Abstract
Aim: To explore whether the aberrant DNA methylation status in plasma could be used as a biomarker for hepatocellular carcinoma (HCC) screening among high-risk individuals. Methods: The promoter methylation status of ELF, RASSF1A, p16, and GSTP1 was investigated by methylation-specific polymerase chain reaction (PCR) in 34 paired HCC and nontumor liver tissue from HCC patients and 10 tissues from patients with liver cirrhosis (LC). Plasma samples from 31 HCC patients, 10 LC patients as well as 7 patients with benign hepatic conditions were also collected and characterized using the same method. Results: Among liver specimens, HCC tissues displayed a significantly higher methylation frequency of each gene compared with nontumor tissue (p<0.05). Moreover, the frequency was much higher in tumor tissues than in nontumor tissue, when the data from two or three genes were combined (p=0.001 and p<0.001, respectively). Among plasma samples, either the frequency of at least one methylated gene (p<0.001) or the average number of methylated genes (p<0.05) demonstrated a stepwise increase in patients with benign lesions, LC, and HCC. Furthermore, when positive results, that is, plasma methylation status of at least one gene were combined with the elevated AFP400 level (serum alpha-fetoprotein [AFP] level at a cutoff of 400 ng/mL), the diagnostic sensitivity of HCC could increase to 93.55%. Conclusions: These results suggested that the methylation of tumor suppressor genes may participate in the development and progression of HCC. Additionally, it may be useful to combine the plasma DNA methylation status of a panel of gene markers and the serum AFP for HCC screening.
Introduction
Primary hepatocellular carcinoma (HCC), one of the most common malignant tumors around the world, is regarded as the third leading cause of cancer-related deaths due to its poor prognosis (Jemal et al., 2011; Siegel et al., 2013). Similar to the multistep carcinogenesis of other cancers, most HCCs in china have undergone a progression from HBV hepatitis, then liver cirrhosis (LC), and finally to carcinoma (Robinson, 1992; Merican et al., 2000).
Despite the unclear molecular pathogenesis of HCC, it has been confirmed that epigenetic aberrance, especially DNA methylation, plays an important role in the progression of carcinoma (Chen et al., 2013; Anwar and Lehmann, 2014; Dong and Wang, 2014). The silence of many tumor suppressor genes (TSGs), caused by the aberrant CpG island methylation of gene promoter, is considered as an early event for HCC pathogenesis (Chang et al., 2008; Nishida et al., 2008; Moribe et al., 2009). TSGs in tissues generally show an increased methylation frequency and different methylation status during the multistep carcinogenesis, which includes normal liver tissues, chronic hepatitis, LC, and HCC (Lee et al., 2003).
Delayed diagnosis was a fatal obstacle to timely and successful treatment, leading to the low survival rate of HCC patients (Singal et al., 2013). Therefore, early diagnosis of HCC is particularly important. Nowadays, serum alpha-fetoprotein (AFP) and ultrasonography (US) are clinically adopted for HCC screening (Masuzaki et al., 2012; Zhu et al., 2013). Nevertheless, serum AFP in about 40% of patients always remains at a low level (<400 ng/mL) (Farinati et al., 2006; Zhang et al., 2010), and US also has limitations in further detection of the features of liver nodules (Ward and Robinson, 2002).
Given that an ideal biomarker should be noninvasively detectable in the early stage of the disease, some studies have been working on the DNA epigenetic abnormalities in peripheral blood plasma or serum (Zhang et al., 2007; Chan et al., 2008; Iyer et al., 2010). As for HCC, during each step of carcinogenesis, the systemic information about the gene methylation status in serum/plasma remains largely unclear. Considering that the liver biopsy, although a gold standard for HCC diagnosis, brings relatively severe trauma and risks (Bongiovanni and Casana, 2008), some more noninvasive markers with high sensitivity for HCC screening are in urgent demand.
In this study, nested methylation-specific PCR (MSP) was performed among plasma and tissue specimens in HCC and LC patients to detect the promoter methylation status of genes, including embryonic liver fodrin (ELF), RASSF1A, p16, and GSTP1. ELF, a β-spectrin, is a stem cell adaptor protein that was recently found to play a pivotal role in TGF-β signaling and markedly decreased in tumor tissues of HCC (Kitisin et al., 2007). While the hypermethylation of the TSGs like RASSF1A, p16, and GSTP1 have already been reported in HCC (Lee et al., 2003; Kimura et al., 2009; Jain et al., 2014), no studies about ELF promoter methylation have ever been reported. This study aimed to investigate the DNA methylation status during multistep hepatocarcinogenesis among tissues and plasma samples, and to seek for noninvasive biomarkers with high sensitivity for HCC screening among high-risk individuals.
