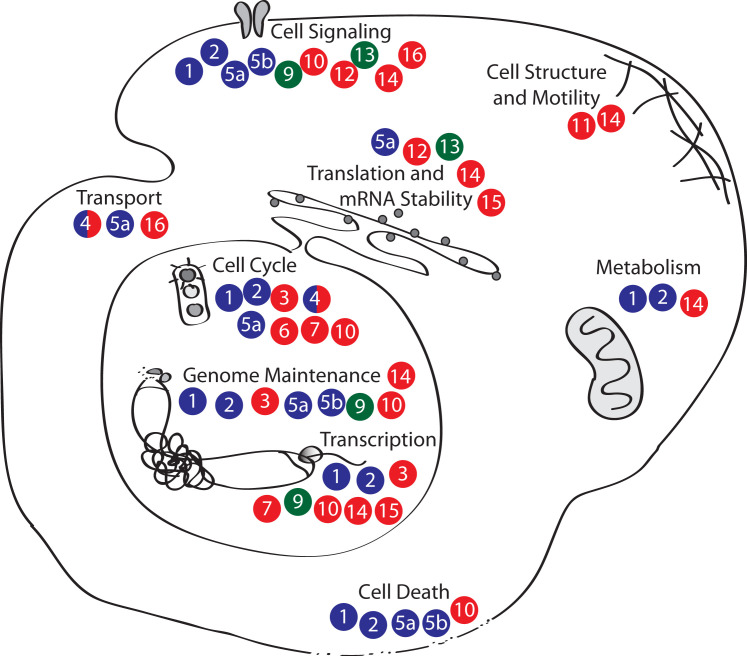
Figure 1.
The PARP Family
PARPs have been linked to nearly all major cellular processes. Juxtaposition of protein identifiers (e.g., 1 = PARP-1) indicates the involvement of the protein in the regulation or execution of the cellular process. Enzymatic activity is indicated by the bubble color: blue = poly(ADP-ribosyl)transferase, red = mono(ADP-ribosyl)transferase, green = no transferase activity. For references, see Table S1.
MS-based proteomics offers three types of data that genomics and transcriptomics cannot: protein-protein interaction mapping (interactomics), identification of protein post-translational modifications, and quantitative information at the protein level (for an in-depth overview of the potential held by MS-based proteomics, we recommend Cox and Mann, 2011). A complete map of the ADP-ribosylated proteome will include all three elements, providing insights into how ADP-ribosylated substrates are regulated via recruitment of MAR/PAR-binding proteins, their sites of modification, and abundance in cells. While the ADP-ribosylated interactome has been explored in the last decade, it is only recently that MS-based techniques have been available for the identification of ADP-ribosylated sites at the proteome scale. In this Perspective, we will explore how MS-based proteomics can help address several important questions in the field of ADP-ribosylation. (1) What is the significance of the many potential amino acid attachment sites? Which attachments are regulated by which enzymes? (2) How can we distinguish between sites of MAR and PAR, and between the many possible structures of PAR, including length and branch variants? How important are these distinctions? (3) What does an increase in cellular PARylation levels mean? Does it reflect an increase in the number of amino acid site modifications, an increase in the number of ADP-ribose units at existing sites, or an increase in unconjugated PAR levels? (4) Are all ADP-ribosylation sites physiologically significant? In the following sections, we will discuss the inherent challenges, existing solutions, and future needs to address these critical questions for a complete, functional understanding of the ADP-ribosylated proteome.
Investigating the ADP-Ribosylated Proteome by Mass Spectrometry: Challenges
Mapping of MARylated and PARylated (collectively, ADP-ribosylated) proteomes requires robust protocols to overcome the dynamic, heterogeneous, and labile nature of these modifications. An initial challenge is the variable PAR attachment sites, which can be found on acidic and basic residues, a list that expands when MARylation sites are also considered (see later sections). This variability results in a wide range of chemical and enzymatic sensitivities (Cervantes-Laurean et al., 1997), greatly hindering the identification of an intact, complete ADP-ribosylated proteome. Second, the modification itself is typically found at low levels in cells and exhibits very fast attachment/removal kinetics (Wielckens et al., 1982), making robust enrichment methods a critical component for elucidating the ADP-ribosylated proteome. Third, the structure of the PAR polymer poses a practical challenge, as it is heterogeneous (between 2 and 200 subunits in vivo, can be branched or linear [Hassa et al., 2006]) and highly charged, characteristics incompatible with most MS methods. Here, we will consider the methods that have addressed and overcome subsets of these challenges and the potential for further progress on those that remain.
A Draft of the ADP-Ribosylated Interactome
Molecular interactions can serve as an early indicator of molecular functions, and a sizeable contribution has already been made to the field of ADP-ribosylation by several large-scale proteomics studies that identify proteins associated with MAR and/or PAR, which are summarized in Figure 2 A (Gagné et al., 2008, Gagné et al., 2012, Isabelle et al., 2012, Jungmichel et al., 2013). These studies used a common experimental design: human cells were exposed to DNA damaging agents, a classical stimulant of PARP-1 PARylation activity, before being lysed and subjected to enrichment of ADP-ribosylated proteins, followed by MS-based protein identification. Because the enrichment is performed under a range of non-denaturing conditions in all of these studies, the proteins identified include not only ADP-ribosylated proteins but also ADP-ribose binding proteins and the larger non-covalent interaction networks, thereby providing an aggregate picture of the ADP-ribosylated interactome. Using all 832 proteins identified in these studies (Table S2), a draft map of biological processes enriched in the DNA damage-induced ADP-ribosylated interactome is presented in Figure 2B and Figure S1 in detail. While the DNA damage response is the canonical role for PARylation in cells, it is clear that additional roles for ADP-ribosylation are present even following genomic insult. In particular, there is a significant enrichment of RNA processing factors (purple boxes), a trend that was noted individually by each group. Such enrichment may be linked to the similarity of the chemical and electrostatic properties of PAR and RNA—cellular biopolymers that are able to share binding partners (e.g., Murawska et al., 2011). Another noted enrichment is seen for cellular macromolecular complex assembly, exemplified in mitotic spindles (Chang et al., 2004), nucleoli (Boamah et al., 2012), stress granules (Leung et al., 2011), DNA repair complexes (Okano et al., 2003), and nuclear matrices (Cardenas-Corona et al., 1987), possibly owing to the polymeric nature of PAR and the plethora of PAR binding domains that may target this polymer as a structural scaffold. Such proteome-wide views of the biological processes regulated by ADP-ribosylation sends researchers and clinicians a key message: a reduction in ADP-ribosylation by PARP inhibitors impacts many aspects of cellular function and should not be seen as a simple block to DNA repair.
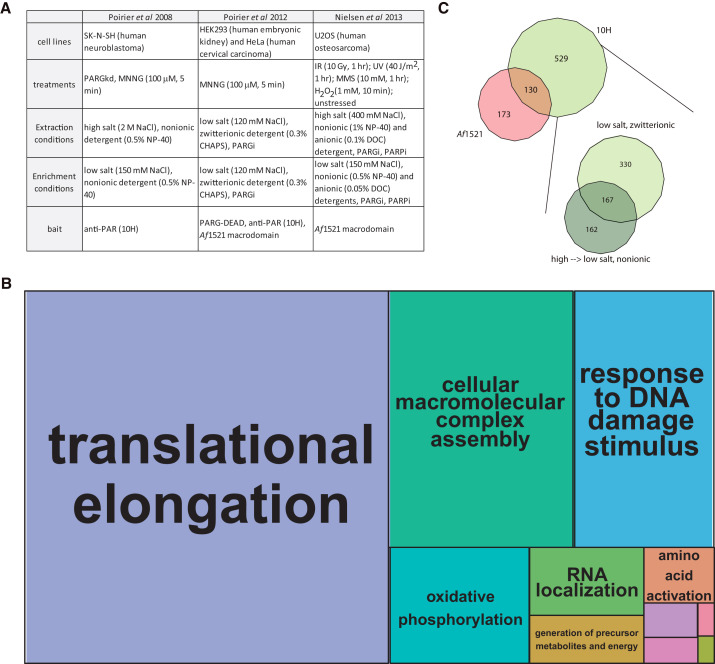
Figure 2.
Processes Enriched in the ADP-Ribosylated Interactome
(A) Experimental design for the interactome studies used for this meta-analysis. PARGi, PARG inhibitor; PARPi, PARP inhibitor; PARGkd, PARG knockdown.
(B) The pooled DNA-damaged induced ADP-ribosylated interactome depicted as a treemap of enriched biological processes. The most enriched biological processes (based on statistical likelihood) are shown as larger components within the map and grouped according to common cellular functions. See Figure S1 for the detailed version of this treemap. Gene ontology determined using DAVID (Huang et al., 2009), treemap constructed using REViGO (Supek et al., 2011) and R (R Development Core Team, 2011).
(C) A compilation of the proteins identified in response to DNA damage can be broken out by enrichment methods (bait) or cell lysis conditions. For comparison of lysis conditions, the 10H enriched proteins were analyzed. Euler diagrams created in VennMaster (Kestler et al., 2005). Source data available in Table S2.
In light of the similarities in experimental design, the methods chosen for cell lysis and enrichment have proven to be critical determinants of the interactome observed by each group. Variations in the enrichment method for ADP-ribosylated proteins produced two nearly distinct sets of proteins (see Figure 2C), partly resulting from biased affinity of the 10H antibody for PAR polymers longer than 20 subunits (Kawamitsu et al., 1984), while the Af1521 macrodomain enriches for both MARylated and PARylated proteins (Dani et al., 2009). Such biased affinity may help explain why the Af1521 macrodomain-enriched interactome contains more known ADP-ribosylated substrates (as determined by their inclusion in site identification studies [Daniels et al., 2014, Zhang et al., 2013]) than the 10H antibody-enriched interactome (see Figure S2), as the longer polymers targeted by the antibody may serve as bait for PAR binding proteins and their interactors. The 10H-derived interactome can be separated into unique networks based on lysis buffer composition (Figure 2C), supporting that many of these protein-protein and protein-PAR interactors are non-covalent and subject to charge disruption. Of note, Nielsen and co-workers emphasized the inclusion of PARP inhibitors in cell lysis buffer to prevent the DNA sheared during the cell lysis procedure from activating the DNA damage-responsive PARPs in vitro; prevention of this activation cuts down on non-physiological PAR-dependent interactions formed in cell lysate (Jungmichel et al., 2013), an observation that may further explain the unique identifications in the studies shown in Figure 2C. While these pioneering studies highlight the importance for the consideration of lysis conditions and enrichment methods, it is clear that we have yet to approach saturation in probing the complete ADP-ribosylated interactome; we expect that a more complete interactome will be obtained using complementary strategies to induce and enrich ADP-ribose. Besides DNA damage, it is equally important to characterize the ADP-ribosylated interactomes under other cellular stress as well as the interactomes within various PAR-enriched cellular macromolecular complexes. Healthy, unstressed cells have also been shown to maintain low basal levels of PAR, with cellular PARylation patterns distinct at different stages of the cell cycle and in different cellular compartments (Vyas et al., 2013). Though it is quite common to increase the amount of endogenous ADP-ribosylated substrates by long-term knockdown of the PAR degradative enzyme PARG, such treatment will likely cause non-physiological changes (as shown in the PARG110 knockout mouse [Min et al., 2010]). While the recent development of cell-permeable PARG inhibitors may provide an alternative to increase the amount of substrates without requiring long-term treatment (Finch et al., 2012), it is a priority to improve the existing methods for enriching ADP-ribosylated substrates (reviewed in Vivelo and Leung, 2015) and increase the sensitivity of MS to detect them from native cell conditions.
