Abstract
Whereas several mitochondrial/chloroplast pentatricopeptide repeat (PPR) proteins have been reported to regulate plant responses to abiotic stresses, no nucleus-localized PPR protein has been found to play role in these processes. In the present experiment, we provide evidence that a cytosol-nucleus dual-localized PPR protein SOAR1, functioning to negatively regulate abscisic acid (ABA) signaling in seed germination and postgermination growth, is a crucial, positive regulator of plant response to abiotic stresses. Downregulation of SOAR1 expression reduces, but upregulation of SOAR1 expression enhances, ABA sensitivity in ABA-induced promotion of stomatal closure and inhibition of stomatal opening, and plant tolerance to multiple, major abiotic stresses including drought, high salinity and low temperature. Interestingly and importantly, the SOAR1-overexpression lines display strong abilities to tolerate drought, salt and cold stresses, with surprisingly high resistance to salt stress in germination and postgermination growth of seeds that are able to potentially germinate in seawater, while no negative effect on plant growth and development was observed. So, the SOAR1 gene is likely useful for improvement of crops by transgenic manipulation to enhance crop productivity in stressful conditions. Further experimental data suggest that SOAR1 likely regulates plant stress responses at least partly by integrating ABA-dependent and independent signaling pathways, which is different from the ABI2/ABI1 type 2C protein phosphatase-mediated ABA signaling. These findings help to understand highly complicated stress and ABA signalling network.
Electronic supplementary material
The online version of this article (doi:10.1007/s11103-015-0327-9) contains supplementary material, which is available to authorized users.
Keywords: Abscisic acid signaling, Arabidopsis thaliana, Pentatricopeptide repeat (PPR) protein, SOAR1, Salinity stress, Drought stress, Cold stress
Introduction
Terrestrial plant may suffer from various abiotic stresses during the whole life cycle, among which, salinity, drought and low temperature are major factors that restrict productivity. Plants have developed a series of resistance mechanisms to adverse environmental factors (Xiong and Zhu 2001; Zhu 2002, 2003). The phytohormone abscisic acid (ABA) plays central roles in modulating plant adaptation to various adverse conditions through regulating a set of stress response genes, which form a gene regulatory network to allow plants to cope with environmental stresses (Xiong and Zhu 2001; Zhu 2002; Finkelstein et al. 2002; Jakoby et al. 2002; Shinozaki et al. 2003; Adie et al. 2007; Cutler et al. 2010; Golldack et al. 2011; Fujita et al. 2011; Qin et al. 2011).
The pentatricopeptide repeat (PPR) superfamily proteins are encoded by one of the largest gene families in plants, which include about 450 members in Arabidopsis thaliana and more than 600 members in rice (Oryza sativa) (Small and Peeters 2000; Lurin et al. 2004; Rivals et al. 2006; Schmitz-Linneweber and Small 2008). PPR proteins are mostly targeted to mitochondria or chloroplasts, and involved in many aspects of RNA processing in these organelles, such as RNA splicing, editing, 5′/3′ end processing, stability, cleavage, and translation (Meierhoff et al. 2003; Williams and Barkan 2003; Lurin et al. 2004). The mitochondrial/chloroplast PPR proteins play diverse and important roles in plant developmental processes and responses to environmental stresses. An Arabidopsis chloroplast PPR protein SVR7 (Lv et al. 2014) was reported to be involved in photosynthesis and oxidative stress tolerance. Six Arabidopsis mitochondrial PPR proteins, PPR40 (Zsigmond et al. 2008), ABO5 (Liu et al. 2010), AHG11 (Murayama et al. 2012), SLG1 (Yuan and Liu 2012), PGN (Laluk et al. 2011), and SLO2 (Zhu et al. 2014), were reported to regulate ABA signaling and salt or drought stress responses. The pgn mutant and PGN-overexpression lines (Laluk et al. 2011), the ppr40-1 (Zsigmond et al. 2008), ahg11 (Murayama et al. 2012), slg1 (Yuan and Liu 2012), and slo2 (Zhu et al. 2014) mutants showed hypersensitivity to salt or osmotic stress during germination and/or postgermination growth, while adult plants of the slo2 or slg1 mutants showed increased drought and/or salt tolerance (Yuan and Liu 2012; Zhu et al. 2014). These data suggest a highly complicated mechanism by which these mitochondrial/chloroplast PPRs regulate plant response to abiotic stresses, though it was proposed that they may regulate reactive oxygen species (ROS) homeostasis to be involved in stress responses or ABA signaling.
Whereas numerous PPR proteins are found to be localized to the mitochondrial or chloroplast intracellular space, few PPR proteins have been found to reside in other cellular compartments such as cytosol or nucleus. To our knowledge, so far, only two PPR proteins were found in the nucleus, which regulate embryogenesis (Ding et al. 2006; Hammani et al. 2011). Up to date, however, no nucleus- or cytosol-localized PPR protein has been found to regulate plant responses to abiotic stresses. Most recently, we identified a cytosol-nucleus dual-localized PPR protein, SOAR1 (for suppressor of the ABAR-overexpressor 1), which functions negatively in ABA signaling in seed germination and seedling growth downstream of the Mg-chelatase H subunit/putative ABA receptor (CHLH/ABAR) and upstream of an important ABA-responsive bZIP transcription factor ABI5 (Jiang et al. 2014; Mei et al. 2014; Wang and Zhang 2014). However, it remains still unknown whether and how SOAR1 regulates plant response to abiotic stresses. In the present experiment, we show that SOAR1 is a crucial, positive regulator of plant response to multiple, major abiotic stresses including drought, high salinity and low temperature. The SOAR1-overexpression lines display strong abilities to tolerate drought, salt and cold stresses, which is likely useful for improvement of crops. Further experimental data suggest that SOAR1 likely regulates plant stress responses at least partly by integrating ABA-dependent and independent signaling pathways. These findings help to understand highly complicated stress and ABA signalling network.
Materials and methods
Plant materials and growth conditions
The T-DNA insertion mutants soar1-2 (stock no. FLAG_546D07) and soar1-3 (stock no. FLAG_500B04) in SOAR1 gene (At5g11310, see Mei et al. 2014) were obtained from Versailles Genetics and Plant Breeding Laboratory, Arabidopsis thaliana Resource Centre (INRA; http://dbsgap.versailles.inra.fr/portail/) with the Col ecotype as background. The abi1-3 abi2-2 double mutant seeds are a generous gift from Dr. Y. Guo (China Agricultural University, Beijing, China), where abi1-3 (stock no. SALK_076309) and abi2-2 (stock no. SALK_015166) mutants, two T-DNA insertion knockout mutants in the ABI1 (At4g26080) and ABI2 (At5g57050) genes, respectively, were obtained from the Arabidopsis Biological Resource Centre (ABRC) with the Col ecotype as background. These mutants were identified previously as we described (Mei et al. 2014).
The Arabidopsis ecotype Col-0 was used to generate transgenic plants. The SOAR1-overexpression lines and ABI2-overexpression line ABI2-OE were generated as described previously (Sun et al. 2011; Mei et al. 2014). Plants were grown on Murashige and Skoog (MS) medium (Murashige and Skoog 1962; PhytoTechnology, Shawnee Mission, KS, USA) containing 3 % (w/v) sucrose and 0.8 % (w/v) agar or in compost soil under a 16 h photoperiod in the growth chamber or phytotron at about 20 °C.
Seed germination and postgermination growth
Seeds were surface sterilized in 4 % (v/v) sodium hypochlorite, and rinsed five times with sterile water. The seeds were sown on MS basal medium as mentioned above, with addition of NaCl or D-mannitol (Amresco, Solon, OH, USA) at indicated concentrations for assaying salt and osmotic stress responses. The seeds were stratified at 4 °C for 3 day, and transferred to 20 °C under long-day cycle (16 h/8 h light/dark) for phenotypic analysis of germination and post-germination growth.
Stomatal movement
Mature rosette leaves from about 4-week-old plants were used for assays of stomatal movement. The stomatal apertures on the abaxial surface of leaves were measured. To observe ABA-induced stomatal closure, leaves were immersed in a buffer consisting of 50 mM KCl and 10 mM MES-KOH with pH 6.15, and then exposed to cold light source (Chongqing Optec Instrument Co., Lt, Chongqing, China) for 2.5 h. Subsequently, the leaves were transferred into the fresh buffer as described above but supplemented with 0 (control) or 20 μM (±) ABA (Sigma, Saint Louis, MO, USA) for an incubation of 2.5 h before stomatal apertures were measured. To study ABA-inhibited stomatal opening, leaves were kept in the dark for 2.5 h, and then exposed to light and incubated for 2.5 h in the buffer as mentioned above and supplemented with 0 (control) or 20 μM (±) ABA before stomatal apertures were measured. The assays were performed with three independent repetitions (n ≥ 80 apertures per experiment).
Water loss from detached leaves
Mature rosette leaves of similar size were sampled from about 4-week-old plants and placed in culture dishes under light at room temperature (about 24 °C) with the relative humidity of air about 40 %. Water loss was evaluated by weighing leaves at the indicated time points. The assays were performed with three independent repetitions.
Drought treatment
Ten-d-old seedlings were transplanted to 7-cm pots filled with compost soil with the same water content. Plants were grown at 22 °C under long-day cycle (light/dark: 16 h/8 h) for 1 week, and drought was imposed on the plants by withholding water for about 3 weeks until the lethal effect was observed on the mutant plants, while the control plants were well watered. The growth status and survival rates of different genotypes were recorded 3 days after the plants were re-watered. The entire experiment was replicated three times with similar results.
Salt treatment
To investigate germination and postgermination growth of different genotypes under salt stress, seeds were sown directly on MS medium containing 130, 150, 175, or 200 mM NaCl and grown 18 days after stratification, and then root length and fresh weight of seedlings were recorded. To test the extremity of the NaCl concentrations which the SOAR1-overexpression lines tolerate, seeds were sown in the MS medium containing 250, 300, 350, 422, and 513 mM NaCl, and germination and postgermination growth were investigated 2 weeks after stratification. To assay of salt response of the whole mature plants, NaCl treatment was imposed with irrigation as described previously (Shi et al. 2003) with some modifications. Ten-day-old seedlings were transferred to soil and continued to grow 1 week before the NaCl treatment. NaCl solution was imposed with three increasing concentrations (100, 150, and 200). With each NaCl concentration the plants were irrigated one time at an interval of 4 days, and for a total period of 12 days plants were irrigated three times with corresponding concentrations of NaCl, and then they continued to be irrigated with 200 mM NaCl solution tow times at an interval of 4 days, and were investigated for salt tolerance 7 days after the last NaCl irrigation. The control plants were irrigated with water.
