Abstract
Background
The Na+/Ca2+ Exchanger (NCX) protein family is a member of the Cation/Ca2+ exchanger superfamily and its members play important roles in cellular Ca2+ homeostasis. While the functions of NCX family of proteins is well understood in humans, not much is known about the total complement of Na+/Ca2+ exchangers in plants and their role in various physiological and developmental processes. In the present study, we have identified all the NCX proteins encoded in the genomes of rice and Arabidopsis and studied their phylogeny, domain architecture and expression profiles across different tissues, at various developmental stages and under stress conditions.
Results
Through whole genome investigation, we identified twenty-two NCX proteins encoded by fifteen genes in rice and sixteen NCX proteins encoded by thirteen genes in Arabidopsis. Based on phylogenetic reconstruction, these could be classified into five clades, members of most of which were found to possess distinct domain architecture. Expression profiling of the identified NCX genes using publicly available MPSS and microarray data showed differential expression patterns under abiotic stresses, and at various development stages. In rice, OsNCX1, OsNCX8, OsNCX9 and OsNCX15 were found to be highly expressed in all the plant parts and various developmental stages. qRT-PCR based expression analysis revealed that OsNCX3, OsNCX10 and OsNCX15 were highly induced by salt and dehydration stress. Besides, expression profiling showed differential regulation of rice NCX genes in response to calcium and EGTA. Interestingly, expression of none of the NCX genes was found to be co-regulated by NaCl and calcium.
Conclusions
Together, our results present insights into the potential role of NCX family of proteins in abiotic stresses and development. Findings of the present investigation should serve as a starting point for future studies aiming functional characterization of plant NCX family proteins.
Electronic supplementary material
The online version of this article (doi:10.1186/s12284-015-0054-5) contains supplementary material, which is available to authorized users.
Keywords: Sodium calcium exchanger, Abiotic stress, Development, MPSS, Microarray, qRT-PCR
Background
Systemic and cellular homeostasis is very important for proper functioning of living organisms, including plants. The alkali (Na+, K+) and alkaline (Ca2+, Mg2+) earth metals play very critical roles in living system, but their optimum cellular concentration must be maintained for proper action (Jon et al. 2008). Calcium is one of the most important and ubiquitous signaling molecules and perturbed Ca2+ homeostasis may induce cellular death (McLean et al. 1965). The role of Ca2+ as a secondary messenger in signal transduction in response to external and internal stimuli like plant growth nutrients, light, fungal elicitors and various abiotic stresses is well known (Hepler and Wayne 1985; Evans et al. 2001). Intracellular Ca2+ concentration in resting cells is maintained at 10–100 nM and it may go up to 10 μM during external or internal perturbations (Peretz et al. 1994; Bootman and Berridge 1995; Hardie 1996). As Ca2+ controls a number of physiological processes both at molecular and cellular levels in plants (Hepler and Wayne 1985), it is very crucial to regulate the Ca2+ level inside the cell.
There are two classes of membrane proteins involved in cellular Ca2+ homeostasis, the ATP-driven calcium pumps or plasma membrane Ca2+ ATPase (PMCA), and sodium/calcium exchangers (Na+/Ca2+ exchanger or NCX) (Haug-Collet et al. 1999). These two pumps complement each other in their functions. Though NCX has a low affinity towards Ca2+, it can transport Ca2+ at very high speed within short duration i.e. up to 5000 calcium ions per second (Carafoli et al. 2001). On the other hand, PMCA has very high affinity towards Ca2+, which normally helps the cell in maintaining the very low concentration of cytosolic Ca2+ in resting cell (Siegel et al. 1999).
NCXs are antiporter membrane proteins and maintain optimum cellular Ca2+ levels. The energy stored in the electrochemical gradient of such proteins is used to exchange Ca2+ with its counterpart Na+, which moves down its electrochemical gradient across the plasma membrane. NCX is considered as one of the most important cellular component for removing Ca2+ (Dipolo and Beauge 2006). NCX functions on the principle of removal of a single calcium ion and import of three sodium ions in exchange (Yu and Choi 1997). NCX also show reversibility in their mode of action i.e. when cellular Na+ concentration increases beyond critical level these proteins start importing calcium ions inside the cell (Bindokas and Miller 1995; Yu and Choi 1997; Wolf et al. 2001). Therefore, NCX can work in both directions depending upon the gradient generated by Ca2+ and Na+ concentrations inside the cell. NCX proteins have been found in many different cell types of animal and plant species. Wide ranges of animal and plant species have highly conserved structural properties of NCX. Amongst animals, invertebrates possess only single NCX gene, whereas vertebrates have multiple NCX genes due to gene duplication (On et al. 2008).
The availability of high quality whole genome sequence of Arabidopsis and rice and their expression data in the form of Microarray and MPSS (Massively Parallel Signature Sequencing) has enabled the study of different gene families which play important roles in plants. Different gene families in Arabidopsis and rice genomes, such as Cation/H+ exchanger family (Kamiya et al. 2005), histidine kinase family (Pareek et al. 2006), SAUR gene family (Jain et al. 2006), protein phosphatase 2C family (Xue et al. 2008), basic leucine zipper transcription factor family in rice (Nijhawan et al. 2008), CBS domain containing gene family (Kushwaha et al. 2009), nucleocytoplasmic transport receptors family (Huang et al. 2010) and glyoxalase family (Mustafiz et al. 2011) etc. have been analyzed. Recently genome wide analysis of gene families has been extended to other plant species like NAC transcription factor family in potato (Singh et al. 2013), Family‑1 UDP glycosyltransferases in maize (Li et al. 2014), and AP2/ERF family genes from Lotus corniculatus (Sun et al. 2014).
Some of the NCX genes are well studied in animal and human systems. Phylogenetic analysis of NCX gene family in fish species has identified 13 NCX genes and revealed that two serial NCX gene duplication events have occurred around the time vertebrates and invertebrates diverged (Marshall et al. 2005). In case of plants, the only characterized NCX protein is AtNCL (an NCX-like protein) from Arabidopsis which was found to play a role in salt stress and maintaining Ca2+ homeostasis (Wang et al. 2012). However, other members of the NCX gene family in plants have not been comprehensively identified and characterized. Considering the importance of this protein family in maintaining Ca2+-homeostasis and the pivotal roles that Ca2+ plays as a second messenger in plants, it is imperative to comprehensively dissect the complement of sodium-calcium exchangers in plants paving the way for functional characterization studies.
Therefore, to gain insights into the structural and functional attributes of NCX family of proteins in plants, in the present study, we have carried out whole genome analyses to identify NCX domain containing proteins in Arabidopsis and rice genomes using TAIR (The Arabidopsis Information Resource) version 10 and RGAP (Rice Genome Annotation Project) release 7, respectively. We have also suggested nomenclature, and provided chromosomal distribution and phylogenetic analysis of NCX genes in Arabidopsis and rice. For a comprehensive phylogenetic analysis, we have additionally identified NCX proteins from two monocotyledonous plant species, Brachypodium (Brachypodium distachyon) and foxtail millet (Setaria italica) and two dicotyledonous plant species, poplar (Populus trichocarpa) and potato (Solanum tuberosum). Detailed in silico expression analysis of NCX genes during developmental and stress conditions provided important clue regarding their possible functions. Quantitative real time PCR (qRT-PCR) analysis of fifteen rice NCX genes under six different abiotic stresses viz. salinity, dehydration, heat, cold, oxidative and UV and under exogenous treatment with CaCl2 and a Ca2+ chelator, EGTA (ethylene glycol tetraacetic acid) was performed.
Results and discussion
Identification and nomenclature of NCX proteins in Arabidopsis and rice genomes
Genes encoding NCX proteins were identified in Arabidopsis (TAIR version 10) and rice (TIGR version 7) genomes by employing HMM (Hidden Markov Model) profile retrieved from Pfam database, keyword search and domain search functions. NCX domain (PF01699) containing proteins having putative sodium/calcium exchanger function have been classified as NCX proteins. Our analysis has identified thirteen distinct chromosomal loci encoding for sixteen NCX proteins in Arabidopsis and fifteen chromosomal loci encoding for twenty-two NCX proteins in rice; and hence are reported to undergo alternative splicing. In Arabidopsis, only two genes (AtNCX5 and AtNCX6), whereas in rice, seven genes (OsNCX1, OsNCX2, OsNCX4, OsNCX5, OsNCX7, OsNCX9 and OsNCX14,) are reported to undergo alternative splicing. In order to confirm the presence of NCX domain and identification of additional domains, all the NCX proteins were analyzed using Pfam and InterProScan. These analyses confirmed the presence of NCX domain in all the identified proteins. In addition, CAX and EF-hand domains were also found in some of the proteins (Table 1). Previously, some NCX family members have been named ambiguously, which has led to confusion in literature. For example, five Arabidopsis proteins which were named as CAX7 to CAX11 by Maser et al. (2001) were later found to be proteins showing very high sequence similarity with mammalian K+-dependent Na+/Ca2+ antiporter NCKX6 (Shigaki et al. 2006). Thus, to avoid any ambiguity and to maintain uniformity, we have suggested new nomenclature for NCX family members. In case where prior information was present in literature regarding any gene, the old name is given along with the new name (Table 1). ‘At’ and ‘Os’ have been used as prefix for the nomenclature in case of Arabidopsis thaliana and Oryza sativa respectively, followed by NCX. The genes were numbered according to their chromosomal location on chromosome 1–5 in case of Arabidopsis and on chromosome 1–12 in case of rice and from top to bottom. Similar criteria have been adapted for nomenclature of NAC proteins in soybean (Le et al. 2011) and potato (Singh et al. 2013) and WRKY proteins in maize (Wei et al. 2012). Details of each NCX member are shown in Table 1, representing their locus numbers, CDS and protein length, chromosomal co-ordinates and domain architecture.
