Abstract
The prefrontal cortex (PFC), seat of the highest-order cognitive functions, constitutes a conglomerate of highly specialized brain areas and has been implicated to have a role in the onset and installation of various neurodevelopmental disorders. The development of a properly functioning PFC is directed by transcription factors, guidance cues and other regulatory molecules and requires the intricate and temporal orchestration of a number of developmental processes. Disturbance or failure of any of these processes causing neurodevelopmental abnormalities within the PFC may contribute to several of the cognitive deficits seen in patients with neurodevelopmental disorders. In this review, we elaborate on the specific processes underlying prefrontal development, such as induction and patterning of the prefrontal area, proliferation, migration and axonal guidance of medial prefrontal progenitors, and their eventual efferent and afferent connections. We furthermore integrate for the first time the available knowledge from genome-wide studies that have revealed genes linked to neurodevelopmental disorders with experimental molecular evidence in rodents. The integrated data suggest that the pathogenic variants in the neurodevelopmental disorder-associated genes induce prefrontal cytoarchitectonical impairments. This enhances our understanding of the molecular mechanisms of prefrontal (mis)development underlying the four major neurodevelopmental disorders in humans, that is, intellectual disability, autism spectrum disorders, attention deficit hyperactivity disorder and schizophrenia, and may thus provide clues for the development of novel therapies.
The prefrontal cortex in neurodevelopmental disorders
Neurodevelopmental disorders affect a large percentage of the population worldwide. Although the available drugs can alleviate some of the symptoms associated with these disorders, they are not curative and adverse drug reactions are often observed. In addition, many neurodevelopmental disorder-associated symptoms, especially cognitive symptoms, still cannot be treated effectively. To improve the prognosis of a given neurodevelopmental disorder, the effectiveness of existing therapies and the potential for finding new treatment strategies, detailed knowledge of the development and pathophysiology of the disorders is mandatory.1, 2 Neurodevelopmental disorders such as intellectual disability (ID), autism spectrum disorders (ASDs), attention deficit (hyperactivity) disorder (AD(H)D) and schizophrenia share particular cytoarchitectonical, connectional and functional features suggesting a similar neurodevelopmental origin. Unfortunately, for the most part, detailed molecular studies of developmental events within brain areas that are involved in the etiology of these neurodevelopmental disorders are still lacking.
A wealth of data indicates that the prefrontal cortex (PFC) contributes to the cognitive deficits or endophenotypes of many, if not all, neurodevelopmental disorders.3, 4, 5, 6, 7, 8, 9, 10, 11, 12 As a conglomerate of individually unique subareas, the PFC has a key role in the execution of higher-order cognitive functions, for example, language comprehension and cognitive functions involved in decision making such as planning and reasoning.13, 14, 15, 16 In this respect, the different subareas within the PFC mediate various processes including response inhibition, working memory, attention or autonomic control.17, 18, 19, 20 Furthermore, the medial regions of the PFC, the mPFC, such as the infralimbic, prelimbic and cingulated areas, have a role in the cognitive deficits of many neurodevelopmental disorders.7, 11
The main neurodevelopmental disorders—ID, ASDs, AD(H)D and schizophrenia—have a complex etiology involving a large number of genes and environmental factors that also affect prefrontal brain regions, including those of the mPFC. Although multiple genes have been found to be associated with each of these disorders, the actual function and involvement of individual genes in the developmental aspects of mPFC formation in particular are largely unknown. Abnormalities in the expression of these genes often lead to impaired or deviant functioning of several brain structures, including the mPFC, affecting behavior as previously shown in animal studies.21, 22
In the following, we will give an overview of the main neurodevelopmental disorders with a particular focus on the defects in the development of the mPFC, bearing in mind that areas other than the mPFC may also contribute to the etiology of the disorders.
ID
The diagnostic category mental retardation groups a number of syndromes with severe ID that are associated with chromosomal abnormalities such as Down Syndrome (trisomy of chromosome 21), Prader–Willi and Angelman Syndromes, Williams–Beuren Syndrome, Smith–Magenis Syndrome, DiGeorge Syndrome and monosomy of chromosome 1p36.1.23, 24, 25, 26 Other ID syndromes show mild-to-moderate phenotypes and are associated with mutations, small insertions/deletions or copy number variations affecting a single gene, for example, fragile X syndrome, caused by a mutation in the FMR1 gene27, 28 and Kleefstra syndrome, caused by a functional loss of the EHMT1 gene.29 Most ID syndromes are associated with developmental deficits in general, including distorted development of the mPFC.23, 24, 26, 30 In this respect, during the development of the mPFC of ID patients, molecular/cellular defects have been shown to occur in (a) the proliferation of neuronal progenitor cells,31, 32 (b) migration of cortical neurons33, 34, 35, 36, 37 and (c) synaptogenesis.32, 38, 39
ASDs
The ASDs include autism, Asperger's syndrome and ‘pervasive developmental disorder not otherwise specified' Diagnostic and Statistical Manual of Mental Disorders-5th edition (DSM-V). They constitute a group of wide-ranging neurodevelopmental disorders that are characterized by variable impairments in three core symptom domains, that is, reciprocal social interaction, (verbal and nonverbal) communication, and restricted, repetitive and stereotyped patterns of behavior, interests and activities.40, 41, 42, 43 Although many of these behavioral impairments are driven by deficits in basal ganglia and amygdala functioning, cognitive dysfunctions such as memory deficits and deficits in social interaction and perception are integrated by the mPFC.44 The neurodevelopmental basis underlying the defects in language and speech, which are often part of the diagnosis in ASDs relates to abnormalities in fronto-striatal functioning.45, 46, 47, 48, 49 Regarding the development of the mPFC of ASD patients, molecular/cellular defects have been reported to occur in (a) the proliferation of neuronal progenitor cells50, 51 resulting in macrocephalic and minicolumn pathology in several brain areas including the PFC,3, 40, 42, 52, 53, 54 (b) migration and differentiation of GABA ergic parvalbumin+ (PV+) interneurons toward the PFC,36, 55, 56 (c) axon guidance, as there seems to be a disconnection of long-distance axonal pathways57, 58 and (d) synaptogenesis, particularly of GABAergic synapses.59, 60, 61 Deficits in integration and early information processing can be explained by hyperconnectivity combined with slower synapses.62 Furthermore, there is evidence for amplified activation and density of microglia within the PFC of ASD patients.57, 63, 64
AD(H)D
Inattention, hyperactivity/impulsivity and motivational/emotional dysregulation are the core symptom domains in AD(H)D. In AD(H)D patients, the mPFC-directed cognitive functions are affected and frequently of early onset.65, 66, 67 A delay in cortical maturation specifically in the most prefrontal areas and its connections to other brain areas has often been observed68 and there is increasing evidence that glutamate signaling is affected.69 During development, the PFC of patients with AD(H)D shows molecular/cellular defects in (a) the white matter, suggesting axon guidance deficits70, 71, 72 (b) dopaminergic and noradrenergic connectivity with the cerebellum and striatum65, 67, 73, 74, 75, 76 and (c) synaptogenesis influencing the electrophysiological properties and functioning of PFC neurons.77, 78, 79
Schizophrenia
Schizophrenia is thought to affect mainly (social) cognition, but it usually is also associated with chronic problems of behavioral and emotional regulation.80 Schizophrenia is characterized by a breakdown of thought processes manifested as delusions and hallucinations (positive symptoms) and by poor emotional responsiveness, and disorganized thinking and speech (negative symptoms). People with schizophrenia are likely to have co-morbidities such as major depression and anxiety disorders. Furthermore, working and long-term memory, attention, executive functioning and speed of processing are often affected.80 All of these symptoms can at least to some extent be linked to (impaired) PFC functioning.5, 12, 81, 82, 83, 84 During development of the mPFC in schizophrenia patients, molecular/cellular defects may occur in the (a) proliferation of neuronal progenitor cells, as reflected by the observed severely decreased gray-matter volume,85 as well as of GABAergic PV+ interneurons,86, 87 (b) postnatal pruning of dendritic trees and synapse loss,88, 89, 90, 91 (c) general connectivity of various neurotransmitter systems such as the glutamate, GABA and dopamine systems together with a reduced connectivity with other cortical areas.92, 93, 94, 95, 96, 97, 98, 99
Rodent models of neurodevelopmental disorders
Before one can start to develop better and more target-specific therapies for patients with neurodevelopmental disorders, it is necessary to first unravel elementary processes of brain development in adequate animal models and to understand subsequent developmental processes in those areas associated with the endophenotypes of neurodevelopmental disorders. In this way, fundamental hypotheses can be created and tested in relation to the etiology of these disorders. Such parallel approaches are crucial to eventually design optimal treatment strategies.
As mentioned before, although the PFC is often referred to as a single brain region, many subdivisions into distinct areas can be made, each of which possesses its own specific cytoarchitecture, cytochemistry, connectivity and functional properties. Defining these areas across species suffers from the fact that large interspecies differences exist in the layering per area, fueling the debate on whether or not rodents possess a region equivalent to the human PFC as they lack a granular zone in this area.100, 101 However, it should be noted that the formation of the general laminar pattern in the PFC shows a relation with phylogenesis: in ‘higher' mammalian species, such as primates and humans, PFC regions can be granular, that is, they possess a granular layer IV, as well as an agranular layer. The ‘lower' the species, the smaller the proportion of granular PFC regions (for reviews, see refs 100, 101). Thus the concept of homologous structures with similar functions may apply.
In this review, we will focus on the rodent mPFC and its structure–function relationships with connected brain areas in the context of neurodevelopmental disorders.102, 103 One example of a well-defined rodent model for neurodevelopmental disorders is the apomorphine-susceptible and apomorphine-unsusceptible Wistar rat. The behavioral impairments seen in the apomorphine-susceptible rats resemble features of schizophrenia.104, 105, 106 At least part of this phenotype can be attributed to the differences in the mesocorticolimbic projections.107
Furthermore, mouse models are ideally suited to study targeted molecular alterations.102, 108, 109, 110, 111, 112, 113, 114 In this way, genetic variants identified through association studies can be tested for their biological function and correlated with cognitive endophenotypes of human neurodevelopmental disorders. However, the traditional techniques of targeted mutation used in these kinds of model systems are systemic in nature and often result in inducing compensation mechanisms. Cre-Lox and knock-in systems still affect a large part of the brain, but can offer cell-type selective and temporally controlled strategies to achieve targeted mutations at different pre- and postnatal ages.115 Although in utero electroporation-mediated gene transfer spatially restrict gene repression or genetic rescues to early developmental time-points (app. E10-E17), virally mediated gene transfer can be performed pre- as well as postnatally.116 Furthermore, intersectional genetics (Flpe/Cre) to selectively mutate genes of interest in overlapping areas between a Cre and a Flpe allele (for example, Dlx5 Flpe and a region-specific Cre to selectively target GABAergic interneurons in a region of interest) increases the spatial selectivity of such approaches. Using these techniques, it is possible to knock down or rescue a particular gene in a specific part of the brain (for example, PFC) and at a specific time during brain development.
By employing various behavioral tasks, it is now possible to specifically test endophenotypes associated with mPFC function in rodent models, such as working memory, conditioned associative learning, attentional set shifting and reversal learning.117, 118, 119, 120, 121, 122 Consequently, by combining the targeted mutation with specific behavioral tests and instead of having to study a particular disease as a whole, one can now molecularly unravel the individual cognitive endophenotypes.21, 22 A further advantage of such an approach is that a causal inference can be made between the expression of a particular gene in a specific brain locus and one or more cognitive (endo)phenotypes, which is not yet possible in humans.
Developmental aspects of PFC formation
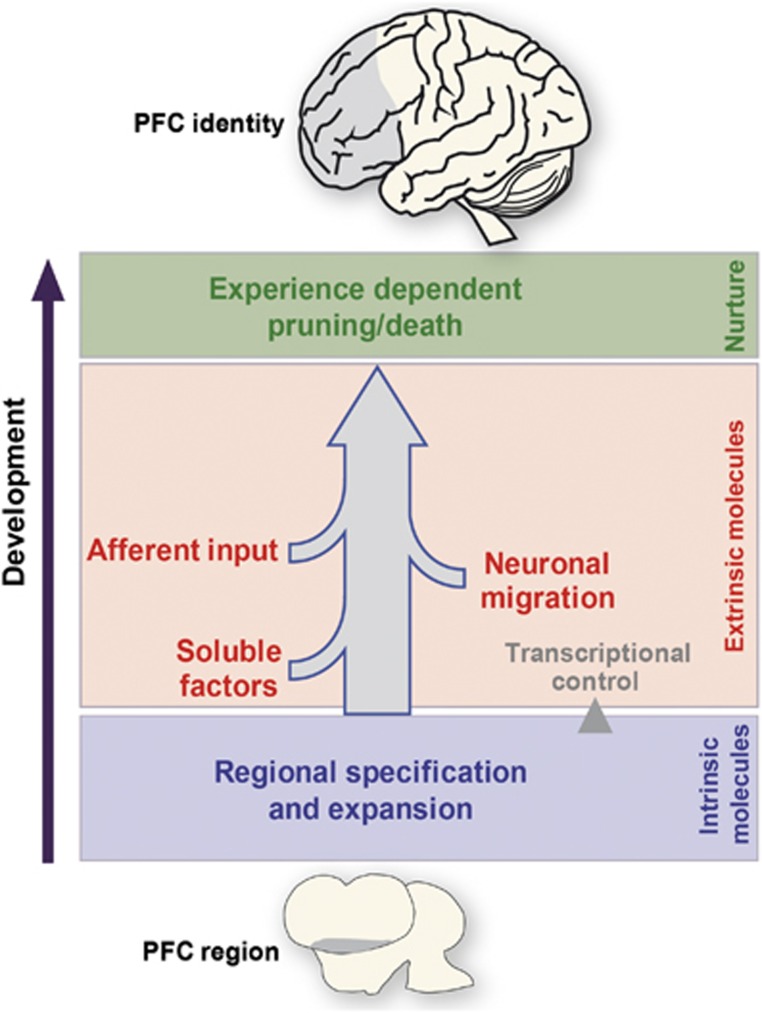
The PFC represents the functionally most advanced brain area with the longest period of maturation. This maturation includes proliferation and migration of neurons, growth of dendrites, the formation of neural micro- and macro-circuits through efferent/afferent axonal projections, and the fine-tuning of synaptic contacts and neuronal density steered by experience. This maturation process starts with an initial phase of cell division within an intrinsically specified PFC region, in which specific transcription factors (TFs) have a timing-critical role (Figure 1). Developmental events such as induction, migration and axon guidance are under the control of extrinsic cues and sculpt the identity of frontal areas. Appropriate cognitive behavior is fine-tuned over time by activity-dependent processes including sensory stimuli and social interactions, which in turn leads to pruning and cell death of unused connections.123 As a result, intricate convergence of connections with various other brain areas occurs, eventually creating the unique identity of the PFC and the subareas it encompasses (Figure 1). Here, the initial focus will be on the early developmental events of the (fore)brain as a whole and the molecules that are relevant during this phase. Although little is known about the early developmental characteristics of the PFC, many early principles and main mechanisms of forebrain compartmentalization and maturation are also applicable to PFC development. Important to keep in mind is the influence of external stimuli (for example, stress, drugs and hormones) that, if excessive, can lead to an altered development of the PFC and its connected areas.123 Thus, the knowledge about the genes that are involved in the structural and functional development of the (fore)brain and in particular the PFC is important for a better understanding of the molecular mechanisms underlying (disturbed) cognitive functions. Eventually, this knowledge may enable us to therapeutically intervene when this ‘developmental balance' is shifted toward neuropsychiatric disorder.
Figure 1.
Bird's eye view of developmental events required for prefrontal cortex (PFC) formation. The identity of the PFC is sculpted over time by intrinsic developmental mechanisms such as expansion by proliferation and regional specification by the differential expression of intrinsic factors (e.g., transcription factors), indicated in blue. These intrinsic factors can control genes (transcriptional control) that affect other developmental events such as the expression and release of soluble morphogens, migration of neurons or guidance molecules that direct axons from other brain areas towards the PFC and vice versa to establish appropriate connectivity. These extrinsic factors are depicted in red. Pruning of appropriate connections and neuron death are under the control of external stimuli (green).
Induction of (pre)frontal boundaries
The developmental progression of the forebrain starts with regional expansion through division of neuronal progenitor cells in proliferative zones lining the embryonic ventricles of the brain. The most anterior part of the neural tube develops into three primary vesicles even before the posterior section of the tube has formed: the prosencephalon (forebrain), mesencephalon (midbrain) and rhombencephalon (hindbrain).124 After closure, the neural tube is characterized by a sequence of swellings and constrictions along the anteroposterior axis, some of which subsequently develop into strict boundaries.125
Except for the specific boundary compartment, the zona limitans intrathalamica (ZLI), no unique set of boundary markers has been identified for regions of the forebrain and most of the telencephalon develops in an unsegmented way.125 Anterior of the midbrain–hindbrain border (MHB) or isthmus, the diencephalon consists of three neuromeres (p1–p3) according to the so-called prosomeric model.125, 126, 127 The more anterior prosomeres (p4–p6) subdivide the secondary prosencephalon (hypothalamus and telencephalon).128 The boundaries that are created function to arrange and stabilize local signaling centers or ‘organizers' important for the early patterning of the embryonic brain (Figures 2a and b). Gradually, gradients of soluble morphogens and growth factors (Fgfs, BMPs, SHH and Wnts)129, 130 are secreted from signaling centers and regulate the graded expression of certain intrinsic TFs, a process that is called induction131 (Figures 2a and b).
