Abstract
In settings of resource constraint, an understanding of HIV drug resistance can guide antiretroviral therapy (ART) at switch to second-line therapy. To determine the prevalence of such HIV drug resistance mutations (HIV DRM), we used an in-house sequencing assay in the pol gene (protease and partial reverse transcriptase) in a cohort of patients suspected of failing a first-line regimen, which in Zambia comprises two nucleoside/nucleotide reverse transcriptase inhibitors (NRTIs) and one non-nucleoside reverse transcriptase inhibitor (NNRTI). Our analysis cohort (n=68) was referred to the University Teaching Hospital in Lusaka from November 2009 to October 2012. Median duration on first-line ART to suspected treatment failure was 3.2 years (IQR 1.7–4.7 years). The majority of patients (95%) harbored HIV-1 subtype C virus. Analysis of reverse transcriptase revealed M184V (88%), K103N/S (32%), and Y181C/I/V (41%) DRMs, with the latter conferring reduced susceptibility to the salvage therapy candidates etravirine and rilpivirine. Three patients (5%) had major protease inhibitor (PI) resistance mutations: all three had the V82A mutation, and one patient (Clade J virus) had a concurrent M46I, Q58E, and L76V DRM. HIV-1 genotyping revealed major and minor DRMs as well as high levels of polymorphisms in subtype C isolates from patients failing first-line antiretroviral therapy. Closer monitoring of DRM mutations at first-line failure can inform clinicians about future options for salvage therapy.
Keywords: HIV drug resistance, pol, diversity, NNRTI, Zambia
Introduction
The administration of combination antiretroviral therapy (ART) in patients with HIV is effective in suppressing viral replication (Egger et al., 2002), prolonging AIDS-free years of life (Berrey et al., 2001), preventing horizontal transmission (Cohen et al., 2011), and vertical transmission (Connor et al., 1994). Development of HIV-1 drug resistance mutations (HIV DRMs) and polymorphisms to the viral proteins protease and reverse transcriptase, however, can undermine the effectiveness of treatment programs, particularly in resource-limited settings where treatment options beyond a “second line” are limited (Gupta et al., 2012). Consequently, monitoring of clinical isolates for HIV DRMs is critical not only for the management of patients, but also for policy makers forecasting drug needs following initial treatment failure. The World Health Organization (WHO) recommends that resistance testing should be performed for public health drug resistance surveillance in resource-constrained settings (Bennett et al., 2008). In this study, the HIV pol region (protease and reverse transcriptase) was sequenced at the time of suspected first-line treatment failure in a cohort of patients referred to the University Teaching Hospital in Lusaka, Zambia. The prevalence of HIV pol DRMs and polymorphisms were determined using a locally developed in-house HIV DRM genotyping assay.
Materials and Methods
Patient Characteristics
We conducted a retrospective analysis of HIV-infected patients (>15 years old) suspected of treatment failure during first-line ART who were referred to the University Teaching Hospital's Advanced Treatment Centre. In Zambia, first-line ART regimens comprise two nucleoside/nucleotide reverse transcriptase inhibitors (NRTIs)—among them tenofovir (TDF), zidovudine (ZDV), stavudine (d4T), abacavir (ABC), lamivudine (3TC), and emtricitabine (FTC) during the period of this analysis—along with a non-nucleoside reverse transcriptase inhibitor (NNRTI), either efavirenz (EFV) or nevirapine (NVP). Second line regimens included two NRTIs and a boosted protease inhibitor (Zambian Ministry of Health, 2010). HIV DRM testing was ordered at the discretion of the attending clinician, and the national guidelines specifies that a viral load >1,000 copies/ml after 6 months of therapy is considered virological failure and advises a change to a second line regimen (Zambian Ministry of Health, 2010). We included demographic information and medical history (including current treatment course) to characterize our population, including pharmacy refill-based medication possession ratio (MPR) as a measure of drug adherence (Chi et al., 2009; Kauf et al., 2012; Roth et al., 2012; Vinikoor et al., 2013). Ethics boards at the University of Zambia (Lusaka, Zambia), the University of Alabama at Birmingham (Birmingham, AL, USA), and the University of North Carolina in Chapel Hill (Chapel Hill, NC, USA) approved the use of programmatic data for outcomes analysis.
