Abstract.
Optical tissue properties limit visible light depth penetration in tissue. Because of this, the recent development of optogenetic tools was quickly followed by the development of light delivery devices for in vivo optogenetics applications. We summarize the efforts made in the last decade to design neural probes that combine conventional electrophysiological recordings and optical channel(s) for optogenetic activation, often referred to as optodes or optrodes. Several aspects including challenges for light delivery in living brain tissue, the combination of light delivery with electrophysiological recordings, probe designs, multimodality, wireless implantable system, and practical considerations guiding the choice of configuration depending on the questions one seeks to address are presented.
Keywords: fiber optics, neuroscience, genetically-encoded sensors, opsins, light-tissue interactions
1. Introduction
In the last decade, the field of neurosciences witnessed the development of several optogenetic tools used to probe and modulate brain activity with a specificity never attained before.1 Optogenetic control of brain activity is achieved with the help of light-sensitive proteins such as ion pumps or channels and, more recently, through light-activated enzymes and G-protein-coupled receptors.2,3 When such receptors or ion channels are expressed at the cell membrane, for example, a simple flash of light can trigger activation or inhibition of cellular activity. The beauty of the technique lies in the fact that protein expression can be targeted to a very specific subpopulation of neurons, enabling selective activation or inhibition of that subpopulation regardless of the light confinement. Thus, for the first time, specificity in sensing or stimulating in the brain is not only determined by spatial factors, i.e., by how precisely the device used to probe is positioned and how limited in space its influence is. In fact, before optogenetics, in many cases, specific activation or sensing was simply impossible because different neuronal populations (often subserving opposite roles) are literally intermingled within the same area.
Neurons, with their colossal axonal and dendritic extensions, and their high levels of transport and secretion, are among the most transcriptionally active cells in the body. Therefore, in contrast to other systems, like the immune system for example, the complexity of neural circuits results in large part from the presence of an enormous diversity of cell types. The transcriptional programs that confer neuron identity enable these cells to maintain highly specialized phenotypes defined by their morphological, biochemical, and physiological properties.4 Given this complexity, an optogenetic strategy becomes essential to any effort to crack the neural code.
Multiple iterations of protein engineering have cascaded in recent years to refine and diversify sensing and actuating with light. The field is booming and we will briefly review some of the recent developments in Sec. 2. But the core objective of the present review is to summarize the strategies used to illuminate brain tissue and, in turn, simultaneously record brain activity, allowing real time readout of light effect on brain activity. Indeed, any efforts to develop novel light-responsive tools are undissociable from those to deliver (and collect) light. It may seem simple to shine light on a brain, but visible light can only poorly penetrate the cranium and light penetration in tissue, especially brain tissue, is limited by tissue optical properties (scattering and absorption). Red-shifted optogenetic actuators have been used with success for stimulation through the skull.5,6 Nevertheless, as a general rule, light delivery in in vivo experiments represents a significant challenge.
To achieve light delivery and electrographic recording from the intact central nervous system, researchers designed dual optical and electrical probes. The combination of optical and electrical elements in one single probe is referred to here as an optrode (the terms “optode” and “optitrode” are sometimes used in the literature). Some of the earliest optrode designs involved the use of standard commercial optical fibers and metal electrodes.7 Later, more sophisticated designs incorporated multiple light delivery and electrical recording sites.8 In other cases, the need to minimize tissue damage led to the development of new micrometric or flexible optrodes made from custom optical waveguides that incorporate electrical recording elements in their structure.9 Finally, recent advances in the developments of micro-LEDs (light-emitting diodes) made them an element of choice for the fabrication of optrodes. Given their small size and low-power consumption, they can be positioned directly at the desired light delivery site. This eliminates the use of optical fibers and raises possibilities for completely wireless systems and freely behaving animal experimentation.
2. Light to Control and Probe the Brain: a Brief Summary
