Summary
One of the exceptional properties of the brain is its ability to acquire new knowledge through learning and to store that information through memory. The epigenetic mechanisms linking changes in neuronal transcriptional programs to behavioral plasticity remain largely unknown. Here, we identify the epigenetic signature of the neuronal enhancers required for transcriptional regulation of synaptic plasticity genes during memory formation, linking this to Reelin signaling. The binding of Reelin to its receptor, LRP8, triggers activation of this cohort of LRP8-Reelin-regulated-Neuronal (LRN) enhancers that serve as the ultimate convergence point of a novel synapse-to-nucleus pathway. Reelin simultaneously regulates NMDA-receptor transmission, which reciprocally permits the required, γ-secretase-dependent cleavage of LRP8, revealing an unprecedented role for its intracellular domain in the regulation of synaptically generated signals. These results uncover an in vivo enhancer code serving as a critical molecular component of cognition and relevant to psychiatric disorders linked to defects in Reelin signaling.
Introduction
Throughout life, in response to environmental cues, the nervous system is required to make appropriate changes at the level of neural circuitries that might be governed by changes in gene expression. Indeed, precise temporal and spatial control of gene transcription is required for the development of the complex brain architecture consisting of hundreds of cell types with highly specialized functions (Bernard et al., 2012; Zeng et al., 2012). Although remarkable progress has been made toward generating genome-wide atlases of transcriptional profiles across the adult brain (Hawrylycz et al., 2012; Lein et al., 2007), the global contributions of gene expression patterns to in vivo behavioral processes remain largely unknown. Advances in the past two decades have unambiguously shown that epigenetic mechanisms leading to finely-tuned gene expression patterns are essential for regulating individual neuronal activity and for sustaining function of neuronal circuits involved in higher-level cognitive behaviors (Telese et al., 2013; West and Greenberg, 2011). A sophisticated pattern of epigenetic control of cognition is emerging from studies of DNA methylation (Guo et al., 2011; Kaas et al., 2013; Miller et al., 2010; Rudenko et al., 2013), histone modifications (Graff et al., 2014; Graff et al., 2012; Gupta et al., 2010; Peleg et al., 2010), and RNA metabolism (Gao et al., 2010; Rajasethupathy et al., 2012; Shirayama et al., 2012).
Studies of genome-wide maps of enhancers in the mammalian genome have estimated that there are 7×104- 105 enhancers/cell (Consortium et al., 2012; Thurman et al., 2012). Interestingly, a widespread association of disease-linked DNA variations in these regulatory elements has been reported (Grossman et al., 2013; Hah et al., 2013; Kim et al., 2010; Li et al., 2013; Maurano et al., 2012; Ripke et al., 2013). Enhancers are themselves regulated transcription units that act to potentiate transcription, putatively by delivering important regulatory factors to the promoter through long-range physical interactions with coding genes promoters (Hah et al., 2013; Kim et al., 2010; Li et al., 2013; Sanyal et al., 2012; Shlyueva et al., 2014). Distal enhancer elements control cell-type transcriptional specificity and orchestrate the extraordinary array of functional diversity in developing tissues in controlled spatiotemporal dynamics (Consortium et al., 2012; Pennacchio et al., 2006; Shen et al., 2012; Visel et al., 2013). Based on the critical roles of enhancers in virtually every aspect of regulated transcriptional programs, it becomes clear that uncovering enhancer regulatory strategies will be important to explicate brain functions.
The Reelin pathway has been well documented to regulate synaptic plasticity (Qiu et al., 2006b; Rogers et al., 2011; Weeber et al., 2002). Reduction in Reelin expression is detected in many psychiatric disorders, including schizophrenia, major depression, epilepsy and autism (Folsom and Fatemi, 2013), thus making it a key candidate pathway for investigation of neuronal function. The extracellular protein, Reelin, has been shown to be a critical factor for the proper development of the laminated neocortex (D'Arcangelo et al., 1995; Rice and Curran, 2001). Recent findings indicate that in the mature brain Reelin promotes long-term potentiation through modulation of glutamatergic synaptic transmission (Beffert et al., 2005; Chen et al., 2005; Qiu et al., 2006a; Qiu et al., 2006b; Rogers et al., 2011; Trotter et al., 2013; Weeber et al., 2002). The emerging literature suggests a predominant role of the Reelin receptor, LRP8, in mediating neuroplasticity in the adult brain, where it is suggested to be a functional component of a multiprotein complex containing the NMDA receptor (NMDA-R) at excitatory synapses (Beffert et al., 2006; Hoe et al., 2006; Weeber et al., 2002). The cytoplasmic domain of LRP8 acts as a signal transducer for the cascade of events initiated by Reelin through a protein-protein interaction network based on its conserved NPTY motif recognized by several adaptor proteins, including DAB1 (Trommsdorff et al., 1999). Therefore, it is of particular interest to uncover the molecular mechanisms by which the Reelin-LRP8 pathway modulates synaptic plasticity events central to learning and memory.
Here, by integrating behavioral paradigms with genomic and transcriptomic strategies, we provide evidence that the Reelin pathway controls learning and memory through activation of specific cohort of cis-regulatory enhancer elements. Because these enhancers are activated by Reelin-LRP8 signaling, we refer to these as the LRP8-Reelin-regulated Neuronal (LRN) enhancers. We elucidate the molecular signature of the LRN enhancers, which involves combinatorial actions of the transcription factors (TFs) CREB and MEF2, a switch in occupancy between the NCoR co-repressor and the CBP co-activator complexes, and precise three-dimensional patterns of enhancer-promoter interactions. CRISPR-mediated interference of enhancer activity documents that LRN enhancers are required for activation of synaptic plasticity genes that constitute a core component of the transcriptional events observed in a fear conditioning paradigm. Furthermore, we report a novel crosstalk between NMDA-R activity and γ-secretase-dependent release of the LRP8 intracellular domain as a necessary component of this epigenomic pathway. Collectively, our results indicate that alterations of the Reelin pathway lead to misregulation of a specific enhancer-driven transcriptional program that could underlie impairment of cognitive functions in a variety of mental illnesses.
RESULTS
Reelin signaling induces transcriptional programs linked to learning and memory
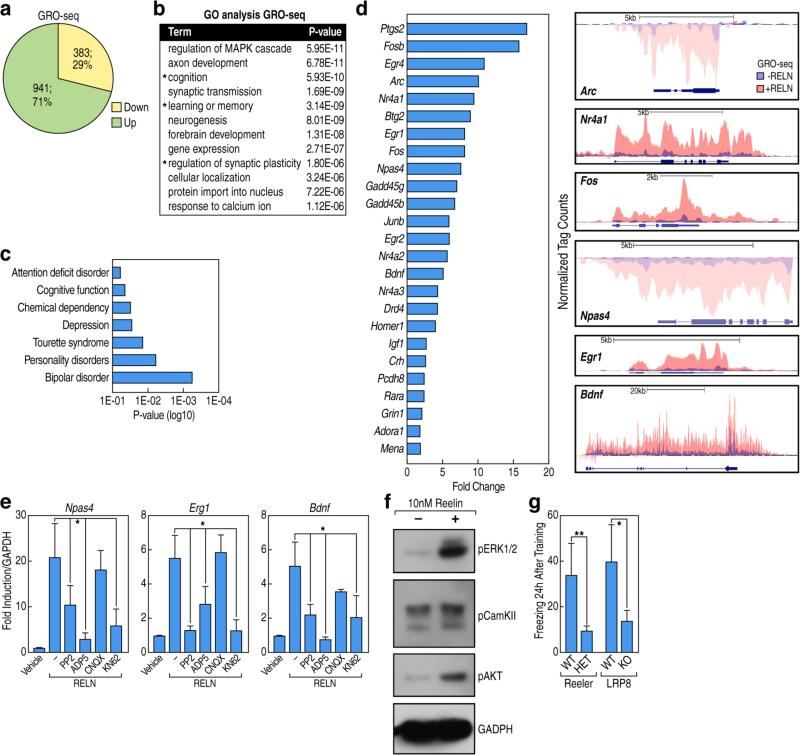
To evaluate whether specific transcriptional programs are initiated by Reelin signaling, we generated transcriptomic profiles via global-run on sequencing (GRO-seq) using primary cortical neurons treated for 1 h with 10nM purified recombinant Reelin protein (RELN) (Figure S1A-B). Using a cut-off threshold of 1.5-fold change, and a false discovery rate (FDR) of 0.001, we detected dynamic changes in the nascent transcripts of 1,324 coding genes (Table S1). The majority of differentially expressed genes (941; 71%) were up-regulated (hereafter referred to as Reelin target genes) (Figure 1A). Gene functional annotations related to regulation of neuroplasticity processes were over-represented in the Reelin-regulated transcriptome (Figure 1B). Moreover, Reelin target genes were positively associated with genes implicated in neuropsychiatric disorders, suggesting a biological relevance of this pathway to disease phenotypes (Figure 1C). Reelin robustly induced the expression of synaptic activity-dependent genes, the regulation of which is a candidate mechanism for NMDA-R-dependent long-term potentiation (LTP) during memory formation (Flavell and Greenberg, 2008; Kandel et al., 2014), as shown in a representative UCSC browser image in Figure 1D and confirmed by qRT-PCR (Figure S1C). Given that Reelin modulates NMDA-R activity in a Src-dependent manner (Chen et al., 2005), we examined the extent of co-regulation of transcriptional events triggered by Reelin and NMDA-R signaling pathways. Therefore, we generated transcriptional profiles by RNA-seq using cortical neurons stimulated by the GABAA receptor blocker bicuculline (50uM) that increases NMDA-R-dependent excitatory synaptic transmission (Figure S1D). Roughly 30% of Reelin-dependent genes were synergistically co-regulated by NMDA-R activation (Figure S1E-F), suggesting that NMDA-R activation contributes to the Reelin-dependent nuclear signaling. In support of our hypothesis, we found by qRT-PCR that the activation of Reelin responsive genes is dramatically impaired by pharmacological inhibition of Src tyrosine kinases (PP2) and by antagonists of NMDA-R (D-AP5), but not antagonists of AMPA receptors (CNQX). It is also impaired by inhibitors of calcium/calmodulin-dependent protein kinase (KN62) that has been implicated in the induction of LTP (Lucchesi et al., 2011; Shalin et al., 2004) (Figure 1E). Consistent with this, Reelin treatment induces phosphorylation of active forms of ERK1/2 and CaMKII, along with phosphorylation of AKT, which serves as a hallmark of activation of the Reelin signaling pathway (Figure 1F) (Beffert et al., 2002). Although a large body of evidence has clearly demonstrated the role of Reelin in the regulation of synaptic plasticity at the cellular level, the requirement of Reelin signaling for cognitive behaviors has been a controversial debate due to a variety of behavioral paradigms used in distinct laboratories. To examine the potential relevance of our findings in vivo, we used a classical form of robust associative learning (contextual fear paradigm) in the Heterozygous Reeler Mice (HRM) and in LRP8-KO mice (Rudy et al., 2004). We observed a severe impairment in freezing behaviors in these mutant mice that reflect a loss of long-term memory formation in absence of intact Reelin signaling (Figure 1G). These studies corroborate the role of a novel synapse-to-nucleus signaling triggered by Reelin-LRP8 signaling connecting transcriptional control to learning and memory behaviors.