HIV-1 Viral Load Measurement and Genotyping
HIV-1 viral load was measured by the Roche Amplicor HIV-1 RNA Monitor kit (version 1.5; Roche Molecular Diagnostics, Pleasanton, CA). CD4+ lymphocyte counts were performed using a Beckman Coulter flow cytometer (Beckman Coulter, Inc., Fullerton, CA). For genotype, viral RNA was extracted, amplified and sequenced using a modified in-house assay adapted from prior reports (Wallis et al., 2010). A 1,200 base pair (bp) amplification fragment was generated from patient virus isolated from 500 ml of blood plasma using the QIAamp viral RNA isolation kit (Qiagen Corporation, Venlo, The Netherlands, performed according to the manufacturer's protocol). Complementary DNA (cDNA) was first generated using a forward strand synthesis (FSS) primer: (CWR: 5′- GCATACTTYCCTGTTTTCAG-3′; HXB2 position 3594–3613) using the Superscript III Reverse Transcriptase (Invitrogen Corporation, Life Technologies, Inc., Carlsbad, CA). At the completion of the cDNA synthesis, the remaining RNA was treated with RNaseOUT (Invitrogen Corp., Life Technologies), and the synthesis mixture was purified using a GeneJet DNA (Thermo Scientific Corporation, Waltham, MA) purification kit. A nested PCR reaction was performed on the cDNA eluate using Platinum Taq DNA polymerase (Invitrogen Corporation, Life Technologies) according to previously published amplification methods (Seu et al., 2014): a 1,200 bp sequence was first amplified using forward primer (CWF: 5′-GAAGGACACCAAATGAAAGAYTG-3′; HXB2 nucleotide position 2,044–2,066) and reverse primer (LSR1: 5′-ACTGTTTTACATCATTAGTGTGGG -3′; HXB2 nucleotide position 3,651–3,628) and a second round PCR amplification using a forward primer (LSF1: 5′-TCAGAGCAGACCAGAGCCAACAGCCCCA-3′; HXB2 nt position 2,136–2,163) and reverse primer (Rev7: 5′-ATCCCTGGGTAAATCTGACTTGCCCA-3′; HXB2 position 3,370–3,345). Amplicons were directly analyzed via population-based Sanger Sequencing using BigDye Terminator chemistry and protocols recommended by the manufacturer (Applied Biosystems, Foster City, CA), and sequencing reaction products were analyzed with an ABI 3130XL genetic analyzer (Applied Biosystems). Bi-directional DNA strands were sequenced using overlapping fragments, and individual sequence fragments for each amplicon were assembled, inspected, and edited using Sequencher software version 5.0 (Gene Codes Corporation, Ann Arbor, MI)—capable of resolving down to 20% of minority variations at a nucleotide position. Fasta sequences of the concatenated amplicons were submitted to the Stanford Database (http://hivdb.stanford.edu/index.html) to generate an HIV-1 drug resistance report. The pol gene encompassing reverse transcriptase and protease was analyzed. HIV DRM and polymorphisms were analyzed according to both the drug resistance profile on the curated website as well as 2013 IAS–USA guidelines (Johnson et al., 2013).
HIV-1 Protease Phylogenetic Analysis
Protease sequences (amino acids 1–99) were aligned using the ClustalX multiple sequence alignment tool using the Clustal method and phylogenetic trees were generated by using the neighbor-joining method with the PHYLIP software package (version 3.52c; Joseph Felsenstein, University of Washington, Felsenstein, 1985). The SEQBOOT program was carried out to generate 100 data sets that represent randomly re-sampled versions of the input-aligned sequences, to test the reliability of the final tree topology. Evolutionary distances were estimated by the character-based maximum likelihood method DNAML using the transition/transversion ratio of 2, with global rearrangements and randomized input order of sequences. The results from the random datasets were then summarized by constructing a majority rule consensus tree with CONSENSE. The phylogenetic relationships were determined by the NJ-plot program (version 2.3; M. Gouy, University of Lyon). Representative sequences from selected HIV-1 clades were obtained from Los Alamos National Laboratories (http://www.hiv.lanl.gov/content/index) in order to represent the 68 isolates within the global group M HIV-1 clades. Brackets were used to indicate the clustering of clades (A–J), and branch numbers to represent bootstrap values >75. The GenBank accession numbers of the 68 sequences of the pol region (nucleotide position 2,253–2,550 relative to HXB2) presented in this study are as follows: KM513662–KM513729.
Results
Patient Characteristics
Demographic and clinical characteristics of the patients are described in Table I. 68 HIV-positive patients on PI sparing first-line therapy (46% male, median age 34.6, interquartile range [IQR] 26.9–41.7) who had viral loads above 1,000 copies/ml had HIV DRM testing performed from November 2009 to October 2012. Among these patients, the median viral load was 29,050 copies/ml (IQR 8,652–79,680) and the median CD4+ T cell counts were 144.5 (IQR 44.5–241.7) assessed at the time of the HIV DRM test. Adherence was generally poor, with only 52% shown to have an MPR >95% during the preceding 12 months.
