Abstract
The small molecule biosynthetic potential of most filamentous fungi remains largely unexplored and represents an attractive source for new compound discovery. Genome sequencing of Calcarisporium arbuscula, a mushroom endophytic fungus, revealed 68 core genes that are involved in natural products biosynthesis. This is in sharp contrast to predominant production of the ATPase inhibitors aurovertin B (1) and D (2). Inactivation of a histone H3 deacetylase HdaA led to pleiotropic activation and overexpression of more than 75% of the biosynthetic genes. Sampling of the overproduced compounds led to isolation of ten compounds of which four contain new structures, including the cyclic peptides arbumycin (3) and arbumelin (4); the diterpenoid arbuscullic acid A (11); and a meroterpenoid arbuscullic acid B (12). Such epigenetic modification therefore provides a rapid and global approach to mine the chemical diversity of endophytic fungi.
Keywords: Endophytic fungus, Genome mining, Epigenetic regulation, Gene expression, Natural products
Filamentous fungi are prolific producers of bioactive natural products,[1] exemplified by the antibiotic penicillin[2] and the anti-hypercholesteromia lovastatin.[3] Recent genome sequencing efforts of many fungal species have revealed significant biosynthetic potential, as represented by a large number of cryptic and diverse biosynthetic pathways.[4] Synthetic biological efforts to activate these silent pathways in well-studied fungal species, mostly in the Penicillium and Aspergillus genera, have led to the discovery of new natural products.[5] In particular, epigenetic approaches that lead to chromatin remodeling have resulted in activation of individual gene clusters.[6]
Endophytic fungi are increasingly recognized as significant underachievers in natural product biosynthesis.[7] For example, recent genome sequencing of entomophytic fungus Metarhizium anisopliae and endophytic fungus Beauveria bassiana revealed each encodes a large number of potential secondary metabolite gene clusters (85 and 45, respectively), significantly more than the number of compounds that has been reported for each strain.[8] Such high biosynthetic potential is a reflection of the complex natural ecological environment,[9] which is difficult to replicate in the laboratory, resulting in most gene clusters being silent in axenic cultures. Therefore, genetically modifying endophytic fungi to activate, ideally globally, the plethora natural product pathways can be particularly fruitful in accessing new chemical scaffolds.
Calcarisporium arbuscula is an endophytic fungus living in the fruiting bodies of mushrooms.[10] Fungi in the Calcarisporium genus have been noted for production of a few antimicrobials and mycotoxins.[11] C. arbuscula predominantly produces F1-ATPase inhibitors 1 and 2, which are polyketides containing an unusual 2,6-dioxabicyclo[3.2.1]octane core connected to an α-pyrone through a triene linker (Scheme 1).[12] Bioinformatics analysis of the draft genome sequence of C. arbuscula showed 68 gene clusters encoding potential natural product biosynthetic pathways, among which 41 contain polyketide synthases (PKSs) (including 4 PKS-nonribosomal peptide synthetase (NRPS) hybrids), 18 contain NRPSs and 9 contain terpene synthases (TS) (Table S1–S3). This large collection of biosynthetic gene clusters is in sharp contrast to the two predominant metabolites 1 and 2 (Figure 1).[12a] This organism therefore represents a prime resource for genome mining using globally effective approaches. However, initial attempts to culture the fungus on different media, such as MEPA, CYA, YMEG or YG, did not lead to production of new compounds. The nongenetic strategy of addition DNA methyltransferase inhibitor 5-azacytidine or histone deacetylase (HDAC) inhibitor suberoyl bis-hydroxamic acid[13] also did not change the metabolomic profile.
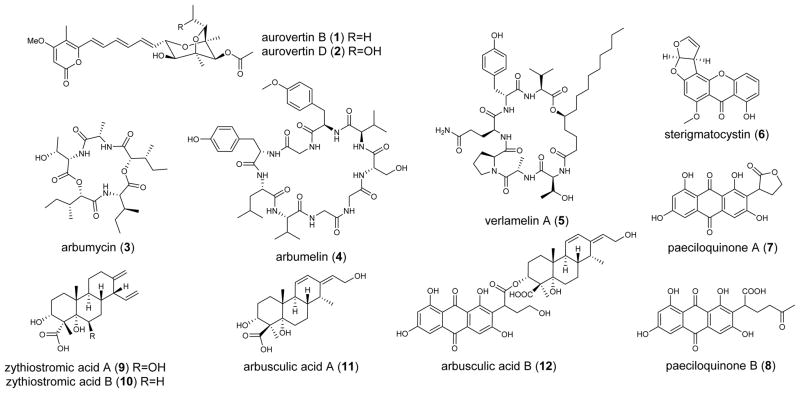
Scheme 1.
