Abstract
The localization and specialized function of Ras-like proteins are largely determined by posttranslational processing events. In a highly regulated process, palmitoyl groups may be added to C-terminal cysteine residues, targeting these proteins to specific membranes. In the human fungal pathogen Cryptococcus neoformans, Ras1 protein palmitoylation is essential for growth at high temperature but is dispensable for sexual differentiation. Ras1 palmitoylation is also required for localization of this protein on the plasma membrane. Together, these results support a model in which specific Ras functions are mediated from different subcellular locations. We therefore hypothesize that proteins that activate Ras1 or mediate Ras1 localization to the plasma membrane will be important for C. neoformans pathogenesis. To further characterize the Ras1 signaling cascade mediating high-temperature growth, we have identified a family of protein S-acyltransferases (PATs), enzymes that mediate palmitoylation, in the C. neoformans genome database. Deletion strains for each candidate gene were generated by homogenous recombination, and each mutant strain was assessed for Ras1-mediated phenotypes, including high-temperature growth, morphogenesis, and sexual development. We found that full Ras1 palmitoylation and function required one particular PAT, Pfa4, and deletion of the PFA4 gene in C. neoformans resulted in altered Ras1 localization to membranes, impaired growth at 37°C, and reduced virulence.
INTRODUCTION
S-palmitoylation involves the reversible attachment of palmitic acid moieties to proteins. This process of posttranslational modification is catalyzed by the protein S-acyltransferase (PAT) enzymes through the thioesterification of a cysteine side chain group. By the addition of this hydrophobic palmitoyl group, proteins often become associated with membranes. However, in contrast to constitutive protein modifications such as prenylation, palmitoylation is often regulated and reversible, allowing for dynamic changes in patterns of subcellular protein localization (1, 2).
PAT genes were first identified in the budding yeast Saccharomyces cerevisiae as the enzymes responsible for the palmitoylation of Ras2 (Erf2) and yeast casein kinase (Akr1) (3, 4). The PAT family is characterized by several molecular features, including multiple transmembrane domains, as well as a DHHC motif (Asp-His-His-Cys) within an approximately 50-amino-acid cysteine-rich domain (CRD) (5). Seven PAT family members have now been identified in yeast, while 24 of these proteins have been identified in mammals. Interestingly, nine PATs have been associated with human disease ranging from neuropsychiatric conditions such as Alzheimer's and Huntington's disease to developmental conditions (Golz syndrome) and cancer (6).
Unlike other types of lipidation, such as prenylation or N-myristoylation, there is no specific consensus sequence for protein palmitoylation. Therefore, simple bioinformatic prediction of palmitoylated proteins has been challenging. However, a comprehensive analysis of palmitoylated proteins in yeast found that there were conserved features among many of the cysteine residues that are likely to undergo this type of modification: most are located near prenylated/myristoylated residues, most are surrounded by basic or hydrophobic amino acids, and many lie in the cytoplasmic regions of membrane-spanning proteins (7).
Lipidation of proteins by palmitoylation mediates membrane localization. The importance of protein localization on function has been extensively explored, especially for Ras-like proteins. In mammalian cells, palmitoylation of Ras dictates not only localization to particular membranes, such as the Golgi complex, plasma membrane (PM), and microdomains within the PM, but also downstream signaling from Ras (8, 9). In the fission yeast Schizosaccharomyces pombe, two distinct Ras protein signaling programs are largely determined by palmitoylation and localization of Ras1 on different cellular membranes (10).
In C. neoformans the Ras1 (CnRas1) protein is required for pathogenesis, morphogenesis, and sexual differentiation. Recently, we demonstrated that palmitoylation and subsequent plasma membrane attachment of Ras1 are essential for the maintenance of normal morphology and pathogenicity. However, protein palmitoylation does not appear to be required for other Ras-related phenotypes such as mating and hyphal differentiation (11).
To elucidate the PAT responsible for C. neoformans Ras1 palmitoylation, we have identified seven genes predicted to encode DHHC motif-containing proteins. Targeted deletion of most of these genes does not affect Ras-dependent cellular functions. However, the pfa4Δ mutant shares some phenotypic similarities with an ras1Δ mutant, such as impaired growth at 37°C and attenuated virulence. Moreover, PM association of a green fluorescent protein (GFP)-Ras1 fusion protein is altered in the pfa4Δ mutant strain compared to that in the wild type, and palmitoylation of Ras1 is reduced in this mutant background. We also found that Pfa4 was required for Ras1-independent phenotypes, indicating that this protein clearly has other targets than Ras proteins. Also, our genetic and biochemical analyses suggest some degree of overlapping functions among the PATs in C. neoformans, underscoring the importance of this process in the growth, differentiation, and virulence of this microbial pathogen.
MATERIALS AND METHODS
Strains and media.
C. neoformans strains used in this study are listed in Table 1. Strains were maintained on YPD (yeast extract, peptone, dextrose) medium (12). To analyze cell morphology in response to temperature stress, strains incubated overnight in liquid YPD cultures with shaking were diluted 1:10 in fresh YPD medium and incubated at either 30°C or 37°C with shaking for either 4 h or 24 h prior to imaging. To examine chitin morphology in response to temperature stress, samples were collected and resuspended in phosphate-buffered saline (PBS) containing 25 μg/ml of calcofluor white and incubated for 10 min. To analyze growth sensitivity in response to temperature and cell wall stress, strains incubated overnight in shaking YPD cultures were serially diluted and spotted onto YPD plates prepared with the indicated concentrations of Congo red, calcofluor white, SDS, and caffeine (per Fig. 4 legend). To examine mating morphology, strains of opposite mating types were mixed together in water and then spotted onto MS mating medium plates (13). Cocultures were incubated in the dark prior to imaging.
TABLE 1.
C. neoformans strains
| Strain | Genotype | Source or reference |
|---|---|---|
| H99 | MATα | 44 |
| KN99a | MATa | 45 |
| CBN336 | MATα ras1::neo | 11 |
| CBN96 | MATα pHIS3-GFP-RAS1-nat | 11 |
| CBN116 | MATα pHIS3-mCherry-RAS1-neo | 46 |
| CBN167 | MATα pHIS3-GFP-ras1C203S,C204S-nata | 11 |
| CBN134 | MATα akr1::neo | This study |
| CBN198 | MATα pfa3::neo | This study |
| CBN124 | MATα pfa5::neo | This study |
| CBN201 | MATα pfa4::neo | This study |
| DPG1 | MATα erf2::neo | This study |
| JLCN437 | MATα cku80::neo | 16 |
| SKC7 | MATα cku80::neo cpt6::nat | This study |
| CBN414 | MATα cpt7::neo | This study |
| CBN389 | MATα pfa3::neo pfa4::neo | This study |
| CBN403 | MATα pfa3::neo pfa4::neo pHIS3-GFP-RAS1-nat | This study |
| CBN255 | MATa pfa4::neo | This study |
| KMP1 | MATα erf4::neo | This study |
| CBN324 | MATα pfa4::neo pPFA4-nat | This study |
| CBN241 | MATα pfa4::neo pHIS3-GFPRAS1-nat | This study |
| CBN370 | MATα pHIS3-mCherry-RAS1-neo pHIS3-GFP-PFA4-nat | This study |
| CBN451 | MATα pHIS3-mCherry-PFA4-neo | This study |
ras1C203S,C204S, the C203S and C204S substitutions encoded by the ras1 gene.
FIG 4.