Materials and Methods
Clinical samples
With informed consent, 82 surgically resected liver specimens and 48 peripheral plasma samples were collected in West China Hospital, Sichuan University, between November 2008 and May 2009. The tissue specimens comprised 34 paired HCC and nontumor liver tissue from HCC patients (mean age, 47.94 years; 28 males and 6 females; 33 HBV-positive and 16 serum AFP >400 ng/mL), 10 from LC patients without concurrent HCC (mean age, 45.40 years; 7 males and 3 females; 8 HBV-positive but serum AFP <400 ng/mL), and 4 from healthy individuals who died of sudden accidents. The diagnosis of all specimens was pathologically confirmed. Plasma samples were obtained from 31 HCC patients (mean age, 45.90 years; 25 males and 6 females; 29 HBV-positive and 15 serum AFP >400 ng/mL), 10 LC patients without concurrent HCC (mean age, 46.50 years; 7 males and 3 females; 7 HBV-positive but serum AFP <400 ng/mL), and 7 patients with benign lesions (6 with liver angioma and 1 with lipid metabolic disorder). Among these specimens, 34 plasma samples were matched with the liver tissues (26 from HCC patients and 8 from LC patients).
Cell line preparation
In this study, cell lines, including Huh-7, T47D, and MCF-7, which have been confirmed to contain promoter hypermethylation of CpG island of RASSF1A, p16, and GSTP1, respectively, were chosen as positive controls (Yang et al., 2001; Di Gioia et al., 2006). Cell lines of MCF-7 and Huh-7 were cultured in PRMI1640, while T47D was cultured in DMEM. Both media contained 10% fetal bovine serum. Well-grown passages were collected for genomic DNA extraction.
DNA extraction
According to the manufacturer's instruction, DNA was extracted from tissues and cell lines with the TIANamp kit and dissolved in 150 μL Tris-EDTA buffer (TE). Likewise, plasma DNA was isolated from 400 μL sample using the Body Fluid Viral DNA/RNA Miniprep Kit (Axygen) and then dissolved in 50 μL TE.
Bisulfite conversion of DNA and MSP
The bisulfite modification was carried out as described previously (Herman et al., 1996; Wang et al., 2006). Nested-MSP was performed to investigate the methylation status of CpG islands of ELF, RASSF1A, p16, and GSTP1. The nested-MSP primers of ELF were designed using MethPrimer and the other three were as described previously (Table 1) (Guo et al., 2006; Kawamoto et al., 2007). During the first round of polymerase chain reaction (PCR), the regions containing short CpG-rich stretches flanked by the universal primers (PNA) were amplified. The 25 μL volume reaction system consisted of 50 ng bisulfate-modified DNA, 1× PCR buffer, 0.2 mM of each deoxynucleotide triphosphates, 0.4 μM of each primer, 2 mM MgCl2, and 1.5 U of Taq DNA polymerase (TaKaRa). Amplification was carried out as follows: initial denaturation at 95°C for 5 min, 35 cycles of 95°C for 30 s, annealing for 30 s and 72°C for 30 s, with a final extension of 10 min at 72°C. One microliter of diluted (1:50) sample of the first round of PCR products was then used as the template for the second round of MSP, while the other 5 μL for unmethylation-specific PCR (USP). The PCR condition was as follows: denaturation at 95°C for 5 min, 30 cycles of 95°C for 30 s, annealing for 30 s and 72°C for 30 s, with a final extension of 5 min at 72°C. Finally, the obtained MSP and USP PCR products were electrophoresed on 20 g/L agarose gels, stained with ethidium bromide, and visualized under UV illumination.
Table 1.
Primer Sequences and Polymerase Chain Reaction Conditions for MSP Analysis
| Primer name | Sense primer (5′-3′) | Antisense primer (5′-3′) | Annealing temp (°C) | PCR cycles | Product size (bp) |
|---|---|---|---|---|---|
| RASSF1A | |||||
| PNAa | GGAGGGAAGGAAGGGTAAG | CAACTCAATAAACTCAAACTCCC | 66 | 35 | 260 |
| MSP | GGGTTTTGCGAGAGCGCG | GCTAACAAACGCGAACCG | 63 | 30 | 169 |
| USP | GGTTTTGTGAGAGTGTGTTTAG | CACTAACAAACACAAACCAAAC | 63 | 30 | 169 |
| p16 | |||||
| PAN | AGAAAGAGGAGGGGTTGGTTGG | ACGCCCACACCTCCTCTACC | 65 | 35 | 193 |
| MSP | TTATTAGAGGGTGGGGCGGATCGC | GACCCCGAACCGCGACCGTAA | 62 | 30 | 150 |
| USP | TTATTAGAGGGTGGGGTGGATTGT | CAACCCCAAACCACAACCATAA | 62 | 30 | 151 |
| GSTP1 | |||||
| PAN | GGGATTTTAGGGTGTTTTTTTG | ACCTCCGAACCTTATAAAAATAATCCC | 62 | 35 | 159 |
| MSP | TTCGGGGTGTAGCGGTCGTC | GCCCCAATACTAAATCACGACG | 59 | 30 | 91 |
| USP | GATGTTTGGGGTGTAGTGGTTGTT | CCACCCCAATACTAAATCACAACA | 59 | 30 | 97 |
| E-cadherin | |||||
| PAN | GTGTTTTCGGGGTTTATTTGGTTGT | TACAACTCCAAAAACCCATAACTAACTAACC | 56 | 35 | 186 |
| MSP | TGTAGTTACGTATTTATTTTTAGTGGCGTC | CGAATACGATCGAATCGAACCG | 60 | 30 | 112 |
| USP | TGGTTGTAGTTATGTATTTATTTTTAGTGGTGTT | ACACCAAATACAATCAAATCAAACCAAA | 60 | 30 | 120 |
| ELF | |||||
| PAN | AATATAATGTTGTTATGTTAATTAGGTGAT | CAAAACTTTTAAATATAACCAATCCTAAAC | 57 | 35 | 182 |
| MSP | GGCGAGTGGTTTTTGATAAAATTATAAAT | ACATACCACGATTACCGAACCCGAA | 63 | 30 | 107 |
| USP | GGTGAGTGGTTTTTGATAAAATTATAAATT | ACATACCACAATTACCAAACCCAAA | 61 | 30 | 107 |
PNA, the first universal primer.