Table 1.
Details of putative NCX proteins encoded in the Arabidopsis and rice genomes. Table enlists NCX genes in Arabidopsis and rice along with their corresponding proteins, their existing nomenclature in TAIR, locus identifiers, CDS, protein length, chromosomal locations and domain architecture
| Gene | Protein | Nomenclature in TAIR/RGAP | Locus | CDS length (bp) | Protein length (aa) | Coordinates (5′-3′) | Domain architecture |
|---|---|---|---|---|---|---|---|
| Arabidopsis thaliana | |||||||
| AtNCX1 | AtNCX1 | AtCAX11 | AT1G08960.1 | 1248 | 416 | 2879642 - 2882231 |
|
| AtNCX2 | AtNCX2 | Sodium/calcium exchanger | AT1G53210.1 | 1758 | 586 | 19844632 - 19847836 |
|
| AtNCX3 | AtNCX3 | AtCCX4 | AT1G54115.1 | 1935 | 645 | 20202118 - 20204177 |
|
| AtNCX4 | AtNCX4 | AtCAX6 | AT1G55720.1 | 1449 | 467 | 20828118 - 20830640 |
|
| AtNCX5 | AtNCX5.1 | AtCAX5 | AT1G55730.1 | 1326 | 442 | 20831052 - 20834519 |
|
| AtNCX5.2 | AT1G55730.2 | 1326 | 442 | 20831052-20834512 | |||
| AtNCX6 | AtNCX6.1 | AtCAX1 | AT2G38170.1 | 1392 | 464 | 15989083 - 15993278 |
|
| AtNCX6.2 | AT2G38170.2 | 1179 | 393 | 15989083 - 15993276 | |||
| AtNCX6.3 | AT2G38170.3 | 1428 | 476 | 15990045 - 15993278 | |||
| AtNCX7 | AtNCX7 | AtMHX1 | AT2G47600.1 | 1620 | 540 | 19524160 - 19527413 |
|
| AtNCX8 | AtNCX8 | AtCAX2 | AT3G13320.1 | 1326 | 442 | 4314529 - 4318351 |
|
| AtNCX9 | AtNCX9 | AtCCX3 | AT3G14070.1 | 1932 | 644 | 4661143 - 4663074 |
|
| AtNCX10 | AtNCX10 | AtCAX3 | AT3G51860.1 | 1380 | 460 | 19239427 - 19242913 |
|
| AtNCX11 | AtNCX11 | AtCAX4 | AT5G01490.1 | 1365 | 455 | 195541 - 198524 |
|
| AtNCX12 | AtNCX12 | AtCAX8 | AT5G17850.1 | 1680 | 560 | 5899253 - 5900932 |
|
| AtNCX13 | AtNCX13 | AtCAX7 | AT5G17860.1 | 1713 | 571 | 5902394 - 5904380 |
|
| Oryza sativa | |||||||
| OsNCX1 | OsNCX1.1 | Sodium/calcium exchanger | LOC_Os01g11414.1 | 1755 | 584 | 6134202 -6139819 |
|
| OsNCX1.2 | LOC_Os01g11414.2 | 1284 | 427 | 6136830-6139189 | |||
| OsNCX2 | OsNCX2.1 | OsCAX1a | LOC_Os01g37690.1 | 1356 | 451 | 21076730 - 21071370 |
|
| OsNCX2.2 | LOC_Os01g37690.2 | 1065 | 354 | 21073919-21072230 | |||
| OsNCX3 | OsNCX3 | Sodium/calcium exchanger | LOC_Os02g04630.1 | 1089 | 362 | 2070492 - 2075369 |
|
| OsNCX4 | OsNCX4.1 | Sodium/calcium exchanger | LOC_Os02g14980.1 | 1728 | 575 | 8358769 - 8362960 |
|
| OsNCX4.2 | LOC_Os02g14980.2 | 1281 | 426 | 8359318-8362959 | |||
| OsNCX5 | OsNCX5.1 | OsCAX1c | LOC_Os02g21009.1 | 1353 | 450 | 12432287 - 12451812 |
|
| OsNCX5.2 | LOC_Os02g21009.2 | 1353 | 451 | 12432287 - 12448557 | |||
| OsNCX6 | OsNCX6 | Sodium/calcium exchanger | LOC_Os03g08230.1 | 1920 | 639 | 4196467 - 4194283 |
|
| OsNCX7 | OsNCX7.1 | Sodium/calcium exchanger | LOC_Os03g27960.1 | 1317 | 437 | 16061184 - 16065169 |
|
| OsNCX7.2 | LOC_Os03g27960.2 | 1314 | 437 | 16061151 - 16065169 | |||
| OsNCX8 | OsNCX8 | Sodium/calcium exchanger | LOC_Os03g45370.1 | 1728 | 575 | 25613825 - 25616214 |
|
| OsNCX9 | OsNCX9.1 | OsCAX3 | LOC_Os04g55940.1 | 1254 | 417 | 33314630 - 33319375 |
|
| OsNCX9.2 | LOC_Os04g55940.2 | 1254 | 418 | 33315199-33319375 | |||
| OsNCX10 | OsNCX10 | OsCAX1b | LOC_Os05g51610.1 | 1362 | 453 | 29593841 - 29588646 |
|
| OsNCX11 | OsNCX11 | Sodium/calcium exchanger | LOC_Os10g30070.1 | 1794 | 597 | 15614210 - 15616127 |
|
| OsNCX12 | OsNCX12 | Sodium/calcium exchanger | LOC_Os11g01580.1 | 1068 | 355 | 335287 – 334094 |
|
| OsNCX13 | OsNCX13 | Sodium/calcium exchanger | LOC_Os11g05070.1 | 798 | 265 | 2220748 - 2217645 |
|
| OsNCX14 | OsNCX14.1 | Sodium/calcium exchanger | LOC_Os11g43860.1 | 1572 | 523 | 26492383 - 26488372 |
|
| OsNCX14.2 | LOC_Os11g43860.2 | 1305 | 434 | 26492229 - 26488372 | |||
| OsNCX15 | OsNCX15 | Sodium/calcium exchanger | LOC_Os12g42910.1 | 1764 | 587 | 26669155 - 26671200 |
|
 NXC,
NXC,  EF-hand,
EF-hand,  CAX domian
CAX domian
Based on the presence of conserved domains, it was found that in Arabidopsis all the NCX proteins contain a pair of NCX domains, except AtNCX2 (At1g53210) that contains only one NCX domain and one EF-hand domain (Table 1). Whereas in rice, eleven NCX genes (OsNCX2, OsNCX3, OsNCX5, OsNCX6, OsNCX7, OsNCX8, OsNCX9, OsNCX10, OsNCX11, OsNCX14 and OsNCX15) code for proteins with a pair of NCX domains, the rest four genes (OsNCX1, OsNCX4, OsNCX12 and OsNCX13) code for proteins with a single NCX domain. Out of four proteins with a single NCX domain, two (OsNCX1 and OsNCX4) contain additional EF-hand domain. Recently, functional characterization of Arabidopsis NCX-like protein, AtNCL (At1g53210; designated as AtNCX2 in present study) was carried out. AtNCL was found to have the ability to bind Ca2+ and its loss of function mutants were less sensitive to salt stress than WT or AtNCL overexpressing lines (Wang et al. 2012). Thus, the presence of EF-hand domain in OsNCX1 and OsNCX4 suggest that these proteins may also bind Ca2+ and regulate cellular Na+ and Ca2+ homeostasis.
Among NCX proteins with a pair of NCX domains, six AtNCXs (AtNCX4, AtNCX5, AtNCX6, AtNCX8, AtNCX10 and AtNCX11) and five OsNCXs (OsNCX2, OsNCX5, OsNCX7, OsNCX9 and OsNCX10) also contain a CAX (Ca2+/H+ exchanger) domain (TIGR00378). Interestingly, both the NCX domains in these proteins were found to lie within the CAX domain. CAX domain is a dominant feature of Cation/H+ exchanger proteins, which transport and regulates Ca2+ homeostasis. Previous characterization of Arabidopsis CAX1 and CAX2 showed that these genes encode for high efficiency and low efficiency H+/Ca2+ exchangers, respectively (Hirschi et al. 1996). Since, these CAX genes were characterized before release of Arabidopsis genome sequence, there appears to be some ambiguity. For example, CAX1 and CAX2 were reported to encode for proteins of 459 and 399 amino acids, respectively. However, in the present study we show that CAX1 and CAX2 (designated as AtNCX6 and AtNCX8, respectively) encode for proteins of 463 and 441 amino acids, respectively. A phylogenetic study of cation transporter families showed that Arabidopsis contains eleven CAX genes named AtCAX1-11 (Mäser et al. 2001). However, later it was shown that AtCAX7 to AtCAX11 have limited amino acid sequence homology with any CAX. In fact, these proteins share a very high sequence similarity with the mammalian K+-dependent Na+/Ca2+ antiporter NCKX6 (Cai and Lytton 2004; Shigaki et al. 2006). Therefore, in the present study all the NCX domain containing proteins were renamed as NCX proteins irrespective of the presence of additional domain(s). The presence of NCX domain along with the CAX domain in these proteins suggests that these proteins may also have dual role as CAX and NCX. To ascertain the relevance of co-occurrence of these domains in a single protein, their functional characterization would be required in future.