By definition, the ADP-ribosylated interactome is composed of covalently ADP-ribosylated substrates, ADP-ribose binding proteins, and their interacting proteins. With the ability to synthesize MAR or PAR with a defined number of ADP-ribose groups (Kistemaker et al., 2015, Lambrecht et al., 2015, Tan et al., 2012), it is foreseeable to further refine the mapping of the proteome that binds to single or multiple ADP-ribose groups non-covalently. Parallel development of techniques to identify the attachment sites of ADP-ribosylation has already allowed for definitive identification of ADP-ribosylated substrates at the proteome level (Daniels et al., 2014, Zhang et al., 2013; see the next section). Combination of these complementary sets of proteomic data will allow researchers more precision in mapping the connections within the ADP-ribosylated interactome.
Characterizing ADP-Ribosylation at the Level of the Amino Acid Attachment Sites
While MARylation and PARylation have long been considered two classes of PTMs, it is useful, and perhaps more accurate, to consider their attachment sites together as a single modification. The first reason for this consideration stems from knowledge of the PAR degradative enzyme PARG (Slade et al., 2011), which is capable of transforming PARylated substrates into MARylated ones, effectively blurring the lines between sites of mono and poly(ADP-ribose). Second, there is evidence of cooperative efforts between enzymes capable of adding mono and poly(ADP-ribose) to proteins (Mao et al., 2011), which may result in a PARP adding polymer to an existing MAR initiation site—an occurrence which has also been shown in vitro through PARP-1 elongation of MARylated agarose beads (Panzeter et al., 1992). This notion of shared sites for MAR and PAR synthesis is taken further by the demonstration that PARP-4 exhibits MARylating activity in isolation but has PARylating activity in its native vault protein complex; this change in activity presumably arises through cooperation with other members of the complex, none of which are known PARPs (Kickhoefer et al., 1999, Vyas et al., 2014). For these reasons, characterization of ADP-ribosylation attachment sites remains distinct from characterization of the heterogeneous molecule (mono/poly, linear/branched) occupying these sites. Accordingly, the MS-based methods for ADP-ribosylation site identification discussed in this section are restricted to identifying the site of the PTM attachment following removal of any subunits beyond the protein–proximal monomer, offering no information with respect to the original size or structure of the corresponding PTM.
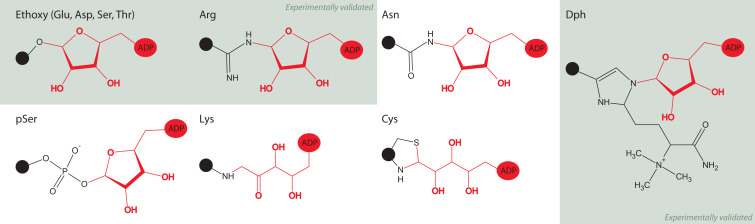
A major analytical challenge in identifying ADP-ribosylation attachment sites comes from the wide variety of amino acids that can be ADP-ribosylated, including glutamic and aspartic acids, serines, threonines (Cervantes-Laurean et al., 1995), phosphoserines (Smith and Stocken, 1975), cysteines (McDonald and Moss, 1994), asparagines (Manning et al., 1984), arginines (Laing et al., 2011), lysines (Altmeyer et al., 2009), and diphthamides (Oppenheimer and Bodley, 1981). This large collection of ADP-ribose acceptors provides a number of unique attachment structures (Figure 3 ) that differ in chemical and enzymatic sensitivities, e.g., acidic, but not basic, amino acids lose ADP-ribose in the presence of high pH, hydroxylamine quickly releases ADP-ribose groups from modified glutamate, aspartate, and, less readily, from arginine, and ADP-ribose is exclusively removed from arginine in the presence of the ADP-ribosyl hydrolase ARH1 (Cervantes-Laurean et al., 1997, Moss et al., 1983, Moss et al., 1992). Though the majority of ADP-ribosylated sites are sensitive to hydroxylamine (Adamietz and Hilz, 1976), hydroxylamine-insensitive sites, such as lysine, may also serve important biological roles (Messner and Hottiger, 2011). Phosphorylated tyrosine sites are relatively rare in comparison to phosphoserine and phosphothreonine, yet they play indispensable roles in cellular biology (Olsen et al., 2006); it would be important, therefore, to study all intracellular protein residue-ADP-ribose attachments to understand the significance of each site of modification.
Figure 3.
ADP-Ribosylation Attachment Sites
Known and predicted structures linking amino acids to ADP-ribose, grayed out boxes show structures that have been validated. See text for references.
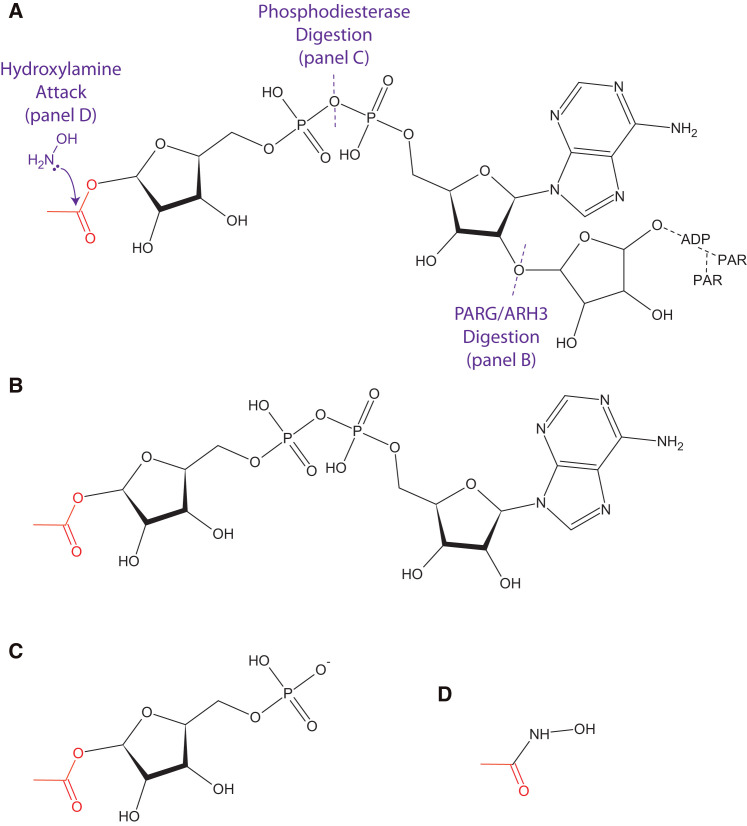
Several sample preparation methods have been developed to study ADP-ribosylation sites by MS. The first relies on the unambiguous identification of MARylated sites, which can be identified as a 541.06 Dalton mass shift above the unmodified form of the peptide (Figures 4A and 4B). A distinct advantage of this method comes from the reliable fragmentation of the modification itself during standard peptide fragmentation, providing diagnostic ions that can confirm the ADP-ribosylation state of the modified peptide (Hengel et al., 2009, Oetjen et al., 2009, Tao et al., 2009). This method has also been utilized for the identification of PARylation sites following treatment of defined substrates in vitro with PARG or ARH3, both of which leave MAR at the otherwise mass variant PAR attachment site (Messner et al., 2010, Rosenthal et al., 2011). It should be noted, however, that an inherent uncertainty underlies a subset of site identifications following PARG/ARH3 treatment as these enzymes release free ADP-ribose—a molecule that has been shown to spontaneously ADP-ribosylate the N terminus of proteins and peptides as well as lysine, arginine, and cysteine residues in vitro (Cervantes-Laurean et al., 1996, Kharadia and Graves, 1987, McDonald and Moss, 1994). As such modifications have the potential to form in any environment rich in free ADP-ribose (e.g., in the vicinity of PARG/ARH3 digestion), the occurrence and significance of these non-enzymatic modifications in cells remain important unanswered questions (see Supplemental Information for further discussion). As such, their presence cannot currently be attributed exclusively to either sample preparation or intracellular biology, particularly when PARG (or any enzyme capable of producing free ADP-ribose) is present in both scenarios, and the field would greatly benefit from performing a series of experiments to clearly establish or dispel whether non-enzymatic ADP-ribosylation should be a concern for proteomics studies.
Figure 4.
ADP-Ribosylation Tags
(A) Poly(ADP-ribose) can be simplified to mono(ADP-ribose) as in (B) by the glycohydrolase activity of PARG/ARH3, (C) to phosphoribose through digestion by phosphodiesterase, or (D) to a hydroxamic acid derivative though exposure to hydroxylamine. Of note, hydroxylamine treatment on ADP-ribosylated arginine results in the formation of the hydroxyamate of ADP-ribose (Moss et al., 1983) and therefore will likely not leave the 15.01 Da signature on formerly modified arginine residues as in glutamate/aspartate residues (Zhang et al., 2013). A representative acidic attachment site (red) is used for illustration.
