Abstract
Efficient HIV transcription requires P-TEFb, an essential co-factor for Tat. In actively replicating cells, P-TEFb is incorporated into the 7SK snRNP complex together with the repressor protein HEXIM1. Using an affinity purification-tandem mass spectrometry approach to identify modification sites on HEXIM1 that regulate the sequestration of P-TEFb by 7SK snRNP, we found that HEXIM1 can be phosphorylated on adjacent residues in a region immediately upstream of the coiled-coil dimerization domain (Ser268, Thr270, Tyr271, and Tyr274). Phosphomimetic mutations of Tyr271 and Tyr274 disrupted the assembly of P-TEFb and HEXIM1 into the 7SK snRNP complex. Although Y271E/Y274E did not adversely affect the nuclear localization pattern of HEXIM1, it induced the redistribution of the CDK9 subunit of P-TEFb into the cytoplasm. By contrast, the Y271F/Y274F HEXIM1 mutant assembled normally with P-TEFb within the 7SK snRNP complex but severely reduced proviral gene expression in T cells in response to activation signals and caused a severe growth defect of Jurkat T cells. Thus Y271F/Y274F, which cannot be phosphorylated on these residues, appears to block the exchange of active P-TEFb from the 7SK complex, thereby limiting the level of P-TEFb below the threshold required to support transcription elongation of the HIV provirus and cellular genes.
Keywords: HIV Tat, 7SK RNP complex, HEXIM1 phosphorylation, P-TEFb
1 Introduction
Defining the RNA/protein interactions that hold the essential Tat co-factor P-TEFb within the repressive 7SK snRNP complex and the distinct mechanisms by which P-TEFb is released from 7SK snRNP by Tat after T-cell activation is an essential step for understanding how HIV transcription and latency is regulated in T-cells. Full length transcription of integrated proviral HIV and the majority of mammalian protein-encoding genes require stimulation of RNA polymerase II (RNAP II) processivity by P-TEFb kinase [1-3]. The major functional P-TEFb complex in actively dividing mammalian cells is a heterodimer complex of the CDK9 serine/threonine kinase and Cyclin T1 (CycT1) [4]. HIV Tat binds directly to CDK9/CycT1 and is recruited to transcriptionally paused RNAP II complexes situated on nascent TAR RNA hairpins at the proviral 5’ LTR [4, 5]. TAR-bound Tat/P-TEFb complexes function to hyperphosphorylate serine residues of the heptad repeats at the C-terminal tail of promoter-proximally paused RNA polymerase II and also phosphorylate the negative elongation factors NELF and DSIF that are responsible for stalling the polymerase and preventing HIV RNA synthesis [6]. Thus, the overall effect of P-TEFb activity on the paused polymerase complex is to stimulate efficient promoter clearance and processive elongation.
Entry of HIV into latency in infected T cells is due to restrictions imposed on both transcription initiation and transcription elongation [1]. Epigenetic restrictions at the viral LTR block initiation and reduce the levels of the viral transactivator protein, Tat, to below its threshold levels. In resting T-cells P-TEFb is nearly absent due to a block in the synthesis of CycT1. In actively replicating cells, a substantial fraction of cellular P-TEFb (~50-80%) is reversibly associated with a nuclear ribonucleoprotein complex 7SK snRNP that inhibits the kinase and suppresses its transcriptional activity on HIV and cellular genes [7, 8]. Central to this inhibitory complex is 7SK snRNA, a highly-expressed and ubiquitous RNA polymerase III transcript 330-332 nucleotides in length [2]. 7SK snRNA folds into a highly structured three-dimensional conformer that provides a structural scaffold that coordinates the binding of two molecules of P-TEFb to a homodimer of the P-TEFb inhibitory partner HEXIM1 [9, 10]. Association of HEXIM1 with 7SK snRNA is critical for P-TEFb incorporation into 7SK snRNP, and HEXIM1 cannot interact with and inhibit P-TEFb in the absence of 7SK snRNA [11]. The 5’ and 3’ ends of 7SK snRNA are bound and protected from nuclease degradation by the 5’ methylphosphate capping enzyme MEPCE and the La-related protein LARP7, respectively [12, 13].
7SK snRNP provides an exchangeable pool from which the catalytically competent and transcriptionally active form of P-TEFb can be readily extracted and directed to genes [14]. For instance, we have previously demonstrated that T-cell activation of latently infected T cells via ligation of the T-cell receptor (TCR) or challenge with a protein kinase C (PKC) agonist, results in a partial but rapid release of P-TEFb and HEXIM1 from 7SK snRNP that is coupled with recruitment of P-TEFb to the HIV LTR and stimulation of processive proviral transcription [15].
Using an affinity purification and tandem mass spectrometry (AP-MS/MS) approach to isolate and characterize P-TEFb complexes, we found that the S175 residue on CDK9 is specifically phosphorylated following T-cell activation [16]. P-Ser175 is only found on P-TEFb that has dissociated from the 7SK snRNP inhibitory complex. Signal-dependent phosphorylation of Ser175, although not required for Tat binding to P-TEFb, favors binding of Tat over the bromodomain extra-terminal protein BRD4, which is used to direct P-TEFb to cellular genes, thus establishing a molecular link between T-cell activation and the availability of P-TEFb for HIV transcriptional control.