Figure1. Reelin activates a transcriptional program linked to learning and memory.
(A) Pie chart showing genes regulated by Reelin assessed by GRO-Seq analysis (FDR <0.001, FC >1.5) in cortical neurons treated 1 h with 10nM Reelin.
(B) Gene Ontology analysis of Reelin-regulated genes showing the top-enriched terms. Asterisks (*) indicate annotations that are related to synaptic plasticity.
(C) Genetic association analysis of all Reelin-regulated genes using DAVID.
(D) Top Reelin-regulated genes are ranked by fold change. UCSC genome browser images showing Reelin-induced transcription by GRO-Seq of selected target genes linked to synaptic plasticity.
(E) qRT-PCR results indicating that Reelin-induced genes are inhibited by specific inhibitors (1uM PP2, 100uM D-AP5 and 40uM CNQX, 10uM KN62). RNA was analyzed 6 h post-treatment. Data are normalized against Gapdh. Data are shown as mean ± SD; *p<0.05 (Two-tailed students T-test).
(F) Western blots analysis of cortical neurons stimulated with Reelin (10nM, 0.5 h), using antibodies against the phosphorylated form of ERK1/2, CaMKII and Akt. GAPDH antibody was used as a loading control.
(G) Heterozygous Reeler (HET) and LRP8-KO mice demonstrate impaired associative fear memory acquisition in contextual paradigms (auditory cue: 30 s, 85 dB tone; foot shock: 2 s, 0.5 mA, constant current). Histograms represent the measurements of freezing behaviors (error bars ± SD; *p < 0.05 or **p<0.001 by Two-tailed students T-test).
See also Fig S1
Reelin signaling activates specific cohorts of enhancer elements in the genome
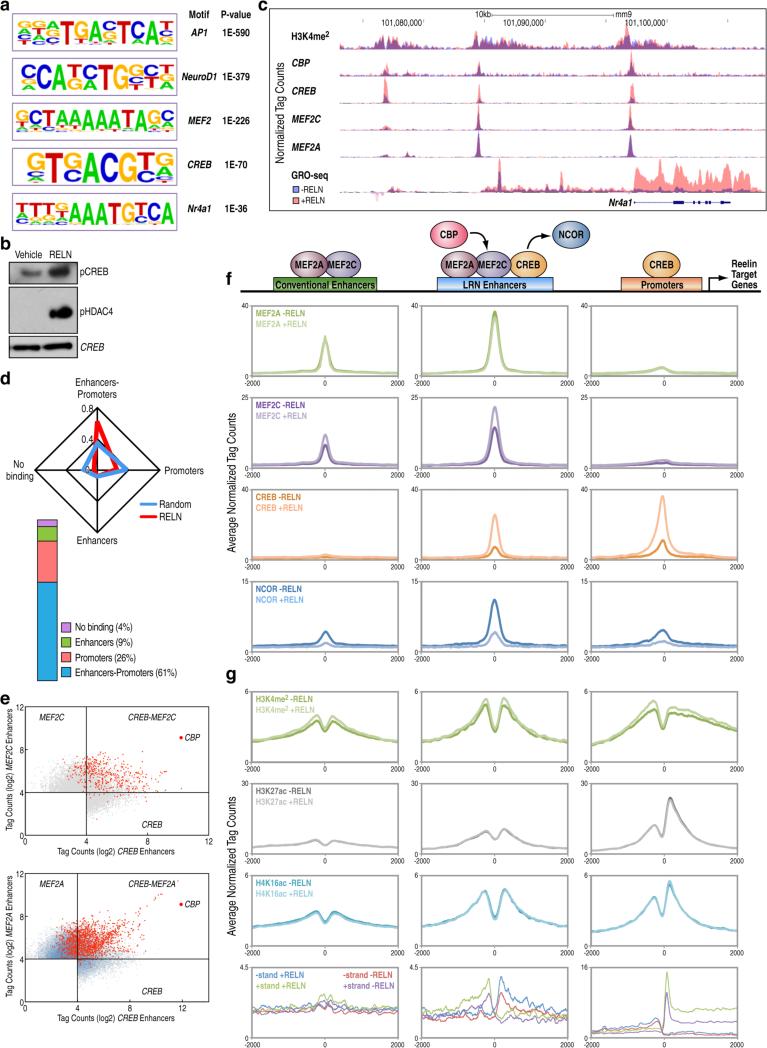
To explore the epigenetic mechanisms underlying the Reelin-dependent transcriptional program, we surveyed the chromatin landscape in cortical neurons by chromatin immunoprecipitation (ChIP-Seq) of histone tail modifications associated with active chromatin states (H3K4me2, K3K27Ac, H4K16Ac) (Heintzman et al., 2009). We identified several thousand putative distal enhancers elements (Figure S2A). To examine how these chromatin states relate to enhancer activity in a signaling-dependent fashion, we profiled the genome-wide distribution of the histone acetyl transferase, CBP, upon Reelin treatment. CBP has served as a predictor of enhancers with tissue-specific activity and it has been reported to label activated enhancers in depolarized neurons (Kim et al., 2010; May et al., 2012). We observed that Reelin stimulation results in the induction of CBP recruitment to its target loci (Figure S2B). The H3K4me2-marked enhancers co-occupied by CBP upon Reelin treatment (n=4,175) are highly enriched for motifs recognized by neuronal TFs, including MEF2 and CREB, which have been described as key regulators of synaptic plasticity (Figure 2A) (Akhtar et al., 2012; Deisseroth et al., 1996; Dietrich, 2013; Flavell et al., 2008). Western blot analysis documented that Reelin robustly induced phosphorylation of CREB at serine 133 (Gonzalez and Montminy, 1989) and phosphorylation of HDAC4 at serine 632, consistent with HDAC4 nuclear export that is required for MEF2 transcriptional activation (Wang et al., 2000) (Figure 2B). Therefore, we carried out ChIP-Seqs in cortical neurons exposed to Reelin for 1 h using antibodies against CREB and two different isoforms of MEF2 TFs (MEF2A and MEF2C), which are highly expressed in cortical neurons (Figure 2SC). A representative UCSC browser image depicting the epigenomic changes induced by Reelin is shown in Figure 2C for the Nr4a1 locus. Reelin substantially increased CREB and MEF2C binding to their genomic loci, but did not alter the recruitment of MEF2A, suggesting a specific involvement of MEF2C, which is particularly intriguing given that MEF2C is the only isoform required for positive regulation of hippocampal associative learning (Barbosa et al., 2008) (Figure S2B). We observed a largely conserved distribution of MEF2A and MEF2C binding patterns (Figure S2D), and a widespread association with distal enhancers sites, suggesting that they may function as lineage-specific components of neuronal enhancers (Figure S2E). Conversely, the CREB binding occurs at both promoter (57%) and enhancer regions (43%) (Figure S2E). The majority of Reelin targets (60%) strongly correlated with CREB binding simultaneously at promoters and enhancers (Figure 2D, Figure S2F), indicating that promoter binding is generally not sufficient to engage transcriptional changes, while the presence of CREB on enhancers is required. Comparative genomic analysis of different TFs in a variety of cell types has suggested that a typical signature of enhancers is the presence of multiple TFs (Chen et al., 2008). To gain insight into in the functional enhancer signature underlying the Reelin-induced epigenomic events, we analyzed different genomic regions based on the interplay of CREB and MEF2 TFs. Scatter plots of the tag counts around genomic coordinates of MEF2 and CREB-occupied enhancers sites show that enhancers co-bound by both factors predominantly coincide with the most active enhancers labeled by CBP upon Reelin treatment (Figure 2E).
Figure 2. Definition of LRP8-Reelin-regulated Neuronal enhancers (LRN enhancers).
(A) De novo motif analysis of CBP-occupied H3K4me2-enhancers showing the top enriched transcription factor motifs after Reelin treatment (10nM, 1 h), with associated P values as indicated.
(B) Western blots of cortical neurons stimulated with 10nM Reelin for 30 minutes using antibodies for phosphorylated CREB and HDAC4; CREB antibody is used as a loading control.
(C) A representative genome browser image depicting ChIP-Seq binding patterns at distal H3K4me2-enhancers at the Nr4a1 gene locus, the transcription of which was enhanced by Reelin (GRO-Seq).