Chromosomal organization of NCX genes
In case of Arabidopsis, NCX genes are distributed on chromosome I, II, III and V, none of the NCX gene is located on chromosome IV (Fig. 1a). Five NCX genes are located on chromosome I, while two NCX genes are located on chromosome II. Chromosome III and chromosome V possess three NCX genes each. It has been previously reported that two rounds of duplication events have occurred in Arabidopsis genome during the course of evolution, followed by gene loss (Bowers et al. 2003; Ermolaeva et al. 2003). This is evident by the observation that only one NCX gene from Arabidopsis, AtNCX6 on chromosome II, seems to be duplicated along with inversion on chromosome III as AtNCX10. These duplicated genes share more than 75 % homology at protein level (data not shown). Similarly, a few genes of various protein families such as, CDCP (Kushwaha et al. 2009) and glyoxalase (Mustafiz et al. 2011) have been reported to be duplicated in Arabidopsis.
Fig. 1.
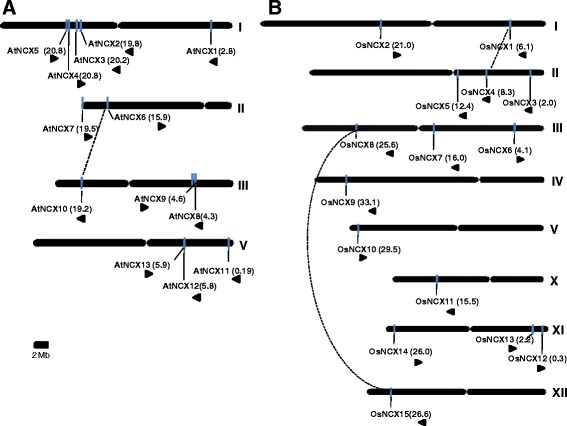
Genomic distribution of NCX genes. a Chromosomal localization of NCX family genes from Arabidopsis. Four out of the five Arabidopsis chromosomes (all except chromosome IV) were found to possess one or more gene(s) encoding NCX family proteins. Bars on chromosomes denote the genes and arrows below the gene names indicate the direction of transcription. b Genomic distribution of NCX family genes on rice chromosomes. Out of the 12 chromosomes of rice, 8 chromosomes possess genes encoding NCX proteins. Chromosome numbers are shown at the right of the chromosomes and the centromeric regions are indicated by ellipse in both rice and Arabidopsis. Events of duplication between AtNCX6 and AtNCX10 in Arabidopsis; and between OsNCX1 and OsNCX4 and OsNCX8 and OsNCX15 in rice are represented by dotted lines. Scale at the bottom represents 2 Mb for both the karyograms
In case of rice, all fifteen NCX genes are distributed on eight out of twelve chromosomes. The chromosome II, III, and XI each possess three NCX genes, while, two NCX genes are located on chromosome I. Rest of the chromosome viz. IV, V, X and XII each contains only single NCX gene (Fig. 1b). Gene duplication plays a central role in plant diversification, a key process that generates the raw material necessary for adaptive evolution (Flagel and Wendel 2009). In case of rice, it has been reported that large-scale losses of duplicated genes might have occurred shortly after the genome duplication (Wang et al. 2005). OsNCX1 localized on chromosome I seems to be duplicated as OsNCX4 on chromosome II. These duplicated genes share 65 % sequence homology at protein level (data not shown). Similarly OsNCX8 on chromosome III seems to be duplicated on chromosome XII as OsNCX15 and they share 58 % sequence homology at protein level with each other (data not shown).
Phylogenetic analysis of NCX proteins of Arabidopsis and rice
To explore the evolutionary relationship between Arabidopsis and rice NCX proteins, multiple sequence alignment of protein sequences was performed using ClustalW2 program and phylogenetic tree was generated using MEGA6 software (Additional file 1: Figure S1). All the members of Arabidopsis and rice NCX family were divided into five major clades. Interestingly, NCX proteins with similar domain architecture were clustered in same clades. Proteins such as AtNCX2, OsNCX1, and OsNCX4, which have calcium binding EF hand-like domain in addition to NCX domain were clustered together in clade I and share 46 to 58 % sequence homology with each other. In clade II, AtNCX4, AtNCX5, AtNCX8, OsNCX3, OsNCX7 and OsNCX9 were clustered together and share 46 to 87 % sequence homology with each other. All the proteins clustered in clade II contain a CAX domain in addition to a pair of NCX domains, except OsNCX3 that contains only a pair of NCX domains. Clade III is the second largest clade, which contains AtNCX6, AtNCX10, AtNCX11, OsNCX2, OsNCX5 and OsNCX10. All the proteins in this clade contain CAX domain in addition to a pair of NCX domains and share 46 to 76 % sequence homology with each other. The clade IV is the smallest clade, having only AtNCX7 and OsNCX14, which share 67.5 % sequence homology with each other. The largest clade V contains eleven proteins (AtNCX3, AtNCX9, OsNCX6, AtNCX12, AtNCX13, OsNCX8, OsNCX15, OsNCX11, OsNCX12, AtNCX1, and OsNCX13) which share sequence similarity ranging from 42 to 80 %. In order to study the phylogenetic relationship of Arabidopsis and rice NCX family members with that of other plants, NCX proteins were identified from the Brachypodium and foxtail millet as representatives of monocotyledonous and from poplar and potato as representatives of dicotyledonous plants. We have identified 18, 16, 18 and 16 NCX proteins from Brachypodium, foxtail millet, poplar and potato, respectively (Additional file 2: Table S1). Orthologs of rice NCX proteins in Brachypodium and foxtail millet and that of Arabidopsis NCX proteins in poplar and potato were also identified (Additional file 2: Table S1). The presence of NCX domain in all these proteins was verified using Pfam and InterProScan. However, in some of the proteins CAX and EF hand domains were also identified (Additional file 3: Table S2). Phylogenetic tree comprising NCX proteins from Arabidopsis and rice with those from Brachypodium, foxtail millet, poplar and potato was constructed using MEGA6 (Fig. 2). Interestingly, NCX proteins from all the plant species studied (except potato) were divided into five clades as found in the case of Arabidopsis and rice NCX proteins, shown in Additional file 1: Figure S1. NCX proteins from potato were found to be distributed in four phylogenetic clades (all except clade IV). Clade V consists of maximum number of NCX proteins (36), followed by clade III, II, I and IV, which contain 24, 23, 16 and 7 NCX proteins, respectively. Similar distribution of NCX proteins of all the six species in five clades was found to be strikingly similar (Fig. 2) as in case of Arabidopsis and rice NCXs (Additional file 1: Figure S1), which was found to be based on domain architecture. Cai and Lytton (2004) have previously performed phylogenetic analysis of the cation/Ca2+ exchangers super family of 147 proteins from different genomes like bacteria, archaea, and eukaryotes, and classified this super family into the YBRG, CAX, NCX, NCKX and CCX families. Phylogenetic relationship of five CAX genes of rice was suggested earlier by Kamiya et al. (2005) which are part of NCX family also. Recently, Emery et al. 2012 have also suggested the evolutionary hierarchy of Ca2+/cation antiporter families in flowering plants which also includes NCX family. The present study provides a comprehensive phylogenetic analysis of all NCX proteins identified from A. thaliana, O. sativa, B. distachyon, S. italica, P. trichocarpa and S. tuberosum.
Fig. 2.