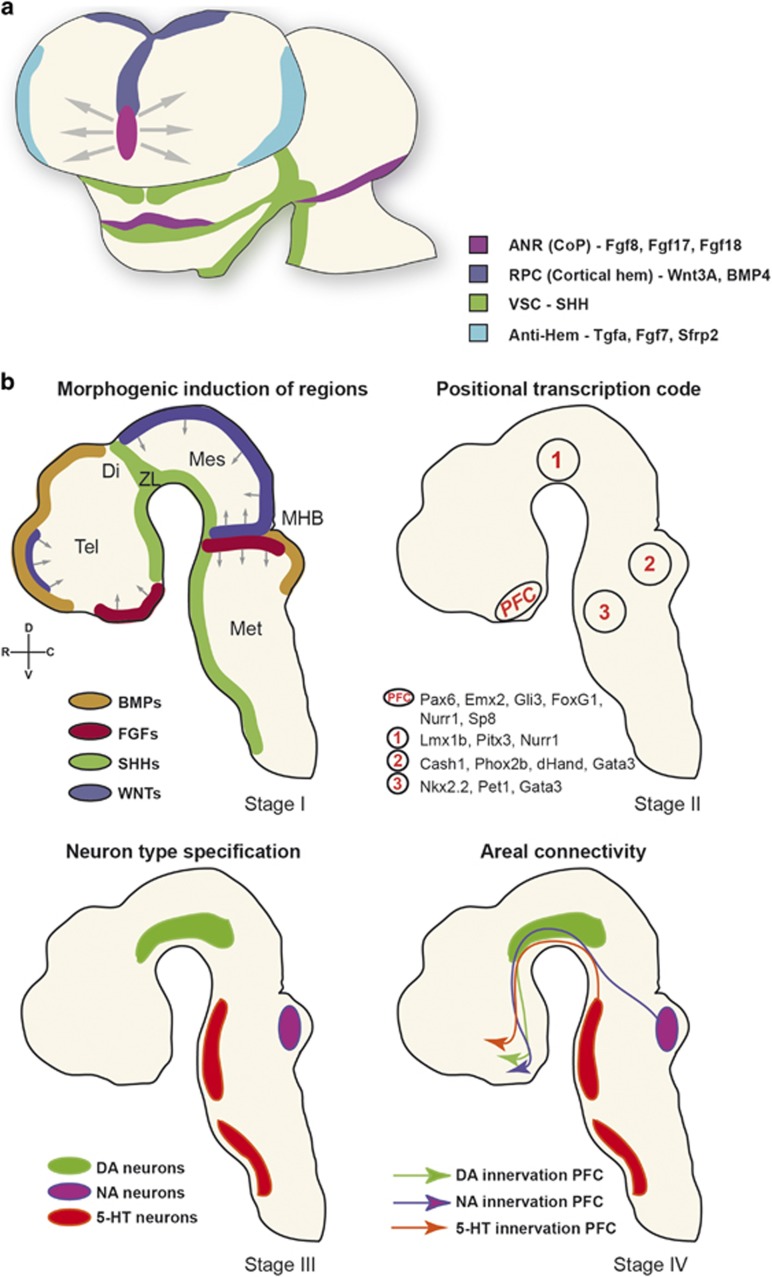
Figure 2.
Molecular stages in the development of the PFC. (a) Schematic representation of the frontal view of a young (E11.5) mouse forebrain showing inductive influences (morphogens such as Fgfs, Wnts, SHH and BMPs; stage I). (b) Sagittal schematic views. These morphogens (stage I) have an effect on regional specification through intrinsic expression of transcription factors (stage II). This combinatorial code will have its effect on the cell-type specification of the major neurotransmitter systems (stage III). The neurotransmitter systems will connect to the PFC, shaping it and establishing the respective neural networks (stage IV). ANR, anterior neural ridge; DA, dopaminergic; DI, diencephalon; MES, mesencephalon; MET, metencephalon; MHB, mid-hindbrain border; NA, noradrenergic; PFC, prefrontal cortex; RPC, rostral patterning center; SHH, sonic hedgehog; Tel, telencephalon; VSC, ventral signaling center; ZL, zona limitans; 5-HT, serotonergic.
Fgfs, especially Fgf8, Fgf17 and Fgf18 from the rostral patterning center (also called anterior neural ridge) provide, apart from their role in other areas, positional information on the presumptive prefrontal region along the rostro-caudal axis of the forebrain.132, 133 The dorsal patterning center or cortical hem secretes Bmp4/Wnt3A, which has a role in medial and dorsal pallium patterning,134, 135, 136 but in combination with SHH also steers prefrontal formation (Figures 2a and b). SHH is expressed by the ventral signaling center and regulates Fgf8 expression through the transcriptional repressor Gli3.137, 138, 139, 140 Absence of Fgf17 leads to a reduced PFC size and abnormal social behavior.141, 142 Thus, Bmp, Wnt and Fgf proteins all work coordinately to pattern the most rostral telencephalon.139, 143 Interference with each of the three Fgf receptor subtypes results in reduced numbers of either excitatory or inhibitory neurons, specifically in the prefrontal area and often resulting in altered behavior.144, 145, 146, 147, 148, 149
Regional identity of the PFC through intrinsic patterning
The gradients of morphogens and signaling molecules from the early patterning centers impart positional information influencing the expression of intrinsic TFs (Figure 2b). These have a crucial role in the regionalization of the forebrain and correlate with morphologic boundaries, the so-called regional specification underlying the spatio-temporal control of postnatal arealization.131, 150, 151, 152 The regional identity that is created by the expression of TFs includes the final cell-type specification.153 The inductive signals provided by morphogens and signaling molecules regulate the combinatorial expression of TFs and other regulatory factors, resulting in the generation of specific neuronal subtypes154, 155 (Figure 2a and b).
The interaction between extrinsic growth factors and intrinsic TFs during the early developmental events evolves through rostral patterning by the factors Fgf8 and Fgf17 through the Fgf receptors. This Fgf-signaling promotes the expression of the TFs Foxg1, Six3, Sp8, Pax6, Erm (etv5), Er81 (etv1), Nkx2.1 and Pea3, and represses the expression of Coup-tf1 and Emx2 more caudally.131, 133, 156 Although it is most likely the expression of a combination of multiple TFs that underlies the identity of an area, there are a few individual TFs that are specifically linked to the development of the most rostral part of the cortex. The expression of the TFs Pax6 and Emx2, for example, is known to have a role in cortical identity in general.131, 157, 158 Yet, very few TFs are specifically expressed in and linked to early PFC development.
During the course of development, distinct neuronal cell types will express a variety of proteins that are involved in migration, targeting (for example, axon guidance) and specific neurotransmitter release. This set of proteins is unique for each cell type, thereby regulating the formation of functional areas.159 The expression of the respective genes (extrinsic genes) is under the control of a distinct combinatorial code of TFs generating neuronal diversity160(Figure 1 and Figure 2). Other TFs such as Rest4 and Nurr1 display increased expression in the PFC and are involved in various aspects of cognitive behavior.161, 162 Although an abundance of genome-wide expression data shows that specific TFs are expressed in later stages of PFC development, their downstream targets and functional relevance are largely unknown.163, 164, 165, 166 In fact, the existing data are now congruent with a model in which each neuronal cell type within the PFC (but also other areas) most likely uses an exclusive code of intrinsic genes to control the expression of extrinsic genes. This code is unique to each particular cell type essential for the sequential steps in development. The next level of complexity starts off when extrinsic mechanisms such as migration and afferent input begin to have a role in the development of the prefrontal areas.
Proliferation and migration of PFC neurons
The PFC, like other cortical areas, expands by generating new neurons through (a)symmetric divisions of radial glia cells in the (sub)ventricular zone lining the ventricles.167, 168 During this process, reduction of the extrinsic morphogen Fgf8 results in less proliferation and more apoptosis, which ultimately changes the identity of the cortex.132, 169, 170 In particular Fgf has a determining role in the production of excitatory glutamatergic pyramidal neurons in the most anterior part of the cortex with deletion of the gene resulting in a reduced number of excitatory cortical neurons.171 Many TFs controlling the cell cycle, including cyclinD1, drive prefrontal expansion.39 Some newborn progenitors or intermediate progenitor cells expressing Tbr2 migrate to the subventricular zone to generate neurons. Lack of Tbr2 expression results in reduced cortical surface and thickness.172, 173, 174, 175 It is furthermore widely accepted that classical neurotransmitters such as dopamine and serotonin have an early role in controlling the neuron numbers within the PFC.176, 177, 178
The differential expression of TFs but also of adhesion and axon guidance molecules reflects a signage map for migrating neurons. The expression patterns are graded along the anterior–posterior and medial–lateral axes of the embryonic brain instructing neurons to establish functionally distinct lamina. During embryogenesis, most brain areas deploy radial migration in multiple waves as their major route to establish lamination within the structure.167, 179, 180 Radial glia cells, with their cell body within the ventricular zone, send out their glial processes toward the pial surface where they attach to the basal membrane. Newborn neurons that become (excitatory) projection neurons use the glial scaffold to migrate to their final place in the brain by using either somal translocation or locomotion.167, 180, 181 The ventricular zone generates the deeper layer neurons, including the subplate, layer VI and subsequently layer V projection neurons. Additionally, Cajal–Retzius neurons are generated within the cortical hem and to a lesser extent at other sites in the subpallium and septum. These layer I neurons express Reelin, a large secreted glycoprotein intricately involved in the inside-out laminar patterning of cortical neurons.182, 183 At later stages, the subventricular zone gives birth to neurons which migrate radially into the cortical plate past the deep layer neurons and form layers IV, III and II of the PFC, creating an inside-out pattern. Most of the projection neurons (80%) use glutamate as their neurotransmitter projecting to distant cortical and subcortical targets. The basic molecular developmental mechanisms that have been elucidated in rodent studies are in principle similar to those in humans, even though the human brain has gone through a series of additional evolutionary steps, including size, shape and gyrification modifications.184, 185, 186
Migration of GABAergic interneurons towards the PFC
A small proportion of neurons, which includes the majority of GABAergic (GAD65/67+) interneurons originating from the ganglionic eminences, migrate tangentially to the cortical plate, then radially to reach their target lamina.187 The subpallial interneurons migrate via a lengthy route towards the PFC using directional cues to eventually position themselves between pyramidal projection neurons on which they synapse.167, 188 Medial ganglionic eminence-derived interneurons will generate PV and somatostatin interneurons that populate all cortical structures (as well as hippocampus, striatum, amygdala, etc). These interneurons are specified in the medial ganglionic eminence by the expression of Nkx2.1 and Lhx6 followed by Sox6 expression as they start migrating. In contrast, caudal ganglionic eminence-derived interneurons encompass all 5-HT3A-expressing interneurons of various morphology and physiology.188 The homeobox TFs Dlx1 and Dlx2 mainly regulate the maturation of GABAergic (inter)neurons within the ganglionic eminences, having the TF Arx as a downstream target.133 However, the combinatorial expression of TFs such as Olig2, Dlx5, Arx, Lhx6, Cux2, NPAS1 and MafB define the various subpopulations of interneurons within the subpallium that end up in the (prefrontal) cortex.188, 189 As development progresses, interneurons within the (prefrontal) cortex start to express transporters (GAT-1 and -3), VGAT and components of GABAergic synapses190 making them highly adaptive to the maturing PFC.
Axon guidance, target selection and synapse formation of PFC neurons
The assembly of neuronal circuits during embryonic development relies upon the guidance of growing axons to their synaptic targets. To help them find their synaptic partners, developing axons are tipped with a highly motile sensory structure, the growth cone. Growth cones are instructed to follow predetermined trajectories by heterogeneously distributed guidance molecules in the extracellular environment. Binding of axon guidance molecules to receptor complexes on the growth cone surface initiates intracellular signaling events, which modulate growth cone morphology and directionality through local modifications of the cytoskeleton. Axon guidance molecules can act as attractants or repellents, that is, either directing growth cones toward a specific structure or preventing them from entering inappropriate regions. Furthermore, these cues exist as membrane-associated molecules acting at short ranges or as soluble agents with long-distance effects.191, 192, 193, 194 The responses of growing axons to particular cues, however, may change as they grow toward their final targets.176 For example, Semaphorin 3F is such a bidirectional guidance cue that, through binding with Neuropilin-2, initially repels dopaminergic axons from the rostral ventral tegmental area on their way to the mPFC, and later attracts and orients them within the mPFC.176 When the axonal growth cone has been guided to the proper target, synaptic contacts can be formed that are mediated by adhesion molecules such as the cadherins.195, 196 Newly formed synaptic contacts change their functional properties as development progresses and contribute to the maturation and functioning of an area.197, 198 Furthermore, the immature afferent projections are refined via the same guidance molecules in topography (pruning of branches), convergence (less efferent projections onto one cell) as well as postsynaptic compartment (less afferent dendritic innervation) in specific brain areas.197, 198, 199 Changes occurring in pyramidal morphology in terms of expansion of dendritic complexity are specifically apparent in layer III.200 Furthermore, during the first four postnatal weeks the local inhibitory interneuron networks in the mPFC undergo an extensive process of maturation, both at the level of intrinsic functional as well as network properties.201, 202 Given that inhibitory network activity is thought to contribute to the proper construction of cortical networks, the refinement of synaptic connectivity in inhibitory and excitatory networks leads to developmental plasticity and fine-tuning of complex behavior.
Topographic map formation in PFC connectivity: parcellation versus lamination
As mentioned above, in rodents and other phylogenetically ‘higher' species, the PFC is not one homogeneous cortical region but is compartmentalized into a number of structurally and functionally distinct prefrontal areas, each of which is thought to possess characteristic input–output profiles. In general, the rodent PFC can be subdivided into medial, lateral and ventral sections. Within the medial portion, the anterior cingulate (Cg), prelimbic (PL) and infralimbic (IL) cortices (Figure 3) and dorsal peduncular cortex can be distinguished from dorsal to ventral.203 The lateral and ventral PFC consists of the orbitofrontal cortex and the agranular insular cortices.204 The different areas of the PFC are connected to various other brain regions through highly organized projections controlling decision-directed behavior.205, 206, 207
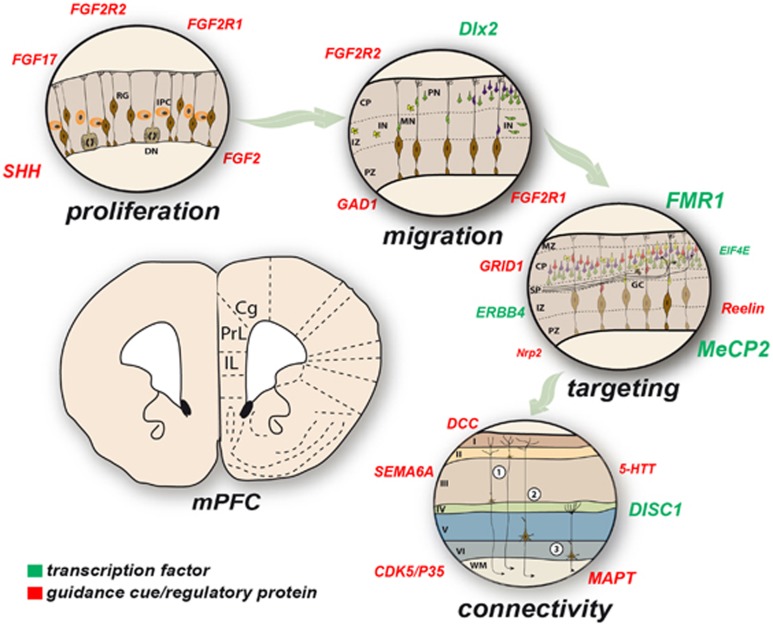
Figure 3.
Neurodevelopmental disorder-associated genes that are involved in mPFC development. Various genes are associated with neurodevelopmental events in the mPFC (proliferation, migration, guidance targeting and connectivity) of which some can also be found in association studies with the four major neurodevelopmental disorders ID, ASDs, AD(H)D, schizophrenia. The letter size in the ‘cloud' of genes is indicative of the frequency of the gene associated with the various neurodevelopmental disorders connected to that particular neurodevelopmental event. Cg, cingulate cortex; CP, cortical plate; DN, dividing neuroblast; GC, growth cone; IL, infralimbic cortex; IN, interneuron; IPC, intermediate progenitor; IZ, intermediate zone; MN, migrating neuron, PN, post-mitotic neuron; PrL, prelimbic cortex; PZ, proliverative zone; RG, radial glia; (1) Commissural and corticocortical projection neurons, respectively; (2) subcerebral projection neurons to basal ganglia, diencephalon, midbrain, hindbrain and spinal cord; (3) corticothalamic projection neurons to mediodorsal thalamic targets; (2) and (3)=corticofugal.