Table I. Clinical and Biological Characteristics of 68 Zambian Patients Screened for HIVDR at First Line Failure.
| Characteristic | N | Value |
|---|---|---|
| Age (years [IQR]) | 52 | 34.6 (26.9–41.7) |
| Male (No. [%]) | 68 | 31 (46%) |
| WHO stage (No. [%]) | 66 | |
| I | 9 (13%) | |
| II | 19 (28%) | |
| III | 31 (46%) | |
| IV | 7 (10%) | |
| BMI (kg/m2) median [IQR]) | 48 | 21.8 (19.3–24.6) |
| CD4 count (cells/mm3) (Value [IQR]) | 60 | 144.5 (44.5–241.7) |
| Hemoglobin (g/dl) value [IQR] | 60 | 10.9 (10.0–12.8) |
| Time on antiretroviral treatment (years [IQR]) | 68 | 3.2 (1.7–4.7 years) |
| Most recent antiretroviral therapy | 68 | |
| ZDV + 3TC + EFV | 1 (1.5%) | |
| ZDV + 3TC + NVP | 26 (38%) | |
| D4T + 3TC + EFV | 3 (4%) | |
| D4T + 3TC + NVP | 18 (27%) | |
| ABC + 3TC + NVP | 1 (1.5%) | |
| TDF + 3TC/FTC+ EFV | 8 (12%) | |
| TDF + 3TC/FTC + NVP | 11 (16%) | |
| Medication Possession Ratio >0.95 (value, %) | ||
| Previous 3 months | 67 | 46 (69%) |
| Previous 6 months | 64 | 38 (59%) |
| Previous 12 months | 62 | 32 (52%) |
| Viral Load at time of HIVDR testing-copies/ml (Interquartile Range) | 68 | 29,050 (8,652–79,680) |
AIDS, Acquired immunodeficiency syndrome; HAART, Highly Active Antiretroviral therapy; HIV, Human immunodeficiency virus; NNRTI, Non-Nucleoside/Nucleotide reverse transcriptase inhibitor; NRTI, Nucleoside/Nucleotide reverse transcriptase inhibitor; PI, Protease inhibitor. Antiretroviral therapy (ART) regimens taken during this course of follow-up are depicted on the graph as either combination (commas) or fixed dose (slash) drugs. 3-letter abbreviations for ARTs are as follows: Nucleoside reverse transcriptase inhibitors (NRTIs) [abacavir- ABC; emtricitabine- FTC; lamivudine- 3TC; stavudine- D4T; tenofovir- TDF; zidovudine -ZDV], non-nucleoside reverse transcriptase inhibitors (NNRTIs) [efavirenz- EFV, nevirapine- NVP].
Prevalence of NRTI and NNRTI Drug Resistance Mutations
Genetic analysis of the 5′ half of reverse transcriptase (RT), amino acids 1–265 (HXB2 nucleotide position 2,550–3,345), demonstrated prevalent NRTI and NNRTI DRMs (Johnson et al., 2013). Assessment of the overall prevalence of first line therapy patients with suspected treatment failure for at least one DRM (n = 67, 98%), one NRTI DRM (n = 61, 90%), or NNRTI DRM (n = 67, 98%) is shown (Fig. 1A). Within this cohort, a summary is provided for the prevalence of drug resistance mutations according to first line regimens prescribed: ZDV + 3TC + NNRTIs, D4T + 3 TC + NNRTIs, ABC + 3TC + NVP, and TDF + 3TC/FTC + NNRTIs (Table I). Notably, the prevalence of NNRTI DRM prevalence was high (at or near 100%) in all first line therapy in this population (Fig. 1B).
Fig. 1.

Sequences were uploaded onto the Stanford Drug Database program (http://sierra2.stanford.edu/sierra/servlet/JSierra), and both Drug Resistance Mutations (DRM) and polymorphisms were analyzed according to both the drug resistance profile on the curated website as well as the 2013 IAS guidelines. (A) Assessment of the overall prevalence of first line therapy patients with suspected treatment failure. (B) Prevalence of drug resistance mutations according to first line regimens prescribed.