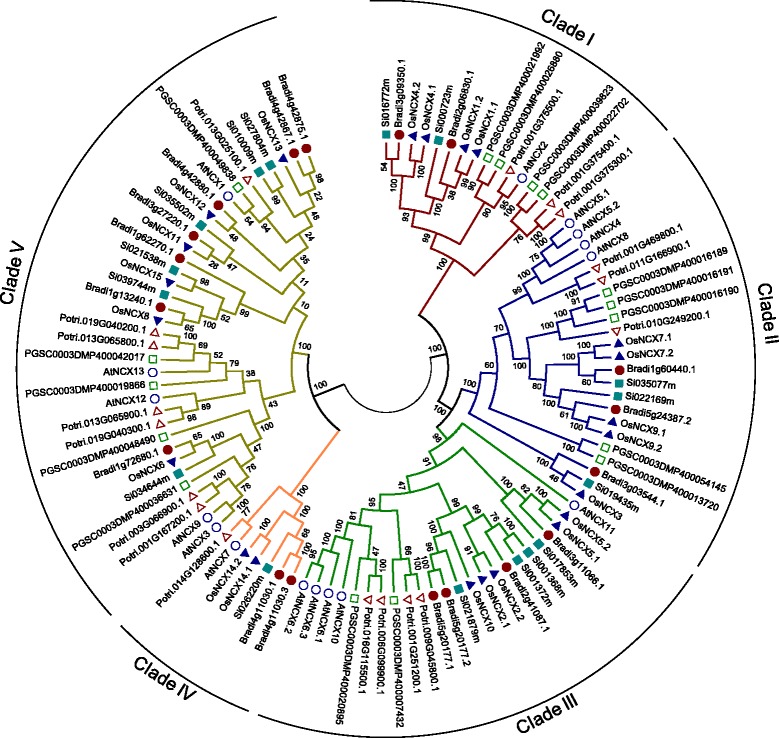
Phylogenetic relationships among Arabidopsis, rice, Brachypodium, foxtail millet, poplar and potato NCX proteins. Multiple sequence alignment of NCX proteins was done using ClustalW2 and the phylogenetic tree was generated using MEGA6 software by the Neighbor-joining method with 1000 bootstrap replicates. NCX proteins were categorized into five different clades depending upon the relative sequence homology of each member of the clades. NCX proteins from Arabidopsis, poplar, potato, rice, Brachypodium and foxtail millet are denoted by different shapes viz. circle, triangle, rectangle, solid triangle, solid circle and solid rectangle, respectively
Topological model prediction for NCX proteins
Topological structures of all the NCX proteins of Arabidopsis and rice (for the longest ORFs among the spliced variants) were predicted using Protter software (Fig. 3). Presence of transmembrane segments (TMSs) indicated that all the NCX proteins are transmembrane proteins. In general, proteins clustered in same clade in phylogenetic tree (Additional file 1: Figure S1) share similar membrane topologies. Thus, topological models were arranged as per the phylogenetic clustering of NCX proteins. The number of TMSs was found to be variable in various NCX proteins and ranged from 5 to 14. In many NCX proteins from both rice and Arabidopsis, a large hydrophilic loop intruding into the cytoplasm apparently separated the TMSs into two groups joined together by the large hydrophilic loop. However, as an exception, in case of OsNCX15, the TMSs were found to be separated by an extracellular loop. It was observed that generally lesser number of TMSs lie towards the N-terminal side of the loop and more were present towards C-terminal side (Fig. 3). This loop comprises sites important for calcium regulation, Na+ dependent inactivation and alternative splicing (Iwamoto et al. 1999; Marshall et al. 2005). Some proteins such as AtNCX2, OsNCX1 and OsNCX4 also have EF hand loop in between two spans of TMS (marked as clade I in Fig. 3). Two spans of TMS have also been reported earlier in case of NCX domain containing calcium/proton exchangers (CAX), calcium/cation exchangers (CCX) and Magnesium/cation exchangers (MHX) in Arabidopsis, but they differ in function with little modification in the N- and C terminal residues of TMS (Kamiya and Maeshima 2004; Shigaki et al. 2006). Based on experimental evidences, NCX proteins are modeled to have nine putative TMSs (Iwamoto et al. 1999; Nicoll et al. 1999) in animals. Five TMSs were reported to be present in the N-terminal domain and 4 in the C-terminal domain. These NCX proteins comprised ~30 residues long signal peptide that is cleaved during initial processing. Interestingly, proteins with signal peptide on their N-terminal were clustered in clade I and clade V (Fig. 3).
Fig. 3.
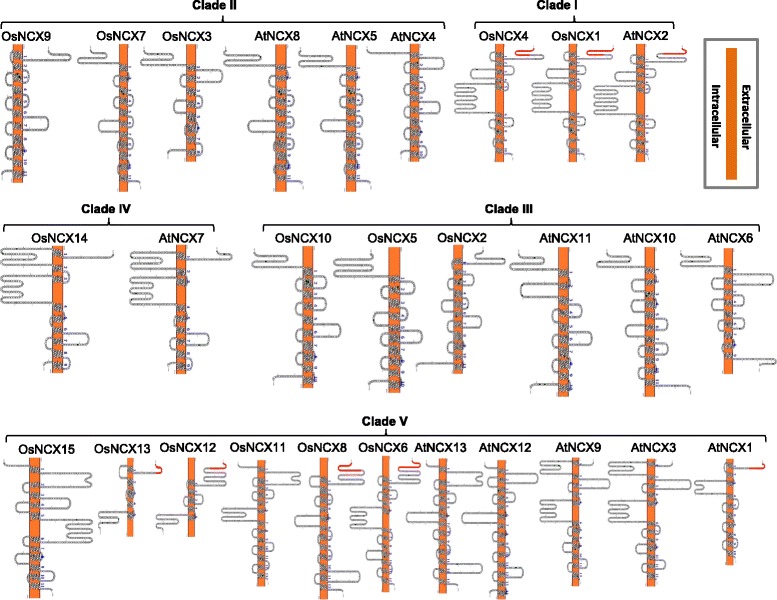
Predicted topological structure of each of the NCX family members in Arabidopsis and rice. Here, all the members are divided into clades according to phylogenetic relationship among Arabidopsis and rice NCX families (as shown in Additional file 1: Figure S1 and in Fig. 3). Topological structures were generated using Protter v1.0 (see Methods). For splice variants possessing a similar predicted topological architecture, only a single model for the topology has been provided. Red colored N-terminal regions in a few NCX proteins represent putative signal peptides. Finger-like projections are loops joining N- and C-terminus groups of TMS, these loops may act as EF hand. The box at the top-left is a schematic representation of the plasma membrane (shown as orange bar) with the extra- and the intra-cellular regions specified
Expression profiling of NCX family genes using Massive parallel signature sequencing (MPSS)-based expression data
Expression analysis of NCX gene family members using Arabidopsis MPSS data (Fig. 4a) revealed that AtNCX2 and AtNCX6 have high transcript level in all the tissues except callus (CAF and CAS) and untreated root tissue (ROF), suggesting their role in most of the development stages. The expression of AtNCX7 seems to be higher only in inflorescence and silique, and remained lower in all other tissues suggesting its specific role in flowering and silique development. Earlier, characterization of AtMHX (a Mg2+/H+ exchanger also found to possess NCX domain; named as AtNCX7 in present study) has been done by Shaul et al. (1999) and it was found to have highest expression in inflorescence, which correspond to our findings (Fig. 4a). Similarly, AtNCX12 has high expression only in case of silique (SIF) and callus (CAS). AtNCX11 and AtNCX4 maintain constitutively medium level of expression in almost all the plant tissues. Expression of AtNCX13 was high in inflorescence tissues. AtNCX10 exhibited higher transcript level in inflorescence (INF), root (ROF) and silique (SIF) tissues.
Fig. 4.
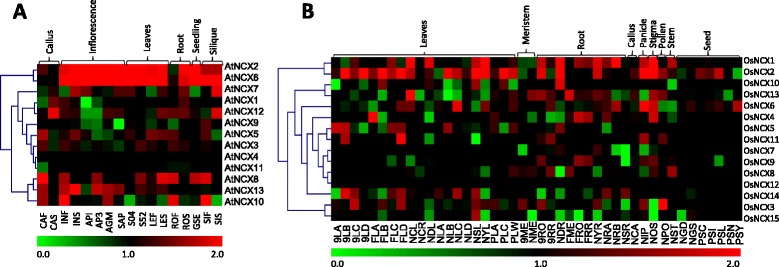
Expression profile of NCX genes as obtained from analysis of MPSS-based expression data. a Expression profile of NCX genes from MPSS data in different tissues/organs of Arabidopsis, b Expression profile of NCX genes from MPSS data in different tissues/organs in rice. The heat maps have been generated using TIGR MeV software package and represent hierarchical clustering of log-transformed signal values of all NCX genes in various tissue/organs (indicated at the top). The scale for relative expression values is represented by color bars below the heat maps, thereby green color representing lowest expression levels, black medium expression and red denotes highest expression level. Names of the MPSS libraries are mentioned below each heat map
Further, we have analyzed the expression pattern of all the rice NCX genes in different tissues using MPSS data (Fig. 4b). Transcript of OsNCX2 was predominantly high in all the tissues, which possibly indicates its important role throughout the plant. OsNCX12 has uniformly intermediate level of expression in all the plant tissues. Previously, the transcript levels of rice CAX genes i.e. OsCAX1a (OsNCX2), OsCAX1b (OsNCX10), OsCAX1c (OsNCX5), OsCAX2 (OsNCX7) and OsCAX3 (OsNCX9), were analyzed in several tissues like, embryo, endosperm, shoot, root, flower, leaf blade, leaf sheath, node, internode, root and callus (Kamiya et al. 2005). Expression profile of these genes reported earlier corresponds to our MPSS expression profile. For example, expression of OsNCX2 (OsCAX1a) was found to be high in all the tissues analyzed by Kamiya et al. (2005), which corresponds to the MPSS data presented in our study. Expression of OsNCX10 (OsCAX1b) was high in case of root (NDR) in MPSS data, which also corresponds to the observations by Kamiya et al. (2005). Expression of OsNCX5 (OsCAX1c) was observed only in case of leaf blade (Kamiya et al. 2005), similarly in MPSS data, its expression was observed in case of leaf only. These observations indicate towards possible function of various NCX genes in different tissues and developmental stages. Further, we identified orthologous genes from Arabidopsis and rice to correlate their expression profiles (Additional file 4: Table S3). Interestingly, AtNCX2 and its ortholog OsNCX1; and AtNCX6 and its ortholog OsNCX2 were found to have high expression in leaf, root and inflorescence tissues (Fig. 4a and b).