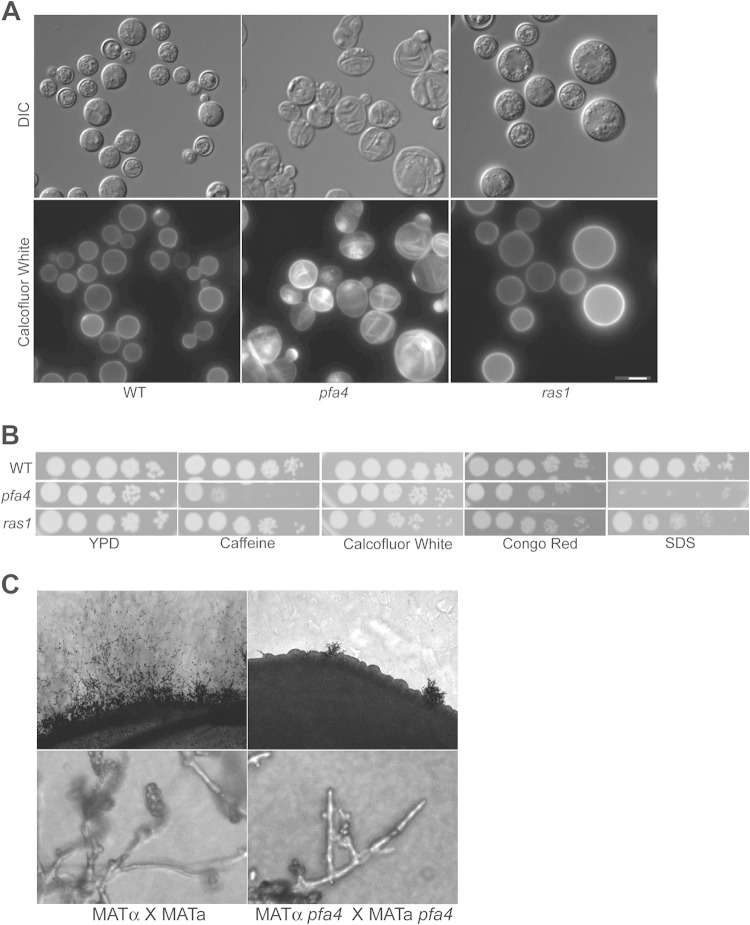
A pfa4 strain exhibits ras1-independent phenotypes. (A) pfa4 mutant cells exhibit abnormal chitin localization. The wild-type (H99) strain and pfa4Δ (CBN201) and ras1Δ (CBN336) mutant strains were incubated overnight in YPD medium, diluted 10-fold in YPD medium, and then grown for an additional 24 h at 37°C. Cells were stained with calcofluor white prior to imaging (magnification, ×100) by fluorescence microscopy. Scale bar, 5 μm. (B) The pfa4Δ mutant strain is sensitive to cell wall stress. The wild-type (H99) strain and pfa4Δ (CBN201) and ras1Δ (CBN336) mutant strains were grown overnight in YPD medium, serially diluted, spotted onto YPD medium containing 1 mg/ml caffeine, 1 mg/ml calcofluor white, 0.5% Congo red, or 0.02% SDS. Plates were incubated at 30°C for 48 h. (C) Pfa4 is required for mating filament formation and differentiation during sexual development in a dose-dependent manner. Bilateral mating mixtures were prepared by mixing overnight cultures of MATα and MATa wild-type cells (H99 and KN99a) and MATα and MATa pfa4 mutant cells (CBN201 and CBN255). Mixtures were spotted onto MS mating medium and incubated for 5 days in the dark. Images were taken at the periphery of the mating mixtures at magnifications of ×100 (top panels) and ×400 (bottom panels).
Molecular biology.
C. neoformans genes identified in this study are listed in Table 2. To generate C. neoformans PAT deletion mutants using a dominant selectable marker, three-piece PCR overlap extension was used to replace the AKR1, PFA3, PFA5, and PFA4 open reading frames with the neomycin (neo) resistance cassette to generate strains CBN134 (akr1::neo), CBN198 (pfa3::neo), CBN124 (pfa5::neo), and CBN201 (pfa4::neo) (14). The cpt7::neo and cpt6::nat constructs were generated using three-piece In-Fusion HD cloning (Clontech). The erf2::neo deletion construct was generated by split-marker double-joint PCR (15). These constructs were used to generate mutant strains DGP1 (erf2::neo), SHC7 (cpt6::nat), and CBN414 (cpt7::neo). All of the PAT mutant strains except for SHC7 were generated in the type H99 background. We were not able to readily generate a cpt6::nat deletion mutant strain in the H99 strain, instead requiring a strain with a deletion of CKU80. Ku80 is required for nonhomologous end joining, and deletion of CKU80 increases homologous recombination in C. neoformans (16). The PFA4 gene was subcloned into the nat resistance plasmid pCH233 and transformed into the pfa4::neo mutant strain CBN201 to create a PFA4 reconstituted strain (CBN324). Deletion strains and the PFA4 reconstituted strain were confirmed by PCR, Southern blot analysis, and reverse transcription-PCR (RT-PCR).
TABLE 2.
C. neoformans genes
| Gene name(s) | Locus tag | GenBank accession no. |
|
|---|---|---|---|
| Chromosome | Protein | ||
| CPT1/AKR1 | CNAG_00436 | CP003828.1 | AFR97092.1 |
| CPT2/PFA3 | CNAG_02481 | CP003825.1 | AFR95354.1 |
| CPT3/PFA5 | CNAG_04849 | CP003829.1 | AFR97366.2 |
| CPT4/PFA4 | CNAG_03981 | CP003821.1 | AFR93481.1 |
| CPT5/ERF2 | CNAG_00274 | CP003820.1 | AFR92407.2 |
| CPT6 | CNAG_06862 | CP003824.1 | AFR94617.2 |
| CPT7 | CNAG_04167 | CP003828.1 | AFR96897.1 |
| ERF4 | CNAG_05730 | CP003825.1 | AFR95328.2 |
To examine the localization of the Pfa4 protein, the PFA4 gene and terminator sequence were amplified by PCR from wild-type C. neoformans genomic DNA and subcloned into the nat resistance plasmid pCN19 containing GFP under the HIS3 promoter and the neo resistance plasmid pCN52 containing mCHERRY under the HIS3 promoter (11).
Genomic integration of the PAT deletion constructs, PFA4 reconstitution construct, and epitope-tagged constructs was performed using the biolistic transformation method as described previously (17, 18).
Microscopy and imaging.
Differential interference contrast (DIC) microscopy and fluorescent images were captured with a Zeiss Axio Imager A1 fluorescence microscope equipped with an AxioCam MRM digital camera. Images were acquired and analyzed by ZEN (black edition) software (Zeiss). High-resolution fluorescent images were captured using a GE DeltaVision Elite deconvolution microscope equipped with a CoolSnap HQ2 high-resolution charge-coupled-device (CCD) camera. Images were acquired and analyzed by softWoRx software (GE). Images were colorized and converted to JPEG format by ImageJ (Fiji) software (19).
Golgi apparatus localization.
The green-emitting fluorescent stain C6-NBD-ceramide (6-{N-[(7-nitrobenzo-2-oxa-1,3-diazol-4-yl)amino]caproylsphingosine}) (Invitrogen) was used to visualize the Golgi apparatus. C6-NBD-ceramide has been used to visualize the Golgi complex in yeasts, including S. cerevisiae and C. neoformans (20, 21). Wild-type cells expressing red fluorescent protein (mCherry)-tagged Ras1 (mCh-Ras1; CBN116) and mCh-Pfa4 (CBN451) were grown overnight in YPD medium, diluted 10-fold, and grown for 4 h at 30°C. Cells were fixed in 3.7% formaldehyde for 10 min, washed three times in PBS, and incubated from 4 h to overnight in 5 μM NBD C6-ceramide.
Palmitoylation assay.