Here, we extend our studies of post-translational modifications that regulate P-TEFb to include modifications on HEXIM1. Specifically, we have identified a cluster of phosphorylation sites at the C-terminal acidic domain and an unstructured region immediately upstream of the coiled-coil dimerization domain. Mutagenesis and biochemical studies strongly suggest that the phosphorylation of two adjacent tyrosine residues (Tyr271/Tyr274) prevents the sequestration of P-TEFb by 7SK snRNP. By contrast, mutant Y271F/Y274F appears to stabilize the 7SK RNP and limit the level of transcriptionally active P-TEFb much below the threshold required to support processive transcription elongation of proviral HIV and cellular genes. These data are consistent with the hypothesis that signal-dependent post-translational modifications of 7SK snRNP components allow sub-threshold levels of Tat to efficiently extract P-TEFb and deliver it to the HIV LTR and drive viral replication.
2 Materials and Methods
2.1 Cell culture and latent HIV reactivation studies
The Jurkat 2D10 T-cell line model of HIV latency, has been extensively characterized [6]. The provirus in 2D10 cells carries a d2EGFP reporter in place of Nef to facilitate measurement of transcription activation by flow cytometry. Jurkat and HEK293T cells were cultured at 37°C in complete media (RPMI 1640 medium (Hyclone), 5% fetal bovine serum, 100 I.U/ml penicillin, 100 μg/ml streptomycin, and 25 mM Hepes, pH 7.2). Jurkat 2D10 cells stably expressing Y271F/Y274F FLAG-HEXIM1 were cultured in the complete medium described above but containing 20% fetal bovine serum. Proviral reactivation was for 18 hr with TNF-α, SAHA, PMA, or a combination of anti-CD3/anti-CD28 antibodies.
2.2 Stable expression of FLAG-HEXIM1 in Jurkat 2D10 and HEK 293T cells
The MSCV retroviral expression system (Clontech) was used to generate Jurkat 2D10 or HEK 293T cells that stably express FLAG-HEXIM1 WT or the mutant versions. Briefly, VSVG-pseudotyped packaged retroviruses carrying MSCV FLAG-HEXIM1 were prepared by co-transfecting HEK 293T cells with pMSCV FLAG-HEXIM1, and expression vectors for retroviral gag-pol and VSVG using Lipofectamine 2000 (Invitrogen). MSCV-infected Jurkat cells were selected in complete media containing 3 μg/ml puromycin. Expression of FLAG-HEXIM1 WT and mutants was confirmed by Western blotting using both anti-FLAG (Sigma) and anti-HEXIM1 antibodies.
2.3 Purification of HEXIM1 for tandem mass spectrometry
5.0 × 108 Jurkat 2D10 cells expressing FLAG-HEXIM1 WT were harvested before and after 1 hr activation by 50 ng/ml PMA. Whole cell extracts (WCEs) were prepared using cell lysis buffer A (150 mM NaCl, 10 mM KCl, 1.5 mM MgCl2, 0.5% NP-40, 1 mM DTT, 10 mM Hepes, [pH 8.0]) containing a protease and phosphatase inhibitor cocktail (Roche). WCEs were cleared by centrifugation at 2000 rpm for 5 min followed by incubation for 2 h with protein A sepharose beads. Cleared WCEs were then incubated with anti-FLAG M2 affinity resin (Sigma) overnight at 4 °C on a gentle rocker (30 μl packed beads for 1 ml of cleared WCEs). The beads were isolated by centrifugation at 2000 rpm for 2 min followed by five 30-min washes of the resin with 10 ml cell lysis buffer A. Bound HEXIM1 protein complexes were eluted with 200 μg/ml FLAG peptide in 2 ml cell lysis buffer A at 4 ¼ on a gentle rocker.
2.4 Preparation of protein samples for LC-MS/MS
Protein eluates were concentrated by methanol-chloroform precipitation and rehydrated with 1X LDS loading buffer containing 50 mM DTT before being resolved by 1D SDS-PAGE on a 4-12% Bis-Tris gel. SYPRO Ruby (Invitrogen) stained gel bands containing HEXIM1 protein were subjected to in-gel digestion with sequencing grade modified trypsin (Promega, WI) according to the procedure described previously [16] or with the enzyme pepsin A. Briefly, gel pieces excised from a SDS-PAGE were first washed with 50% acetonitrile in 50 mM ammonium bicarbonate, and then dehydrated with acetonitrile. Before an overnight proteolytic digestion, proteins were reduced with 20 mM DTT at room temperature for 1 hr and alkylated with 55 mM iodoacetamide in 50 mM ammonium bicarbonate for 30 min in the dark. After proteolytic digestion, peptides were extracted from the gel with 5% formic acid in 50% acetonitrile and then resuspended in 0.1% formic acid after being dried completely under the Speed Vacuum.
