Abstract
The genetic code allows most amino acids a choice of optimal and nonoptimal codons. We report that synonymous codon choice is tuned to promote interaction of nascent polypeptides with the signal recognition particle (SRP), which assists in protein translocation across membranes. Cotranslational recognition by the SRP in vivo is enhanced when mRNAs contain nonoptimal codon clusters 35–40 codons downstream of the SRP-binding site, the distance that spans the ribosomal polypeptide exit tunnel. A local translation slowdown upon ribosomal exit of SRP-binding elements in mRNAs containing these nonoptimal codon clusters is supported experimentally by ribosome profiling analyses in yeast. Modulation of local elongation rates through codon choice appears to kinetically enhance recognition by ribosome-associated factors. We propose that cotranslational regulation of nascent-chain fate may be a general constraint shaping codon usage in the genome.
Understanding how newly synthesized proteins fold in the cell remains a fundamental problem in biology. Nascent polypeptides must interact cotranslationally with ribosome-associated chaperones and factors assisting in folding, translocation and quality control1. The determinants establishing cotranslational specificity in ribosome nascent-chain recognition by these factors are poorly understood. The roles of translation itself and of the mRNA sequences are particularly unexplored. The genetic code allows most amino acids a choice of optimal and nonoptimal codons, which are translated at different speeds2–4. Here we examine whether synonymous codon choice is tuned to promote interaction of nascent polypeptides with the SRP, which assists in protein translocation across membranes.
Up to a third of the eukaryotic proteome is translocated into the endoplasmic reticulum (ER). Most translocation in eukaryotes involves the cotranslational action of the SRP, a key protein-biogenesis factor acting on nascent chains to ensure their efficient delivery to the secretory pathway5–8. SRP recognizes hydrophobic N-terminal signal sequences (SSs) or transmembrane (TM) segments in the translating polypeptides after they emerge from the ribosome exit tunnel9,10. SRP-bound ribosome nascent chain complexes (RNCs) are then transferred via the SRP receptor to a translocon channel in the ER membrane5,6. Although SRP-independent post-translational translocation can also occur11, cotranslational translocation advantageously limits the cytoplasmic exposure of aggregation-prone, hydrophobic SS and TM segments12, thereby coupling protein synthesis and export.
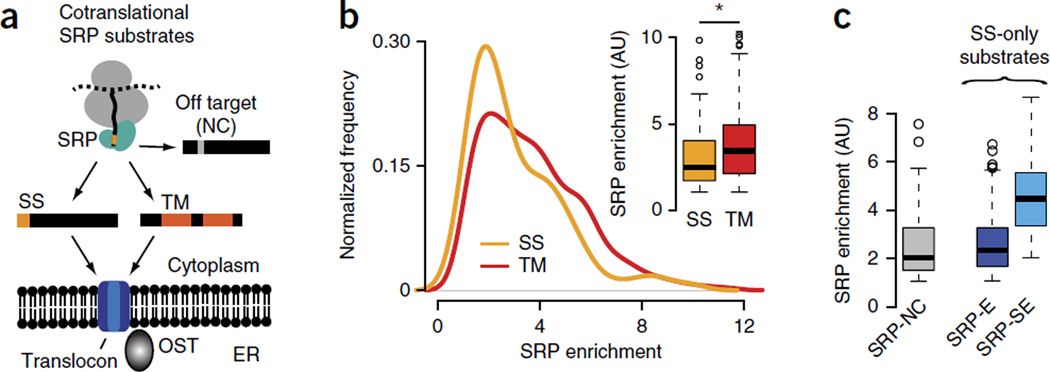
The SRP offers a unique opportunity to understand the determinants of cotranslational recognition of RNCs by ribosome-associated factors, given the extensive understanding of SRP-recognition sites in nascent polypeptides. All SSs share a short, positively charged N-terminal region followed by a tract of ~8–15 hydrophobic amino acids and capped by a cleavage site13; membrane-spanning TM segments contain longer hydrophobic stretches of ~20 amino acids. The hydrophobic groove in SRP54 (ref. 9) binds a minimum of 8 or 9 hydrophobic amino acids in the substrate14. Despite this understanding, the determinants establishing the specificity of the SRP in vivo remain enigmatic in light of several puzzling observations. First, beyond a minimal hydrophobicity threshold15, SRP binding is quite tolerant to sequence variation. SSs share little sequence similarity and are often highly divergent16. Strikingly, ~20% of random sequences can act in vivo as SSs when fused to the N terminus of invertase17. Second, the SRP can bind to secretory nascent polypeptides without canonical SSs18 while ignoring other substrates with canonical SSs11. Third, the SRP can bind in vitro with nanomolar affinity to ribosomes translating cytoplasmic proteins without SSs or TM helices19. Accordingly, the SRP could in principle interact with many cytoplasmic proteins in vivo. Despite these observations, analysis of the cotranslational specificity of the SRP in vivo has indicated a high degree of selectivity for bona fide substrates containing SSs and TM helices20. Indeed, most cotranslational SRP substrates in vivo are secretory and membrane proteins (Fig. 1a), and few off-target noncognate proteins bind the SRP.
Figure 1.
The SRP binds nascent chains in vivo with a broad distribution of specificities. (a) Schematics of selective, cotranslational SRP interaction with proteins bearing signal sequences (SS) or transmembrane (TM) segments. The SRP also binds few noncognate (NC) cytosolic and nuclear off-target proteins that do not get translocated. (b) Enrichment scores of cotranslational SRP binding for SS proteins (i.e., proteins with an N-terminal SS) and for TM proteins (i.e., proteins with TM segments as SRP-binding sites). Inset, significantly higher enrichment of SRP binding in TM proteins (n = 190) compared to SS proteins (n = 107) (P = 0.0007 by Wilcoxon rank-sum test). (c) Classification of SS proteins into noncognate (SRP-NC; n = 109), enriched (SRP-E; n = 78) and strongly enriched (SRP-SE; n = 29) substrates, on the basis of increasing SRP enrichment scores. Box plots show the data distribution through median (center line), first and third quartiles (filled box), 1.5× interquartile range (dashed line) and extreme values (circles). AU, arbitrary units. *P < 0.05.
The above observations suggest that the presence of an SS or TM segment may not be the only determinant driving SRP recognition. A quantitative analysis of cotranslational SRP-substrate interactions in vivo20 in yeast cells revealed that some secretory and transmembrane proteins are more enhanced in SRP binding than others (Fig. 1b; data set from ref. 20). Although RNCs containing the longer TM segments as the first putative SRP-binding site are, in general, significantly more enriched in SRP binding than nascent chains with an SS (P = 0.0007 by Wilcoxon rank-sum test; Fig. 1b; data set from ref. 20), there is a wide range in the observed enrichment of SRP interaction within proteins with either SS or TM domains (Fig. 1b). Analysis of this unique in vivo interaction data set offers an opportunity to identify global determinants governing nascent-chain recognition by the SRP and may offer general insight into the recognition of nascent chains by cotranslationally acting factors.