We developed a modified version of the acyl-biotinyl switch assay developed by Wan et al. (22) to assess the palmitoylation status of C. neoformans Ras1 (11). Briefly, wild-type and pfa4Δ mutant strains expressing GFP-RAS1 were grown overnight in YPD medium to an optical density at 600 nm (OD600) of 1.0. Twenty-milliliter samples were centrifuged and resuspended in 0.5 ml of lysis buffer containing 10 mM N-ethylmaleimide (NEM; Thermo Scientific) and 2× protease inhibitors (Complete Mini, EDTA-free; Roche). After lysis, protein concentration was equalized using Precision Red (Cytoskeleton), and the samples were processed as previously described (11) with the exception that GFP-Trap resin (ChromoTek) was used to precipitate GFP-Ras1 in the control samples. Bound proteins were eluted from NeutrAvidin resin (palmitoylated Ras1; ThermoScientific) and GFP-Trap resin (control Ras1) into 30 μl of 5× Laemmli sample buffer and analyzed by Western blotting.
Comparative quantification of Ras1 palmitoylation in the pfa4Δ mutant strain compared to that of the wild type was performed by densitometric analysis of digitized Western blot signals using ImageJ (Fiji) software (gel analysis function) (19). Quantified Western blot signals for palmitoylated Ras1 were normalized to total Ras1 signal for each sample and expressed as a percentage of the value for the wild-type strain.
Western blot analysis.
Samples were heated to 95°C for 4 min, and half of each sample (15 μl) was loaded and separated on a NuPAGE 4 to 12% Bis-Tris gel (Invitrogen) using morpholinepropanesulfonic acid (MOPS) running buffer. Samples were electrophoretically transferred to Invitrolon polyvinylidene difluoride (PVDF) membrane (Invitrogen). The membrane blot was blocked with Starting Block T20 (Pierce) for 1 h, incubated with anti-GFP antibody (1/1,000 dilution; Roche) for 1 h, washed three times for 10 min each with Tris-buffered saline plus Tween (TBST), incubated with an anti-mouse peroxidase-conjugated secondary antibody (1/50,000 dilution; Jackson Laboratory), and washed three times for 10 min each with TBST. Reactive proteins were detected by enhanced chemiluminescence (ECL Prime Western blotting detection reagent; GE Healthcare).
Animal studies.
Using the murine inhalation model of systemic cryptococcosis, we inoculated female A/J (Jackson labs) mice intranasally with 5 × 105 C. neoformans cells as previously described (23). Groups of 10 mice were inoculated with one of three strains: the wild type (H99), pfa4Δ (CBN201) mutant, and reconstituted pfa4Δ/PFA4 (CBN321) strain. Mice were monitored daily for signs of infection and sacrificed at predetermined clinical endpoints correlating with an imminently lethal infection. The statistical significance in the difference between the survival curves of the animals inoculated with each strain was evaluated using a log rank test (JMP software; SAS Institute, Cary, NC). All studies were performed in compliance with Duke University institutional guidelines for animal experimentation.
RESULTS
C. neoformans protein S-acyltransferases.
We used two approaches to identify an Ras1-specific PAT in C. neoformans. First, we probed the C. neoformans var. grubii genome database with a BLAST search for homologs of S. cerevisiae Erf2, the primary protein S-acyltransferase for S. cerevisiae Ras2 (ScRas2). Second, we queried the C. neoformans genome databases (Cryptococcus neoformans var. grubii H99 Sequencing Project, Broad Institute of Harvard and MIT [24] and Fungi DB [25]) for proteins predicted to contain a DHHC domain (Pfam PF01529). Seven sequences were selected for further analysis and were initially designated Cryptococcus protein S-acyltransferases (CPT genes) (Table 2; see also Fig. S1 in the supplemental material). Two of the sequences, CNAG_04361 (CPT1) and CNAG_03981 (CPT4), have been previously named AKR1 and PFA4 based on similarity to S. cerevisiae sequences. In a phylogenic alignment and tree construction (26) comprised of PAT amino acid sequences from C. neoformans, S. cerevisiae, and Homo sapiens, we also found that Cpt1 clustered with S. cerevisiae Akr1 and Ark2 while Cpt4 clustered with S. cerevisiae Pfa4 and human ZDHC6 (see Fig. S2 in the supplemental material). Also, three other C. neoformans PATs (CNAG_02481, CNAG_04849, and CNAG_00274) each clustered with individual S. cerevisiae PATs (Pfa3, Pfa5, and Erf2). Based on these data, we redesignated the C. neoformans CPT1 to CPT5 genes AKR1, PFA3, PFA5, PFA4, and ERF2, respectively, while CPT6 and CPT7 retain their initial designations. However, our functional analysis suggests that, at least in the case of CnPfa4, the C. neoformans PATs are not true orthologs of the S. cerevisiae proteins and appear to have overlapping substrates.
We also found that CNAG_03981 (PFA4) did not initially appear to encode a protein with an intact DHHC domain as predicted by automated algorithms. However, manual inspection of likely splice junctions revealed errors in automated intron-exon border prediction. Amplification and sequencing of the cDNA associated with this gene confirmed this misannotation, indicating that this sequence actually encoded a DHHC domain-containing protein.
In addition we identified a putative Erf4 homolog in C. neoformans (Table 2). In S. cerevisiae, Erf2 palmitoyltransferase activity requires Erf4, a nonenzymatic accessory protein (27). This association is conserved in mammalian cells, with Gcp16 acting as a cofactor for the Ras PAT DHHC9 (28). In S. cerevisiae, Erf4 acts to stabilize Erf2 against degradation and is also involved in stabilization of the Erf2-palmitoyl intermediate and transfer of the group to the substrate protein Ras2 (27). Erf4 and Gcp16 share a conserved domain (Erf4 domain; Pfam 10256) that was used to identify CNAG_02504 in C. neoformans.
Mutation of Pfa4 and Pfa3 results in various temperature sensitivities.
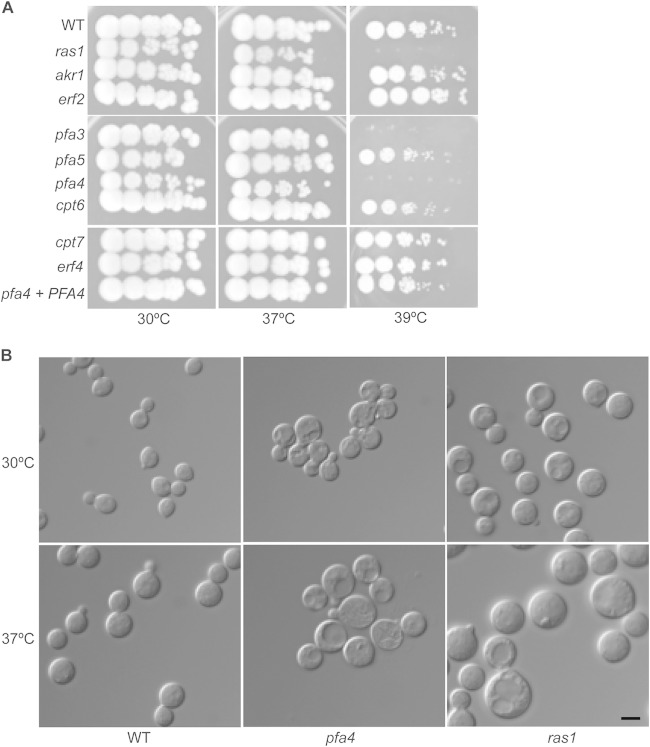
To evaluate the role of these putative protein acyltransferases in Ras-related cellular processes, we examined strains with targeted mutations in each PAT gene, including ERF4. None of the these genes was essential for growth, and initial phenotypic testing showed that only the pfa4Δ mutant strain was temperature sensitive at 37°C, similar to the ras1Δ mutant (Fig. 1A). In addition, the pfa3Δ deletion strain exhibited a subtle temperature sensitivity at 39°C (Fig. 1A). These results were confirmed by testing the phenotypes for a subset of these mutants (akr1Δ, pfa3Δ, pfa4Δ, and erf2Δ strains) that were present in an independent collection of C. neoformans mutant strains (29).