2.5 LC-MS/MS analysis of PTMs
Liquid chromatography-tandem mass spectrometry analysis was performed by using a LTQ Velos-Orbitrap mass spectrometer (Thermo Fisher Scientific, Waltham, MA) interfaced with Waters nano Acquity UPLC (Waters, MA). The full MS scan was performed with 120,000 resolution in the Orbitrap detector, while the MS/MS scan was performed in ion trap detector on the top twenty most abundant precursor ions in CID mode at the normalized collision energy of 35%. The data were analyzed by Mascot Daemon (version 2.4.0, Matrix Science, Boston, MA) using an IPI human database (version June 2010) with the setting of 10 ppm for parent ions and 0.8 Da for product ions. For HEXIM1 mutant samples, MASCOT search used a customized database including WT, as well as the mutant HEXIM1 proteins in which the specific amino acids were replaced with the corresponding mutants. Carbamidomethylation of Cys, oxidation of Met, phosphorylation of Ser, Thr and Tyr, as well as the acetylation of Lys were set as variable modifications. Candidate sites of post-translational modification initially identified with Mascot software were further verified by manual interpretation of the obtained MS/MS spectra.
2.6 Immunofluorescence microscopy
Cells were allowed to adhere to plastic cover slips coated with poly-L-lysine for 10 min at 37 °C. The cover slips were washed once with 1X PBS and bound cells were fixed for 15 min. in 4% formaldehyde solution at room temperature. Fixed cells were permeabilized with 1X BD Perm/Wash buffer (BD Biosciences) for 30 min. at room temperature and incubated with blocking solution containing 10% Normal Donkey Serum (The Jackson Laboratory) for 15 min. at room temperature. Immunostaining with primary antibodies was performed for 1 hr at room temperature in 200 μl blocking buffer. For the detection of total HEXIM1 and CDK9 cells were separately incubated with 9 μg anti-HEXIM1 (custom synthesized by Covance Research Products) and 2 μg anti-CDK9 (Santa Cruz Biotechnology), respectively. Afterwards the cells were washed three times with 1X PBS and then incubated with a 1:100 dilution of AF488-conjugated anti-Rabbit IgG secondary antibody in 200 μl blocking buffer (Jackson ImmunoResearch) for 1 h. For the detection of FLAG-HEXIM1, cells were incubated with 2 μg of anti-FLAG M2 antibody that had been directly conjugated to AF594 using the Zenon mouse labeling kit (Life Technologies). All cover slips were counterstained with 1 μg/ml DAPI for 3 min. at room temperature, mounted onto glass slides using ProLong Antifade (Life Technologies), and viewed using a Deltavision epifluorescent microscope (Applied Precision). Images were captured in z series, deconvolved, and processed using the Softworx analysis program (Applied Precision).
3 Results
3.1 Tandem mass spectrometry of HEXIM1 affinity isolated from Jurkat 2D10 T cells
HEXIM1 is required to recruit P-TEFb into the 7SK RNP complex, which then provides an exchangeable pool of P-TEFb used to sustain efficient gene expression [14]. Several groups, including ours, have demonstrated that the exchange of P-TEFb from the 7SK RNP complex is regulated by cellular signaling pathways that induce post-translational modifications (PTMs) on P-TEFb subunits [15-19]. In the current study, we focused on PTMs on HEXIM1 that are associated with the assembly and disassembly of the 7SK RNP complex.
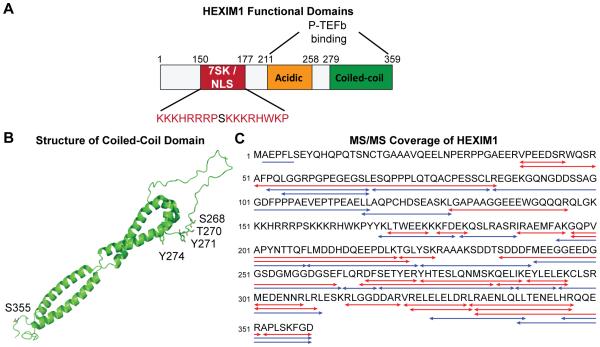
Whole cell extracts were prepared from Jurkat 2D10 T cells stably expressing N-terminally FLAG-tagged HEXIM1 either before or after cellular activation with 50 ng/ml PMA for 1 hr. The HEXIM1-containing complexes were then affinity purified by anti-FLAG immunoprecipitation followed by elution with FLAG peptide and resolved by 1D SDS-PAGE. In order to maximize the portion of the protein sequence of HEXIM1 that would be detected in the LC-MS/MS analysis, gel bands containing HEXIM1 were subjected to in-gel digestion with either trypsin or pepsin A. As shown in Fig. 1, the combined MS/MS sequence coverage of HEXIM1 from unambiguously identified peptides cleaved with trypsin or pepsin A was 83%. The unidentified sequences were located mainly at the N-terminus and the basic 7SK snRNA binding region.
Figure 1.
Identification of PTMs on HEXIM1 by tandem mass spectrometry. (A) Cartoon based on the published NMR structure of the coiled-coil dimeric region of HEXIM1 [26] showing the locations of the phospho-sites S268, T270, Y271, Y274, and S355 relative to the coiled-coil dimer. (B) Combined MS/MS sequence coverage of trypsin (in red) and pepsin A (in blue) cleaved HEXIM1 affinity purified from Jurkat T cells.