(D) Radar plot showing the frequency of occupancy of CREB at enhancers and/or promoter sites of Reelin-regulated genes, compared to Reelin-independent genes.
(E) CREB and MEF2 co-bound genomic sites are visualized by their respective normalized ChIP-Seq tag counts (log2) within 200 bp of a given peak in Reelin-treated condition; top-enriched peaks overlapping with CBP are visualized in red.
(F) Profile plots of normalized ChIP-Seq tag intensities for the indicated transcriptional factors/cofactors. Each tag density profile is centered on TSS of Reelin-regulated genes or MEF2A binding peaks on distal H3K4me2-marked elements, including conventional enhancers and LRN enhancers. Additional 2kb from the center of each plot is shown. Schematic diagram of distinct regulatory regions around Reelin-induced genes is depicted in the top panel.
(G) Tag density profiles based on the same regulatory regions as in Fig 2F show the enrichment of active enhancer marks by ChIP-Seq and the induction of eRNA transcripts by GRO-Seq across the LRN enhancers.
See also Fig S2 and Fig S3
While long-term gene repression is maintained mainly by histone modifications associated with heterochromatic regions, signal-dependent regulation of gene transcription can be mediated by dynamic exchange of corepressor and coactivator complexes (Baek et al., 2002; Lee et al., 2001; Perissi et al., 2004; Perissi et al., 2010). We examined the possible role of the NCoR corepressor complex, a known component of the HDAC complexes, including HDAC4, which specifically regulates MEF2 TFs activation and is involved in learning and memory (Fitzsimons et al., 2013; Sando et al., 2012). NCoR ChIP-Seq showed that approximately 60% of NCoR binding sites colocalized with distal enhancers elements and, strikingly, Reelin treatment triggered a substantial dismissal of the NCoR corepressor machinery from these binding sites (Figure 2SB, Figure S2E). Accordingly, NCoR ability to bind to MEF2C is greatly impaired by Reelin treatment (Figure 2SG). Furthermore, ChIP-Seq tag distribution profiles of proximal or distal cis-regulatory elements revealed that these dynamic changes occur predominately at specific cohorts of enhancer sites activated by Reelin, as compared to most of the conventional neuronal enhancers that recruit MEF2 TFs (Figure 2F). Similar results were obtained when H3K27Ac-marked enhancers were analyzed (data not shown). These results permitted us to identify a signature of the Reelin-regulated enhancer code that we refer to as LRP8-Reelin-regulated neuronal (LRN) enhancers. We identified 738 distal sites showing the LRN enhancer signature based on CBP, CREB and MEF2 recruitment and NCoR dismissal (Figure S2H). Consistent with these results, the LRN enhancers correlated with the highest level of active histone marks (Figure 2G, Figure S3A) and with the most induced eRNA transcripts (Figure 2G). Recent studies demonstrated that eRNAs are locally generated by Pol II as bidirectional non-coding transcripts important in establishing enhancers function and in regulating gene transcription through various mechanisms (Kim et al., 2010; Lam et al., 2013; Li et al., 2013; Wang et al., 2011). These results provide evidence that combinatorial action of CREB and MEF2 TFs, together with a signaling-dependent switch of CBP and NCoR complexes, are hallmarks of LRN enhancers, which are themselves signal-responsive transcription units.
Reelin-induced γ-secretase-dependent release of LRP8 intracellular domain and NMDA-R activity
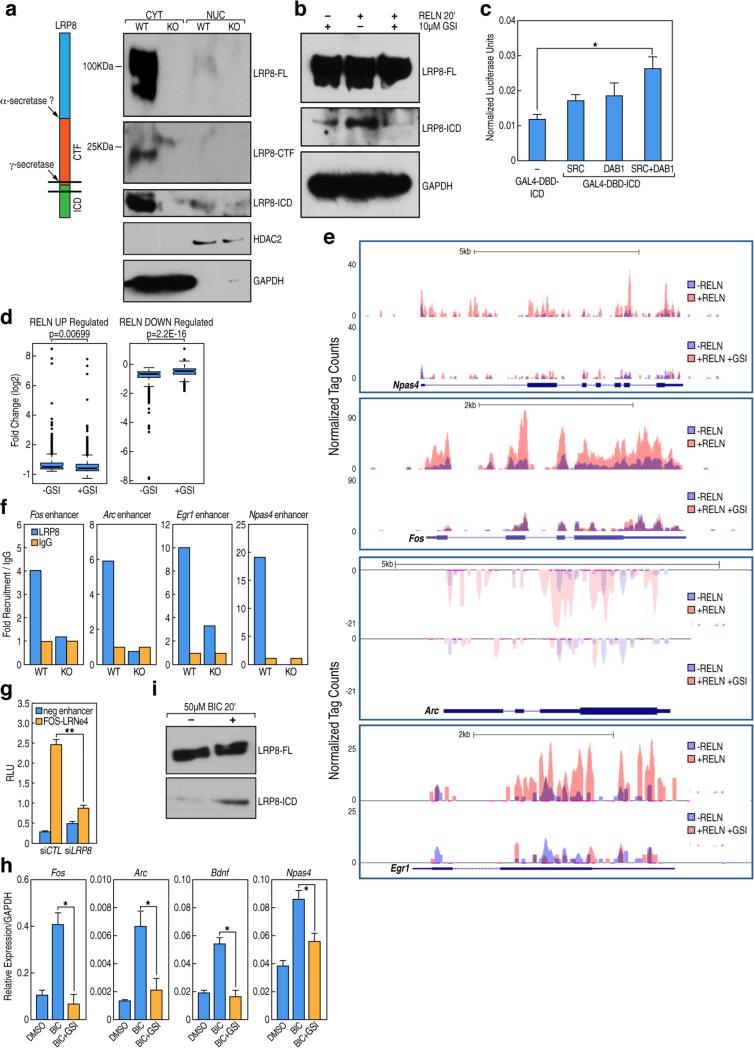
In neurons, synapse-to-nucleus communication is a widely used process for relaying extracellular stimuli to the nucleus. Various mechanisms of nucleo-cytoplasmic shuttling of synaptically enriched components have been reported, including the regulated intramembrane proteolysis of γ-secretase substrates that results in the release of an intracellular domain, which accumulates in the nucleus and acts as an intermediary in the transmission of extracellular stimuli (Selkoe and Wolfe, 2007). Because it is known that the Reelin receptor, LRP8, is a substrate of γ-secretase activity in vitro (Hoe and Rebeck, 2005; May et al., 2003), we investigated whether LRP8 undergoes this process in vivo in a signaling-dependent manner. We confirmed that the proteolytic processing of LRP8 occurs in vivo by using fractionated protein extracts of cortical brain dissections obtained from WT and LRP8-KO mice (Figure 3A, Figure S3B). We detected three major proteolytic products of LRP8, including the full-length mature form (105 kDa), the carboxy-terminal fragment generated by extracellular shedding (25 kDa) and the intracellular domain (ICD) released by intramembranous cleavage by γ-secretase activity (14 kDa), which translocates into the nuclear fraction. Treatment of cortical neurons with recombinant Reelin protein enhanced the release of LRP8-ICD that was specifically blocked by γ–secretase inhibitors (GSI) (Figure 3B). To begin to evaluate the involvement of LPR8-ICD in the transcriptional regulation, we used GAL4-based transactivation assays. When fused to the GAL4 DNA binding domain, LRP8-ICD showed a transactivation activity that was significantly enhanced by the co-expression of SRC and DAB1 (Figure 3C), suggesting that the core components of Reelin signaling are required to promote LRP8-ICD function in the nucleus. To further explore the involvement of γ-secretase activity in the regulation of Reelin-dependent transcriptomes, we performed RNA-Seq profiling of cortical neurons treated with Reelin in presence or absence of GSI. Blocking the γ-secretase activity was sufficient to abolish the Reelin-induced transcriptional changes (Figure 3D, Figure 3E). These results indicate a direct involvement of γ-secretase-mediated cleavage of LRP8 as an upstream step in the synapse-to-nucleus communication promoted by Reelin. Furthermore, LRP8-ICD was recruited to LRN enhancers (Figure 3F) and transient knockdown of LRP8 by siRNA in cortical neurons lead to substantial reduction of a reporter gene transcription driven by a FOS enhancer carrying the LRN enhancer signature (Figure 3G). Intriguingly, we observed that the activation of NMDA-R by bicuculline treatment does not overcome the lack of regulated intramembrane proteolysis of LRP8, as demonstrated by qRT-PCR experiments in which the inhibition of γ-secretase activity was sufficient to block NMDA-receptor-dependent transcriptional regulation (Figure 3H). Accordingly, bicuculline treatment enhanced the release of the LRP8-ICD (Figure 3I), indicating that there is a mutual interdependence between the LRP8 and NMDA receptors, both of which functionally interact at post-synaptic densities (Hoe et al., 2006). Our data suggest that the processing of LRP8 by γ-secretase activity represents a shared component of two signaling cascades initiated at the membrane level by ligand-induced stimulation. This crosstalk might ultimately serve as reinforcing mechanism to achieve optimal gene expression regulation.
Figure 3. Bidirectional regulation of Reelin-dependent transcriptome by γ-secretase-mediated release of LRP8-ICD and NMDA-R signaling.
(A) Western blot results of immunoprecipitation with anti-LRP8 antibody from brain cortex of WT and mutant LRP8-KO mice showing the proteolytical processing of LRP8; only the ICD fragment is present in the nuclear fraction. A diagram of the cleavage events is depicted on the left.
(B) Western blot data from protein extracts of cortical neurons showing that the processing of LRP8 is enhanced in the presence Reelin (10nM, 0.5 h) and abolished in the presence of 1uM GSI.