Two alternatives have been demonstrated in recent studies to identify ADP-ribosylation sites at the proteome level. The first method—digestion of MAR and PAR down to their phosphoribose attachment sites (Figures 4A and 4C) (Chapman et al., 2013, Daniels et al., 2014, Hengel and Goodlett, 2012, Palazzo et al., 2015)—relies upon the pyrophosphatase activity of either snake venom phosphodiesterase (Matsubara et al., 1970), a standard enzyme for in vitro PAR digestion, or human NudT16, a recently discovered ADP-ribosyl phosphodiesterase that is ∼100-fold less efficient than SVP but can be synthesized as a recombinant protein (Palazzo et al., 2015). Similar to the PARG/ARH3 method, the chemistry of the attachment site is maintained; however, the iso-ADP-ribose fragments released by phosphodiesterase do not allow for formation of the reactive aldehyde group, which has shown to be responsible for spontaneous ADP-ribosylation (Cervantes-Laurean et al., 1996). The apparent unbiased digestion of PAR and MAR by SVP suggests that this method will be amenable to all forms of amino acid attachments and has indeed produced acidic, basic, and nucleophilic site identifications from endogenously modified proteins (Daniels et al., 2014, Vyas et al., 2014). The second method relies upon the release of ADP-ribose from acidic (glutamic and aspartic) amino acid residues by hydroxylamine, a standard method for distinguishing between amino acid acceptors of ADP-ribose (Cervantes-Laurean et al., 1997). The utility of this method lies in the alteration of the acidic group following hydroxylamine release of ADP-ribose (Figure 4A); the resultant hydroxamic acid derivative produces a mass shift of 15.01 Daltons, which is easily distinguishable by MS (Figure 4D) (Zhang et al., 2013). Though limited to identifying only acidic ADP-ribosylation sites, this method has provided a list of 1,048 sites on 340 proteins from the acidic ADP-ribosylated proteome, highlighting for the first time the widespread modification of substrate proteins in cells (Zhang et al., 2013).
With the ability to definitively identify ADP-ribosylation sites, it is now possible to begin addressing the roles of protein ADP-ribosylation. The functional impact of such modified sites can, to some extent, be addressed by mutagenesis studies using recombinant proteins or by targeted genome editing techniques in cells. However, unique difficulties accompany these classic means of characterizing PTM effects, as point mutations are limited by the large number of amino acids that can be ADP-ribosylated (Figure 3). For example, mutation of a glutamic acid to an aspartic acid will not guarantee a lack of ADP-ribosylation, requiring researchers to, in the interest of blocking ADP-ribosylation, mutate acidic sites to non-acidic residues. The requirement of such mutagenesis strategies further complicates the interpretation of molecular or cellular effects—is ADP-ribosylation of the residue important, or has the loss of an acidic residue changed the structure or interaction network of the protein? As an alternative to blocking ADP-ribosylation by mutational means, chemical strategies have been developed to introduce ADP-ribose groups at specific residues on purified peptides (Kistemaker et al., 2013, Moyle and Muir, 2010, van der Heden van Noort et al., 2010); this technique could allow researchers to mimic the ADP-ribosylated form of a protein by conjugating the modified peptide of interest to the terminus/termini of the parent protein, a technique (termed semisynthesis), which has allowed for functional analysis of phosphorylated proteins in vitro (Szewczuk et al., 2009). Another way to ascertain functional roles of these sites in cells involves following their modification status temporally upon treatment that induces or inhibits ADP-ribosylation. For example, quantitative proteomics techniques have already been utilized to map out the temporal coordination of ADP-ribose related protein complexes in response to DNA damage—a necessary step toward understanding the mechanism of ADP-ribose-dependent DNA damage repair (Gagné et al., 2012, Isabelle et al., 2012). With these newly developed site identification techniques, we can further define the temporal changes of the ADP-ribosylated substrates at the site level, potentially indicating which particular sites are of physiological significance. Additionally, it has been shown that only a subset of ADP-ribosylation sites within the proteome are sensitive to treatment by chemotherapeutic PARP inhibitors currently in Phase III clinical trials (Zhang et al., 2013). These variable responses to PARP inhibition may indicate the mechanism of action of these drugs, providing the molecular basis of the clinical benefits and side effects observed in patients.
Defining Target Specificity for Addition and Removal of ADP-Ribosylation
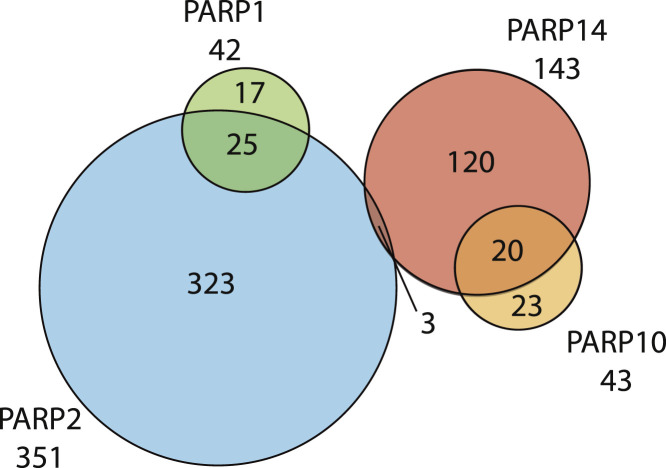
Given the large number of cellular processes regulated by PARPs (see Figure 1), it will be interesting to determine the shared and unique substrates of each of the enzymatically active family members. Using a protein microarray that consists of 8,000 proteins, two groups have identified the sub-proteomes that can be modified by PARP-2, PARP-10, and PARP-14 in vitro (Feijs et al., 2013a, Troiani et al., 2011). Alternatively, the Cohen group has engineered PARP-1 and PARP-2 mutants that specifically use a bio-orthogonal NAD+ analog for the identification of their respective PARP-specific substrates from nuclear extracts in vitro (Carter-O’Connell et al., 2014). The majority of proteins modified by individual PARPs are distinct, suggesting that each PARP exhibits unique substrate specificity (Figure 5 ; Table S3). When coupled with site identification techniques, it is now possible to determine whether there is a defined motif surrounding the ADP-ribosylation sites modified by each PARP. For example, whether any of the PARPs are responsible for the consensus sequence of PXE∗, E∗P, PXXE, or E∗XXG surrounding the modified glutamate (E∗) residue, as identified by the Yu group recently (Zhang et al., 2013). Similar experimental designs may allow us to deduce whether there are specific motifs for modification by individual PARPs, such as those identified for PARP-5a substrates (Guettler et al., 2011).
Figure 5.
PARP Substrate Specificity
Substrates for PARP-1, PARP-2, PARP-10, and PARP-14 were identified in three studies using protein arrays or analog-sensitive mutant protein identification (see text). Euler diagrams created in VennMaster. Source data available in Table S3.
One puzzling piece of data from the current studies is that ADP-ribosylation sites auto-modified by each PARP are found at diverse amino acids, such as acidic (Glu/Asp), basic (Lys, Arg), and nucleophilic (Cys) residues; this apparent lack of specificity is true for PARPs that add multiple (PARP-1) or single ADP-ribose groups (PARP-3, PARP-6, PARP-10, PARP-11, PARP-12, and PARP-16) (Daniels et al., 2014, Vyas et al., 2014). This flexibility in amino-acid acceptor residues argues against the amino acid specificity of these enzymes, at least during in vitro auto-modification. One possible explanation is that these PARPs are acting as NADases, which hydrolyze NAD+ in vitro (Desmarais et al., 1991), and the released ADP-ribose groups non-enzymatically conjugate to reactive amino acid residues. Though no studies have yet to investigate such non-enzymatic modification on PARPs, Cervantes-Laurean et al. showed that histones can be modified non-enzymatically by incubation with ADP-ribose in vitro and deduced that lysines are the primary sites (Cervantes-Laurean et al., 1996). On the other hand, only cysteine residues were identified in auto-modified PARP-8 in vitro, suggesting that certain PARPs may have defined amino acid specificity (Vyas et al., 2014). It will therefore be of interest to examine whether there are any amino acid preferences on endogenous protein substrates of each PARP at a proteome-wide scale. One major drawback of the current techniques to identify proteome-wide enzyme-substrate relationships is that these experiments were all performed in vitro, thus losing the proper physiological context (e.g., cellular localization, enzyme concentration, protein modification states). Therefore, techniques are urgently needed to identify PARP-specific proteomes in cells.
So far, hydrolases that remove the single ADP-ribose groups from arginine and glutamate have been identified (Table 1 ), but it is not clear whether modifications at other amino acids are reversible. Do hydrolases exhibit amino acid specificity with regard to ADP-ribose removal? Similarly, would the biological modules that bind ADP-ribose groups, such as a macrodomain, have substrate or amino acid binding specificity? Notably, the specificities of macrodomains have been shown to be dependent on the amino acids surrounding the modified sites (Forst et al., 2013, Moyle and Muir, 2010). Thus, these macrodomains will likely enrich for a restricted set of endogenous ADP-ribosylated proteins. Recently, by comparing the ADP-ribosylated proteome from human and mouse cells before and after enrichment by the Af1521 macrodomain, our group found that the macrodomain-enriched proteome selects against ADP-ribosylated glutamate residues globally (Daniels et al., 2014), consistent with the earlier findings that this macrodomain bears hydrolase activity against acidic MARylated amino acids of a single substrate (Jankevicius et al., 2013, Rosenthal et al., 2013). It can be postulated, then, that the glutamate sites identified following enrichment by Af1521 macrodomain were PARylated prior to enrichment, as MAR would have been hydrolyzed off. Using this same line of reasoning, binding and hydrolase specificity (for both the targeted ADP-ribosylated residues and neighboring amino acids) of all ADP-ribose binding modules can be systematically defined. Table 1 summarizes the binding affinity and substrate specificity of some of the most-studied ADP-ribose binding domains and hydrolases. While the primary aim of these characterization studies is often to elucidate the role these protein domains play in cell biology, they have also provided a much-needed expansion of a “biological toolbox” for distinguishing between classes of ADP-ribosylated substrates, an effort which began 20 years ago with the ARH1-aided classification of substrates carrying MARylation on arginine residues (Ohno et al., 1995). This toolbox should provide the means for enriching targeted groups of ADP-ribosylated proteins to expand our knowledge of the ADP-ribosylated proteome.
Table 1.