FIG 1.
The pfa4Δ mutant strain exhibits ras1-like defects at high temperature. (A) Overnight cultures of wild-type (WT; H99), akr1Δ (CBN134), pfa3Δ (CBN198), pfa5Δ (CBN124), pfa4Δ (CBN201), erf2Δ (DPG1), cpt6Δ (SKC7), cpt7Δ (CBN414), ras1Δ (CBN336), and erf4Δ (KMP1) strains and the reconstituted pfa4Δ/PFA4 (CBN324) mutant strain were serially diluted, spotted onto YPD medium, and incubated at 30°C, 37°C, and 39°C for 48 h. (B) Overnight cultures of the wild-type (H99) strain and ras1Δ (CBN336) and pfa4Δ (CBN201) mutant strains were diluted in YPD liquid medium and shifted to either 30°C or 37°C for 4 h. Cells were imaged (magnification, ×100) with DIC optics to assess temperature-dependent alterations in cell size and morphology. Scale bar, 5 μm.
Strains with ras1Δ mutations have pronounced morphology defects in both the yeast and hyphal forms. For the asexual yeast form, ras1 cells are larger than those of wild-type strains, resulting from defects in cell polarity and actin cytoskeleton dynamics (30). This cell size difference is noted at 30°C, but it is most apparent at higher growth temperatures. Among the PAT mutant strains, only the pfa4Δ mutant exhibited a temperature-dependent size increase although this was not as dramatic as that of the ras1Δ mutant strain (Fig. 1B; also data not shown).
To ensure that any phenotypes in the pfa4Δ mutant strain were appropriately attributed to this specific mutation, an intact copy of the PFA4 gene was reintroduced into this mutant strain. The resulting pfa4Δ/PFA4 reconstituted strain demonstrated complete restoration of wild-type levels of growth at 37°C, as well as normal morphogenesis at all temperatures (Fig. 1A).
Ras1 localization is impaired in the pfa4Δ mutant strain.
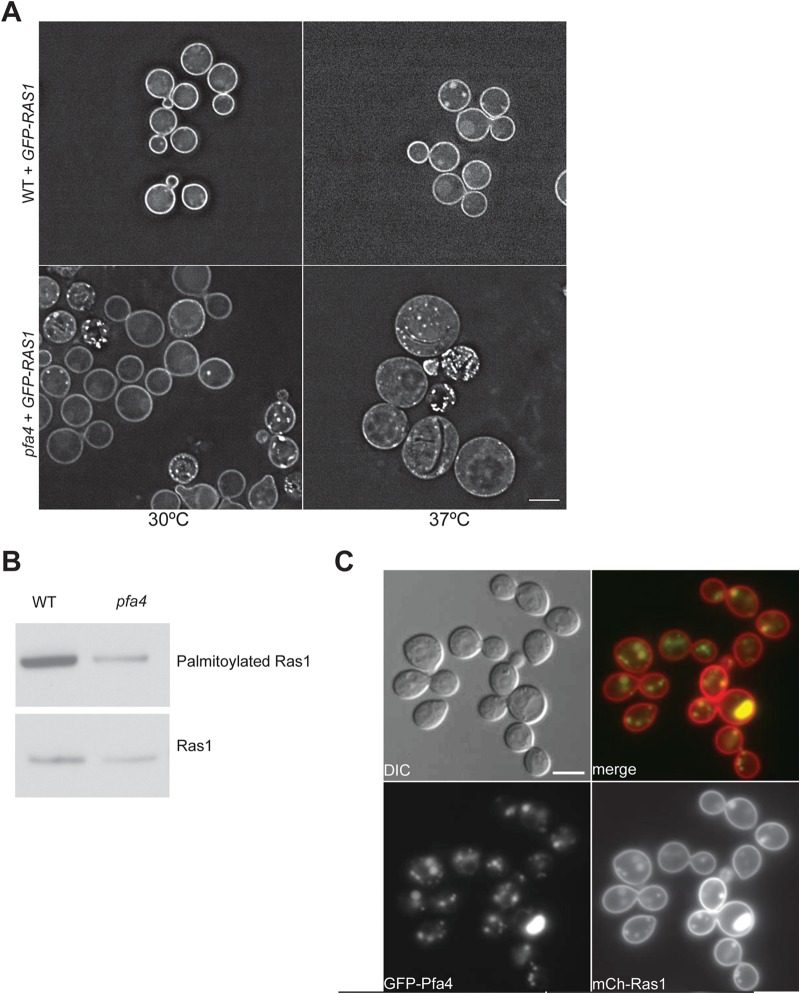
Palmitoylation of Ras proteins promotes plasma membrane localization. Previously, we demonstrated that the palmitoylation of amino acid residue C203 or C204 is sufficient for Ras1 function and localization to the plasma membrane (11). To determine if Ras1 localization was impaired in the pfa4Δ mutant strain, we generated a pfa4Δ mutant strain constitutively expressing a GFP-Ras1 fusion protein. In wild-type cells, GFP-Ras1 localizes to the plasma membrane and endomembrane structures resembling punctate dots. In the pfa4Δ mutant there was an overall decrease in GFP-Ras1 localization to the plasma membrane in addition to an increase of punctate dots in some cells (Fig. 2A). The altered PM association of GFP-Ras1 in the pfa4Δ mutant was even more apparent at 37°C, in contrast to observations in the wild-type strain background that demonstrated equally intense patterns of GFP-Ras1 PM localization at both temperatures (Fig. 2A). We observed no alteration of GFP-Ras1 localization in the other PAT mutant strains or in the erf4Δ mutant strain (data not shown).
FIG 2.
Ras1 palmitoylation is dependent on Pfa4.(A) Ras1 localization to the PM is reduced in the pfa4Δ mutant strain. The wild-type strain and pfa4Δ mutant strain expressing GFP-RAS1 under the control of the histone H3 promoter (CBN96 and CBN241) were grown overnight in YPD medium, diluted 10-fold, and grown for 24 h at 30°C or 37°C. Cells were imaged (magnification, ×63) using DeltaVision microscopy to assess the pattern and intensity of fluorescent fusion protein localization. All images were obtained using identical exposure times. Scale bar, 5 μm. (B) Ras1 palmitoylation is reduced in the pfa4Δ mutant strain. Cell lysates obtained from the wild-type and pfa4 mutant strains expressing GFP-Ras1 (CBN96 and CBN241) were accessed for palmitoylation using a modified version of the acyl-biotinyl switch assay. Lysate samples were split between experimental samples (top panel) and loading control samples (bottom panel) to quantify palmitoylated Ras1 and total Ras1 levels, respectively. Proteins in the experimental samples were labeled with biotin and precipitated using NeutrAvidin beads. In this assay, biotinylation occurs specifically on proteins that contain a free sulfhydryl group generated by hydroxylamine cleavage of a thioester bond (palmitoylated proteins). GFP-Ras1 proteins in the control samples were immunoprecipitated with GFP-Trap resin. Samples were separated by NuPAGE electrophoresis, transferred to PVDF membranes, and immunoblotted with anti-GFP antibodies. Digital images were assessed using densitometry (ImageJ) for signal intensity. Palmitoylated-Ras1 signal was normalized relative to that of total Ras1 signal for each sample. Representative images from multiple independent experiments are demonstrated. (C) Pfa4 and Ras1 colocalize at punctate dots in the cytoplasm. A wild-type strain coexpressing GFP-Pfa4 and mCh-Ras1 (CBN370) was grown overnight in YPD medium, diluted 10-fold, and grown for 4 h at 30°C. Images (magnification, ×100) were taken using the appropriate filter and merged. Scale bar, 5 μm.