Prevalence of Drug Resistance Mutations to Salvage Therapy Options
As expected, the most common NRTI DRM was M184V (selected by 3TC and FTC) present in 60 patient isolates (88%), followed by K65R (selected by TDF, d4T, abacavir [ABC], didanosine [DDI]) (n = 21, 31%). A wide range of mutations conferring multi-NRTI resistance were also observed, including M41L (n = 7, 10%), A62V (n = 15, 22%), D67N (n = 10, 15%), K70R (n = 10, 15%), V75I (n = 2, 3%), F77L (n = 1, 1%), Y115F (n = 6, 9%), F116Y (n = 1, 1%), Q151M (n = 1, 1%), L210W (n = 3, 4%), T215Y/F (n = 7, 10%) and (n = 5, 7%), and K219Q/E (n = 4, 6%) and (n = 7, 10%). The mean number of thymidine analog mutations (TAMs) per patient currently prescribed thymidine analogs was 0.9 out of the 46 patients (range 0–4 TAMs per patient) (Fig. 2A). The most prevalent NNRTI mutations were Y181C/I/V (n = 24, 35%), (n = 3, 4%), and (n = 1 (1%), respectively, K103N/S (n = 21 (32%) and n = 1 [1%]), G190A/S/E (n = 20 (29%), n = 1 (1%), and (n = 2, 3%), respectively, V108I (n = 13, 19%), and V106A/M (n = 2, 3%) and (n = 10, 15%) (Fig. 2B). Assessment of NNRTI DRMs to rilpivirine (Fig. 2C) and etravirine (Fig. 2D) revealed the prevalence of several mutations conferring reduced susceptibility to these potential options for salvage therapy. Of note, there was a high prevalence of Y181C/I/V (n = 28, 41% cumulatively) that confers reduced susceptibility to both rilpivirine (RPV) and etravirine (ETR), and Y188L (n = 5, 7%) which leads to reduced susceptibility to RPV.
Fig. 2.
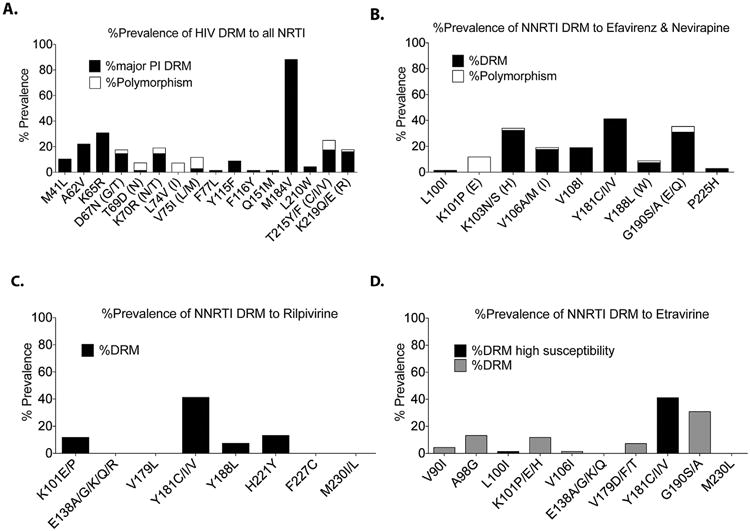
(A) All NRTI DRMs. (B) NNRTI-Efavirenz and Nevaripine DRMs. (C) NNRTI-Rilpivirine DRMs. (D) NNRTIEtravirine DRMs. Alphanumerical symbols refer to the amino acid position of the drug resistance mutation. The first letter represents the wild type residue, followed by the amino acid position, followed by the major/minor drug resistance mutation. In parentheses are polymorphisms at this genetic locus.
Protease Inhibitor Drug Resistance Mutations
Despite a lack of prior exposure to protease inhibitors, three patients (5%) had one or more major PI DRM (full length protease 1–99 amino acids). V82A was detected in all three patients, with concurrent M46I, Q58E, and L76V in one of the three patients harboring the clade J HIV-1 virus. (Supplementary Fig. 1A) Genotypic assessment of prevalence of PI-specific DRM are also shown: lopinavir/r, indinavir/r, and darunavir/r (Supplementary Fig. 1B–D).
Phylogenetic Analysis
The majority of isolates (n = 65, 95%) were identified as HIV-1 subtype C during phylogenetic analysis of pol amino acid positions 1–99 of protease (HXB2 nucleotide [nt] position 2253–2550). Sequences representative of HIV-1 clades were obtained from Los Alamos National Laboratories (http://www.hiv.lanl.gov/content/index) and represent global group M HIV-1 subtypes. Brackets indicate the clustering of clades, and branch numbers represent bootstrap values >75. Non-clade C protease sequences were identified in three individuals and were HIV-1 subtypes A, B, and J (Fig. 3). Phylogenetic review showed two main branches of clade C virus among generated HIV sequences from patients. One main group (n = 24, 35%) clustered around sequences from viruses isolated in Zambia (ZM) and Malawi (MW); another group (n = 19, 28%) clustered around sequences from isolates from South Africa (ZA) Somalia (SO), Tanzania (TZ), and Botswana (BZ). Lastly, another group (n = 22, 32%) clustered around Clade C sequences from Israel (IL) and Georgia (GE). This phylogenetic analysis indicates the broad sequence diversity of the HIV protease gene from a sample population in Lusaka, Zambia.