MSP, methylation-specific PCR; USP, unmethylation-specific polymerase chain reaction.
Bisulfite-modified DNA from cell lines with methylated genes mentioned above served as positive controls of RASSF1A, p16, and GSTP1, respectively; positive control of ELF utilized the completely methylated plasmid DNA containing promoter of ELF (ELF prom). In addition, bisulfite-treated DNA from healthy liver tissues served as negative control.
Statistical analyses
The differences and associations of the promoter methylation status between varied tissue specimens and plasma samples were analyzed by the Pearson's χ2 test or Fisher's exact test according to the absolute numbers included in the analysis. The concordances of methylation status for each gene between tissues and matched plasma samples were evaluated using the simple kappa coefficient. Associations between clinicopathologic variables and promoter methylation status were analyzed using Fisher's exact test. One-way ANOVA was used to evaluate the significance of the differences observed between three or more means. A value of p<0.05 (two sided) was considered significant. Statistical analyses were carried out with SPSS software (version 13.0).
Results
Methylation frequency of individual TSG and cumulative methylation patterns in HCC, Nontumor, and LC tissues
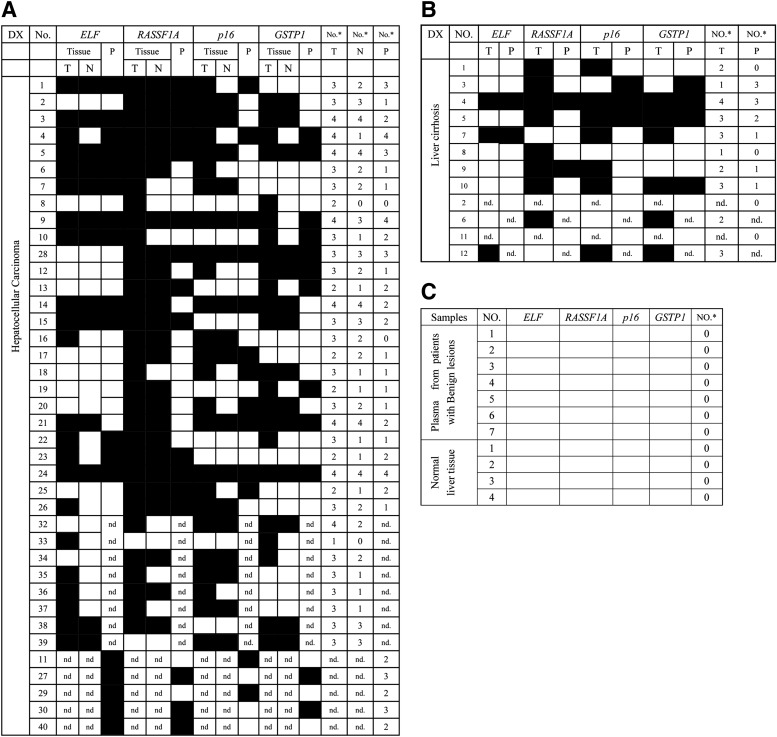
In this study, the methylation status of the CpG islands of ELF, RASSF1A, p16, and GSTP1 in different tissue specimens is shown in Figure 1. The detailed specific MSP results of each specimen are given in Figure 2. As shown in Table 2, the methylation frequency of each TSG was significantly higher in tumor than in nontumor tissue (p<0.05, χ2 test), whereas no aberrant methylation was found in the healthy liver tissues.
FIG. 1.