In S. cerevisiae, there is a clear overlap of PAT functions. ScRas2 palmitoylation is not completely abrogated in an erf2 mutant strain (31). Additionally, the yeast PATs Akr1, Pfa2, Pfa4, and Pfa3 each exhibit in vitro PAT activity toward Ras2 (32). To begin to explore whether other PAT proteins contribute to Ras1 palmitoylation, we generated a pfa3Δ pfa4Δ double mutant. The Pfa3 protein was chosen for specific inclusion, given its modest but measurable role in thermotolerance. The pfa3Δ pfa4Δ double mutant strain displayed no additive phenotypic changes compared with the pfa4Δ mutant in terms of thermotolerance or yeast cell morphogenesis. In addition, localization of GFP-Ras1 in the pfa3Δ pfa4Δ double mutant strain was similar to that in the single pfa4Δ mutant strain (data not shown).
Ras1 palmitoylation is decreased in the pfa4Δ mutant strain.
In addition to assessing the localization of Ras1 in the pfa4Δ mutant strain, we directly assessed the palmitoylation status of Ras1 using a modified version of the acyl-biotinyl switch assay (22). In this assay, palmitoyl groups are captured by biotinylation, and relative Ras protein palmitoylation activity can be assessed in different strain backgrounds. In two independent experiments, Ras1 palmitoylation was decreased by 50 and 59% in the pfa4Δ mutant compared to the level in the wild type but was not eliminated (Fig. 2B). Similar reductions of Ras1 palmitoylation were observed in the pfa3Δ pfa4Δ double mutant strain (data not shown). These measurements were consistent with the degree of altered GFP-Ras1 PM localization observed in each of these strain backgrounds (Fig. 2A). Together, these results suggest that the Pfa4 protein serves a major role in the palmitoylation of the CnRas1 protein under these growth conditions.
Ras1 and Pfa4 colocalize on endomembrane structures.
In current models, palmitoylation promotes the localization of Ras proteins from the Golgi compartment to the PM. If Pfa4 functions as an Ras PAT, we would expect Pfa4 to be localized on endomembrane structures (Golgi compartment or endoplasmic reticulum [ER]) to catalyze the palmitoylation of Ras1. In fact, other yeast Ras PATs localize to the Golgi compartment (S. pombe) or ER (S. cerevisiae) (31, 33). To assess the localization of Pfa4, we generated a GFP-tagged version of this protein. As predicted, we observed that GFP-Pfa4 localized prominently to distinct punctate regions in the cytoplasm as well as in the perinuclear region, consistent with Golgi compartment/ER localization (Fig. 2C).
To probe the association of Pfa4 and Ras1 localization patterns, we examined the localization of GFP-Pfa4 and mCh-Ras1 in a wild-type strain. C. neoformans Ras1 localized primarily to the PM but also to punctate dots in the cytoplasm and the vacuole, consistent with its predicted progression from endomembranes to the PM (Fig. 2C) (11). However, mCh-Ras1 was not observed in the perinuclear region, nor was Gfp-Pfa4 observed at the PM. Instead, both fluorescent fusion proteins colocalized to the cytoplasmic punctate structures, consistent with previously reported patterns of Golgi compartment localization. We also observed larger regions of colocalization in a subset of cells that appeared to be in vacuole-like regions (Fig. 2C).
To determine the precise sites of Ras1 and Pfa4 colocalization, we independently examined the localization of mCh-Ras1 and mCh-Pfa4 in fixed cells incubated with the fluorescent Golgi complex stain C6-NBD-ceramide. The C6-NBD-ceramide-stained Golgi complex strongly colocalized with mCh-Ras1 in punctate dots in the cytoplasm (see Fig. S3 in the supplemental material). Additionally, we observed colocalization between mCh-Pfa4 and C6-NBD-ceramide in a subset of cytoplasmic puncta, suggesting that a fraction of Pfa4 also localizes to the Golgi compartment (see Fig. S3 in the supplemental material). Taken together, these data suggest that Pfa4 exists on several endomembrane structures including the ER (perinuclear) and Golgi complex (cytoplasmic puncta). Additionally, Pfa4 and Ras1 colocalize at the Golgi compartment to facilitate PM association of the Ras proteins.
Pfa4 is required for pathogenesis.
Impaired growth of C. neoformans strains at elevated temperatures often correlates with reduced virulence in animal models of cryptococcosis. We therefore tested the ability of the pfa4Δ mutant strain to survive and cause lethal disease in a murine inhalational model of C. neoformans infection. Female C57BL/6 mice were sedated and inoculated by the inhalation of yeast cells. The infected mice were monitored for clinical signs (weight loss or neurological symptoms) consistent with progressive, disseminated cryptococcal infection. Infections with the wild-type or pfa4Δ/PFA4 reconstituted strain resulted in a similar progression of lethal disease within approximately 3 weeks, with all infected mice demonstrating neurological symptoms consistent with meningoencephalitis, the prominent form of this infection. In contrast, none of the mice infected with the pfa4Δ mutant strain succumbed to the infection; these mice continued to gain weight and appear healthy through the 6-week course of the experiment with no pulmonary or neurological symptoms (Fig. 3). Together, these results indicate that the Pfa4 protein is required for the full virulence potential of C. neoformans and that it is likely required for the establishment of the initial infection and/or dissemination to the central nervous system. Therefore, in spite of the presence of multiple genes encoding putative PATs in the C. neoformans genome, the single Pfa4 protein is required for full Ras1 palmitoylation, high-temperature growth, and virulence in an animal model of cryptococcal infection.
FIG 3.
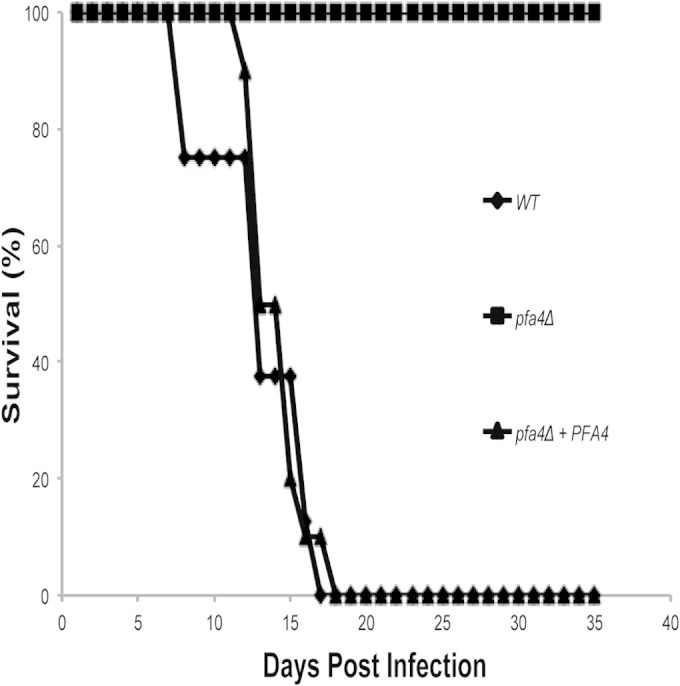
PFA4 is required for pathogenesis in a murine model of cryptococcosis. A/Jcr mice were intranasally inoculated with the wild-type H99, pfa4Δ mutant (CBN201), and pfa4Δ/PFA4 reconstituted (CBN324) strains. Animal survival was monitored for 40 days.
The pfa4Δ mutant strain exhibits ras1-independent associated phenotypes.
Ras1 is unlikely to be the only target of Pfa4 in C. neoformans. In S. cerevisiae, Erf2 has several confirmed substrates, including Ras2, Rho2, and Gpa1 (3, 7, 31). Consistent with this hypothesis, our analysis of the C. neoformans pfa4Δ mutant strain revealed several ras1-independent phenotypes. During overnight incubations at 37°C, pfa4Δ mutant cells resemble irregular, deflated balloons, in contrast to the more uniform, large, unbudded phenotype of ras1Δ mutant cells (Fig. 4A). In addition, these cells stain abnormally with the fluorescent chitin stain calcofluor white. In both wild-type and ras1Δ mutant cells, calcofluor white uniformly stains the cell wall and septa. In contrast, calcofluor white staining is concentrated in rings and patches over the surface of pfa4Δ mutant cells when they are grown at 37°C (Fig. 4A).