This section will briefly review some of the main optogenetics tools developed over the last decade to control and probe brain activity with high-temporal resolution. Here, we focus primarily on control (actuators), but the congruently rapidly growing “probing” (sensors) aspect of optogenetics is equally important, especially in an effort to deploy multimodal interrogation of brain function.
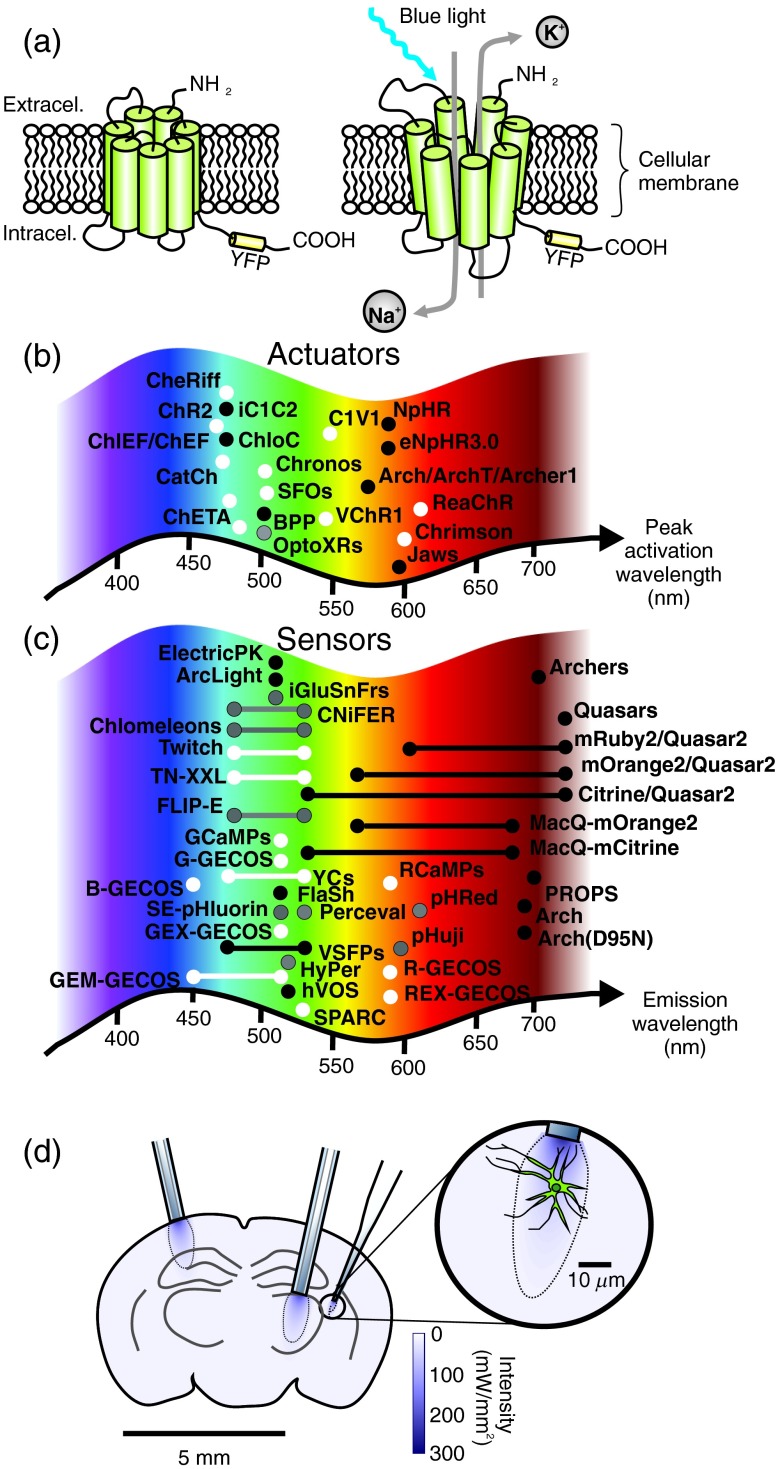
Well before optogenetic manipulation, light was used to induce photolysis of a compound,10,11 but the use or development of light-responsive proteins was especially attractive and became a real quest. In the early 2000’s, Miesenböck proposed the insertion of exogenous opsins in neurons to control their activity.12 Soon after, other successful neuronal insertions of light-responsive proteins were reported, such as the engineering of light-sensitive channels.13 This work was quickly followed by the development of light-sensitive membrane neurotransmitter receptors.14 The latter approach has the advantage of allowing one to study the contribution of specific ion channels or receptors to information coding by neurons, but these multicomponent approaches, requiring exogenous agents, remain complex to use. The development of single-component optogenetics, using channelrhodopsin15,16 has been the main catalyst for the growth of the field of optogenetics. Its main advantages are the combined simplicity of the system—a single component approach—and fast kinetics, allowing millisecond time scale optical control of neurons.17 Equally important is the subsequent development of single component inhibitory control of neural activity with halorhodopsin and archeorhodopsin.18–20 Figure 1(a) shows a schematic representation of channelrhodopsin-2(ChR2) before and after light activation.
Fig. 1.
Optogenetic tools and light tissue penetration: (a) schematic representation of a transmembrane channelrhodopsin protein in its closed (left) and opened (right) configurations following blue light illumination; (b) and (c) schematic representation of the peak activation wavelength of the actuator (white)/silencing (black) proteins listed in Table 1(b) and the fluorescent sensor proteins [voltage (black), calcium (white), and other sensors (gray)] listed in Table 2. Proteins for intracellular signaling control in panel b are represented in gray; (d) schematic representation of a coronal cross-section of a mouse brain illustrating the typical light penetrance achieved (distribution of irradiance, calculated using the equation in the Appendix) at 473 nm using a diameter optical fiber () and a micro-optrode.
The last decade witnessed an extraordinarily fast development of many light-sensitive proteins bioengineered to control brain activity, with refinements to modify conductance, ion selectivity, kinetics, spectral properties, and intracellular trafficking.1 Table 1 lists some of them. Originally, these proteins were activate by blue light, like ChR2, but today light-sensitive proteins absorption spectra covers much of the visible spectrum. Photocurrent characteristics of the different optogenetic actuators are also important to consider and are continuously being improved. Each variant comes with different intrinsic photosensitivity, but photocurrents are also dependent on the protein expression levels, the targeted cell type and the irradiance. Kleinlogel et al.21 compared photocurrents amplitude, rise time, and decay kinetics of different channel rhodopsin variants. Figure 1(b) represents the peak activation wavelength of the main optogenetic actuators designed to control cellular activity. It is important to keep in mind that opsin activation spectra are relatively large and are not limited to the precise activation peaks represented in Fig. 1(b). Hence, achieving two-color optical drives can be challenging due to the activation spectra overlaps.
Table 1.
Some of the main optogenetic actuators to control cellular activity.
| Protein | Absorption wavelength range (nm) | Action | References |
|---|---|---|---|
| (a) Excitatory tools | |||
| CHR2 | 400 to 500 | Cation channel | 17 |
| VChR1 | 500 to 550 | Cation channel | 22 |
| C1V1 | (peak) | Cation channel | 23 |
| ChETA | (peak) | Cation channel | 24 |
| ChEF | 450 to 500 | Cation channel | 25 |
| ChiEF | 450 to 500 | Cation channel | 25 |
| CheRiff | 450 to 500 | Cation channel | 26 |
| ReaChR | 590 to 630 | Cation channel | 6 |
| Chrimson | (peak) | Cation channel | 27 |
| Chronos | (peak) | Cation channel | 27 |
| CatCh | (peak) | -permeable channelrhodopsin | 21 |
| SFOsa |
450 to 590 |
Cation channel |
28 |
| (b) Intracellular signaling control | |||
| OptoXRs |
500 |
Intracellular signaling control (G protein) |
29 |
| (c) Inhibitory tools | |||
| NpHR | 550 to 620 | Chloride pump | 19 |
| eNpHR3.0 | 550 to 620 | Chloride pump | 20 |
| Arch | 500 to 600 | Proton pump | 30,31 |
| ArchT | 500 to 600 | Proton pump | 31 |
| Blue-shifted proton pump (BPP) | (peak) | Proton pump | 32 |
| Archer1 | 500 to 600 | Proton pump | 33 |
| ChloC | (peak) | Chloride channel | 34 |
| iC1C2 | 450 to 500 | Chloride channel | 35 |
| Jaws | 550 to 625 | Chloride pump | 36 |
Requires two activation wavelengths, one to initiate the current and a second to terminate it. There are also red-shifted versions of step function opsins (SFOs, see Ref. 37).