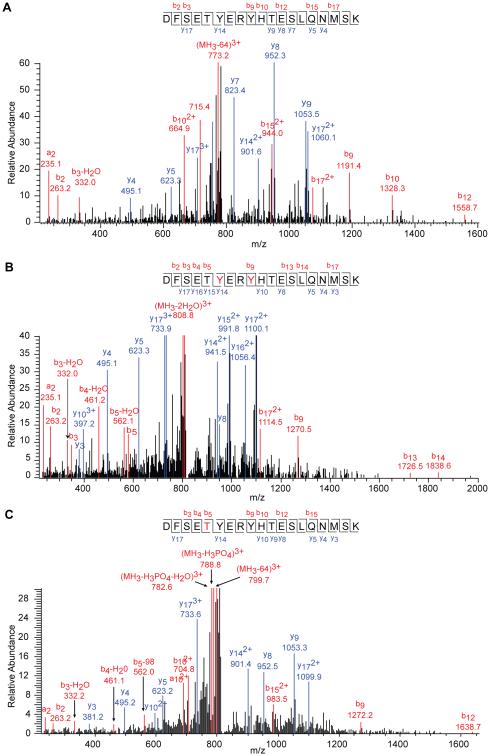
Data that was generated following MS/MS were searched against an IPI human database with programmed settings to identify phosphorylation as well Lys acetylation sites (Supplementary Table 1). Candidate PTMs were further verified by manual interpretation of the obtained MS/MS spectra (Fig. 2 and Supplementary Fig. 1). Fig. 2 presents spectra covering the HEXIM1 266-284 peptide, which is phosphorylated at Y271 or Y274 after PMA treatment. It is important to note that each of the identified peptides could also be captured in their unmodified states, reflecting the heterogeneity inherent in the dynamic 7SK RNP regulatory system.
Figure 2.
Manually annotated MS/MS fragmentation spectra. Unmodified (upper), phospho-Tyr HEXIM1 266-284 (middle), and phospho-Thr270 (lower) tryptic precursor peptides. The observed b and y series fragment ions are marked in red and blue, respectively.
3.2 Phosphomimetic mutation of Y271 and Y274 block 7SK snRNP assembly
A striking result of the MS/MS analysis was that a significant number of HEXIM1 phosphorylation sites were clustered at both the C-terminal acidic domain (S233, T236, and S237) and an unstructured peptide region immediately upstream of the coiled-coil homodimerization domain (S268, T270, Y271, and Y274). Because multiple closely spaced phosphorylation sites were present on the same peptides, and the population of molecules was intrinsically heterogeneous, we introduced unmodifiable residues and phosphomimetic mutants at each of the potential sites of phosphorylation. Specifically S233, T236, and S237 were each mutated to A or D, S268 and T270 were each converted to A or E, and the Y residues were each mutated to F or E. The mutants were evaluated by stable infection of Jurkat cell lines (Fig. 3) and HEK293T cells (Supplementary Fig. 2) using retroviral vectors expressing the FLAG HEXIM1 wild-type or the mutant constructs. Whole cell extracts from these cells were prepared as detailed in the Methods section and anti-FLAG immunoprecipitation followed by Western blotting was performed to assess the effects of these mutations on the ability of HEXIM1 to co-precipitate with P-TEFb subunits and the 7SK snRNP integral component LARP7 [20] (Fig. 3).
Figure 3.
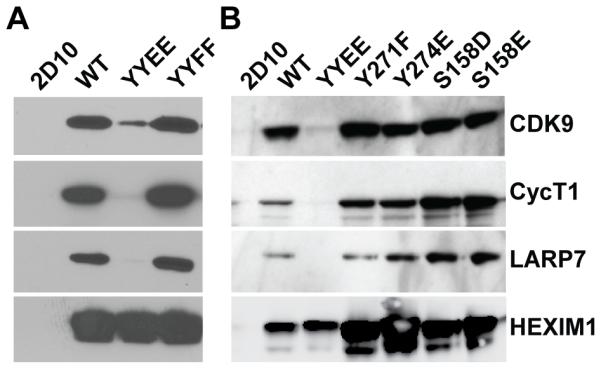
Y271E/Y274E disrupts sequestration of P-TEFb by 7SK snRNP. (A) Co-Immunoprecipitation of FLAG-HEXIM1, CDK9, Cyclin T1, and LARP7 from Jurkat T cells stably transduced with FLAG HEXIM1, FLAG Y271E/Y274E (YYEE), or FLAG Y271F/Y271F (YYFF). (B) Phosphomimetic mutations of Ser158 have no adverse effect on the assembly of 7SK snRNP.
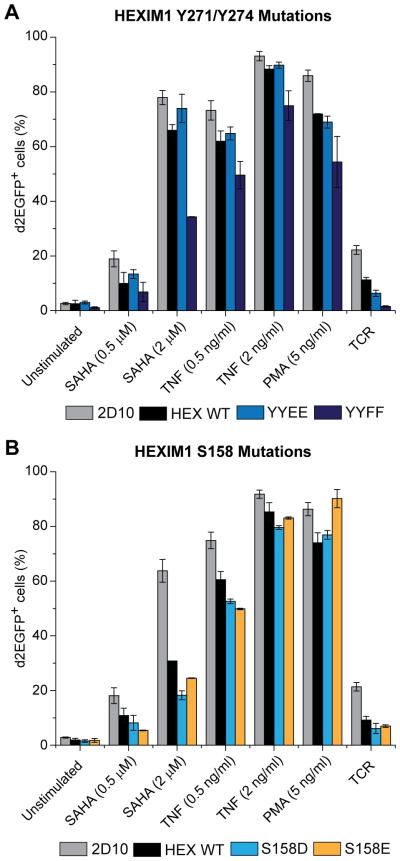
The major phenotypes were seen with mutants at Y271 and Y274. As shown in Fig. 3A the Y271E/Y274E mutation potently blocked assembly of the 7SK snRNP complex and only minimal levels of CDK9, CycT1 and LARP7 co-precipitated with HEXIM. By contrast, Y271F/Y274F mutation formed stable associations. The mutagenesis results therefore strongly suggest that phosphorylation of the unstructured peptide upstream of the coiled-coil (HEXpep258-279) could be involved in the dissociation of the 7SK snRNP.