The pfa4Δ mutant strain is also sensitive to reagents that perturb the cell wall, showing sensitivity to SDS, caffeine, and Congo red. In comparison, in these assays the ras1Δ mutant strain exhibited only a slight sensitivity to SDS (Fig. 4B). Taken together with the unusual morphology of the pfa4Δ mutant cells, these results suggest that Pfa4 palmitoylates substrates involved in cell wall structure and integrity.
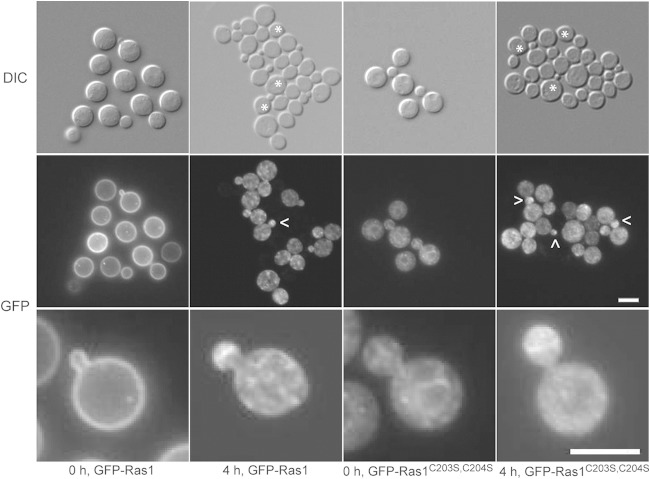
Although Ras1 is required for sexual differentiation in C. neoformans, our previous results indicate that C. neoformans mating is a palmitoylation-independent process. The Ras1 protein with the C203S and C204S substitutions (Ras1C203S, C204S) is unable to undergo C-terminal palmitoylation, and it is restricted to endomembranes. However, this palmitoylation-defective mutant ras1 strain fully supports all stages of sexual differentiation in C. neoformans (11). To further characterize the role of palmitoylation and Ras protein localization in the mating process, we assessed the localization of the N-terminally tagged Ras1 alleles during a mating reaction and found a dynamic, palmitoylation-independent alteration of Ras1 localization in cells undergoing fusion. GFP-Ras1 initially primarily localized to the PM in MATα wild-type cells. However, after several hours of incubation with a MATa mating partner, the GFP-Ras1 localization to the PM was lost, and GFP-Ras1 was instead observed as puncta in the cytoplasm and at the cell bud (Fig. 5). Interestingly, we observed a similar pattern of mating-dependent localization of GFP-Ras1 in cells expressing the mutant form of GFP-Ras1C203S, C204S that cannot be palmitoylated, consistent with the intact mating phenotype in this strain (Fig. 5). Previously we found that GFP-Ras1C203S, C204S is unable to localize to the PM and is restricted to the endomembranes of nonmating yeast cells. Taken together, these observations suggest that while the localization of CnRas1 during mating is independent of palmitoylation, it is very dynamic, moving from the PM to endomembrane structures and polarizing to the emerging bud.
FIG 5.
Ras1 is repolarized in a mating-dependent manner. Mating mixtures were prepared from overnight cultures of MATα wild-type cells expressing either GFP-Ras1 (CBN96) or the palmitoylation-defective GFP-Ras1C203S, C204S (CBN167) with wild-type MATa cells (KN99a). Mixtures were spotted onto MS medium plates in 100-μl aliquots. At regular intervals, cells were scraped off the plate, resuspended in PBS, and examined for GFP-Ras localization. No fluorescence was detected in wild-type MATa cells (representative cells are marked by asterisks in the DIC image). Top panel, DIC images (magnification, ×100). Middle and bottom panels, fluorescent images (magnification, ×100). Arrowheads specify cells containing GFP-Ras1 polarized to emerging buds. Bar, 5 μm.
Interestingly, we observed that Pfa4 is required for mating. The pfa4Δ mutant exhibited a significant mating defect when both partners contained the pfa4 deletion (Fig. 4C, bilateral mating assay). In these assays very few mating foci were observed, mating hyphae were short in comparison to those of the wild type, and few basidiospores were produced. No defects were observed in unilateral pfa4Δ mutant mating assays between a pfa4Δ mutant strain and a wild-type strain. In comparison, ras1Δ mutant strains were unable to undergo cell-cell fusion or form filaments in either unilateral or bilateral crosses (34, 35). These results, together with the prior observation that ras1 palmitoylation mutant strains are completely mating competent (11), suggest that the mating defect of the pfa4Δ mutant strain likely involves the mislocalization of Pfa4 substrates other than Ras1.
DISCUSSION
Protein palmitoylation is a reversible process, allowing a dynamic temporal/spatial movement of targeted proteins to various cellular membranes. In this study, we sought to identify proteins involved in the palmitoylation of C. neoformans Ras1. Similar to other organisms, C. neoformans contains a family of putative protein S-acyltransferases identified by a highly conserved DHHC domain.
Based on sequence analysis and phylogeny, C. neoformans Erf2 is the closest homolog to S. cerevisiae Erf2, the primary Ras PAT protein in this species. Additionally, S. cerevisiae Erf2 activity is dependent on Erf4 (3, 31, 33). However, our experiments suggest that CnPfa4, which is most similar to S. cerevisiae Pfa4, is the primary PAT for Ras1. C. neoformans erf2Δ and erf4Δ mutant strains do not exhibit any ras1-like phenotypes. In contrast, pfa4Δ mutant cells exhibit temperature sensitivity and a temperature-dependent cell size increase, as would be expected with altered CnRas1 function. In addition, the localization of GFP-Ras1 at the PM was perturbed in the pfa4Δ mutant but localized normally in the other PAT mutant strains. Although CnRas1 palmitoylation and PM localization were not completely eliminated in this single-mutant background, the observed level of reduction of Ras palmitoylation is similar to that in S. cerevisiae and S. pombe erf2 mutant strains (31, 33).
S. cerevisiae PATs and PAT substrates fall into groups. For example, Erf2 substrates have additional lipid modifications (prenylation or myristoylation) that occur prior to palmitoylation. In contrast, Pfa4 substrates include proteins such as amino acid permeases that contain transmembrane repeats (7). Our results demonstrate that Pfa4 in C. neoformans has activity on Ras1, offering a different paradigm from that in S. cerevisiae. Whether CnPfa4 has additional activity on transmembrane protein substrates has yet to be determined.
Interestingly, S. cerevisiae Pfa4 also has a non-transmembrane protein target, Chs3 (chitin synthase 3), and palmitoylation of Chs3 is required for this protein to exit the ER (36). In C. neoformans Chs3 is required for cell wall integrity (37, 38). chs3Δ and pfa4Δ strains exhibit similar sensitivities to cell wall stressors Congo red, caffeine, and SDS, suggesting that CnPfa4 may function as a PAT to Chs3 (37). It is possible that PAT substrate specificity has been altered or expanded in C. neoformans so that Pfa4 serves as the primary PAT for substrates that are targeted by PATs in other species.
Our characterization of the pfa4Δ mutant strain identified ras1-independent phenotypes, including altered susceptibility to cell wall stress, suggesting additional targets of Pfa4. Both the ras1Δ and pfa4Δ mutant strains have mating defects, but the nature of the altered mating differs between the two strains. The pfa4Δ mutant strain has a bilateral mating defect in which both mating partners must be mutants for mating to be altered. In contrast, the ras1Δ mutant exhibits a unilateral mating defect, and this mating phenotype is independent of palmitoylation (11). In addition to Ras proteins, S. cerevisiae Erf2 substrates include G-alpha proteins such as Gpa1 and Gpa2 and the G-gamma subunit Ste18 (7). In C. neoformans, G-alpha Gpa1 and G-gamma Gpg1 contain putative palmitoylation sites, and the gpa1 and gpg1 mutant strains exhibit bilateral mating defects similar to those of the pfa4Δ mutant strain (39, 40).