Freezing treatment
The freezing treatment was performed as described previously (Shi et al. 2012) with some modifications. Two-week-old seedlings grown in petri dishes at 20 °C were used for the cold-acclimation (CA) treatment and non-acclimated (NA) treatment. Seedlings of the NA group were subjected directly to a freezing treatment, while seedlings of the CA group were acclimated at 4 °C for 7 days before the freezing treatment. Seedlings were placed in a freezing chamber (RuMED4001, Stuttgart, Germany) set at 0 °C programmed to cool at 1 °C per hour until the minimum temperature. Petri dishes of plants were removed at the indicated time or temperature points. After the freezing treatment, the plants were incubated at 4 °C in the dark for 12 h and then transferred to light at 20 °C in the growth chamber. The survival rates of the seedlings were scored visually after recovering for about 2 days. In addition, the electrolyte leakage and proline content of the plants treated by freezing were measured according to the procedures described previously (Bates et al. 1973; Lee et al. 2001; Yang et al. 2010).
Quantitative real-time PCR
Quantitative real-time PCR for the osmosis-, salt- and cold-responsive genes (see Supplementary Table S1 online for the gene-specific primers) was performed essentially as described previously (Shang et al. 2010; Mei et al. 2014), with the Bio-Rad Real-Time System CFX96TM C1000 thermal cycler (Bio-Rad, Hercules, CA, USA) and following the manufacture’s instructions. Total RNA was isolated with plant total RNA extraction kit (BioTeke Corporation, Beijing, China) supplemented with DNA digestion (New England Biolabs RNase-Free DNase I, Beijing, China), and then the RNA sample was reverse-transcribed with the Transcriptor First Strand cDNA Synthesis Kit (Roche, Mannheim, Germany) according to the manufacturer’s instructions. Amplification of ACTIN2/8 genes was used as an internal control. The cDNA was amplified using SYBR Premix Ex Taq (TaKaRa, Dalian, China) with the CFX96TM C1000 thermal cycler in a 10 ml volume. The relative expression levels were calculated as described (Mei et al. 2014). It is noteworthy that we used two soar1 mutant and two SOAR1-overexpresssion lines to carry this experiment, and got the similar results. We presented the data of one mutant and one OE line as a representative.
Results
Downregulation of SOAR1 expression reduces, but upregulation of SOAR1 expression enhances, dehydration tolerance
ABI2 is a member of the clade-A type-2C protein phosphatases (PP2Cs) that regulates negatively ABA signaling and stress responses (Leung et al. 1997; Schweighofer et al. 2004; Cutler et al. 2010; Liang and Zhang 2014). We observed that the stomata of an ABI2-overexpression line ABI2-OE that we generated previously (Sun et al. 2011; Mei et al. 2014) kept open even in the dark and exhibited strong ABA-insensitive phenotypes in ABA-induced promotion of stomatal closure and inhibition of stomatal opening (Fig. 1a), revealing that guard cell signaling in response to ABA was seriously lesioned in the ABI2-OE line. Similar to this ABI2-overexpression line, the two SOAR1-knockdown mutant alleles soar1-2 and soar1-3 showed ABA-insensitive phenotypes in ABA-induced promotion of stomatal closure and inhibition of stomatal opening, of which the intensity was slightly weaker than that of the ABI2-OE line (Fig. 1a). In contrast to the soar1-2 and soar1-3 mutants, the SOAR1-overexpression lines OE1 and OE6 showed ABA-hypersensitive phenotypes in ABA-induced promotion of stomatal closure and inhibition of stomatal opening (Fig. 1a). These data show that SOAR1 positively regulates guard cell signaling in response to ABA.
Fig. 1.
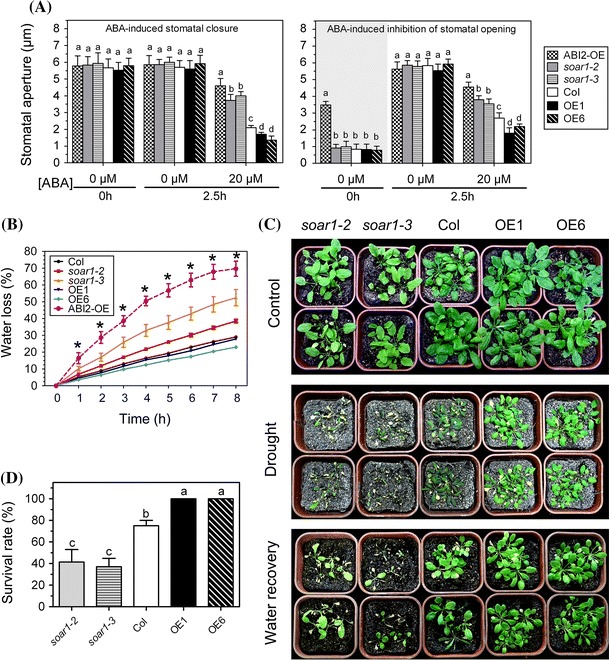
SOAR1 positively regulates plant resistance to drought stress. a ABA-induced stomatal closure (left panel) and inhibition of stomatal opening (right panel) of the wild-type Col, two SOAR1-knockdown mutant alleles soar1-2 and soar1-3, SOAR1-overexpressing lines OE1 and OE6, and an ABI2-overexpressing line ABI2-OE. Mature rosette leaves from 4-week-old seedlings were used for the assays. Values are the mean ± SE from three independent experiments (n ≥ 80 apertures per experiment), and different letters indicate significant differences at P < 0.05 (Duncan’s multiple range test) when comparing values within the same ABA concentration. b Water loss rates during a 6-h period from the detached leaves of the different genotypes described in (a). Values are the mean ± SE of five independent experiments. Star indicates that significant differences at P < 0.05 (Duncan’s multiple range test) exist when comparing values within the same time point. The entire experiment was replicated five times with similar results. c Plant growth status in the drought assays. Drought was imposed on the wild-type Col, soar1-2 and soar1-3 mutants, as well as OE1 and OE6, by withholding water for about 3 weeks until the lethal effect was observed on the mutant plants, while the control plants were well watered. The growth status was recorded 3 days after the plants were re-watered. The entire experiment was replicated three times with similar results. d Survival rate of different genotypes as mentioned in (c). Drought was imposed on the plants by withholding water until the lethal effect was observed on the mutant plants, then survival rate was recorded 3 days after the plants were re-watered. Values are the mean ± SE from three independent experiments (n ≥ 50 plants per line for each experiment) and different letters indicate significant differences at P < 0.05 (Duncan’s multiple range test)
Essentially consistent with the ABA-related phenotypes in stomatal movement, the detached leaves of soar1-2 and soar1-3 mutants, as well as those of the ABI2-OE line, showed higher rates, but the SOAR1-overexpression line OE6 showed lower rates, of water loss than those of the wild-type plants under dehydration conditions (Fig. 1b, Supplementary Table S2). It was noted, however, that the SOAR1-overexpression line OE1 showed no significant difference from the wild-type plants in relation to the water loss from detached leaves (Fig. 1b, Supplementary Table S2). Interestingly and importantly, the soar1-2 and soar1-3 mutants showed significantly more sensitive to drought than the wild-type plants, while both SOAR1-overexpression lines showed higher capacity to conserve water and to tolerate drought stress condition (Fig. 1c, d). It is noteworthy, however, that the amplitude of change in water loss from detached leaves of the SOAR1-overexpression lines was relatively small compared with wild types, while this change resulted in significant increase in their drought tolerance. We did not observe differences in stomata number between wild-type plants and SOAR1-overexpression lines. So, overexpression of SOAR1 may induce additional mechanism, such as enhanced ability of cells to tolerate osmotic stress, to allow plants to cope with drought environment.
Downregulation of SOAR1 expression reduces, but upregulation of SOAR1 expression enhances, osmotic and salt tolerance in seed germination and postgermination growth
We investigated phenotypes of the soar1 mutants and the SOAR1-overexpression lines subjected to both the osmotic stress induced by application of 300-mM mannitol in the medium and salt stress in the 175-mM NaCl-containing medium where the seeds of the different genotypes were directly sown. We observed that, compared with the wild-type Col seeds, the germination rates of seeds, survival rates of the germinating seeds, and the subsequent early-postgermination growth (7 d after stratification) of soar1-2 and soar1-3 decreased, but those of the SOAR1-overexpression lines OE1, OE3 and OE6 increased under both stressful conditions (Fig. 2a–e, Supplementary Tables S3-S5). The ABI2-overexpressing line ABI2-OE showed similar mannitol/NaCl-insensitive phenotypes to those of the SOAR1-overexpression lines (Fig. 2a–e, Supplementary Tables S3-S5).
Fig. 2.