Fig. 3.

The protease sequences obtained were aligned using the ClustalX multiple sequence alignment tool using the Clustal method. Phylogenetic trees were generated by using the neighbor-joining method with the PHYLIP software package (version 3.52c; Joseph Felsenstein, University of Washington). Sequences representative of HIV-1 clades were obtained from Los Alamos National Laboratories (www.LANL.gov) andrepresent global group M HIV-1 subtypes. Brackets indicate the clustering of clades, and branch numbers represent bootstrap values >75. Subtype nomenclature are denoted as the following: A.BC.xx, where “A” represents the HIV clade, “BC” represents the two letter country abbreviation, and “xx” represents the first two numbers of the sequence accession numbers on LANL. Country codes are represented as Ghana (GH), Nigeria (NG), Kenya (KE), Tanzania (TZ), Uganda (UG), Rwanda (RW), South Africa (ZA), Cameroon (CM), Democratic Republic of Congo (CD), Senegal (SN), Botswana (BW), Zambia (ZM), Malawi (MW), Ethiopia (ET), Somalia (SO), Central African Republic (CF), China (CN), Canada (CA), United States of America (US), Brazil (BR), Argentina (AR), Spain (ES), Cyprus (CY), Georgia (GE), India (IN), and Israel (IL). Patients within the study cohort are denoted as numerical values from 1 to 68.
Discussion
In this retrospective study, we sequenced the HIV-1 pol gene in ART patients suspected of first-line failure using an in-house genotyping assay in Zambia, thus providing critical HIV drug resistance mutation (HIV DRM) information. We observed a high prevalence of the Y181C/I/V (41%) and L100I (2%), an important consideration as these NNRTI DRMs confers reduced susceptibility to the third line salvage candidate etravirine. We performed a genetic analysis of HIV protease sequences that revealed the level of genetic diversity of HIV clades in Zambia, with sequences deposited to Genbank (accession IDs: KM513662–KM513729). Lastly, we performed all virological sequencing assays for HIV DRM analysis at the clinical diagnostic laboratory in the same urban township as the tertiary care referral center in Lusaka, Zambia.
Our study confirms preliminary reports that subtype C is the predominant clade in Zambia (Tang et al., 2002). Phylogenetic analysis of HIV-1 protease showed three main branches of clade C virus among generated HIV sequences from patients, from southern and eastern Sub-Saharan Africa as well as beyond. Some clades, such as clades C and D, are linked with faster disease progression, and differing subtypes can have differences in their phenotypic drug susceptibilities (Harrigan et al., 2001; Pant Pai et al., 2012). The overall goal of the phylogenetic analysis was to capture the variability of circulating native HIV-1 genetic strains from this cohort of patients in Lusaka, Zambia. We see that despite the incomplete assessment, we already see a wide range of genetic variability of Clade C viruses that share sequence concordance with other regional Clade C viruses (Fig. 3). Our sequencing analysis reveals many naturally occurring polymorphisms in HIV-1 subtype C isolates from PI drug-naive individuals. Because of this, careful attention needs to be paid to these patients when switching to second-line therapy, such as ritonavir-boosted lopinavir-based ART. Such polymorphisms could lead to rapid treatment failure and development of drug-resistant HIV-1 mutants in individuals initiating lopinavir/ritonavir-based second-line regimens (Wainberg and Brenner 2012). The breadth of sequence diversity underscores the need for further HIV-1 genotyping studies, particularly in light of different drug susceptibilities to HIV-1 nucleotide polymorphisms at these loci.