Previously our lab demonstrated that Ras1 localization to the PM was not required for sexual differentiation. This was surprising since many of the signaling molecules involved in mating reside on the PM. Here, we observed that Ras1 localization shifts during sexual differentiation from uniform PM localization to cytoplasmic patches and bud tips in a palmitoylation-independent manner. These results provide an explanation of why palmitoylation is not required for Ras1 mating competence in C. neoformans. Through a mechanism distinct from palmitoylation, Ras1 is delivered from the endomembranes to specific sites on the PM (bud). This shift of localization is not a result of lipid remodeling or relocalization of Ras1 within the PM since it occurs even with completely palmitoylation-defective mutant forms of Ras1.
One of the most striking consequences that we observed among the C. neoformans PAT mutations was the dramatically altered virulence of the single pfa4Δ mutant strain. This virulence attenuation is likely multifactorial, given the observed defects in several phenotypes required for survival in vivo. The pfa4Δ mutant is unable to grow at mammalian body temperature, and several C. neoformans mutant strains with altered thermotolerance are hypovirulent (30, 34, 41, 42). Moreover, the prominent cell wall alteration in the pfa4Δ mutant strain also likely contributes to its reduced virulence. Interestingly, expression of the inducible virulence-associated phenotypes of capsule and melanin remains intact with a pfa4Δ mutation. Together, these data suggest that Pfa4 is the primary PAT for the C. neoformans Ras1 protein and that compensatory palmitoylation by other PATs is not complete in the absence of Pfa4. Also, the Pfa4 protein has substrates in addition to Ras1 that mediate cellular processes involved in mating and cell wall integrity. Therefore, targeting single S-acyltransferases as an antifungal drug target remains a viable strategy despite the presence of a large family of potentially functionally related proteins.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by PHS grant 1R01-AI050128. Duke Light Microscopy Core Facility DeltaVision instrumentation was funded by grant 1S10RR027528-01.
The C. neoformans mutant strain collection created by Hiten Madhani (29) was obtained through the Fungal Genetics Stock Center (43). We also acknowledge the Fungal Genomes Initiative at the Broad Institute and Fungi DB (24, 25). We thank Sam Johnson at Duke Light Microscopy Core Facility for instruction. We also thank Sandra Breeding for assistance with lab strain curation.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00010-15.
REFERENCES
- 1.Baekkeskov S, Kanaani J. 2009. Palmitoylation cycles and regulation of protein function. Mol Membr Biol 26:42–54. doi: 10.1080/09687680802680108. [DOI] [PubMed] [Google Scholar]
- 2.Salaun C, Greaves J, Chamberlain LH. 2010. The intracellular dynamic of protein palmitoylation. J Cell Biol 191:1229–1238. doi: 10.1083/jcb.201008160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lobo S, Greentree WK, Linder ME, Deschenes RJ. 2002. Identification of a Ras palmitoyltransferase in Saccharomyces cerevisiae. J Biol Chem 277:41268–41273. doi: 10.1074/jbc.M206573200. [DOI] [PubMed] [Google Scholar]
- 4.Roth AF, Feng Y, Chen L, Davis NG. 2002. The yeast DHHC cysteine-rich domain protein Akr1p is a palmitoyl transferase. J Cell Biol 159:23–28. doi: 10.1083/jcb.200206120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mitchell DA, Vasudevan A, Linder ME, Deschenes RJ. 2006. Protein palmitoylation by a family of DHHC protein S-acyltransferases. J Lipid Res 47:1118–1127. doi: 10.1194/jlr.R600007-JLR200. [DOI] [PubMed] [Google Scholar]
- 6.Hornemann T. 2015. Palmitoylation and depalmitoylation defects. J Inherit Metab Dis 38:179–186. doi: 10.1007/s10545-014-9753-0. [DOI] [PubMed] [Google Scholar]
- 7.Roth AF, Wan J, Bailey AO, Sun B, Kuchar JA, Green WN, Phinney BS, Yates JR, Davis NG. 2006. Global analysis of protein palmitoylation in yeast. Cell 125:1003–1013. doi: 10.1016/j.cell.2006.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prior IA, Hancock JF. 2012. Ras trafficking, localization and compartmentalized signalling. Semin Cell Dev Biol 23:145–153. doi: 10.1016/j.semcdb.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eisenberg S, Laude AJ, Beckett AJ, Mageean CJ, Aran V, Hernandez-Valladares M, Henis YI, Prior IA. 2013. The role of palmitoylation in regulating Ras localization and function. Biochem Soc Trans 41:79–83. doi: 10.1042/BST20120268. [DOI] [PubMed] [Google Scholar]
- 10.Chang EC, Philips MR. 2006. Spatial segregation of Ras signaling: new evidence from fission yeast. Cell Cycle 5:1936–1939. doi: 10.4161/cc.5.17.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nichols CB, Ferreyra J, Ballou ER, Alspaugh JA. 2009. Subcellular localization directs signaling specificity of the Cryptococcus neoformans Ras1 protein. Eukaryot Cell 8:181–189. doi: 10.1128/EC.00351-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sherman F. 1991. Getting started with yeast. Methods Enzymol 194:3–21. doi: 10.1016/0076-6879(91)94004-V. [DOI] [PubMed] [Google Scholar]
- 13.Xue C, Tada Y, Dong X, Heitman J. 2007. The human fungal pathogen Cryptococcus can complete its sexual cycle during a pathogenic association with plants. Cell Host Microbe 1:263–273. doi: 10.1016/j.chom.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 14.Fraser JA, Subaran RL, Nichols CB, Heitman J. 2003. Recapitulation of the sexual cycle of the primary fungal pathogen Cryptococcus neoformans var. gattii: implications for an outbreak on Vancouver Island, Canada. Eukaryot Cell 2:1036–1045. doi: 10.1128/EC.2.5.1036-1045.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim MS, Kim SY, Jung KW, Bahn YS. 2012. Targeted gene disruption in Cryptococcus neoformans using double-joint PCR with split dominant selectable markers. Methods Mol Biol 845:67–84. doi: 10.1007/978-1-61779-539-8_5. [DOI] [PubMed] [Google Scholar]
- 16.Goins CL, Gerik KJ, Lodge JK. 2006. Improvements to gene deletion in the fungal pathogen Cryptococcus neoformans: absence of Ku proteins increases homologous recombination, and co-transformation of independent DNA molecules allows rapid complementation of deletion phenotypes. Fungal Genet Biol 43:531–544. doi: 10.1016/j.fgb.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 17.Toffaletti DL, Rude TH, Johnston SA, Durack DT, Perfect JR. 1993. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J Bacteriol 175:1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davidson RC, Cruz MC, Sia RA, Allen B, Alspaugh JA, Heitman J. 2000. Gene disruption by biolistic transformation in serotype D strains of Cryptococcus neoformans. Fungal Genet Biol 29:38–48. doi: 10.1006/fgbi.1999.1180. [DOI] [PubMed] [Google Scholar]