Analysis of the complete set of mutations in HEK293T cell lines (Supplementary Fig. 2A and 2B), confirmed that Y271E blocked 7SK RNP complex assembly and a further reduction was seen when Y271E was combined with Y274E (which did not have a strong individual phenotype). By contrast, HEXIM1 association with P-TEFb subunits and LARP7 were maintained in Y271F and Y271F/Y274F mutant cells. The remaining mutations had no significant effects on 7SK RNP assembly. In a complementary approach we also showed that 7SK RNA is selectively lost from immunoprecipitates prepared with the Y271E/Y274E mutation in HEXIM1 (Supplementary Fig. 3)
HEXIM1 also possesses a consensus protein kinase C modification site, S158, which is situated at the center of its bipartite 7SK snRNA binding motif. Based on findings from in vitro RNA binding experiments and both in vitro and in vivo HEXIM1 phosphorylation assays, Fujinaga et al. [17] have postulated that phosphorylation of HEXIM1 at Ser158 by PKC-theta, leads to the dissociation of P-TEFb from 7SK snRNP. We were unable to verify the PKC-dependent phosphorylation of S158 by tandem mass spectrometry because peptides belonging to the 7SK snRNA binding motif could not be resolved (Fig. 1B). However, when we mutated S158 to A, D, and E (Fig. 3B and Supplementary Fig. 2C) we did not observe significant loss of HEXIM1 association with P-TEFb or LARP7 with the phosphomimetic mutations S158D or S158E.
In summary, the mutagenesis results strongly support the hypothesis that phosphorylation of Y271 and Y274 is an important signaling mechanism regulating the dissociation of P-TEFb and HEXIM1 from the 7SK snRNP complex in T cells.
3.3 Y271F/Y274F inhibits proviral gene expression
Since Y271E/Y274E HEXIM1 prevented the incorporation of P-TEFb into 7SK snRNP, we anticipated that the 7SK-associated pool of P-TEFb might be reduced in T cells expressing this mutant. We also expected that the 7SK-free pool of P-TEFb in T cells would be severely limited in cells expressing Y271F/Y274F HEXIM1 since the inability to phosphorylate these tyrosine sites might hinder the signal-dependent dissociation of P-TEFb from 7SK snRNP. The impact of these mutations on proviral gene expression in Jurkat 2D10 T cells was studied by flow cytometry before and after cellular activation by 18 h stimulation with TNF-α, the HDAC inhibitor SAHA, the phorbol ester PMA, or T-cell receptor signaling. Ectopic expression of HEXIM1 wild-type and Y271E/Y274E caused modest declines in the percentage of EGFP-positive cells in response to SAHA, TNF-α, and PMA relative to control 2D10 T cells (Fig. 4A). T-cell receptor stimulation of proviral expression was reduced 2-fold and 3.5-fold due to ectopic expression of HEXIM1 wild-type and Y271E/Y274E, respectively. Expression of the Y271F/Y274F mutant led to more severe inhibiton of proviral expression. There was a 14.35-fold decrease in EGFP-positive cells after T-cell receptor stimulation, a 2-fold decrease under basal conditions and 2.75-fold and 2.27-fold decreases following stimulation with SAHA at 0.5 μM and 2 μM, respectively. In cells treated with 0.5 ng/mL TNF-α and 5 ng/mL PMA the Y271F/Y274F mutant caused 1.48-fold, and 1.58-fold reductions in EGFP-positive cells, respectively. Taken together, these data indicate that Y271F/Y274F HEXIM1 can severely restrict the reactivation of latent proviral HIV well beyond the modest inhibition observed with the wild-type protein or the Y271E/Y274E phosphomimetic mutation. Consistent with these results, ectopic expression of HEXIM1 wild-type or the Y271E/Y274E mutant did not affect Tat protein expression from TNF-activated proviruses, while the Y271F/Y274F mutant caused a 12-fold decrease in Tat protein levels (Supplementary Figure 4)
Figure 4.
Effect of mutations in HEXIM1 on HIV transcription. (A) Jurkat 2D10 cells carrying stably expressed HEXIM1 were cultured in 20% FBS complete medium and activated as shown for 18 h and then analyzed by flow cytometry for proviral EGFP reporter gene expression due toHEXIM WT, YYEE, or YYFF. (B) Effect of mutating Ser158 on HIV reactivation. Cells were cultured in 5% FBS and treated as indicated. Error bars denote +/− standard error of the mean from at least 3 independent replicates.
During the course of establishing the Y271F/Y274F HEXIM1 stable Jurkat T-cell line, we also found unexpectedly that expression of Y271F/Y274F HEXIM1 resulted in a severe growth defect in 5% fetal bovine serum (FBS) (Supplementary Figure 5) which can be reversed by addition of 20% FBS.