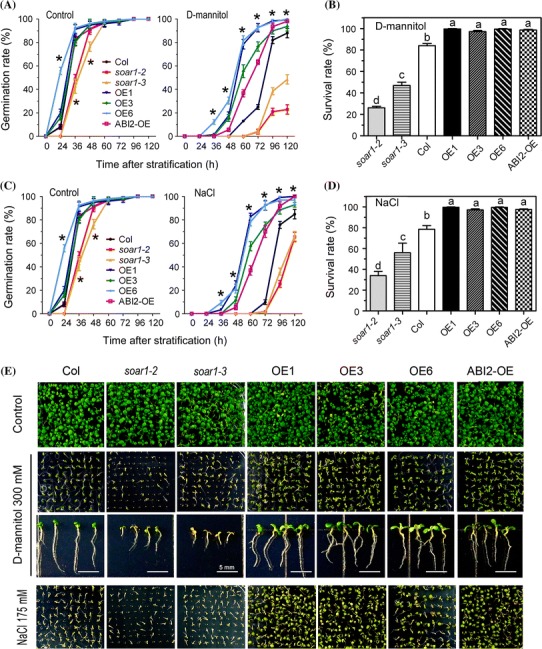
Seed germination and postgermination growth of different genotypes under D-mannitol-induced osmotic stress and salt stress. a Seed germination rates of the wild-type Col, two SOAR1-knockdown mutant alleles soar1-2 and soar1-3, three SOAR1-overexpression lines (OE1, OE3 and OE6), and the ABI2-overexpression line (ABI2-OE). Seeds were sown in the mannitol-free (control) and D-mannitol-containing (300 mM) MS-medium, and the germination rates were recorded from 24 to 120 h after stratification at 4 °C for 3 days. b Survival rate of different genotypes as mentioned in (A) grown on the mannitol-containing medium 7 days after stratification. c Seed germination rates of the genotypes as described in A. Seeds were sown in the NaCl-free (control) and NaCl-containing (175 mM) MS-medium, and the germination rates were recorded from 24 to 120 h after stratification at 4 °C for 3 days. d Survival rate of different genotypes as mentioned in (a) grown on the NaCl-containing medium (175 mM) 7 days after stratification. e Postgermination growth of the different genotypes described in (a) in the mannitol/NaCl-free (control) and 300 mM-D-mannitol- or 175 mM-NaCl-containing medium 7 days after stratification. Bars indicate 5 mm. Each value in A-D is the mean ± SE of three independent experiments, and stars in A and C indicate that significant differences at P < 0.05 (Duncan’s multiple range test) exist when comparing values within the same time point, while different letters in B and D indicate significant differences at P < 0.05 (Duncan’s multiple range test) between different genotypes
Further, NaCl was applied at different concentrations in the medium where the seeds were directly sown to test the responses of these genotypes to salt stress in postgermination growth during a relatively prolonged period (18 d after stratification). We observed that, in the medium containing 130, 175 or 200 mM NaCl, the postgermination growth of the soar1-2 and soar1-3 mutants and abi1-3 abi2-2 double mutant (knockout mutant of ABI1 and ABI2 genes) was significantly reduced, while that of the SOAR1-overexpression lines OE1, OE3 and OE6 as well as the ABI2-OE line was significantly enhanced in comparison of wild-type seedlings (Fig. 3a–c, Supplementary Fig. S1, Supplementary Table S6). These data are essentially consistent with the above-mentioned observations (Fig. 2).
Fig. 3.
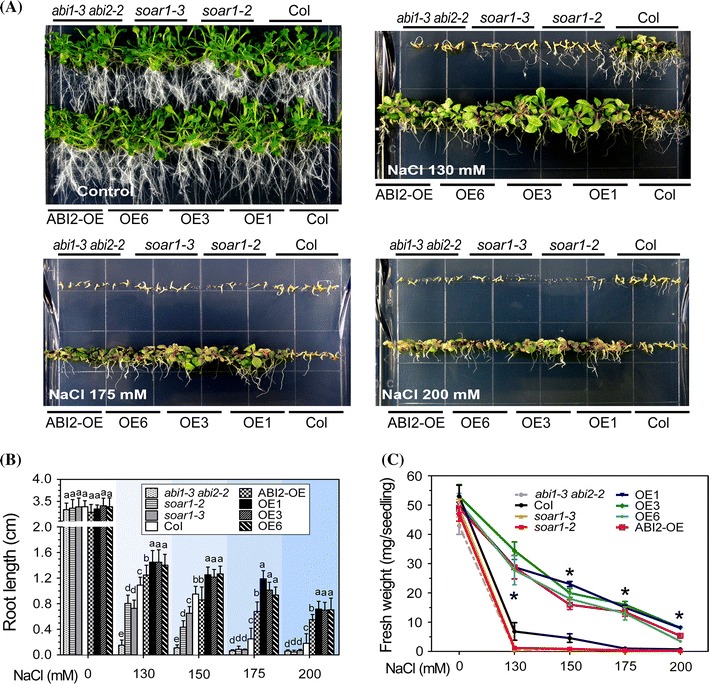
Postgermination growth of different genotypes under salt stress. a Postgermination growth of the wild-type Col, two SOAR1-knockdown mutant alleles soar1-2 and soar1-3, three SOAR1-overexpression lines (OE1, OE3 and OE6), abi1-3 abi2-2 double knockout-mutant, and the ABI2-overexpression line (ABI2-OE) in the MS medium containing 0 (control), 130, 175, and 200 mM NaCl (18 days after stratification). The entire experiment was replicated three times with similar results. b, c Statistical values of the root length (b) and fresh weight (c) of the different genotypes described in (A) grown in the medium supplemented with 0, 130, 150, 175, and 200 mM NaCl. Each value is the mean ± SE of five biological determinations with different letters (b) or stars (c) indicating significant differences at P < 0.05 (Duncan’s multiple range test) when comparing values within the same NaCl concentration
SOAR1-overexpression results in resistance of seed germination to extremely high salinity, and in salt insensitivity of mature plants, in contrast to salt hypersensitivity resulting from SOAR1 down-expression
We tested the extremity of NaCl concentrations under which the SOAR1-expression lines germinate and continue to grow by applying NaCl at different concentrations (250, 300, 350, 422, and 513 mM) in the medium in which the seeds were directly sown. Surprisingly, we observed that the seeds of the SOAR1-expression lines OE1, OE3 and OE6 germinated even in the medium containing higher than 500 mM of NaCl, and continued postgermination growth in the medium containing higher than 350 mM of NaCl, whereas the wild-type seeds scarcely germinated at the medium containing 250 mM of NaCl (Fig. 4, Supplementary Fig. S2). The phenotypes of the ABI2-overexpressing line ABI2-OE showed similar to, but weaker than, those of the SOAR1-overexpression lines (Fig. 4, Supplementary Fig. S2). The NaCl concentrations of 422 mM and 513 mM approximate those of artificial seawater (Lyman and Fleming 1940; Kester et al. 1967; Veerman et al. 2009), suggesting that the seeds of the SOAR1-expression lines may germinate in seawater.
Fig. 4.
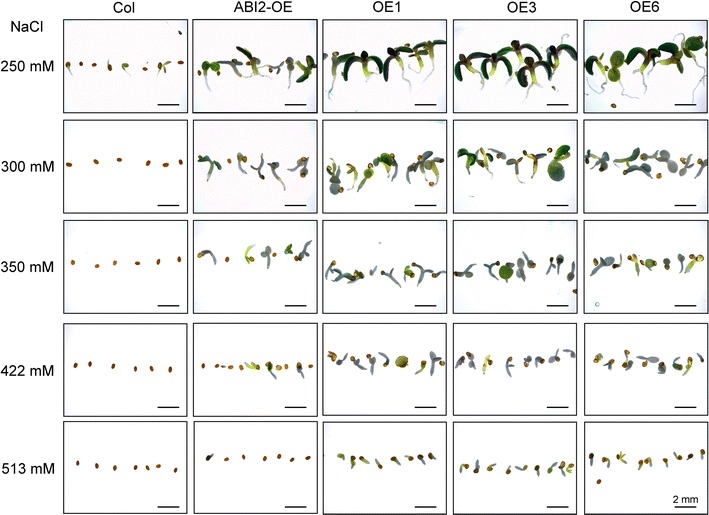
Test of the extremity of the NaCl concentrations under which seeds of the SOAR1-overexpression lines germinate and continue to grow. Germination and postgermination growth of the wild-type Col, the ABI2-overexpression line (ABI2-OE) and three SOAR1-overexpression lines OE1, OE3 and OE6 in the MS medium containing 250, 300, 350, 422, and 513 mM NaCl, were investigated 2 weeks after stratification. The entire experiment was replicated three times with similar results. Bars indicate 2 mm
Interestingly, further experiments of salt stress, imposed on the mature, whole plants by irrigation with NaCl solution, showed that the mature plants of the SOAR1-expression lines OE1, OE3 and OE6 were insensitive, but those of the soar1-2 and soar1-3 mutants were hypersensitive, to salt stress in comparison with wild-type plants (Fig. 5). It is particularly noteworthy that, in contrast to the mannitol/NaCl-insensitive phenotypes of their seeds sown directly in the mannitol or NaCl-containing medium, the mature plants of the ABI2-overexpressing line ABI2-OE showed highly sensitive to NaCl (Fig. 5). However, the mature plants of the abi1-3 abi2-2 double mutant showed NaCl-insensitive phenotype, which is similar to, but weaker than, that of the SOAR1-expression lines (Fig. 5). These data suggest that SOAR1 regulates plant response against abiotic stress by mechanisms at least partly different from those used by PP2Cs like ABI2.
Fig. 5.

Growth status of the whole mature plants of different genotypes under salt stress. Growth of wild-type Col, soar1-2 and soar1-3 mutants, abi1-3 abi2-2 double knockout-mutant, the ABI2-overexpression line ABI2-OE, and SOAR1-overexpression lines OE1 and OE6 is shown after NaCl treatment. Ten-day-old seedlings were transferred to soil and continued to grow 1 week before the NaCl treatment. NaCl solution was imposed with three increasing concentrations (100, 150, and 200). With each NaCl concentration the plants were irrigated one time at an interval of 4 days, and for a total period of 12 days plants were irrigated three times with corresponding concentrations of NaCl, and then they continued to be irrigated with 200 mM NaCl solution two times at an interval of 4 days, and the pictures were taken 7 days after the last NaCl irrigation. The control plants were irrigated with water. The entire experiment was replicated three times with similar results
Downregulation of SOAR1 expression reduces, but upregulation of SOAR1 expression enhances, freezing tolerance
For the freezing assays, 2-week-old seedlings of the non-cold-acclimated group were subjected directly to a progressive freezing process, but those of the cold-acclimated group were treated with the progressive freezing process after a pretreatment of so-called cold-acclimation (see Materials and methods), and then growth status and survival rates were recorded to estimate the freezing consequences. We observed that, in comparison with wild-type plants, the soar1-2 and soar1-3 mutants showed freezing-sensitive phenotypes, while the SOAR1-overexpression line OE1 showed freezing-tolerant phenotypes, as evidenced by the data of growth status (Fig. 6a) and especially survival rates after freezing (Fig. 6b), which is true regardless of the cold- or non-cold- acclimation pretreatment (Fig. 6a, b), suggesting that SOAR1 positively regulates both the basal and acquired freezing tolerance. Consistent with these observations, the soar1-2 and soar1-3 mutants showed higher electrolyte leakage and lower level of proline, and the SOAR1-overexpression line showed lower electrolyte leakage and higher level of proline particularly following the freezing treatments (Fig. 6c, d), indicating that the cell membranes of the soar1-2 and soar1-3mutants may be damaged by the freezing treatment, and the cells of the SOAR1-overexpression line may be protected from freezing partly by a lower level of electrolyte leakage from cells and a higher level of proline in cells.