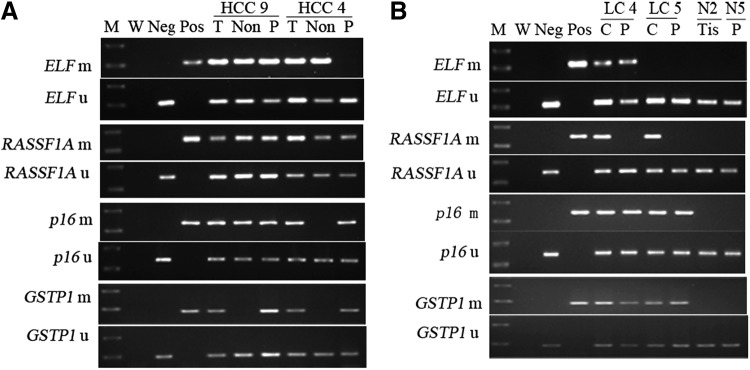
MSP analysis of ELF, RASSF1A, p16, and GSTP1 in liver tissue and plasma samples of hepatocellular carcinoma (HCC) and liver cirrhosis (LC) patients. (A) Representative examples of tumor, nontumor tissue, and corresponding plasma samples from two HCC patients. (B) Methylation status in cirrhosis and corresponding plasma from two LC patients and one normal liver tissue and one plasma sample from patients without liver disease. Bisulfite-treated DNA was amplified with primers specific to the methylated (m) or the unmethylated (u) CpG islands of each gene. MSP products were stained with ethidium bromide after 2.0% agarose gel electrophoresis. C, cirrhosis; M, molecular weight standard; MSP, methylation-specific PCR; m, methylated; W, water contamination control; Pos, MSP product from positive plasma or cell line DNA used as positive control (ELF prom for ELF, Huh-7 for RASSF1A, T47D for p16, and MCF-7 for GSTP1); Neg, unmethylated product of normal liver DNA as negative control; Non, nontumor; N, normal cases; P, plasma samples; PCR, polymerase chain reaction; T, tumor; Tis, tissue; u, unmethylated.
FIG. 2.
Summary of methylation analysis of ELF, RASSF1A, p16, and GSTP1 in liver tissue and corresponding plasma samples. Filled boxes indicate the presence of methylation and open boxes indicate the absence of methylation. (A) MSP in tumor, nontumor, and plasma samples of HCC patients. (B) MSP in cirrhosis and corresponding plasma samples of LC patients. (C) MSP in normal liver tissues and plasma samples from patients with benign lesions. T, tumor tissue; N, paired nontumorous liver tissue; No., case number; No.*, number of genes methylated; nd., not detected.
Table 2.
Methylation Frequency for Four Genes in Samples of HCC and LC Cases
| Gene | Tumor (n=34) (%) | Nontumor (n=34) (%) | HCC plasma (n=31) (%) | LC tissue (n=10) (%) | LC plasma (n=10) (%) | Tumor vs. nontumor, p-value | Tumor vs. LC, p-value | HCC plasma vs. LC plasma p-value |
|---|---|---|---|---|---|---|---|---|
| ELF | 22 (64.70) | 13 (38.24) | 18 (58.06) | 3 (30) | 2 (20) | 0.029 | 0.113 | 0.084 |
| RASSF1A | 32 (94.12) | 25 (73.52) | 16 (51.61) | 8 (80.0) | 2 (20) | 0.021 | 0.460 | 0.244 |
| p16 | 25 (73.52) | 17 (50) | 13 (41.94) | 7 (70) | 3 (30) | 0.046 | 1.000 | 0.919 |
| GSTP1 | 23 (67.65) | 14 (41.17) | 12 (38.70) | 6 (60) | 4 (40) | 0.028 | 0.945 | 1.000 |
p-Value, analyzed by χ2 test.
HCC, hepatocellular carcinoma; LC, liver cirrhosis.
Moreover, to investigate whether accumulation of epigenetic aberrance is involved in the carcinogenesis of HCC, cumulative methylation patterns of the four TSGs were analyzed (Table 3). We discovered that, with the combination of at least two (p=0.001, χ2 test) or three genes, the methylation frequency was even higher in tumor than in nontumor (p<0.001, χ2 test).
Table 3.
The Frequency of CpG Island Hypermethylation in Tumor and Nontumorous Liver Samples from HCC, Cirrhotic Liver Samples Without Concurrent HCC, and Normal Liver Samples
| Diagnosis | No. of cases | No. of cases methylated for at least one gene (%)a | No. of cases methylated for at least two genes (%)a | No. of cases methylated for at least three genes (%)a | Average no. of methylated geneb |
|---|---|---|---|---|---|
| Tumor tissue | 34 | 34 (100) | 33 (97.06)c | 27 (79.41)d | 3.00±0.74e |
| Nontumor tissue | 34 | 32 (94.12) | 22 (64.71)c | 12 (35.30)d | 2.03±1.17e |
| LC | 10 | 10 (100) | 8 (80) | 5 (50) | 2.40±0.97 |
| Normal liver | 4 | 0 | 0 | 0 | 0e |
p-Value results for other tests are not significant.
Analyzed by χ2 test or Fisher's exact test, as appropriate.
Analyzed by one-way ANOVA test.
p=0.001, tumor versus nontumorous tissue.
p<0.001, tumor versus nontumorous tissue.
p<0.001, tumor versus nontumorous tissue, nontumorous versus normal liver tissue, LC versus normal liver tissue.
ANOVA, analysis of variance.