3.4 Effect of Y271E/Y274E on the subcellular localization of HEXIM1 and P-TEFb
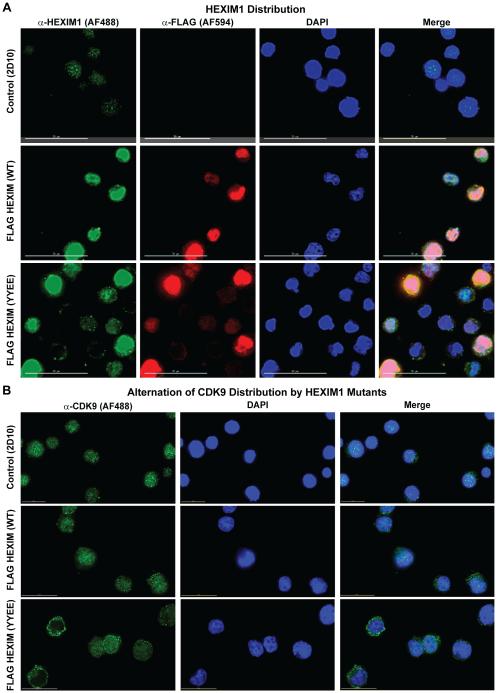
Immunofluorescence deconvolution microscopy was employed to examine the sub-cellular distribution patterns of HEXIM1 and the P-TEFb subunit CDK9 in Jurkat 2D10 T cells belonging to control, FLAG HEXIM1 wild-type, and FLAG Y271E/Y274E HEXIM1. HEXIM1 protein normally exhibits a punctate nucleoplasmic immunostaining pattern characteristic of nuclear speckles [21], and has been identified as a functional component of other nuclear non-7SK complexes belonging to the steroid hormone nuclear receptor superfamily [22-24]. Immunostaining of control Jurkat 2D10 T cells for HEXIM1 and CDK9 displayed the expected distribution patterns (Fig. 5A). Both FLAG HEXIM1 wild-type and Y271E/Y274E cells largely maintained a nuclear localization of HEXIM1, as detected by both anti-HEXIM1 and anti-FLAG immunostaining, but lost its punctate distribution pattern due to the overexpression of the protein in these cells. In the Y271E/Y274E cells, a perinuclear cytoplasmic staining with anti-CDK9 antibodies was observed (Fig. 5B and Supplementary Fig. 6) suggesting that the inability to incorporate P-TEFb into the nucleoplasmic 7SK snRNP complex in Y271E/Y274E cells releases the majority of the CDK9 from the nucleus.
Figure 5. Effect of Y271E/Y274E on the subcellular localization of HEXIM1 and P-TEFb.
(A) The Y271E/Y274E mutation does not adversely interfere with the nuclear localization pattern of HEXIM1 in Jurkat T cells. Uninfected Jurkat T cells or cells stably expressing FLAG HEXIM1 WT or the YYEE mutant from the integrated MSCV were fixed in 4% formaldehyde, permeabilized and then immunostained with fluorochrome-conjugated antibodies to HEXIM1 or the FLAG epitope. Images were obtained using a DeltaVision deconvolution microscope at 60X magnification. Scale bar represents a length of 30 μM. (B) Immunofluorescence microscopy to examine effect of the YYEE mutation on the subcellular localization of the CDK9 subunit of P-TEFB. Uninfected Jurkat T cells or cells stably expressing FLAG HEXIM1 WT or the YYEE mutant from the integrated MSCV were fixed in 4% formaldehyde, permeabilized and then immunofluorescently stained for CDK9. Images were obtained using a DeltaVision deconvolution microscope at 60X magnification. Scale bar represents a length of 15 μM.
4 Discussion
HEXIM1 is required to form a ribonucleoprotein complex with 7SK snRNA and stabilize the nuclear pool of P-TEFb [10]. The sequestration of P-TEFb by HEXIM1 and 7SK snRNA is regulated by cellular signaling pathways resulting in post-translational modifications (PTMs) on P-TEFb subunits and HEXIM1 following their dissociation from the 7SK RNP complex [15-18].
In the current study, we focused on identifying PTMs on HEXIM1 that regulate the sequestration of P-TEFb by 7SK snRNP in T cells. Combined MS/MS analysis of trypsin and pepsin A cleaved HEXIM1 that was affinity purified from Jurkat T cells led to >80% capture of the proteins amino acid sequence. Many of the identified phosphorylation sites were found to be clustered at the C-terminal acidic domain and an unstructured region immediately upstream of the coiled-coil dimerization domain.
We focused our studies on two tyrosines that become phosphorylated after PMA stimulation of cells. A phosphomimetic mutation introduced at two adjacent tyrosine residues (Y271E/Y274E) completely disrupted the sequestration of P-TEFb by 7SK snRNP. While this mutation, Y271E/Y274E, did not affect the nuclear localization of HEXIM1 it did cause a noticeable redistribution of the CDK9 subunit of P-TEFb into the cytoplasm. Stable expression of Y271E/Y274E HEXIM1 in Jurkat 2D10 cells slightly inhibited proviral gene expression, contrary to what had been previously reported [9, 25]. It seems likely that in this case endogenous HEXIM1 remained at sufficient levels to form a 7SK RNP complex that could support gene expression. The complementary phosphorylation-defective mutant, Y271F/Y274F, was still able to assemble normally with P-TEFb within the 7SK snRNP but strongly inhibited proviral gene expression, presumably because this mutation prevented dissociation of the 7SK RNP complex. Interestingly, the Y271F/Y274F HEXIM1 mutation also resulted in a severe defect in the growth of Jurkat T cells, implying that it restricts the level of transcriptionally active P-TEFb needed to support proviral HIV and cellular gene expression.