A Biological Toolbox of ADP-Ribose Binding and Hydrolysis Protein Domains
 |
Our current understanding of the most well-studied ADP-ribose binding domains and hydrolases. Green = Yes, Red = No, E/R = hydrolysis shown specifically for glutamate or arginine residues, respectively. MD = macrodomain, N/A = not applicable, blank = possible but currently unknown. SARS-CoV, Severe Acute Respiratory Syndrome-Coronavirus; HEV, Hepatitis E Virus; SFV, Semliki Forest Virus.
gmouse PARP-14 macrodomain 2; Forst et al., 2013, Rosenthal et al., 2013
lTARG1 removes the complete PAR chain from modified glutamate residues, rather than hydrolyzing glycosidic bonds between subunits of PAR as in PARG and ARH3; Rosenthal et al., 2013, Sharifi et al., 2013
∗ARH3 showed no hydrolase activities against MARylated arginine, cysteine, diphthamide, and asparagine
Distinguishing between Sites of MAR- and PARylation
While it is advisable—and at this point only possible—to study the attachment sites of all forms of ADP-ribosylation together, the distinction between MAR and PAR, as well as the many subclasses of PAR, will likely prove critical for interpretation of the role played by the modified residue of interest. For example, five out of the 15 enzymatically active human PARPs are responsible for PARylation activity, with the other 10 restricted to MARylation (Figure 1), meaning that a change in the PARylation status of a residue can only be attributed to the enzymatic activity of those five PARPs. A similar analysis could be employed for the ADP-ribosyl hydrolases: two can only remove MAR (macroD1 and macroD2), two can turn PAR into MAR (PARG and ARH3), and one can remove both PAR and MAR (TARG1; see Table 1) (Barkauskaite et al., 2013). Therefore, understanding how an ADP-ribosylation site is changing between an unmodified state and carrying MAR or PAR can suggest the enzymes responsible for its regulation. The clinical implications of understanding the distinction between PAR versus MAR is exemplified in the PARP inhibitor classification performed by Wahlberg et al., where 185 PARP inhibitors were assayed for their abilities to bind members of the human PARP family; many of these inhibitors bind to MARylating as well as PARylating members of the family (Wahlberg et al., 2012). Such potential off-target inhibition of MARylation would not be revealed by the typical assay for monitoring the effectiveness of PARP inhibitors, which only measures changes in PARylation level. Knowing which ADP-ribosylation sites are affected by these inhibitors (or in disease states) and how those ADP-ribosylation sites are changing between unmodified, MARylated, and PARylated will be predictive of the PARPs targeted in cells. Finally, multiple ADP-ribose groups in PAR may define functional roles distinct from MAR. For example, while wild-type, PARylation-capable PARP-1 is able to fully rescue DNA repair in PARP-1−/− MEFs, a PARP-1 mutant that is only capable of MARylation activity cannot (Mortusewicz et al., 2007). Such detrimental changes brought on by converting PARylation to MARylation sites may be because the structure of PAR is similar to that of polynucleic acids (e.g., DNA) and thus could compete for, or modulate the functions of, factors that bind nucleic acids. For these reasons, we will now examine potential methods for classifying sites of ADP-ribosylation based on the structure of their PTM.
As diagrammed in Figure 4B, MAR is a homogenous modification with a predictable mass of 541.06 Daltons, allowing MARylation site localization by MS. Given that MARylated peptides can be captured by phosphopeptide enrichment techniques (Laing et al., 2011), it is feasible to globally enrich MARylated peptides from protease-digested cell lysates. In fact, re-analysis of phosphoproteomic data uncovered 79 MARylated proteins (Matic et al., 2012). However, this re-analysis likely underestimates the global level of MARylation due to the high pH (pH 10) phosphopeptide elution employed (Huttlin et al., 2010), a condition that results in loss of ADP-ribose groups conjugated to acidic sites (Cervantes-Laurean et al., 1997). Consistently, all but one of the MARylated sites identified in the re-analysis were arginine, an observation otherwise attributed to the increased stability of ADP-ribosylated arginine as opposed to ADP-ribosylated glutamate in the conditions employed for their study (Matic et al., 2012). For non-biased detection of MARylated proteomes, the labile bond between ADP-ribose groups and acidic amino acids must be preserved, e.g., by choosing a neutral phosphate buffer for eluting the phosphopeptide enrichment matrices (as in Daniels et al., 2014).
Another possibility to distinguish MARylated substrates from PARylated substrates is to exploit the distinct properties of protein domains that specifically recognize them (see biological toolbox, Table 1). For example, the WWE domain recognizes iso-ADP-ribose—the molecular structure spanning consecutive ADP ribose subunits of PAR (Wang et al., 2012); therefore, this domain could be an ideal tool for enriching PARylated, but not MARylated, targets. Alternatively, MacroD2 can be engineered to abrogate its inherent ADP-ribose hydrolase activity but retain its binding specificity toward MARylated substrates (Jankevicius et al., 2013). However, most of these domains were tested with single MARylated or PARylated substrates. Use of this biological toolbox for proteome-wide investigation warrants systematic analyses of these ADP-ribose binding modules to fully characterize their substrate specificities for both binding and hydrolysis.
Free/Conjugated, Branched/Linear: The Many Forms of Poly(ADP-ribose)
Besides identifying the ADP-ribosylation sites, MS can also be used to accurately quantitate PAR levels with femtomole sensitivity (Martello et al., 2013). Assuming an average chain length of 10 ADP-ribose units per PAR molecule, the Bürkle group estimated that there are about 3,000 PAR molecules/cell in native cellular conditions, which can be induced to >150,000 molecules/cell upon DNA damage, with a branching frequency of 1%–2% (Martello et al., 2013). Combining this methodology with site identification could allow researchers to deduce whether the increase in PARylation is a result of new PARylation sites and/or substrates, or simply elongation of existing sites on existing substrates. However, one should be aware of an alternative source of PAR—the soluble PAR that is not attached to target proteins. The existence of soluble PAR in vivo has been inferred from mounting evidence that PARG has both endo- and exo-glycosidic activity, allowing this enzyme to produce and regulate levels of free PAR (Barkauskaite et al., 2013). Additionally the ADP-ribosyl hydrolase TARG1 has been shown to reduce PARylation levels on auto-modified PARP-1 without releasing free ADP-ribose in vitro (Sharifi et al., 2013), indicating that the entire PAR chain could be released as a single unit in cells. The cellular implications of free PAR were demonstrated by the release of apoptosis-inducing factor (AIF) following exposure of cells to free PAR, an effect that was not observed in the presence of digested PAR (Yu et al., 2006). Finally, the ADP-ribosyl hydrolase ARH3, which degrades PAR, regulates the release of AIF in cells, hypothetically through its ability to degrade free PAR (Mashimo et al., 2013). Notably, cellular PAR levels are an important clinical parameter to measure the effectiveness of PARP inhibitors and/or chemotherapeutic agents in clinical trials (as in NCI standard operating procedure #340505) as well as a predictive biomarker proposed for PARP inhibitor sensitivity (Gottipati et al., 2010, Oplustilova et al., 2012). An understanding of the conjugation state of cellular PAR is necessary for accurate interpretation of changing PARylation levels.
Current approaches do not account for another important parameter—the structural subclasses of PAR. These subclasses include length variants (Hottiger et al., 2010, Vyas et al., 2014) as well as branching variants (PARPs −1 and −2 make branched polymers while −5a makes linear polymers [Alvarez-Gonzalez and Jacobson, 1987, Amé et al., 1999, Rippmann et al., 2002]). These differences could functionally impact PAR’s role as a scaffold, where different lengths of the polymer have already been shown to recruit distinct populations of proteins (e.g., Fahrer et al., 2007)—a potential mechanism for temporal coordination of cellular processes (Leung, 2014, Realini and Althaus, 1992). The development of proteomic tools to determine polymer length and structure in cells could shed light on the unique roles played by the many forms of PAR. The recent development of a purification scheme for large amounts of PAR standards of defined length (Kistemaker et al., 2015, Lambrecht et al., 2015, Tan et al., 2012) could potentially pave the way for characterizing the length of the polymer on PARylated substrates. Ultimately, the goal is to use MS to simultaneously identify both the sites of ADP-ribosylation and the number of ADP-ribose groups that are attached to those modified sites. Such technical challenges bear remarkable similarity to the problem of the site-specific microheterogeneity observed in N-linked glycosylation, where structures of sugar polymer attached to the modified sites could be of different lengths and varied degrees of branching (An et al., 2009). Recent advances in search algorithms have been able to map simultaneously the glycosylation sites, the number of sugar moieties and the branch points of the sugar polymer attached at the modified site of single proteins (Chandler et al., 2013). Though an ADP-ribose moiety carries more negative charge and generally 2-fold more mass than sugar moieties, it is perhaps feasible to map both the modified sites and short oligomers (< 5 mers) attached on single PARylated proteins in the future.
Assessing the Physiological Relevance of ADP-Ribosylation Sites
Site Occupancy
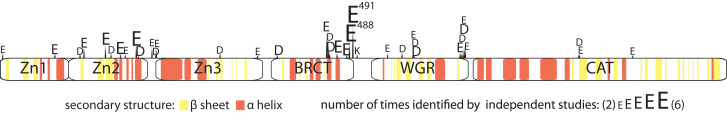
Complete characterization of a single ADP-ribosylation site will include accurate identification of four factors: (1) amino acid conjugation site, (2) enzymes responsible for addition and removal of the modification, (3) structural make-up of the modification (mono? poly? branched?), and (4) site occupancy/stoichiometry. While progress has been made in the first three endeavors as discussed above, it is the last aim that will most aid in the determination of functionally and physiologically relevant sites of ADP-ribosylation; functional (and therefore regulated) sites will likely exhibit a defined stoichiometric change in response to stimulus, while non-functional sites will show no change or changes that cannot be associated consistently with the biological stimulus applied. Quantifying a change in site occupancy, however, is much more challenging than quantifying a change in protein levels as the measurement may track the changing intensity of a single peptide as opposed to many peptides from a single protein (Wu et al., 2011). Additionally many of the modifications may exist at very low stoichiometries, making quantification extra sensitive to variability introduced during sample preparation, a challenge which has been mitigated by the use of internal, stable-isotope-labeled standards (Kettenbach et al., 2011, Olsen et al., 2010). Investigation of site occupancies (and the identification of robust, reproducible changes at determined sites) has the potential to test two hypotheses: (1) that some protein/peptide N-terminal, lysine, arginine, and cysteine modifications may be non-functional (and therefore represent biological noise), as they have the potential to be formed non-enzymatically by ADP-ribose groups that are released from PAR degradation by PARG/ARH3 and/or NADase activity of PARPs (Cervantes-Laurean et al., 1996, Desmarais et al., 1991, Kharadia and Graves, 1987, McDonald and Moss, 1994) and (2) that ADP-ribosylation of proteins is not always residue-specific, and may occasionally be mapped to a protein region as opposed to an amino acid. This latter hypothesis has been proposed to explain PARP-1 PARylation of BRCA1, wherein regions of BRCA1 were identified as PARylation acceptors as opposed to sites (Hu et al., 2014). This observation stands in contrast to PARP-1-mediated PARylation of the tumor suppressor p53, of which mutational analysis has yielded three p53 PARylation sites that account for nearly all of the PARylation present on the substrate (Kanai et al., 2007). Mutating all three residues to alanine resulted in cytoplasmic accumulation of p53 and further biochemical experiments indicated that this site-specific PARylation on p53 blocked its interaction with the nuclear export receptor Crm1 (Kanai et al., 2007). Both region-specific as well as site-specific mechanisms appear to be at play following PARP-1 auto-modification, an event that has been carefully characterized by a number of MS studies in recent years, resulting in a large number of site identifications (see Figure 6 , source data in Table S4) (Chapman et al., 2013, Daniels et al., 2014, Gagné et al., 2015, Sharifi et al., 2013, Tao et al., 2009, Zhang et al., 2013). While several defined modification sites such as E488 and E491 have been identified by all studies, there are also regions—such as the C terminus of the WGR domain stretching from E642–E650—that show regional, but not necessarily site-specific, overlap between studies. The ability to monitor whether sites or protein regions exhibit the regulatory patterns associated with cellular changes will provide essential data for determining their relative importance.