(C) Luciferase reporter assay showing the effect of the LRP8-ICD when fused to the Gal4 DNA-binding domain in the presence of SRC and/or DAB1. Relative luminescence units (RLU) were measured 24 h after transfection and values normalized to Firefly/Renilla. RLU are shown as means ±SD based on 3 individual experiments; *p<0.005 (Two-tailed students T-test).
(D) Box-and-whisker plots of expression values for genes up- or down-regulated by Reelin treatment showing the effect of the GSI. The y axis shows expression value in log2 of fold changes. The central horizontal bar indicates the median fold change.
(E) UCSC genome browser images showing the normalized RNA-Seq tag densities for Npas4, Fos, Arc and Egr1 genes in Reelin-stimulated neurons (10nM, 6h) in absence or presence of 1uM GSI.
(F) ChIP-qPCR showing the recruitment of LRP8 on specific enhancer elements in hippocampi from WT and mutant LRP8-KO mice. y axis represents fold recruitment values compared to control IgG ChIPs. Each sample represents a pool of hippocampal dissections from 4 individual animals.
(G) Effect of siLRP8 on the activity of enhancers driving a luciferase reporter gene compared to a control siRNA. Bars represent mean normalized values from three independent experiments ± SD, *P < 0.05 (two-tailed Student's t-test).
(H) Fold change in gene expression of representative coding genes quantified by RT-qPCR from cortical neurons stimulated with 50uM bicuculline for 6 h in presence or absence of GSI. Data represent mean ± SD *P < 0.05 (two-tailed Student's t-test).
(I) Western blot results from total protein extracts immunoprecipitated with anti-LRP8 antibodies in bicuculline-stimulated neurons (50uM, 0.5 h) in presence or absence of 1uM GSI.
See also Fig S3
LRN enhancers are required for the activation of Reelin target genes
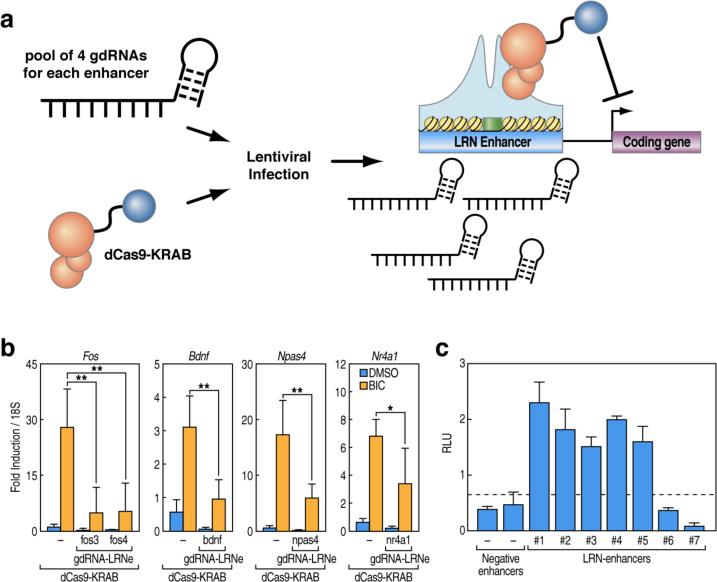
To gain further insights into the contribution of LRN enhancers in the regulation of Reelin target genes, we took advantage of a recently developed Cas9/CRISPR (clustered regularly interspaced short palindromic repeats) methodology (Cong et al., 2013), in which a catalytically inactive or “dead” Cas9 (dCas9) can be exploited as a platform for site-selective RNA-guided transcriptional regulation (Qi et al., 2013). We designed small guide RNAs (sgRNAs) targeting DNA sequences encoded in the LRN enhancers in proximity of key Reelin-dependent genes. For each enhancer, we multiplexed four different sgRNAs directed to the core region encompassing CREB and MEF2 binding sites. We co-expressed the sgRNAs by lentiviral infection of cortical neurons together with a version of dCAS9 protein fused to a transcriptional repression domain (the Krüppel-associated box (KRAB) domain), which efficiently triggers gene expression silencing if guided by a specific sgRNA (Gilbert et al., 2013; Maeder et al., 2013) (Figure 4A, Figure S4A). Using qRT-PCR experiments, we demonstrated that blocking the activity of LRN enhancers was sufficient to abolish the activation of Reelin-dependent genes (Figure 4B). Furthermore, using a luciferase reporter assay, we found that LRN enhancers confer an intrinsically higher activity compared to other enhancers (Figure 4C). These results provide strong evidence that the activation of LRN enhancers directly plays a pivotal role in orchestrating signal-dependent transcriptional regulation and is a crucial step in the synapse-to-nuclear signaling triggered by Reelin via NMDA-R.
Figure 4. LRN enhancers are required for transcriptional activation of target coding genes.
(A) Schematic describing the experimental CRISPR interference approach to inhibit LRN enhancer activity in cortical neurons. We hypothesize that the catalytically inactive CAS9 fused to the repression domain, KRAB, can interfere with enhancer activity and abolish the expression of cognate coding genes.
(B) dCas9-KRAB fusion protein in presence of gdRNAs directed against specific LRN enhancers efficiently silences expression of coding genes in transduced cortical neurons upon stimulation with bicuculline (50uM, 6h). qRT-PCR experiments show fold induction calculated based on three independent experiments as mean ± SD, *P<0.05.
(C) Assessment of enhancer activity by cloning several enhancer genomic regions (1kb) upstream of the luciferase reporter gene driven by the Fos minimal promoter. Bars represent mean normalized values from three independent experiments ± SD, *P < 0.05 (two-tailed Student's t-test).
See also Fig S4
LRN enhancers engage in long-range interactions with target promoters
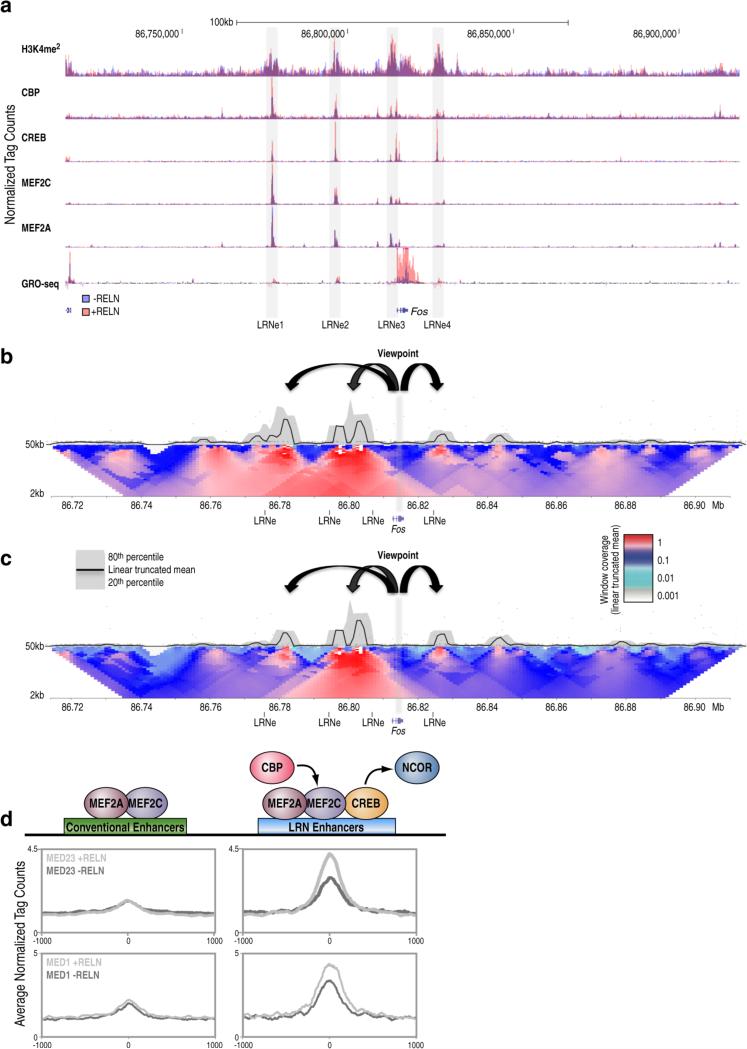
Physical communication between distal cis-regulatory sequences in the genome utilizes chromatin looping and serves as a three-dimensional platform for fine-tuned regulation of several cellular processes, including gene expression (Jin et al., 2013; Sanyal et al., 2012; Zhang et al., 2013). To further investigate the possibility that LRN enhancers engage in long-range interactions with target promoters of Reelin-induced genes we carried out 4C-sequencing (4C-Seq) experiments in cortical neurons. We took advantage of two cutter 4C-Seq methodology to identify specific looping interactions in an unbiased manner after defining a “view point” of interest positioned near a promoter site (van de Werken et al., 2012). We focused on the Fos gene locus due to its complex genomic architecture characterized by 4 distinct enhancers displaying the LRN enhancer signature (Figure 5A). Analysis of 4C contact profiles from duplicate experiments revealed that LRN enhancers engage in long-range interactions with target coding gene promoters (Figure 5B-S5A). Importantly, the chromatin architecture of the Fos locus was conserved in vivo, as shown by 4C contact profiles detected in the hippocampal regions of WT mice (Figure 5C-S5B), suggesting that precise three-dimensional architecture may be involved in modulation of brain function in vivo. Given that the 4C method does not allow for accurate measurement of looping dynamics, we hypothesized that mapping the mediator complex (Kagey et al., 2010) could begin to answer the question of whether activation of LRN enhancers relies on dynamic long-range interactions. Therefore, we carried out ChIP-Seq experiments for mediator components MED1 and MED23, which have been linked to intellectual disability (Hashimoto et al., 2011). The ChIP-seq analysis revealed that mediator complex is specifically recruited to LRN enhancers upon Reelin signaling (Figure 5D), suggesting that looping interactions of LRN enhancers and promoters serve as a three-dimensional platform that controls the actions of enhancers on their specific targets and integrate an additional layer of regulation to the molecular signature of Reelin-regulated cistromes.