Sequencing of RT revealed genotypes with multiple TAMs from patients with exposure to ZDV and d4T, reflecting the influence of these historical agents for first-line treatment in Zambia. The most prevalent NNRTI mutations were Y181C/I/V (41%), G190A/S/E (34%), K103N/S (32%), and Y188L (7%). These results are of particular relevance as Y181C/I/V and Y188L (and to a lesser extent, G190A/S/E) renders second-generation NNRTI drugs (e.g., ETR, RPV) ineffectual for potential salvage therapies (Johnson et al., 2013). Our study also included an assessment of minor DRMs as well as documented HIV-1 polymorphisms. Minor DRMs emerge later than major mutations and in isolation do not have a substantial effect on phenotype. Some may improve replication of viruses containing major mutations (Johnson et al., 2013). Furthermore, polymorphisms not conferring drug resistance were also seen, especially at RT codon positions 67, 69, 70, 74, 75, and 215, and protease positions 63 and 89, which may indicate viruses on the pathway toward additional mutations that could ultimately result in HIVDR if selected under drug pressure (Wainberg and Brenner, 2012).
A prior study of antiretroviral drug-näve HIV-1-infected Zambian adults (n=28) demonstrated a high frequency of pre-existing minor mutations in the protease gene—including I93L (92%), L89M (79%), and M36I (79%)—which corroborates results from our current study (Handema et al., 2003). Another Zambian study compared therapy-näve patients before (n = 30) and after (n = 66) the availability of free-of-charge ART, showing no statistically significant difference in major PI DRMs (V82I: 10% vs. 3%, P = 0.20), NNRTIs (K103N: 0% vs. 1%, P = 1.0), or NRTIs (M184V: 0% vs. 1%, P = 1.0). However, authors observed an increase in the number of minor, borderline, or partial resistance mutations in the later cohort (Gonzalez et al., 2010). These findings are in line with the data presented in our study. Preexisting minor mutations as well as naturally occurring HIV polymorphisms may have an impact on drug susceptibility; as such the monitoring of HIV genotypes in patients is warranted to guide the switching of ART regimens.
The major limitation of our study is its limited external validity. This population, referred to the University Teaching Hospital's Advance Treatment Centre, is a subset of those receiving care in Lusaka. Furthermore, viral load and HIV drug resistance testing were requested at the discretion of the clinical staff based on medical history; however, we did not collect information about criteria used for each individual. Because of this fact, the cohort represented in this report may represent a group of patients who have severe complications in HIV treatment, which in turn may explain the high prevalence of those with ≥1 NRTI or ≥1 NNRTI DRM. This finding is consistent with a recent study in Thailand, South Africa, India, Malawi, and Tanzania showing that, of 148 patients with genotypic resistance testing after first-line antiretroviral therapy (ART) failure, 93% (n = 138) and 96% (n = 142) had at least one NRTI and NNRTI DRM, respectively. (Wallis et al., 2014) Additionally, only 16% in our cohort reported use of the TDF-based regimens that have been incorporated into first-line treatment for most Sub-Saharan African countries (World Health Organization, 2010). In fact, Zambia was one of the first in the region to roll out TDF-based ART (Chi et al., 2010). Given the time that elapsed between the implementation of TDF (2007) and our analysis cohort (2009–2012), the relatively low prevalence of its use in our study could suggest better patient outcomes with TDF-based ART. Large-scale studies of patient outcomes, ones that include virologic outcomes and HIV resistance testing, are needed to confirm this finding. Finally, we recognize that the small sample size may lead to imprecision to our estimates. A follow-up study of HIV DRM within a larger and better-defined group of first-line ART failing patients may necessary to confirm our findings here.
In conclusion, this study documents the HIV-1 drug resistance profile and genetic diversity in patients within the public health system in Zambia. Our major finding was the high frequency of NNRTI DRMs that may compromise the effectiveness of third-line salvage candidates such as etravirine and rilpivirine. Closer monitoring of DRM mutations at first-line failure can better inform clinicians and healthy policy advisors about what future ART regimens should comprise at a regional level. Ongoing surveillance for population antiretroviral drug resistance—using lower cost testing assays that can be performed in local settings—is urgently needed.
Supplementary Material
Acknowledgments
The programmatic work described herein was supported by the U.S. Centers for Disease Control and Prevention, through separate awards to the University Teaching Hospital (U2G GH000078), the University of Zambia (1U2G GH000109–01), and the Centre for Infectious Disease Research in Zambia (U2G GH000226). Additional trainee support (L.S.) was provided by the NIH Fogarty International Center (R24 TW007988). The authors would like to thank Dr. Ranjit Warrier and Thabiso M. Phiri for transferring the original in-house HIV DRM assay to CIDRZ Central Laboratory. They would also like to acknowledge the clinical and administrative staff of the Adult Infectious Disease Center (AIDC) at the University Teaching Hospital as well as the laboratory personnel at the CIDRZ Central Laboratory in Lusaka, Zambia.