Compounds purified from C. arbuscula. Compounds 1 and 2 are from wild type, and 3-12 are from the ΔhdaA mutant.
Figure 1.
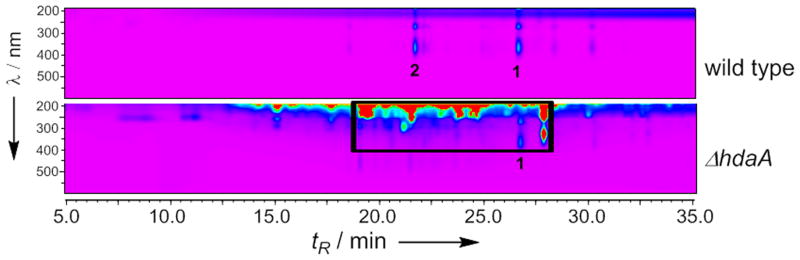
HPLC contour plot of extracts from WT and the ΔhdaA C. arbuscula strains. The boxed region contains compounds 3-12 isolated in this work.
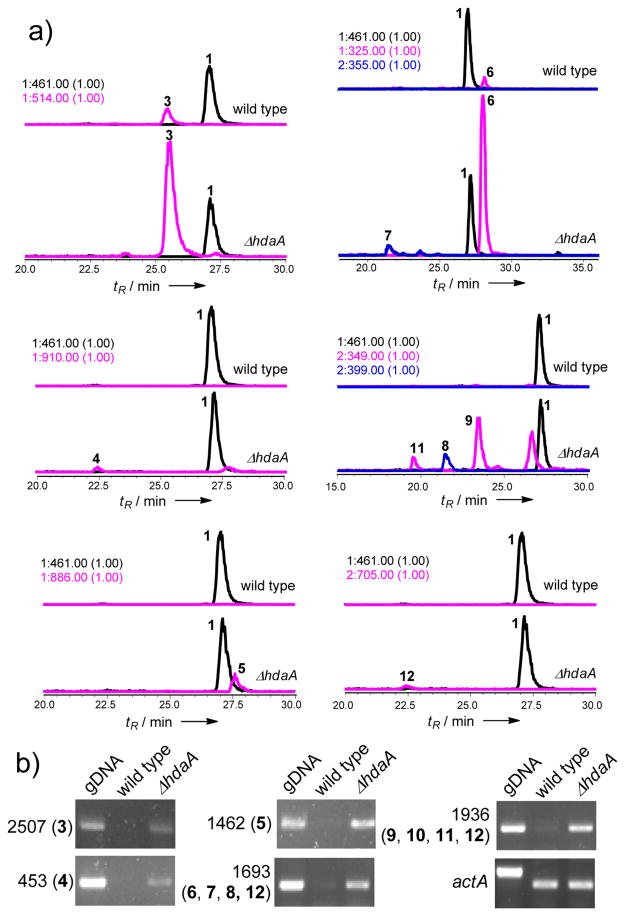
In filamentous fungi, many silenced gene clusters are located within the heterochromatic regions and subsequently transcriptionally repressed.[14] HDACs remove acetyl groups from the amino-tails of histones and maintain the chromatin in an inaccessible state for the transcriptional machinery.[15] Keller and coworkers showed that fungal HDACs negatively regulate production of sterigmatocystin and penicillin in A. nidulans; and attenuate transcription of NRPS gene clusters in A. fumigatus.[16] To investigate the roles of HDACs in C. arbuscula, we deleted hdaA (Figure S3), which encodes the histone H3 lysine 14 (K14) deacetylase. Removal of hdaA resulted in slower growth, shorter mycelia and defective sporulation in C. arbuscula (Figure S4). Metabolite extraction and LC/MS analysis revealed production of significantly more compounds compared to the wild type (Figure 1). Comprehensive RT-PCR analysis of all 68 core biosynthetic genes showed that while weak to no expression of most core genes were observed in wild type, deletion of hdaA led to increased expression of 75% (31/41) of the PKS genes, 78% (14/18) of the NRPS genes and 78% (7/9) of the TS genes (Figure S5). Hence, C. arbuscula HdaA globally suppresses biosynthetic genes under axenic growth conditions and its deletion leads to pleiotropic activation of secondary metabolism.