Fig. 6.
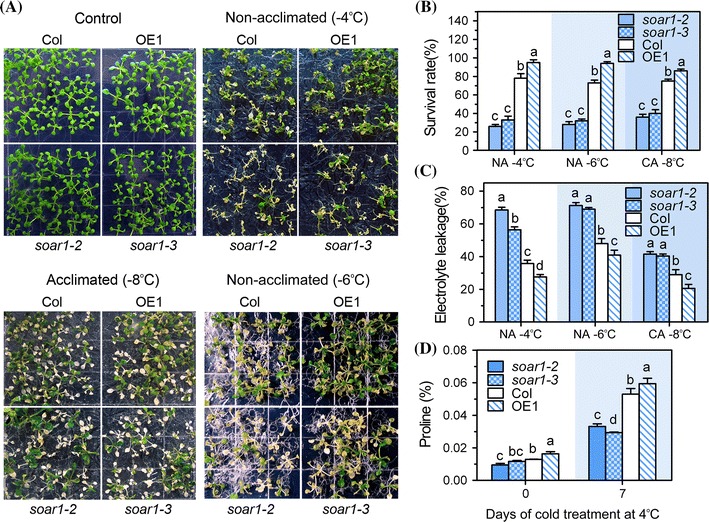
Changes in growth, electrolyte leakage and proline contents of different genotypes under freezing condition. a, b Freezing assay of wild-type Col, soar1-2, soar1-3, and SOAR1-overexpression line (OE1). Two-week-old seedlings were placed directly under freezing condition (non-acclimated plants) at −4 °C (top, right in A, and NA −4 °C in B) or −6 °C (bottom, right in A, and NA −6 °C in B) for 1 h, while other two-week-old seedlings were firstly acclimated at 4 °C for 7 days and then placed under a freezing condition at −8 °C for 1 h (cold-acclimated plants, bottom, left in A, and CA −8 °C in B). The growth and survival rates of different genotypes were recorded after a 2-days recovery at 20 °C and are shown in A and B, respectively. The control plants were grown at 20 °C. Each value in B is the mean ± SE of three biological determinations and different letters indicate significant differences at P < 0.05 (Duncan’s multiple range test) within the same treatment. c Ion leakage of different genotypes described in A and B after freezing treatment. Each value is the mean ± SE of three independent determinations and different letters indicate significant differences at P < 0.05 (Duncan’s multiple range test) within the same treatment. (D) Proline content in different genotypes described in A. The plants were grown at 20 °C for 2 weeks, acclimated at 4 °C for 7 days, and sampled for analysis. Each value is the mean ± SE of three independent determinations and different letters indicate significant differences at P < 0.05 (Duncan’s multiple range test) within the same time point
Changes in SOAR1 expression alter expression of a subset of genes involved in osmotic, salt and cold responses
We tested SOAR1 expression in wild-type plants under salt and cold stresses before assessing a series of known stress-responsive genes, and found that the SOAR1 expression levels increased with salt stress, which was similar to those of SOS1 and SOS2, two well-characterized genes positively involved in plant response to salt (Liu et al. 2000; Qiu et al. 2002; Shi et al. 2003) (Supplementary Fig. S3). Cold treatment at 4 °C induced a transient, but that at 0 °C induced a constant, increase of SOAR1 expression (Supplementary Fig. S4). These data reveal that SOAR1 is a salt- and cold-induced gene.
The expression of ABI2 was strongly induced by SOAR1 overexpression (Fig. 7a, b), which confirms our previous observation (Mei et al. 2014), and the ABI2 expression levels were even much higher in the SOAR1 overexpression line than in the wild-type plants with salt and mannitol treatments (Fig. 7a, b). Both the salt and mannitol treatments promoted ABI1 expression while its expression levels of both soar1-2 and OE1 remained lower than those of wild type, noting scarcely detectable levels of the ABI1 gene in the soar1-2 mutant (Fig. 7a, b). The expression levels of the ABA-responsive gene OST1 (Mustilli et al. 2002) and abiotic stress/ABA-responsive gene DREB2A (Liu et al. 1998), were repressed in the soar1-2 mutant, while these two genes and the ABA-responsive genes ABF4 (Choi et al. 2000), RD29A (Yamaguchi-Shinozaki and Shinozaki 1994) and KIN1 (Gilmour et al. 1998) were up-regulated in the OE1 line (Fig. 7a). In response to the mannitol-induced osmotic stress, the expression of OST1, DREB2A, ABF4, P5CS1, RD29A, RD29B (Yamaguchi-Shinozaki and Shinozaki 1994), and KIN1 was repressed in the soar1-2 mutant, while that of OST1, DREB2A and ABF4 was significantly up-regulated in the OE1 line (Fig. 7a). All these genes encodes positive ABA/stress-signaling regulators except for ABI1 and ABI2, so these gene expression data, partly explaining the earlier-mentioned stress-response observations of the different genotypes, suggest a highly complex mechanism of osmotic/salt stress response by which SOAR1 functions.
Fig. 7.
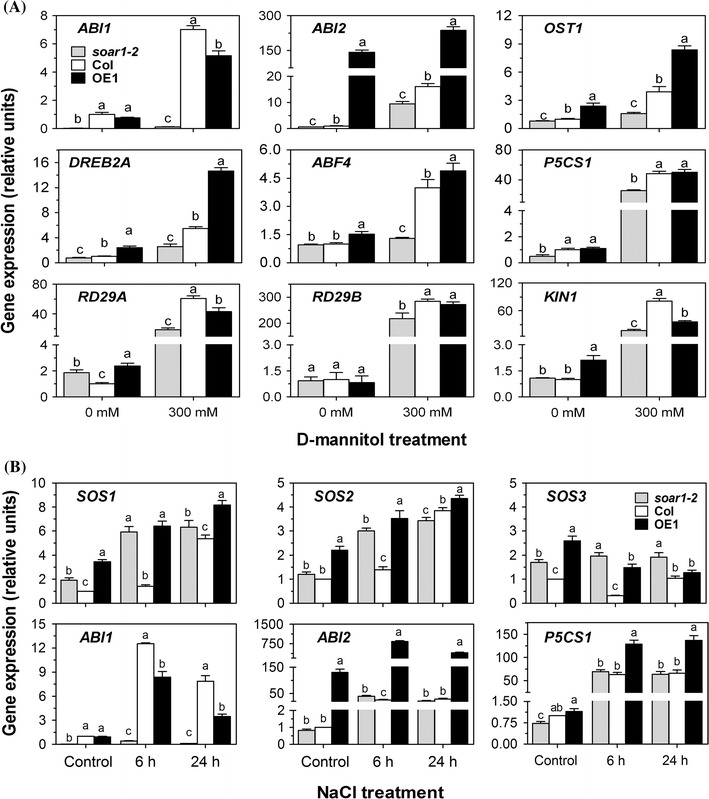
Changes in SOAR1 expression alter expression of a subset of genes involved in osmotic and salt stress responses. The mRNA levels in the seedlings of wild-type Col, soar1-2 mutant and the SOAR1-overexpression line OE1 were determined by real time RT-PCR. Two-week-old seedlings grown at 20 °C were treated with mannitol-free (0 mM) (as a control) or mannitol-containing (300 mM) solution for 24 h (a), or with the NaCl-free (0 mM) (as a control) or NaCl-containing (200 mM) solution for 6 or 24 h (b). The expression of the osmotic and salt stress responsive genes (as indicated in the figures) was analyzed. The gene expression levels were relative units normalized relative to the value from the sample of the wild-type Col plants (as 1). Each value is the mean ± SE of three independent determinations and different letters indicate significant differences at P < 0.05 (Duncan’s multiple range test) within the same treatment or at the same time point
Down- or up-regulation of SOAR1 expression in the soar1-2 mutant and the SOAR1-overexpression OE1 line, respectively, both enhanced the expression levels of SOS1, SOS2 (Liu et al. 2000; Qiu et al. 2002; Shi et al. 2003) and SOS3 (Liu and Zhu 1998) only with no statistically difference between the wild-type and soar1-2 plants for SOS3 with 24-h salt treatment (Fig. 7b), and the salt treatment promoted the expression levels of SOS1 and SOS2 in all the genotypes, noting a significant highest level of the two gene expression in the OE1 line with salt treatment of a 24-h duration (Fig. 7b). However, the salt treatment decreased the expression levels of SOS3 in OE1 line with a transient decrease in wild-type and a slight stimulation in soar1-2 (Fig. 7b). These data suggest that the positive regulatory roles of SOAR1 in response to salt stress is likely difficult to be fully explained by the SOS-mediated mechanism.
P5CS1 is a gene involved in proline biosynthesis (Verbruggen et al. 1993; Kavi Kishor et al. 1995; Hare and Cress 1997), of which the expression levels increased with the mannitol and salt treatment, and the highest levels were observed in the SOAR1 overexpression line with salt treatment (Fig. 7a, b). These findings are consistent with tolerance to salt stress of the SOAR1 overexpression lines.