Previous reports have implicated Ser158 phosphorylation in the control of 7SK RNP complex formation [17]. However, we found that Ser158 phosphomimetic mutations neither disrupted the assembly of 7SK snRNP nor affected the basal or induced proviral gene expression levels. Therefore, although Ser158 of HEXIM1 is probably phosphorylated in vivo in response to PKC activation, our mutagenesis analyses suggest that its phosphorylation, unlike that of Tyr271 and Tyr274, may not be sufficient to prevent HEXIM1 assembly with 7SK snRNA.
In summary, the 7SK snRNP complex creates an exchangeable pool of P-TEFb that is delivered to genes in response to cellular signaling. Our results strongly suggest that a key regulatory event in this pathway is tyrosine phosphorylation of HEXIM1. It remains to be determined which kinases are responsible for these modifications and how HEXIM1 phosphorylation is coordinated with PTMs on the P-TEFb subunits, such as the phosphorylation of CDK9 on S175 [16] and the acetylation of CycT1 [19], that also regulate 7SK RNP formation and HIV gene expression.
Supplementary Material
Significance.
The release of the transcription elongation factor P-TEFb from the 7SK RNP complex and its binding to the HIV Tat transactivator protein enables the efficient transcription of HIV proviruses. In resting memory T-cells, which carry the bulk of the latent HIV viral pool, limiting the cellular levels of P-TEFb ensures that the provirus remains silenced unless the host cell is activated. Here we demonstrate that activation of T cells induces phosphorylation of HEXIM1, a repressor protein required for the formation of the 7SK RNP complex, at a cluster of phosphorylation sites at the C-terminal acidic domain and an unstructured region immediately upstream of the coiled-coil dimerization domain. Mutagenesis and biochemical studies demonstrated that phosphomimetic substitutions at two adjacent tyrosine residues (Tyr271/Tyr274) prevents the sequestration of P-TEFb by 7SK snRNP. By contrast, mutant Y271F/Y274F appears to stabilize the 7SK RNP and limit the level of transcriptionally active P-TEFb much below the threshold required to support transcription proviral HIV and cellular genes. Thus, signal dependent post-translational modifications that alter the assembly of the 7SK snRNP allow sub-threshold levels of Tat to efficiently extract P-TEFb and deliver it to the HIV LTR, thereby initiating a positive feedback loop that drives viral replication.
Acknowledgments
This work was supported by grants from the National Institutes of Health, R01-AI067093 and DP1-DA028869 to JK and the Martin Delaney CARE Collaboratory, U19-AI096113. UM was supported by 108266-51-RFRL from amfAR (The Foundation For AIDS Research). We also thank the CWRU/UH Center for AIDS Research (P30-AI036219) for provision of flow cytometry services and clinical samples.
Abbreviations
- 7SK snRNP
7SK small nuclear ribonucleoprotein
- HIV
Human immunodeficiency virus
- P-TEFb
Positive transcription elongation factor b
- HEXIM1
Hexamethylene-bis-acetamide inducible protein 1
- HIV LTR
HIV long terminal repeat
- TAR
Transactivation-responsive element
- HDAC inhibitor
Histone deacetylase inhibitor
- TNF-α
Tumor necrosis factor α
- CID
Collision-induced dissociation
References
- [1].Mbonye U, Karn J. Transcriptional control of HIV latency: Cellular signaling pathways, epigenetics, happenstance and the hope for a cure. Virology. 2014;454-455C:328–339. doi: 10.1016/j.virol.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Peterlin BM, Brogie JE, Price DH. 7SK snRNA: a noncoding RNA that plays a major role in regulating eukaryotic transcription. Wiley Interdiscip Rev RNA. 2012;3:92–103. doi: 10.1002/wrna.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bartkowiak B, Greenleaf AL. Phosphorylation of RNAPII: To P-TEFb or not to P-TEFb? Transcription. 2011;2:115–119. doi: 10.4161/trns.2.3.15004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Wei P, Garber ME, Fang SM, Fischer WH, Jones KA. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- [5].Tahirov TH, Babayeva ND, Varzavand K, Cooper JJ, et al. Crystal structure of HIV-1 Tat complexed with human P-TEFb. Nature. 2010;465:747–751. doi: 10.1038/nature09131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Jadlowsky JK, Wong JY, Graham AC, Dobrowolski C, et al. The negative elongation factor (NELF) is required for the maintenance of proviral latency but does not induce promoter proximal pausing of RNAP II on the HIV LTR. Mol Cell Biol. 2014;34:1911–1928. doi: 10.1128/MCB.01013-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Nguyen VT, Kiss T, Michels AA, Bensaude O. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature. 2001;414:322–325. doi: 10.1038/35104581. [DOI] [PubMed] [Google Scholar]
- [8].Yang Z, Zhu Q, Luo K, Zhou Q. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature. 2001;414:317–322. doi: 10.1038/35104575. [DOI] [PubMed] [Google Scholar]