Yet, as mentioned earlier, optogenetics tools are not limited to actuators (activators and silencers), with their more recent incursion into light-controllable enzymatic reactions and even transcriptional processes, but also include many sensors: proteins developed so that their fluorescence is environment-dependent.1,38,39 For example, fluorescent voltage-sensitive proteins can be used to isolate the electrical activity of a subpopulation of cells.39,40 Until very recently, the temporal response and/or the signal-to-noise ratio of these proteins were unsatisfactory. However, the novel generation of fluorescent voltage sensitive proteins displays promising properties.41 Similarly, the use of calcium-sensitive proteins, such as the GCaMP or the GECO (genetically encoded calcium indicator for optical imaging) families,42,43 helps to report population-specific cellular activity and intracellular signaling events. Table 2 lists some of the main optogenetic sensors; their emission peak is also illustrated graphically in Fig. 1(c). Note that optogenetic sensing is not limited to transmembrane potential and calcium; proteins sensors for other ionic species, and now even for neurotransmitters, are becoming available (see Table 2). Akin to optogenetics actuators and other fluorescent dyes, efforts are being made to develop a red-shifted version of these proteins. Beyond spectral properties, challenges include optimization of the brightness, the temporal response, affinity, size, trafficking, and targeting of these proteins.
Table 2.
Some of the main optogenetic tools for sensing cellular activity.
| Protein | Excitation/emission wavelengths (nm) | Action | References |
|---|---|---|---|
| (a) Voltage sensors | |||
| FlaSh | 450/510 | Voltage sensor | 44 |
| VSFP1 | 440/480 and 530 | Voltage sensor | 45 |
| VSFP2.3 | 440/480 and 530 | Voltage sensor | 46 |
| SPARC | 470/500 to 550 | Voltage sensor | 47 |
| hVOS | 480/520 | Voltage sensor | 48 |
| ElectricPK | 485/510 | Voltage sensor | 49 |
| ArcLight | 485/510 | Voltage sensor | 50 |
| PROPS | 550/700 | Voltage sensor | 51 |
| Arch | 560/690 | Voltage sensor | 41 |
| Arch(D95N) | 585/690 | Voltage sensor | 41 |
| QuasAr1/QuasAr2 | 590/715 | Voltage sensor | 26 |
| Archer1/Archer2 | Voltage sensor | 33 | |
| MacQ-mCitrine | 500/530 and 580 | Voltage sensor | 52 |
| MacQ-mOrange2 | 530/560 and 580 | Voltage sensor | 52 |
| Citrine eFRET GEVI (QuasAr2) | 500/530 and 715 | Voltage sensor | 53 |
| mOrange2 eFRET GEVI (QuasAr2) | 530/560 and 715 | Voltage sensor | 53 |
| mRuby2 eFRET GEVI (QuasAr2) |
560/600 and 715 |
Voltage sensor |
53 |
| (b) Calcium sensors | |||
| GCaMPs | 490/510 | Calcium sensor | 43,54 |
| RCaMPs | 570/585 | Calcium sensor | 55 |
| G-Gecos | 490/510 | Calcium sensor | 42,56 |
| B-Gecos | 375/450 | Calcium sensor | 42 |
| R-Gecos | 565/585 | Calcium sensor | 42 |
| Twitch | 435/470 and 530 | Calcium sensor | 57 |
| GEM-GECO1 | 400/450 and 510 | Ratiometric calcium sensor | 42 |
| GEX-GECO1 | 400 and 480/510 | Ratiometric calcium sensor | 42 |
| REX-GECO1 | 480 and 570/585 | Ratiometric calcium sensor | 58 |
| Cameleon derivatives (YCs) | 435/470 and 530 | Ratiometric calcium sensor | 59 |
| TN-XXL |
435/480 and 530 |
Calcium sensor |
60 |
| (c) Others | |||
| FLIP-E | 435/480 and 530 | Glutamate sensor | 61 |
| iGluSnFrs | 485/510 | Glutamate sensor | 62 |
| Chlomeleon/Super Chlomeleon | 435/470 and 530 | Ratiometric chloride sensor | 63,64 |
| CNiFER | 435/480 and 530 | Acetylcholine sensor | 65 |
| pHRed | 440 and 580/610 | Ratiometric pH sensor | 66 |
| SE-pHluorin | 400 and 475/510 | pH sensor | 67 |
| pHuji | 570/600 | pH sensor | 67 |
| Perceval | 405 and 490/530 | ATP/ADP sensor | 68 |
| HyPer | 430 and 500/520 | sensor | 69 |
3. Light Delivery in Living Tissue
Brain tissue is not transparent to visible light and one of the main challenges for the use of optogenetics in vivo is the limited light penetration and confinement in deeper structures. Indeed, due to light scattering and absorption, a usable amount of visible light cannot easily reach deep brain structures and illumination from an external source is, in practice, limited to the cortex [Fig. 1(d)]. The Beer–Lambert equation can be used to estimate light attenuation after propagation in tissue:
| (1) |
In Eq. (1), stands for the light intensity after a traveled distance of , is the initial intensity, and is the extinction coefficient. This coefficient can be calculated from the absorption and scattering coefficient in biological tissue, and , respectively:
| (2) |
Both and depend on the wavelength, . Note that light scattering in tissue is anisotropic, meaning that light will scatter with preferential angles. The value thus incorporates an anisotropic factor, :
| (3) |
Extinction coefficients and anisotropic factors for brain tissues (white and gray matters) are listed in Table 3.