Input connectivity of the mPFC
In terms of the afferent connectivity of the mPFC, a comprehensive and detailed comparison of area-specific input connectivity is still lacking. The mPFC is known to receive long ascending projections from the ventral hippocampus,208, 209 from cholinergic neurons of the basal forebrain,210, 211 from dopaminergic neurons of the rostral part of the medial ventral tegmental area176, 212, 213 and from serotonergic/cholinergic neurons of the brainstem along a highly defined trajectory.214, 215 Functionally, the connection with the ventral hippocampus is thought to be of particular importance for the functioning of the mPFC during cognitive tasks.216, 217 The cholinergic and dopaminergic systems are considered to modulate mPFC activity and attentional performance.218, 219 Interestingly, the dopaminergic projections from the ventral tegmental area show strong laminar and cell-type specificity. They form dense contacts exclusively with interneurons in layers V and VI,176, 213, 220, 221 while for example projections from limbic and thalamic regions innervate both PV+ interneurons and pyramidal cells throughout layers II–VI.222, 223, 224 Furthermore, connections of the mPFC with both the basolateral amygdala209, 225 and the striatum are implicated in motivated behavior.226, 227 Interestingly, the long-range connections originating from the basolateral amygdala have been shown to not only be layer- but also cell-type specific. Neurons in the basolateral amygdala preferentially target layer II pyramidal neurons in the mPFC, such as PL, and amygdala, with which they can form reciprocal connections.225, 228
Output connectivity of the mPFC
As in other cortical areas, the long-range efferent connections of the PFC are mediated by excitatory projection neurons, that is, glutamatergic pyramidal cells. Depending on the PFC area, the pyramidal cells project to many structures such as the basal forebrain, olfactory and cortical structures, amygdala, striatum, (hypo)thalamus and the brainstem.204, 215, 225, 226, 229 In addition, prefrontal pyramidal neurons project to various subcortical areas thereby modulating dopaminergic, adrenergic, cholinergic and serotonergic projection systems.101, 204 The targets of the projection neurons show distinct layer specificity. Layer III pyramidal neurons connect the mPFC mainly to other cortical areas, whereas layers V and VI pyramidal cells project primarily to subcortical targets.230, 231 Furthermore, there is evidence for layer specificity of projections onto individual subcompartments of single brain structures. In terms of the nucleus accumbens, mPFC layer II pyramidal neurons preferentially innervate the core region, whereas neurons of deep layers V and VI innervate the core as well as the shell region.232
In contrast to the input connectivity, there is ample data demonstrating that the output connectivity properties of the mPFC are area dependent, which supports the notion that prefrontal areas are involved in modulating various aspects of cognitive behavior,203, 204, 229 not only in rodents but also in a number of other species.220, 229, 230 The dorsomedial areas of the PFC establish connections with the sensorimotor and association cortex, which are lacking in the ventral parts of the PFC. The ventral parts, however, establish relatively strong connections with the amygdaloid complex and limbic association cortices. Furthermore, the IL has been shown to mainly project to autonomic/visceral related sites, supporting its role in visceromotor activity,204 whereas the PL primarily innervates limbic sites that are thought to affect cognition.
Future translational avenues of research
In summary, substantial progress has been made in the past decades toward understanding the etiology of neurodevelopmental disorders at the molecular, cellular and systems levels. Nevertheless, we have only just begun to thoroughly study the development of a conglomerate of specific brain areas that as a group define the PFC and that are involved in the etiology of these disorders. In this context, it is remarkable that the exact molecular orchestration of the development of the PFC is still largely unknown. What are the molecular mechanisms that create a correctly parcellated and layered PFC? How are the extensive and highly specific interactions between various signaling pathways that are connecting the individual areas fine-tuned and how can we manipulate these? We are also only beginning to shed light on the large variety of neuronal cells and their integration in prefrontal local and global networks, let alone that we would know all the molecules that guide their differentiation and projections.
To test targeted molecular variations, rodents have emerged as an excellent model. Animal models and functional assays are invaluable as it comes to decipher the exact functions of the large number of genes that are involved in the various aspects of PFC development, that is, induction of prefrontal boundaries, intrinsic patterning of the PFC, proliferation and migration of (pyramidal) PFC neurons, migration of GABAergic interneurons toward the PFC, axon guidance, target selection and synapse formation of PFC neurons, and PFC connectivity formation. Slowly, the view is emerging that some of these genes are identical to the susceptibility genes of neurodevelopmental disorders (Table 1). However, up to now only a few of the genes could be directly linked to one or more of the developmental events within the PFC as well as one or more of the four major neurodevelopmental disorders, that is, ID, ASDs, AD(H)D and/or schizophrenia.
Table 1. Commonalities in gene association between PFC developmental events and the four major neurodevelopmental disorders.
| Gene | Involvement in PFC development | ID | ASDs | AD(H)D | Schizophrenia |
|---|---|---|---|---|---|
| Induction of prefrontal boundaries | |||||
| FGF17 | Fgf17 is secreted by the the rostral patterning center (RSC) and is involved in the induction of prefrontal boundaries.141, 142, 233 | Fgf17 knockout mice display deficits in specific social interactions that have been linked to ASDs.142 | |||
| SHH | Shh is secreted by the VSC and regulates the expression of Fgf8, which is involved in the induction of prefrontal boundaries.137, 138, 139 | Mutations in SHH cause holoprosencephaly, a common forebrain malformation associated with craniofacial anomalies and MR.234 | Significantly higher levels of serum SHH protein were found in children with autism.235 | A mutation in SHH was found in two boys with ADHD.236 | |
| Proliferation and migration of PFC neurons | |||||
| FGF2 | Fgf2 has an important role in the production of glutamatergic pyramidal neurons in the (pre)frontal cortex.237 | Fgf2 knockout mice show hyperactivity.238 | Serum FGF2 levels were found to be increased in people with schizophrenia.239 | ||
| FGFR1 | Fgfr1 is required for the proper number of glutamatergic pyramidal neurons in the frontal cortex.144 | Dominant or recessive FGFR1 mutations are responsible for Hartsfield syndrome.240 | Dysfunctional Fgfr1 signalling is associated with spontaneous hyperactivity.144 | FGFR1 levels are higher in schizophrenia241 and th-fgfr1(tk-) transgenic mice exhibit behavior resembling human schizophrenia.242 | |
| FGFR2 | Fgfr2 is involved in generating excitatory glutamatergic neurons in the mPFC.147 | Mutations in FGFR2 cause Crouzon's or Apert syndrome, which can be associated with MR.243, 244 | Deletions of FGFR2 are associated with ASD.245 | Some Fgfr2 deficient mice display hyperactive behavior.246 | A SNP flanking the FGFR2 gene is associated with schizophrenia.247 |
| Migration of GABAergic interneurons into the PFC | |||||
| DLX2 | Dlx2 controls interneurons migration toward frontal forebrain.248 | Deletions of DLX2 are associated with MR.249 | DLX2 shows genetic association with autism.250 | ||
| GAD1 | Gad1 regulates the migration of GABA-ergic interneurons to the PFC.251, 252 | Gad1 is an ASD susceptibility gene.253, 254, 255, 256 | GAD1 expression is altered in schizophrenia patients and is considered a risk gene.257, 258, 259 Review: ref 260. | ||
| Axon guidance, target selection and synapse formation of PFC neurons | |||||
| ERBB4 | Erbb4 regulates dendritic spine formation and density of PV+ interneurons in the PFC.261, 262, 263, 264 | ERBB4 is associated with ID.265 | Numerous studies implicate ERBB4 as schizophrenia risk genes.266, 267 For reviews, see refs 268,269. | ||
| EIF4E | Eif4e has a role in synaptic function, dendritic spine density and synaptic plasticity of PFC neurons.61 | EIF4E shows genetic association with autism.270, 271, 272 Eif4e transgenic mice display autism-like behaviors.61, 273 | |||
| FMR1 | Fmr1 functions in synaptogenesis of dendritic spines of PFC neurons.62, 274, 275, 276, 277 | Mutations/deletions of FMR1 cause Fragile X Syndrome, most common known hereditary cause of MR/ID and autism. Reviews: refs 28,30,278. | Mutations/deletions of FMR1 cause Fragile X Syndrome, most common known hereditary cause of MR/ID and autism. Reviews: refs 279,280,281. | Human and animal models carrying the FMR1 mutation display ADHD symptoms.282, 283, 284, 285 | Reduced levels of FMR1 and mutations of associated genes in schizophrenia patients.286, 287, 288 |
| GRID1 | Grid1 has a role in synaptogenesis of PFC neurons.289 | Genetic association290 and Grid1 knockout mice show autism-like behavior.289 | GRID1 shows genetic association with schizophrenia and gray-matter reduction in patients.291, 292 | ||
| NRP2 | Nrp2 is involved in regulating axon guidance of PFC neurons.293 | NRP2 mutations are associated with autism.294, 295 | |||
| RELN | Reln is involved in regulating spine density and network formation.296 | Disruption of RELN is associated with MR.297 | RELN shows genetic association with autism. Reviews: refs 298,299,300. | RELN shows genetic association with schizophrenia. Reviews: refs 301,302,303. | |
| MECP2 | MeCP2 plays a critical role in the regulation of GABAergic transmission and cortical excitability of PFC pyramidal.304 | MECP2 is associated with MR/ID and especially linked to Rett syndrome. Reviews: refs 305,306. | MECP2 is genetically linked to ASD.307, 308 Review: ref 309. | De novo mutations of MECP2 found in schizophrenia patients.310, 311 | |
| PFC connectivity | |||||
| DCC | DCC influences the prefrontal maturation and network formation with the dopaminergic midbrain.312, 313 | Association between schizophrenia and genetic variation in DCC.314 | |||
| DISC1 | Disc1 KD is associated with dendritic abnormalities and affected cAMP signalling and hampers the mesocortical dopaminergic network formation.21, 315 | DISC1 shows genetic association with autism.316, 317, 318, 319 | DISC1 shows genetic association with ADHD in adults.320 | DISC1 is a strong candidate gene for schizophrenia (recent reviews: refs 321,322,323. | |
| CDK5/P35 | Cdk5r1 knockout mice display improper mesolimbic circuitry of the PFC.324 | Cdk5/P35 knockout mice display ADHD- like behavior.324 | Lower levels of CDK5/P35 in people with schizophrenia.325, 326 | ||
| MAPT | Mutations in MAPT are associated with altered functional connectivity in the human PFC.327 | MAPT CNVs and microdeletions in patients with MR.328, 329, 330, 331 | |||
| SEMA6A | Loss of Sema6a causes prefrontal loss of connectivity.332 | Sema6a mutant mice display ASD-like behaviors.332 | Sema6a mutant mice display schizophrenia-like behaviors.332 | ||
| 5-HTT | 5-HTT is involved in proper raphe-prefrontal network formation.215 | 5-HTT is associated with schizophrenia.333, 334 | |||
Abbreviations: AD(H)D, attention deficit hyperactivity disorder; ASD, autism spectrum disorder; GABA, γ-aminobutyric acid; ID, intellectual disability; PFC, prefrontal cortex; PV+, parvalbumin+; RSC, rostral spinal cord; VSC, ventral signaling center.
Synopsis of the most cited genes that have been directly linked—through rodent studies—to one or more of the developmental events of PFC development (indicated in italics) and that have been directly genetically linked to the etiology of ID/MR, ASDs, AD(H)D and/or schizophrenia. Notes: (1) focus was on only those genes that were proven to be involved in prefrontal developmental events and not just expressed or involved in cortical development in general (e.g., Reelin); (2) A selection of references was made when more than three references were found.
Especially the availability of in utero electroporation-mediated gene transfer and other genetic approaches and hence the possibility to locally knock down or rescue particular genes will hopefully enable us to unravel the exact orchestration of brain areas such as those within the PFC in the near future. Such knowledge will assist in developing early intervention approaches by altering the susceptibility genes at a particular time and place, such that we deviate from the predetermined developmental path, even before the onset of the neurodevelopmental disorder(s) in question. Considering that individual susceptibility genes of neurodevelopmental disorders have often been found to be associated with multiple disorders, we can assume that several disorders share a common neurodevelopmental origin. It will be a challenge to dissect the individual genetic (and possibly even epigenetic) contributions to a disorder by using functional studies combined with behavioral tasks. For example, gene-environment interactions are crucial to distinguish between risk and vulnerability.
It is to be expected that in the coming years many more genes regulating developmental processes in the PFC and other brain structures will be linked to neurodevelopmental disorders and vice versa. Animal models, in which we can specifically alter gene expression in the PFC, can be instrumental for the understanding of the aetiopathological aspects of the disorder(s), as we can monitor the early disturbances that will eventually lead to defects in brain maturation and behavior. In order to move toward better and more preventive treatment of the neurodevelopmental disorders, bridges need to be built between disciplines such as combining genetic analyses of patients suffering from neurodevelopmental disorders with structural and functional brain imaging and in-depth molecular in vitro and in vivo approaches with cell and animal models. Exploring the molecular and cellular aspects during the progression of the disease process in animal models will clarify the pathological mechanisms, which in turn may provide clues to develop novel treatments for these disorders. The earlier during life and the more personalized the treatment strategies are applied, the better, alleviating symptoms at an early stage and reducing medical costs dramatically.
Acknowledgments
This work was supported by grants from the Donders Centre for Neuroscience, Radboud University Nijmegen (DS, SMK). The authors thank Prof B. Franke, Dr W. Scheenen, Prof H. van Bokhoven and Prof A. Kriegstein for critically reviewing the manuscript and the anonymous reviewers for their comments. Also, we apologize for those primary works not referenced here due to space limitations.