Microarray-based expression analysis of NCX family genes in Arabidopsis and rice
NCX proteins can alter cytosolic Ca2+ levels, which is an important second messenger during various physiological and developmental signals. However, most of the plant NCX proteins are uncharacterized and their physiological function remains to be studied. Studying the expression profiling of the uncharacterized genes can provide important clues regarding their function (Mustafiz et al. 2011; Tripathi et al. 2015). Therefore, to gain preliminary insight into the possible function of plant NCX proteins in stress response and during development, we have exploited publicly available microarray data for Arabidopsis and rice. Arabidopsis Microarray data in response to different abiotic stresses (Fig. 5a and b) and different developmental and reproductive stages (Fig. 6) was retrieved from AtGenExpress (http://jsp.weigelworld.org/expviz/expviz.jsp). As shown in Fig. 5, AtNCX7, AtNCX9, AtNCX10, AtNCX12 and AtNCX13, exhibited upregulation under salt and osmotic stress conditions in both root and shoot. Expression of AtNCX2 in root largely remained unchanged in response to all the stresses. While in shoot, its expression was upregulated in response to salt, osmotic, cold and wounding stresses. Previously, Wang et al. (2012) have also shown its induction in response to salt, ABA, cold stress and heat stress. AtNCX1 and AtNCX3 are the only two genes which did not show any significant change in expression in roots. Expression of AtNCX6 and AtNCX11 was found to be upregulated in response to salt, cold and heat stresses in roots, while in shoots their expression was largely unaltered. The expression data for AtNCX4 (At1g55720) was not available in the microarray database and hence could not be included in expression analysis under different stresses.
Fig. 5.
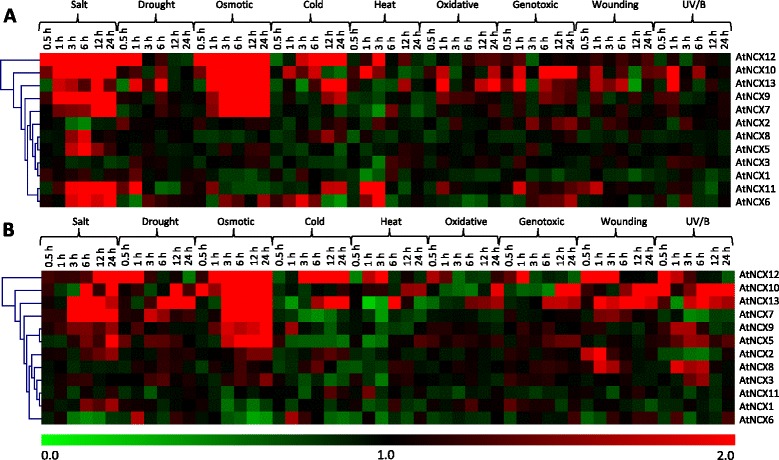
Microarray-based expression profile of Arabidopsis NCX genes under various abiotic stress conditions. Heat maps show the fold changes in expression of Arabidopsis NCX genes in a root and b shoot tissues under different abiotic stress conditions such as salt, drought, osmotic, cold, heat, oxidative, genotoxic, wounding and UV/B stress. Heat maps were generated using microarray data obtained from AtGenExpress. Microarray data was obtained for different time points for different stresses viz 0.5 h, 1 h, 3 h, 6 h, 12 h and 24 h for both root and shoot tissues and analyzed with respect to control. Relative signal values are represented by color bar shown at the bottom of heat map; thereby green color representing downregulation, black signifies no change in expression and red shows upregulation
Fig. 6.

Expression analysis of Arabidopsis NCX genes at different developmental stages using microarray data. Heat map and hierarchical cluster displays differential expression profile of NCX genes across various developmental stages. Color bar at the left represents log10 expression values; thereby green color representing lowest expression levels, black medium expression and red signifies highest expression level. Developmental stages used for expression profiling are mentioned on the right side of the heat map
We also performed the expression profiling of NCX genes during different vegetative and reproductive stages using Microarray data (Fig. 6). Microarray data contain more tissues and developmental stages as compared to MPSS data. Largely, expression profile of NCX genes obtained from microarray data corroborate with MPSS data. For instance, AtNCX2 and AtNCX6 were found to be highly expressed in all the developmental stages, which is in agreement to their expression profile observed in MPSS data. On the other hand expression of AtNCX11 was low at almost all the developmental stages. Specific high transcript of AtNCX9 in stage 8, 9 and 10 of seed development, indicates toward its particular role in seed development.
Gene Expression Omnibus (GEO) database of the NCBI (http://www.ncbi.nlm.nih.gov/geo/) was used to obtain transcriptional data of NCX genes under different developmental stages of rice (Fig. 7). Based on the studies by Itoh et al. (2005) and information from Oryzabase (http://www.shigen.nig.ac.jp/rice/oryzabase/top/top.jsp), rice panicle and seed developmental stages were divided into 6 and 5 major categories, respectively. We found that some of the NCX genes like, OsNCX1, OsNCX6, OsNCX8 and OsNCX9 were expressed at high level in almost all the developmental stages, suggesting their broad role in plant development. The expression of OsNCX10 and OsNCX12 was low in all the developmental stages. Expression of OsNCX2 and OsNCX7 was also found to be intermediate in all the developmental stages. Earlier using GUS-reporter assay the expression of OsCAX1a (named as OsNCX2 in the present study) has been found in stomata, trichomes and vascular cells (Kamiya et al. 2006). Our development stage specific microarray data also complements the expression profile of CAX genes as reported previously (Kamiya et al. 2005), which shows high transcript levels of OsCAX1a (OsNCX2), OsCAX2 (OsNCX7) and OsCAX3 (OsNCX9) while OsCAX1b (OsNCX10) and OsCAX1c (OsNCX5) show very low transcript level in different plant tissues. On comparing the expression profiles of orthologous genes in Arabidopsis and rice (orthologous genes are shown in Additional file 4: Table S3), it was found that AtNCX2 and its ortholog OsNCX1; AtNCX3 and its ortholog OsNCX6; and AtNCX6 and its ortholog OsNCX2 have constitutive expression in all the tissues analyzed in Figs. 6 and 7.
Fig. 7.
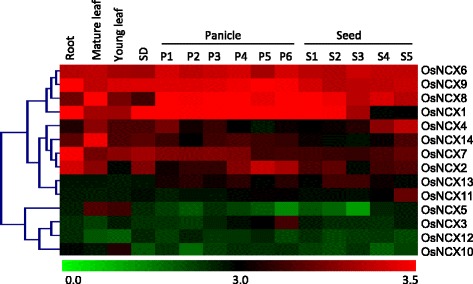
Expression analysis of rice NCX genes at different developmental stages using microarray data. Heat map and hierarchical clustering displays differential expression profile of rice NCX genes across various developmental stages. Color bar at the bottom represents log2 expression values, thereby green color representing lowest expression levels, black medium expression and red signifies highest expression level. Developmental stages used for expression analysis are mentioned on the top of each column. Various stages used for the expression profiling (from left to right in the heat map) are: Roots of 7- day old seedling, Mature leaf (collected before pollination), young leaf, SD-7-day old seedling, P1 (0–3 cm panicle), P2 (3–5 cm panicle), P3 (5–10 cm panicle), P4 (10–15 cm panicle), P5 (15–22 cm panicle), P6 (22–30 cm panicle), S1 (0–2 DAP), S2 (3–4 DAP), S3 (5–10 DAP), S4 (11–20 DAP) and S5 (21–29 DAP)
Expression profiling of rice NCX genes using quantitative real time PCR under various abiotic stress and Ca2+ stimuli conditions
Calcium is one of the most important secondary messengers in plant system and its role in various adverse environmental conditions like biotic and abiotic stresses has been well studied. NCX proteins play a pivotal role in Ca2+-homeostasis and hence can potentially participate in the diverse physiological processes involving Ca2+ as the second messenger. To examine the role of rice NCX family members in abiotic stress and calcium homeostasis and to validate the expression profile obtained via analysis of microarray-based expression data, we have carried out expression analysis using quantitative real-time reverse transcription-PCR (qRT-PCR) in rice. The relative transcript abundance of all the OsNCX genes under various abiotic stresses viz. salinity, dehydration, oxidative, heat, cold and UV stress and treatments with calcium and EGTA has been presented as bar graphs and heat maps (Fig. 8a–i). Previous reports have shown that stress related genes are often differentially expressed in rice (Kumari et al. 2009). Expression of OsNCX3, OsNCX9 and OsNCX10 was either upregulated or unaltered under all the stresses imposed in present study which suggests that these NCX proteins might have some important role in response to abiotic stress. In contrast, expression of OsNCX5 was repressed under all the stresses except under UV stress. Interestingly, we found that relative transcript abundance of all the NCX genes was upregulated in response to UV stress. Particularly, OsNCX9, OsNCX10, OsNCX11 and OsNCX12 were highly up regulated by 100, 300, 56 and 50 fold, respectively in response to UV. OsNCX3 seems to be highly responsive to oxidative stress as it showed 122-fold upregulation in response to MV. In response to cold stress transcript level of OsNCX1 was specifically high, suggesting it might have some role in maintaining calcium level during cold stress. The expression of OsNCX3, OsNCX10 and OsNCX15 was predominantly up-regulated in response to salinity and dehydration stress. Transcript levels of OsNCX4 were very low, thus we were not able to detect it under any of the conditions tested in the present study.