Table 3.
Brain tissue absorption and scattering coefficient and anisotropic factor for .70
| Parameters | Tissue type | Values |
|---|---|---|
| White matter | 0.8 | |
| Gray matter | 0.88 | |
| White matter | ||
| Gray matter | ||
| White matter | 1 to | |
| Gray matter | 0.17 to |
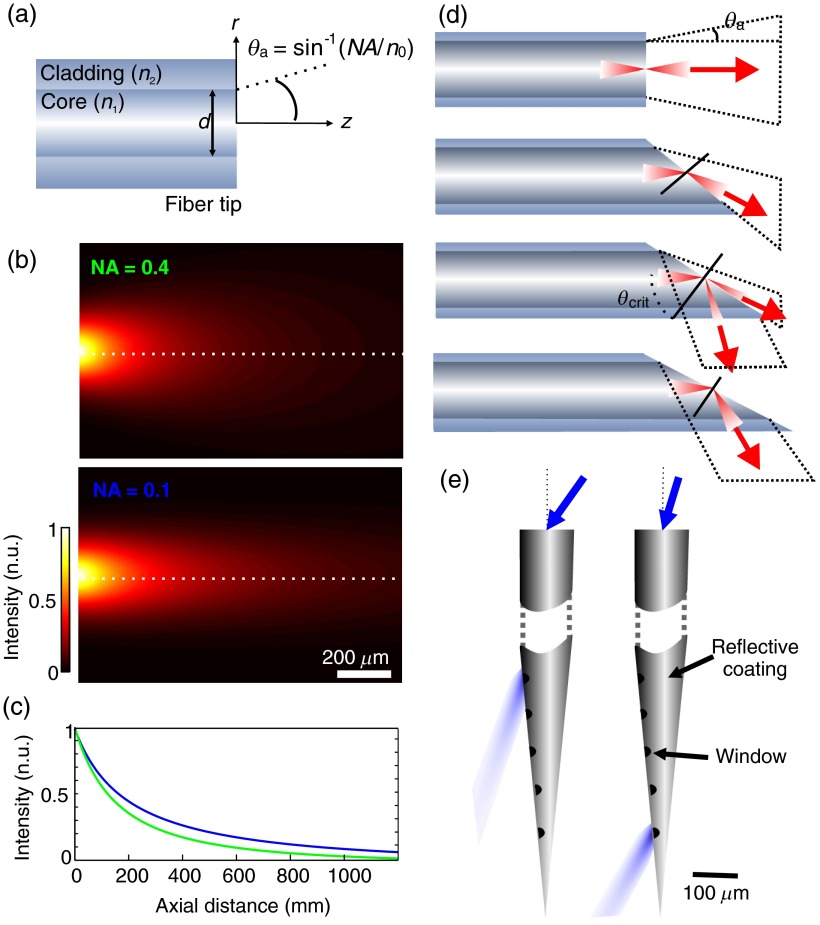
Optical fibers thus offer a convenient alternative to reach deeper structures. Two main aspects are important when choosing the right fiber: the optical fiber core size, which should be in the same order of magnitude than the targeted area, and the optical fiber numerical aperture (NA). The later influences the transversal spread of the illumination volume and is defined as
| (4) |
where and are the refractive indices of the fiber core and cladding, respectively [Fig. 2(a)].
Fig. 2.
Tissue irradiance profiles and strategies for controlled light delivery: (a) schematic representation of an optical fiber tip and the calculated irradiance profile (b) at its tip for and ; the diameter of the fiber core used in the calculation was and values of absorption and scattering were chosen for a wavelength of 473 nm; (c) axial normalized intensity profile at fiber center, corresponding to the dashed lines in (b); (d) different irradiance profile obtained from different tip geometries (adapted from Vo-Dinh70); (e) flexible illumination location from a coated tapered fiber (adapted from Ref. 71). Optical windows ( wide) were made along the tapered shaft. Using different light input angles, different zones are illuminated.
Knowing these values and those of tissue optical properties (extinction coefficients and refractive index), one can easily estimate the effective excitation volume at the fiber tip. For neuron activation, the outer limit of this volume that can be defined has the minimal intensity value for action potential generation, which is reported to be approximately in the case of ChR2.72 As stated earlier, this value will differ from one protein to another and from one subject to another since the membrane expression level of the protein may vary. The volume of activation can always be enlarged by increasing light intensity at the fiber tip (increasing the number of recruited cells), but this intensity must be kept below tissue damage threshold. This threshold will be different for the different protocols used and is dependent on the stimulation duration and repetition rate, but in most cases an intensity at the fiber tip around was reported to be safe.73 However, for sustained stimulation, damages were reported at levels above .74 Using this input intensity at the tip of an optical fiber of core diameter with an NA of 0.2, the limit for ChR2 activation () is reached at a distance of . A schematic representation of the irradiance profiles at different fiber tips are represented in Fig. 1(d) (also see Ref. 75 for an illustration of blue light penetration in cortical tissue). The dashed line represents the activation threshold. A number of papers describe methods for calculating illumination profile or volume at optical fiber tips, mainly for other purposes (spectroscopy, fluorescence or phototherapy).8,9,76–78 In most cases a simple geometric propagation and extinction coefficient is sufficient to have a rough estimate of the illumination profile. Figure 2(a) shows a schematic representation of an optical fiber tip and its acceptance or irradiance angle (). Figures 2(b) and 2(c) show a two-dimensional representation of an irradiance profile calculated from a simplified MATLAB® script (see Appendix) for two different values of NA. Profiles were calculated from the model described by Foutz et al.78 The Stanford University Optogenetics webpage offers a helpful tool for irradiance estimation.79 It is important to mention that the irradiance profile can be shaped using different fiber tip geometries and coatings.71,80 Figure 2(d) shows example of different fiber tips and the illumination they provide. This can be extremely useful to limit the light irradiance to a defined layer or region within the tissue.81 Recently, Pisanello et al.71 proposed an innovative method to achieve multipoint illumination from a single tapered waveguide. Its design takes advantage of the angular dependence of the losses in a tapered waveguide [see Fig. 2(e)]. Using different injection angle, they could control the illumination location.71 This approach is promising to vary the simulation site, and can be used, for example, to alternatively stimulate different brain layers via the same optical route. However, the precision of the light injection angle at the fiber input must be firmly regulated and the total input power requirements are much higher.