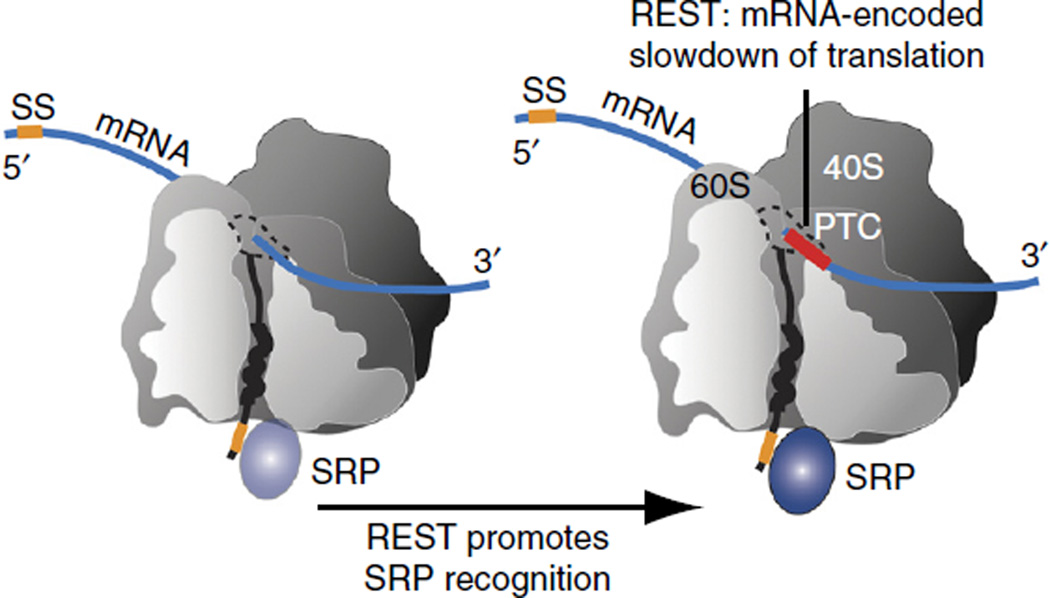
Here we show that in vivo cotranslational recognition by the SRP is enhanced when the mRNA encoding the secretory polypeptide contains a cluster of nonoptimal codons (~35–40 codons) downstream of the SRP-binding site, a distance long enough to span the ribosomal exit tunnel21. Ribosome profiling analyses corroborate that preferential SRP recognition is linked to nonoptimal codons causing a local slowdown of translation upon ribosomal exposure of SRP-binding elements. Our results thus suggest that mRNAs contain sequence elements, which we propose to name ‘REST’, for mRNA-encoded slowdown of translation, that modulate local elongation rates to enhance recognition by ribosome-associated factors and regulate nascent chain fate.
RESULTS
SRP nascent-chain enrichment in vivo is not explained by the SS
To analyze substrates with SRP-binding sites of comparable length, we first focused on SS-containing proteins (Fig. 1 and Supplementary Fig. 1). We defined two sets of SS proteins on the basis of their in vivo SRP binding enrichment (Fig. 1c and Supplementary Fig. 1): those enriched (SRP-E) and those strongly enriched (SRP-SE) in SRP. We also analyzed noncognate interactors (SRP-NC), which are cytosolic, mitochondrial and nuclear proteins that lack an SS or TM domain but nonetheless bind SRP in vivo, albeit at lower enrichment (Fig. 1c). For SRP-NC proteins, we examined the longest N-terminal hydrophobic stretch in their sequences. The decreasing preference for SRP association in these curated sets, ranging from SRP-SE to SRP-E to SRP-NC, allowed the systematic analysis of determinants promoting SRP recognition. We examined the overall hydrophobicity, maximum hydrophobicity and sequence information content in SSs with distinctly higher enrichment scores (Supplementary Fig. 2). Strikingly, SS hydrophobicity, putative binding motifs and distinct sequence patterns could not explain the in vivo differences in SRP interaction observed for SRP-SE and SRP-E proteins, thus suggesting that the properties of the SS do not play a major part in the observed SRP enrichment (Supplementary Fig. 2).
SRP-SE OST subunits contain a translational-efficiency dip
Because individual codons are translated at different speeds2–4, and the SRP acts cotranslationally, we considered whether local elongation rates have a role in SRP recognition. Modulation of local translation kinetics through optimal (faster translated) and nonoptimal (more slowly translated) codons has been linked to cotranslational protein folding22–25. We computed mRNA translational efficiency (TE) profiles for all SRP substrates, using a scale that incorporates the competition between tRNA supply and demand25,26 (Supplementary Fig. 3 and Supplementary Table 1).
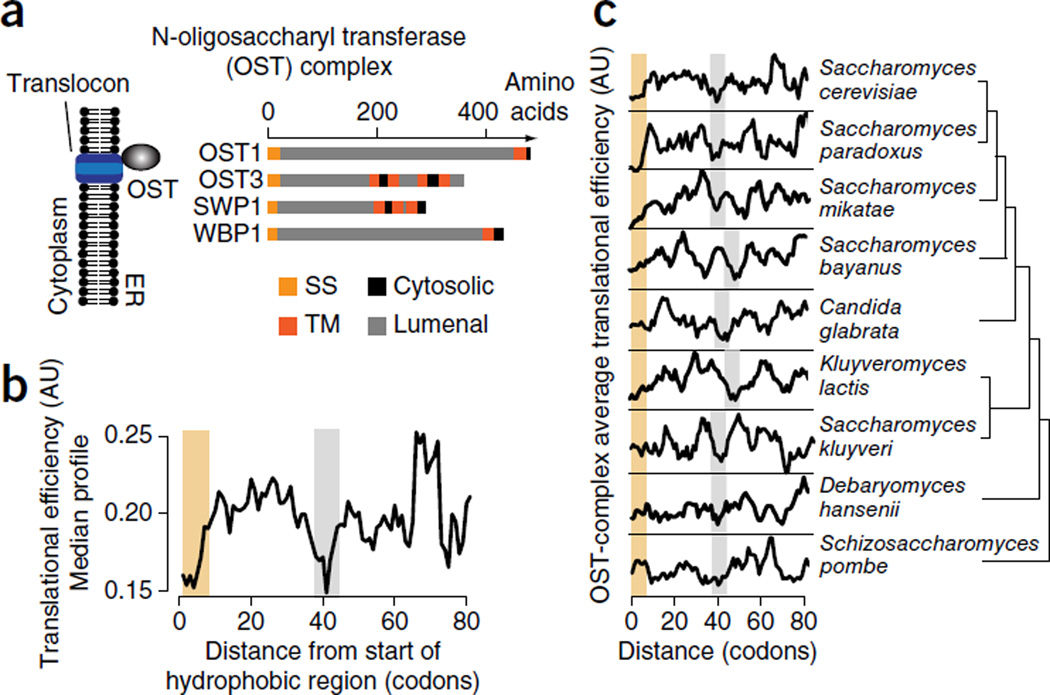
We first examined individual TE profiles for the strongest SRP interactors, namely subunits of the essential oligosaccharyl transferase (OST) complex (Supplementary Fig. 4a). OST subunits OST1, OST3, SWP1 and WBP1 have a unique topology consisting of an SS followed by a large N-terminal lumenal domain (Fig. 2a and Supplementary Fig. 4b). Accordingly, their SSs may be under strong selective pressure for efficient cotranslational recognition to ensure translocation of the N-terminal domains. The median TE profile of the mRNAs encoding these OST subunits revealed two regions surrounding the SS enriched in nonoptimal codons, which would be slowly translated (Fig. 2b). The beginning of the coding sequence contained a previously described low-TE ramp linked to facilitating translation initiation27. The second region of low TE occurred ~40 codons downstream of the start of the SS (Fig. 2b). Given the ribosomal tunnel length of ~30–35 amino acids21, this low-TE region seems optimally positioned to slow elongation just as the hydrophobic SS emerges from the ribosome exit tunnel. Of note, this local dip in TE is evolutionarily conserved across closely related yeasts, thus suggesting an important functional role (Fig. 2c). This led us to hypothesize that enhanced SRP interaction responds to a genetically programmed slowdown in elongation when the SS emerges from the ribosome. We also examined the TE of invertase, the model substrate that could accommodate many random sequences as a functional SS17. Intriguingly, we also found a distinct region of low TE within its coding sequence, ~34–40 codons downstream from the SS (Supplementary Fig. 4c); this could account for the puzzling ability of many diverse sequences with suboptimal binding sites to mediate SRP interaction.