Figure 6.
PARP-1 Auto-modification Sites
Schematic of PARP-1 includes protein domains and secondary structure; α helices are shown in red, β sheets in yellow. Auto-modification sites identified by at least two independent studies are shown. Size of annotated residues is based on the number of studies that have identified the modification sites. E488 and E491 located at the C terminus of the BRCT domain are identified by all MS studies and are shown as the two major auto-modification sites. Source data available in Table S4.
Top-Down Proteomics
A necessary step forward will come from linking ADP-ribosylation into the established network of integrated PTMs (Woodsmith et al., 2013). Some work has already been done to link PARylation and ubiquitination (Kang et al., 2011, Zhang et al., 2011), as well as ADP-ribosylation and acetylation (Kowieski et al., 2008), elucidating important cellular mechanisms. Future findings will be brought on by the constant development of MS analysis software, a critical component in PTM identification, as well as the increasing availability of liquid chromatography methods and mass analyzers that are compatible with top-down proteomics. As top-down proteomics analyzes intact proteins (rather than the peptides which result from proteolysis), this method can often distinguish between protein proteoforms, i.e., gene products that are post-translationally processed in multiple ways, often with functional implications (Smith et al., 2013, Tran et al., 2011). This technique has proven powerful in the analysis of complex proteoforms such as histone variants, enabling the simultaneous characterization of the 14 H2A proteoforms (Boyne et al., 2006), and more recently, whole-protein kinetics of acetylation turnover on histones H3, H4, and H2A (Zheng et al., 2013). In the same way, top-down proteomics could facilitate the identification of groups of temporally or spatially correlated ADP-ribosylation sites, as well as other protein modifications. Integration of ADP-ribosylation into the growing network of PTMs has the potential to reveal novel regulatory roles for ADP-ribosylation and provide context for the physiological changes brought on by its modulation.
Conclusions
The power to monitor and interpret proteome-wide changes in ADP-ribosylation states promises to advance the fundamental understanding of ADP-ribosylation biology and facilitate further connections between cellular and patient responses to therapeutic PARP inhibition (Box 1 ). The depth of the proteome will clearly be advanced with the invention of better tools to enrich ADP-ribosylated proteomes—MAR/PAR-binding proteomes, MAR/PARylated proteomes and PARP-specific proteomes from cells in different cellular conditions, particularly native conditions that are understudied due to their low levels of ADP-ribosylation. However, such procurement of vast amounts of data must be coupled with the urgency to address basic questions such as whether the site of the PTM attachment matters, whether the PTM is always added enzymatically, and what the functional consequences are of adding single versus multiple ADP-ribose residues onto the attachment site. In light of the promise shown by these new proteomic tools for the study of ADP-ribosylation, it is high time to investigate this therapeutically important, yet enigmatic, protein modification at a detailed mechanistic level.
Box 1. How Proteomics Can Push the Field of ADP-Ribosylation Forward.
-
(1)
Identifying sites of ADP-ribosylation: knowing the amino acid attachment informs a researcher about the potential impact of the modification; for example, whether the PTM is switching the charge state of modified amino acids, and what class of enzymes is most likely responsible for its attachment and removal. This information allows further study through mutation of the amino acid attachment site. Knowing the stoichiometry of these modifications, and how they respond to stimuli, will provide clues as to which sites or protein regions are important regulatory switches.
-
(2)
Distinguishing between MAR and the many structures of PAR. A single ADP-ribosylation site may represent any number of PTMs, including MAR, linear PAR, and branched PAR, with polymers ranging between 2 and 200 subunits in vivo. Knowing the structure of the PARylation in question will allow researchers to speculate on the consequences resulting from the buildup of negative charges and the potential for this PTM to serve as a scaffold for recruiting PAR-binding partners.
-
(3)
Profiling the cellular response to PARP inhibitors. A proteome-wide view of the ADP-ribosylation state of a cell or tissue may reveal the molecular basis for chemotherapeutic responses, informing the design and development of PARP inhibitors for effective therapy.
Acknowledgments
We thank Drs. Phil Sharp, Paul Chang, Rhoel Dinglasan, Michael Cohen, Andrew Holland, Ted Dawson, and Michael Matunis for critical reading of the manuscript. The proteomics work in the A.K.L.L. and S.-E.O. Laboratories have been supported by a DOD Breast Cancer Research Program Idea Award #BC101881 (A.K.L.L.), an NIH grant R01-GM104135 (A.K.L.L.), the Safeway Research Foundation (A.K.L.L), the Patrick C. Walsh Prostate Cancer Research Fund (A.K.L.L.), the Allegheny Health Network–Johns Hopkins Cancer Research Fund (A.K.L.L.), a Journal of Cell Science Travelling Fellowship (C.M.D.), a Joy Cappel Young Investigator Award (C.M.D.), an NCI training grant 5T32CA009110 (C.M.D.), and an NIDA grant P30-DA028846 (S.-E.O.).
Footnotes
Supplemental Information includes two figures and four tables and can be found with this article online at http://dx.doi.org/10.1016/j.molcel.2015.06.012.
Supplemental Information
Sourced data used to generate the ADP-ribosylated interactomes shown in Figure 2. See Supplemental Text for criteria of data inclusion.
Sourced data used to generate the Euler diagrams showing PARP-1, PARP-2, PARP-10, and PARP-14 specific substrates in Figure 5.
Sourced data used to generate the PARP-1 auto-modification site map from Figure 6.
References
- Adamietz P., Hilz H. Poly(adenosine diphosphate ribose) is covalently linked to nuclear proteins by two types of bonds. Hoppe Seylers Z. Physiol. Chem. 1976;357:527–534. doi: 10.1515/bchm2.1976.357.1.527. [DOI] [PubMed] [Google Scholar]
- Ahel I., Ahel D., Matsusaka T., Clark A.J., Pines J., Boulton S.J., West S.C. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008;451:81–85. doi: 10.1038/nature06420. [DOI] [PubMed] [Google Scholar]
- Ahel D., Horejsí Z., Wiechens N., Polo S.E., Garcia-Wilson E., Ahel I., Flynn H., Skehel M., West S.C., Jackson S.P. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 2009;325:1240–1243. doi: 10.1126/science.1177321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmeyer M., Messner S., Hassa P.O., Fey M., Hottiger M.O. Molecular mechanism of poly(ADP-ribosyl)ation by PARP1 and identification of lysine residues as ADP-ribose acceptor sites. Nucleic Acids Res. 2009;37:3723–3738. doi: 10.1093/nar/gkp229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Gonzalez R., Jacobson M.K. Characterization of polymers of adenosine diphosphate ribose generated in vitro and in vivo. Biochemistry. 1987;26:3218–3224. doi: 10.1021/bi00385a042. [DOI] [PubMed] [Google Scholar]
- Amé J.C., Rolli V., Schreiber V., Niedergang C., Apiou F., Decker P., Muller S., Höger T., Ménissier-de Murcia J., de Murcia G. PARP-2, A novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J. Biol. Chem. 1999;274:17860–17868. doi: 10.1074/jbc.274.25.17860. [DOI] [PubMed] [Google Scholar]
- An H.J., Froehlich J.W., Lebrilla C.B. Determination of glycosylation sites and site-specific heterogeneity in glycoproteins. Curr. Opin. Chem. Biol. 2009;13:421–426. doi: 10.1016/j.cbpa.2009.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aredia F., Scovassi A.I. Poly(ADP-ribose): a signaling molecule in different paradigms of cell death. Biochem. Pharmacol. 2014;92:157–163. doi: 10.1016/j.bcp.2014.06.021. [DOI] [PubMed] [Google Scholar]
- Barkauskaite E., Jankevicius G., Ladurner A.G., Ahel I., Timinszky G. The recognition and removal of cellular poly(ADP-ribose) signals. FEBS J. 2013;280:3491–3507. doi: 10.1111/febs.12358. [DOI] [PubMed] [Google Scholar]
- Boamah E.K., Kotova E., Garabedian M., Jarnik M., Tulin A.V. Poly(ADP-Ribose) polymerase 1 (PARP-1) regulates ribosomal biogenesis in Drosophila nucleoli. PLoS Genet. 2012;8:e1002442. doi: 10.1371/journal.pgen.1002442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyne M.T., 2nd, Pesavento J.J., Mizzen C.A., Kelleher N.L. Precise characterization of human histones in the H2A gene family by top down mass spectrometry. J. Proteome Res. 2006;5:248–253. doi: 10.1021/pr050269n. [DOI] [PubMed] [Google Scholar]
- Cardenas-Corona M.E., Jacobson E.L., Jacobson M.K. Endogenous polymers of ADP-ribose are associated with the nuclear matrix. J. Biol. Chem. 1987;262:14863–14866. [PubMed] [Google Scholar]
- Carter-O’Connell I., Jin H., Morgan R.K., David L.L., Cohen M.S. Engineering the substrate specificity of ADP-ribosyltransferases for identifying direct protein targets. J. Am. Chem. Soc. 2014;136:5201–5204. doi: 10.1021/ja412897a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes-Laurean D., Loflin P.T., Minter D.E., Jacobson E.L., Jacobson M.K. Protein modification by ADP-ribose via acid-labile linkages. J. Biol. Chem. 1995;270:7929–7936. doi: 10.1074/jbc.270.14.7929. [DOI] [PubMed] [Google Scholar]