4. Combining Light Delivery with Electrographic Recordings: Optrodes Designs
4.1. Preliminary Designs
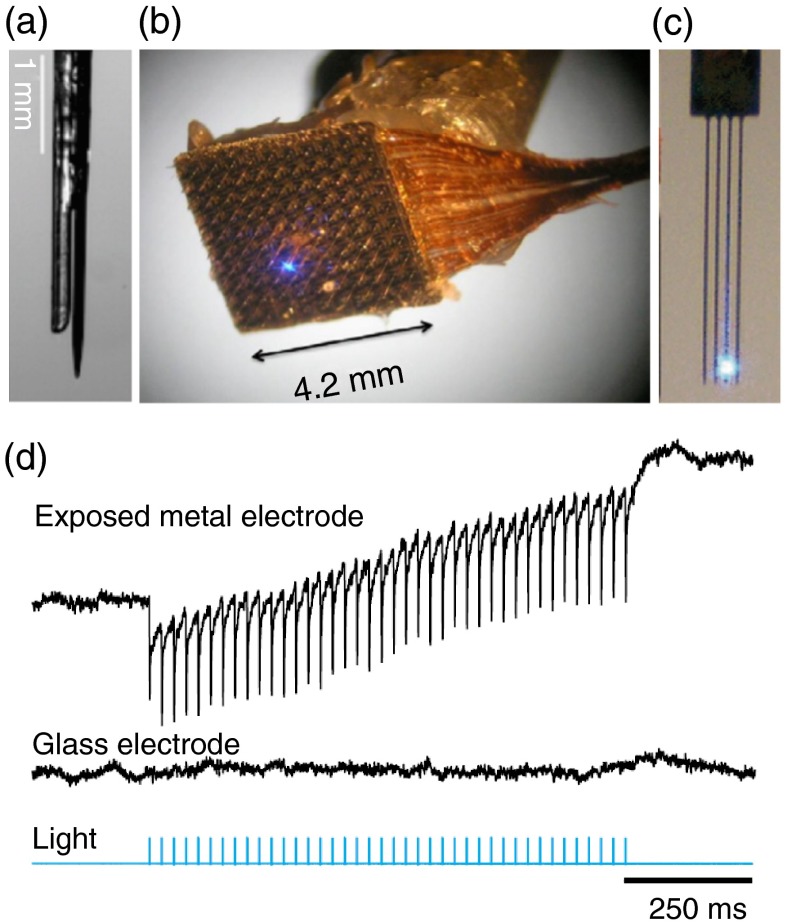
Monitoring cellular activity at the stimulation site is necessary to confirm the illumination effect on the targeted population of neurons. Brain activity can be monitored in various ways. Optical functional imaging, functional magnetic resonance imaging, and behavioral assessments are examples of possible readout strategies.7 However, one of the most popular methods used in neurosciences research remains electrophysiology. It offers a high-temporal resolution and can probe activity at the cellular or network levels. Electrodes can be implanted and used for several weeks or months. Unlike fluorescent measurements using calcium or voltage fluorescent sensors, electrophysiological recordings come at virtually no cost; no dye introduction or transfection is needed. Extracellular recording is noninvasive to the cell, minimally fade with time (unlike photobleaching of fluorescent dyes), and is nontoxic (no photochemistry involved). It is thus, not surprising that this modality is often preferred over others. However, it is important to note that electrode implantation causes a certain amount of tissue and vascular damage, and they are susceptible to photo-electric artifacts (discussed in Sec. 4.2), to cross-talk between recording channels and signal degradation over long periods of time.82 Therefore, the right electrode assembly is primordial. By varying electrode types and frequency bandwidth, one may choose to record single units, multiunits, local field potentials, corticograms or even electroencephalograms.83 The need for combined optical light guides and electrodes (here referred to as optrodes) was present in the field of neurosciences before the beginning of optogenetics and preliminary designs of optrodes were made in the past for different applications.84,85 A major application for these early optrodes was simultaneous electrophysiological recordings and fluorescence collection. For example, Duff Davis, and Schmidt84 combined extracellular recordings to calcium-sensitive fluorescence measurement in awake animals using optrodes. Original optrode designs simply combined a commercial optical fiber to a commercial extracellular electrode.7,72 Figure 3(a) shows a micrograph of such an optrode. This is a flexible, simple, and low-cost design that can easily be integrated in any electrophysiological recordings setup. In most cases, researchers choose conventional recording modes such as extracellular single-or multiunit or local field potential recordings, but other types of recording electrodes, such as ion-sensitive electrodes, can be used.86 An alternative to optical fibers for light delivery is to use a glass pipette recording electrode as the light guide. This is obviously not an optimal waveguide, but optical losses can be compensated by a higher input power. The pipette can thus serve simultaneously as an electrode and an illumination channel. Note that optical losses will occur along the shaft of the tapered electrode and a metal coating should be used to confine illumination at the electrode tip. Along this line, recently, Katz et al.87 presented the “optopatcher” that could potentially be used for in vivo patch clamp.
Fig. 3.