References
- Patel V, Boyce N, Collins PY, Saxena S, Horton R. A renewed agenda for global mental health. Lancet. 2011;378:1441–1442. doi: 10.1016/S0140-6736(11)61385-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belfer ML. Child and adolescent mental disorders: the magnitude of the problem across the globe. J Child Psychol Psychiatry. 2008;49:226–236. doi: 10.1111/j.1469-7610.2007.01855.x. [DOI] [PubMed] [Google Scholar]
- Mitchell SR, Reiss AL, Tatusko DH, Ikuta I, Kazmerski DB, Botti JA, et al. Neuroanatomic alterations and social and communication deficits in monozygotic twins discordant for autism disorder. Am J Psychiatry. 2009;166:917–925. doi: 10.1176/appi.ajp.2009.08101538. [DOI] [PubMed] [Google Scholar]
- Shenton ME, Dickey CC, Frumin M, McCarley RW. A review of MRI findings in schizophrenia. Schizophr Res. 2001;49:1–52. doi: 10.1016/s0920-9964(01)00163-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesh TA, Niendam TA, Minzenberg MJ, Carter CS. Cognitive control deficits in schizophrenia:mechanisms and meaning. Neuropsychopharmacology. 2011;36:316–338. doi: 10.1038/npp.2010.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova MF. Functional and anatomical aspects of prefrontal pathology in schizophrenia. Schizophrenia bulletin. 1997;23:517–519. doi: 10.1093/schbul/23.3.517. [DOI] [PubMed] [Google Scholar]
- Arnsten AF. Stress signalling pathways that impair prefrontal cortex structure and function. Nat Rev. 2009;10:410–422. doi: 10.1038/nrn2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnsten AF. Prefrontal cortical network connections: key site of vulnerability in stress and schizophrenia. Int J Dev Neurosci. 29:215–223. doi: 10.1016/j.ijdevneu.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins PY, Patel V, Joestl SS, March D, Insel TR, Daar AS, et al. Grand challenges in global mental health. Nature. 2011;475:27–30. doi: 10.1038/475027a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Neale MC. Endophenotype: a conceptual analysis. Mol Psychiatry. 2010;15:789–797. doi: 10.1038/mp.2010.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamo NJ, Arnsten AF. Molecular modulation of prefrontal cortex: rational development of treatments for psychiatric disorders. Behav Neurosci. 2012;125:282–296. doi: 10.1037/a0023165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DA, Curley AA, Glausier JR, Volk DW. Cortical parvalbumin interneurons and cognitive dysfunction in schizophrenia. Trends Neurosci. 2012;35:57–67. doi: 10.1016/j.tins.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson-Schill SL, Bedny M, Goldberg RF. The frontal lobes and the regulation of mental activity. Curr Opin Neurobiol. 2005;15:219–224. doi: 10.1016/j.conb.2005.03.006. [DOI] [PubMed] [Google Scholar]
- Egner T, Hirsch J. Cognitive control mechanisms resolve conflict through cortical amplification of task-relevant information. Nat Neurosci. 2005;8:1784–1790. doi: 10.1038/nn1594. [DOI] [PubMed] [Google Scholar]
- Etkin A, Egner T, Peraza DM, Kandel ER, Hirsch J. Resolving emotional conflict: a role for the rostral anterior cingulate cortex in modulating activity in the amygdala. Neuron. 2006;51:871–882. doi: 10.1016/j.neuron.2006.07.029. [DOI] [PubMed] [Google Scholar]
- Miller EK. The prefrontal cortex and cognitive control. Nat Rev. 2000;1:59–65. doi: 10.1038/35036228. [DOI] [PubMed] [Google Scholar]
- Miller EK, Cohen JD. An integrative theory of prefrontal cortex function. Ann Rev Neurosci. 2001;24:167–202. doi: 10.1146/annurev.neuro.24.1.167. [DOI] [PubMed] [Google Scholar]
- Summerfield C, Egner T, Greene M, Koechlin E, Mangels J, Hirsch J. Predictive codes for forthcoming perception in the frontal cortex. Science. 2006;314:1311–1314. doi: 10.1126/science.1132028. [DOI] [PubMed] [Google Scholar]
- Duncan J. An adaptive coding model of neural function in prefrontal cortex. Nat Rev. 2001;2:820–829. doi: 10.1038/35097575. [DOI] [PubMed] [Google Scholar]
- Mansouri FA, Tanaka K, Buckley MJ. Conflict-induced behavioural adjustment: a clue to the executive functions of the prefrontal cortex. Nat Rev. 2009;10:141–152. doi: 10.1038/nrn2538. [DOI] [PubMed] [Google Scholar]
- Niwa M, Kamiya A, Murai R, Kubo K, Gruber AJ, Tomita K, et al. Knockdown of DISC1 by in utero gene transfer disturbs postnatal dopaminergic maturation in the frontal cortex and leads to adult behavioral deficits. Neuron. 2010;65:480–489. doi: 10.1016/j.neuron.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loos M, Mueller T, Gouwenberg Y, Wijnands R, van der Loo RJ, Birchmeier C, et al. Neuregulin-3 in the mouse medial prefrontal cortex regulates impulsive action. Biol Psychiatry. 2014;76:648–655. doi: 10.1016/j.biopsych.2014.02.011. [DOI] [PubMed] [Google Scholar]
- Antonarakis SE, Lyle R, Dermitzakis ET, Reymond A, Deutsch S. Chromosome 21 and down syndrome: from genomics to pathophysiology. Nat Rev Genet. 2004;5:725–738. doi: 10.1038/nrg1448. [DOI] [PubMed] [Google Scholar]
- Contestabile A, Benfenati F, Gasparini L. Communication breaks-down: from neurodevelopment defects to cognitive disabilities in Down syndrome. Prog Neurobiol. 2010;91:1–22. doi: 10.1016/j.pneurobio.2010.01.003. [DOI] [PubMed] [Google Scholar]
- Gardiner K, Herault Y, Lott IT, Antonarakis SE, Reeves RH, Dierssen M. Down syndrome: from understanding the neurobiology to therapy. J Neurosci. 2010;30:14943–14945. doi: 10.1523/JNEUROSCI.3728-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bokhoven H. Genetic and epigenetic networks in intellectual disabilities. Annu Rev Genet. 2011;45:81–104. doi: 10.1146/annurev-genet-110410-132512. [DOI] [PubMed] [Google Scholar]
- Chelly J, Mandel JL. Monogenic causes of X-linked mental retardation. Nat Rev Genet. 2001;2:669–680. doi: 10.1038/35088558. [DOI] [PubMed] [Google Scholar]
- Kim M, Ceman S. Fragile X mental retardation protein: past, present and future. Curr Protein Pept Sci. 2012;13:358–371. doi: 10.2174/138920312801619420. [DOI] [PubMed] [Google Scholar]
- Kleefstra T, Smidt M, Banning MJ, Oudakker AR, Van Esch H, de Brouwer AP, et al. Disruption of the gene euchromatin histone methyl transferase1 (Eu-HMTase1) is associated with the 9q34 subtelomeric deletion syndrome. J Med Genet. 2005;42:299–306. doi: 10.1136/jmg.2004.028464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercaldo V, Descalzi G, Zhuo M. Fragile X mental retardation protein in learning-related synaptic plasticity. Mol Cells. 2009;28:501–507. doi: 10.1007/s10059-009-0193-x. [DOI] [PubMed] [Google Scholar]
- Cheng A, Haydar TF, Yarowsky PJ, Krueger BK. Concurrent generation of subplate and cortical plate neurons in developing trisomy 16 mouse cortex. Dev Neurosci. 2004;26:255–265. doi: 10.1159/000082142. [DOI] [PubMed] [Google Scholar]
- Chakrabarti L, Galdzicki Z, Haydar TF. Defects in embryonic neurogenesis and initial synapse formation in the forebrain of the Ts65Dn mouse model of Down syndrome. J Neurosci. 2007;27:11483–11495. doi: 10.1523/JNEUROSCI.3406-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen MH, Vissers LE, Willemsen MA, van Bon BW, Kroes T, de Ligt J, et al. Mutations in DYNC1H1 cause severe intellectual disability with neuronal migration defects. J Med Genet. 49:179–183. doi: 10.1136/jmedgenet-2011-100542. [DOI] [PubMed] [Google Scholar]
- Pilz DT, Matsumoto N, Minnerath S, Mills P, Gleeson JG, Allen KM, et al. LIS1 and XLIS (DCX) mutations cause most classical lissencephaly, but different patterns of malformation. Hum Mol Genet. 1998;7:2029–2037. doi: 10.1093/hmg/7.13.2029. [DOI] [PubMed] [Google Scholar]
- Rafalowska J, Dziewulska D, Podlecka A, Maslinska D. Early ontogenic disturbances in cell migration in mentally disabled adult. Clin Neuropathol. 2001;20:13–18. [PubMed] [Google Scholar]
- Penagarikano O, Abrahams BS, Herman EI, Winden KD, Gdalyahu A, Dong H, et al. Absence of CNTNAP2 leads to epilepsy, neuronal migration abnormalities, and core autism-related deficits. Cell. 2011;147:235–246. doi: 10.1016/j.cell.2011.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JS. Molecular genetics of neuronal migration disorders. Curr Neurol Neurosci Rep. 11:171–178. doi: 10.1007/s11910-010-0176-5. [DOI] [PubMed] [Google Scholar]
- Alarcon JM, Malleret G, Touzani K, Vronskaya S, Ishii S, Kandel ER, et al. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: a model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron. 2004;42:947–959. doi: 10.1016/j.neuron.2004.05.021. [DOI] [PubMed] [Google Scholar]
- Nobs L, Baranek C, Nestel S, Kulik A, Kapfhammer J, Nitsch C, et al. Stage-specific requirement for cyclin D1 in glial progenitor cells of the cerebral cortex. Glia. 2014;62:829–839. doi: 10.1002/glia.22646. [DOI] [PubMed] [Google Scholar]
- DiCicco-Bloom E, Lord C, Zwaigenbaum L, Courchesne E, Dager SR, Schmitz C, et al. The developmental neurobiology of autism spectrum disorder. J Neurosci. 2006;26:6897–6906. doi: 10.1523/JNEUROSCI.1712-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grafodatskaya D, Chung B, Szatmari P, Weksberg R. Autism spectrum disorders and epigenetics. J Am Acad Child Adolesc Psychiatry. 2010;49:794–809. doi: 10.1016/j.jaac.2010.05.005. [DOI] [PubMed] [Google Scholar]
- Amaral DG, Schumann CM, Nordahl CW. Neuroanatomy of autism. Trends Neurosci. 2008;31:137–145. doi: 10.1016/j.tins.2007.12.005. [DOI] [PubMed] [Google Scholar]
- Vorstman JA, Staal WG, van Daalen E, van Engeland H, Hochstenbach PF, Franke L.Identification of novel autism candidate regions through analysis of reported cytogenetic abnormalities associated with autism Mol Psychiatry 200611118–28. [DOI] [PubMed] [Google Scholar]
- Etkin A, Gyurak A, O'Hara R. A neurobiological approach to the cognitive deficits of psychiatric disorders. Dialogues Clin Neurosci. 2013;15:419–429. doi: 10.31887/DCNS.2013.15.4/aetkin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop DV. Genes, cognition, and communication: insights from neurodevelopmental disorders. Ann NY Acad Sci. 2009;1156:1–18. doi: 10.1111/j.1749-6632.2009.04419.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folia V, Udden J, Forkstam C, Ingvar M, Hagoort P, Petersson KM. Implicit learning and dyslexia. Ann NY Acad Sci. 2008;1145:132–150. doi: 10.1196/annals.1416.012. [DOI] [PubMed] [Google Scholar]
- Orban P, Lungu O, Doyon J. Motor sequence learning and developmental dyslexia. Ann NY Acad Sci. 2008;1145:151–172. doi: 10.1196/annals.1416.016. [DOI] [PubMed] [Google Scholar]
- Pennington BF, Bishop DV. Relations among speech, language, and reading disorders. Annu Rev Psychol. 2009;60:283–306. doi: 10.1146/annurev.psych.60.110707.163548. [DOI] [PubMed] [Google Scholar]
- Abrams DA, Lynch CJ, Cheng KM, Phillips J, Supekar K, Ryali S, et al. Underconnectivity between voice-selective cortex and reward circuitry in children with autism. Proc Natl Acad Sci USA. 2013;110:12060–12065. doi: 10.1073/pnas.1302982110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccarino FM, Grigorenko EL, Smith KM, Stevens HE. Regulation of cerebral cortical size and neuron number by fibroblast growth factors: implications for autism. J Autism Dev Disord. 2009;39:511–520. doi: 10.1007/s10803-008-0653-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler MG, Dasouki MJ, Zhou XP, Talebizadeh Z, Brown M, Takahashi TN, et al. Subset of individuals with autism spectrum disorders and extreme macrocephaly associated with germline PTEN tumour suppressor gene mutations. J Med Genet. 2005;42:318–321. doi: 10.1136/jmg.2004.024646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courchesne E, Campbell K, Solso S. Brain growth across the life span in autism: age-specific changes in anatomical pathology. Brain Res. 2011;1380:138–145. doi: 10.1016/j.brainres.2010.09.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courchesne E, Pierce K, Schumann CM, Redcay E, Buckwalter JA, Kennedy DP, et al. Mapping early brain development in autism. Neuron. 2007;56:399–413. doi: 10.1016/j.neuron.2007.10.016. [DOI] [PubMed] [Google Scholar]
- Schumann CM, Bloss CS, Barnes CC, Wideman GM, Carper RA, Akshoomoff N, et al. Longitudinal magnetic resonance imaging study of cortical development through early childhood in autism. J Neurosci. 2010;30:4419–4427. doi: 10.1523/JNEUROSCI.5714-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatemi SH, Halt AR, Stary JM, Kanodia R, Schulz SC, Realmuto GR. Glutamic acid decarboxylase 65 and 67 kDa proteins are reduced in autistic parietal and cerebellar cortices. Biol Psychiatry. 2002;52:805–810. doi: 10.1016/s0006-3223(02)01430-0. [DOI] [PubMed] [Google Scholar]
- Zhang ZW, Zak JD, Liu H. MeCP2 is required for normal development of GABAergic circuits in the thalamus. J Neurophysiol. 2010;103:2470–2481. doi: 10.1152/jn.00601.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zikopoulos B, Barbas H. Changes in prefrontal axons may disrupt the network in autism. J Neurosci. 2010;30:14595–14609. doi: 10.1523/JNEUROSCI.2257-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zikopoulos B, Barbas H. Altered neural connectivity in excitatory and inhibitory cortical circuits in autism. Front Hum Neurosci. 2013;7:609. doi: 10.3389/fnhum.2013.00609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medrihan L, Tantalaki E, Aramuni G, Sargsyan V, Dudanova I, Missler M, et al. Early defects of GABAergic synapses in the brain stem of a MeCP2 mouse model of Rett syndrome. J Neurophysiol. 2008;99:112–121. doi: 10.1152/jn.00826.2007. [DOI] [PubMed] [Google Scholar]
- Radyushkin K, Hammerschmidt K, Boretius S, Varoqueaux F, El-Kordi A, Ronnenberg A, et al. Neuroligin-3-deficient mice: model of a monogenic heritable form of autism with an olfactory deficit. Genes Brain Behav. 2009;8:416–425. doi: 10.1111/j.1601-183X.2009.00487.x. [DOI] [PubMed] [Google Scholar]
- Santini E, Huynh TN, MacAskill AF, Carter AG, Pierre P, Ruggero D, et al. Exaggerated translation causes synaptic and behavioural aberrations associated with autism. Nature. 2013;493:411–415. doi: 10.1038/nature11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa-Silva G, Loebel A, Giugliano M, de Kock CP, Mansvelder HD, Meredith RM. Hyperconnectivity and slow synapses during early development of medial prefrontal cortex in a mouse model for mental retardation and autism. Cereb Cortex. 2012;22:1333–1342. doi: 10.1093/cercor/bhr224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JT, Chana G, Pardo CA, Achim C, Semendeferi K, Buckwalter J, et al. Microglial activation and increased microglial density observed in the dorsolateral prefrontal cortex in autism. Biol Psychiatry. 2010;68:368–376. doi: 10.1016/j.biopsych.2010.05.024. [DOI] [PubMed] [Google Scholar]
- Morgan JT, Chana G, Abramson I, Semendeferi K, Courchesne E, Everall IP. Abnormal microglial-neuronal spatial organization in the dorsolateral prefrontal cortex in autism. Brain Res. 2012;1456:72–81. doi: 10.1016/j.brainres.2012.03.036. [DOI] [PubMed] [Google Scholar]
- Biederman J. Attention-deficit/hyperactivity disorder: a selective overview. Biol Psychiatry. 2005;57:1215–1220. doi: 10.1016/j.biopsych.2004.10.020. [DOI] [PubMed] [Google Scholar]
- Liston C, Malter Cohen M, Teslovich T, Levenson D, Casey BJ. Atypical prefrontal connectivity in attention-deficit/hyperactivity disorder: pathway to disease or pathological end point. Biol Psychiatry. 2011;69:1168–1177. doi: 10.1016/j.biopsych.2011.03.022. [DOI] [PubMed] [Google Scholar]
- Brennan AR, Arnsten AF. Neuronal mechanisms underlying attention deficit hyperactivity disorder: the influence of arousal on prefrontal cortical function. Ann NY Acad Sci. 2008;1129:236–245. doi: 10.1196/annals.1417.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw P, Eckstrand K, Sharp W, Blumenthal J, Lerch JP, Greenstein D, et al. Attention- deficit/hyperactivity disorder is characterized by a delay in cortical maturation. Proc Natl Acad Sci USA. 2007;104:19649–19654. doi: 10.1073/pnas.0707741104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltezos S, Horder J, Coghlan S, Skirrow C, O'Gorman R, Lavender TJ, et al. Glutamate/glutamine and neuronal integrity in adults with ADHD: a proton MRS study. Transl Psychiatry. 2014;4:e373. doi: 10.1038/tp.2014.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Agati E, Casarelli L, Pitzianti MB, Pasini A. Overflow movements and white matter abnormalities in ADHD. Prog Neuropsychopharmacol Biol Psychiatry. 2010;34:441–445. doi: 10.1016/j.pnpbp.2010.01.013. [DOI] [PubMed] [Google Scholar]