Intriguingly, when comparing the average number of aberrantly methylated genes in each step of the multistep carcinogenesis, we found that the average number showed a stepwise increase during the progression of the lesion, that is, 0 for healthy liver tissues, 2.03 for nontumor tissues, and 3.00 for HCC tissues, and the difference between each step was statistically significant (p<0.001, one-way ANOVA test). However, the average number of methylated genes for LC tissues (2.40) did not show any significant difference compared with either tumor or nontumor tissue.
Methylation frequency of individual TSG and cumulative methylation patterns in plasma samples
To study whether plasma DNA methylation status during the multistep carcinogenesis could be monitored, the plasma samples from 31 HCC patients, 10 LC patients, and 7 patients with benign lesions were analyzed (Fig. 1). Details of each sample are illustrated in Figure 2. The aberrant plasma DNA methylation of the four TSGs was detected in patients with HCC and LC (Table 2), whereas no significant results were found in patients with benign lesions.
In our study, 26 HCC tissue specimens along with their matched plasma samples were set up, and the methylation profiles are shown in Figure 2. In these cases, aberrant RASSF1A methylation occurred in all those tumor tissues and 16 plasma samples. Among 22 patients with ELF methylation in HCC, an identical epigenetic alteration was detected in plasma of 82% (18 of 22) patients in association (p<0.001, Fisher's exact test) and in concordance (κ=0.761) with tumor methylation status, respectively. Similarly, when estimate the consistence of TSG methylation between plasma and tissue, 11 of 19 for p16 methylation showed that the frequency in plasma was associated (p=0.01, Fisher's exact test) and in concordance with that in tumor tissues (κ=0.425); Also, aberrant GSTP1 methylation was detected in 18 tumor tissues and 10 plasma samples, with significant association (p=0.009, Fisher's exact test) and concordance (κ=0.435) between tissues and plasma samples.
To evaluate whether or not the cumulative methylation pattern of the four TSGs could be applied in HCC screening among populations of high risk, all plasma samples were reanalyzed on the basis of the combination of these aberrantly methylated genes (Table 4). The plasma DNA methylation frequency with at least one methylated gene showed a stepwise increase during the progression of the lesions, that is, 0 of 7 for patients with benign lesions, 6 of 10 for LC patients, and 29 of 31 for HCC, and the differences between each step were statistically significant (p<0.05, Fisher's exact test). Accordingly, the plasma status with at least one of these TSGs being methylated could be considered as a biomarker for HCC screening among populations of high risk, for its high sensitivity of 93.55% in HCC patients. Moreover, the average number of methylated genes in plasma samples from patients with benign lesions (0), LC (1.10), and HCC (1.90) also showed a stepwise increase (p<0.05, one-way ANOVA test).
Table 4.
The Frequency of CpG Island Hypermethylation in Plasma Samples from HCC and LC Patients
| Diagnosis of patients | No. of cases | No. of cases methylated for at least one gene (%)a | Average no. of methylated geneb |
|---|---|---|---|
| HCC | 31 | 29 (93.55)c | 1.90±1.08d |
| LC | 10 | 6 (60)c | 1.10±1.20d |
| Benign lesionse | 7 | 0c | 0d |
Analyzed by χ2 test or Fisher's exact test, as appropriate.
Analyzed by one-way ANOVA test.
p=0.024, HCC plasma versus LC plasma; p=0.035, LC plasma versus normal plasma.
p=0.037, HCC plasma versus LC plasma; p=0.035, LC plasma versus normal plasma.
Benign lesions, patients with benign lesions.
Analysis of diagnostic sensitivity of HCC with combination of serum AFP level and plasma DNA methylation status
Among the 31 HCC plasma samples, it was of great importance to discover that when referring to the plasma DNA methylation frequency with at least one gene being methylated, the diagnostic sensitivity became significantly higher than the well-known biomarker AFP400 (93.55% vs. 48.39%, p<0.001, χ2 test). Furthermore, the diagnostic sensitivity of HCC could increase to 93.55% with the combination of these two markers.
Association between TSG methylation profile and clinicopathological features of HCC
Based on the number of methylated genes, the 34 HCC patients were categorized into group 1 (more methylated genes in tumor than in ANLT) and group 2 (equal number of methylated genes in tumor and ANLT). Among these cases, 24 belonged to group 1, while 10 belonged to the other. The clinicopathological features of HCC, including patients' age, gender, HBsAg, status of LC, serum AFP level, tumor size, tumor quantity, grade of differentiation, portal vein embolus, metastasis and recurrence, 1-year postoperative mortality, were analyzed in association with the methylation of individual TSG or the cumulative patterns. Portal vein embolus and paracirrhosis occurred more frequently in group 1 (p=0.015 and p=0.031, respectively, Fisher's exact test). No significant association was observed between the TSG methylation profile and the other clinicopathological features of HCC.