Fig. 8.
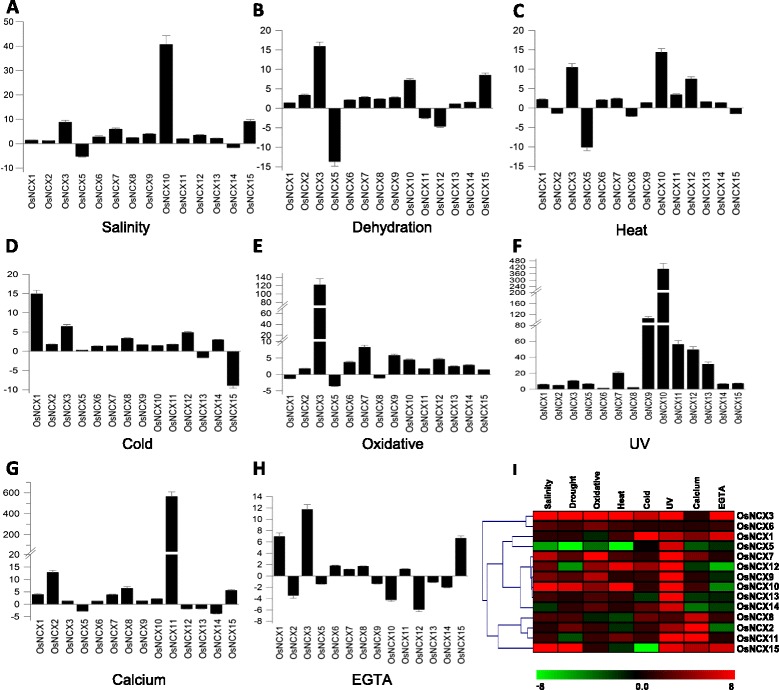
Expression profiling of rice NCX genes under different abiotic stresses using qRT-PCR. Bar graphs show fold change in expression of rice NCX genes under various abiotic stresses, viz. a salinity, b dehydration, c heat, d cold, e oxidative (MV), f UV-B, g calcium (calcium chloride) and h calcium chelator EGTA i Heat map generated from the above qRT-PCR data representing the expression profile of rice NCX genes under different abiotic stresses and calcium and EGTA treatment. Color bars at the base represent fold change in expression; green, black and red colors represent downregulated, unaltered and upregulated expression of genes relative to the untreated control, respectively
Sodium calcium exchangers are involved in intracellular regulation of calcium levels. If Ca2+ concentration is low inside the cell, NCX proteins help the cell to intake calcium from outside and vice-versa if calcium concentration is high inside the cell. To study the calcium mediated- regulation of NCX genes at transcriptional level, we examined the transcript abundance of OsNCX genes in a moderately stress-sensitive rice cultivar – IR64, in response to calcium and its chelator (EGTA). The transcript level of most of the NCX genes was upregulated in response to exogenous calcium, with some exceptions like OsNCX5, OsNCX12, OsNCX13 and OsNCX14, which were downregulated under Ca2+ treatment. OsNCX11 was highly upregulated in response to calcium. Interestingly, EGTA treatment repressed the transcript level of OsNCX genes. However, expression of OsNCX1, OsNCX3, OsNCX6, OsNCX7, OsNCX11 and OsNCX15 was upregulated in response to EGTA. OsNCX12 showed extremely low expression in response to both calcium and EGTA implying its restricted role in calcium transportation across or inside the cell. This data indicates that NCX genes are under tight control of intracellular calcium levels. However, it is also apparent from the expression profile obtained in our study that expression pattern of NCX genes is different in response to NaCl and calcium treatments, suggesting their unique role in specific conditions. Further characterization of putative OsNCX family members, at both molecular and chemical level, needs to be carried out in order to ascertain their role in specific abiotic stress conditions.
Conclusions
Plants require various transporters for exchange of different ions. NCX proteins are one of them which maintain the intracellular and extracellular Ca2+ concentrations. However, not much has been studied about plant NCX proteins, and their entire repertoire remains to be discovered. The current study has comprehensively identified genes encoding putative NCX proteins in Arabidopsis, rice, Brachypodium, foxtail millet, poplar and potato based on NCX domain. We have also proposed new nomenclature to NCX family members of Arabidopsis and rice, which could be helpful for future studies. The differential expression pattern of NCX members under various developmental stages and stress conditions suggest their role in associated physiological processes. qRT-PCR based expression analysis of rice NCX family members has suggested many new genes involved in abiotic stress response and future studies are needed to validate their function. Taken together, this study has imparted new insights into the putative functions of NCX proteins in plants. Detailed functional characterization of individual NCX family member would be a future challenge and it could help in understanding the role(s) of NCX proteins in various aspects of the life cycle of plants.
Methods
Identification of Na+/Ca2+ exchanger proteins from various dicots and monocots
In order to identify Sodium/calcium exchanger (NCX) proteins in Arabidopsis and rice, hidden markov model (HMM) search was performed in TAIR 10 and RGAP 7 databases, respectively with an e-value cut off 0.001. Similarly, HMM search was performed to identify NCX proteins in Brachypodium distachyon, Setaria italica, Populus trichocarpa and Solanum tuberosum using phytozome version 10.1 database (http://phytozome.jgi.doe.gov/pz/portal.html; Goodstein et al. 2012). All the putative NCX proteins identified using HMM search were subjected to Pfam (http://Pfam.sanger.ac.uk/) and InterProScan (http://www.ebi.ac.uk/Tools/pfa/iprscan5/) analysis to verify the presence of NCX domains. BLASTn search program was used to determine the position of each NCX gene on Arabidopsis and rice chromosomes available at TAIR (ver.10) and RGAP (ver.7.0), respectively. For nomenclature prefix “At” or “Os” for Arabidopsisthaliana and Oryza sativa, respectively was added followed by NCX and numbered according to its position from top to bottom on the respective chromosome. Alternatively spliced forms were represented by Arabic numbers after “.” sign. For identification of orthologous proteins of O. sativa NCXs in B. distachyon and S. italica; and for A. thaliana NCX proteins in P. trichocarpa and S. tuberosum, Blastp search was performed in respective proteome database in phytozome version 10.1.
Prediction of putative transmembrane topology of NCX proteins from rice and Arabidopsis
The putative transmembrane topology for longest ORF of each of the NCX proteins was predicted with the help of PROTTER version 1.0 (Omasits et al. 2014; http://wlab.ethz.ch/protter/start/).
Phylogenetic analysis
Multiple sequence alignment was performed using ClustalW2 (Larkin et al. 2007) and phylogenetic tree for NCX proteins was plotted using MEGA6 software (Tamura et al. 2013) using Neighbour-joining method (Saitou and Nei 1987) with 1000 bootstrap replicates. Rest of the parameters used in MEGA 6 were set to default.
Expression analysis using MPSS database
Expression data from MPSS tags was retrieved from the Arabidopsis and rice MPSS project websites (http://mpss.udel.edu). The signature was considered to be significant if it uniquely identifies an individual gene and shows perfect match (100 % identity over 100 % length of the tag). The normalized abundance (tags per million, tpm) of these signatures for a given gene in a given library represents a quantitative estimate of expression of that gene. Descriptions of the MPSS libraries of A. thaliana and O. sativa are provided in Additional file 5: Table S4 and Additional file 6: Table S5, respectively. Heatmaps were generated using expression values obtained from MPSS database for respective NCX genes with the help MeV software (http://www.tm4.org/mev.html, Eisen et al. 1998).
Expression analysis using microarray data
Expression profile of Arabidopsis NCX genes was analyzed using microarray data generated through AtGenExpress (https://www.arabidopsis.org/portals/expression/microarray/ATGenExpress.jsp) under different abiotic stress conditions such as, salt, drought, osmotic, cold, heat, oxidative, genotoxic, wounding and UV/B stress (Kilian et al. 2007) and various developmental stages (Schmid et al. 2005). Microarray data was obtained for different stresses at different time points viz 0.5 h, 1 h, 3 h, 6 h, 12 h and 24 h for both root and shoot tissues. Fold change at transcript level of different genes under stress was calculated with respect to their controls. For the developmental stage data, Affymetrix values were log10 transformed, heat maps generated and hierarchical clustering done using the MeV software package (Eisen et al. 1998). For microarray analysis of rice NCX genes, Affymetrix GeneChip rice genome arrays (http://www.ncbi.nlm.nih.gov/geo/; Gene Expression Omnibus platform accession nos. GSE6893) was used. The Affymetrix values were log2 transformed and heatmaps generated using TIGR MeV software (Eisen et al. 1998). Microarray dataset in rice is described as: Roots of 7- day old seedling, Mature leaf (collected before pollination), young leaf, SD-7-day old seedling, P1 (0-3 cm panicle), P2 (3–5 cm panicle), P3 (5–10 cm panicle), P4 (10–15 cm panicle), P5 (15–22 cm panicle), P6 (22–30 cm panicle), S1 (0–2 DAP), S2 (3–4 DAP), S3 (5–10 DAP), S4 (11–20 DAP) and S5 (21–29 DAP).