Figure 2.
An evolutionarily conserved local dip in translational efficiency characterizes the SRP-dependent translocation of the essential N-oligosaccharyl transferase (OST). (a) Four of eight OST subunits, namely OST1, OST3, SWP1 and WBP1, have a distinct topology of an SS followed by a long lumenal domain. (b) Median translational efficiency (TE) profile of the OST subunits OST1, OST3, SWP1 and WBP1, showing a distinct region of low TE (gray bar) ~40 codons downstream of the start of the hydrophobic region of the SS (yellow bar). (c) Evolutionary conservation of the region of low TE (gray bars) ~40 codons downstream of the start of the hydrophobic region of the SS (yellow bars) in OST subunits across closely related yeast species. TE profiles show predicted relative translation speeds and are in arbitrary units (AU).
mRNA-encoded translation slowdown promotes SS recognition
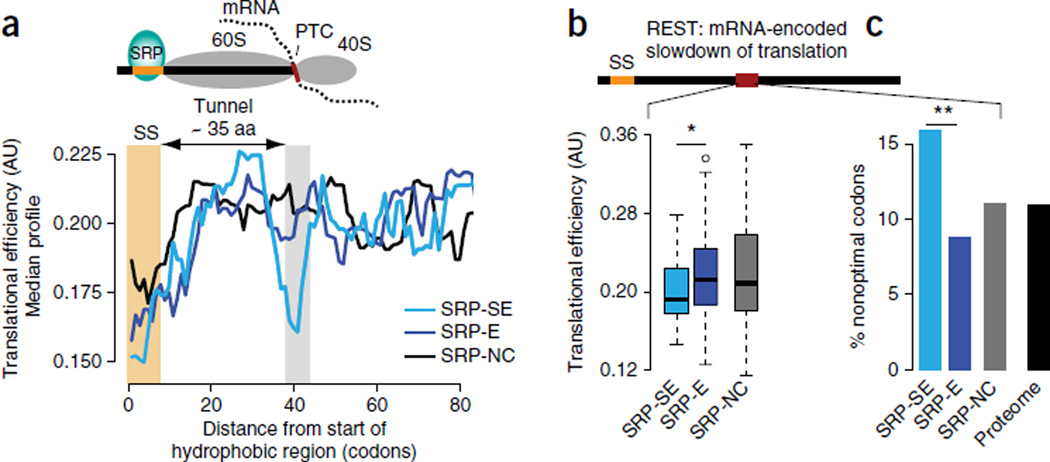
We next calculated the median TE for all SRP-SE proteins and compared it to the median TEs of SRP-E and SRP-NC substrates. Remarkably, strongly enriched SRP substrates systematically had a low-TE region located ~38–43 codons downstream of the SS, which we did not observe in SRP-E or SRP-NC substrates (Fig. 3a). Thus, this region of lower codon optimality translated when the SS emerges from the ribosome is a shared feature of SRP-enriched substrates. Correspondingly, individual mRNAs encoding SRP-SE substrates, but not SRP-E interactors, contained, on average, regions downstream of the SS with significantly lower TE (P = 0.02 by Wilcoxon rank-sum test; Fig. 3b). Of note, the lower TE in SRP-SE substrates can be directly traced to a local enrichment in nonoptimal codons (P = 0.002 by Fisher’s exact test; Fig. 3c). The cytosolic SRP-NC substrates show, on average, an intermediate TE in this region, which is absent from cytosolic proteins that have a long hydrophobic stretch but do not bind to the SRP (Fig. 3b and Supplementary Fig. 4d,e); this may contribute to their nonspecific interactions with the SRP. Our results suggest that mRNAs encoding nascent chains that bind strongly to the SRP in vivo contain a strategically placed sequence element that slows the elongation rate to coordinate translation with nascent-chain recognition by the ER translocation factor SRP. We propose calling this type of element REST, for mRNA-encoded slowdown of translation. It is noteworthy that SRP-SE substrates exhibit lower local translational efficiencies downstream of the signal sequences (Supplementary Fig. 4) even though they are more highly expressed and thus have overall higher translational efficiencies26. This suggests that for these mRNAs the lower TE fulfills a regulatory function, thus further supporting the importance of the REST elements in strongly enriched SRP substrates.
Figure 3.
Slowdown of translation by nonoptimal codons promotes SRP recognition. (a) Top, schematic. Bottom, median TE profiles for each class of SRP substrates, showing a dip in TE in the downstream region (gray bar) in the mRNA sequences of all SRP-SE (n = 29) substrates, but not those of SRP-E (n = 78) and SPR-NC (n = 109), just after emergence of the hydrophobic SRP-binding site from the ribosome exit tunnel. Aa, amino acids. (b) Statistical analysis showing significantly lower average TE for individual SRP-SE substrates in the translational-slowdown element encoded in the mRNA sequences downstream of the SS, compared to SRP-E substrates (P = 0.02 by Wilcoxon rank-sum test; n values as in a). (c) Enrichment in nonoptimal codons. The translational-slowdown element in the mRNA sequences of SRP-SE substrates is enriched in nonoptimal codons (P = 0.002 by Fisher’s exact test on counts of optimal and nonoptimal codons; two sided, with n values as in a). Box plots show the data distribution through median (center line), first and third quartiles (filled box), 1.5× interquartile range (dashed line) and extreme values (circles). *P < 0.05; **P < 0.01.
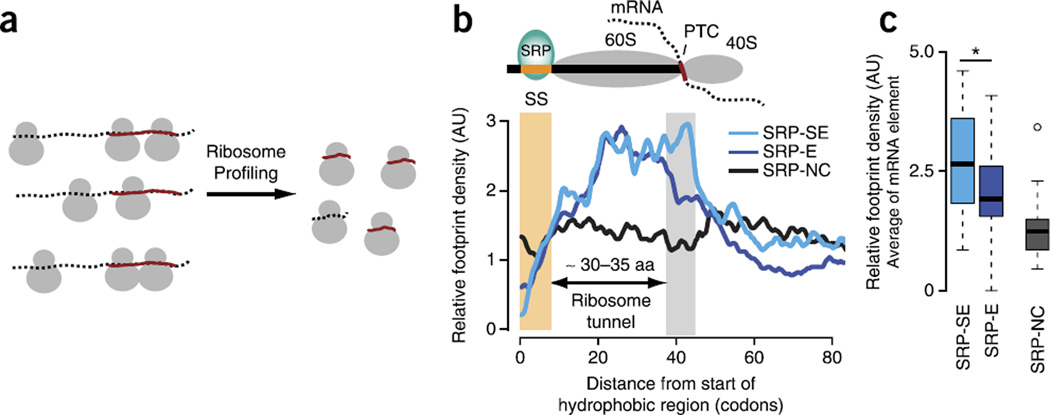
Translation elongation kinetics can be experimentally measured at high resolution by ribosome profiling, wherein slower local translation produces higher densities of ribosome-protected footprints28 (Fig. 4a and Supplementary Fig. 5a,b). We assessed the presence of a local translation slowdown occurring in mRNAs encoding SRP-SE but not in SRP-E substrates, by using ribosome profiling data29. Indeed, the SRP-SE substrates exhibited higher average ribosome footprint densities ~35–40 codons downstream of the SS (Fig. 4b), the region predicted to be more slowly translated by TE analysis. The footprint densities for individual proteins in this downstream region were, on average, significantly higher for SRP-SE than for SRP-E (P = 0.042 by Wilcoxon rank-sum test; Fig. 4c and Supplementary Fig. 5c,d) and SRP-NC substrates, results also consistent with our TE analysis. An additional buildup of increased ribosome footprint densities from around position 20, shared by all substrates with cognate SSs, may arise from interactions between hydrophobic and helical signal anchor sequences30 including their N-terminal positive charges31 with the ribosome exit tunnel. Thus, we find good agreement between the low TE predicted in the mRNA region downstream of the SS and the observed slow translation kinetics in this region of the mRNA. It appears that a strategic enrichment in nonoptimal codons in the mRNA through a REST element slows down translation upon ribosomal exit of the SS, thereby kinetically increasing the time window for SRP binding and enhancing recognition of SRP-SE proteins.