- Cervantes-Laurean D., Jacobson E.L., Jacobson M.K. Glycation and glycoxidation of histones by ADP-ribose. J. Biol. Chem. 1996;271:10461–10469. doi: 10.1074/jbc.271.18.10461. [DOI] [PubMed] [Google Scholar]
- Cervantes-Laurean D., Jacobson E.L., Jacobson M.K. Preparation of low molecular weight model conjugates for ADP-ribose linkages to protein. Methods Enzymol. 1997;280:275–287. doi: 10.1016/s0076-6879(97)80119-x. [DOI] [PubMed] [Google Scholar]
- Chandler K.B., Pompach P., Goldman R., Edwards N. Exploring site-specific N-glycosylation microheterogeneity of haptoglobin using glycopeptide CID tandem mass spectra and glycan database search. J. Proteome Res. 2013;12:3652–3666. doi: 10.1021/pr400196s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang P., Jacobson M.K., Mitchison T.J. Poly(ADP-ribose) is required for spindle assembly and structure. Nature. 2004;432:645–649. doi: 10.1038/nature03061. [DOI] [PubMed] [Google Scholar]
- Chapman J.D., Gagné J.P., Poirier G.G., Goodlett D.R. Mapping PARP-1 auto-ADP-ribosylation sites by liquid chromatography-tandem mass spectrometry. J. Proteome Res. 2013;12:1868–1880. doi: 10.1021/pr301219h. [DOI] [PubMed] [Google Scholar]
- Cox J., Mann M. Quantitative, high-resolution proteomics for data-driven systems biology. Annu. Rev. Biochem. 2011;80:273–299. doi: 10.1146/annurev-biochem-061308-093216. [DOI] [PubMed] [Google Scholar]
- Curtin N.J., Szabo C. Therapeutic applications of PARP inhibitors: anticancer therapy and beyond. Mol. Aspects Med. 2013;34:1217–1256. doi: 10.1016/j.mam.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dani N., Stilla A., Marchegiani A., Tamburro A., Till S., Ladurner A.G., Corda D., Di Girolamo M. Combining affinity purification by ADP-ribose-binding macro domains with mass spectrometry to define the mammalian ADP-ribosyl proteome. Proc. Natl. Acad. Sci. USA. 2009;106:4243–4248. doi: 10.1073/pnas.0900066106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels C.M., Ong S.E., Leung A.K. Phosphoproteomic approach to characterize protein mono and poly(ADP-ribosyl)ation sites from whole cell lysate. J. Proteome Res. 2014;3:3510–3522. doi: 10.1021/pr401032q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmarais Y., Ménard L., Lagueux J., Poirier G.G. Enzymological properties of poly(ADP-ribose)polymerase: characterization of automodification sites and NADase activity. Biochim. Biophys. Acta. 1991;1078:179–186. doi: 10.1016/0167-4838(91)99007-f. [DOI] [PubMed] [Google Scholar]
- Egloff M.P., Malet H., Putics A., Heinonen M., Dutartre H., Frangeul A., Gruez A., Campanacci V., Cambillau C., Ziebuhr J. Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains. J. Virol. 2006;80:8493–8502. doi: 10.1128/JVI.00713-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrer J., Kranaster R., Altmeyer M., Marx A., Bürkle A. Quantitative analysis of the binding affinity of poly(ADP-ribose) to specific binding proteins as a function of chain length. Nucleic Acids Res. 2007;35:e143. doi: 10.1093/nar/gkm944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feijs K.L., Kleine H., Braczynski A., Forst A.H., Herzog N., Verheugd P., Linzen U., Kremmer E., Lüscher B. ARTD10 substrate identification on protein microarrays: regulation of GSK3β by mono-ADP-ribosylation. Cell Commun. Signal. 2013;11:5. doi: 10.1186/1478-811X-11-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feijs K.L., Verheugd P., Lüscher B. Expanding functions of intracellular resident mono-ADP-ribosylation in cell physiology. FEBS J. 2013;280:3519–3529. doi: 10.1111/febs.12315. [DOI] [PubMed] [Google Scholar]
- Finch K.E., Knezevic C.E., Nottbohm A.C., Partlow K.C., Hergenrother P.J. Selective small molecule inhibition of poly(ADP-ribose) glycohydrolase (PARG) ACS Chem. Biol. 2012;7:563–570. doi: 10.1021/cb200506t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forst A.H., Karlberg T., Herzog N., Thorsell A.G., Gross A., Feijs K.L., Verheugd P., Kursula P., Nijmeijer B., Kremmer E. Recognition of mono-ADP-ribosylated ARTD10 substrates by ARTD8 macrodomains. Structure. 2013;21:462–475. doi: 10.1016/j.str.2012.12.019. [DOI] [PubMed] [Google Scholar]
- Gagné J.P., Isabelle M., Lo K.S., Bourassa S., Hendzel M.J., Dawson V.L., Dawson T.M., Poirier G.G. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res. 2008;36:6959–6976. doi: 10.1093/nar/gkn771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagné J.P., Pic E., Isabelle M., Krietsch J., Ethier C., Paquet E., Kelly I., Boutin M., Moon K.M., Foster L.J., Poirier G.G. Quantitative proteomics profiling of the poly(ADP-ribose)-related response to genotoxic stress. Nucleic Acids Res. 2012;40:7788–7805. doi: 10.1093/nar/gks486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagné J.P., Ethier C., Defoy D., Bourassa S., Langelier M.F., Riccio A.A., Pascal J.M., Moon K.M., Foster L.J., Ning Z. Quantitative site-specific ADP-ribosylation profiling of DNA-dependent PARPs. DNA Repair (Amst.) 2015;30:68–79. doi: 10.1016/j.dnarep.2015.02.004. [DOI] [PubMed] [Google Scholar]
- Garber K. PARP inhibitors bounce back. Nat. Rev. Drug Discov. 2013;12:725–727. doi: 10.1038/nrd4147. [DOI] [PubMed] [Google Scholar]
- Gibson B.A., Kraus W.L. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat. Rev. Mol. Cell Biol. 2012;13:411–424. doi: 10.1038/nrm3376. [DOI] [PubMed] [Google Scholar]
- Gottipati P., Vischioni B., Schultz N., Solomons J., Bryant H.E., Djureinovic T., Issaeva N., Sleeth K., Sharma R.A., Helleday T. Poly(ADP-ribose) polymerase is hyperactivated in homologous recombination-defective cells. Cancer Res. 2010;70:5389–5398. doi: 10.1158/0008-5472.CAN-09-4716. [DOI] [PubMed] [Google Scholar]
- Gottschalk A.J., Trivedi R.D., Conaway J.W., Conaway R.C. Activation of the SNF2 family ATPase ALC1 by poly(ADP-ribose) in a stable ALC1·PARP1·nucleosome intermediate. J. Biol. Chem. 2012;287:43527–43532. doi: 10.1074/jbc.M112.401141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guettler S., LaRose J., Petsalaki E., Gish G., Scotter A., Pawson T., Rottapel R., Sicheri F. Structural basis and sequence rules for substrate recognition by Tankyrase explain the basis for cherubism disease. Cell. 2011;147:1340–1354. doi: 10.1016/j.cell.2011.10.046. [DOI] [PubMed] [Google Scholar]
- Hassa P.O., Haenni S.S., Elser M., Hottiger M.O. Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol. Mol. Biol. Rev. 2006;70:789–829. doi: 10.1128/MMBR.00040-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F., Tsuda K., Takahashi M., Kuwasako K., Terada T., Shirouzu M., Watanabe S., Kigawa T., Kobayashi N., Güntert P. Structural insight into the interaction of ADP-ribose with the PARP WWE domains. FEBS Lett. 2012;586:3858–3864. doi: 10.1016/j.febslet.2012.09.009. [DOI] [PubMed] [Google Scholar]
- Hengel S.M., Goodlett D.R. A review of tandem mass spectrometry characterization of adenosine diphosphate-ribosylated peptides. Int. J. Mass Spectrom. 2012;312:114–121. doi: 10.1016/j.ijms.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengel S.M., Shaffer S.A., Nunn B.L., Goodlett D.R. Tandem mass spectrometry investigation of ADP-ribosylated kemptide. J. Am. Soc. Mass Spectrom. 2009;20:477–483. doi: 10.1016/j.jasms.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hottiger M.O., Hassa P.O., Lüscher B., Schüler H., Koch-Nolte F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem. Sci. 2010;35:208–219. doi: 10.1016/j.tibs.2009.12.003. [DOI] [PubMed] [Google Scholar]
- Houtkooper R.H., Pirinen E., Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat. Rev. Mol. Cell Biol. 2012;13:225–238. doi: 10.1038/nrm3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Petit S.A., Ficarro S.B., Toomire K.J., Xie A., Lim E., Cao S.A., Park E., Eck M.J., Scully R. PARP1-driven poly-ADP-ribosylation regulates BRCA1 function in homologous recombination-mediated DNA repair. Cancer Discov. 2014;4:1430–1447. doi: 10.1158/2159-8290.CD-13-0891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huttlin E.L., Jedrychowski M.P., Elias J.E., Goswami T., Rad R., Beausoleil S.A., Villén J., Haas W., Sowa M.E., Gygi S.P. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143:1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isabelle M., Gagné J.P., Gallouzi I.E., Poirier G.G. Quantitative proteomics and dynamic imaging reveal that G3BP-mediated stress granule assembly is poly(ADP-ribose)-dependent following exposure to MNNG-induced DNA alkylation. J. Cell Sci. 2012;125:4555–4566. doi: 10.1242/jcs.106963. [DOI] [PubMed] [Google Scholar]
- Jankevicius G., Hassler M., Golia B., Rybin V., Zacharias M., Timinszky G., Ladurner A.G. A family of macrodomain proteins reverses cellular mono-ADP-ribosylation. Nat. Struct. Mol. Biol. 2013;20:508–514. doi: 10.1038/nsmb.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmichel S., Rosenthal F., Altmeyer M., Lukas J., Hottiger M.O., Nielsen M.L. Proteome-wide identification of poly(ADP-Ribosyl)ation targets in different genotoxic stress responses. Mol. Cell. 2013;52:272–285. doi: 10.1016/j.molcel.2013.08.026. [DOI] [PubMed] [Google Scholar]
- Kanai M., Hanashiro K., Kim S.H., Hanai S., Boulares A.H., Miwa M., Fukasawa K. Inhibition of Crm1-p53 interaction and nuclear export of p53 by poly(ADP-ribosyl)ation. Nat. Cell Biol. 2007;9:1175–1183. doi: 10.1038/ncb1638. [DOI] [PubMed] [Google Scholar]