Figure 5. LRN enhancers engage in long-term interactions with the promoter of their target gene.
(A) UCSC browser image of the Fos genomic locus showing normalized tag counts for GRO-Seq and indicated ChIP-Seqs; gray bars indicate the location of distinct LRN enhancers (e1-e4).
(B-C) Contact profiles depicting the genomic interactions of the Fos promoter (viewpoint, gray bar) in cortical neurons (B) or hippocampi (C). The black curve represents the normalized contact frequency calculated based on the linear-truncated mean values with a resolution of 1kb. The expected interactions of enhancers with the promoter (black arrows) are observed. Black bars indicate the coordinates of known LRN enhancers.
(D) Profiles of normalized ChIP-seq tags of MED23 and MED1 before and after Reelin treatment are shown for distinct enhancers regions.
See also Fig S5
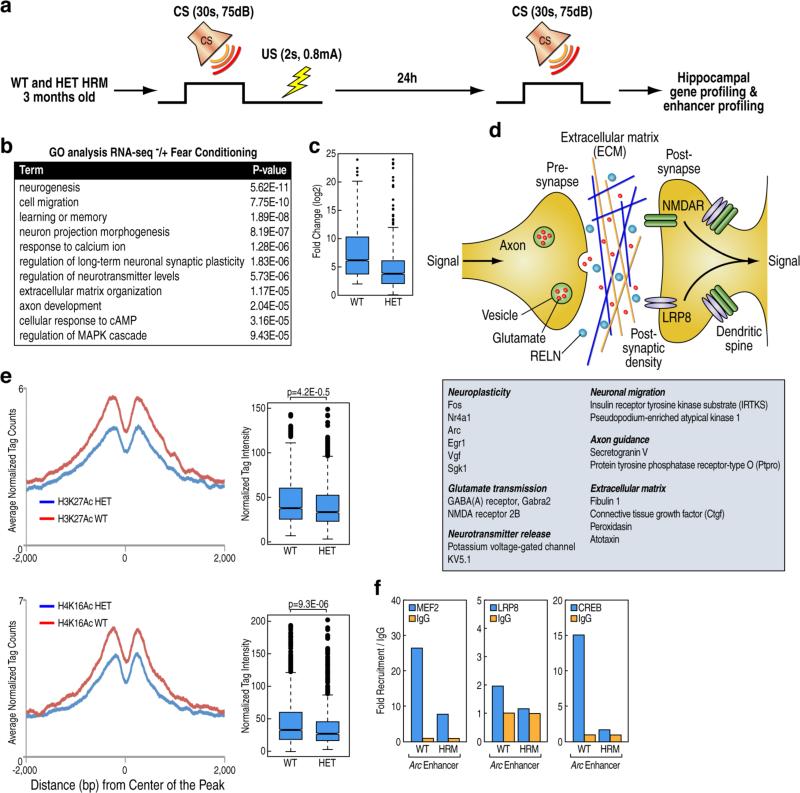
Reelin-dependent epigenomic events are required for hippocampal-dependent associative learning
Based on our results we hypothesized that Reelin-dependent transcriptional programs dictated by LRN enhancer activity may ultimately serve as an epigenetic mechanism underlying learning and memory formation. To trace memories at the molecular level, we examined the gene expression patterns induced by fear conditioning in hippocampal regions of WT and HRM by generating transcriptional profiles after 24 h of contextual training (Figure 6A). Analysis of differentially expressed genes identified 234 regulated genes in WT hippocampi, hereafter referred to as “learning genes” (Figure 6B, Figure S6A, Table S3). We observed a significant reduction of learning genes activation in the HRM, which exhibit a severe impairment in the execution of this behavioral task (Figure 6C, Figure 1G), providing clear evidence that intact Reelin signaling is required for the regulation of specific transcriptional programs during long-term memory formation. Of these induced transcripts, ~30% corresponded to those exhibiting robust transcriptional induction upon Reelin treatment (Table S3) and, intriguingly, corresponded to key genes important for neuroplasticity events and extracellular matrix (ECM). This result provides an intriguing perspective for future investigation given the well-documented function of ECM dysfunction in schizophrenia (Berretta, 2012) (Figure 6D).
Figure 6. Reelin nuclear signaling is misregulated during memory formation in vivo.
(A) Schematic describing the contextual fear conditioning paradigm used to isolate RNA and chromatin from hippocampi for global transcriptomic and epigenomic analysis.
(B) Functional gene ontology annotations associated with genes regulated during memory formation in WT mice.
(C) Box-and-whisker plots of normalized fold changes (log2) of genes in WT and HET Reeler mice 24 h after fear conditioning training. The y axis shows expression values in log2 of fold changes. The central horizontal bar indicates the median normalized tag distribution.
(D) Table shows representative learning-induced genes that are misregulated by the altered Reelin signaling in the heterozygous Reeler mice. A schematic of the synaptic processes linked to the Reelin-learning-dependent genes is depicted on the top panel.
(E) Distribution of normalized tag densities of ChIP-seq of indicated histone marks around LRN enhancers elements in hippocampi from naïve WT and HET Reeler mice. Box-and-whisker plots show ChIP-Seq tag counts (log2, y axis) in WT and HET Reeler mice. The central horizontal bar indicates the median normalized tag distribution. Each sample represents a pool of hippocampal dissections from 4 individual animals.
(F) ChIP-qPCR showing loss of recruitment of MEF2C, LRP8 and CREB at the indicated genomic locations in hippocampi isolated from WT and HET Reeler mice. Each sample represents a pool of hippocampal dissections from 4 individual animals.
See also Fig S6
To further investigate the extent to which LRN enhancers activity contributes to the cognitive deficits of HRM, we mapped the enhancer landscape in the hippocampi of WT and HRM. The chromatin state of LRN enhancers, labeled by active enhancer marks (H3K27Ac and H4K16Ac), was significantly impaired in absence of intact Reelin signaling (Figure 6E). Accordingly, we showed by conventional ChIP that the recruitment of LRN enhancer-associated transcriptional regulators was strongly altered (Figure 6F), suggesting that LRN enhancers activity is crucial for licensing appropriate transcriptional regulation in the brain in response to environmental stimuli.
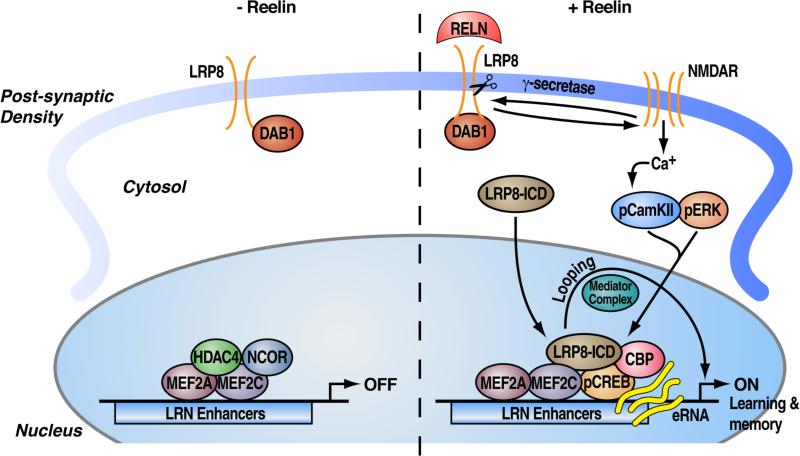
Together our results uncover a novel molecular mechanism that enables the information delivered by Reelin at the synapses to be transferred to the nucleus, where it is deciphered by transcriptional/co-factors complexes at the level of specific cis-regulatory elements, which drive the expression of neuroplasticity genes important for learning and memory in vivo. This long-distance communication signaling involves the γ–secretase-dependent release of the LRP8-ICD, which is required for calcium influx through NMDA receptor and is essential to drive gene expression within the nucleus (Figure 7).
Figure 7. Synapse-to-nucleus signaling triggered by Reelin converges on the activation of LRN enhancers.
(Model) Reelin activates a synapse-to-nuclear signaling by triggering the proteolytical processing of its receptor LRP8 by γ–secretase activity. The regulated intramembrane proteolysis of LRP8 is bidirectionally regulated by the activation of the NMDA-R signaling. The LRP8-ICD is released from the membrane and translocates into the nucleus where it takes part to the regulation of epigenomic events centered on a specific cohort of enhancers that recruit the neuronal transcription factors MEF2 and CREB. LRN enhancer activity depends on the switch of a CBP co-activator complex and NCoR corepressor complex. Long-range interactions between LRN enhancer elements and target promoters involve the mediator complex. LRN enhancers are required for the activation of synaptic plasticity genes that are regulated during memory formation in vivo.
Discussion
Reelin signaling promotes activation of transcriptional programs linked to learning and memory
In the past three decades, groundbreaking studies from Aplysia, and subsequently from mammalian models have clearly demonstrated that long-term plasticity critically depends on transcriptional activation and protein synthesis of activity-dependent genes, which are robustly turned on by the activity of CREB (Kaang et al., 1993; Schacher et al., 1988; Sheng et al., 1990). However, the underpinning epigenomic mechanisms and their link to behavioral outputs have yet to be resolved. Here, we expand our understanding of activity-dependent transcriptional activation by utilizing the power of genomic technologies, which enable new perspectives in the understanding of the molecular mechanisms of neuroplasticity. We provide clear evidence that the Reelin-LRP8 pathway is required for hippocampal-dependent associative learning and is directly linked to epigenomic events that support memory formation. We used GRO-Seq to reveal that stimulation of cortical neurons with Reelin results in the rapid activation of a transcriptome linked to learning and memory. Furthermore, the genetic ablation of Reelin signaling in vivo was strongly correlated with lack of specific transcriptional responses in contextual fear conditioning paradigms. Particularly interesting is the connection with genes involved in the extracellular matrix that controls a variety of neuronal processes that are altered in schizophrenia. While our study has predominantly focused on hippocampus-dependent plasticity, we do not exclude the possibility that Reelin signaling might be involved in similar and/or distinct mechanisms in other areas of the brain implicated in the induction of memory traces, such as the amydgala or the cortical regions. While our data revealed an extensive hippocampal transcriptional response to conditioned learning stimuli, it will be of particular interest in the future to elucidate the contribution of functionally distinct neuronal subtypes to the regulation of specific gene networks. Indeed, due to the technical obstacles imposed by tissue heterogeneity, only substantial changes would be recorded in our hippocampal transcriptomes, while changes of cell-type-specific transcripts might be more difficult to detect.