Optrode designs: (a) optrode design made from a core multimode fiber and a metal recording electrode;7 (b) integrated device that combines a multielectrode array and a fine illumination site;88 (c) multiarray silicon probes with integrated optical fibers for spatiotemporal brain control and recording;89 and (d) simultaneous local field potential recordings from mice tissue not expressing opsins with light-exposed metal (upper trace) and glass microelectrodes (middle trace). Light stimulation is represented in blue. Note the photo-electric artifacts in the upper trace (modified from Ref. 74).
More sophisticated optrode designs included several recording sites90,91 or several light stimulation sites or wavelengths.92 Zhang et al.,88 for example, developed an integrated device that combines a multielectrode array and a fine illumination site. This site was made from an etched multimode fiber on which gold was deposited. The fiber tip was left bare providing an optical aperture diameter of . The gold thin film served as an extracellular electrode. This etched optical fiber was integrated into a recording sites silicon-based multielectrode array fabricated by Blackrock Microsystems. Figure 3(b) shows a micrograph of this device. This design allows for spatio-temporal analysis of the electrical activity in optogenetics experiments. The illumination, though, was limited to a single restricted area. The device was recently used for in vivo chronic experimentation.93 However, given its geometry, it is not really amenable for recording from deeper brain structures. Other optrode designs incorporate several illumination sites and wavelengths; in some cases, to help shape the illumination profile94 and in other cases to provide neuronal control at multiple sites.8,89,95–99 The latter is of particular interest for local circuitry dissection. Figure 3(c) shows a representation of an opto-electronic array manufactured from multipolar silicon electrodes and fibers.8,89
Optrodes, such as those described here, were used for several neurosciences applications. For example, they were used to help deconstruct Parkinson neural circuitry,72 to demonstrate the role of pyramidal cells and interneurons interaction for the generation of ripple oscillations,100 and to demonstrate that parvalbumin interneurons mediate inhibition-induced theta-resonance in cortical networks.92
It is important to stress that, in addition to the geometry, the irradiance profile and the electrical properties of optrodes, light sources must be chosen with care for an optimized optogenetic stimulation setup. Important factors to keep in mind when choosing the right light source are: emission wavelength, emission power, and the possibility to modulate emission intensity with high-temporal resolution. Optical fiber can be coupled to a lamp, a LED or a laser depending on the light stimulation needs. Not all laser sources can be modulated by an external signal. However, mechanical shutters, filter wheels, and/or acousto-optic modulators can be used to tune illumination as needed. These must be chosen considering the needs of the experiments in mind. Mechanical shutters are slower than acousto-optic modulators and do not provide switching capabilities with temporal precision comparable to that of the latter. Yet, acousto-optic modulators are expensive and optimized for a given wavelength, limiting their use in multimodal systems. In addition, factors such as the divergence of the light source, the size of the optical fiber, its NA, the presence of a taper or any optical components in the light path likely to induce optical losses must be considered when choosing the minimal source output power needed.
4.2. Photo-Electric Artifacts
An important factor to keep in mind when designing and fabricating optrodes is the generation of photo-electric artifacts. When metal electrodes are exposed to light with the right frequency or wavelength, the photo-electric effect frees electrons generating measurable currents. This results in sharp artifacts in the electrophysiological recordings. Beltramo et al.101 proposed a method to subtract these artifacts. They reproduced their light stimulation protocol postmortem, evaluated the photo-electric artifacts and subtracted them from their recordings. Others used sinusoidal light stimuli to prevent spike waveforms distortion.102 These artifacts can be reduced by minimizing the electrode exposure to light, by choosing the right material or by using nonmetal electrodes, such as glass electrodes.74 Indeed, optrodes made from all-glass electrodes9,87 are free of photo-electric artifacts. Figure 3(d) shows simultaneous local field potential recordings from mice tissue not expressing opsins with light-exposed metal and glass microelectrodes. Unlike the signal recorded from the metal electrode, the signal recorded from the glass microelectrode is free of photo-electric artifacts. Efforts are also on-going to develop metal-based electrode free of photoartifacts.103
4.3. Specialty Fibers and Waveguides
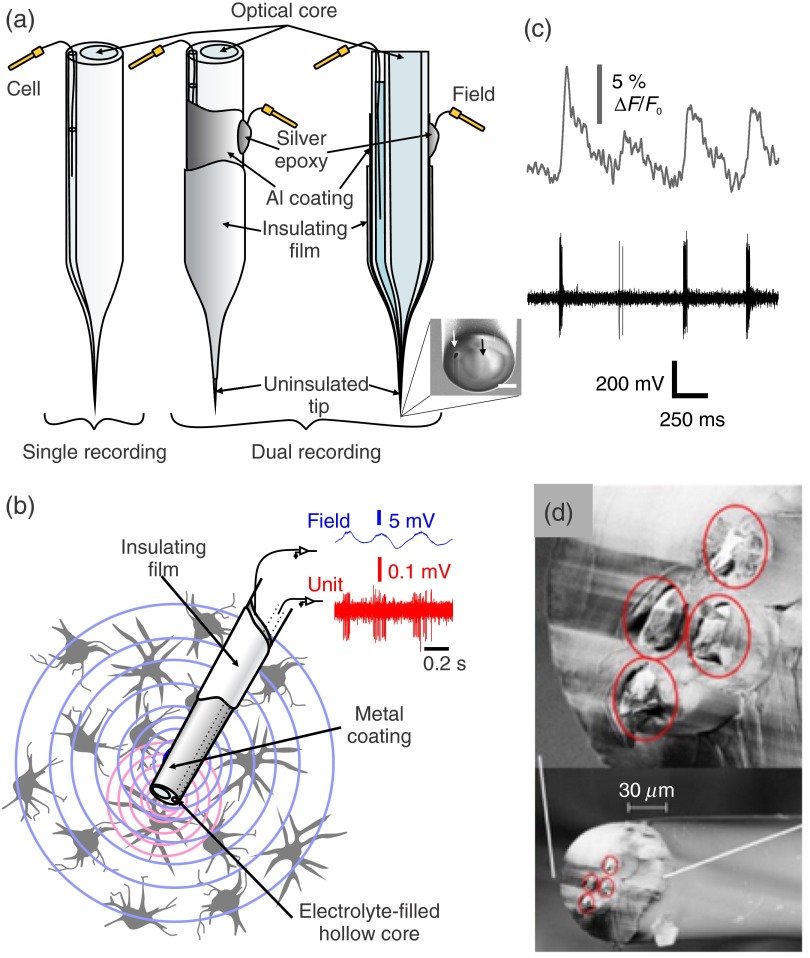
Most optrode designs are based on commercially available multimode optical fibers, but researchers have also customized fibers to embed electrodes directly in their structure. LeChasseur et al.9 developed a dual core optical fiber for simultaneous fluorescence collection and extracellular recordings. Their fiber structure incorporates two parallel cores: a standard optical multimode core and a hollow core which, once filled with an electrolyte solution, can be used as a regular glass microelectrode. They demonstrated that this customized fiber could also be used for optogenetics purposes.9,104 Figure 4(a) shows a schematic representation of the dual core fiber. The structure can be tapered and the size of the final micro-optrode diameter can be chosen to achieve very precise cellular level optical stimulation (and/or monitoring) and electrical recordings. Metal coatings can be added on the external wall