To characterize the newly produced compounds in the ΔhdaA strain, the organic extracts were partitioned with n-hexane and methanol. Then the methanol phase was fractionated by reversed phase chromatography. Four fractions particularly rich in metabolites were selected for further purification, which led to the isolation of 10 compounds, including three peptides (3, 4, 5), three polyketides (6, 7, 8), three diterpenes (9, 10, 11) and one polyketide-diterpene hybrid (12). Among them, 3, 4, 11 and 12 are new (Scheme 1).
Two new cyclic peptides arbumycin (3) (10 mg/L) and arbumelin (4) (0.17 mg/L), and a lipopeptide verlamelin A (5) (0.25 mg/L) were isolated from the ΔhdaA mutant. When selective ion monitoring was performed, a ten-fold decrease in the level of 3 can be detected in the wild type, and no trace of 4 or 5 could be found (Figure 3a). The structure of 3 was elucidated as a cyclic penta-depsipeptide based on NMR spectra (Table S11, Figure S13–S18) and X-ray crystallography (Figure S6). The asymmetric peptide is derived from three amino acids (L-Ala, L-Thr, L-Ile) and two identical hydroxyl acids (2S,3R)-2-hydroxy-3-methylpentanoic acid 13 (Figure 2c). Most fungal depsipeptides characterized to date are symmetrical and are catalyzed by the iterative action of a two module NRPS depsi-peptide synthase.[17] Therefore, the asymmetrical nature of 3 presents an interesting case of structure variation in this family. The (2S,3R) stereochemistry in 13 was an unexpected finding from the crystal structure, and indicates possible epimerization of the 3R methyl group from (2S,3S)-isoleucine.
Figure 3.
Over-production of compounds in the ΔhdaA mutant. a) Higher yields of compounds 3-9, 11 and 12 in the ΔhdaA mutant based on EIC compared to 1. b) RT-PCR analysis of gene expression of the selected core genes. The housekeeping actA served as the internal control. The contig numbers (with corresponding compounds in the parentheses) are also shown.
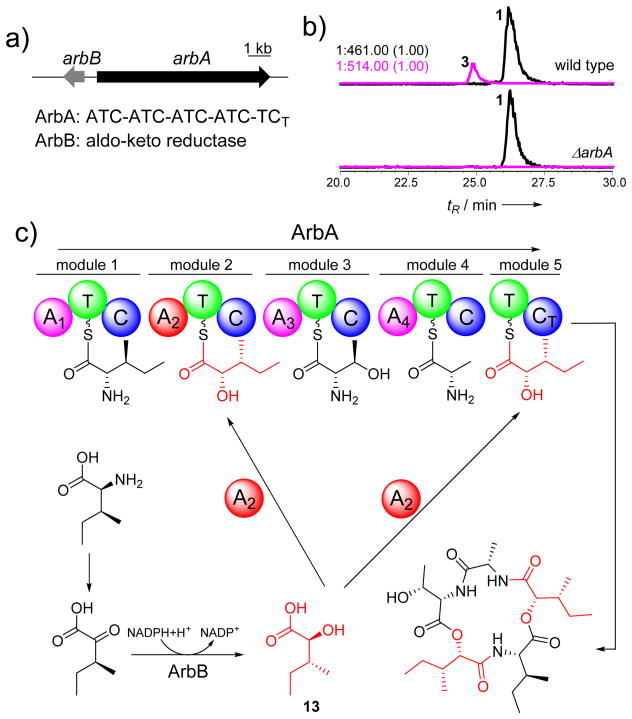
Figure 2.
Characterization of arbumycin (3) and its gene cluster in C. arbuscula. a) The putative gene cluster for the biosynthesis of 3; b) Genetic verification of the gene cluster for the biosynthesis of 3; c) A proposed pathway for the biosynthesis of 3.