Crosstalk between LRP8 regulated intramembrane proteolysis and NMDA receptor-dependent nuclear signaling
Unexpectedly, we found that proteolytical cleavage of LRP8 is a crucial component of the synapse-to-nuclear signaling triggered by Reelin. Further investigation is needed to elucidate which nucleo-cytoplasmic shuttling mechanism is utilized by LRP8-ICD. Spatial and temporal constraints of intracellular trafficking represent future challenges to be addressed, given the current technical difficulties in tracking few molecules across long distances dictated by the unique neuronal morphology. However, we have begun to elucidate the mechanism by which synapses targeted by Reelin communicate with the nuclear compartment. We report that the LRP8 proteolysis and NMDA-R activation are reciprocally regulated in a signaling-dependent manner. This intriguing result confers on Reelin signaling a pivotal role in the regulation of excitatory glutamatergic transmission. The sustained hypofunction of glutamatergic signaling via NMDA receptor is considered one of the major mechanistic hypotheses of schizophrenia (Veerman et al., 2014). Our results provide a candidate mechanism explaining how the reduction in Reelin expression observed in schizophrenic patients could contribute to the development of cognitive impairment of the disorder.
LRN enhancers as a novel epigenomic signature underlying the synaptic plasticity events triggered by Reelin
By combining genome-wide epigenetic profiling with de novo motif analysis, measurement of eRNA transcripts and assessment of three-dimensional architecture, we identified enhancer regulatory elements that serve as a signal-responsive genomic signature for learning and memory. Those LRN enhancers are functionally linked to neuroplasticity events initiated by Reelin and participate in rapid and robust activation of key target genes. Recent studies have estimated that a striking fraction of loci identified in genome-wide association studies are enriched in non-coding sequences (Maher, 2012; Visel et al., 2009). These large-scale projects aim to elucidate the genetic architecture of human disease and corroborate our appreciation of the importance of cis-regulatory elements, particularly enhancers, as a widespread feature of common and complex diseases susceptibility, but will require not only the global mapping of enhancers, but also the elucidation of the precise molecular mechanisms that regulate their function. In this context, our study provides an example of a biological pathway relevant to cognition that is directly linked to a genomic regulatory network of enhancer elements, which is required for proper transcriptional regulation of key synaptic plasticity genes. The link of LRN enhancers to human sequence variations identified in neuropsychiatric disorders remains to be explored.
EXPERIMENTAL PROCEDURES
All animal care and experimental procedures were in accordance with the University of California, San Diego research guidelines for the care and use of laboratory animals. See also Supplemental Information.
Primary Cortical Neurons and Cell Cultures
Mixed cortical cultures were established by dissecting E15.5 mouse embryos as previously described (Jossin et al., 2007). Cultures were maintained for 10-11 in vitro before treatment. N2A, HEK-293-T and HEK-293 lacking N-acetylglucosaminyltransferase I (GnTI) activity (HEK293S GnTI) were obtained from ATCC. Neuronal cultures were treated with recombinant Reelin that was purified as described in Supplemental Information. Luciferase assay, immunoprecipitation and fractionated cell extracts were performed as described in Extended Experimental Procedures.
Reagents and Antibodies
The antibodies used include: LRP8 (ab108208), CBP (ab2832), H3K27Ac (ab4729), MEF2C (ab79436) from Abcam; H3K4me2 (07-030), H4K16Ac (07-329), CREB (17-600), phopho-CREB-(Ser133) (17-10131) from Millipore; MED23 (NB200-337) from Novus; p-CaMKII (sc-32289), GAPDH (sc-365062), MEF2 (sc-313) from Santa Cruz Biotechnology; p-HDAC4-Ser632 (#3424), from CellSignaling Technologies; MED1 (A300-793A) from Bethyl Laboratories. The rabbit anti-N-CoR antibody was generated as described previously (Heinzel et al., 1997). The chemical reagents used include: CNQX (#0190), D-AP5 (#0106), Bicuculline (#0130) from Tocris; PP2 (#529573), KN62 (#422706), γ-secretase Inhibitor II (#565755) from Millipore.
RNA purification and RNA-seq
RNA was isolated using Trizol (Life Technologies) for hippocampal dissections or RNeasy column (Qiagen) for cortical neurons. RNA was analyzed for purity using the Agilent 2100 Bioanalyser (Agilent). RNA samples from 4 littermates for each treatment and genetic background were pooled and used for library preparations. RNA-seq library preparations were performed using Ribo-Zero™ rRNA Removal Kits and ScriptSeq™ v2 RNA-Seq Library Preparation Kit, according to manufacturing procedures (Epicentre). The ScriptSeq™ Index PCR Primers (Epicentre) were used to add an Illumina® barcode to an RNA-Seq libraries during PCR amplification using the FailSafe PCR Polymerase (Epicentre).
GRO-Seq libraries preparation
GRO-Seq experiments were performed as previously reported (Core et al., 2008; Li et al., 2013). Briefly, nuclei were prepared form ~20 millions of cortical neurons. For the run-on assay, nuclear-run-on-RNA (NRO-RNA) was extracted with TRIzol LS reagent (Life Technologies) following manufacturer's instructions. NRO-RNA was fragmented to ~300-500nt by alkaline base hydrolysis and followed by treatment with DNase I and antarctic phosphatase. At this step, anti-BrdU argarose beads (Santa Cruz Biotechnology) were utilized to purify the Br-UTP labeled nascent RNA. Fragmented RNA was end-repaired by T4 PNK and purified by acidic phenol-chloroform extraction. cDNA synthesis was performed as previously described (Ingolia et al., 2009) with few modifications. The RNA fragments were subjected to poly-A tailing, followed by reverse transcription using oNTI223 primer and superscript III RT kit (Life Technologies). The cDNA products were size-selected by gel excision using a 10% polyacrylamide TBE-urea gel (~100-500bp). The first-strand cDNA was circularized by CircLigase (Epicentre) and re-linearized by APE1 (NEB) (sscDNA) and then separated/excised by a 10% polyacrylamide TBE gel as described above (~120-320bp). Finally, sscDNA template was amplified by PCR using the Phusion High-Fidelity enzyme (NEB). The oligonucleotide primers oNTI200 and oNTI201 were used to generate DNA for deep sequencing (GRO-seq primer sequences are included in Table S2).
ChIP, ChIP-seq library preparations
ChIP was performed as previously described with some modifications (Johnson et al., 2007). Briefly, ~20 millions neurons were double cross-linked with 2mM DSG (ProteoChem) for 45 minutes and then for another 10 minutes with 1% formaldehyde. Soluble chromatin was fragmented using a Bioruptor® sonicator (~200-400bp). Soluble chromatin was incubated at 4C overnight with 2-5μg antibodies pre-bound to 20μl Dynabeads Protein G (Life Technologies). After washing, ChIP-ed DNA was eluted/de-crosslinked at 65C for 4 hours and purified using iPureLink kit (Diagenode). For ChIP-seq, the DNA libraries were constructed following Illumina's ChIP-seq Sample prep kit (following manufacturer's instructions). For ChIPs conducted on hippocampal microdissections, a similar protocol described for the nuclear/cytosolic protein extraction was performed to obtain single cell suspensions before cross-linking. For ChIP-qPCR experiments from LRP8-KO or HRM mice, chromatin extracted from hippocampal microdissections of 4 individual animals was pooled to perform ChIP for each antibody.
4C-seq libraries preparation
Chromosome confirmation capture was performed as described previously (Lieberman-Aiden et al., 2009). Briefly, 20 million cells were used for each 4C. For 4C-Seqs conducted on hippocampal microdissections, a similar protocol described for the nuclear/cytosolic protein extraction was performed to obtain single cell suspensions before cross-linking. Cells were crosslinked with 1% formaldehyde for 10 minutes. Soluble chromatin was subjected to restriction endonuclease digestion overnight, using 400 units of HindIII (NEB). Further, intra-molecular ligations were supported using 1000 units of T4 DNA ligase (NEB) for 4 hours at 16°C. Then chromatin was de-crosslinked at 65°C and purified. Second restriction digestion was performed overnight, using 50 units of DpnII (NEB). Subsequently, intra-molecular interactions were ligated overnight. Chromatin was subjected to RNaseA digestion, purified using Qiagen columns and subjected to PCR (Expand long range PCR system, Roche diagnostics) using a first primer designed on the viewpoint and a second outer primer designed beside the DpnII site (Table S2). Both primers contained illumina sequencing adapters and Barcodes for multiplexing. Approximately 16 reactions were performed for each viewpoint to generate complex libraries.