- [9].Li Q, Price JP, Byers SA, Cheng D, et al. Analysis of the large inactive P-TEFb complex indicates that it contains one 7SK molecule, a dimer of HEXIM1 or HEXIM2, and two P-TEFb molecules containing Cdk9 phosphorylated at threonine 186. J. Biol. Chem. 2005;280:28819–28826. doi: 10.1074/jbc.M502712200. [DOI] [PubMed] [Google Scholar]
- [10].Yik JH, Chen R, Nishimura R, Jennings JL, et al. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol. Cell. 2003;12:971–982. doi: 10.1016/s1097-2765(03)00388-5. [DOI] [PubMed] [Google Scholar]
- [11].Michels AA, Fraldi A, Li Q, Adamson TE, et al. Binding of the 7SK snRNA turns the HEXIM1 protein into a P-TEFb (CDK9/cyclin T) inhibitor. EMBO J. 2004;23:2608–2619. doi: 10.1038/sj.emboj.7600275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Markert A, Grimm M, Martinez J, Wiesner J, et al. The La-related protein LARP7 is a component of the 7SK ribonucleoprotein and affects transcription of cellular and viral polymerase II genes. EMBO Rep. 2008;9:569–575. doi: 10.1038/embor.2008.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Xue Y, Yang Z, Chen R, Zhou Q. A capping-independent function of MePCE in stabilizing 7SK snRNA and facilitating the assembly of 7SK snRNP. Nucleic Acids Res. 2010;38:360–369. doi: 10.1093/nar/gkp977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zhou Q, Li T, Price DH. RNA polymerase II elongation control. Annu Rev Biochem. 2012;81:119–143. doi: 10.1146/annurev-biochem-052610-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kim YK, Mbonye U, Hokello J, Karn J. T-Cell Receptor Signaling Enhances Transcriptional Elongation from Latent HIV Proviruses by Activating P-TEFb through an ERK-Dependent Pathway. Journal of molecular biology. 2011;410:896–916. doi: 10.1016/j.jmb.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Mbonye UR, Gokulrangan G, Datt M, Dobrowolski C, et al. Phosphorylation of CDK9 at Ser175 enhances HIV transcription and is a marker of activated P-TEFb in CD4(+) T lymphocytes. PLoS Pathog. 2013;9:e1003338. doi: 10.1371/journal.ppat.1003338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Fujinaga K, Barboric M, Li Q, Luo Z, et al. PKC phosphorylates HEXIM1 and regulates P-TEFb activity. Nucleic Acids Res. 2012;40:9160–9170. doi: 10.1093/nar/gks682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Chen R, Liu M, Li H, Xue Y, et al. PP2B and PP1alpha cooperatively disrupt 7SK snRNP to release P-TEFb for transcription in response to Ca2+ signaling. Genes Dev. 2008;22:1356–1368. doi: 10.1101/gad.1636008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Cho S, Schroeder S, Kaehlcke K, Kwon HS, et al. Acetylation of cyclin T1 regulates the equilibrium between active and inactive P-TEFb in cells. EMBO J. 2009;28:1407–1417. doi: 10.1038/emboj.2009.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Krueger BJ, Jeronimo C, Roy BB, Bouchard A, et al. LARP7 is a stable component of the 7SK snRNP while P-TEFb, HEXIM1 and hnRNP A1 are reversibly associated. Nucleic Acids Res. 2008;36:2219–2229. doi: 10.1093/nar/gkn061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Dow EC, Liu H, Rice AP. T-loop phosphorylated Cdk9 localizes to nuclear speckle domains which may serve as sites of active P-TEFb function and exchange between the Brd4 and 7SK/HEXIM1 regulatory complexes. J. Cell. Physiol. 2010;224:84–93. doi: 10.1002/jcp.22096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wittmann BM, Fujinaga K, Deng H, Ogba N, Montano MM. The breast cell growth inhibitor, estrogen down regulated gene 1, modulates a novel functional interaction between estrogen receptor alpha and transcriptional elongation factor cyclin T1. Oncogene. 2005;24:5576–5588. doi: 10.1038/sj.onc.1208728. [DOI] [PubMed] [Google Scholar]
- [23].Yeh IJ, Song K, Wittmann BM, Bai X, et al. HEXIM1 plays a critical role in the inhibition of the androgen receptor by anti-androgens. Biochem J. 2014;462:315–327. doi: 10.1042/BJ20140174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Shimizu N, Ouchida R, Yoshikawa N, Hisada T, et al. HEXIM1 forms a transcriptionally abortive complex with glucocorticoid receptor without involving 7SK RNA and positive transcription elongation factor b. Proc. Natl. Acad. Sci. U S A. 2005;102:8555–8560. doi: 10.1073/pnas.0409863102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Fujinaga K, Luo Z, Peterlin BM. Genetic analysis of the structure and function of 7SK small nuclear ribonucleoprotein (snRNP) in cells. J Biol Chem. 2014;289:21181–21190. doi: 10.1074/jbc.M114.557751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Dames SA, Schonichen A, Schulte A, Barboric M, et al. Structure of the Cyclin T binding domain of Hexim1 and molecular basis for its recognition of P-TEFb. Proc. Natl. Acad. Sci. U S A. 2007;104:14312–14317. doi: 10.1073/pnas.0701848104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.