of the fiber to provide additional extracellular local field potential recordings. This offers the advantage of recording multiple different electrophysiological signals, larger field potentials, and single unit activity at the same time as calcium sensor fluorescence, in parallel [Figs. 4(b) and 4(c)]. Since extracellular electrophysiological recordings cannot directly impact membrane potential, single cell optogenetics can be valuable to probe membrane properties via photo-stimulation from the extracellular medium. Lin et al.105 chose to embed an electrical recording element (a twisted-wire tetrode) in their waveguide structure [Fig. 4(d)]. A more recent fiber design allowed for the combination of optical stimulation, electrical recordings and drug delivery.106 Canales et al.106 described several strategies to combine multiple electrical recording sites and drug delivery channels in single probes with diameter ranging from 416 to 85 μm. This technology uses polymers and presents a higher flexibility than a glass optrode. Note that configuration with optical fibers are not limited to photo-stimulation; the light waveguide can also serve for fluorescence detection.9 In the latter case, the micro-optrode presents several advantages over other approaches with sparse labeling because it enables placing the collection tip very close to the cell, providing ideal signal sensitivity to monitor intracellular events reported by optogenetic sensors9 [Fig. 4(c)]. For fluorescence detection, high-quality glasses should be chosen over polymers in the fabrication of optrodes to avoid the collection of intrinsic fluorescence and background signal.
Fig. 4.
Micro-optrodes made from customized optical fibers: (a) schematic representation of a tapered dual-core fiber (left). One core (optical-core) serves as an optical channel and the second, a hollow core, is filled with an electrolyte to serve as an extracellular single unit recording electrode.9 Metal coatings can be added further on the external wall of the fiber to provide additional extracellular local field potential recording capabilities. The middle panel shows a three-dimensional view of the coated probe and the right panel a transverse cut view.104 The inset is a scanning electron micrograph of the fiber tip (scale bas is ). The white arrow points at the hollow core used for electrical recording and the black arrow points at the optical core. (b) Schematic representation of the electrophysiological signals recorded with the metal coated fiber (wide field potentials) versus the electrolyte-filled hollow core (single unit spikes) of the same fiber.104 (c) Example of simultaneous cellular level electrophysiological and calcium monitoring (adapted from Ref. 9). (d) Micrograph of a flexible optitrode made from polymer and embedded twisted-wire tetrode (modified from Ref. 105).
4.4. Optrodes in Freely Behaving Animals
When choosing optrodes made from optical fibers in experiments with freely moving animals, one has to prevent possible tangling and animal mobility restriction due to optical and electrical cabling. One way to achieve this is to use rotary joints to connect the optical implanted optrodes to the light source.107,108 A schematic representation of an optical patch cord and an electrical cable connected to rotary joints is shown in Fig. 5(a). Rotary joints serve to ease the animal motility and prevent torsion on the animal head and the implanted devices. They are now available commercially in industries specialized in fiber optics components or optogenetics supplies and they can even be adapted to accommodate multichannel connections.109 An alternative is to use light-weight cables and head mounted light sources.8 In some cases, optogenetics is used to decrypt brain macrocircuitry and electrode implantation will be necessary not only in the activated zone, but also in a remote postsynaptic zone. Designing completely wireless optogenetics systems [see Fig. 5(b), Ref. 110] is attractive for freely behaving experiments and a major challenge for using optical fiber in optrode designs for freely behaving animals lies in the fact that they are not easily adaptable to wireless recordings. A fiber optic patch cord has to be connected to the light source. This is where micro-LEDs open new avenues. Recent advances in power consumption, output power, size, heat generation, and dissipation of blue micro-LEDs make them promising candidates for wireless implantable optogenetics devices.8,111–113 Wireless implantable electrophysiological recording chips is a common theme in biomedical and electrical engineering;114 and now, a rising portion of these implantable chips offer micro-LEDs for optogenetic stimulation.115–117 Although miniaturization remains a challenge, efforts are now being made to print optoelectronic devices on thin malleable substrates, ideal for chronic implants. Figure 5(c) shows a schematic representation of the layer composition of a multifunctional, implantable optoelectronic device incorporating various components that can be used for wireless optogenetics and electrophysiology.117 Figure 6 represents a flow chart intended to help in choosing the appropriate tools for in vivo experimentation. In short, any optrode type could be used for acute experimentation in anesthetized or restrained animals. However, for experiments in freely behaving and presumably long term recording, optrodes with solid-based electrodes (e.g., metal, silicon, and polymer) should be employed. The optrode illumination field must be optimized for the size of the targeted cell population. Micro-optrodes (diameter ) must be used for single cell activation. In principle, all fiber-optic sensors can be used for fluorescence detection using proper fluorescence optical beam splitters and filters, but again the size of the fiber tip must be adapted to the size of the population targeted. For LED-based optrodes, an optical waveguide or a photodetector could be added for fluorescence detection.