Grant sponsor: U.S. Centers for Disease Control and Prevention; Grant number: U2G GH000078; Grant sponsor: University Teaching Hospital; Grant number: U2G GH000078; Grant sponsor: University of Zambia; Grant number: 1U2G GH000109-01; Grant sponsor: Centre for Infectious Disease Research in Zambia; Grant number: U2G GH000226; Grant sponsor: NIH Fogarty International Center; Grant number: R24 TW007988
Footnotes
Supporting Information: Additional supporting information may be found in the online version of this article at the publisher's web-site.
References
- Bennett DE, Myatt M, Bertagnolio S, Sutherland D, Gilks CF. Recommendations for surveillance of transmitted HIV drug resistance in countries scaling up antiretroviral treatment. Antivir Ther. 2008;13:S25–S36. [PubMed] [Google Scholar]
- Berrey MM, Schacker T, Collier AC, Shea T, Brodie SJ, Mayers D, Coombs R, Krieger J, Chun TW, Fauci A, Self SG, Corey L. Treatment of primary human immunodeficiency virus type 1 infection with potent antiretroviral therapy reduces frequency of rapid progression to AIDS. J Infect Dis. 2001;183:1466–1475. doi: 10.1086/320189. [DOI] [PubMed] [Google Scholar]
- Chi BH, Cantrell RA, Zulu I, Mulenga LB, Levy JW, Tambatamba BC, Reid S, Mwango A, Mwinga A, Bulterys M, Saag MS, Stringer JS. Adherence to first-line antiretroviral therapy affects non-virologic outcomes among patients on treatment for more than 12 months in Lusaka, Zambia. Int J Epidemiol. 2009;38:746–756. doi: 10.1093/ije/dyp004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi BH, Mwango A, Giganti M, Mulenga LB, Tambatamba-Chapula B, Reid SE, Bolton-Moore C, Chintu N, Mulenga PL, Stringer EM, Sheneberger R, Mwaba P, Stringer JS. Early clinical and programmatic outcomes with tenofovir-based anti-retroviral therapy in Zambia. J Acquir Immune Defic Syndr. 2010;54:63–70. doi: 10.1097/QAI.0b013e3181c6c65c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen MS, Chen YQ, McCauley M, Gamble T, Hosseinipour MC, Kumarasamy N, Hakim JG, Kumwenda J, Grinsztejn B, Pilotto JH, Godbole SV, Mehendale S, Chariyalertsak S, Santos BR, Mayer KH, Hoffman IF, Eshleman SH, Piwowar-Manning E, Wang L, Makhema J, Mills LA, de Bruyn G, Sanne I, Eron J, Gallant J, Havlir D, Swindells S, Ribaudo H, Elharrar V, Burns D, Taha TE, Nielsen-Saines K, Celentano D, Essex M, Fleming TR HPTN 052 Study Team. Prevention of HIV-1 infection with early antiretroviral therapy. N Engl J Med. 2011;365:493–505. doi: 10.1056/NEJMoa1105243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor EM, Sperling RS, Gelber R, Kiselev P, Scott G, O'Sullivan MJ, VanDyke R, Bey M, Shearer W, Jacobson RL. Reduction of maternal-infant transmission of human immunodeficiency virus type 1 with zidovudine treatment. Pediatric AIDS Clinical Trials Group Protocol 076 Study Group. N Engl J Med. 1994;331:1173–1180. doi: 10.1056/NEJM199411033311801. [DOI] [PubMed] [Google Scholar]
- Egger M, May M, Chêne G, Phillips AN, Ledergerber B, Dabis F, Costagliola D, D'Arminio Monforte A, de Wolf F, Reiss P, Lundgren JD, Justice AC, Staszewski S, Leport C, Hogg RS, Sabin CA, Gill MJ, Salzberger B, Sterne JA Cohort Collaboration ART. Prognosis of HIV-1-infected patients starting highly active antiretroviral therapy: A collaborative analysis of prospective studies. Lancet. 2002;360:119–129. doi: 10.1016/s0140-6736(02)09411-4. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Gonzalez S, Gondwe C, Tully DC, Minhas V, Shea D, Kankasa C, M'soka T, Wood C. Short communication: Antiretroviral therapy resistance mutations present in the HIV type 1 subtype C pol and env regions from therapy-naive patients in Zambia. AIDS Res Hum Retroviruses. 2010;26:795–803. doi: 10.1089/aid.2009.0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RK, Jordan MR, Sultan BJ, Hill A, Davis DH, Gregson J, Sawyer AW, Hamers RL, Ndembi N, Pillay D, Bertagnolio S. Global trends in antiretroviral resistance in treatmentnaive individuals with HIV after rollout of antiretroviral treatment in resource-limited settings: A global collaborative study and meta-regression analysis. Lancet. 2012;380:1250–1258. doi: 10.1016/S0140-6736(12)61038-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handema R, Terunuma H, Kasolo F, Kasai H, Sichone M, Yamashita A, Deng X, Harrigan PR, Montaner JS, Wegner SA, Verbiest W, Miller V, Wood R, Larder BA. World-wide variation in HIV-1 phenotypic susceptibility in untreated individuals: Biologically relevant values for resistance testing. AIDS. 2001;15:1671–1677. doi: 10.1097/00002030-200109070-00010. [DOI] [PubMed] [Google Scholar]