- Xia S, Li X, Kimball AE, Kelly MS, Lesser I, Branch C. Thalamic shape and connectivity abnormalities in children with attention-deficit/hyperactivity disorder. Psychiatry Res. 2012;204:161–167. doi: 10.1016/j.pscychresns.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart H, Marquand AF, Smith A, Cubillo A, Simmons A, Brammer M, et al. Predictive neurofunctional markers of attention-deficit/hyperactivity disorder based on pattern classification of temporal processing. J Am Acad Child Adolesc Psychiatry. 2014;53:569–78, e1. doi: 10.1016/j.jaac.2013.12.024. [DOI] [PubMed] [Google Scholar]
- Arnsten AF. Fundamentals of attention-deficit/hyperactivity disorder: circuits and pathways. J Clin Psychiatry. 2006;67 (Suppl 8:7–12. [PubMed] [Google Scholar]
- Rommelse NN, Geurts HM, Franke B, Buitelaar JK, Hartman CA. A review on cognitive and brain endophenotypes that may be common in autism spectrum disorder and attention- deficit/hyperactivity disorder and facilitate the search for pleiotropic genes. Neurosci Biobehav Rev. 2011;35:1363–1396. doi: 10.1016/j.neubiorev.2011.02.015. [DOI] [PubMed] [Google Scholar]
- Leo D, Sorrentino E, Volpicelli F, Eyman M, Greco D, Viggiano D, et al. Altered midbrain dopaminergic neurotransmission during development in an animal model of ADHD. Neurosci Biobehav Rev. 2003;27:661–669. doi: 10.1016/j.neubiorev.2003.08.009. [DOI] [PubMed] [Google Scholar]
- Miller EM, Pomerleau F, Huettl P, Gerhardt GA, Glaser PE. Aberrant glutamate signaling in the prefrontal cortex and striatum of the spontaneously hypertensive rat model of attention- deficit/hyperactivity disorder. Psychopharmacology. 2014;231:3019–3029. doi: 10.1007/s00213-014-3479-4. [DOI] [PubMed] [Google Scholar]
- Fossella JA, Sommer T, Fan J, Pfaff D, Posner MI. Synaptogenesis and heritable aspects of executive attention. Ment Retard Dev Disabil Res Rev. 2003;9:178–183. doi: 10.1002/mrdd.10078. [DOI] [PubMed] [Google Scholar]
- Kenar AN, Ay OI, Herken H, Erdal ME. Association of VAMP-2 and Syntaxin 1A genes with adult attention deficit hyperactivity disorder. Psychiatry Investig. 2014;11:76–83. doi: 10.4306/pi.2014.11.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawi Z, Matthews N, Wagner J, Wallace RH, Butler TJ, Vance A, et al. DNA variation in the SNAP25 gene confers risk to ADHD and is associated with reduced expression in prefrontal cortex. PloS One. 2013;8:e60274. doi: 10.1371/journal.pone.0060274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy SE, McCombie WR, Corvin A. Unlocking the treasure trove: from genes to schizophrenia biology. Schizophr Bull. 2014;40:492–496. doi: 10.1093/schbul/sbu042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DA, Cruz D, Eggan S, Erickson S. Postnatal development of prefrontal inhibitory circuits and the pathophysiology of cognitive dysfunction in schizophrenia. Ann NY Acad Sci. 2004;1021:64–76. doi: 10.1196/annals.1308.008. [DOI] [PubMed] [Google Scholar]
- Connor CM, Guo Y, Akbarian S. Cingulate white matter neurons in schizophrenia and bipolar disorder. Biol Psychiatry. 2009;66:486–493. doi: 10.1016/j.biopsych.2009.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beneyto M, Lewis DA. Insights into the neurodevelopmental origin of schizophrenia from postmortem studies of prefrontal cortical circuitry. Int J Dev Neurosci. 2011;29:295–304. doi: 10.1016/j.ijdevneu.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnsten AF. Prefrontal cortical network connections: key site of vulnerability in stress and schizophrenia. Int J Dev Neurosci. 2011;29:215–223. doi: 10.1016/j.ijdevneu.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zierhut KC, Schulte-Kemna A, Kaufmann J, Steiner J, Bogerts B, Schiltz K. Distinct structural alterations independently contributing to working memory deficits and symptomatology in paranoid schizophrenia. Cortex. 2012;49:1063–1072. doi: 10.1016/j.cortex.2012.08.027. [DOI] [PubMed] [Google Scholar]
- Arnold SE, Talbot K, Hahn CG. Neurodevelopment, neuroplasticity, and new genes for schizophrenia. Prog Brain Res. 2005;147:319–345. doi: 10.1016/S0079-6123(04)47023-X. [DOI] [PubMed] [Google Scholar]
- Bernstein HG, Smalla KH, Durrschmidt D, Keilhoff G, Dobrowolny H, Steiner J, et al. Increased density of prohibitin-immunoreactive oligodendrocytes in the dorsolateral prefrontal white matter of subjects with schizophrenia suggests extraneuronal roles for the protein in the disease. Neuromolecular Med. 2012;14:270–280. doi: 10.1007/s12017-012-8185-y. [DOI] [PubMed] [Google Scholar]
- Glantz LA, Lewis DA. Decreased dendritic spine density on prefrontal cortical pyramidal neurons in schizophrenia. Arch Gen Psychiatry. 2000;57:65–73. doi: 10.1001/archpsyc.57.1.65. [DOI] [PubMed] [Google Scholar]
- Bennett MR. Schizophrenia: susceptibility genes, dendritic-spine pathology and gray matter loss. Prog Neurobiol. 2011;95:275–300. doi: 10.1016/j.pneurobio.2011.08.003. [DOI] [PubMed] [Google Scholar]
- Glausier JR, Fish KN, Lewis DA. Altered parvalbumin basket cell inputs in the dorsolateral prefrontal cortex of schizophrenia subjects. Mol Psychiatry. 2014;19:30–36. doi: 10.1038/mp.2013.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glausier JR, Lewis DA. Dendritic spine pathology in schizophrenia. Neuroscience. 2013;251:90–107. doi: 10.1016/j.neuroscience.2012.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson A. The neurochemical circuitry of schizophrenia. Pharmacopsychiatry. 2006;39:S10–S14. doi: 10.1055/s-2006-931483. [DOI] [PubMed] [Google Scholar]
- Seamans JK, Yang CR. The principal features and mechanisms of dopamine modulation in the prefrontal cortex. Prog Neurobiol. 2004;74:1–58. doi: 10.1016/j.pneurobio.2004.05.006. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Burgos G, Hashimoto T, Lewis DA. Alterations of cortical GABA neurons and network oscillations in schizophrenia. Curr Psychiatry Rep. 2010;12:335–344. doi: 10.1007/s11920-010-0124-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DA, Gonzalez-Burgos G. Neuroplasticity of neocortical circuits in schizophrenia. Neuropsychopharmacology. 2008;33:141–165. doi: 10.1038/sj.npp.1301563. [DOI] [PubMed] [Google Scholar]
- Volk DW, Lewis DA. Prefrontal cortical circuits in schizophrenia. Curr Top Behav Neurosci. 2010;4:485–508. doi: 10.1007/7854_2010_44. [DOI] [PubMed] [Google Scholar]
- Rolls ET, Loh M, Deco G, Winterer G. Computational models of schizophrenia and dopamine modulation in the prefrontal cortex. Nat Rev. 2008;9:696–709. doi: 10.1038/nrn2462. [DOI] [PubMed] [Google Scholar]
- Chen L, Perez SM, Lodge DJ. An augmented dopamine system function is present prior to puberty in the methylazoxymethanol acetate rodent model of schizophrenia. Dev Neurobiol. 2014;74:907–917. doi: 10.1002/dneu.22172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deserno L, Sterzer P, Wustenberg T, Heinz A, Schlagenhauf F. Reduced prefrontal-parietal effective connectivity and working memory deficits in schizophrenia. J Neurosci. 2012;32:12–20. doi: 10.1523/JNEUROSCI.3405-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elston GN. Cortex, cognition and the cell: new insights into the pyramidal neuron and prefrontal function. Cereb Cortex. 2003;13:1124–1138. doi: 10.1093/cercor/bhg093. [DOI] [PubMed] [Google Scholar]
- Ongur D, Price JL. The organization of networks within the orbital and medial prefrontal cortex of rats, monkeys and humans. Cereb Cortex. 2000;10:206–219. doi: 10.1093/cercor/10.3.206. [DOI] [PubMed] [Google Scholar]
- Kellendonk C, Simpson EH, Kandel ER. Modeling cognitive endophenotypes of schizophrenia in mice. Trends Neurosci. 2009;32:347–358. doi: 10.1016/j.tins.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaaro-Peled H. Gene models of schizophrenia: DISC1 mouse models. Prog Brain Res. 2009;179:75–86. doi: 10.1016/S0079-6123(09)17909-8. [DOI] [PubMed] [Google Scholar]
- van Loo KM, Martens GJ. Genetic and environmental factors in complex neurodevelopmental disorders. Curr Genomics. 2007;8:429–444. doi: 10.2174/138920207783591717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coolen MW, Van Loo KM, Van Bakel NN, Pulford DJ, Serneels L, De Strooper B, et al. Gene dosage effect on gamma-secretase component Aph-1b in a rat model for neurodevelopmental disorders. Neuron. 2005;45:497–503. doi: 10.1016/j.neuron.2004.12.054. [DOI] [PubMed] [Google Scholar]
- Van Schijndel JE, Van Zweeden M, Van Loo KM, Martens GJ. Gene expression profiling in brain regions of a rat model displaying schizophrenia-related features. Behav Brain Res. 2010;207:476–479. doi: 10.1016/j.bbr.2009.10.042. [DOI] [PubMed] [Google Scholar]
- van der Elst MC, Roubos EW, Ellenbroek BA, Veening JG, Cools AR. Apomorphine-susceptible rats and apomorphine-unsusceptible rats differ in the tyrosine hydroxylase-immunoreactive network in the nucleus accumbens core and shell. Exp Brain Res. 2005;160:418–423. doi: 10.1007/s00221-004-2025-8. [DOI] [PubMed] [Google Scholar]
- Moron JA, Brockington A, Wise RA, Rocha BA, Hope BT. Dopamine uptake through the norepinephrine transporter in brain regions with low levels of the dopamine transporter: evidence from knock-out mouse lines. J Neurosci. 2002;22:389–395. doi: 10.1523/JNEUROSCI.22-02-00389.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roullet FI, Crawley JN. Mouse models of autism: testing hypotheses about molecular mechanisms. Curr Top Behav Neurosci. 2011;7:187–212. doi: 10.1007/7854_2010_113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moy SS, Nadler JJ. Advances in behavioral genetics: mouse models of autism. Mol Psychiatry. 2008;13:4–26. doi: 10.1038/sj.mp.4002082. [DOI] [PubMed] [Google Scholar]
- Desbonnet L, Waddington JL, Tuathaigh CM. Mice mutant for genes associated with schizophrenia: common phenotype or distinct endophenotypes. Behav Brain Res. 2009;204:258–273. doi: 10.1016/j.bbr.2009.04.001. [DOI] [PubMed] [Google Scholar]
- Johnstone M, Thomson PA, Hall J, McIntosh AM, Lawrie SM, Porteous DJ. DISC1 in schizophrenia: genetic mouse models and human genomic imaging. Schizophr Bull. 2011;37:14–20. doi: 10.1093/schbul/sbq135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa A. Genetic animal models for schizophrenia: advantages and limitations of genetic manipulation in drosophila, zebrafish, rodents, and primates. Prog Brain Res. 2009;179:3–6. doi: 10.1016/S0079-6123(09)17901-3. [DOI] [PubMed] [Google Scholar]
- Chen J, Lipska BK, Weinberger DR. Genetic mouse models of schizophrenia: from hypothesis-based to susceptibility gene-based models. Biol Psychiatry. 2006;59:1180–1188. doi: 10.1016/j.biopsych.2006.02.024. [DOI] [PubMed] [Google Scholar]
- Baek ST, Kerjan G, Bielas SL, Lee JE, Fenstermaker AG, Novarino G, et al. Off-target effect of doublecortin family shRNA on neuronal migration associated with endogenous microRNA dysregulation. Neuron. 2014;82:1255–1262. doi: 10.1016/j.neuron.2014.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolk SM, de Mooij-Malsen AJ, Martens GJ. Spatiotemporal molecular approach of in utero electroporation to functionally decipher endophenotypes in neurodevelopmental disorders. Front Mol Neurosci. 2011;4:37. doi: 10.3389/fnmol.2011.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissonette GB, Powell EM. Reversal learning and attentional set-shifting in mice. Neuropharmacology. 2011;62:1168–1174. doi: 10.1016/j.neuropharm.2011.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissonette GB, Martins GJ, Franz TM, Harper ES, Schoenbaum G, Powell EM. Double dissociation of the effects of medial and orbital prefrontal cortical lesions on attentional and affective shifts in mice. J Neurosci. 2008;28:11124–11130. doi: 10.1523/JNEUROSCI.2820-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigman JL, Rothblat LA. Stimulus specific deficit on visual reversal learning after lesions of medial prefrontal cortex in the mouse. Behav Brain Res. 2008;187:405–410. doi: 10.1016/j.bbr.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Kellendonk C, Simpson EH, Polan HJ, Malleret G, Vronskaya S, Winiger V, et al. Transient and selective overexpression of dopamine D2 receptors in the striatum causes persistent abnormalities in prefrontal cortex functioning. Neuron. 2006;49:603–615. doi: 10.1016/j.neuron.2006.01.023. [DOI] [PubMed] [Google Scholar]
- Ben Abdallah NM, Fuss J, Trusel M, Galsworthy MJ, Bobsin K, Colacicco G, et al. The puzzle box as a simple and efficient behavioral test for exploring impairments of general cognition and executive functions in mouse models of schizophrenia. Exp Neurol. 2011;227:42–52. doi: 10.1016/j.expneurol.2010.09.008. [DOI] [PubMed] [Google Scholar]
- Chudasama Y. Animal models of prefrontal-executive function. Behav Neurosci. 2011;125:327–343. doi: 10.1037/a0023766. [DOI] [PubMed] [Google Scholar]
- Kolb B, Mychasiuk R, Muhammad A, Li Y, Frost DO, Gibb R. Experience and the developing prefrontal cortex. Proc Natl Acad Sci USA. 2012;109:17186–17193. doi: 10.1073/pnas.1121251109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira C, Pombero A, Garcia-Lopez R, Gimeno L, Echevarria D, Martinez S. Molecular mechanisms controlling brain development: an overview of neuroepithelial secondary organizers. Int J Dev Biol. 2010;54:7–20. doi: 10.1387/ijdb.092853cv. [DOI] [PubMed] [Google Scholar]
- Puelles L, Rubenstein JL. Forebrain gene expression domains and the evolving prosomeric model. Trends Neurosci. 2003;26:469–476. doi: 10.1016/S0166-2236(03)00234-0. [DOI] [PubMed] [Google Scholar]
- Andersson E, Tryggvason U, Deng Q, Friling S, Alekseenko Z, Robert B, et al. Identification of intrinsic determinants of midbrain dopamine neurons. Cell. 2006;124:393–405. doi: 10.1016/j.cell.2005.10.037. [DOI] [PubMed] [Google Scholar]
- Pombero A, Martinez S. Telencephalic morphogenesis during the process of neurulation: an experimental study using quail-chick chimeras. J Comp Neurol. 2009;512:784–797. doi: 10.1002/cne.21933. [DOI] [PubMed] [Google Scholar]
- Rubenstein JL, Martinez S, Shimamura K, Puelles L. The embryonic vertebrate forebrain: the prosomeric model. Science (New York, NY. 1994;266:578–580. doi: 10.1126/science.7939711. [DOI] [PubMed] [Google Scholar]
- Borello U, Pierani A. Patterning the cerebral cortex: traveling with morphogens. Curr Opin Genet Dev. 2010;20:408–415. doi: 10.1016/j.gde.2010.05.003. [DOI] [PubMed] [Google Scholar]
- Lander AD. Morpheus unbound: reimagining the morphogen gradient. Cell. 2007;128:245–256. doi: 10.1016/j.cell.2007.01.004. [DOI] [PubMed] [Google Scholar]
- O'Leary DD, Chou SJ, Sahara S. Area patterning of the mammalian cortex. Neuron. 2007;56:252–269. doi: 10.1016/j.neuron.2007.10.010. [DOI] [PubMed] [Google Scholar]
- Fukuchi-Shimogori T, Grove EA. Neocortex patterning by the secreted signaling molecule FGF8. Science (New York, NY. 2001;294:1071–1074. doi: 10.1126/science.1064252. [DOI] [PubMed] [Google Scholar]
- Rubenstein JL. Research Review: Development of the cerebral cortex: implications for neurodevelopmental disorders. J Child Psychol Psychiatry. 2010;52:339–355. doi: 10.1111/j.1469-7610.2010.02307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert JM, Hayhurst M, Marks ME, Kulessa H, Hogan BL, McConnell SK. BMP ligands act redundantly to pattern the dorsal telencephalic midline. Genesis. 2003;35:214–219. doi: 10.1002/gene.10183. [DOI] [PubMed] [Google Scholar]
- Fernandes M, Gutin G, Alcorn H, McConnell SK, Hebert JM. Mutations in the BMP pathway in mice support the existence of two molecular classes of holoprosencephaly. Development (Cambridge, England) 2007;134:3789–3794. doi: 10.1242/dev.004325. [DOI] [PubMed] [Google Scholar]
- Hebert JM, Mishina Y, McConnell SK. BMP signaling is required locally to pattern the dorsal telencephalic midline. Neuron. 2002;35:1029–1041. doi: 10.1016/s0896-6273(02)00900-5. [DOI] [PubMed] [Google Scholar]
- Rash BG, Grove EA. Patterning the dorsal telencephalon: a role for sonic hedgehog. J Neurosci. 2007;27:11595–11603. doi: 10.1523/JNEUROSCI.3204-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuschel S, Ruther U, Theil T. A disrupted balance between Bmp/Wnt and Fgf signaling underlies the ventralization of the Gli3 mutant telencephalon. Dev Biol. 2003;260:484–495. doi: 10.1016/s0012-1606(03)00252-5. [DOI] [PubMed] [Google Scholar]
- Ohkubo Y, Chiang C, Rubenstein JL. Coordifnate regulation and synergistic actions of BMP4, SHH and FGF8 in the rostral prosencephalon regulate morphogenesis of the telencephalic and optic vesicles. Neuroscience. 2002;111:1–17. doi: 10.1016/s0306-4522(01)00616-9. [DOI] [PubMed] [Google Scholar]