To understand the biosynthesis of 3, we identified a potential cluster on scaffold 82 that consists of a five module NRPS (ArbA) and an aldo-ketoreductase (ArbB) (Figure 2a). The transcription level of arbA is increased in the ΔhdaA strain (Figure 3b). While ArbA contains a C-terminal condensation domain (CT) consistent with formation of a cyclic product,[18] the last module (module 5) did not contain an adenylation (A) domain. This indicates module 5 may use the same substrate (13 is used twice in 3) as a previous module and an A domain may be shared between the two modules. Sequence analysis of the 10-amino acid specificity codes of A domains revealed that A2 should be responsible for incorporation of 13, as the highly conserved Asp235 that anchors the amino groups of amino acids is replaced by Ala235; while a threonine (Thr330) conserved in hydroxyacid-activating A domains is present (Table S4).[17] Indeed, inactivation of arbA in wild type C. arbuscula (Figure S7) completely abolished the production of 3 (Figure 2b). The proposed biosynthesis of 3 from ArbA and ArbB is shown in Figure 2c. We suggest that ArbB may be responsible for the ketoreduction and epimerization of 2-keto-3S-pentanoic acid to yield 13. This is highly reminiscent of the epimerizing ketoreductase (KR) domains found in bacterial type I PKSs, in which the KR domains are able to control the stereochemistry at vicinal α-methyl and β-hydroxyl substituted carbons starting from α-methyl-β-ketone substrates.[19]
Arbumelin 4 is a new cyclic nonapeptide containing three Gly and two D-amino acids as characterized by NMR (Table S12–S13, Figure S19–S27). The stereochemical configurations of the constituent amino acids were determined through hydrolysis, derivatization and HPLC analysis (Supporting Information). One nine-module NRPS on contig 453 is proposed to be the candidate for the biosynthesis of 4. Sequence analysis showed that the 10-amino acid specificity code of A domains in module 3, 7 and 8 are nearly identical and consistent with three Gly positions in 4 (Table S5). The E domains in modules 4 and 5 should be responsible for the epimerization of the consecutive DO-methyl-Tyr and D-Val. Taken together, the linear sequence of 4 is likely to be L-Leu-L-Tyr-Gly-D-O-methyl-Tyr-D-Val-L-Ser-Gly-Gly-L-Val, which is macrocyclized by the terminal CT domain in the NRPS (Scheme S1).[18] Immediately adjacent to the NRPS gene is a gene encoding a methyltransferase (Figure S8). It may be responsible for formation of the O-methyl-Tyr residue prior to activation by module 4. 5 was solved by extensive NMR spectroscopy (Figure S27–S31) and is the same as a recently reported cyclic hexa-lipopeptide verlamelin, an antifungal compound isolated from Lecanicillium sp. HF627.[20] The corresponding gene cluster is located on contig 1462 with the same set of genes (such as NRPS, fatty acyl ligase, etc) as reported for verlamelin (Figure S8).[20] The NRPS-encoding genes on contig 453 and 1462 were verified to be overexpressed in the ΔhdaA mutant (Figure 3b).
Deletion of hdaA led to a twenty-fold increase in the level of the mycotoxin sterigmatocystin (6) (Figure 3a). The activated gene cluster on contig 1693 is readily identified based on >70% sequence identity to that from Aspergillus ochraceoroseus (Figure S1).[21] Overexpression of this pathway (Figure 3b) also led to the isolation of the protein tyrosine kinase inhibitors paeciloquinone A (7) and B (8) (Figure S33–S34), which are shunt products of the biosynthetic pathway of 6 (Scheme S2).[22] Bioinformatic analysis of remaining PKS gene clusters also revealed the potential to produce other mycotoxins such as citrinin (contig921) (Figure S2).[23]