Discussion
Current evidence indicates that a number of major pathways are implicated in HCC, including the TGF-β −Smad pathway, RAF-MKK1-MAPK, Pl3K–AKT1–mTO, WNT–catenin β-1, IGF-I, hepatocyte growth factor–c-MET, and growth-factor-regulated angiogenic signaling. These pathways are involved in the progression of hepatocarcinogenesis and development, such as epithelial mesenchymal transition, self-renewal of liver cancer stem cell, and tumor invasion and metastasis, and in regulation of general cellular process such as proliferation, differentiation, and apoptosis (Majumdar et al., 2012; Gedaly et al., 2014). Effectively, many TSGs play important roles in controlling the pathways to prevent hepatocarcinogenesis. Unfortunately, epigenetic silencing of TSGs by CpG island hypermethylation commonly occurs in HCCs (Neumann et al., 2012; Nishida et al., 2014).
For instance, RASSF1A, a number of the Ras association domain family, as a candidate of TSG plays a crucial function in cell cycle regulation, apoptosis, and microtubule stability by regulating the Raf-Ras pathway; p16, as an inhibitor of cyclin-dependent CDK4 and CDK6, arrests cell cycle by preserving activation of Rb. In addition, GSTP1, a type of GSTs that catalyze the conjugation of carcinogens, natural toxins, and exogenous drugs, is involved in attenuating hepatocarcinogenesis due to exposure to toxins (e.g., alcohol or aflatoxin), although it does not interfere with the pathways. It has been reported that hypermethylation of each of the three TSGs is an early event in HCC (Wong et al., 1999; Wang et al., 2006; Chan et al., 2008).
The most interesting TSG is ELF, which is a crucial modulator of TGF-β signaling. ELF, as an adaptor of Smad3/4, associates with TGFR, facilitating the complex translocation to the nucleus. Recent study has shown that ELF expresses in the normal human liver progenitor cells but not in STAT3/Oct4-positive cancer stem cells of HCC, implying the inhibition effect of ELF on hepatocarcinogenesis (Lin et al., 2009). However, what leads ELF to lose expression in HCC stem cells is not yet understood in detail. Thus, we supposed that ELF might be silenced by its promoter methylation similar to the inactivation mechanism of most TSGs.
Since silence of ELF, RASSF1A, p16, and GSTP1 is intimately implicated in hepatocarcinogenesis, we selected promoter methylation of the four genes as a biomarker for screening HCC among high-risk populations.
To the best of our knowledge, this was the first study to demonstrate that a high frequency of DNA methylation of ELF gene promoter was detected in HCC, which gave a new sight to study the mechanisms of ELF involving in the pathogenesis of HCC and its application in prognosis. In the meantime, the present study confirmed that the methylation frequency of ELF, RASSF1A, p16, and GSTP1, the average number of methylated genes or methylation frequencies with combination of at least two or three genes in HCC tissues were significantly higher than in nontumor, which is in consistence with previous reports (Lee et al., 2003; Yang et al., 2003; Wang et al., 2006; Li et al., 2010). All these results suggest that events of DNA methylation had taken place even early in the liver injury stages, and a trend of increase of methylated genes occurred during progression of HCC.
In our experiment, the aberrant hypermethylation of four genes ELF, RASSF1A, p16, and GSTP1 was observed in plasma samples with the positive results of the matched HCC tissues, which is concordant with the previous study regarding p16 and GSTP1 (Wong et al., 1999, 2000, 2003; Wang et al., 2006), indicating the refection of the epigenetic changes of corresponding tumors. It should be noted that p16 and GSTP1 were found to be methylated in the plasma sample of one LC case, but not in the matched liver lesion. A possible explanation for the discordance is that the methylation of a given gene might occur in some nodules, which were missed being collected and analyzed.
The combination between aberrant plasma DNA methylation status and serum AFP may offer a promising approach to improve the sensitivity of the noninvasive diagnosis of early HCC, since in our study, the plasma status with at least one methylated gene and the serum AFP400 were taken into account together, and the diagnostic sensitivity in HCCs could improve to 93.55%. These results imply that once studies with a larger sample size confirm our idea, a better way of screening the high-risk population will come into use, benefiting further the monitoring, diagnosis, and treatment of early HCC in LC patients.
It has been shown that methylation of TSGs is intimately involved in the development and progression of tumor (Zhang, 2015). In this study, the clinicopathological features of HCC patients were analyzed in association with methylation of either an individual gene or cumulative patterns, respectively. Portal vein embolus was found to be more frequent in group 1 (with more methylated genes in tumor than nontumor) than in group 2 (with equal number of methylated genes in tumor and nontumor). In the previous study, the portal vein embolus was a major determinant of the outcome of HCC patients, and the survival rate was lower for those with portal vein embolus (Shirabe et al., 2009). In addition, it was reported that HCC patients with methylation of GSTP1 or E-cadherin showed a poorer survival than those without (Lee et al., 2003). In view of the follow-up data of only 1 year in our study, this could be a limitation for the prognosis evaluation, which prompted us to investigate more TSGs and to collect more follow-up data in further studies.