- Blaess S, Stephen D, Joyner AL. Gli3 coordinates three-dimensional patterning and growth of the tectum and cerebellum by integrating Shh and Fgf8 signaling. Development (Cambridge, England) 2008;135:2093–2103. doi: 10.1242/dev.015990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cholfin JA, Rubenstein JL. Frontal cortex subdivision patterning is coordinately regulated by Fgf8, Fgf17, and Emx2. J Comp Neurol. 2008;509:144–155. doi: 10.1002/cne.21709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scearce-Levie K, Roberson ED, Gerstein H, Cholfin JA, Mandiyan VS, Shah NM, et al. Abnormal social behaviors in mice lacking Fgf17. Genes Brain Behav. 2008;7:344–354. doi: 10.1111/j.1601-183X.2007.00357.x. [DOI] [PubMed] [Google Scholar]
- Shimogori T, Banuchi V, Ng HY, Strauss JB, Grove EA. Embryonic signaling centers expressing BMP, WNT and FGF proteins interact to pattern the cerebral cortex. Development (Cambridge, England) 2004;131:5639–5647. doi: 10.1242/dev.01428. [DOI] [PubMed] [Google Scholar]
- Shin DM, Korada S, Raballo R, Shashikant CS, Simeone A, Taylor JR, et al. Loss of glutamatergic pyramidal neurons in frontal and temporal cortex resulting from attenuation of FGFR1 signaling is associated with spontaneous hyperactivity in mice. J Neurosci. 2004;24:2247–2258. doi: 10.1523/JNEUROSCI.5285-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson RE, Kind PC, Graham NA, Etherson ML, Kennedy J, Fernandes AC, et al. Fgf receptor 3 activation promotes selective growth and expansion of occipitotemporal cortex. Neural Dev. 2009;4:4. doi: 10.1186/1749-8104-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson RE, Pellicano F, Iwata T. Fibroblast growth factor receptor 3 kinase domain mutation increases cortical progenitor proliferation via mitogen-activated protein kinase activation. J Neurochem. 2007;100:1565–1578. doi: 10.1111/j.1471-4159.2006.04285.x. [DOI] [PubMed] [Google Scholar]
- Stevens HE, Smith KM, Maragnoli ME, Fagel D, Borok E, Shanabrough M, et al. Fgfr2 is required for the development of the miedial prefrontal cortex and its connections with limbic circuits. J Neurosci. 2010;30:5590–5602. doi: 10.1523/JNEUROSCI.5837-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller Smith K, Fagel DM, Stevens HE, Rabenstein RL, Maragnoli ME, Ohkubo Y, et al. Deficiency in inhibitory cortical interneurons associates with hyperactivity in fibroblast growth factor receptor 1 mutant mice. Biol Psychiatry. 2008;63:953–962. doi: 10.1016/j.biopsych.2007.09.020. [DOI] [PubMed] [Google Scholar]
- Inglis-Broadgate SL, Thomson RE, Pellicano F, Tartaglia MA, Pontikis CC, Cooper JD, et al. FGFR3 regulates brain size by controlling progenitor cell proliferation and apoptosis during embryonic development. Dev Biol. 2005;279:73–85. doi: 10.1016/j.ydbio.2004.11.035. [DOI] [PubMed] [Google Scholar]
- Hoch RV, Rubenstein JL, Pleasure S. Genes and signaling events that establish regional patterning of the mammalian forebrain. Semin Cell Dev Biol. 2009;20:378–386. doi: 10.1016/j.semcdb.2009.02.005. [DOI] [PubMed] [Google Scholar]
- Sur M, Rubenstein JL. Patterning and plasticity of the cerebral cortex. Science (New York, NY. 2005;310:805–810. doi: 10.1126/science.1112070. [DOI] [PubMed] [Google Scholar]
- Mason I. Initiation to end point: the multiple roles of fibroblast growth factors in neural development. Nat Rev. 2007;8:583–596. doi: 10.1038/nrn2189. [DOI] [PubMed] [Google Scholar]
- Schuurmans C, Guillemot F. Molecular mechanisms underlying cell fate specification in the developing telencephalon. Curr Opin Neurobiol. 2002;12:26–34. doi: 10.1016/s0959-4388(02)00286-6. [DOI] [PubMed] [Google Scholar]
- Jessell TM. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat Rev Genet. 2000;1:20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- Goridis C, Rohrer H. Specification of catecholaminergic and serotonergic neurons. Nat Rev. 2002;3:531–541. doi: 10.1038/nrn871. [DOI] [PubMed] [Google Scholar]
- Hasegawa H, Ashigaki S, Takamatsu M, Suzuki-Migishima R, Ohbayashi N, Itoh N, et al. Laminar patterning in the developing neocortex by temporally coordinated fibroblast growth factor signaling. J Neurosci. 2004;24:8711–8719. doi: 10.1523/JNEUROSCI.3070-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou SJ, Perez-Garcia CG, Kroll TT, O'Leary DD. Lhx2 specifies regional fate in Emx1 lineage of telencephalic progenitors generating cerebral cortex. Nat Neurosci. 2009;12:1381–1389. doi: 10.1038/nn.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangale VS, Hirokawa KE, Satyaki PR, Gokulchandran N, Chikbire S, Subramanian L, et al. Lhx2 selector activity specifies cortical identity and suppresses hippocampal organizer fate. Science (New York, NY. 2008;319:304–309. doi: 10.1126/science.1151695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polleux F, Ince-Dunn G, Ghosh A. Transcriptional regulation of vertebrate axon guidance and synapse formation. Nat Rev. 2007;8:331–340. doi: 10.1038/nrn2118. [DOI] [PubMed] [Google Scholar]
- Flames N, Pla R, Gelman DM, Rubenstein JL, Puelles L, Marin O. Delineation of multiple subpallial progenitor domains by the combinatorial expression of transcriptional codes. J Neurosci. 2007;27:9682–9695. doi: 10.1523/JNEUROSCI.2750-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida S, Hara K, Kobayashi A, Funato H, Hobara T, Otsuki K, et al. Early life stress enhances behavioral vulnerability to stress through the activation of REST4-mediated gene transcription in the medial prefrontal cortex of rodents. J Neurosci. 2010;30:15007–15018. doi: 10.1523/JNEUROSCI.1436-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas P, Joodmardi E, Hong Y, Perlmann T, Ogren SO. Adult mice with reduced Nurr1 expression: an animal model for schizophrenia. Mol Psychiatry. 2007;12:756–766. doi: 10.1038/sj.mp.4001993. [DOI] [PubMed] [Google Scholar]
- Johnson MB, Kawasawa YI, Mason CE, Krsnik Z, Coppola G, Bogdanovic D, et al. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron. 2009;62:494–509. doi: 10.1016/j.neuron.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Wang K, Valladares O, Hannenhalli S, Bucan M. Genome-wide expression profiling and bioinformatics analysis of diurnally regulated genes in the mouse prefrontal cortex. Genome Biol. 2007;8:R247. doi: 10.1186/gb-2007-8-11-r247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erraji-Benchekroun L, Underwood MD, Arango V, Galfalvy H, Pavlidis P, Smyrniotopoulos P, et al. Molecular aging in human prefrontal cortex is selective and continuous throughout adult life. Biol Psychiatry. 2005;57:549–558. doi: 10.1016/j.biopsych.2004.10.034. [DOI] [PubMed] [Google Scholar]
- Zhang YE, Landback P, Vibranovski MD, Long M. Accelerated recruitment of new brain development genes into the human genome. PLoS Biol. 2011;9:e1001179. doi: 10.1371/journal.pbio.1001179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegstein AR, Noctor SC. Patterns of neuronal migration in the embryonic cortex. Trends Neurosci. 2004;27:392–399. doi: 10.1016/j.tins.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Flint AC, Weissman TA, Dammerman RS, Kriegstein AR. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409:714–720. doi: 10.1038/35055553. [DOI] [PubMed] [Google Scholar]
- Storm EE, Garel S, Borello U, Hebert JM, Martinez S, McConnell SK, et al. Dose-dependent functions of Fgf8 in regulating telencephalic patterning centers. Development (Cambridge, England) 2006;133:1831–1844. doi: 10.1242/dev.02324. [DOI] [PubMed] [Google Scholar]
- Garel S, Huffman KJ, Rubenstein JL. Molecular regionalization of the neocortex is disrupted in Fgf8 hypomorphic mutants. Development (Cambridge, England) 2003;130:1903–1914. doi: 10.1242/dev.00416. [DOI] [PubMed] [Google Scholar]
- Raballo R, Rhee J, Lyn-Cook R, Leckman JF, Schwartz ML, Vaccarino FM. Basic fibroblast growth factor (Fgf2) is necessary for cell proliferation and neurogenesis in the developing cerebral cortex. J Neurosci. 2000;20:5012–5023. doi: 10.1523/JNEUROSCI.20-13-05012.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa A, Mao CA, Hadjantonakis AK, Klein WH, Broccoli V. Tbr2 directs conversion of radial glia into basal precursors and guides neuronal amplification by indirect neurogenesis in the developing neocortex. Neuron. 2008;60:56–69. doi: 10.1016/j.neuron.2008.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa A, Mao CA, Colasante G, Nini A, Klein WH, Broccoli V. Tbr2-positive intermediate (basal) neuronal progenitors safeguard cerebral cortex expansion by controlling amplification of pallial glutamatergic neurons and attraction of subpallial GABAergic interneurons. Genes Dev. 2010;24:1816–1826. doi: 10.1101/gad.575410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold SJ, Huang GJ, Cheung AF, Era T, Nishikawa S, Bikoff EK, et al. The T-box transcription factor Eomes/Tbr2 regulates neurogenesis in the cortical subventricular zone. Genes Dev. 2008;22:2479–2484. doi: 10.1101/gad.475408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney NT, Tierney H, Feldheim DA. Tbr2 is required to generate a neural circuit mediating the pupillary light reflex. J Neurosci. 2014;34:5447–5453. doi: 10.1523/JNEUROSCI.0035-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolk SM, Gunput RA, Tran TS, van den Heuvel DM, Prasad AA, Hellemons AJ, et al. Semaphorin 3F is a bifunctional guidance cue for dopaminergic axons and controls their fasciculation, channeling, rostral growth, and intracortical targeting. J Neurosci. 2009;29:12542–12557. doi: 10.1523/JNEUROSCI.2521-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinast K, Peeters D, Kolk SM, Schubert D, Homberg JR. Genetic and pharmacological manipulations of the serotonergic system in early life: neurodevelopmental underpinnings of autism-related behavior. Front Cell Neurosci. 2013;7:72. doi: 10.3389/fncel.2013.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popolo M, McCarthy DM, Bhide PG. Influence of dopamine on precursor cell proliferation and differentiation in the embryonic mouse telencephalon. Dev Neurosci. 2004;26:229–244. doi: 10.1159/000082140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegstein A, Noctor S, Martinez-Cerdeno V. Patterns of neural stem and progenitor cell division may underlie evolutionary cortical expansion. Nat Rev. 2006;7:883–890. doi: 10.1038/nrn2008. [DOI] [PubMed] [Google Scholar]
- Ayala R, Shu T, Tsai LH. Trekking across the brain: the journey of neuronal migration. Cell. 2007;128:29–43. doi: 10.1016/j.cell.2006.12.021. [DOI] [PubMed] [Google Scholar]
- Rakic P. The radial edifice of cortical architecture: from neuronal silhouettes to genetic engineering. Brain Res Rev. 2007;55:204–219. doi: 10.1016/j.brainresrev.2007.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuesel I. Reelin-mediated signaling in neuropsychiatric and neurodegenerative diseases. Prog Neurobiol. 2010;91:257–274. doi: 10.1016/j.pneurobio.2010.04.002. [DOI] [PubMed] [Google Scholar]
- Gaiano N. Strange bedfellows: Reelin and Notch signaling interact to regulate cell migration in the developing neocortex. Neuron. 2008;60:189–191. doi: 10.1016/j.neuron.2008.10.009. [DOI] [PubMed] [Google Scholar]
- Lui JH, Hansen DV, Kriegstein AR. Development and evolution of the human neocortex. Cell. 2011;146:18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMonica BE, Lui JH, Wang X, Kriegstein AR. OSVZ progenitors in the human cortex: an updated perspective on neurodevelopmental disease. Curr Opin Neurobiol. 2012;22:747–753. doi: 10.1016/j.conb.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertz CC, Lui JH, LaMonica BE, Wang X, Kriegstein AR. Diverse behaviors of outer radial glia in developing ferret and human cortex. J Neurosci. 2014;34:2559–2570. doi: 10.1523/JNEUROSCI.2645-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi G, Butt SJ, Takebayashi H, Fishell G. Physiologically distinct temporal cohorts of cortical interneurons arise from telencephalic Olig2-expressing precursors. J Neurosci. 2007;27:7786–7798. doi: 10.1523/JNEUROSCI.1807-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southwell DG, Nicholas CR, Basbaum AI, Stryker MP, Kriegstein AR, Rubenstein JL, et al. Interneurons from embryonic development to cell-based therapy. Science (New York, NY. 2014;344:1240622. doi: 10.1126/science.1240622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobos I, Long JE, Thwin MT, Rubenstein JL. Cellular patterns of transcription factor expression in developing cortical interneurons. Cereb Cortex. 2006;16:i82–i88. doi: 10.1093/cercor/bhk003. [DOI] [PubMed] [Google Scholar]
- Le Magueresse C, Monyer H. GABAergic interneurons shape the functional maturation of the cortex. Neuron. 2013;77:388–405. doi: 10.1016/j.neuron.2013.01.011. [DOI] [PubMed] [Google Scholar]
- Tessier-Lavigne M, Goodman CS. The molecular biology of axon guidance. Science (New York, NY. 1996;274:1123–1133. doi: 10.1126/science.274.5290.1123. [DOI] [PubMed] [Google Scholar]
- Dickson BJ. Molecular mechanisms of axon guidance. Science (New York, NY. 2002;298:1959–1964. doi: 10.1126/science.1072165. [DOI] [PubMed] [Google Scholar]
- O'Donnell M, Chance RK, Bashaw GJ. Axon growth and guidance: receptor regulation and signal transduction. Annu Rev Neurosci. 2009;32:383–412. doi: 10.1146/annurev.neuro.051508.135614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudanova I, Klein R. Integration of guidance cues: parallel signaling and crosstalk. Trends Neurosci. 2013;36:295–304. doi: 10.1016/j.tins.2013.01.007. [DOI] [PubMed] [Google Scholar]
- Kiryushko D, Berezin V, Bock E. Regulators of neurite outgrowth: role of cell adhesion molecules. Ann NY Acad Sci. 2004;1014:140–154. doi: 10.1196/annals.1294.015. [DOI] [PubMed] [Google Scholar]
- Stoya G, Redies C, Schmid-Hertel N. Inversion of layer-specific cadherin expression profiles and maintenance of cytoarchitectonic areas in the allocortex of the reeler mutant mouse. J Comp Neurol. 2014. [DOI] [PubMed]
- Lai KO, Ip NY. Synapse development and plasticity: roles of ephrin/Eph receptor signaling. Curr Opin Neurobiol. 2009;19:275–283. doi: 10.1016/j.conb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- Tessier CR, Broadie K. Activity-dependent modulation of neural circuit synaptic connectivity. Front Mol Neurosci. 2009;2:8. doi: 10.3389/neuro.02.008.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderhaeghen P, Cheng HJ. Guidance molecules in axon pruning and cell death. Cold Spring Harbor Perspect Biol. 2010;2:a001859. doi: 10.1101/cshperspect.a001859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petanjek Z, Judas M, Kostovic I, Uylings HB. Lifespan alterations of basal dendritic trees of pyramidal neurons in the human prefrontal cortex: a layer-specific pattern. Cereb Cortex. 2008;18:915–929. doi: 10.1093/cercor/bhm124. [DOI] [PubMed] [Google Scholar]
- de Almeida J, Jourdan I, Murer MG, Belforte JE. Refinement of neuronal synchronization with gamma oscillations in the medial prefrontal cortex after adolescence. PloS One. 2013;8:e62978. doi: 10.1371/journal.pone.0062978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JM, Zhang J, Yu YQ, Duan S, Li XM. Postnatal development of 2 microcircuits involving fast-spiking interneurons in the mouse prefrontal cortex. Cereb Cortex. 2014;24:98–109. doi: 10.1093/cercor/bhs291. [DOI] [PubMed] [Google Scholar]
- Hoover WB, Vertes RP. Anatomical analysis of afferent projections to the medial prefrontal cortex in the rat. Brain Struct Funct. 2007;212:149–179. doi: 10.1007/s00429-007-0150-4. [DOI] [PubMed] [Google Scholar]
- George O, Koob GF. Individual differences in prefrontal cortex function and the transition from drug use to drug dependence. Neurosci Biobehav Rev. 2010;35:232–247. doi: 10.1016/j.neubiorev.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisboa SF, Stecchini MF, Correa FM, Guimaraes FS, Resstel LB. Different role of the ventral medial prefrontal cortex on modulation of innate and associative learned fear. Neuroscience. 2010;171:760–768. doi: 10.1016/j.neuroscience.2010.09.048. [DOI] [PubMed] [Google Scholar]