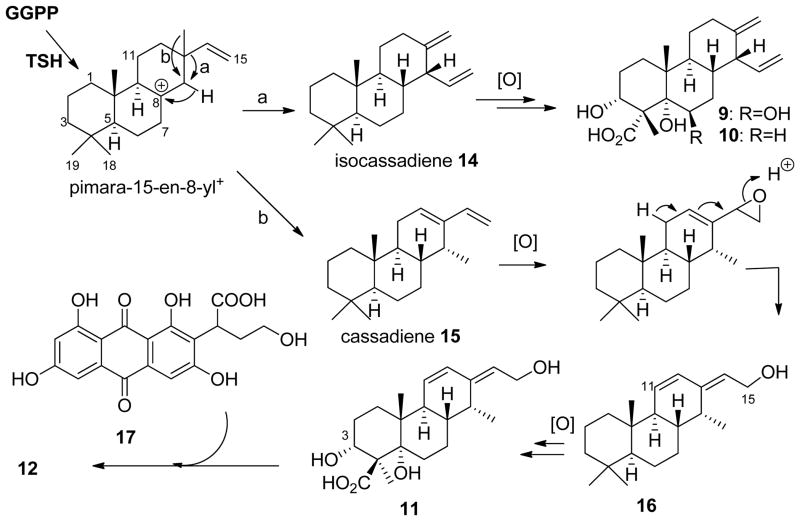
The ΔhdaA strain produced three labdane-related diterpenoid compounds (9-11) as characterized by NMR (Table S14, Figure S35–S44). Upon structure elucidation, 9 (0.67 mg/L) and 10 (0.23 mg/L) were confirmed to be the isocassadienes zythiostromic acid A and B, respectively.[24] Both compounds are heavily oxidized derivatives of isocassadiene 14.[25] The new diterpene arbusculic acid A (11) is structurally related to 9 and 10, and most likely derives from cassadiene 15 (Scheme 2). 11 contains a dienol functional group that has not been observed in fungal diterpenoids. Because of their structural similarities, we propose that compounds 9-11 are synthesized from the same diterpene biosynthetic gene cluster. BLAST search against pimaradiene synthase from A. nidulans (AN1594)[26] and ent-kaurene synthase from the fungus Phaeosphaeria sp. L487[27] revealed a diterpene synthase with the highest identity on contig 1936 (Figure S9a). Based on structural modeling against the taxadiene synthase from Taxus brevifolia,[28] this C. arbuscula diterpene cyclase is organized in three α-helix domains, including two class I and one class II domains (Figure S10b, c).[28] This gene was highly transcribed in the mutant strain compared to the wild type (Figure 3b). Also found in the gene cluster are two adjacent P450 with sequence similarity to ent-kaurene oxidases (Figure S10).[27] The proposed mechanism of formation of 11 is shown in Scheme 2, in which 15 is first epoxidized at the terminal olefin followed by acid-catalyzed ring-opening hydrolysis with transposition of the diene to yield 16. Both 15 and 16 can be subjected to multiple hydroxylations at C3 and C5, as well as six-electron oxidation at C19 to yield 9 and 11, respectively. Intriguingly, we also isolated a meroterpenoid compound arbuscullic acid B (12) (0.2 mg/L) from the ΔhdaA mutant (Table S15, Figure S45–S49). 12 is derived from the esterification of 11 to the carboxylic acid moiety of paeciloquinone D (17),[22a] which is the free acid form of the lactone 7, through the C3 hydroxyl group. The regioselectivity of the ester linkage was confirmed by extensive 2D NMR spectroscopy (Table S15), while no alternatively esterification regioselectivity between 11 and 17 was detected. This suggests the coupling of 11 and 17 may be enzyme catalyzed instead of from a nonenzymatic reaction.
Scheme 2.
Proposed pathways for the biosynthesis of 9-12.
In summary, deletion of a single HDAC gene in the endophytic C. arbuscula led to the global alteration of secondary metabolism and pleiotropic production of new natural products of different chemotypes and biosynthetic origins. Continued chemical isolation from this strain, along with additional regulatory engineering efforts, can lead to the enhanced realization of the biosynthetic potential of this and other endophytic fungi.
Supplementary Material
Acknowledgments
This work was supported by the National Institute of Health (1DP1GM106413). Dr. Y. C. Hu was supported by PUMC Youth Fund (33320140175) and State Key Laboratory Fund for Excellent Young Scientists (GTZB201401). Dr. X.M. Mao was funded by the fellowship of New Star Project from Zhejiang University. We thank Prof. David E. Cane for his helpful discussions.
Footnotes
Supporting information for this article is given via a link at the end of the document.
Contributor Information
Dr. Xu-Ming Mao, Department of Chemical and Biomolecular Engineering, Department of Chemistry and Biochemistry, University of California, University of California, Los Angeles, CA 90095 (USA). College of Life Sciences, Zhejiang University, Hangzhou 310058 (China).