In conclusion, this study revealed human methylation status of ELF, RASSF1A, p16, and GSTP1 in both plasma and liver tissues along the progression of hepatocarcinogenesis, including benign lesion, LC, and HCC. We demonstrated that the methylation of TSGs tend to accumulate during a multistep hepatocarcinogenesis. Our present data also showed that the combination between plasma methylation of a panel of gene markers and the serum AFP may be useful for HCC screening for high-risk individuals and for the monitoring of LC. However, further large studies are still required to confirm our current findings and to identify new noninvasive biomarkers with higher accuracy for screening populations of high risk and for the diagnosis of HCC.
Acknowledgments
We sincerely thank Prof. Tianfu Wen and Wentao Wang for assistance with the samples. This work was supported by a grant from the National Natural Science Foundation of China (No. 30870957) and a grant from the Sichuan Province Science and Technology Department, China (No. 2008SZ0028).
Author Disclosure Statement
No competing financial interests exist.
References
- Anwar SL, Lehmann U. (2014) DNA methylation, microRNAs, and their crosstalk as potential biomarkers in hepatocellular carcinoma. World J Gastroenterol 20:7894–7913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongiovanni M, Casana M. (2008) Non-invasive markers of liver fibrosis in HCV mono-infected and in HIV/HCV co-infected subjects. Med Chem 4:513–519 [DOI] [PubMed] [Google Scholar]
- Chan KC, Lai PB, Mok TS, et al. (2008) Quantitative analysis of circulating methylated DNA as a biomarker for hepatocellular carcinoma. Clin Chem 54:1528–1536 [DOI] [PubMed] [Google Scholar]
- Chang H, Yi B, Li L, et al. (2008) Methylation of tumor associated genes in tissue and plasma samples from liver disease patients. Exp Mol Pathol 85:96–100 [DOI] [PubMed] [Google Scholar]
- Chen K, Huang W, Huang B, et al. (2013) BORIS, brother of the regulator of imprinted sites, is aberrantly expressed in hepatocellular carcinoma. Genet Test Mol Biomarkers 17:160–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Gioia S, Bianchi P, Destro A, et al. (2006) Quantitative evaluation of RASSF1A methylation in the non-lesional, regenerative and neoplastic liver. BMC Cancer 6:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Wang A. (2014) Aberrant DNA methylation in hepatocellular carcinoma tumor suppression (Review). Oncol Lett 8:963–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farinati F, Marino D, De Giorgio M, et al. (2006) Diagnostic and prognostic role of alpha-fetoprotein in hepatocellular carcinoma: both or neither? Am J Gastroenterol 101:524–532 [DOI] [PubMed] [Google Scholar]
- Gedaly R, Galuppo R, Daily MF, et al. (2014) Targeting the Wnt/beta-catenin signaling pathway in liver cancer stem cells and hepatocellular carcinoma cell lines with FH535. PLoS One 9:e99272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Ren J, House MG, et al. (2006) Accumulation of promoter methylation suggests epigenetic progression in squamous cell carcinoma of the esophagus. Clin Cancer Res 12:4515–4522 [DOI] [PubMed] [Google Scholar]
- Herman JG, Graff JR, Myohanen S, et al. (1996) Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A 93:9821–9826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer P, Zekri AR, Hung CW, et al. (2010) Concordance of DNA methylation pattern in plasma and tumor DNA of Egyptian hepatocellular carcinoma patients. Exp Mol Pathol 88:107–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Xie L, Boldbaatar B, et al. (2014) Differential methylation of the promoter and first exon of the RASSF1A gene in hepatocarcinogenesis. Hepatol Res doi: 10.1111/hepr.12449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemal A, Bray F, Center MM, et al. (2011) Global cancer statistics. CA Cancer J Clin 61:69–90 [DOI] [PubMed] [Google Scholar]
- Kawamoto K, Okino ST, Place RF, et al. (2007) Epigenetic modifications of RASSF1A gene through chromatin remodeling in prostate cancer. Clin Cancer Res 13:2541–2548 [DOI] [PubMed] [Google Scholar]
- Kimura N, Moribe T, Iizuka N, et al. (2009) Rapid and quantitative detection of CpG-methylation status using TaqMan PCR combined with methyl-binding-domain polypeptide. Clin Biochem 42:1113–1122 [DOI] [PubMed] [Google Scholar]
- Kitisin K, Ganesan N, Tang Y, et al. (2007) Disruption of transforming growth factor-beta signaling through beta-spectrin ELF leads to hepatocellular cancer through cyclin D1 activation. Oncogene 26:7103–7110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Lee HJ, Kim JH, et al. (2003) Aberrant CpG island hypermethylation along multistep hepatocarcinogenesis. Am J Pathol 163:1371–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Liu W, Wang L, et al. (2010) CpG island methylator phenotype associated with tumor recurrence in tumor-node-metastasis stage I hepatocellular carcinoma. Ann Surg Oncol 17:1917–1926 [DOI] [PubMed] [Google Scholar]