- Kang H.C., Lee Y.I., Shin J.H., Andrabi S.A., Chi Z., Gagné J.P., Lee Y., Ko H.S., Lee B.D., Poirier G.G. Iduna is a poly(ADP-ribose) (PAR)-dependent E3 ubiquitin ligase that regulates DNA damage. Proc. Natl. Acad. Sci. USA. 2011;108:14103–14108. doi: 10.1073/pnas.1108799108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karras G.I., Kustatscher G., Buhecha H.R., Allen M.D., Pugieux C., Sait F., Bycroft M., Ladurner A.G. The macro domain is an ADP-ribose binding module. EMBO J. 2005;24:1911–1920. doi: 10.1038/sj.emboj.7600664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamitsu H., Hoshino H., Okada H., Miwa M., Momoi H., Sugimura T. Monoclonal antibodies to poly(adenosine diphosphate ribose) recognize different structures. Biochemistry. 1984;23:3771–3777. doi: 10.1021/bi00311a032. [DOI] [PubMed] [Google Scholar]
- Kestler H.A., Müller A., Gress T.M., Buchholz M. Generalized Venn diagrams: a new method of visualizing complex genetic set relations. Bioinformatics. 2005;21:1592–1595. doi: 10.1093/bioinformatics/bti169. [DOI] [PubMed] [Google Scholar]
- Kettenbach A.N., Rush J., Gerber S.A. Absolute quantification of protein and post-translational modification abundance with stable isotope-labeled synthetic peptides. Nat. Protoc. 2011;6:175–186. doi: 10.1038/nprot.2010.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharadia S.V., Graves D.J. Relationship of phosphorylation and ADP-ribosylation using a synthetic peptide as a model substrate. J. Biol. Chem. 1987;262:17379–17383. [PubMed] [Google Scholar]
- Kickhoefer V.A., Siva A.C., Kedersha N.L., Inman E.M., Ruland C., Streuli M., Rome L.H. The 193-kD vault protein, VPARP, is a novel poly(ADP-ribose) polymerase. J. Cell Biol. 1999;146:917–928. doi: 10.1083/jcb.146.5.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistemaker H.A., van Noort G.J., Overkleeft H.S., van der Marel G.A., Filippov D.V. Stereoselective ribosylation of amino acids. Org. Lett. 2013;15:2306–2309. doi: 10.1021/ol400929c. [DOI] [PubMed] [Google Scholar]
- Kistemaker H.A., Lameijer L.N., Meeuwenoord N.J., Overkleeft H.S., van der Marel G.A., Filippov D.V. Synthesis of well-defined adenosine diphosphate ribose oligomers. Angew. Chem. Int. Ed. Engl. 2015;54:4915–4918. doi: 10.1002/anie.201412283. [DOI] [PubMed] [Google Scholar]
- Konczalik P., Moss J. Identification of critical, conserved vicinal aspartate residues in mammalian and bacterial ADP-ribosylarginine hydrolases. J. Biol. Chem. 1999;274:16736–16740. doi: 10.1074/jbc.274.24.16736. [DOI] [PubMed] [Google Scholar]
- Kowieski T.M., Lee S., Denu J.M. Acetylation-dependent ADP-ribosylation by Trypanosoma brucei Sir2. J. Biol. Chem. 2008;283:5317–5326. doi: 10.1074/jbc.M707613200. [DOI] [PubMed] [Google Scholar]
- Laing S., Koch-Nolte F., Haag F., Buck F. Strategies for the identification of arginine ADP-ribosylation sites. J. Proteomics. 2011;75:169–176. doi: 10.1016/j.jprot.2011.07.003. [DOI] [PubMed] [Google Scholar]
- Lambrecht M.J., Brichacek M., Barkauskaite E., Ariza A., Ahel I., Hergenrother P.J. Synthesis of dimeric ADP-ribose and its structure with human poly(ADP-ribose) glycohydrolase. J. Am. Chem. Soc. 2015;137:3558–3564. doi: 10.1021/ja512528p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung A.K. Poly(ADP-ribose): an organizer of cellular architecture. J. Cell Biol. 2014;205:613–619. doi: 10.1083/jcb.201402114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung A.K., Vyas S., Rood J.E., Bhutkar A., Sharp P.A., Chang P. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol. Cell. 2011;42:489–499. doi: 10.1016/j.molcel.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord C.J., Tutt A.N., Ashworth A. Synthetic lethality and cancer therapy: lessons learned from the development of PARP inhibitors. Annu. Rev. Med. 2015;66:455–470. doi: 10.1146/annurev-med-050913-022545. [DOI] [PubMed] [Google Scholar]
- Manning D.R., Fraser B.A., Kahn R.A., Gilman A.G. ADP-ribosylation of transducin by islet-activation protein. Identification of asparagine as the site of ADP-ribosylation. J. Biol. Chem. 1984;259:749–756. [PubMed] [Google Scholar]
- Mao Z., Hine C., Tian X., Van Meter M., Au M., Vaidya A., Seluanov A., Gorbunova V. SIRT6 promotes DNA repair under stress by activating PARP1. Science. 2011;332:1443–1446. doi: 10.1126/science.1202723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martello R., Mangerich A., Sass S., Dedon P.C., Bürkle A. Quantification of cellular poly(ADP-ribosyl)ation by stable isotope dilution mass spectrometry reveals tissue- and drug-dependent stress response dynamics. ACS Chem. Biol. 2013;8:1567–1575. doi: 10.1021/cb400170b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashimo M., Kato J., Moss J. ADP-ribosyl-acceptor hydrolase 3 regulates poly (ADP-ribose) degradation and cell death during oxidative stress. Proc. Natl. Acad. Sci. USA. 2013;110:18964–18969. doi: 10.1073/pnas.1312783110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matic I., Ahel I., Hay R.T. Reanalysis of phosphoproteomics data uncovers ADP-ribosylation sites. Nat. Methods. 2012;9:771–772. doi: 10.1038/nmeth.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara H., Hasegawa S., Fujimura S., Shima T., Sugimura T. Studies on poly (adenosine diphosphate ribose). V. Mechanism of hydrolysis of poly (adenosine diphosphate ribose) by snake venom phosphodiesterase. J. Biol. Chem. 1970;245:3606–3611. [PubMed] [Google Scholar]
- McDonald L.J., Moss J. Enzymatic and nonenzymatic ADP-ribosylation of cysteine. Mol. Cell. Biochem. 1994;138:221–226. doi: 10.1007/BF00928465. [DOI] [PubMed] [Google Scholar]
- Messner S., Hottiger M.O. Histone ADP-ribosylation in DNA repair, replication and transcription. Trends Cell Biol. 2011;21:534–542. doi: 10.1016/j.tcb.2011.06.001. [DOI] [PubMed] [Google Scholar]
- Messner S., Altmeyer M., Zhao H., Pozivil A., Roschitzki B., Gehrig P., Rutishauser D., Huang D., Caflisch A., Hottiger M.O. PARP1 ADP-ribosylates lysine residues of the core histone tails. Nucleic Acids Res. 2010;38:6350–6362. doi: 10.1093/nar/gkq463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min W., Cortes U., Herceg Z., Tong W.M., Wang Z.Q. Deletion of the nuclear isoform of poly(ADP-ribose) glycohydrolase (PARG) reveals its function in DNA repair, genomic stability and tumorigenesis. Carcinogenesis. 2010;31:2058–2065. doi: 10.1093/carcin/bgq205. [DOI] [PubMed] [Google Scholar]
- Mortusewicz O., Amé J.C., Schreiber V., Leonhardt H. Feedback-regulated poly(ADP-ribosyl)ation by PARP-1 is required for rapid response to DNA damage in living cells. Nucleic Acids Res. 2007;35:7665–7675. doi: 10.1093/nar/gkm933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss J., Yost D.A., Stanley S.J. Amino acid-specific ADP-ribosylation. J. Biol. Chem. 1983;258:6466–6470. [PubMed] [Google Scholar]
- Moss J., Stanley S.J., Nightingale M.S., Murtagh J.J., Jr., Monaco L., Mishima K., Chen H.C., Williamson K.C., Tsai S.C. Molecular and immunological characterization of ADP-ribosylarginine hydrolases. J. Biol. Chem. 1992;267:10481–10488. [PubMed] [Google Scholar]
- Moyle P.M., Muir T.W. Method for the synthesis of mono-ADP-ribose conjugated peptides. J. Am. Chem. Soc. 2010;132:15878–15880. doi: 10.1021/ja1064312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller-Dieckmann C., Kernstock S., Lisurek M., von Kries J.P., Haag F., Weiss M.S., Koch-Nolte F. The structure of human ADP-ribosylhydrolase 3 (ARH3) provides insights into the reversibility of protein ADP-ribosylation. Proc. Natl. Acad. Sci. USA. 2006;103:15026–15031. doi: 10.1073/pnas.0606762103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murawska M., Hassler M., Renkawitz-Pohl R., Ladurner A., Brehm A. Stress-induced PARP activation mediates recruitment of Drosophila Mi-2 to promote heat shock gene expression. PLoS Genet. 2011;7:e1002206. doi: 10.1371/journal.pgen.1002206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuvonen M., Ahola T. Differential activities of cellular and viral macro domain proteins in binding of ADP-ribose metabolites. J. Mol. Biol. 2009;385:212–225. doi: 10.1016/j.jmb.2008.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberoi J., Richards M.W., Crumpler S., Brown N., Blagg J., Bayliss R. Structural basis of poly(ADP-ribose) recognition by the multizinc binding domain of checkpoint with forkhead-associated and RING Domains (CHFR) J. Biol. Chem. 2010;285:39348–39358. doi: 10.1074/jbc.M110.159855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oetjen J., Rexroth S., Reinhold-Hurek B. Mass spectrometric characterization of the covalent modification of the nitrogenase Fe-protein in Azoarcus sp. BH72. FEBS J. 2009;276:3618–3627. doi: 10.1111/j.1742-4658.2009.07081.x. [DOI] [PubMed] [Google Scholar]
- Ohno T., Tsuchiya M., Osago H., Hara N., Jidoi J., Shimoyama M. Detection of arginine-ADP-ribosylated protein using recombinant ADP-ribosylarginine hydrolase. Anal. Biochem. 1995;231:115–122. doi: 10.1006/abio.1995.1510. [DOI] [PubMed] [Google Scholar]
- Oka S., Kato J., Moss J. Identification and characterization of a mammalian 39-kDa poly(ADP-ribose) glycohydrolase. J. Biol. Chem. 2006;281:705–713. doi: 10.1074/jbc.M510290200. [DOI] [PubMed] [Google Scholar]