Dr. Wei Xu, Department of Chemical and Biomolecular Engineering, Department of Chemistry and Biochemistry, University of California, University of California, Los Angeles, CA 90095 (USA)
Dr. Dehai Li, Department of Chemical and Biomolecular Engineering, Department of Chemistry and Biochemistry, University of California, University of California, Los Angeles, CA 90095 (USA). Key Laboratory of Marine Drugs, Chinese Ministry of Education, School of Medicine and Pharmacy, Ocean University of China, Qingdao, 266003 (China)
Dr. Wen-Bing Yin, Department of Chemical and Biomolecular Engineering, Department of Chemistry and Biochemistry, University of California, University of California, Los Angeles, CA 90095 (USA)
Dr. Yit-Heng Chooi, Department of Chemical and Biomolecular Engineering, Department of Chemistry and Biochemistry, University of California, University of California, Los Angeles, CA 90095 (USA)
Dr. Yong-Quan Li, College of Life Sciences, Zhejiang University, Hangzhou 310058 (China)
Dr. Yi Tang, Email: yitang@ucla.edu, Department of Chemical and Biomolecular Engineering, Department of Chemistry and Biochemistry, University of California, University of California, Los Angeles, CA 90095 (USA)
Dr. Youcai Hu, Email: huyoucai@imm.ac.cn, Department of Chemical and Biomolecular Engineering, Department of Chemistry and Biochemistry, University of California, University of California, Los Angeles, CA 90095 (USA). State Key Laboratory of Bioactive Substance and Function of Natural Medicines, Institute of Materia Medica, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing 100050 (China).
References
- 1.Schueffler A, Anke T. Nat Prod Rep. 2014;31:1425–1448. doi: 10.1039/c4np00060a. [DOI] [PubMed] [Google Scholar]
- 2.Ozcengiz G, Demain AL. Biotech Adv. 2013;31:287–311. doi: 10.1016/j.biotechadv.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 3.Mulder KC, Mulinari F, Franco OL, Soares MS, Magalhaes BS, Parachin NS. Biotech Adv. 2015 doi: 10.1016/j.biotechadv.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 4.Chiang YM, Lee KH, Sanchez JF, Keller NP, Wang CC. Nat Prod Commun. 2009;4:1505–1510. [PMC free article] [PubMed] [Google Scholar]
- 5.a) Brakhage AA, Schroeckh V. Fungal Genet Biol. 2011;48:15–22. doi: 10.1016/j.fgb.2010.04.004. [DOI] [PubMed] [Google Scholar]; b) Zabala AO, Xu W, Chooi YH, Tang Y. Chem Biol. 2012;19:1049–1059. doi: 10.1016/j.chembiol.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cichewicz RH. Nat Prod Rep. 2010;27:11–22. doi: 10.1039/b920860g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chandra S. Appl Microbiol Biotechnol. 2012;95:47–59. doi: 10.1007/s00253-012-4128-7. [DOI] [PubMed] [Google Scholar]
- 8.Gibson DM, Donzelli BG, Krasnoff SB, Keyhani NO. Nat Prod Rep. 2014;31:1287–1305. doi: 10.1039/c4np00054d. [DOI] [PubMed] [Google Scholar]
- 9.Kusari S, Hertweck C, Spiteller M. Chem Biol. 2012;19:792–798. doi: 10.1016/j.chembiol.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 10.Watson P. Trans Br Mycol Soc. 1955;38:409–414. [Google Scholar]
- 11.a) Silber J, Ohlendorf B, Labes A, Erhard A, Imhoff JF. Mar Drugs. 2013;11:3309–3323. doi: 10.3390/md11093309. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Silber J, Ohlendorf B, Labes A, Nather C, Imhoff JF. J Nat Prod. 2013;76:1461–1467. doi: 10.1021/np400262t. [DOI] [PubMed] [Google Scholar]; c) Chrisp P, Dewick PM, Boyle FT. Z Naturforsch C. 1990;45:179–186. doi: 10.1515/znc-1990-3-407. [DOI] [PubMed] [Google Scholar]; d) Yu NJ, Guo SX, Lu HY. J Asian Nat Prod Res. 2002;4:179–183. doi: 10.1080/10286020290011387. [DOI] [PubMed] [Google Scholar]