Plant material and stress treatment for qRT-PCR analysis
Seeds of IR64 rice cultivar were germinated in hydroponic system. The seedlings were grown under control conditions in Yoshida medium (Yoshida et al. 1972) in a growth chamber at 28 ± 2 °C and 16 h photoperiod. After 10 days, various stress treatments were given to seedlings viz. salinity stress (200 mM NaCl), dehydration (air dry), oxidative stress (10 μM methyl viologen), heat stress (42 °C), cold stress (4 °C), UV stress (UV-B exposure), calcium (100 mM CaCl2) or calcium chelator (10 mM EGTA). The duration of all the stress treatments was 6 h and untreated seedlings were taken as the experimental control. After 6 h, shoots were cut, weighed and frozen in liquid nitrogen for further use.
Total RNA was isolated from the frozen stressed and control shoot samples using RaFlex™ solution I and solution II (Bangalore Genei, India) according to the manufacturer’s protocol. Total RNA was used for first strand cDNA synthesis using RevertAidTM RNAse H minus cDNA synthesis kit as per manufacturer’s protocol (Fermentas Life Sciences, USA). Primers were designed from 3′ UTR unique region for each of the rice NCX gene using Primer3 software. Sequences of the primers have been provided as Additional file 7: Table S6. For qRT- PCR, the reaction mixture comprised 5 μl of 10 times diluted cDNA, 12.5 μl of 2X SYBR Green PCR Master Mix (Applied Biosystems, USA) and 200 nM of each gene specific primer in a final volume of 25 μl reaction mixture in 48 well optical reaction plates (Applied Biosystems, USA). Applied Biosystems Step OneTM Real time PCR machine was used to perform qRT-PCR. The PCR conditions for all the genes were kept same i.e. 10 min at 95 °C and 35 cycles of 15 s at 95 °C, 30 s at 55 °C and 30 s at 72 °C. Melt curve analysis was performed for each experiment for testing the specificity of the amplification. The relative expression value was calculated by using REST 2009 software (Vandesompele et al. 2002). At least two biological replicates and three technical replicates were analyzed for qRT-PCR.
Acknowledgements
SLS-P acknowledges financial support received from ICGEB, DBT and CSIR. AKT thanks the Department of Biotechnology, Government of India for awarding Senior Research Fellowship.
Abbreviations
- NCX
Sodium/Calcium exchanger
- CAX
Calcium/proton exchangers
- EF
EF hand
- EGTA
Ethylene glycol tetraacetic acid
- MV
Methyl viologen
- UV
Ultra violet
- TMS
Transmembrane segments
Additional files
Phylogenetic relationships among Arabidopsis and rice NCX proteins. Unrooted tree was generated using MEGA6 software using the Neighbor-joining method with 1000 bootstrap replicates. NCX proteins are categorized into five different clades depending upon the relative sequence homology of each member of the clades. Each clade is denoted by different color shading.
List of orthologous proteins of Arabidopsis NCX proteins in poplar and potato and rice NCX proteins in Brachypodium and foxtail millet. Orthologous proteins were identified in Phytozome 10.1 against database of respective species using Blastp search function.
Position of CAX, NCX and EF hand domains in NCX proteins of Brachypodium, foxtail millet, poplar and potato.
Orthologous proteins of Arabidopsis NCXs in rice alongwith locus IDs of their respective genes.
Description of Arabidopsis MPSS libraries.
Description of rice MPSS libraries.
List of primers used for qRT-PCR analysis and their sequence, and the expected amplicon size.
Footnotes
Anil Kumar Singh and Ritesh Kumar contributed equally to this work.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
SLS-P and AP conceived the study, planned the experiments and finalized the manuscript. AKS and RK carried out the experiments and took part in drafting of the manuscript. AKT and BKG participated in data interpretation and in writing of the manuscript. All authors read and approved the final manuscript.
Contributor Information
Anil Kumar Singh, Email: anils13@gmail.com.
Ritesh Kumar, Email: kumarritesh1983@gmail.com.
Amit K. Tripathi, Email: akt@icgeb.res.in
Brijesh K. Gupta, Email: brijgene@gmail.com
Ashwani Pareek, Email: ashwanip@mail.jnu.ac.in.
Sneh L. Singla-Pareek, Phone: +91-11-26741358, Email: sneh@icgeb.res.in
References
- Bindokas VP, Miller RJ. Excitotoxic degeneration is initiated at non random sites in cultured rat cerebellar neurons. J Neurosci. 1995;15:6999–7011. doi: 10.1523/JNEUROSCI.15-11-06999.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bootman MD, Berridge MJ. The elemental principles of calcium signaling. Cell. 1995;83:675–678. doi: 10.1016/0092-8674(95)90179-5. [DOI] [PubMed] [Google Scholar]
- Bowers JE, Chapman BA, Rong J, Paterson AH. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature. 2003;422:433–438. doi: 10.1038/nature01521. [DOI] [PubMed] [Google Scholar]
- Cai X, Lytton J. The cation/Ca2+ exchanger superfamily: Phylogenetic analysis and structural implications. Mol Biol Evol. 2004;21:1692–1703. doi: 10.1093/molbev/msh177. [DOI] [PubMed] [Google Scholar]
- Carafoli E, Santella L, Branca D, Brini M. Generation, control, and processing of cellular calcium signals. Crit Rev Biochem Mol Biol. 2001;36:107–260. doi: 10.1080/20014091074183. [DOI] [PubMed] [Google Scholar]
- DiPolo R, Beauge L. Sodium/calcium exchanger: Influence of metabolic regulation on ion carrier interactions. Physiol Rev. 2006;86:155–203. doi: 10.1152/physrev.00018.2005. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery L, Whelan S, Hirschi KD, Pittman JK. Protein phylogenetic analysis of Ca(2+)/cation antiporters and insights into their evolution in plants. Front Plant Sci. 2012;3:1. doi: 10.3389/fpls.2012.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermolaeva MD, Wu MM, Eisen JA, Salzberg SL. The age of the Arabidopsis thaliana genome duplication. Plant Mol Biol. 2003;51:859–866. doi: 10.1023/A:1023001130337. [DOI] [PubMed] [Google Scholar]
- Evans NH, McAinsh MR, Hetherington AM. Calcium oscillations in higher plants. Curr Opin Plant Biol. 2001;4:415–420. doi: 10.1016/S1369-5266(00)00194-1. [DOI] [PubMed] [Google Scholar]
- Flagel LE, Wendel JF. Gene duplication and evolutionary novelty in plants. New Phytol. 2009;183:557–564. doi: 10.1111/j.1469-8137.2009.02923.x. [DOI] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, Rokhsar DS. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie RC. INDO-1 measurements of absolute resting and light induced calcium concentration in Drosophila photoreceptors. J Neurosci. 1966;16:2924–2933. doi: 10.1523/JNEUROSCI.16-09-02924.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haug-Collet K, Pearson B, Webel B, Szerencsei RT, Winkfein RJ, Schnetkamp PPM, Colley NJ. Cloning and characterization of a potassium-dependent sodium/calcium exchanger in Drosophila. J Cell Biol. 1999;147:659–669. doi: 10.1083/jcb.147.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepler PK, Wayne RO. Calcium and plant development. Annu Rev Plant Physiol. 1985;36:395–439. doi: 10.1146/annurev.pp.36.060185.002145. [DOI] [Google Scholar]
- Hirschi K, Zhen R, Cunningham KW, Rea PA, Fink GR. CAX1, a H+/Ca2+ antiporter from Arabidopsis. Proc Natl Acad Sci U S A. 1996;93:8782–8786. doi: 10.1073/pnas.93.16.8782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JG, Yang M, Liu P, Yang GD, Wu CA, Zheng CC. Genome-wide profiling of developmental, hormonal or environmental responsiveness of the nucleocytoplasmic transport receptors in Arabidopsis. Gene. 2010;451:38–44. doi: 10.1016/j.gene.2009.11.009. [DOI] [PubMed] [Google Scholar]
- Itoh H, Sasaki A, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Hasegawa Y, Minami E, Ashikari M, Matsuoka M. Dissection of the phosphorylation of rice DELLA protein, SLENDER RICE1. Plant Cell Physiol. 2005;46:1392–1399. doi: 10.1093/pcp/pci152. [DOI] [PubMed] [Google Scholar]
- Iwamoto T, Nakamura TY, Pan Y, Uehara A, Imanaga I, Shigekawa M. Unique topology of the internal repeats in the cardiac Na+/Ca2+ exchanger. FEBS Lett. 1999;446:264–268. doi: 10.1016/S0014-5793(99)00218-5. [DOI] [PubMed] [Google Scholar]
- Jain M, Tyagi AK, Khurana JP. Genome-wide analysis, evolutionary expansion, and expression of early auxin-responsive SAUR gene family in rice (Oryza sativa) Genomics. 2006;88:360–371. doi: 10.1016/j.ygeno.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Jon K, Pittman CE, Paul AS, Clifford MB. A cation-regulated and proton gradient-dependent cation transporter from Chlamydomonas reinhardtii has a role in calcium and sodium homeostasis. J Biol Chem. 2008;284:525–533. doi: 10.1074/jbc.M807173200. [DOI] [PubMed] [Google Scholar]