- 19.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levine TP, Wiggins CA, Munro S. 2000. Inositol phosphorylceramide synthase is located in the Golgi apparatus of Saccharomyces cerevisiae. Mol Biol Cell 11:2267–2281. doi: 10.1091/mbc.11.7.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kmetzsch L, Joffe LS, Staats CC, de Oliveira DL, Fonseca FL, Cordero RJ, Casadevall A, Nimrichter L, Schrank A, Vainstein MH, Rodrigues ML. 2011. Role for Golgi reassembly and stacking protein (GRASP) in polysaccharide secretion and fungal virulence. Mol Microbiol 81:206–218. doi: 10.1111/j.1365-2958.2011.07686.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wan J, Roth AF, Bailey AO, Davis NG. 2007. Palmitoylated proteins: purification and identification. Nat Protoc 2:1573–1584. doi: 10.1038/nprot.2007.225. [DOI] [PubMed] [Google Scholar]
- 23.Cox GM, Mukherjee J, Cole GT, Casadevall A, Perfect JR. 2000. Urease as a virulence factor in experimental cryptococcosis. Infect Immun 68:443–448. doi: 10.1128/IAI.68.2.443-448.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Janbon G, Ormerod KL, Paulet D, Byrnes EJ III, Yadav V, Chatterjee G, Mullapudi N, Hon CC, Billmyre RB, Brunel F, Bahn YS, Chen W, Chen Y, Chow EW, Coppee JY, Floyd-Averette A, Gaillardin C, Gerik KJ, Goldberg J, Gonzalez-Hilarion S, Gujja S, Hamlin JL, Hsueh YP, Ianiri G, Jones S, Kodira CD, Kozubowski L, Lam W, Marra M, Mesner LD, Mieczkowski PA, Moyrand F, Nielsen K, Proux C, Rossignol T, Schein JE, Sun S, Wollschlaeger C, Wood IA, Zeng Q, Neuveglise C, Newlon CS, Perfect JR, Lodge JK, Idnurm A, Stajich JE, Kronstad JW, Sanyal K, Heitman J, Fraser JA, et al. 2014. Analysis of the genome and transcriptome of Cryptococcus neoformans var. grubii reveals complex RNA expression and microevolution leading to virulence attenuation. PLoS Genet 10:e1004261. doi: 10.1371/journal.pgen.1004261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stajich JE, Harris T, Brunk BP, Brestelli J, Fischer S, Harb OS, Kissinger JC, Li W, Nayak V, Pinney DF, Stoeckert CJ Jr, Roos DS. 2012. FungiDB: an integrated functional genomics database for fungi. Nucleic Acids Res 40:D675–D681. doi: 10.1093/nar/gkr918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitchell DA, Hamel LD, Ishizuka K, Mitchell G, Schaefer LM, Deschenes RJ. 2012. The Erf4 subunit of the yeast Ras palmitoyl acyltransferase is required for stability of the acyl-Erf2 intermediate and palmitoyl transfer to a Ras2 substrate. J Biol Chem 287:34337–34348. doi: 10.1074/jbc.M112.379297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Swarthout JT, Lobo S, Farh L, Croke MR, Greentree WK, Deschenes RJ, Linder ME. 2005. DHHC9 and GCP16 constitute a human protein fatty acyltransferase with specificity for H- and N-Ras. J Biol Chem 280:31141–31148. doi: 10.1074/jbc.M504113200. [DOI] [PubMed] [Google Scholar]
- 29.Liu OW, Chun CD, Chow ED, Chen C, Madhani HD, Noble SM. 2008. Systematic genetic analysis of virulence in the human fungal pathogen Cryptococcus neoformans. Cell 135:174–188. doi: 10.1016/j.cell.2008.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nichols CB, Perfect ZH, Alspaugh JA. 2007. A Ras1-Cdc24 signal transduction pathway mediates thermotolerance in the fungal pathogen Cryptococcus neoformans. Mol Microbiol 63:1118–1130. doi: 10.1111/j.1365-2958.2006.05566.x. [DOI] [PubMed] [Google Scholar]
- 31.Bartels DJ, Mitchell DA, Dong X, Deschenes RJ. 1999. Erf2, a novel gene product that affects the localization and palmitoylation of Ras2 in Saccharomyces cerevisiae. Mol Cell Biol 19:6775–6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Budde C, Schoenfish MJ, Linder ME, Deschenes RJ. 2006. Purification and characterization of recombinant protein acyltransferases. Methods 40:143–150. doi: 10.1016/j.ymeth.2006.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Young E, Zheng Z-Y, Wilkins AD, Jeong H-T, Li M, Lichtarge O, Chang EC. 2014. Regulation of Ras localization and cell transformation by evolutionarily conserved palmitoyltransferases. Mol Cell Biol 34:374–385. doi: 10.1128/MCB.01248-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alspaugh JA, Cavallo LM, Perfect JR, Heitman J. 2000. RAS1 regulates filamentation, mating and growth at high temperature of Cryptococcus neoformans. Mol Microbiol 36:352–365. doi: 10.1046/j.1365-2958.2000.01852.x. [DOI] [PubMed] [Google Scholar]
- 35.Waugh MS, Nichols CB, DeCesare CM, Cox GM, Heitman J, Alspaugh JA. 2002. Ras1 and Ras2 contribute shared and unique roles in physiology and virulence of Cryptococcus neoformans. Microbiology 148:191–201. [DOI] [PubMed] [Google Scholar]
- 36.Lam KK, Davey M, Sun B, Roth AF, Davis NG, Conibear E. 2006. Palmitoylation by the DHHC protein Pfa4 regulates the ER exit of Chs3. J Cell Biol 174:19–25. doi: 10.1083/jcb.200602049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Banks IR, Specht CA, Donlin MJ, Gerik KJ, Levitz SM, Lodge JK. 2005. A chitin synthase and its regulator protein are critical for chitosan production and growth of the fungal pathogen Cryptococcus neoformans. Eukaryot Cell 4:1902–1912. doi: 10.1128/EC.4.11.1902-1912.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baker LG, Specht CA, Donlin MJ, Lodge JK. 2007. Chitosan, the deacetylated form of chitin, is necessary for cell wall integrity in Cryptococcus neoformans. Eukaryot Cell 6:855–867. doi: 10.1128/EC.00399-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alspaugh JA, Perfect JR, Heitman J. 1997. Cryptococcus neoformans mating and virulence are regulated by the G-protein alpha subunit GPA1 and cAMP. Genes Dev 11:3206–3217. doi: 10.1101/gad.11.23.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li L, Shen G, Zhang ZG, Wang YL, Thompson JK, Wang P. 2007. Canonical heterotrimeric G proteins regulating mating and virulence of Cryptococcus neoformans. Mol Biol Cell 18:4201–4209. doi: 10.1091/mbc.E07-02-0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Odom A, Muir S, Lim E, Toffaletti DL, Perfect J, Heitman J. 1997. Calcineurin is required for virulence of Cryptococcus neoformans. EMBO J 16:2576–2589. doi: 10.1093/emboj/16.10.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ballou ER, Nichols CB, Miglia KJ, Kozubowski L, Alspaugh JA. 2010. Two CDC42 paralogues modulate Cryptococcus neoformans thermotolerance and morphogenesis under host physiological conditions. Mol Microbiol 75:763–780. doi: 10.1111/j.1365-2958.2009.07019.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCluskey K, Wiest A, Plamann M. 2010. The Fungal Genetics Stock Center: a repository for 50 years of fungal genetics research. J Biosci 35:119–126. doi: 10.1007/s12038-010-0014-6. [DOI] [PubMed] [Google Scholar]
- 44.Perfect JR, Lang SD, Durack DT. 1980. Chronic cryptococcal meningitis: a new experimental model in rabbits. Am J Pathol 101:177–194. [PMC free article] [PubMed] [Google Scholar]
- 45.Nielsen K, Cox GM, Wang P, Toffaletti DL, Perfect JR, Heitman J. 2003. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and α isolates. Infect Immun 71:4831–4841. doi: 10.1128/IAI.71.9.4831-4841.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Selvig K, Ballou ER, Nichols CB, Alspaugh JA. 2013. Restricted substrate specificity for the geranylgeranyltransferase-I enzyme in Cryptococcus neoformans: implications for virulence. Eukaryot Cell 12:1462–1471. doi: 10.1128/EC.00193-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.