Figure 4.
Ribosome profiling confirms a translational slowdown that associates with enhanced SRP binding. (a) Schematic for ribosome profiling. Ribosome profiling experimentally measures local translation kinetics through selective sequencing of ribosome-protected footprints. Higher footprint densities indicate slower local translation kinetics. (b) Median ribosome footprint density profiles (arbitrary units, AU) for SRP-SE (n = 16), SRP-E (n = 33) and SRP-NC (n = 20) substrates. Only SRP-SE substrates exhibit higher footprint densities in the translational-slowdown element (gray bar) downstream of the SS (yellow bar). Aa, amino acids. (c) Distributions of average relative footprint densities in the downstream translational-slowdown element for individual SRP-SE, SRP-E and SRP-NC proteins. SRP-SE substrates are locally translated more slowly compared to SRP-E and SRP-NC substrates just when the hydrophobic SS is exposed outside the ribosome (P = 0.042 by Wilcoxon rank-sum test; n values as in b). Box plots show the data distribution through median (center line), first and third quartiles (filled box), 1.5× interquartile range (dashed line) and extreme values (circles). *P < 0.05.
REST sequence elements in TM proteins
We next examined whether an mRNA-encoded slowdown of translation also promotes SRP binding to TM proteins. For mRNAs encoding TM domains, we examined both their codon optimality (Fig. 5a) and their in vivo footprint densities from ribosome profiling experiments (Fig. 5b). Similarly to what we observed for SS proteins, there was a characteristic region of average lower TE (Fig. 5a) and a corresponding region of higher footprint density (Fig. 5b) downstream of the first TM segment. Surprisingly, the TM segments themselves also exhibited lower local translation rates evident in both the predicted lower TE and the higher ribosome footprint densities (Fig. 5a,b). The slow elongation of the TM domain, which extends its residence time within the exit tunnel, resonates with in vitro experiments suggesting that contacts between TM helices and the tunnel wall may help recruit the SRP from within the ribosome30. Such intraribosomal signaling could promote SRP interaction with TM domains.
Figure 5.
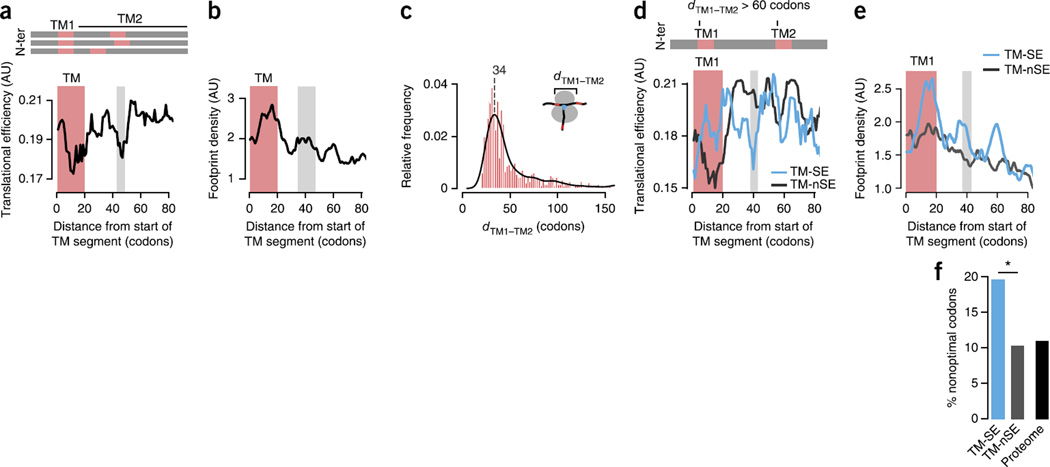
A local slowdown of translation promotes TM-helix recognition by the SRP. (a,b) Median TE (a) and median footprint density (n = 177) (b) profiles for TM proteins (n = 37), showing slower translation of TM segments (red bars) and the region ~40 codons downstream (gray bars). N-ter, N terminus. (c) Distribution of the distance dTM1–TM2 between the start of the first and second TM segments in proteins with multiple TM helices. The most frequent distance is ~34 codons. (d) Median TE profiles for strongly enriched (TM-SE; n = 46) and not strongly enriched (TM-nSE; n = 165) TM proteins with dTM1–TM2 > 60 codons, showing that nonoptimal codons promote SRP recognition independent of the presence of a second TM successive segment. (e) Median ribosome footprint density profiles confirming locally slower translation kinetics for TM-SE (n = 14) substrates compared to TM-nSE (n = 27) substrates. (f) Enrichment in nonoptimal codons at the translational-slowdown element in TM-nSE substrates (P = 0.030 by Fisher’s exact test on counts of optimal and nonoptimal codons; two sided). *P < 0.05.
Proteins often have several TM domains. In light of the lower TEs of TM domains themselves, we examined the average distances between successive TM segments in membrane proteins. Notably, the most frequent distance between the start sites of successive TM helices in TM proteins is ~32–35 codons (Fig. 5c), i.e., the length of the ribosomal tunnel. The spacing may be favored in evolution to exploit the slow translation of the second TM helix and to promote binding of the SRP to the first TM upon emerging from the ribosome, particularly where the first TM segment constitutes the SRP-binding site.
To further examine whether nonoptimal codons promote SRP binding to TM segments without the confounding effect of a second downstream TM helix, we next extracted all TM proteins with a minimum distance of 60 codons between the first and the second TM segment. Because most TM proteins in this data set are strongly enriched in SRP binding (TM-SE; Fig. 1b), we compared them to all TM proteins that were not strongly enriched (TM-nSE). Similarly to what we observed for SS proteins, the TM-SE substrates exhibited a lower predicted TE downstream of the TM segment both in representative median TE profiles (Fig. 5d) and upon analysis of individual mRNAs (P = 0.045 by Wilcoxon rank-sum test; Supplementary Fig. 6a). Analysis of ribosome profiling data for TM-SE substrates also indicated a higher ribosome footprint density ~40 codons downstream of the first TM segment (Fig. 5e and Supplementary Fig. 6b), in agreement with the predicted TE elongation slowdown. Furthermore, the lower TE is linked to the increased presence of nonoptimal codons (P = 0.030 by Fisher’s exact test; Fig. 5f). On average, increased SRP enrichment correlates with the strength of predicted translation slowdown (Supplementary Fig. 6c). Individual examples illustrate these findings. For instance, α-1,6-mannosyltransferase MNN11 has a signal-anchor TM followed by a long lumenal domain (Supplementary Fig. 6d). Efficient and reliable recognition by the SRP is required to avoid aggregation of the lumenal domain in the cytosol. We found a distinct region of low TE (Supplementary Fig. 6e) downstream of the TM, which also results in a higher footprint density in ribosome profiling experiments (Supplementary Fig. 6f). Of note, this low-TE region is also evolutionarily conserved across closely related yeasts (Supplementary Fig. 6g), thus suggesting a functional role that is under selection. Both the dip in predicted TE and the peak in the ribosome footprint density profile are ~55 codons downstream of the start of the TM domain. This distance, longer than that observed for the SS, would allow the longer TM domain to fully emerge from the ribosome exit tunnel (Fig. 5e and Supplementary Fig. 6h–j). Generally, TM domains comprise stronger SRP-binding sites than SSs, and this may weaken the requirement for a REST element for recognition by SRP. Thus, whereas the shorter SSs often span nearly the length of the SRP-binding pocket, the much longer TM helices may provide an extended time window for SRP binding, spanning the full hydrophobic region emerging from the ribosome.