- 12.a) Osselton MD, Baum H, Beechey RB. Biochem Soc Trans. 1974;2:200–202. [Google Scholar]; b) van Raaij MJ, Abrahams JP, Leslie AG, Walker JE. Proc Natl Acad Sci U S A. 1996;93:6913–6917. doi: 10.1073/pnas.93.14.6913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yakasai AA, Davison J, Wasil Z, Halo LM, Butts CP, Lazarus CM, Bailey AM, Simpson TJ, Cox RJ. J Am Chem Soc. 2011;133:10990–10998. doi: 10.1021/ja204200x. [DOI] [PubMed] [Google Scholar]
- 14.a) Palmer JM, Keller NP. Curr Opin Microbiol. 2010;13:431–436. doi: 10.1016/j.mib.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Gacek A, Strauss J. Appl Microbiol Biotechnol. 2012;95:1389–1404. doi: 10.1007/s00253-012-4208-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bulger M. J Biol Chem. 2005;280:21689–21692. doi: 10.1074/jbc.R500004200. [DOI] [PubMed] [Google Scholar]
- 16.a) Shwab EK, Bok JW, Tribus M, Galehr J, Graessle S, Keller NP. Eukaryot Cell. 2007;6:1656–1664. doi: 10.1128/EC.00186-07. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lee I, Oh JH, Shwab EK, Dagenais TR, Andes D, Keller NP. Fungal Genet Biol. 2009;46:782–790. doi: 10.1016/j.fgb.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sussmuth R, Muller J, von Dohren H, Molnar I. Nat Prod Rep. 2011;28:99–124. doi: 10.1039/c001463j. [DOI] [PubMed] [Google Scholar]
- 18.Gao X, Haynes SW, Ames BD, Wang P, Vien LP, Walsh CT, Tang Y. Nat Chem Biol. 2012;8:823–830. doi: 10.1038/nchembio.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.a) Valenzano CR, Lawson RJ, Chen AY, Khosla C, Cane DE. J Am Chem Soc. 2009;131:18501–18511. doi: 10.1021/ja908296m. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) You YO, Khosla C, Cane DE. J Am Chem Soc. 2013;135:7406–7409. doi: 10.1021/ja4014776. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Garg A, Khosla C, Cane DE. J Am Chem Soc. 2013;135:16324–16327. doi: 10.1021/ja408944s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishidoh K, Kinoshita H, Nihira T. Appl Microbiol Biotechnol. 2014;98:7501–7510. doi: 10.1007/s00253-014-5803-7. [DOI] [PubMed] [Google Scholar]
- 21.Cary JW, Ehrlich KC, Beltz SB, Harris-Coward P, Klich MA. Mycologia. 2009;101:352–362. doi: 10.3852/08-173. [DOI] [PubMed] [Google Scholar]
- 22.a) Petersen F, Fredenhagen A, Mett H, Lydon NB, Delmendo R, Jenny HB, Peter HH. J Antibiot (Tokyo) 1995;48:191–198. doi: 10.7164/antibiotics.48.191. [DOI] [PubMed] [Google Scholar]; b) Minto RE, Townsend CA. Chem Rev. 1997;97:2537–2556. doi: 10.1021/cr960032y. [DOI] [PubMed] [Google Scholar]; c) Townsend CA. Nat Prod Rep. 2014;31:1260–1265. doi: 10.1039/c4np00092g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li YP, Xu Y, Huang ZB. Biotechnol Lett. 2012;34:131–136. doi: 10.1007/s10529-011-0745-y. [DOI] [PubMed] [Google Scholar]
- 24.Ayer WA, Khan AQ. Phytochemistry. 1996;42:1647–1652. [Google Scholar]
- 25.Peters RJ. Nat Prod Rep. 2010;27:1521–1530. doi: 10.1039/c0np00019a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bromann K, Toivari M, Viljanen K, Vuoristo A, Ruohonen L, Nakari-Setala T. PloS One. 2012;7:e35450. doi: 10.1371/journal.pone.0035450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawaide H, Imai R, Sassa T, Kamiya Y. J Biol Chem. 1997;272:21706–21712. doi: 10.1074/jbc.272.35.21706. [DOI] [PubMed] [Google Scholar]
- 28.a) Koksal M, Jin Y, Coates RM, Croteau R, Christianson DW. Nature. 2011;469:116–120. doi: 10.1038/nature09628. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Koksal M, Hu H, Coates RM, Peters RJ, Christianson DW. Nat Chem Biol. 2011;7:431–433. doi: 10.1038/nchembio.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.