- Diamond A. Biological and social influences on cognitive control processes dependent on prefrontal cortex. Prog Brain Res. 2011;189:319–339. doi: 10.1016/B978-0-444-53884-0.00032-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson BM, Baratta MV, Biedenkapp JC, Rudy JW, Watkins LR, Maier SF. Activation of the infralimbic cortex in a fear context enhances extinction learning. Learn Mem. 2010;17:591–599. doi: 10.1101/lm.1920810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laroche S, Davis S, Jay TM. Plasticity at hippocampal to prefrontal cortex synapses: dual roles in working memory and consolidation. Hippocampus. 2000;10:438–446. doi: 10.1002/1098-1063(2000)10:4<438::AID-HIPO10>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Little JP, Carter AG. Subcellular synaptic connectivity of layer 2 pyramidal neurons in the medial prefrontal cortex. J Neurosci. 2012;32:12808–12819. doi: 10.1523/JNEUROSCI.1616-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golmayo L, Nunez A, Zaborszky L. Electrophysiological evidence for the existence of a posterior cortical-prefrontal-basal forebrain circuitry in modulating sensory responses in visual and somatosensory rat cortical areas. Neuroscience. 2003;119:597–609. doi: 10.1016/s0306-4522(03)00031-9. [DOI] [PubMed] [Google Scholar]
- Henny P, Jones BE. Projections from basal forebrain to prefrontal cortex comprise cholinergic, GABAergic and glutamatergic inputs to pyramidal cells or interneurons. Eur J Neurosci. 2008;27:654–670. doi: 10.1111/j.1460-9568.2008.06029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauber W. Dopamine release in the prefrontal cortex and striatum: temporal and behavioural aspects. Pharmacopsychiatry. 43:S32–S41. doi: 10.1055/s-0030-1248300. [DOI] [PubMed] [Google Scholar]
- van Schouwenburg M, Aarts E, Cools R. Dopaminergic modulation of cognitive control:distinct roles for the prefrontal cortex and the basal ganglia. Curr Pharm Des. 2010;16:2026–2032. doi: 10.2174/138161210791293097. [DOI] [PubMed] [Google Scholar]
- Fitzgerald PJ. A neurochemical yin and yang: does serotonin activate and norepinephrine deactivate the prefrontal cortex. Psychopharmacology. 2011;213:171–182. doi: 10.1007/s00213-010-1856-1. [DOI] [PubMed] [Google Scholar]
- Witteveen JS, Middelman A, van Hulten JA, Martens GJ, Homberg JR, Kolk SM. Lack of serotonin reuptake during brain development alters rostral raphe-prefrontal network formation. Front Cell Neurosci. 2013;7:143. doi: 10.3389/fncel.2013.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto Y, O'Donnell P. Altered prefrontal cortex-nucleus accumbens information processing in a developmental animal model of schizophrenia. Ann NY Acad Sci. 2003;1003:398–401. doi: 10.1196/annals.1300.035. [DOI] [PubMed] [Google Scholar]
- Goto Y, Grace AA. Limbic and cortical information processing in the nucleus accumbens. Trends Neurosci. 2008;31:552–558. doi: 10.1016/j.tins.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarter M, Hasselmo ME, Bruno JP, Givens B. Unraveling the attentional functions of cortical cholinergic inputs: interactions between signal-driven and cognitive modulation of signal detection. Brain Res. 2005;48:98–111. doi: 10.1016/j.brainresrev.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Del Arco A, Mora F. Neurotransmitters and prefrontal cortex-limbic system interactions: implications for plasticity and psychiatric disorders. J Neural Transm. 2009;116:941–952. doi: 10.1007/s00702-009-0243-8. [DOI] [PubMed] [Google Scholar]
- Zhang ZW, Burke MW, Calakos N, Beaulieu JM, Vaucher E. Confocal analysis of cholinergic and dopaminergic inputs onto pyramidal cells in the prefrontal cortex of rodents. Front Neuroanat. 2010;4:21. doi: 10.3389/fnana.2010.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otani S, Daniel H, Roisin MP, Crepel F. Dopaminergic modulation of long-term synaptic plasticity in rat prefrontal neurons. Cereb Cortex. 2003;13:1251–1256. doi: 10.1093/cercor/bhg092. [DOI] [PubMed] [Google Scholar]
- Gabbott P, Headlam A, Busby S. Morphological evidence that CA1 hippocampal afferents monosynaptically innervate PV-containing neurons and NADPH-diaphorase reactive cells in the medial prefrontal cortex (Areas 25/32) of the rat. Brain Res. 2002;946:314–322. doi: 10.1016/s0006-8993(02)02487-3. [DOI] [PubMed] [Google Scholar]
- Gabbott PL, Warner TA, Busby SJ. Amygdala input monosynaptically innervates parvalbumin immunoreactive local circuit neurons in rat medial prefrontal cortex. Neuroscience. 2006;139:1039–1048. doi: 10.1016/j.neuroscience.2006.01.026. [DOI] [PubMed] [Google Scholar]
- Rotaru DC, Barrionuevo G, Sesack SR. Mediodorsal thalamic afferents to layer III of the rat prefrontal cortex: synaptic relationships to subclasses of interneurons. J Comp Neurol. 2005;490:220–238. doi: 10.1002/cne.20661. [DOI] [PubMed] [Google Scholar]
- Little JP, Carter AG. Synaptic mechanisms underlying strong reciprocal connectivity between the medial prefrontal cortex and basolateral amygdala. J Neurosci. 2013;33:15333–15342. doi: 10.1523/JNEUROSCI.2385-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst M, Fudge JL. A developmental neurobiological model of motivated behavior: anatomy, connectivity and ontogeny of the triadic nodes. Neurosci Biobehav Reviews. 2009;33:367–382. doi: 10.1016/j.neubiorev.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst M, Pine DS, Hardin M. Triadic model of the neurobiology of motivated behavior in adolescence. Psychol Med. 2006;36:299–312. doi: 10.1017/S0033291705005891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senn V, Wolff SB, Herry C, Grenier F, Ehrlich I, Grundemann J, et al. Long-range connectivity defines behavioral specificity of amygdala neurons. Neuron. 2014;81:428–437. doi: 10.1016/j.neuron.2013.11.006. [DOI] [PubMed] [Google Scholar]
- Heidbreder CA, Groenewegen HJ. The medial prefrontal cortex in the rat: evidence for a dorso-ventral distinction based upon functional and anatomical characteristics. Neurosci Biobehav Rev. 2003;27:555–579. doi: 10.1016/j.neubiorev.2003.09.003. [DOI] [PubMed] [Google Scholar]
- Gabbott PL, Warner TA, Jays PR, Salway P, Busby SJ. Prefrontal cortex in the rat: projections to subcortical autonomic, motor, and limbic centers. J Comp Neurol. 2005;492:145–177. doi: 10.1002/cne.20738. [DOI] [PubMed] [Google Scholar]
- Dembrow NC, Chitwood RA, Johnston D. Projection-specific neuromodulation of medial prefrontal cortex neurons. J Neurosci. 2010;30:16922–16937. doi: 10.1523/JNEUROSCI.3644-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding DC, Gabbott PL, Totterdell S. Differences in the laminar origin of projections from the medial prefrontal cortex to the nucleus accumbens shell and core regions in the rat. Brain Res. 2001;917:81–89. doi: 10.1016/s0006-8993(01)02912-2. [DOI] [PubMed] [Google Scholar]
- Cholfin JA, Rubenstein JL.Genetic regulation of prefrontal cortex development and function Novartis Found Symp. 2007288165–173.discussion 73–7, 276–81. [PubMed] [Google Scholar]
- Schell-Apacik C, Rivero M, Knepper JL, Roessler E, Muenke M, Ming JE. SONIC HEDGEHOG mutations causing human holoprosencephaly impair neural patterning activity. Hum Genet. 2003;113:170–177. doi: 10.1007/s00439-003-0950-4. [DOI] [PubMed] [Google Scholar]
- Al-Ayadhi LY. Relationship between Sonic hedgehog protein, brain-derived neurotrophic factor and oxidative stress in autism spectrum disorders. Neurochem Res. 2012;37:394–400. doi: 10.1007/s11064-011-0624-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heussler HS, Suri M, Young ID, Muenke M. Extreme variability of expression of a Sonic Hedgehog mutation: attention difficulties and holoprosencephaly. Arch Dis Child. 2002;86:293–296. doi: 10.1136/adc.86.4.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raballo R, Rhee J, Lyn-Cook R, Leckman JF, Schwartz ML, Vaccarino FM. Basic fibroblast growth factor (Fgf2) is necessary for cell proliferation and neurogenesis in the developing cerebral cortex. J Neurosci. 2000;20:5012–5023. doi: 10.1523/JNEUROSCI.20-13-05012.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwisscha van Scheltinga AF, Bakker SC, Kahn RS, Kas MJ. Fibroblast growth factors in neurodevelopment and psychopathology. Neuroscientist. 2013;19:479–494. doi: 10.1177/1073858412472399. [DOI] [PubMed] [Google Scholar]
- Hashimoto K, Shimizu E, Komatsu N, Nakazato M, Okamura N, Watanabe H, et al. Increased levels of serum basic fibroblast growth factor in schizophrenia. Psychiatry Res. 2003;120:211–218. doi: 10.1016/s0165-1781(03)00186-0. [DOI] [PubMed] [Google Scholar]
- Simonis N, Migeotte I, Lambert N, Perazzolo C, de Silva DC, Dimitrov B, et al. FGFR1 mutations cause Hartsfield syndrome, the unique association of holoprosencephaly and ectrodactyly. J Med Genet. 2013;50:585–592. doi: 10.1136/jmedgenet-2013-101603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaughran F, Payne J, Sedgwick PM, Cotter D, Berry M. Hippocampal FGF-2 and FGFR1 mRNA expression in major depression, schizophrenia and bipolar disorder. Brain Res Bull. 2006;70:221–227. doi: 10.1016/j.brainresbull.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Stachowiak MK, Kucinski A, Curl R, Syposs C, Yang Y, Narla S, et al. Schizophrenia: a neurodevelopmental disorder – integrative genomic hypothesis and therapeutic implications from a transgenic mouse model. Schizophr Res. 2013;143:367–376. doi: 10.1016/j.schres.2012.11.004. [DOI] [PubMed] [Google Scholar]
- Mahieu-Caputo D, Sonigo P, Amiel J, Simon I, Aubry MC, Lemerrer M, et al. Prenatal diagnosis of sporadic Apert syndrome: a sequential diagnostic approach combining three-dimensional computed tomography and molecular biology. Fetal Diagn Ther. 2001;16:10–12. doi: 10.1159/000053872. [DOI] [PubMed] [Google Scholar]
- Padmanabhan V, Hegde AM, Rai K. Crouzon's syndrome: a review of literature and case report. Contemp Clin Dent. 2011;2:211–214. doi: 10.4103/0976-237X.86464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentz E, Vujic M, Karrstedt EL, Erlandsson A, Gillberg C. A case report of two male siblings with autism and duplication of Xq13-q21, a region including three genes predisposing for autism. European Child Adolesc Psychiatry. 2014;23:329–336. doi: 10.1007/s00787-013-0455-1. [DOI] [PubMed] [Google Scholar]
- Kaga Y, Shoemaker WJ, Furusho M, Bryant M, Rosenbluth J, Pfeiffer SE, et al. Mice with conditional inactivation of fibroblast growth factor receptor-2 signaling in oligodendrocytes have normal myelin but display dramatic hyperactivity when combined with Cnp1 inactivation. J Neurosci. 2006;26:12339–12350. doi: 10.1523/JNEUROSCI.3573-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donovan MC, Norton N, Williams H, Peirce T, Moskvina V, Nikolov I, et al. Analysis of 10 independent samples provides evidence for association between schizophrenia and a SNP flanking fibroblast growth factor receptor 2. Mol Psychiatry. 2009;14:30–36. doi: 10.1038/mp.2008.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens HE, Su T, Yanagawa Y, Vaccarino FM. Prenatal stress delays inhibitory neuron progenitor migration in the developing neocortex. Psychoneuroendocrinology. 2013;38:509–521. doi: 10.1016/j.psyneuen.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theisen A, Rosenfeld JA, Shane K, McBride KL, Atkin JF, Gaba C, et al. Refinement of the region for split hand/foot malformation 5 on 2q31.1. Mol Syndromol. 2010;1:262–271. doi: 10.1159/000328405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Novosedlik N, Wang A, Hudson ML, Cohen IL, Chudley AE, et al. The DLX1and DLX2 genes and susceptibility to autism spectrum disorders. Eur J Hum Genet. 2009;17:228–235. doi: 10.1038/ejhg.2008.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida T, Furukawa T, Iwata S, Yanagawa Y, Fukuda A. Selective loss of parvalbumin-positive GABAergic interneurons in the cerebral cortex of maternally stressed Gad1-heterozygous mouse offspring. Transl Psychiatry. 2014;4:e371. doi: 10.1038/tp.2014.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarus MS, Krishnan K, Huang ZJ.GAD67 Deficiency in parvalbumin interneurons produces deficits in inhibitory transmission and network disinhibition in mouse prefrontal cortex Cereb Cortex 2013(e-pub ahead of print). [DOI] [PMC free article] [PubMed]
- Bacchelli E, Blasi F, Biondolillo M, Lamb JA, Bonora E, Barnby G, et al. Screening of nine candidate genes for autism on chromosome 2q reveals rare nonsynonymous variants in the cAMP-GEFII gene. Mol Psychiatry. 2003;8:916–924. doi: 10.1038/sj.mp.4001340. [DOI] [PubMed] [Google Scholar]
- Rabionet R, Jaworski JM, Ashley-Koch AE, Martin ER, Sutcliffe JS, Haines JL, et al. Analysis of the autism chromosome 2 linkage region: GAD1 and other candidate genes. Neurosci Lett. 2004;372:209–214. doi: 10.1016/j.neulet.2004.09.037. [DOI] [PubMed] [Google Scholar]
- Buttenschon HN, Lauritsen MB, El Daoud A, Hollegaard M, Jorgensen M, Tvedegaard K, et al. A population-based association study of glutamate decarboxylase 1 as a candidate gene for autism. J Neural Transm. 2009;116:381–388. doi: 10.1007/s00702-008-0142-4. [DOI] [PubMed] [Google Scholar]
- Chang SC, Pauls DL, Lange C, Sasanfar R, Santangelo SL. Common genetic variation in the GAD1 gene and the entire family of DLX homeobox genes and autism spectrum disorders. Am J Med Genet B Neuropsychiatr Genet. 2011;156:233–239. doi: 10.1002/ajmg.b.31148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbarian S, Huang HS. Molecular and cellular mechanisms of altered GAD1/GAD67 expression in schizophrenia and related disorders. Brain Res Rev. 2006;52:293–304. doi: 10.1016/j.brainresrev.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Ayalew M, Le-Niculescu H, Levey DF, Jain N, Changala B, Patel SD, et al. Convergent functional genomics of schizophrenia: from comprehensive understanding to genetic risk prediction. Mol Psychiatry. 2012;17:887–905. doi: 10.1038/mp.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub RE, Lipska BK, Egan MF, Goldberg TE, Callicott JH, Mayhew MB, et al. Allelic variation in GAD1 (GAD67) is associated with schizophrenia and influences cortical function and gene expression. Mol Psychiatry. 2007;12:854–869. doi: 10.1038/sj.mp.4001988. [DOI] [PubMed] [Google Scholar]
- Cherlyn SY, Woon PS, Liu JJ, Ong WY, Tsai GC, Sim K. Genetic association studies of glutamate, GABA and related genes in schizophrenia and bipolar disorder: a decade of advance. Neurosci Biobehav Rev. 2010;34:958–977. doi: 10.1016/j.neubiorev.2010.01.002. [DOI] [PubMed] [Google Scholar]
- Cooper MA, Koleske AJ. Ablation of ErbB4 from excitatory neurons leads to reduced dendritic spine density in mouse prefrontal cortex. J Comp Neurol. 2014;522:3351–3362. doi: 10.1002/cne.23615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting AK, Chen Y, Wen L, Yin DM, Shen C, Tao Y, et al. Neuregulin 1 promotes excitatory synapse development and function in GABAergic interneurons. J Neurosci. 2011;31:15–25. doi: 10.1523/JNEUROSCI.2538-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou XJ, Ni KM, Yang JM, Li XM. Neuregulin 1/ErbB4 enhances synchronized oscillations of prefrontal cortex neurons via inhibitory synapses. Neuroscience. 2014;261:107–117. doi: 10.1016/j.neuroscience.2013.12.040. [DOI] [PubMed] [Google Scholar]
- Yang JM, Zhang J, Chen XJ, Geng HY, Ye M, Spitzer NC, et al. Development of GABA circuitry of fast-spiking basket interneurons in the medial prefrontal cortex of erbb4-mutant mice. J Neurosci. 2013;33:19724–19733. doi: 10.1523/JNEUROSCI.1584-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasnauskiene J, Ciuladaite Z, Preiksaitiene E, Utkus A, Peciulyte A, Kucinskas V. A new single gene deletion on 2q34: ERBB4 is associated with intellectual disability. Am J Med Genet. 2013;161A:1487–1490. doi: 10.1002/ajmg.a.35911. [DOI] [PubMed] [Google Scholar]
- Joshi D, Fullerton JM, Weickert CS. Elevated ErbB4 mRNA is related to interneuron deficit in prefrontal cortex in schizophrenia. J Psychiatr Res. 2014;53:125–132. doi: 10.1016/j.jpsychires.2014.02.014. [DOI] [PubMed] [Google Scholar]
- Del Pino I, Garcia-Frigola C, Dehorter N, Brotons-Mas JR, Alvarez-Salvado E, Martinez de Lagran M, et al. Erbb4 deletion from fast-spiking interneurons causes schizophrenia-like phenotypes. Neuron. 2013;79:1152–1168. doi: 10.1016/j.neuron.2013.07.010. [DOI] [PubMed] [Google Scholar]
- Buonanno A. The neuregulin signaling pathway and schizophrenia: from genes to synapses and neural circuits. Brain Res Bull. 2010;83:122–131. doi: 10.1016/j.brainresbull.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang UE, Puls I, Muller DJ, Strutz-Seebohm N, Gallinat J. Molecular mechanisms of schizophrenia. Cell Physiol Biochem. 2007;20:687–702. doi: 10.1159/000110430. [DOI] [PubMed] [Google Scholar]