The cold-responsive genes involved in the C-repeat binding factor/DRE-binding factor (CBF/DREB) transcriptional regulatory cascade, the best characterized cold-signalling pathway, were assayed after 4 °C treatment, which include CBF1/DREB1B, CBF2/DREB1C, CBF3/DREB1A (Yamaguchi-Shinozaki and Shinozaki 1994; Liu et al. 1998; Thomashow 1999), and CBF regulon genes COR15A, COR15B, COR47, COR414, KIN1 (Gilmour et al. 1998), and RD29A (Yamaguchi-Shinozaki and Shinozaki 1994), as well as CBF upstream regulator-encoding genes MYB15 (Agarwal et al. 2006), ICE1 (Chinnusamy et al. 2003) and SIZ1 (Miura et al. 2007). Cold treatment increased the expression levels of CBF1, CBF2 and CBF3 with an attenuation transition from 12 h- to 24 h-treatment, noting higher levels in the OE1 and lower levels in the soar1-2 mutant in most cases in comparison with wild type (Fig. 8, Supplementary Fig. S5). The expression profiles of the CBF downstream regulons genes COR15A, COR15B, COR414 and KIN1 were similar to those of the CBFs with a progressive increase instead of the attenuation transition from12 h- to 24 h-treatment for CBFs (Fig. 8, Supplementary Fig. S5). The data from these genes are globally consistent with the observation that SOAR1 positively regulates plant response to cold stress. The other two CBF downstream regulons genes COR47 and RD29A were also induced by the cold treatment but with higher levels in both the soar1-2 mutant and OE1 line by the 24 h-cold treatment. The cold-induced expression of the gene encoding an upstream, positive regulator of CBFs, ICE1, following an essentially similar profile to that of CBFs, could partly explain the expression profile of CBFs, but the expression of other two genes encoding the CBF-upstream, positive regulator SIZ1 and negative regulator MYB15, could not explain the expression profile of CBFs under the cold condition (Fig. 8).
Fig. 8.
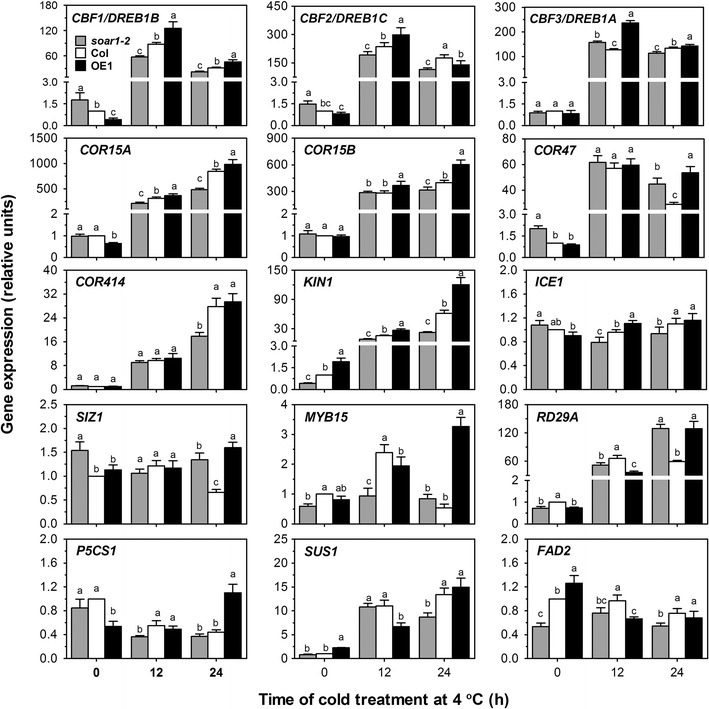
Changes in SOAR1 expression alter expression of a subset of genes involved in cold stress response. The mRNA levels in the seedlings of wild-type Col, soar1-2 mutant and the SOAR1-overexpression line OE1 were determined by real time RT-PCR. Seedlings were grown at 20 °C for 2 weeks, and transferred into a chamber at 4 °C for the indicated time. Expression of CBF1/DREB1B, CBF2/DREB1C, CBF3/DREB1A, COR15A, COR15B, COR47, COR414, KIN1, ICE1, SIZ1, MYB15, RD29A, P5SC1, SUS, and FAD2 were analyzed. The gene expression levels were relative units normalized relative to the value from the sample of the wild-type Col plants (as 1). Each value is the mean ± SE of three independent determinations, and different letters indicate significant differences at P < 0.05 (Duncan’s multiple range test) within the same time point
Additionally, we tested three genes involved in metabolisms potentially to help cells to cope with cold stress. Globally, the P5CS1 gene expression were not significantly induced by cold treatment in most cases, but showed a significantly high level in the SOAR1-overexpression OE1 line following a 24-h cold-treatment, which is consistent with the cold tolerance of the OE1. The expression of the SUS1 gene, involved in solute production (Déjardin et al. 1999; Gilmour et al. 2000) like P5CS1, was strongly induced by the cold treatment; and the FAD2 gene, positively involved in membrane stability (Miquel et al. 1993), was repressed by the cold treatment (Fig. 8). However, the levels of the two genes in the different genotypes (Fig. 8) could not explain the data of plant tolerance to cold stress (Fig. 6).
Previously, we observed that ABA concentration was not affected by down- or up-regulation of SOAR1 expression (Mei et al. 2014). We tested further, in the present experiment, the expression of genes involved in ABA biosynthesis and catabolism. The tested ABA biosynthetic enzyme-encoding genes include AAO1, AAO3, AAO4 (Sekimoto et al. 1998; Seo et al. 2000), ABA1 (Duckham et al. 1991; Xiong et al. 2002), ABA2 (Cheng et al. 2002; González-Guzmán et al. 2002), ABA3 (Bittner et al. 2001; Xiong et al. 2001), ABA4 (North et al. 2007), and NCED3 (Iuchi et al. 2001). The tested ABA catabolic enzyme-encoding genes include AtCYP707A1, AtCYP707A2, AtCYP707A3 (Kushiro et al. 2004; Saito et al. 2004), and UGT71B6 (Lim et al. 2005; Priest et al. 2006). We observed that, in the germinating seeds, the expression of most ABA biosynthetic and catabolic genes was not significantly changed in the OE1 line, while in the soar1-2 mutant, the expression of most of both the ABA biosynthetic and catabolic enzyme-encoding genes was up-regulated as compared with the wild-type plants (Supplementary Fig. S6), suggesting that, in the soar1-2 mutant, both ABA biosynthetic and catabolic processes are likely to be promoted. With exogenous ABA treatment, differences in the expression levels of these genes were attenuated between the soar1-2 mutant and the wild-type plants, while in OE1, ABA-induced effects of these genes observed in wild-type plants decreased (Supplementary Fig. S6). Overall, these data suggest that changes in SOAR1 expression may trigger a mechanism to balance ABA biosynthetic and catabolic processes, globally consistent with the endogenous ABA concentrations, which do not significantly differ among different genotypes as described previously (Mei et al. 2014).
Discussion
SOAR1 is a crucial, positive regulator of plant response to drought, salt and cold stresses, and likely to be useful in crop improvement
We provide genetic evidence that downregulation of SOAR1 expression reduces, but upregulation of SOAR1 expression enhances, ABA sensitivity in ABA-induced promotion of stomatal closure and inhibition of stomatal opening, and plant tolerance to multiple, major abiotic stresses including drought, high salinity and low temperature (Figs. 1, 2, 3, 4, 5, 6, Supplementary Figs. 1 and 2), demonstrating that SOAR1 is a crucial, positive regulator of plant response to abiotic stresses. Whereas several mitochondrial/chloroplast PPR proteins have been reported to regulate plant response to abiotic stresses (Zsigmond et al. 2008; Liu et al. 2010; Laluk et al. 2011; Murayama et al. 2012; Yuan and Liu 2012; Zhu et al. 2014), SOAR1 is the first cytosol-nucleus dual-localized protein, to our knowledge, to be identified as a crucial player in plant response to multiple, major abiotic stresses. The discovery of a cytosolic-nuclear PPR protein to regulate plant stress signaling suggest that PPR proteins may play roles in RNA processing not only in the organelles mitochondrion and chloroplast to regulate ROS homeostasis of cells, but also in the nucleus to modulate a wide range of cellular signaling processes in response to environmental cues of stresses.
Interestingly and importantly, the SOAR1-overexpression lines display strong abilities to tolerate drought, salt and cold stresses, with surprisingly high resistance to salt stress in germination and postgermination growth of seeds that are able to potentially germinate in seawater (Figs. 3, 4, Supplementary Figs. 1 and 2), while no negative impact of SOAR1-overexpression on plant growth and development was observed (Figs. 1, 3, 5). SOAR1 is highly conserved and has homologues in different plant species such as in Vitis vinifera, Ricinus communis, Populus trichocarpa, Sorghum bicolor, and Oryza sativa (Supplementary Fig. S7). Therefore, the SOAR1 gene may likely be useful for improvement of crops by transgenic manipulation to enhance crop productivity in stressful conditions.
How does SOAR1 function in response to abiotic stresses?
ABA is a central stress signal that has been believed to mainly modulates both water balance and osmotic stress/cellular dehydration tolerance to regulate plant adaptation to water deficit and salt stress, where the water balance is mainly controlled through guard cell regulation, the dehydration tolerance is dependent of osmosis-regulation proteins in all cells (Shinozaki et al. 2003; Zhu 2002, 2003). We previously showed that SOAR1 is a negative regulator of ABA signaling in seed germination and postgermination growth (Mei et al. 2014; Jiang et al. 2014). Unexpectedly, in the present experiment, we showed that SOAR1 is positively, but not negatively, involved in ABA-induced promotion of stomatal closure and inhibition of stomatal opening (Fig. 1), which explain in part enhanced tolerance of the SOAR1-overexpression plants to drought stress (Fig. 1). Additionally, the ability to significantly tolerate drought of these SOAR1-overexpression plants may also be linked to their tolerance to osmotic stress.