Detailed bioinformatics methods for the analysis of deep-dequencing experiments were performed as described in Supplemental Information.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Janet Hightower for assistance with figure preparation, to Dr. Amanda Roberts from the Mouse Behavioral Assessment Core at the Scripps Research Institute. We thank Dr Mobley for critical reading of the manuscript. M.G.R. is an investigator with the Howard Hughes Medical Institute. This work was supported by DK018477, NS034934, DK039949, HL065445, REF OF SUH and CA173903 from NIH and NCI to M.G.R; grants from Roche Extending Innovation Network Program to F.T.; and a DOD fellowship to D.N.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
M.G.R. and F.T. conceived the project and wrote the manuscript. F.T conducted the majority of the experiments with participation of D.N for 4C-seq experiments; S.O. and W.Li for GRO-Seq experiments. Q.M. performed the bioinformatic analyses with contributions from F.T. Technical assistance for cloning, luciferase and lentiviral infection experiments was provided by P.M.P. D.C. conducted Reelin recombinant protein purification. Technical assistance for deep-sequencing was provided by K.O., and H.T. contributed to mice breeding and handling.
Accession Numbers
The NCBI Gene Expression Omnibus accession number for the sequencing data reported in this paper is GSE66710.
The authors declare no conflict of interests.
References
- Akhtar MW, Kim MS, Adachi M, Morris MJ, Qi X, Richardson JA, Bassel-Duby R, Olson EN, Kavalali ET, Monteggia LM. In vivo analysis of MEF2 transcription factors in synapse regulation and neuronal survival. PLoS One. 2012;7:e34863. doi: 10.1371/journal.pone.0034863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek SH, Ohgi KA, Rose DW, Koo EH, Glass CK, Rosenfeld MG. Exchange of N-CoR corepressor and Tip60 coactivator complexes links gene expression by NF-kappaB and beta-amyloid precursor protein. Cell. 2002;110:55–67. doi: 10.1016/s0092-8674(02)00809-7. [DOI] [PubMed] [Google Scholar]
- Barbosa AC, Kim MS, Ertunc M, Adachi M, Nelson ED, McAnally J, Richardson JA, Kavalali ET, Monteggia LM, Bassel-Duby R, et al. MEF2C, a transcription factor that facilitates learning and memory by negative regulation of synapse numbers and function. Proc Natl Acad Sci U S A. 2008;105:9391–9396. doi: 10.1073/pnas.0802679105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beffert U, Durudas A, Weeber EJ, Stolt PC, Giehl KM, Sweatt JD, Hammer RE, Herz J. Functional dissection of Reelin signaling by site-directed disruption of Disabled-1 adaptor binding to apolipoprotein E receptor 2: distinct roles in development and synaptic plasticity. J Neurosci. 2006;26:2041–2052. doi: 10.1523/JNEUROSCI.4566-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beffert U, Morfini G, Bock HH, Reyna H, Brady ST, Herz J. Reelin-mediated signaling locally regulates protein kinase B/Akt and glycogen synthase kinase 3beta. J Biol Chem. 2002;277:49958–49964. doi: 10.1074/jbc.M209205200. [DOI] [PubMed] [Google Scholar]
- Beffert U, Weeber EJ, Durudas A, Qiu S, Masiulis I, Sweatt JD, Li WP, Adelmann G, Frotscher M, Hammer RE, et al. Modulation of synaptic plasticity and memory by Reelin involves differential splicing of the lipoprotein receptor Apoer2. Neuron. 2005;47:567–579. doi: 10.1016/j.neuron.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Bernard A, Lubbers LS, Tanis KQ, Luo R, Podtelezhnikov AA, Finney EM, McWhorter MM, Serikawa K, Lemon T, Morgan R, et al. Transcriptional architecture of the primate neocortex. Neuron. 2012;73:1083–1099. doi: 10.1016/j.neuron.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berretta S. Extracellular matrix abnormalities in schizophrenia. Neuropharmacology. 2012;62:1584–1597. doi: 10.1016/j.neuropharm.2011.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Chen Y, Beffert U, Ertunc M, Tang TS, Kavalali ET, Bezprozvanny I, Herz J. Reelin modulates NMDA receptor activity in cortical neurons. J Neurosci. 2005;25:8209–8216. doi: 10.1523/JNEUROSCI.1951-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium EP, Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Arcangelo G, Miao GG, Chen SC, Soares HD, Morgan JI, Curran T. A protein related to extracellular matrix proteins deleted in the mouse mutant reeler. Nature. 1995;374:719–723. doi: 10.1038/374719a0. [DOI] [PubMed] [Google Scholar]
- Deisseroth K, Bito H, Tsien RW. Signaling from synapse to nucleus: postsynaptic CREB phosphorylation during multiple forms of hippocampal synaptic plasticity. Neuron. 1996;16:89–101. doi: 10.1016/s0896-6273(00)80026-4. [DOI] [PubMed] [Google Scholar]
- Dietrich JB. The MEF2 family and the brain: from molecules to memory. Cell Tissue Res. 2013;352:179–190. doi: 10.1007/s00441-013-1565-2. [DOI] [PubMed] [Google Scholar]
- Fitzsimons HL, Schwartz S, Given FM, Scott MJ. The histone deacetylase HDAC4 regulates long-term memory in Drosophila. PLoS One. 2013;8:e83903. doi: 10.1371/journal.pone.0083903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell SW, Greenberg ME. Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu Rev Neurosci. 2008;31:563–590. doi: 10.1146/annurev.neuro.31.060407.125631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell SW, Kim TK, Gray JM, Harmin DA, Hemberg M, Hong EJ, Markenscoff-Papadimitriou E, Bear DM, Greenberg ME. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron. 2008;60:1022–1038. doi: 10.1016/j.neuron.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folsom TD, Fatemi SH. The involvement of Reelin in neurodevelopmental disorders. Neuropharmacology. 2013;68:122–135. doi: 10.1016/j.neuropharm.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Wang WY, Mao YW, Graff J, Guan JS, Pan L, Mak G, Kim D, Su SC, Tsai LH. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature. 2010;466:1105–1109. doi: 10.1038/nature09271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez GA, Montminy MR. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell. 1989;59:675–680. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- Graff J, Joseph NF, Horn ME, Samiei A, Meng J, Seo J, Rei D, Bero AW, Phan TX, Wagner F, et al. Epigenetic priming of memory updating during reconsolidation to attenuate remote fear memories. Cell. 2014;156:261–276. doi: 10.1016/j.cell.2013.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff J, Rei D, Guan JS, Wang WY, Seo J, Hennig KM, Nieland TJ, Fass DM, Kao PF, Kahn M, et al. An epigenetic blockade of cognitive functions in the neurodegenerating brain. Nature. 2012;483:222–226. doi: 10.1038/nature10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman SR, Andersen KG, Shlyakhter I, Tabrizi S, Winnicki S, Yen A, Park DJ, Griesemer D, Karlsson EK, Wong SH, et al. Identifying recent adaptations in large-scale genomic data. Cell. 2013;152:703–713. doi: 10.1016/j.cell.2013.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Ma DK, Mo H, Ball MP, Jang MH, Bonaguidi MA, Balazer JA, Eaves HL, Xie B, Ford E, et al. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat Neurosci. 2011;14:1345–1351. doi: 10.1038/nn.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Kim SY, Artis S, Molfese DL, Schumacher A, Sweatt JD, Paylor RE, Lubin FD. Histone methylation regulates memory formation. J Neurosci. 2010;30:3589–3599. doi: 10.1523/JNEUROSCI.3732-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hah N, Murakami S, Nagari A, Danko CG, Kraus WL. Enhancer transcripts mark active estrogen receptor binding sites. Genome Res. 2013;23:1210–1223. doi: 10.1101/gr.152306.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto S, Boissel S, Zarhrate M, Rio M, Munnich A, Egly JM, Colleaux L. MED23 mutation links intellectual disability to dysregulation of immediate early gene expression. Science. 2011;333:1161–1163. doi: 10.1126/science.1206638. [DOI] [PubMed] [Google Scholar]
- Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, van de Lagemaat LN, Smith KA, Ebbert A, Riley ZL, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzel T, Lavinsky RM, Mullen TM, Soderstrom M, Laherty CD, Torchia J, Yang WM, Brard G, Ngo SD, Davie JR, et al. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- Hoe HS, Pocivavsek A, Chakraborty G, Fu Z, Vicini S, Ehlers MD, Rebeck GW. Apolipoprotein E receptor 2 interactions with the N-methyl-D-aspartate receptor. J Biol Chem. 2006;281:3425–3431. doi: 10.1074/jbc.M509380200. [DOI] [PubMed] [Google Scholar]
- Hoe HS, Rebeck GW. Regulation of ApoE receptor proteolysis by ligand binding. Brain Res Mol Brain Res. 2005;137:31–39. doi: 10.1016/j.molbrainres.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Jin F, Li Y, Dixon JR, Selvaraj S, Ye Z, Lee AY, Yen CA, Schmitt AD, Espinoza CA, Ren B. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- Jossin Y, Gui L, Goffinet AM. Processing of Reelin by embryonic neurons is important for function in tissue but not in dissociated cultured neurons. J Neurosci. 2007;27:4243–4252. doi: 10.1523/JNEUROSCI.0023-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaang BK, Kandel ER, Grant SG. Activation of cAMP-responsive genes by stimuli that produce long-term facilitation in Aplysia sensory neurons. Neuron. 1993;10:427–435. doi: 10.1016/0896-6273(93)90331-k. [DOI] [PubMed] [Google Scholar]