- Yonan AL, Palmer AA, Smith KC, Feldman I, Lee HK, Yonan JM, et al. Bioinformatic analysis of autism positional candidate genes using biological databases and computational gene network prediction. Genes Brain Behav. 2003;2:303–320. doi: 10.1034/j.1601-183x.2003.00041.x. [DOI] [PubMed] [Google Scholar]
- Yonan AL, Alarcon M, Cheng R, Magnusson PK, Spence SJ, Palmer AA, et al. A genomewide screen of 345 families for autism-susceptibility loci. Am J Hum Genet. 2003;73:886–897. doi: 10.1086/378778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves-Pereira M, Muller B, Massie D, Williams JH, O'Brien PC, Hughes A, et al. Deregulation of EIF4E: a novel mechanism for autism. J Med Genet. 2009;46:759–765. doi: 10.1136/jmg.2009.066852. [DOI] [PubMed] [Google Scholar]
- Gkogkas CG, Sonenberg N. Translational control and autism-like behaviors. Cell Logist. 2013;3:e24551. doi: 10.4161/cl.24551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger DD, Osterweil EK, Chen SP, Tye LD, Bear MF. Cognitive dysfunction and prefrontal synaptic abnormalities in a mouse model of fragile X syndrome. Proc Natl Acad Sci USA. 2011;108:2587–2592. doi: 10.1073/pnas.1013855108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhakar AL, Dolen G, Bear MF. The pathophysiology of fragile X (and what it teaches us about synapses) Annu Rev Neurosci. 2012;35:417–443. doi: 10.1146/annurev-neuro-060909-153138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin SA, Galvez R, Greenough WT. Dendritic spine structural anomalies in fragile-X mental retardation syndrome. Cereb Cortex. 2000;10:1038–1044. doi: 10.1093/cercor/10.10.1038. [DOI] [PubMed] [Google Scholar]
- De Rubeis S, Fernandez E, Buzzi A, Di Marino D, Bagni C. Molecular and cellular aspects of mental retardation in the Fragile X syndrome: from gene mutation/s to spine dysmorphogenesis. Adv Exp Med Biol. 2012;970:517–551. doi: 10.1007/978-3-7091-0932-8_23. [DOI] [PubMed] [Google Scholar]
- Maurin T, Zongaro S, Bardoni B.Fragile X Syndrome: from molecular pathology to therapy Neurosci Biobehav Rev 2014(in press). [DOI] [PubMed]
- Rotschafer SE, Razak KA. Auditory processing in fragile x syndrome. Front Cell Neurosci. 2014;8:19. doi: 10.3389/fncel.2014.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oddi D, Crusio WE, D'Amato FR, Pietropaolo S. Monogenic mouse models of social dysfunction: implications for autism. Behav Brain Res. 2013;251:75–84. doi: 10.1016/j.bbr.2013.01.002. [DOI] [PubMed] [Google Scholar]
- Tranfaglia MR. The psychiatric presentation of fragile x: evolution of the diagnosis and treatment of the psychiatric comorbidities of fragile X syndrome. Dev Neurosci. 2011;33:337–348. doi: 10.1159/000329421. [DOI] [PubMed] [Google Scholar]
- Kramvis I, Mansvelder HD, Loos M, Meredith R. Hyperactivity, perseveration and increased responding during attentional rule acquisition in the Fragile X mouse model. Front Behav Neurosci. 2013;7:172. doi: 10.3389/fnbeh.2013.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim L, He L, Maaswinkel H, Zhu L, Sirotkin H, Weng W. Anxiety, hyperactivity and stereotypy in a zebrafish model of fragile X syndrome and autism spectrum disorder. Prog Neuropsychopharmacol Biol Psychiatry. 2014;55:40–49. doi: 10.1016/j.pnpbp.2014.03.007. [DOI] [PubMed] [Google Scholar]
- Kraan CM, Hocking DR, Georgiou-Karistianis N, Metcalfe SA, Archibald AD, Fielding J, et al. Impaired response inhibition is associated with self- reported symptoms of depression, anxiety, and ADHD in female FMR1 premutation carriers. Am J Med Genet B Neuropsychiatr Genet. 2014;165:41–51. doi: 10.1002/ajmg.b.32203. [DOI] [PubMed] [Google Scholar]
- Hunter JE, Epstein MP, Tinker SW, Abramowitz A, Sherman SL. The FMR1 premutation and attention-deficit hyperactivity disorder (ADHD): evidence for a complex inheritance. Behav Genet. 2012;42:415–422. doi: 10.1007/s10519-011-9520-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs T, Kelemen O, Keri S. Decreased fragile X mental retardation protein (FMRP) is associated with lower IQ and earlier illness onset in patients with schizophrenia. Psychiatry Res. 2013;210:690–693. doi: 10.1016/j.psychres.2012.12.022. [DOI] [PubMed] [Google Scholar]
- Fatemi SH, Folsom TD. The role of fragile X mental retardation protein in major mental disorders. Neuropharmacology. 2011;60:1221–1226. doi: 10.1016/j.neuropharm.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–184. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav R, Gupta SC, Hillman BG, Bhatt JM, Stairs DJ, Dravid SM. Deletion of glutamate delta-1 receptor in mouse leads to aberrant emotional and social behaviors. PloS One. 2012;7:e32969. doi: 10.1371/journal.pone.0032969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griswold AJ, Ma D, Cukier HN, Nations LD, Schmidt MA, Chung RH, et al. Evaluation of copy number variations reveals novel candidate genes in autism spectrum disorder-associated pathways. Hum Mol Genet. 2012;21:3513–3523. doi: 10.1093/hmg/dds164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo SZ, Huang K, Shi YY, Tang W, Zhou J, Feng GY, et al. A case-control association study between the GRID1 gene and schizophrenia in the Chinese Northern Han population. Schizophr Res. 2007;93:385–390. doi: 10.1016/j.schres.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Treutlein J, Muhleisen TW, Frank J, Mattheisen M, Herms S, Ludwig KU, et al. Dissection of phenotype reveals possible association between schizophrenia and Glutamate Receptor Delta 1 (GRID1) gene promoter. Schizophr Res. 2009;111:123–130. doi: 10.1016/j.schres.2009.03.011. [DOI] [PubMed] [Google Scholar]
- Kolk SM, Gunput RA, Tran TS, van den Heuvel DM, Prasad AA, Hellemons AJ, et al. Semaphorin 3F is a bifunctional guidance cue for dopaminergic axons and controls their fasciculation, channeling, rostral growth, and intracortical targeting. J Neurosci. 2009;29:12542–12557. doi: 10.1523/JNEUROSCI.2521-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks-Kayal A. Molecular mechanisms of cognitive and behavioral comorbidities of epilepsy in children. Epilepsia. 2011;52:13–20. doi: 10.1111/j.1528-1167.2010.02906.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Yue W, Jia M, Ruan Y, Lu T, Gong X, et al. Association of the neuropilin-2 (NRP2) gene polymorphisms with autism in Chinese Han population. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:492–495. doi: 10.1002/ajmg.b.30495. [DOI] [PubMed] [Google Scholar]
- Iafrati J, Orejarena MJ, Lassalle O, Bouamrane L, Gonzalez-Campo C, Chavis P. Reelin, an extracellular matrix protein linked to early onset psychiatric diseases, drives postnatal development of the prefrontal cortex via GluN2B-NMDARs and the mTOR pathway. Mol Psychiatry. 2014;19:417–426. doi: 10.1038/mp.2013.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki M, Shehab M, El-Aleem AA, Abdel-Salam G, Koeller HB, Ilkin Y, et al. Identification of a novel recessive RELN mutation using a homozygous balanced reciprocal translocation. Am J Med Genet. 2007;143A:939–944. doi: 10.1002/ajmg.a.31667. [DOI] [PubMed] [Google Scholar]
- Folsom TD, Fatemi SH. The involvement of Reelin in neurodevelopmental disorders. Neuropharmacology. 2013;68:122–135. doi: 10.1016/j.neuropharm.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett CW, Gharani N, Millonig JH, Brzustowicz LM. Three autism candidate genes: a synthesis of human genetic analysis with other disciplines. Int J Dev Neurosci. 2005;23:221–234. doi: 10.1016/j.ijdevneu.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Fatemi SH. The role of Reelin in pathology of autism. Mol Psychiatry. 2002;7:919–920. doi: 10.1038/sj.mp.4001248. [DOI] [PubMed] [Google Scholar]
- Fatemi SH. Reelin glycoprotein in autism and schizophrenia. Int Rev Neurobiol. 2005;71:179–187. doi: 10.1016/s0074-7742(05)71008-4. [DOI] [PubMed] [Google Scholar]
- Lakatosova S, Ostatnikova D. Reelin and its complex involvement in brain development and function. Int J Biochem Cell Biol. 2012;44:1501–1504. doi: 10.1016/j.biocel.2012.06.002. [DOI] [PubMed] [Google Scholar]
- Frotscher M. Role for Reelin in stabilizing cortical architecture. Trends Neurosci. 2010;33:407–414. doi: 10.1016/j.tins.2010.06.001. [DOI] [PubMed] [Google Scholar]
- Zhang W, Peterson M, Beyer B, Frankel WN, Zhang ZW. Loss of MeCP2 from forebrain excitatory neurons leads to cortical hyperexcitation and seizures. J Neurosci. 2014;34:2754–2763. doi: 10.1523/JNEUROSCI.4900-12.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francke U. Mechanisms of disease: neurogenetics of MeCP2 deficiency. Nat Clin Pract Neurol. 2006;2:212–221. doi: 10.1038/ncpneuro0148. [DOI] [PubMed] [Google Scholar]
- Samaco RC, Neul JL. Complexities of Rett syndrome and MeCP2. J Neurosci. 2011;31:7951–7959. doi: 10.1523/JNEUROSCI.0169-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanz TA, Guilmette E, Gosink MM, Fischer JE, Fitzgerald LW, Stephenson DT, et al. Transcriptomic analysis of genetically defined autism candidate genes reveals common mechanisms of action. Mol Autism. 2013;4:45. doi: 10.1186/2040-2392-4-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter B, Treadwell-Deering D, Zoghbi HY, Glaze DG, Neul JL. Brief report: MECP2 mutations in people without Rett syndrome. J Autism Dev Disord. 2014;44:703–711. doi: 10.1007/s10803-013-1902-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neul JL. The relationship of Rett syndrome and MECP2 disorders to autism. Dialogues Clin Neurosci. 2012;14:253–262. doi: 10.31887/DCNS.2012.14.3/jneul. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy SE, Gillis J, Kramer M, Lihm J, Yoon S, Berstein Y, et al. De novo mutations in schizophrenia implicate chromatin remodeling and support a genetic overlap with autism and intellectual disability. Mol Psychiatry. 2014;19:652–658. doi: 10.1038/mp.2014.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piton A, Gauthier J, Hamdan FF, Lafreniere RG, Yang Y, Henrion E, et al. Systematic resequencing of X-chromosome synaptic genes in autism spectrum disorder and schizophrenia. Mol Psychiatry. 2011;16:867–880. doi: 10.1038/mp.2010.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Goldman JS, Rymar VV, Forget C, Lo PS, Bull SJ, et al. Critical roles for the netrin receptor deleted in colorectal cancer in dopaminergic neuronal precursor migration, axon guidance, and axon arborization. Neuroscience. 2010;169:932–949. doi: 10.1016/j.neuroscience.2010.05.025. [DOI] [PubMed] [Google Scholar]
- Manitt C, Eng C, Pokinko M, Ryan RT, Torres-Berrio A, Lopez JP, et al. Dcc orchestrates the development of the prefrontal cortex during adolescence and is altered in psychiatric patients. Transl Psychiatry. 2013;3:e338. doi: 10.1038/tp.2013.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant A, Fathalli F, Rouleau G, Joober R, Flores C. Association between schizophrenia and genetic variation in DCC: a case-control study. Schizophr Res. 2012;137:26–31. doi: 10.1016/j.schres.2012.02.023. [DOI] [PubMed] [Google Scholar]
- El-Hassar L, Simen AA, Duque A, Patel KD, Kaczmarek LK, Arnsten AF, et al. Disrupted in schizophrenia 1 modulates medial prefrontal cortex pyramidal neuron activity through cAMP regulation of transient receptor potential C and small-conductance K channels. Biol Psychiatry. 2014;76:476–485. doi: 10.1016/j.biopsych.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpinen H, Ylisaukko-Oja T, Hennah W, Palo OM, Varilo T, Vanhala R, et al. Association of DISC1 with autism and Asperger syndrome. Mol Psychiatry. 2008;13:187–196. doi: 10.1038/sj.mp.4002031. [DOI] [PubMed] [Google Scholar]
- Zheng F, Wang L, Jia M, Yue W, Ruan Y, Lu T, et al. Evidence for association between disrupted-in-Schizophrenia 1 (DISC1) gene polymorphisms and autism in Chinese Han population: a family-based association study. Behav Brain Funct. 2011;7:14. doi: 10.1186/1744-9081-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny EM, Cormican P, Furlong S, Heron E, Kenny G, Fahey C, et al. Excess of rare novel loss-of-function variants in synaptic genes in schizophrenia and autism spectrum disorders. Mol Psychiatry. 2013;19:872–879. doi: 10.1038/mp.2013.127. [DOI] [PubMed] [Google Scholar]
- Kalkman HO. A review of the evidence for the canonical Wnt pathway in autism spectrum disorders. Mol Autism. 2012;3:10. doi: 10.1186/2040-2392-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen KK, Halmoy A, Sanchez-Mora C, Ramos-Quiroga JA, Cormand B, Haavik J, et al. DISC1 in adult ADHD patients: an association study in two European samples. Am J Med Genet B Neuropsychiatr Genet. 2013;162B:227–234. doi: 10.1002/ajmg.b.32136. [DOI] [PubMed] [Google Scholar]
- Narayan S, Nakajima K, Sawa A. DISC1: a key lead in studying cortical development and associated brain disorders. Neuroscientist. 2013;19:451–464. doi: 10.1177/1073858412470168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikida T, Gamo NJ, Sawa A. DISC1 as a therapeutic target for mental illnesses. Expert Opin Ther Targets. 2012;16:1151–1160. doi: 10.1517/14728222.2012.719879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duff BJ, Macritchie KA, Moorhead TW, Lawrie SM, Blackwood DH. Human brain imaging studies of DISC1 in schizophrenia, bipolar disorder and depression: a systematic review. Schizophr Res. 2013;147:1–13. doi: 10.1016/j.schres.2013.03.015. [DOI] [PubMed] [Google Scholar]
- Drerup JM, Hayashi K, Cui H, Mettlach GL, Long MA, Marvin M, et al. Attention-deficit/hyperactivity phenotype in mice lacking the cyclin- dependent kinase 5 cofactor p35. Biol Psychiatry. 2010;68:1163–1171. doi: 10.1016/j.biopsych.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Miguel A, Meana JJ, Garcia-Sevilla JA. Cyclin-dependent kinase-5 and p35/p25 activators in schizophrenia and major depression prefrontal cortex: basal contents and effects of psychotropic medications. Int J Neuropsychopharmacol. 2013;16:683–689. doi: 10.1017/S1461145712000879. [DOI] [PubMed] [Google Scholar]
- Engmann O, Hortobagyi T, Pidsley R, Troakes C, Bernstein HG, Kreutz MR, et al. Schizophrenia is associated with dysregulation of a Cdk5 activator that regulates synaptic protein expression and cognition. Brain. 2011;134:2408–2421. doi: 10.1093/brain/awr155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitwell JL, Josephs KA, Avula R, Tosakulwong N, Weigand SD, Senjem ML, et al. Altered functional connectivity in asymptomatic MAPT subjects: a comparison to bvFTD. Neurology. 2011;77:866–874. doi: 10.1212/WNL.0b013e31822c61f2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregor A, Krumbiegel M, Kraus C, Reis A, Zweier C. De novo triplication of the MAPT gene from the recurrent 17q21.31 microdeletion region in a patient with moderate intellectual disability and various minor anomalies. Am J Med Genet A. 2012;158A:1765–1770. doi: 10.1002/ajmg.a.35427. [DOI] [PubMed] [Google Scholar]
- Rovelet-Lecrux A, Campion D. Copy number variations involving the microtubule-associated protein tau in human diseases. Biochem Soc Trans. 2012;40:672–676. doi: 10.1042/BST20120045. [DOI] [PubMed] [Google Scholar]
- Sapir T, Frotscher M, Levy T, Mandelkow EM, Reiner O. Tau's role in the developing brain: implications for intellectual disability. Hum Mol Genet. 2012;21:1681–1692. doi: 10.1093/hmg/ddr603. [DOI] [PubMed] [Google Scholar]
- Kitsiou-Tzeli S, Frysira H, Giannikou K, Syrmou A, Kosma K, Kakourou G, et al. Microdeletion and microduplication 17q21.31 plus an additional CNV, in patients with intellectual disability, identified by array-CGH. Gene. 2012;492:319–324. doi: 10.1016/j.gene.2011.10.023. [DOI] [PubMed] [Google Scholar]
- Runker AE, O'Tuathaigh C, Dunleavy M, Morris DW, Little GE, Corvin AP, et al. Mutation of Semaphorin-6A disrupts limbic and cortical connectivity and models neurodevelopmental psychopathology. PloS One. 2011;6:e26488. doi: 10.1371/journal.pone.0026488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosia M, Anselmetti S, Pirovano A, Ermoli E, Marino E, Bramanti P, et al. HTTLPR functional polymorphism in schizophrenia: executive functions versus sustained attention dissociation. Prog Neuropsychopharmacol Biol Psychiatry. 2010;34:81–85. doi: 10.1016/j.pnpbp.2009.10.001. [DOI] [PubMed] [Google Scholar]
- Lin C, Tang W, Hu J, Gao L, Huang K, Xu Y, et al. Haplotype analysis confirms association of the serotonin transporter (5-HTT) gene with schizophrenia in the Han Chinese population. Neurosci Lett. 2009;453:210–213. doi: 10.1016/j.neulet.2009.02.023. [DOI] [PubMed] [Google Scholar]