DISCUSSION
Translational elongation kinetics are emerging as an important new but poorly understood mechanism to pervasively control the fate of nascent chains in vivo. Our analyses establish a link between the strength of SRP–nascent chain interaction in vivo and REST elements in the mRNA sequences of both SS and TM proteins. An increased time window for SS or TM-helix recognition at the ribosome exit site systematically distinguishes strongly enriched from enriched SRP substrates. Indeed, strongly enriched SRP substrates contain mRNA elements with a preference for nonoptimal codons positioned to slow down translation upon ribosomal exit of the SRP-binding site, thus kinetically favoring recognition and engagement by the SRP.
Our work adds a kinetic dimension to the regulation of SRP binding in vivo, highlighting the dynamic nature of cotranslational events in the cell. Despite the profound understanding of the interaction of the SRP with SS and TM segments, the preferential in vivo SRP engagement with different substrates20 could not be easily rationalized. Although our analysis clearly confirms that a hydrophobic SS or TM sequence is critical for SRP binding, we also find that, beyond a minimal threshold, increased hydrophobicity in SSs does not lead to stronger SRP recognition. Instead, we uncover a prominent role of synonymous codon choice in cotranslational recruitment of the SRP (Fig. 6). A strategically placed nonoptimal codon cluster in the mRNA sequence ~35–40 codons downstream of the SS or TM segments enhances SRP binding in vivo. Ribosome profiling confirms the slowdown of translation just as the SRP-binding site emerges from the ribosome. The longer TM domains already have an extended association time window, and this, together with possible signaling from within the ribosomal tunnel30, may explain their enhanced SRP recognition. Notably, using a previously published translocation assay32 that relies on SRP recognition and translocation of the signal anchor TM helix of the model substrate Pho8, we find that changing codon optimality at the naturally occurring upstream REST sequence decreases the efficiency of translocation (Supplementary Fig. 7).
Figure 6.
REST coordinates translation elongation and SRP recognition. Translational coordination of local translation rates with in vivo SRP–nascent chain recognition. Strategically positioned nonoptimal codons form an mRNA-encoded slowdown of translation (REST) element downstream of SS or TM segments. The increased time window of the SS or TM segment at the ribosome exit promotes recognition and binding by the SRP.
One corollary of these findings is that evolutionary pressures to enhance SRP binding have shaped mRNA sequences to enhance their efficient cotranslational targeting to membranes. The idea that elongation rates are attuned to cotranslational SRP recruitment explains previous observations that antibiotics affecting translation can enhance SRP binding33. Upon binding RNCs containing SS and TM sites, the SRP itself can further contribute to a kinetic elongation slowdown via its Alu domain9albeit one that would affect all SRP substrates34. Although our analyses focused on nonoptimal codons, it will be interesting to examine whether additional elongation-slowdown mechanisms, including stalling through positive charges30proline residues35 and RNA secondary structure36have roles in regulating nascent-chain interactions.
The availability of global interaction and ribosome profiling datasets provides an unprecedented opportunity to delve into a hitherto-unexplored aspect of cotranslational biology, namely the kinetic determinants of cotranslational folding and recognition. The SRP, being a cotranslational binding factor with a defined recognition site in the emerging nascent chain, serves as a general paradigm for cotranslational nascent-chain recognition by chaperones and other factors. REST elements may generally modulate the fate of nascent polypeptides at the ribosome, coordinating their recognition by chaperones and other factors37,38. Indeed, bacterial SRP appears to rely on Shine-Dalgarno–like sequence elements to coordinate translation and nascent-chain recognition39. Of note, regulation of local translation kinetics through synonymous codons allows for additional levels of dynamic control, e.g., via tRNA regulation or modification40. One intriguing consequence of this idea is the concept that synonymous mutations, and thus genomic coding sequences, are under direct selective pressure to accommodate the complexity of polypeptide folding and translocation in the cell.
ONLINE METHODS
Data sources and classification
The experimental data set on the global profiling of SRP specificity for its cotranslational interactions with nascent polypeptides was obtained from ref. 20. Briefly, del Alamo et al. first isolated SRP-bound ribosome–nascent chain complexes (RNCs) from cells by tandem affinity purification (TAP) of SRP54-TAP. They next identified the protein substrates through their encoding mRNAs by DNA microarray hybridization. Comparison of mRNAs encoding the polypeptide substrates bound by SRP to the total mRNA pool in the cell allowed quantification of which nascent chains selectively interact with SRP cotranslationally. As a control and reference, a pulldown of the ribosomal protein RPL16-TAP served to quantify the amount of ongoing translation, herein referred to as the ‘translatome’.
Microarray data were analyzed with the SAM algorithm41, which statistically tests for differential expression between mRNAs attached to SRP-bound RNCs and all cellular mRNAs. Cotranslational SRP substrates were defined as those proteins whose messages were significantly enriched in the SRP pulldown over the total cellular mRNAs20. We compared the SRP substrates to the translatome by a ‘two-class unpaired’ SAM analysis to identify nascent chains whose interaction with the SRP is enriched over their levels of translation42. Whereas the analysis of the SRP54-TAP pulldown quantifies the fraction of mRNAs of each kind that is attached to SRP-bound RNCs compared to the total cellular mRNA pool, the comparison to the translatome reveals for each gene the fraction of mRNAs attached to SRP-bound RNCs in relation to the number of mRNAs that are being translated.
We retrieved the list of transmembrane (TM) regions across the Saccharomyces cerevisiae proteome from the Saccharomyces Genome Database (http://www.yeastgenome.org/), which are based on predictions by TMHMM 2.0 (ref. 43). We predicted signal sequences (SS) with SignalP44 and Phobius45 and considered the consensus with the annotations in the curated UniProt database (http://www.uniprot.org/). We computed the length of the hydrophobic region in SS with Phobius, as described in ref. 11.
The SRP binds to hydrophobic N-terminal signal sequences or TM segments. TM proteins often lack a SS, in which case the first TM segment acts as a canonical SRP-binding site. Our initial analysis revealed a clear correlation between the length of the hydrophobic stretch in SS or TM segments and SRP enrichment, wherein a longer hydrophobic binding region leads to preferential SRP interaction (Fig. 1b). However, big differences in enrichment are observed even for substrates with SRP-binding sites of comparable lengths, i.e., for SS proteins or TM proteins. To shed light on additional determinants beyond the length of the hydrophobic region that may affect SRP recognition, we split the data set of cotranslational SRP substrates into proteins with and without SSs (Supplementary Fig. 1). We defined SS proteins as those SRP substrates that had a predicted SS that was also listed in the UniProt database and no TM segment in at least the first 100 codons (schematized in Supplementary Fig. 1). In these proteins, the SS is the initial cognate SRP-binding site. We defined all proteins with predicted TM segments but no predicted or annotated SS as TM proteins (Supplementary Fig. 1).