- Kaas GA, Zhong C, Eason DE, Ross DL, Vachhani RV, Ming GL, King JR, Song H, Sweatt JD. TET1 controls CNS 5-methylcytosine hydroxylation, active DNA demethylation, gene transcription, and memory formation. Neuron. 2013;79:1086–1093. doi: 10.1016/j.neuron.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel ER, Dudai Y, Mayford MR. The molecular and systems biology of memory. Cell. 2014;157:163–186. doi: 10.1016/j.cell.2014.03.001. [DOI] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam MT, Cho H, Lesch HP, Gosselin D, Heinz S, Tanaka-Oishi Y, Benner C, Kaikkonen MU, Kim AS, Kosaka M, et al. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature. 2013;498:511–515. doi: 10.1038/nature12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Lee YC, Na SY, Jung DJ, Lee SK. Transcriptional coregulators of the nuclear receptor superfamily: coactivators and corepressors. Cellular and molecular life sciences : CMLS. 2001;58:289–297. doi: 10.1007/PL00000856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Li W, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY, Merkurjev D, Zhang J, Ohgi K, Song X, et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498:516–520. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchesi W, Mizuno K, Giese KP. Novel insights into CaMKII function and regulation during memory formation. Brain research bulletin. 2011;85:2–8. doi: 10.1016/j.brainresbull.2010.10.009. [DOI] [PubMed] [Google Scholar]
- Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK. CRISPR RNA-guided activation of endogenous human genes. Nature methods. 2013;10:977–979. doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher B. ENCODE: The human encyclopaedia. Nature. 2012;489:46–48. doi: 10.1038/489046a. [DOI] [PubMed] [Google Scholar]
- Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May D, Blow MJ, Kaplan T, McCulley DJ, Jensen BC, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, et al. Large-scale discovery of enhancers from human heart tissue. Nat Genet. 2012;44:89–93. doi: 10.1038/ng.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May P, Bock HH, Nimpf J, Herz J. Differential glycosylation regulates processing of lipoprotein receptors by gamma-secretase. J Biol Chem. 2003;278:37386–37392. doi: 10.1074/jbc.M305858200. [DOI] [PubMed] [Google Scholar]
- Miller CA, Gavin CF, White JA, Parrish RR, Honasoge A, Yancey CR, Rivera IM, Rubio MD, Rumbaugh G, Sweatt JD. Cortical DNA methylation maintains remote memory. Nat Neurosci. 2010;13:664–666. doi: 10.1038/nn.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg S, Sananbenesi F, Zovoilis A, Burkhardt S, Bahari-Javan S, Agis-Balboa RC, Cota P, Wittnam JL, Gogol-Doering A, Opitz L, et al. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science. 2010;328:753–756. doi: 10.1126/science.1186088. [DOI] [PubMed] [Google Scholar]
- Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- Perissi V, Aggarwal A, Glass CK, Rose DW, Rosenfeld MG. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell. 2004;116:511–526. doi: 10.1016/s0092-8674(04)00133-3. [DOI] [PubMed] [Google Scholar]
- Perissi V, Jepsen K, Glass CK, Rosenfeld MG. Deconstructing repression: evolving models of co-repressor action. Nat Rev Genet. 2010;11:109–123. doi: 10.1038/nrg2736. [DOI] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu S, Korwek KM, Pratt-Davis AR, Peters M, Bergman MY, Weeber EJ. Cognitive disruption and altered hippocampus synaptic function in Reelin haploinsufficient mice. Neurobiol Learn Mem. 2006a;85:228–242. doi: 10.1016/j.nlm.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Qiu S, Zhao LF, Korwek KM, Weeber EJ. Differential reelin-induced enhancement of NMDA and AMPA receptor activity in the adult hippocampus. J Neurosci. 2006b;26:12943–12955. doi: 10.1523/JNEUROSCI.2561-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasethupathy P, Antonov I, Sheridan R, Frey S, Sander C, Tuschl T, Kandel ER. A role for neuronal piRNAs in the epigenetic control of memory-related synaptic plasticity. Cell. 2012;149:693–707. doi: 10.1016/j.cell.2012.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice DS, Curran T. Role of the reelin signaling pathway in central nervous system development. Annu Rev Neurosci. 2001;24:1005–1039. doi: 10.1146/annurev.neuro.24.1.1005. [DOI] [PubMed] [Google Scholar]
- Ripke S, O'Dushlaine C, Chambert K, Moran JL, Kahler AK, Akterin S, Bergen SE, Collins AL, Crowley JJ, Fromer M, et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet. 2013;45:1150–1159. doi: 10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers JT, Rusiana I, Trotter J, Zhao L, Donaldson E, Pak DT, Babus LW, Peters M, Banko JL, Chavis P, et al. Reelin supplementation enhances cognitive ability, synaptic plasticity, and dendritic spine density. Learn Mem. 2011;18:558–564. doi: 10.1101/lm.2153511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudenko A, Dawlaty MM, Seo J, Cheng AW, Meng J, Le T, Faull KF, Jaenisch R, Tsai LH. Tet1 is critical for neuronal activity-regulated gene expression and memory extinction. Neuron. 2013;79:1109–1122. doi: 10.1016/j.neuron.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudy JW, Huff NC, Matus-Amat P. Understanding contextual fear conditioning: insights from a two-process model. Neuroscience and biobehavioral reviews. 2004;28:675–685. doi: 10.1016/j.neubiorev.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Sando R, 3rd, Gounko N, Pieraut S, Liao L, Yates J, 3rd, Maximov A. HDAC4 governs a transcriptional program essential for synaptic plasticity and memory. Cell. 2012;151:821–834. doi: 10.1016/j.cell.2012.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schacher S, Castellucci VF, Kandel ER. cAMP evokes long-term facilitation in Aplysia sensory neurons that requires new protein synthesis. Science. 1988;240:1667–1669. doi: 10.1126/science.2454509. [DOI] [PubMed] [Google Scholar]
- Selkoe DJ, Wolfe MS. Presenilin: running with scissors in the membrane. Cell. 2007;131:215–221. doi: 10.1016/j.cell.2007.10.012. [DOI] [PubMed] [Google Scholar]
- Shalin SC, Zirrgiebel U, Honsa KJ, Julien JP, Miller FD, Kaplan DR, Sweatt JD. Neuronal MEK is important for normal fear conditioning in mice. Journal of neuroscience research. 2004;75:760–770. doi: 10.1002/jnr.20052. [DOI] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng M, McFadden G, Greenberg ME. Membrane depolarization and calcium induce c-fos transcription via phosphorylation of transcription factor CREB. Neuron. 1990;4:571–582. doi: 10.1016/0896-6273(90)90115-v. [DOI] [PubMed] [Google Scholar]
- Shirayama M, Seth M, Lee HC, Gu W, Ishidate T, Conte D, Jr., Mello CC. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell. 2012;150:65–77. doi: 10.1016/j.cell.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlyueva D, Stampfel G, Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet. 2014;15:272–286. doi: 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- Telese F, Gamliel A, Skowronska-Krawczyk D, Garcia-Bassets I, Rosenfeld MG. “Seq-ing” insights into the epigenetics of neuronal gene regulation. Neuron. 2013;77:606–623. doi: 10.1016/j.neuron.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trommsdorff M, Gotthardt M, Hiesberger T, Shelton J, Stockinger W, Nimpf J, Hammer RE, Richardson JA, Herz J. Reeler/Disabled-like disruption of neuronal migration in knockout mice lacking the VLDL receptor and ApoE receptor 2. Cell. 1999;97:689–701. doi: 10.1016/s0092-8674(00)80782-5. [DOI] [PubMed] [Google Scholar]
- Trotter J, Lee GH, Kazdoba TM, Crowell B, Domogauer J, Mahoney HM, Franco SJ, Muller U, Weeber EJ, D'Arcangelo G. Dab1 is required for synaptic plasticity and associative learning. J Neurosci. 2013;33:15652–15668. doi: 10.1523/JNEUROSCI.2010-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Werken HJ, Landan G, Holwerda SJ, Hoichman M, Klous P, Chachik R, Splinter E, Valdes-Quezada C, Oz Y, Bouwman BA, et al. Robust 4C-seq data analysis to screen for regulatory DNA interactions. Nature methods. 2012;9:969–972. doi: 10.1038/nmeth.2173. [DOI] [PubMed] [Google Scholar]
- Veerman SR, Schulte PF, de Haan L. The Glutamate Hypothesis: A Pathogenic Pathway from which Pharmacological Interventions have Emerged. Pharmacopsychiatry. 2014;47:121–130. doi: 10.1055/s-0034-1383657. [DOI] [PubMed] [Google Scholar]
- Visel A, Rubin EM, Pennacchio LA. Genomic views of distant-acting enhancers. Nature. 2009;461:199–205. doi: 10.1038/nature08451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Taher L, Girgis H, May D, Golonzhka O, Hoch RV, McKinsey GL, Pattabiraman K, Silberberg SN, Blow MJ, et al. A high-resolution enhancer atlas of the developing telencephalon. Cell. 2013;152:895–908. doi: 10.1016/j.cell.2012.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang AH, Kruhlak MJ, Wu J, Bertos NR, Vezmar M, Posner BI, Bazett-Jones DP, Yang XJ. Regulation of histone deacetylase 4 by binding of 14-3-3 proteins. Molecular and cellular biology. 2000;20:6904–6912. doi: 10.1128/mcb.20.18.6904-6912.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474:390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeber EJ, Beffert U, Jones C, Christian JM, Forster E, Sweatt JD, Herz J. Reelin and ApoE receptors cooperate to enhance hippocampal synaptic plasticity and learning. J Biol Chem. 2002;277:39944–39952. doi: 10.1074/jbc.M205147200. [DOI] [PubMed] [Google Scholar]
- West AE, Greenberg ME. Neuronal activity-regulated gene transcription in synapse development and cognitive function. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H, Shen EH, Hohmann JG, Oh SW, Bernard A, Royall JJ, Glattfelder KJ, Sunkin SM, Morris JA, Guillozet-Bongaarts AL, et al. Large-scale cellular-resolution gene profiling in human neocortex reveals species-specific molecular signatures. Cell. 2012;149:483–496. doi: 10.1016/j.cell.2012.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wong CH, Birnbaum RY, Li G, Favaro R, Ngan CY, Lim J, Tai E, Poh HM, Wong E, et al. Chromatin connectivity maps reveal dynamic promoter-enhancer long-range associations. Nature. 2013;504:306–310. doi: 10.1038/nature12716. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.