- Lin L, Amin R, Gallicano GI, et al. (2009) The STAT3 inhibitor NSC 74859 is effective in hepatocellular cancers with disrupted TGF-beta signaling. Oncogene 28:961–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar A, Curley SA, Wu X, et al. (2012) Hepatic stem cells and transforming growth factor beta in hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol 9:530–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuzaki R, Karp SJ, Omata M. (2012) New serum markers of hepatocellular carcinoma. Semin Oncol 39:434–439 [DOI] [PubMed] [Google Scholar]
- Merican I, Guan R, Amarapuka D, et al. (2000) Chronic hepatitis B virus infection in Asian countries. J Gastroenterol Hepatol 15:1356–1361 [DOI] [PubMed] [Google Scholar]
- Moribe T, Iizuka N, Miura T, et al. (2009) Methylation of multiple genes as molecular markers for diagnosis of a small, well-differentiated hepatocellular carcinoma. Int J Cancer 125:388–397 [DOI] [PubMed] [Google Scholar]
- Neumann O, Kesselmeier M, Geffers R, et al. (2012) Methylome analysis and integrative profiling of human HCCs identify novel protumorigenic factors. Hepatology 56:1817–1827 [DOI] [PubMed] [Google Scholar]
- Nishida N, Chishina H, Arizumi T, et al. (2014) Identification of epigenetically inactivated genes in human hepatocellular carcinoma by integrative analyses of methylation profiling and pharmacological unmasking. Dig Dis 32:740–746 [DOI] [PubMed] [Google Scholar]
- Nishida N, Nagasaka T, Nishimura T, et al. (2008) Aberrant methylation of multiple tumor suppressor genes in aging liver, chronic hepatitis, and hepatocellular carcinoma. Hepatology 47:908–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson WS. (1992) The role of hepatitis B virus in the development of primary hepatocellular carcinoma: Part I. J Gastroenterol Hepatol 7:622–638 [DOI] [PubMed] [Google Scholar]
- Shirabe K, Kajiyama K, Harimoto N, et al. (2009) Prognosis of hepatocellular carcinoma accompanied by microscopic portal vein invasion. World J Gastroenterol 15:2632–2637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R, Naishadham D, Jemal A. (2013) Cancer statistics, 2013. CA Cancer J Clin 63:11–30 [DOI] [PubMed] [Google Scholar]
- Singal AG, Waljee AK, Patel N, et al. (2013) Therapeutic delays lead to worse survival among patients with hepatocellular carcinoma. J Natl Compr Canc Netw 11:1101–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Qin Y, Li B, et al. (2006) Detection of aberrant promoter methylation of GSTP1 in the tumor and serum of Chinese human primary hepatocellular carcinoma patients. Clin Biochem 39:344–348 [DOI] [PubMed] [Google Scholar]
- Ward J, Robinson PJ. (2002) How to detect hepatocellular carcinoma in cirrhosis. Eur Radiol 12:2258–2272 [DOI] [PubMed] [Google Scholar]
- Wong IH, Lo YM, Yeo W, et al. (2000) Frequent p15 promoter methylation in tumor and peripheral blood from hepatocellular carcinoma patients. Clin Cancer Res 6:3516–3521 [PubMed] [Google Scholar]
- Wong IH, Lo YM, Zhang J, et al. (1999) Detection of aberrant p16 methylation in the plasma and serum of liver cancer patients. Cancer Res 59:71–73 [PubMed] [Google Scholar]
- Wong IH, Zhang J, Lai PB, et al. (2003) Quantitative analysis of tumor-derived methylated p16INK4a sequences in plasma, serum, and blood cells of hepatocellular carcinoma patients. Clin Cancer Res 9:1047–1052 [PubMed] [Google Scholar]
- Yang B, Guo M, Herman JG, Clark DP. (2003) Aberrant promoter methylation profiles of tumor suppressor genes in hepatocellular carcinoma. Am J Pathol 163:1101–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Yan L, Davidson NE. (2001) DNA methylation in breast cancer. Endocr Relat Cancer 8:115–127 [DOI] [PubMed] [Google Scholar]
- Zhang XF, Qi X, Meng B, et al. (2010) Prognosis evaluation in alpha-fetoprotein negative hepatocellular carcinoma after hepatectomy: comparison of five staging systems. Eur J Surg Oncol 36:718–724 [DOI] [PubMed] [Google Scholar]
- Zhang Y. (2015) Detection of epigenetic aberrations in the development of hepatocellular carcinoma. Methods Mol Biol 1238:709–731 [DOI] [PubMed] [Google Scholar]
- Zhang YJ, Wu HC, Shen J, et al. (2007) Predicting hepatocellular carcinoma by detection of aberrant promoter methylation in serum DNA. Clin Cancer Res 13:2378–2384 [DOI] [PubMed] [Google Scholar]
- Zhu K, Dai Z, Zhou J. (2013) Biomarkers for hepatocellular carcinoma: progression in early diagnosis, prognosis, and personalized therapy. Biomark Res 1:10. [DOI] [PMC free article] [PubMed] [Google Scholar]