We first sought to harness the SS proteins as a paradigm of differential engagement with the SRP. We defined proteins with a SAM enrichment score of at least 2 and defined those within the top 30% of the strongest enrichment of SRP association over the translatome as strongly enriched SRP substrates, herein SRP-SE (n = 29). We classified all other SS proteins that significantly interact with the SRP as enriched (SRP-E) substrates (n = 78). Of note, because both SRP-SE and SRP-E proteins are cotranslational substrates of SRP-dependent protein translocation, this classification allows us to analyze global determinants of SRP specificity. We further included all cotranslational interactors of the SRP that did not have a predicted or annotated SS or TM segment as noncanonical (SRP-NC) substrates (n = 109). To identify the putative SRP-binding site, we searched for the first stretch of at least five consecutive hydrophobic amino acids in the N-terminal region of each amino acid sequence of the SRP-NC proteins.
TM proteins are generally translocated in an SRP-dependent manner5,6,20, as reflected in systematically higher enrichment scores of SRP interaction20 (Fig. 1b). Multipass TM proteins have several potential SRP-binding sites, and this complicates the interpretation of the statistical-enrichment score of SRP interaction. Furthermore, TM proteins that do interact with the SRP cotranslationally, albeit with very low enrichment scores, tend to be weakly expressed; thus, sufficient coverage is lacking in the ribosome profiling data to allow analysis of experimentally measured translation kinetics. We first looked at a set of n = 177 TM proteins with detectable expression levels46, of which n = 37 had ribosome density profiles with enough coverage. Because a second, slowly translated TM helix strategically spaced downstream may also promote SRP recognition of the first TM helix irrespective of nonoptimal codons, we analyzed all TM proteins, and then separately analyzed only those TM proteins with at least 60 codons between the start of the first and the second TM segments. To obtain sufficiently large sample sizes for the latter group, we defined TM proteins with a SAM enrichment score of SRP interaction of at least 3 as strongly enriched (TM-SE) substrates (n = 46) and all other TM proteins as not strongly enriched (TM-nSE) proteins (n = 165).
Sequence hydrophobicity
Hydrophobic interactions are central to intra- and interchain interactions between polypeptides. Hydrophobicity profiles were computed from the amino acid sequences with the Kyte-Doolittle hydrophobicity scale. Individual hydrophobicity profiles were smoothed with sliding windows of size 7. To facilitate interpretation, we used a linearly rescaled version of the Kyte-Doolittle scale that has been normalized to have a zero mean and unitary s.d.47. Herein, hydrophobicity values greater than one are considered very hydrophobic.
To obtain a representative hydrophobicity profile for a group of multiple sequences, we aligned the sequences at the start of the hydrophobic region of the SS and computed the median hydrophobicity at each position. We present the median profiles rather than the mean profiles because the median is more robust toward extreme values, especially at small sample sizes. However, both representations led to the same conclusions. To validate that the median profiles are indeed representative for individual sequences, we computed the average hydrophobicity of the SS (or hydrophobic stretch in the SRP-NC substrates) for each sequence and compared the distributions of the SRP-SE, SRP-E and SRP-NC proteins. Because structural data suggest a minimum binding site of 8 or 9 amino acids in the nascent protein that interact with the hydrophobic groove in SRP54 (ref. 14), we searched for the stretch of length 8 with the highest average hydrophobicity in each signal sequence. We validated that the lack of a global difference in hydrophobicity between SRP-SE and SRP-E substrates is independent of the choice of the window size considered (Supplementary Fig. 2f).
Putative SRP-binding motif
Protein sequence motifs, also referred to as ‘linear motifs’, are defined sequence patterns that mediate protein-protein interactions. Distinct motifs are ubiquitous in facilitating protein binding, targeting, cleavage and post-translational modifications. Whereas SSs have generally well-conserved properties, i.e., a short, positively charged N-terminal region of ~1–4 amino acids followed by a hydrophobic stretch of ~8–15 amino acids and a cleavage site, SSs share little direct sequence similarity and conservation. Here, we asked whether SRP-SE substrates share distinct sequence patterns in their hydrophobic regions that may explain their stronger SRP enrichment compared to SRP-E substrates.
The strength of motifs in biological sequences can be quantified by computing the information content (IC) from sequence alignments of the motif occurrences48. The IC is a theoretical measure based on the Kullback-Leibler divergence or ‘relative entropy’, and it estimates the difference between the probabilities of the observed sequence patterns and the background distribution of the amino acid frequencies. Given a sequence alignment, the IC is calculated as:
where fi,j is the observed frequency of amino acid j at position i in the alignment, pj is the background probability of amino acid j and n is the length of the alignment. We here used the amino acid frequencies in the secretome, i.e., all proteins with either SS or TM segments, as background probabilities.
Because the quality of the alignment strongly influences the discovery of sequence motifs, we tested different approaches. First, we aligned all SSs in the SRP-SE, SRP-E and SRP-NC substrates at the start of their hydrophobic regions as they emerge from the ribosome exit tunnel. Alternatively, we used the constraint-based alignment algorithm COBALT49 with an artificially high gap opening penalty of −100 to generate local sequence alignments of the hydrophobic regions without gaps because any putative motif binding the SRP54 binding pocket would have to be continuous. We generated sequence logos of motifs in the SRP-SE, SRP-E and SRP-NC proteins with the WebLogo webserver (http://weblogo.berkeley.edu/).
We hypothesized that, if there is a distinct sequence pattern that correlates with stronger SRP enrichment, we should observe a more pronounced motif, i.e., a higher IC, in the SRP-SE substrates. In relatively small multiple sequence alignment of low numbers of sequences, the present sequences more strongly affect the observed amino acid frequencies. To avoid any systematic bias toward the smaller group of SRP-SE substrates, we randomly sampled 1,000 times the number of SRP-SE sequences (n = 29) from the pool of all SRP-E substrates and, in a separate analysis, all SRP-E substrates plus SS proteins that do not cotranslationally interact with the SRP, and computed the IC. We then compared the IC of the SRP-SE proteins to that expected from the random distribution. In all these analyses, we found that the SRP-SE proteins do not contain a more defined sequence pattern in their hydrophobic SSs but rather fall onto the mean of the random distribution (Supplementary Fig. 2j).
Translational efficiency predictions
Almost all amino acids can be encoded by multiple codons that are translated at different speeds2–4. To compute translational efficiency (TE) profiles from the protein-coding sequences, we made use of a recently introduced approach that takes into consideration the competition between tRNA supply and demand to predict codon-specific TEs25,26. Herein, tRNA supply is estimated on the basis of the gene copy numbers of tRNAs, which are strongly correlated with cellular tRNA abundance, and selective constraints for wobble base pairs50. The tRNA demand is estimated on the basis of how often each codon is requesting a specific tRNA during translation, i.e., the frequency of codons in the mRNA sequences weighted by the mRNA expression levels25. Because secretory and transmembrane proteins (i.e., the secretome) are translated by ribosomes at or near the ER membrane12,15, we computed a TE scale for all yeasts, analyzed specifically for the secretome (Supplementary Fig. 3 and Supplementary Table 1). We first examined whether the increased demand in the secretome for tRNAs that encode hydrophobic amino acids dramatically alters the TE scale compared to that of the full S. cerevisiae proteome. As expected, codons encoding hydrophobic amino acids become less optimal, and codons encoding charged amino acids become more optimal (Supplementary Fig. 2b). However, the fold changes between codon-specific TE values computed for the secretome compared to the full proteome are relatively small (Supplementary Fig. 2b). Accordingly, the resulting TE scale for the secretome is almost perfectly correlated to the TE scale for the full proteome (R2 = 0.96; n = 61 sense codons; Supplementary Fig. 2c) and does not result in changes in any classification into optimal and nonoptimal codons.