It is well known that ABA accumulates in salt stress as in other abiotic stresses, and increased levels of ABA result in inhibition of seed germination and is required for tolerance of seedling growth to salt (Zhu 2002, 2003; Shinozaki et al. 2003). We previously showed that down-expression of SOAR1 increases, but up-expression of SOAR1 dramatically decreases ABA sensitivity in ABA-induced seed-germination inhibition and postgermination-growth arrest (Mei et al. 2014; Jiang et al. 2014), and the SOAR1-overexpressiing seeds germinate even in 500 μM-(±)ABA-containing medium (Jiang et al. 2014). Therefore, the hypersensitivity resulting from SOAR1 down-expression and resistance against high salinity from SOAR1 up-expression in seed germination and postgermination growth (Figs. 2, 3, 4, Supplementary Figs. 1 and 2) should be attributed to ABA hypersensitivity of the SOAR1 down-expressing mutants and strong ABA insensitivity of the SOAR1 over-expressing lines (see Mei et al. 2014; Jiang et al. 2014) in the situation of the salt-induced high levels of ABA. This point of view may be supported by the observations that the ABI2-overexpressing line, highly insensitive to ABA (Sun et al. 2011; Mei et al. 2014), tolerates, but abi1 abi2 double knockout mutant, overly sensitive to ABA, is hypersensitive, to salt stress in seed germination and postgermination growth, as observed in this experiment (Figs. 2, 3, 4, Supplementary Figs. 1 and 2). The same phenomenon was also observed previously (Achard et al. 2006). The mannitol-induced osmosis-related phenotypes of the SOAR1-down- or—over-expressing plants in seed germination and postgermination growth may be also explained by changes in ABA sensitivity of these genotypes (Fig. 2). Interestingly and importantly, however, the mature plants of the SOAR1-over-expressing lines, similar to those of the abi1 abi2 double knockout mutant, tolerate, but the mature plants of the SOAR-down-expressing mutants, similar to those of the ABI2-overexpressing line, are overly sensitive to, salt stress (Fig. 5), which reveals that SOAR1 positively, but ABI2 negatively, regulates plant response to salt stress in seed germination and seedling growth, whereas both are negative regulators of ABA signaling in these two developmental processes. Therefore, unlike the ABI2-overexpression line, the tolerance to salt stress of the SOAR1-overexpression lines in postgermination growth especially during a prolonged period (18 days after stratification, Fig. 3), observed when their seeds were directly sown in ABA-containing medium, should be ascribed not only to its negative role of SOAR1 in ABA signaling in seed germination (resulting in strong ABA insensitivity of the these lines) but also to its positive role in response to salt stress.
Plant response to cold stress, like that to drought and salt stresses, requires both ABA-dependent and -independent signaling pathways (Shinozaki et al. 2003; Zhu 2002, 2003; Qin et al. 2011; Ma and Qi 2014; Shi and Yang 2014). The CBF-mediated signaling pathway has been believed to be an ABA-independent signaling pathway (Yamaguchi-Shinozaki and Shinozaki 1994; Liu et al. 1998; Thomashow 1999; Gilmour et al. 1998; Chinnusamy et al. 2003, 2007; Agarwal et al. 2006; Miura et al. 2007; Lata and Prasad 2011; Shi and Yang 2014). Like its positive role in plant response to drought and salt stresses as mentioned above, SOAR1 positively regulates plant response to cold stress (Fig. 6), functioning at least partly through the CBF signaling pathway, as evidenced by the analysis of gene expression in the SOAR1-down- and over-expressing plants (Fig. 8).
Taken together, all these findings reveal that SOAR1, a cytosolic-nuclear PPR protein, plays crucial roles in plant response to multiple, major abiotic stresses, likely integrating ABA-dependent and independent pathways. The SOAR1-mediated ABA signaling, functioning positively in ABA-induced inhibition of seed germination and postgermination growth arrest but negatively in ABA-induced promotion of stomatal closure and inhibition of stomatal opening, is likely different from the ABI2/ABI1 PP2C-mediated signaling downstream of PYR/PYL-RCAR receptors for ABA (Mustilli et al. 2002; Yoshida et al. 2006; Fujii et al. 2009; Ma et al. 2009; Park et al. 2009; Santiago et al. 2009; Umezawa et al. 2009; Vlad et al. 2009, 2010; Nishimura et al. 2010; Cutler et al. 2010). SOAR1 has been identified as a downstream player of CHLH/ABAR, a candidate ABA receptor that positively regulates ABA signaling (Shen et al. 2006; Legnaioli et al. 2009; Wu et al. 2009; Shang et al. 2010; Jia et al. 2011; Jiang et al. 2011; Tsuzuki et al. 2011, 2013; Du et al. 2012; Liu et al. 2012; Xu et al. 2012; Liu et al. 2013; Yan et al. 2013; Mei et al. 2014; Jiang et al. 2014; Zhang et al. 2013, 2014; Wang and Zhang, 2014). Elucidation of the mechanism by which SOAR1 functions, especially in the nuclear events, will be of great importance to help to understand highly complicated ABA and stress signalling network.
Electronic supplementary material
Acknowledgments
We thank Drs Shu-Hua Yang and Yan Guo (China Agricultural University, Beijing, China), and Dr Dong Liu (Tsinghua University, Beijing, China) for help with materials and equipment. This research was supported by the National Key Basic Research Program of China (2012CB114300-002), National Natural Science Foundation of China (Grant Nos. 31200213 and 31170268), and the Ministry of Agriculture of China (Grant 2014ZX08009003).
Contributor Information
Xiao-Fang Wang, Email: wangxf@biomed.tsinghua.edu.cn.
Da-Peng Zhang, Email: zhangdp@mail.tsinghua.edu.cn, Email: zhangdp@tsinghua.edu.cn.
References
- Achard P, Cheng H, De Grauwe L, Decat J, Schoutteten H, Moritz T, Van der Straeten D, Peng JR, Harberd NP. Integration of plant responses to environmentally activated phytohormonal signals. Science. 2006;311:91–94. doi: 10.1126/science.1118642. [DOI] [PubMed] [Google Scholar]
- Adie BAT, Pérez-Pérez J, Pérez-Pérez MM, Godoy M, Sánchez-Serrano JJ, Schmelz EA, Solano R. ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell. 2007;19:1665–1681. doi: 10.1105/tpc.106.048041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal M, Hao Y, Kapoor A, Dong CH, Fujii H, Zheng X, Zhu JK. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J Biol Chem. 2006;281:37636–37645. doi: 10.1074/jbc.M605895200. [DOI] [PubMed] [Google Scholar]
- Bates LS, Waldren RP, Teare ID. Rapid determination of free proline for water-stress studies. Plant Soil. 1973;39:205–207. [Google Scholar]
- Bittner F, Oreb M, Mendel RR. ABA3 is a molybdenum cofactor sulfurase required for activation of aldehyde oxidase and xanthine dehydrogenase in Arabidopsis thaliana. J Biol Chem. 2001;276:40381–40384. doi: 10.1074/jbc.C100472200. [DOI] [PubMed] [Google Scholar]
- Cheng WH, Endo A, Zhou L, Penney J, Chen HC, Arroyo A, Leon P, Nambara E, Asami T, Seo M, et al. A unique short-chain dehydrogenase/reductase in arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell. 2002;14:2723–2743. doi: 10.1105/tpc.006494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong XH, Agarwal M, Zhu JK. ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Gene Dev. 2003;17:1043–1054. doi: 10.1101/gad.1077503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Zhu J, Zhu JK. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007;12:444–451. doi: 10.1016/j.tplants.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Choi HI, Hong JH, Ha JO, Kang JY, Kim SY. ABFs, a family of ABA-responsive element binding factors. J Biol Chem. 2000;275:1723–1730. doi: 10.1074/jbc.275.3.1723. [DOI] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- Déjardin A, Sokolov LN, Kleczkowski LA. Sugar/osmoticum levels modulate differential abscisic acid-independent expression of two stress-responsive sucrose synthase genes in Arabidopsis. Biochem J. 1999;344:503–509. [PMC free article] [PubMed] [Google Scholar]
- Ding YH, Liu NY, Tang ZS, Liu J, Yang WC. Arabidopsis GLUTAMINE-RICH PROTEIN23 is essential for early embryogenesis and encodes a novel nuclear PPR motif protein that interacts with RNA polymerase II subunit III. Plant Cell. 2006;18:815–830. doi: 10.1105/tpc.105.039495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du SY, Zhang XF, Lu ZK, Xin Q, Wu Z, Jiang T, Lu Y, Wang XF, Zhang DP. Roles of the different components of magnesium chelatase in abscisic acid signal transduction. Plant Mol Biol. 2012;80:519–537. doi: 10.1007/s11103-012-9965-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckham S, Linforth R, Taylor I. Abscisic-acid-deficient mutants at the aba gene locus of Arabidopsis thaliana are impaired in the epoxidation of zeaxanthin. Plant Cell Environ. 1991;14:601–606. [Google Scholar]
- Finkelstein RR, Gampala SSL, Rock CD. Abscisic acid signaling in seeds and seedlings. Plant Cell. 2002;14:S15–S45. doi: 10.1105/tpc.010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK. In vitro reconstitution of an abscisic acid signalling pathway. Nature. 2009;462:660–664. doi: 10.1038/nature08599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Fujita M, Shinozaki K, Yamaguchi-Shinozaki K. ABA-mediated transcriptional regulation in response to osmotic stress in plants. J Plant Res. 2011;124:509–525. doi: 10.1007/s10265-011-0412-3. [DOI] [PubMed] [Google Scholar]
- Gilmour SJ, Zarka DG, Stockinger EJ, Salazar MP, Houghton JM, Thomashow MF. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 1998;16:433–442. doi: 10.1046/j.1365-313x.1998.00310.x. [DOI] [PubMed] [Google Scholar]
- Gilmour SJ, Sebolt AM, Salazar MP, Everard JD, Thomashow MF. Overexpression of the ArabidopsisCBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 2000;124:1854–1865. doi: 10.1104/pp.124.4.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golldack D, Lüking I, Yang O. Plant tolerance to drought and salinity: stress regulating transcription factors and their functional significance in the cellular transcriptional network. Plant Cell Rep. 2011;30:1383–1391. doi: 10.1007/s00299-011-1068-0. [DOI] [PubMed] [Google Scholar]