- Kamiya T, Akahori T, Ashikari M, Maeshima M. Expression of the vacuolar Ca2+/H+ exchanger, OsCAX1a, in rice: cell and age specificity of expression, and enhancement by Ca2+ Plant Cell Physiol. 2006;47:96–106. doi: 10.1093/pcp/pci227. [DOI] [PubMed] [Google Scholar]
- Kamiya T, Akahori T, Maeshima M. Expression profile of the genes for rice cation/H+ exchanger family and functional analysis in yeast. Plant Cell Physiol. 2005;46:1735–1740. doi: 10.1093/pcp/pci173. [DOI] [PubMed] [Google Scholar]
- Kamiya T, Maeshima M. Residues in internal repeats of the rice cation/H+ exchanger are involved in the transport and selection of cations. J Biol Chem. 2004;279:812–819. doi: 10.1074/jbc.M309726200. [DOI] [PubMed] [Google Scholar]
- Kilian J, Whiteland D, Horak J, Wanke D, Weini S, Batistic O, D’Angelo C, Bornberg-Bauer E, Kudla J, Harter K. The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 2007;50:347–363. doi: 10.1111/j.1365-313X.2007.03052.x. [DOI] [PubMed] [Google Scholar]
- Kumari S, Punjabi-Sabharwal V, Khushwaha HR, Sopory SK, Singla-Pareek SL, Pareek A. Transcriptome map for seedling stage specific salinity response indicates a specific set of genes as candidate for saline tolerance in Oryza sativa L. Funct Integr Genomics. 2009;9:109–123. doi: 10.1007/s10142-008-0088-5. [DOI] [PubMed] [Google Scholar]
- Kushwaha HR, Singh AK, Sopory SK, Singla-Pareek SL, Pareek A. Genome wide expression analysis of CBS domain containing proteins in Arabidopsis thaliana (L.) Heynh and Oryza sativa L. reveals their developmental and stress regulation. BMC Genomics. 2009;10:200. doi: 10.1186/1471-2164-10-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, Mc William H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. ClustalW and ClustalX version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Le DT, Nishiyama R, Watanabe Y, Mochida K, Yamaguchi-Shinozaki K, Shinozaki K, Tran LS. Genome-wide survey and expression analysis of the plant-specific NAC transcription factor family in soybean during development and dehydration stress. DNA Res. 2011;18:263–276. doi: 10.1093/dnares/dsr015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Li P, Wang Y, Dong R, Yu H, Hou B. Genome-wide identification and phylogenetic analysis of Family-1 UDP glycosyltransferases in maize (Zea mays) Planta. 2014;239:1265–1279. doi: 10.1007/s00425-014-2050-1. [DOI] [PubMed] [Google Scholar]
- Marshall CR, Fox JA, Butland SL, Ouellette BF, Brinkman FS, Tibbits GF. Phylogeny of Na+/Ca2+ exchanger (NCX) genes from genomic data identifies new gene duplications and a new family member in fish species. Physiol Genomics. 2005;21:161–173. doi: 10.1152/physiolgenomics.00286.2004. [DOI] [PubMed] [Google Scholar]
- Mäser P, Thomine S, Schroeder JI, Ward JM, Hirschi K, Sze H, Talke IN, Amtmann A, Maathuis FJ, Sanders D, Harper JF, Tchieu J, Gribskov M, Persans MW, Salt DE, Kim SA, Guerinot ML. Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol. 2001;126:1646–1667. doi: 10.1104/pp.126.4.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean AEM, McLean E, Judah JD. Cellular necrosis in liver induced and modified by drugs. Int Rev Exp Pathol. 1965;4:127–157. [PubMed] [Google Scholar]
- Mustafiz A, Singh AK, Pareek A, Sopory SK, Singla-Pareek SL. Genome-wide analysis of rice and Arabidopsis identifies two glyoxalase genes that are highly expressed in abiotic stresses. Funct Integr Genomics. 2011;11:293–305. doi: 10.1007/s10142-010-0203-2. [DOI] [PubMed] [Google Scholar]
- Nicoll DA, Ottolia M, Lu L, Lu Y, Philipson KD. A new topological model of the cardiac sarcolemmal Na+-Ca2+ exchanger. J Biol Chem. 1999;274:910–917. doi: 10.1074/jbc.274.2.910. [DOI] [PubMed] [Google Scholar]
- Nijhawan A, Jain M, Tyagi AK, Khurana JP. Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol. 2008;146:333–350. doi: 10.1104/pp.107.112821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- On C, Marshall CR, Chen N, Moyes CD, Tibbits GF. Gene structure evolution of the Na+-Ca2+ exchanger (NCX) family. BMC Evol Biol. 2008;8:127. doi: 10.1186/1471-2148-8-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omasits U, Ahrens CH, Müller S, Wollscheid B. Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics. 2014;30:884–886. doi: 10.1093/bioinformatics/btt607. [DOI] [PubMed] [Google Scholar]
- Pareek A, Singh A, Kumar M, Kushwaha HR, Lynn AM, Singla-Pareek SL. Whole-genome analysis of Oryza sativa reveals similar architecture of two-component signaling machinery with Arabidopsis. Plant Physiol. 2006;142:380–397. doi: 10.1104/pp.106.086371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peretz A, Suss T, Rom G, Arnon A, Payne R, Minke B. The light response of Drosophila photoreceptors is accompanied by an increase in cellular calcium: effects of specific mutations. Neuron. 1994;12:1257–1267. doi: 10.1016/0896-6273(94)90442-1. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohman JU. A gene expression map of Arabidopsis thaliana development. Nat Genet. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- Shaul O, Hilgemann DW, de-Almeida-Engler J, Van Montagu M, Inz D, Galili G. Cloning and characterization of a novel Mg(2+)/H(+) exchanger. EMBO J. 1999;18:3973–3980. doi: 10.1093/emboj/18.14.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigaki T, Rees I, Nakhleh L, Hirschi KD. Identification of three distinct phylogenetic groups of CAX Cation/Proton antiporters. J Mol Evol. 2006;63:815–825. doi: 10.1007/s00239-006-0048-4. [DOI] [PubMed] [Google Scholar]
- Siegel GJ, Agranoff BW, Albers RW, Fisher SK, Uhler . Basic Neurochemistry: Molecular, Cellular and Medical aspects: 6th ed: Lippincott. Philadelphia: Williams & Wilkins; 1999. [Google Scholar]
- Singh AK, Sharma V, Pal AK, Acharya V, Ahuja PS. Genome-wide organization and expression profiling of NAC transcription factor family in potato (Solanum tuberosum L.) DNA Res. 2013;20:403–423. doi: 10.1093/dnares/dst019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun ZM, Zhou ML, Xiao XG, Tang YX, Wu YM. Genome-wide analysis of AP2/ERF family genes from Lotus corniculatus shows LcERF054 enhances salt tolerance. Funct Integr Genomics. 2014;14:453–466. doi: 10.1007/s10142-014-0372-5. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi AK, Singh K, Pareek A, Singla-Pareek SL. Histone chaperones in Arabidopsis and rice: genome-wide identification, phylogeny, architecture and transcriptional regulation. BMC Plant Biol. 2015;15:42. doi: 10.1186/s12870-015-0414-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, Katleen DP, Filip P, Bruce P, Nadine VR, Anee DP, Frank S. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3. Research. 2002;0034:1–0034.11. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Li Z, Wei J, Zhao Z, Sun D, Cui S. A Na+/Ca2+ exchanger-like protein (AtNCL) involved in salt stress in Arabidopsis. J Biol Chem. 2012;287:44062–44070. doi: 10.1074/jbc.M112.351643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Shi X, Hao B, Ge S, Luo J. Duplication and DNA segmental loss in the rice genome: implications for diploidization. New Phytol. 2005;165:937–946. doi: 10.1111/j.1469-8137.2004.01293.x. [DOI] [PubMed] [Google Scholar]
- Wei KF, Chen J, Chen YF, Wu LJ, Xie DX. Molecular phylogenetic and expression analysis of the complete WRKY transcription factor family in maize. DNA Res. 2012;19:153–164. doi: 10.1093/dnares/dsr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf JA, Stys PK, Lusardi T, Meaney D, Smith DH. Traumatic axonal injury induces calcium influx modulated by tetrodotoxin-sensitive sodium channels. J Neurosci. 2001;21:1923–1930. doi: 10.1523/JNEUROSCI.21-06-01923.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue T, Wang D, Zhang S, Ehlting J, Ni F, Jakab S, et al. Genome-wide and expression analysis of protein phosphatase 2C in rice and Arabidopsis. BMC Genomics. 2008;9:550. doi: 10.1186/1471-2164-9-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Forno DA, Cock JH, Gomez KA. Laboratory Manual for Physiological Studies of Rice. Manila: International Rice Research Institute; 1972. [Google Scholar]
- Yu SP, Choi DW. Na+–Ca2+ exchange currents in cortical neurons: concomitant forward and reverse operation and effect of glutamate. Eur J Neurosci. 1997;9:1273–1281. doi: 10.1111/j.1460-9568.1997.tb01482.x. [DOI] [PubMed] [Google Scholar]