One characteristic of this normalized TE scale is the indication that for most codons, tRNA supply and demand are closely matched; of note, this is directly supported by experimental ribosome profiling data51. The analysis of the cellular tRNA pools also suggests two distinct tails of codons for which supply clearly exceeds demand and vice versa. To gain more discriminative power and to better reflect this balance between tRNA supply and demand, we here define the 20% least efficient codons as nonoptimal (Supplementary Fig. 2). Furthermore, to support an argument of selective codon choice, we separately analyzed only those nonoptimal codons whose corresponding amino acids are encoded by at least three codons, thus omitting three codons (TTT − F; TAT − Y; AAT− N) from the list of nonoptimal codons (Supplementary Fig. 2a). We report the results for the more degenerate nonoptimal codons in the main text. However, our results and their significance are the same for both definitions.
We computed median TE profiles for the SRP-SE, SRP-E and SRP-NC proteins, to represent the different classes of SRP substrates. To validate the presence of low-TE regions, we computed the mean TE in all individual sequences between codons 38 and 45 downstream of the start of the SS. Importantly, although the graph shows the median TE profiles smoothed by averaging over sliding windows of size 7 for clarity of representation, we evaluated the statistical differences between substrate classes from the raw TE profiles without any prior smoothing.
Ribosome profiling analysis
Ribosome profiling globally measures translation kinetics at high resolution by the selective sequencing of ribosome-protected footprints28. By producing a global snapshot of translating ribosomes along mRNAs, higher ribosome footprint densities indicate regions that are translated more slowly. Because unbiased statistical analyses of ribosome protection require sufficient coverage even in regions of low density, and ongoing improvements in NGS sequencing technology have improved the coverage of more-recent ribosome profiling data sets, we use a recent ribosome profiling data set obtained from ref. 29. We computed the correlation coefficient between the ribosome footprint profiles of replica 1 and replica 2 in the profiling data set29 as well as the consensus average footprint density profile for each ORF (Supplementary Fig. 5a). Importantly, analysis of the correlation between replicas as a function of the average coverage (Supplementary Fig. 5b) indicates that the higher the minimum coverage, the higher, on average, the agreement between footprint density profiles between replicas. Accordingly, and following the approach of ref. 52, a minimum coverage of, on average, at least ten sequencing reads per codon was chosen, to yield a curated ribosome profiling data set with good agreement between replicas. ORFs without sufficient coverage or reproducibility were discarded from further analyses. We further normalized the footprint density profiles to have an average coverage of 1.
To assess local translation rates downstream of SS or TM segments, we aligned the sequences at the start of their hydrophobic regions and computed representative median ribosome footprint density profiles for each class of substrates. We confirmed that the median profiles indeed reflect characteristics of individual profiles (Supplementary Fig. 5c,d). To further validate that qualitative differences in median profiles reflect quantitative differences in individual proteins, we calculated the average footprint density of the downstream slowdown region. For SS proteins, we calculated the average footprint density for the region 38 to 45 codons downstream of the start of the hydrophobic stretch and for TM proteins the region 39 to 41 codons downstream of the start of the TM segment.
Several individual biochemically validated examples demonstrate that synonymous codons are indeed translated at different speeds2–4, thus directly affecting protein folding and function53–55. Further evidence for an important role of nonoptimal, more slowly translated codons stems from findings that overexpression of tRNAs corresponding to nonoptimal codons disrupts protein homeostasis and leads to widespread aggregation56. Our analysis found clusters of nonoptimal codons leading to a region of three to seven codons that is translated at significantly lower speed. Furthermore, we found significant differences in ribosome footprint densities that correlated with an enrichment in nonoptimal codons. The increasing availability of data sets with even deeper coverage will increase the statistical power of ribosome profiling data and allow detection of additional signals of translational attenuation that are not apparent in existing data sets with less coverage. This will further illuminate the contribution and hierarchy of additional factors beyond codon optimality that are known to contribute to local slower elongation rates, including positive charges31, polyproline motifs35, tRNA modifications40, RNA secondary structure39 and codon-pair frequencies57, which are also likely to influence local translation rates in vivo.
Statistical analysis
Microarray data were analyzed with the SAM algorithm41. All data analyses were performed in Python and the statistics environment R (http://www.r-project.org/). Differences between distributions were tested for significance by the nonparametric Wilcoxon rank-sum test, and two-sided P values are reported. The enrichment of nonoptimal codons was tested by Fisher’s exact test, and two-sided P values are reported.
Supplementary Material
Acknowledgments
We thank members of the Frydman laboratory for helpful discussions and K. Dalton for comments on the manuscript. We gratefully acknowledge support from a European Molecular Biology Organization Long-Term Fellowship (ALTF 1334-2010) to S.P., US National Institutes of Health (NIH) grant GM108325 to J.C. and NIH grant GM56433 and Human Frontier Science Program Grant RGP0025/2012 to J.F.
Footnotes
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
S.P. and J.F. conceived the research project. S.P. performed all computational analyses. J.C. performed the translocation experiment. S.P. and J.F. wrote the manuscript; all authors discussed the results and commented on the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html
References
- 1.Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU. Molecular chaperone functions in protein folding and proteostasis. Annu. Rev. Biochem. 2013;82:323–355. doi: 10.1146/annurev-biochem-060208-092442. [DOI] [PubMed] [Google Scholar]
- 2.Spencer PS, Siller E, Anderson JF, Barral JM. Silent substitutions predictably alter translation elongation rates and protein folding efficiencies. J. Mol. Biol. 2012;422:328–335. doi: 10.1016/j.jmb.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu Y, et al. Non-optimal codon usage is a mechanism to achieve circadian clock conditionality. Nature. 2013;495:116–120. doi: 10.1038/nature11942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dana A, Tuller T. The effect of tRNA levels on decoding times of mRNA codons. Nucleic Acids Res. 2014;42:9171–9181. doi: 10.1093/nar/gku646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keenan RJ, Freymann DM, Stroud RM, Walter P. The signal recognition particle. Annu. Rev. Biochem. 2001;70:755–775. doi: 10.1146/annurev.biochem.70.1.755. [DOI] [PubMed] [Google Scholar]
- 6.Akopian D, Shen K, Zhang X, Shan S-o. Signal recognition particle: an essential protein-targeting machine. Annu. Rev. Biochem. 2013;82:693–721. doi: 10.1146/annurev-biochem-072711-164732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE. Biological and chemical approaches to diseases of proteostasis deficiency. Annu. Rev. Biochem. 2009;78:959–991. doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- 8.Landry SJ, Gierasch LM. Recognition of nascent polypeptides for targeting and folding. Trends Biochem. Sci. 1991;16:159–163. doi: 10.1016/0968-0004(91)90060-9. [DOI] [PubMed] [Google Scholar]
- 9.Halic M, et al. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature. 2004;427:808–814. doi: 10.1038/nature02342. [DOI] [PubMed] [Google Scholar]
- 10.Noriega TR, et al. Signal recognition particle-ribosome binding is sensitive to nascent chain length. J. Biol. Chem. 2014;289:19294–19305. doi: 10.1074/jbc.M114.563239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ast T, Cohen G, Schuldiner M. A network of cytosolic factors targets SRP-independent proteins to the endoplasmic reticulum. Cell. 2013;152:1134–1145. doi: 10.1016/j.cell.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 12.Rapoport TA. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450:663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- 13.von Heijne G. Signal sequences: the limits of variation. J. Mol. Biol. 1985;184:99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]