- González-Guzmán M, Apostolova N, Bellés JM, Barrero JM, Piqueras P, Ponce MR, Micol JL, Serrano R, Rodríguez PL. The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. Plant Cell. 2002;14:1833–1846. doi: 10.1105/tpc.002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammani K, Gobert A, Hleibieh K, Choulier L, Small I, Giegé P. An Arabidopsis dual-localized pentatricopeptide repeat protein interacts with nuclear proteins involved in gene expression regulation. Plant Cell. 2011;23:730–740. doi: 10.1105/tpc.110.081638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare PD, Cress WA. Metabolic implications of stress-induced proline accumulation in plants. Plant Growth Regul. 1997;21:79–102. [Google Scholar]
- Iuchi S, Kobayashi M, Taji T, Naramoto M, Seki M, Kato T, Tabata S, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 2001;27:325–333. doi: 10.1046/j.1365-313x.2001.01096.x. [DOI] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002;7:106–111. doi: 10.1016/s1360-1385(01)02223-3. [DOI] [PubMed] [Google Scholar]
- Jia HF, Chai YM, Li CL, Qin L, Shen YY. Cloning and characterization of the H subunit of a magnesium chelatase gene (PpCHLH) in peach. J Plant Growth Regul. 2011;30:445–455. [Google Scholar]
- Jiang T, Zhang XF, Wang XF, Zhang DP. Arabidopsis 3-Ketoacyl-CoA Thiolase-2 (KAT2), an enzyme of fatty acid beta-oxidation, is involved in ABA signal transduction. Plant Cell Physiol. 2011;52:528–538. doi: 10.1093/pcp/pcr008. [DOI] [PubMed] [Google Scholar]
- Jiang SC, Mei C, Wang XF, Zhang DP. A hub for ABA signaling to the nucleus: significance of a cytosolic and nuclear dual-localized PPR protein SOAR1 acting downstream of Mg-chelatase H subunit. Plant Signaling and Behavior. 2014;9:e972899. doi: 10.4161/15592316.2014.972899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavi Kishor PB, Hong Z, Miao GH, Hu CAA, Verma DPS. Overexpression of Δ1-pyrroline-5-carboxylate synthetase increases proline production and confers osmotolerance in transgenic plants. Plant Physiol. 1995;108:1387–1394. doi: 10.1104/pp.108.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kester DR, Duedall IW, Connors DN, Pytkowicz RM. Preparation of artificial sea water. Limnol Oceanogr. 1967;12:176–179. [Google Scholar]
- Kushiro T, Okamoto M, Nakabayashi K, Yamagishi K, Kitamura S, Asami T, Hirai N, Koshiba T, Kamiya Y, Nambara E. The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: key enzymes in ABA catabolism. EMBO J. 2004;23:1647–1656. doi: 10.1038/sj.emboj.7600121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laluk K, Abuqamar S, Mengiste T. The Arabidopsis mitochondria-localized pentatricopeptide repeat protein PGN functions in defense against necrotrophic fungi and abiotic stress tolerance. Plant Physiol. 2011;156:2053–2068. doi: 10.1104/pp.111.177501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lata C, Prasad M. Role of DREBs in regulation of abiotic stress responses in plants. J Exp Bot. 2011;62:4731–4748. doi: 10.1093/jxb/err210. [DOI] [PubMed] [Google Scholar]
- Lee HJ, Xiong LM, Gong ZZ, Ishitani M, Stevenson B, Zhu JK. The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleo-cytoplasmic partitioning. Gene Dev. 2001;15:912–924. doi: 10.1101/gad.866801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legnaioli T, Cuevas J, Mas P. TOC1 functions as a molecular switch connecting the circadian clock with plant responses to drought. EMBO J. 2009;28:3745–3757. doi: 10.1038/emboj.2009.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J, Merlot S, Giraudat J. The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell. 1997;9:759–771. doi: 10.1105/tpc.9.5.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S, Zhang DP. Protein kinases and phosphatases involved in ABA signaling. In: Zhang DP, editor. Abscisic acid: metabolism, transport and signaling. Heidelberg: Springer; 2014. pp. 137–175. [Google Scholar]
- Lim EK, Doucet CJ, Hou B, Jackson RG, Abrams SR, Bowles DJ. Resolution of (+)-abscisic acid using an Arabidopsis glycosyltransferase. Tetrahedron Asymmetry. 2005;16:143–147. [Google Scholar]
- Liu JP, Zhu JK. A calcium sensor homolog required for plant salt tolerance. Science. 1998;280:1943–1945. doi: 10.1126/science.280.5371.1943. [DOI] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Ishitani M, Halfter U, Kim CS, Zhu JK. The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc Natl Acad Sci USA. 2000;97:3730–3734. doi: 10.1073/pnas.060034197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, He J, Chen Z, Ren X, Hong X, Gong Z. ABA overly-sensitive 5 (ABO5), encoding a pentatricopeptide repeat protein required for cis-splicing of mitochondrial nad2 intron 3, is involved in the abscisic acid response in Arabidopsis. Plant J. 2010;63:749–765. doi: 10.1111/j.1365-313X.2010.04280.x. [DOI] [PubMed] [Google Scholar]
- Liu ZQ, Yan L, Wu Z, Mei C, Lu K, Yu YT, Liang S, Zhang XF, Wang XF, Zhang DP. Cooperation of three WRKY-domain transcription factors WRKY18, WRKY40, and WRKY60 in repressing two ABA-responsive genes ABI4 and ABI5 in Arabidopsis. J Exp Bot. 2012;63:6371–6392. doi: 10.1093/jxb/ers293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Xu YH, Jiang SC, Lu K, Lu YF, Feng XJ, Wu Z, Liang S, Yu YT, Wang XF, et al. Light-harvesting chlorophyll a/b-binding proteins, positively involved in abscisic acid signalling, require a transcription repressor, WRKY40, to balance their function. J Exp Bot. 2013;64:5443–5456. doi: 10.1093/jxb/ert307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurin C, Andres C, Aubourg S, Bellaoui M, Bitton F, Bruyere C, Caboche M, Debast C, Gualberto J, Hoffmann B, et al. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell. 2004;16:2089–2103. doi: 10.1105/tpc.104.022236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv HX, Huang C, Guo GQ, Yang ZN. Roles of the nuclear-encoded chloroplast SMR domain-containing PPR protein SVR7 in photosynthesis and oxidative stress tolerance in Arabidopsis. J Plant Biol. 2014;57:291–301. [Google Scholar]
- Lyman J, Fleming RH. Composition of sea water. J Mar Res. 1940;3:134–146. [Google Scholar]
- Ma Y, Qi F (2014) ABA regulation of plant responses to drought and salt stresses. In: Zhang DP (ed) Abscisic acid: metabolism, transport and signaling. Springer, Heidelberg, pp 315–336
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- Mei C, Jiang SC, Lu YF, Wu FQ, Yu YT, Liang S, Feng XJ, Portoles S, Lu K, Wu Z, et al. Arabidopsis pentatricopeptide repeat protein SOAR1 plays a critical role in abscisic acid signalling. J Exp Bot. 2014;65:5317–5330. doi: 10.1093/jxb/eru293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meierhoff K, Felder S, Nakamura T, Bechtold N, Schuster G. HCF152, an Arabidopsis RNA binding pentatricopeptide repeat protein involved in the processing of chloroplast psbB-psbT-psbH-petB-petD RNAs. Plant Cell. 2003;15:1480–1495. doi: 10.1105/tpc.010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miquel M, James D, Jr, Dooner H, Browse J. Arabidopsis requires polyunsaturated lipids for low-temperature survival. Proc Natl Acad Sci USA. 1993;90:6208–6212. doi: 10.1073/pnas.90.13.6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Jin JB, Lee J, Yoo CY, Stirm V, Miura T, Ashworth EN, Bressan RA, Yun D, Hasegawa PM. SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell. 2007;19:1403–1414. doi: 10.1105/tpc.106.048397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Murayama M, Hayashi S, Nishimura N, Ishide M, Kobayashi K, Yagi Y, Asami T, Nakamura T, Shinozaki K, Hirayama T. Isolation of Arabidopsisahg11, a weak ABA hypersensitive mutant defective in nad4 RNA editing. J Exp Bot. 2012;63:5301–5310. doi: 10.1093/jxb/ers188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustilli AC, Merlot S, Vavasseur A, Fenzi F, Giraudat J. Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell. 2002;14:3089–3099. doi: 10.1105/tpc.007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N, Sarkeshik A, Nito K, Park SY, Wang A, Carvalho PC, Lee S, Caddell DF, Cutler SR, Chory J, et al. PYR/PYL/RCAR family members are major in vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J. 2010;61:290–299. doi: 10.1111/j.1365-313X.2009.04054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North HM, Almeida AD, Boutin JP, Frey A, To A, Botran L, Sotta B, Marion-Poll A. The Arabidopsis ABA-deficient mutant aba4 demonstrates that the major route for stress-induced ABA accumulation is via neoxanthin isomers. Plant J. 2007;50:810–824. doi: 10.1111/j.1365-313X.2007.03094.x. [DOI] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TFF, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priest DM, Ambrose SJ, Vaistij FE, Elias L, Higgins GS, Ross ARS, Abrams SR, Bowles DJ. Use of the glucosyltransferase UGT71B6 to disturb abscisic acid homeostasis in Arabidopsis thaliana. Plant J. 2006;46:492–502. doi: 10.1111/j.1365-313X.2006.02701.x. [DOI] [PubMed] [Google Scholar]
- Qin F, Shinozaki K, Yamaguchi-Shinozaki K. Achievements and challenges in understanding plant abiotic stress responses and tolerance. Plant Cell Physiol. 2011;52:1569–1582. doi: 10.1093/pcp/pcr106. [DOI] [PubMed] [Google Scholar]