- Okano S., Lan L., Caldecott K.W., Mori T., Yasui A. Spatial and temporal cellular responses to single-strand breaks in human cells. Mol. Cell. Biol. 2003;23:3974–3981. doi: 10.1128/MCB.23.11.3974-3981.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen J.V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- Olsen J.V., Vermeulen M., Santamaria A., Kumar C., Miller M.L., Jensen L.J., Gnad F., Cox J., Jensen T.S., Nigg E.A. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 2010;3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- Oplustilova L., Wolanin K., Mistrik M., Korinkova G., Simkova D., Bouchal J., Lenobel R., Bartkova J., Lau A., O’Connor M.J. Evaluation of candidate biomarkers to predict cancer cell sensitivity or resistance to PARP-1 inhibitor treatment. Cell Cycle. 2012;11:3837–3850. doi: 10.4161/cc.22026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheimer N.J., Bodley J.W. Diphtheria toxin. Site and configuration of ADP-ribosylation of diphthamide in elongation factor 2. J. Biol. Chem. 1981;256:8579–8581. [PubMed] [Google Scholar]
- Palazzo L., Thomas B., Jemth A.S., Colby T., Leidecker O., Feijs K.L., Zaja R., Loseva O., Puigvert J.C., Matic I. Processing of protein ADP-ribosylation by Nudix hydrolases. Biochem. J. 2015;468:293–301. doi: 10.1042/BJ20141554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panzeter P.L., Zweifel B., Althaus F.R. Synthesis of poly(ADP-ribose)-agarose beads: an affinity resin for studying (ADP-ribose)n-protein interactions. Anal. Biochem. 1992;207:157–162. doi: 10.1016/0003-2697(92)90517-b. [DOI] [PubMed] [Google Scholar]
- Perraud A.L., Fleig A., Dunn C.A., Bagley L.A., Launay P., Schmitz C., Stokes A.J., Zhu Q., Bessman M.J., Penner R. ADP-ribose gating of the calcium-permeable LTRPC2 channel revealed by Nudix motif homology. Nature. 2001;411:595–599. doi: 10.1038/35079100. [DOI] [PubMed] [Google Scholar]
- R Development Core Team . R Foundation for Statistical Computing; Vienna: 2011. R: A language and environment for statistical computing. [Google Scholar]
- Realini C.A., Althaus F.R. Histone shuttling by poly(ADP-ribosylation) J. Biol. Chem. 1992;267:18858–18865. [PubMed] [Google Scholar]
- Rippmann J.F., Damm K., Schnapp A. Functional characterization of the poly(ADP-ribose) polymerase activity of tankyrase 1, a potential regulator of telomere length. J. Mol. Biol. 2002;323:217–224. doi: 10.1016/s0022-2836(02)00946-4. [DOI] [PubMed] [Google Scholar]
- Rosenthal F., Messner S., Roschitzki B., Gehrig P., Nanni P., Hottiger M.O. Identification of distinct amino acids as ADP-ribose acceptor sites by mass spectrometry. Methods Mol. Biol. 2011;780:57–66. doi: 10.1007/978-1-61779-270-0_4. [DOI] [PubMed] [Google Scholar]
- Rosenthal F., Feijs K.L., Frugier E., Bonalli M., Forst A.H., Imhof R., Winkler H.C., Fischer D., Caflisch A., Hassa P.O. Macrodomain-containing proteins are new mono-ADP-ribosylhydrolases. Nat. Struct. Mol. Biol. 2013;20:502–507. doi: 10.1038/nsmb.2521. [DOI] [PubMed] [Google Scholar]
- Sharifi R., Morra R., Appel C.D., Tallis M., Chioza B., Jankevicius G., Simpson M.A., Matic I., Ozkan E., Golia B. Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease. EMBO J. 2013;32:1225–1237. doi: 10.1038/emboj.2013.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slade D., Dunstan M.S., Barkauskaite E., Weston R., Lafite P., Dixon N., Ahel M., Leys D., Ahel I. The structure and catalytic mechanism of a poly(ADP-ribose) glycohydrolase. Nature. 2011;477:616–620. doi: 10.1038/nature10404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J.A., Stocken L.A. Chemical and metabolic properties of adenosine diphosphate ribose derivatives of nuclear proteins. Biochem. J. 1975;147:523–529. doi: 10.1042/bj1470523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith L.M., Kelleher N.L., Consortium for Top Down Proteomics Proteoform: a single term describing protein complexity. Nat. Methods. 2013;10:186–187. doi: 10.1038/nmeth.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen J.D., Brody J.R., Armen R.S., Pascal J.M. Structural implications for selective targeting of parps. Front Oncol. 2013;3:301. doi: 10.3389/fonc.2013.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F., Bošnjak M., Škunca N., Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szewczuk L.M., Tarrant M.K., Cole P.A. Protein phosphorylation by semisynthesis: from paper to practice. Methods Enzymol. 2009;462:1–24. doi: 10.1016/S0076-6879(09)62001-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan E.S., Krukenberg K.A., Mitchison T.J. Large-scale preparation and characterization of poly(ADP-ribose) and defined length polymers. Anal. Biochem. 2012;428:126–136. doi: 10.1016/j.ab.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Z., Gao P., Liu H.W. Identification of the ADP-ribosylation sites in the PARP-1 automodification domain: analysis and implications. J. Am. Chem. Soc. 2009;131:14258–14260. doi: 10.1021/ja906135d. [DOI] [PubMed] [Google Scholar]
- Timinszky G., Till S., Hassa P.O., Hothorn M., Kustatscher G., Nijmeijer B., Colombelli J., Altmeyer M., Stelzer E.H., Scheffzek K. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat. Struct. Mol. Biol. 2009;16:923–929. doi: 10.1038/nsmb.1664. [DOI] [PubMed] [Google Scholar]
- Tran J.C., Zamdborg L., Ahlf D.R., Lee J.E., Catherman A.D., Durbin K.R., Tipton J.D., Vellaichamy A., Kellie J.F., Li M. Mapping intact protein isoforms in discovery mode using top-down proteomics. Nature. 2011;480:254–258. doi: 10.1038/nature10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troiani S., Lupi R., Perego R., Depaolini S.R., Thieffine S., Bosotti R., Rusconi L. Identification of candidate substrates for poly(ADP-ribose) polymerase-2 (PARP2) in the absence of DNA damage using high-density protein microarrays. FEBS J. 2011;278:3676–3687. doi: 10.1111/j.1742-4658.2011.08286.x. [DOI] [PubMed] [Google Scholar]
- van der Heden van Noort G.J., van der Horst M.G., Overkleeft H.S., van der Marel G.A., Filippov D.V. Synthesis of mono-ADP-ribosylated oligopeptides using ribosylated amino acid building blocks. J. Am. Chem. Soc. 2010;132:5236–5240. doi: 10.1021/ja910940q. [DOI] [PubMed] [Google Scholar]
- Vivelo C.A., Leung A.K. Proteomics approaches to identify mono-(ADP-ribosyl)ated and poly(ADP-ribosyl)ated proteins. Proteomics. 2015;15:203–217. doi: 10.1002/pmic.201400217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas S., Chesarone-Cataldo M., Todorova T., Huang Y.H., Chang P. A systematic analysis of the PARP protein family identifies new functions critical for cell physiology. Nat. Commun. 2013;4:2240. doi: 10.1038/ncomms3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas S., Matic I., Uchima L., Rood J., Zaja R., Hay R.T., Ahel I., Chang P. Family-wide analysis of poly(ADP-ribose) polymerase activity. Nat. Commun. 2014;5:4426. doi: 10.1038/ncomms5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlberg E., Karlberg T., Kouznetsova E., Markova N., Macchiarulo A., Thorsell A.G., Pol E., Frostell Å., Ekblad T., Öncü D. Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors. Nat. Biotechnol. 2012;30:283–288. doi: 10.1038/nbt.2121. [DOI] [PubMed] [Google Scholar]
- Wang Z., Michaud G.A., Cheng Z., Zhang Y., Hinds T.R., Fan E., Cong F., Xu W. Recognition of the iso-ADP-ribose moiety in poly(ADP-ribose) by WWE domains suggests a general mechanism for poly(ADP-ribosyl)ation-dependent ubiquitination. Genes Dev. 2012;26:235–240. doi: 10.1101/gad.182618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wielckens K., Schmidt A., George E., Bredehorst R., Hilz H. DNA fragmentation and NAD depletion. Their relation to the turnover of endogenous mono(ADP-ribosyl) and poly(ADP-ribosyl) proteins. J. Biol. Chem. 1982;257:12872–12877. [PubMed] [Google Scholar]
- Woodsmith J., Kamburov A., Stelzl U. Dual coordination of post translational modifications in human protein networks. PLoS Comput. Biol. 2013;9:e1002933. doi: 10.1371/journal.pcbi.1002933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R., Haas W., Dephoure N., Huttlin E.L., Zhai B., Sowa M.E., Gygi S.P. A large-scale method to measure absolute protein phosphorylation stoichiometries. Nat. Methods. 2011;8:677–683. doi: 10.1038/nmeth.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S.W., Andrabi S.A., Wang H., Kim N.S., Poirier G.G., Dawson T.M., Dawson V.L. Apoptosis-inducing factor mediates poly(ADP-ribose) (PAR) polymer-induced cell death. Proc. Natl. Acad. Sci. USA. 2006;103:18314–18319. doi: 10.1073/pnas.0606528103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu S., Mickanin C., Feng Y., Charlat O., Michaud G.A., Schirle M., Shi X., Hild M., Bauer A. RNF146 is a poly(ADP-ribose)-directed E3 ligase that regulates axin degradation and Wnt signalling. Nat. Cell Biol. 2011;13:623–629. doi: 10.1038/ncb2222. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Wang J., Ding M., Yu Y. Site-specific characterization of the Asp- and Glu-ADP-ribosylated proteome. Nat. Methods. 2013;10:981–984. doi: 10.1038/nmeth.2603. [DOI] [PubMed] [Google Scholar]
- Zheng Y., Thomas P.M., Kelleher N.L. Measurement of acetylation turnover at distinct lysines in human histones identifies long-lived acetylation sites. Nat. Commun. 2013;4:2203. doi: 10.1038/ncomms3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sourced data used to generate the ADP-ribosylated interactomes shown in Figure 2. See Supplemental Text for criteria of data inclusion.
Sourced data used to generate the Euler diagrams showing PARP-1, PARP-2, PARP-10, and PARP-14 specific substrates in Figure 5.
Sourced data used to generate the PARP-1 auto-modification site map from Figure 6.