- 14.Janda CY, et al. Recognition of a signal peptide by the signal recognition particle. Nature. 2010;465:507–510. doi: 10.1038/nature08870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hegde RS, Kang S-W. The concept of translocational regulation. J. Cell Biol. 2008;182:225–232. doi: 10.1083/jcb.200804157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng N, Gierasch LM. Signal sequences: the same yet different. Cell. 1996;86:849–852. doi: 10.1016/s0092-8674(00)80159-2. [DOI] [PubMed] [Google Scholar]
- 17.Kaiser CA, Preuss D, Grisafi P, Botstein D. Many random sequences functionally replace the secretion signal sequence of yeast invertase. Science. 1987;235:312–317. doi: 10.1126/science.3541205. [DOI] [PubMed] [Google Scholar]
- 18.Kraut-Cohen J, Gerst JE. Addressing mRNAs to the ER: cis sequences act up. Trends Biochem. Sci. 2010;35:459–469. doi: 10.1016/j.tibs.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 19.Flanagan JJ, et al. Signal recognition particle binds to ribosome-bound signal sequences with fluorescence-detected subnanomolar affinity that does not diminish as the nascent chain lengthens. J. Biol. Chem. 2003;278:18628–18637. doi: 10.1074/jbc.M300173200. [DOI] [PubMed] [Google Scholar]
- 20.del Alamo M, et al. Defining the specificity of cotranslationally acting chaperones by systematic analysis of mRNAs associated with ribosome-nascent chain complexes. PLoS Biol. 2011;9:e1001100. doi: 10.1371/journal.pbio.1001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fedyukina DV, Cavagnero S. Protein folding at the exit tunnel. Annu. Rev. Biophys. 2011;40:337–359. doi: 10.1146/annurev-biophys-042910-155338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komar AA. A pause for thought along the co-translational folding pathway. Trends Biochem. Sci. 2009;34:16–24. doi: 10.1016/j.tibs.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 23.Zhang G, Ignatova Z. Folding at the birth of the nascent chain: coordinating translation with co-translational folding. Curr. Opin. Struct. Biol. 2011;21:25–31. doi: 10.1016/j.sbi.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 24.O’Brien EP, Vendruscolo M, Dobson CM. Prediction of variable translation rate effects on cotranslational protein folding. Nat. Commun. 2012;3:868. doi: 10.1038/ncomms1850. [DOI] [PubMed] [Google Scholar]
- 25.Pechmann S, Frydman J. Evolutionary conservation of codon optimality reveals hidden signatures of cotranslational folding. Nat. Struct. Mol. Biol. 2013;20:237–243. doi: 10.1038/nsmb.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gingold H, Pilpel Y. Determinants of translation efficiency and accuracy. Mol. Syst. Biol. 2011;7:481. doi: 10.1038/msb.2011.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tuller T, et al. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell. 2010;141:344–354. doi: 10.1016/j.cell.2010.03.031. [DOI] [PubMed] [Google Scholar]
- 28.Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zinshteyn B, Gilbert WV. Loss of a conserved tRNA anticodon modification perturbs cellular signaling. PLoS Genet. 2013;9:e1003675. doi: 10.1371/journal.pgen.1003675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berndt U, Oellerer S, Zhang Y, Johnson AE, Rospert S. A signal-anchor sequence stimulates signal recognition particle binding to ribosomes from inside the exit tunnel. Proc. Natl. Acad. Sci. USA. 2009;106:1398–1403. doi: 10.1073/pnas.0808584106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Charneski CA, Hurst LD. Positively charged residues are the major determinants of ribosomal velocity. PLoS Biol. 2013;11:e1001508. doi: 10.1371/journal.pbio.1001508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dalley JA, Selkirk A, Pool MR. Access to ribosomal protein Rpl25p by the signal recognition particle is required for efficient cotranslational translocation. Mol. Biol. Cell. 2008;19:2876–2884. doi: 10.1091/mbc.E07-10-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang D, Shan S-o. Translation elongation regulates substrate selection by the signal recognition particle. J. Biol. Chem. 2012;287:7652–7660. doi: 10.1074/jbc.M111.325001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mason N, Ciufo LF, Brown JD. Elongation arrest is a physiologically important function of signal recognition particle. EMBO J. 2000;19:4164–4174. doi: 10.1093/emboj/19.15.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doerfel LK, et al. EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science. 2013;339:85–88. doi: 10.1126/science.1229017. [DOI] [PubMed] [Google Scholar]
- 36.Yang JR, Chen X, Zhang J. Codon-by-codon modulation of translational speed and accuracy via mRNA folding. PLoS Biol. 2014;12:e1001910. doi: 10.1371/journal.pbio.1001910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sherman MY, Qian S-B. Less is more: improving proteostasis by translation slow down. Trends Biochem. Sci. 2013;38:585–591. doi: 10.1016/j.tibs.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 38.Kramer G, Boehringer D, Ban N, Bukau B. The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat. Struct. Mol. Biol. 2009;16:589–597. doi: 10.1038/nsmb.1614. [DOI] [PubMed] [Google Scholar]
- 39.Fluman N, Navon S, Bibi E, Pilpel Y. mRNA-programmed translation pauses in the targeting of E. coli membrane proteins. Elife. 2014;3:e03440. doi: 10.7554/eLife.03440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.El Yacoubi B, Bailly M, De Crécy-Lagard V. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu. Rev. Genet. 2012;46:69–95. doi: 10.1146/annurev-genet-110711-155641. [DOI] [PubMed] [Google Scholar]
- 41.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Willmund F, et al. The cotranslational function of ribosome-associated Hsp70 in eukaryotic protein homeostasis. Cell. 2013;152:196–209. doi: 10.1016/j.cell.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krogh A, Larsson B, Von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 44.Nielsen H, Engelbrecht J, Brunak S, Von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 45.Käll L, Krogh A, Sonnhammer EL. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 2004;338:1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 46.Holstege FC, et al. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 47.Pechmann S, Levy ED, Tartaglia GG, Vendruscolo M. Physicochemical principles that regulate the competition between functional and dysfunctional association of proteins. Proc. Natl. Acad. Sci. USA. 2009;106:10159–10164. doi: 10.1073/pnas.0812414106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.D’haeseleer P. What are DNA sequence motifs? Nat. Biotechnol. 2006;24:423–425. doi: 10.1038/nbt0406-423. [DOI] [PubMed] [Google Scholar]
- 49.Papadopoulos JS, Agarwala R. COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics. 2007;23:1073–1079. doi: 10.1093/bioinformatics/btm076. [DOI] [PubMed] [Google Scholar]
- 50.dos Reis M, Savva R, Wernisch L. Solving the riddle of codon usage preferences: a test for translational selection. Nucleic Acids Res. 2004;32:5036–5044. doi: 10.1093/nar/gkh834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qian W, Yang JR, Pearson NM, Maclean C, Zhang J. Balanced codon usage optimizes eukaryotic translational efficiency. PLoS Genet. 2012;8:e1002603. doi: 10.1371/journal.pgen.1002603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li GW, Oh E, Weissman JS. The anti-Shine–Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature. 2012;484:538–541. doi: 10.1038/nature10965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang F, Saha S, Shabalina SA, Kashina A. Differential arginylation of actin isoforms is regulated by coding sequence–dependent degradation. Science. 2010;329:1534–1537. doi: 10.1126/science.1191701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang G, Hubalewska M, Ignatova Z. Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat. Struct. Mol. Biol. 2009;16:274–280. doi: 10.1038/nsmb.1554. [DOI] [PubMed] [Google Scholar]
- 55.Kimchi-Sarfaty C, et al. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- 56.Fedyunin I, et al. tRNA concentration fine tunes protein solubility. FEBS Lett. 2012;586:3336–3340. doi: 10.1016/j.febslet.2012.07.012. [DOI] [PubMed] [Google Scholar]
- 57.Coleman JR, et al. Virus attenuation by genome-scale changes in codon pair bias. science. 2008;320:1784–1787. doi: 10.1126/science.1155761. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.