- Qiu QS, Guo Y, Dietrich MA, Schumaker KS, Zhu JK. Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc Natl Acad Sci USA. 2002;99:8436–8441. doi: 10.1073/pnas.122224699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivals E, Bruyere C, Toffano-Nioche C, Lecharny A. Formation of the Arabidopsis pentatricopeptide repeat family. Plant Physiol. 2006;141:825–839. doi: 10.1104/pp.106.077826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito S, Hirai N, Matsumoto C, Ohigashi H, Ohta D, Sakata K, Mizutani M. Arabidopsis CYP707As encode (+)-abscisic acid 8′-hydroxylase, a key enzyme in the oxidative catabolism of abscisic acid. Plant Physiol. 2004;134:1439–1449. doi: 10.1104/pp.103.037614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago J, Dupeux F, Round A, Antoni R, Park SY, Jamin M, Cutler SR, Rodriguez PL, Márquez JA. The abscisic acid receptor PYR1 in complex with abscisic acid. Nature. 2009;462:665–668. doi: 10.1038/nature08591. [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Small I. Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci. 2008;13:663–670. doi: 10.1016/j.tplants.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Schweighofer A, Hirt H, Meskiene I. Plant PP2C phosphatases: emerging functions in stress signaling. Trends Plant Sci. 2004;9:236–243. doi: 10.1016/j.tplants.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Sekimoto H, Seo M, Kawakami N, Komano T, Desloire S, Liotenberg S, Marion-Poll A, Caboche M, Kamiya Y, Koshiba T. Molecular cloning and characterization of aldehyde oxidases in Arabidopsis thaliana. Plant Cell Physiol. 1998;39:433–442. doi: 10.1093/oxfordjournals.pcp.a029387. [DOI] [PubMed] [Google Scholar]
- Seo M, Peeters AJM, Koiwai H, Oritani T, Marion-Poll A, Zeevaart JAD, Koornneef M, Kamiya Y, Koshiba T. The Arabidopsis aldehyde oxidase 3 (AAO3) gene product catalyzes the final step in abscisic acid biosynthesis in leaves. Proc Natl Acad Sci USA. 2000;97:12908–12913. doi: 10.1073/pnas.220426197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Yan L, Liu ZQ, Cao Z, Mei C, Xin Q, Wu FQ, Wang XF, Du SY, Jiang T, et al. The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell. 2010;22:1909–1935. doi: 10.1105/tpc.110.073874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen YY, Wang XF, Wu FQ, Du SY, Cao Z, Shang Y, Wang XL, Peng CC, Yu XC, Zhu SY, et al. The Mg-chelatase H subunit is an abscisic acid receptor. Nature. 2006;443:823–826. doi: 10.1038/nature05176. [DOI] [PubMed] [Google Scholar]
- Shi Y, Yang S. ABA regulation of the cold stress response in plants. In: Zhang DP, editor. Abscisic acid: metabolism, transport and signaling. Heidelberg: Springer; 2014. pp. 337–363. [Google Scholar]
- Shi H, Lee BH, Wu SJ, Zhu JK. Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nat Biotechnol. 2003;21:81–85. doi: 10.1038/nbt766. [DOI] [PubMed] [Google Scholar]
- Shi Y, Tian S, Hou L, Huang X, Zhang X, Guo H, Yang S. Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type-A ARR genes in Arabidopsis. Plant Cell. 2012;24:2578–2595. doi: 10.1105/tpc.112.098640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K, Seki M. Regulatory network of gene expression in the drought and cold stress responses. Curr Opin Plant Biol. 2003;6:410–417. doi: 10.1016/s1369-5266(03)00092-x. [DOI] [PubMed] [Google Scholar]
- Small ID, Peeters N. The PPR motif: a TPR-related motif prevalent in plant organellar proteins. Trends Biochem Sci. 2000;25:46–47. doi: 10.1016/s0968-0004(99)01520-0. [DOI] [PubMed] [Google Scholar]
- Sun HL, Wang XJ, Ding WH, Zhu SY, Zhao R, Zhang YX, Xin Q, Wang XF, Zhang DP. Identification of an important site for function of the type 2C protein phosphatase ABI2 in abscisic acid signalling in Arabidopsis. J Exp Bot. 2011;62:5713–5725. doi: 10.1093/jxb/err274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow MF. Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Biol. 1999;50:571–599. doi: 10.1146/annurev.arplant.50.1.571. [DOI] [PubMed] [Google Scholar]
- Tsuzuki T, Takahashi K, Inoue S, Okigaki Y, Tomiyama M, Hossain MA, Shimazaki K, Murata Y, Kinoshita T. Mg-chelatase H subunit affects ABA signaling in stomatal guard cells, but is not an ABA receptor in Arabidopsis thaliana. J Plant Res. 2011;124:527–538. doi: 10.1007/s10265-011-0426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuzuki T, Takahashi K, Tomiyama M, Inoue S, Kinoshita T. Overexpression of the Mg-chelatase H subunit in guard cells confers drought tolerance via promotion of stomatal closure in Arabidopsis thaliana. Frontier Plant Sci. 2013;4:440. doi: 10.3389/fpls.2013.00440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, Yamaguchi-Shinozaki K, Ishihama Y, Hirayama T, Shinozaki K. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci USA. 2009;106:17588–17593. doi: 10.1073/pnas.0907095106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veerman J, Saakes M, Metz SJ, Harmsen GJ. Reverse electrodialysis: performance of a stack with 50 cells on the mixing of sea and river water. J Membrane Sci. 2009;327:136–144. [Google Scholar]
- Verbruggen N, Villarroel R, Van Montagu M. Osmoregulation of a pyrroline-5-carboxylate reductase gene in Arabidopsis thaliana. Plant Physiol. 1993;103:771–781. doi: 10.1104/pp.103.3.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlad F, Rubio S, Rodrigues A, Sirichandra C, Belin C, Robert N, Leung J, Rodriguez PL, Laurière C, Merlot S. Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell. 2009;21:3170–3184. doi: 10.1105/tpc.109.069179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlad F, Droillard MJ, Valot B, Khafif M, Rodrigues A, Brault M, Zivy M, Rodriguez PL, Merlot S, Laurière C. Phospho-site mapping, genetic and in planta activation studies reveal key aspects of the different phosphorylation mechanisms involved in activation of SnRK2s. Plant J. 2010;63:778–790. doi: 10.1111/j.1365-313X.2010.04281.x. [DOI] [PubMed] [Google Scholar]
- Wang XF, Zhang DP. ABA signal perception and ABA receptors. In: Zhang DP, editor. Abscisic acid: metabolism, transport and signaling. Heidelberg: Springer; 2014. pp. 89–116. [Google Scholar]
- Williams PM, Barkan A. A chloroplast-localized PPR protein required for plastid ribosome accumulation. Plant J. 2003;36:675–686. doi: 10.1046/j.1365-313x.2003.01915.x. [DOI] [PubMed] [Google Scholar]
- Wu FQ, Xin Q, Cao Z, Liu ZQ, Du SY, Mei C, Zhao CX, Wang XF, Shang Y, Jiang T, et al. The magnesium-chelatase H subunit binds abscisic acid and functions in abscisic acid signaling: new evidence in Arabidopsis. Plant Physiol. 2009;150:1940–1954. doi: 10.1104/pp.109.140731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Zhu JK. Abiotic stress signal transduction in plants: molecular and genetic perspectives. Physiol Plantarum. 2001;112:152–166. doi: 10.1034/j.1399-3054.2001.1120202.x. [DOI] [PubMed] [Google Scholar]
- Xiong L, Ishitani M, Lee H, Zhu JK. The Arabidopsis LOS5/ABA3 locus encodes a molybdenum cofactor sulfurase and modulates cold stress- and osmotic stress-responsive gene expression. Plant Cell. 2001;13:2063–2083. doi: 10.1105/TPC.010101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Lee H, Ishitani M, Zhu JK. Regulation of osmotic stress-responsive gene expression by the LOS6/ABA1 locus in Arabidopsis. J Biol Chem. 2002;277:8588–8596. doi: 10.1074/jbc.M109275200. [DOI] [PubMed] [Google Scholar]
- Xu YH, Liu R, Yan L, Liu ZQ, Jiang SC, Shen YY, Wang XF, Zhang DP. Light-harvesting chlorophyll a/b-binding proteins are required for stomatal response to abscisic acid in Arabidopsis. J Exp Bot. 2012;63:1095–1106. doi: 10.1093/jxb/err315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell. 1994;6:251–264. doi: 10.1105/tpc.6.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Liu ZQ, Xu YH, Lu K, Wang XF, Zhang DP. Auto- and cross-repression of three Arabidopsis WRKY transcription factors WRKY18, WRKY40, and WRKY60 negatively involved in ABA signaling. J Plant Growth Regul. 2013;32:399–416. [Google Scholar]
- Yang H, Shi Y, Liu J, Guo L, Zhang X, Yang S. A mutant CHS3 protein with TIR-NB-LRR-LIM domains modulates growth, cell death and freezing tolerance in a temperature-dependent manner in Arabidopsis. Plant J. 2010;63:283–296. doi: 10.1111/j.1365-313X.2010.04241.x. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Nishimura N, Kitahata N, Kuromori T, Ito T, Asami T, Shinozaki K, Hirayama T. ABA-hypersensitive germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiol. 2006;140:115–126. doi: 10.1104/pp.105.070128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Liu D. Functional disruption of the pentatricopeptide protein SLG1 affects mitochondrial RNA editing, plant development, and responses to abiotic stresses in Arabidopsis. Plant J. 2012;70:432–444. doi: 10.1111/j.1365-313X.2011.04883.x. [DOI] [PubMed] [Google Scholar]
- Zhang XF, Jiang T, Wu Z, Du SY, Yu YT, Jiang SC, Lu K, Feng XJ, Wang XF, Zhang DP. Cochaperonin CPN20 negatively regulates abscisic acid signaling in Arabidopsis. Plant Mol Biol. 2013;83:205–218. doi: 10.1007/s11103-013-0082-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XF, Jiang T, Yu YT, Wu Z, Jiang SC, Lu K, Feng XJ, Liang S, Lu YF, Wang XF, et al. Arabidopsis co-chaperonin CPN20 antagonizes Mg-chelatase H subunit to derepress ABA-responsive WRKY40 transcription repressor. Science China Life Sciences. 2014;57:11–21. doi: 10.1007/s11427-013-4587-9. [DOI] [PubMed] [Google Scholar]
- Zhu JK. Salt and drought stress signal transduction in plants. Annu Rev Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. Regulation of ion homeostasis under salt stress. Curr Opin Plant Biol. 2003;6:441–445. doi: 10.1016/s1369-5266(03)00085-2. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Dugardeyn J, Zhang C, Mühlenbock P, Eastmond PJ, Valcke R, De Coninck B, Öden S, Karampelias M, Cammue BPA, et al. The Arabidopsisthaliana RNA editing factor SLO2, which affects the mitochondrial electron transport chain, participates in multiple stress and hormone responses. Mol Plant. 2014;7:290–310. doi: 10.1093/mp/sst102. [DOI] [PubMed] [Google Scholar]
- Zsigmond L, Rigó G, Szarka A, Székely G, Ötvös K, Darula Z, Medzihradszky KF, Koncz C, Koncz Z, Szabados L. Arabidopsis PPR40 connects abiotic stress responses to mitochondrial electron transport. Plant Physiol. 2008;146:1721–1737. doi: 10.1104/pp.107.111260. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