- Harrigan PR, Montaner JS, Wegner SA, Verbiest W, Miller V, Wood R, Larder BA. World-wide variation in HIV-1 phenotypic susceptibility in untreated individuals: Biologically relevant values for resistance testing. AIDS. 2001;15:1671–1677. doi: 10.1097/00002030-200109070-00010. [DOI] [PubMed] [Google Scholar]
- Johnson VA, Calvez V, Gunthard HF, Paredes R, Pillay D, Shafer RW, Wensing AM, Richman DD. Update of the drug resistance mutations in HIV-1: March. Top Antivir Med. 2013;21:6–14. [PMC free article] [PubMed] [Google Scholar]
- Kauf TL, Davis KL, Earnshaw SR, Davis EA. Spillover adherence effects of fixed-dose combination HIV therapy. Patient Prefer Adherence. 2012;6:155–164. doi: 10.2147/PPA.S28482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant Pai, Shivkumar N, Cajas S. Does genetic diversity of HIV-1 non-B subtypes differentially impact disease progression in treatment-naive HIV-1-infected individuals? A systematic review of evidence: 1996–2010. J Acquir Immune Defic Syndr. 2012;59:382–388. doi: 10.1097/QAI.0b013e31824a0628. [DOI] [PubMed] [Google Scholar]
- Roth AM, Holmes AM, Stump TE, Aalsma MC, Ackermann RT, Carney TS, Katz BP, Kesterson J, Erdman SM, Balt CA, Inui TS. Can lay health workers promote better medical self-management by persons living with HIV? An evaluation of the Positive Choices program. Patient Educ Couns. 2012;89:184–190. doi: 10.1016/j.pec.2012.06.010. [DOI] [PubMed] [Google Scholar]
- Seu L, Mwape I, Guffey MB. Single genome amplification of proviral HIV-1 DNA from dried blood spot specimens collected during early infant screening programs in Lusaka, Zambia. J Virol Methods. 2014;203:97–101. doi: 10.1016/j.jviromet.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Tang S, Lobashevsky E, Myracle D, Fideli, Aldrovandi G, Allen S, Musonda R, Kaslow A. Favorable and unfavorable hla class i alleles and haplotypes in zambians predominantly infected with clade c human immunodeficiency virus type 1. J Virol. 2002;76:8276–8284. doi: 10.1128/JVI.76.16.8276-8284.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinikoor MJ, Schuttner L, Moyo C, Li M, Musonda P, Hachaambwa LM, Stringer JS, Chi BH. Late refills during the first year of antiretroviral therapy predict mortality and program failure among HIV-infected adults in Urban Zambia. AIDS Res Hum Retroviruses. 2013;30:74–77. doi: 10.1089/aid.2013.0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainberg MA, Brenner BG. The impact of HIV genetic polymorphisms and subtype differences on the occurrence of resistance to antiretroviral drugs. Mol Biol Int. 2012;2012:256982. doi: 10.1155/2012/256982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis CL, Aga E, Ribaudo H, Saravanan S, Norton M, Stevens W, Kumarasamy N, Bartlett J, Katzenstein D A5230 team. Drug susceptibility and resistance mutations after first-line failure in resource limited settings. Clin Infect Dis. 2014;59:706–715. doi: 10.1093/cid/ciu314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis CL, Papathanasopoulos MA, Lakhi S, Karita E, Kamali A, Kaleebu P, Sanders E, Anzala O, Bekker LG, Stevens G. Affordable in-house antiretroviral drug resistance assay with good performance in non-subtype B HIV-1. J Virol Methods. 2010;163:505–508. doi: 10.1016/j.jviromet.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. Antiretroviral therapy for HIV infection in adults and adolescents: Recommendations for a public health approach 2010 revision. Geneva: World Health Organization; 2010. [PubMed] [Google Scholar]
- Zambian Ministry of Health. Adult and adolescent antiretroviral therapy protocols. Lusaka: Printech Press; 2010. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