Fig. 5.
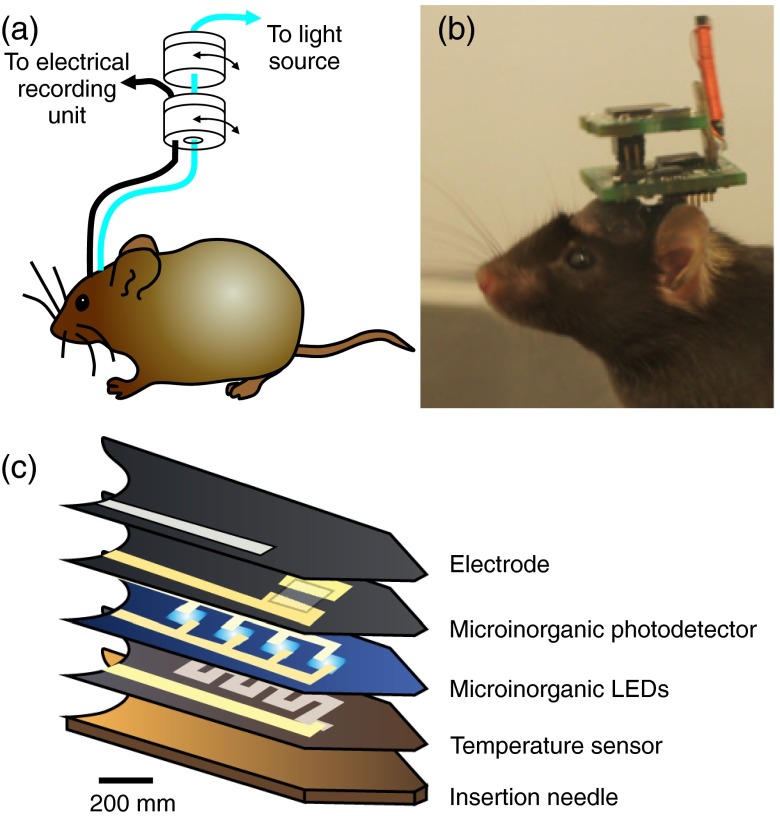
Multimodal optogenetics configurations for experiments in freely moving animals: (a) representation of a mouse implanted with a fiber optic cannula and a chronic electrophysiogical electrode and the rotary joints enabling free movements of the animal; (b) image of a mouse implanted with a wireless optogenetic system (Figure modified from Ref. 110); (c) schematics of the layer composition of multifunctional, implantable optoelectronic device incorporating various elements that can be used for wireless optogenetics and electrophysiology (adapted from Ref. 117).
Fig. 6.
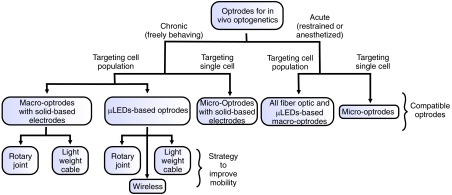
Flow chart of the different possible avenues and associated compatible optrode designs including strategies to improve mobility.
4.5. Combining Light Delivery and Optical Monitoring Modalities
Electrophysiological recordings, providing various possibilities and high-temporal resolution, are extremely popular as a readout strategy. Yet, as mentioned previously, optrodes can also be used for optical fluorescence monitoring.9,84,86,104,118 For example, They can serve to excite and record signals from voltage or calcium sensitive fluorescent probes allowing for an all-optical optogenetics strategy.119 Optrodes can also help to confirm opsin expression at the optrode implantation site9 by detecting the fluorescence of the reporter gene. Fluorescence signal detection is achieved by integrating a fluorescence detection light path at the optical fiber end9 and, again, the fiber size must be adapted to the experiments. For example, large probes are not ideal to detect the location of a sparse, evenly distributed cell population, but they can be used to detect a localized cell population or to monitor bulk voltage or calcium signals originating from specific subpopulations of cells. Fluorescent signal detection via optrodes can be brought down to the single cell level and be used to identify a genetically or retrogradely identified cell9 allowing targeted electrophysiological recording and optogenetic stimulation in deeper brain structures. In contrast to electrophysiological recordings, multicolor optical detection and activation/inhibition can serve to simultaneously probe differently labeled populations of cells and help discriminate their different roles in information processing and signal generation. It also offer the capability to probe intracellular signalling events such as calcium waves [see Fig. 4(c) adapted from Ref. 9], enzymatic reactions through reporter genes, or even molecular interactions through Förster resonance energy transfer, for example. For such multimodal sensing/actuating experiments, proteins must be chosen to avoid overlap between imaging (excitation and emission) and opsin activation wavelengths. A promising avenue for fluorescence detection is the combination of CMOS detector array and micro-LEDs120 or the use of microendoscopy121–123 to allow for combined spatiotemporal segregation of optical recordings. Yet a major challenge in the case of sensing remains the availability of photons, which are much more limited on the emission than on the excitation side. Fore sparse fluorescent labeling, the usability of large probes for optical monitoring purposes is limited. Micro-optrode designs, with micron to few tens of micron-scale tips, allowing local measurements with minimized optical cells-to-probe paths, offer promising avenues for integrating optical control and monitoring in deep brain structures.9
An alternative to using optrodes in anesthetized or restrained animals is in vivo optical imaging which is a readout method of choice for superficial brain regions easily accessible via a cranial window.124–126 Although optical imaging modalities are limited in depth, they offer submicrometer spatial resolution and can be used to discriminate several cells or cell compartments within the same field of view. As with optrodes, optical imaging can be used for multicolor detection, membrane potential monitoring, intracellular calcium concentration measurements, and so on. To overcome depth accessibility, microendoscopes have been developed for in vivo imaging,127,128 but the size of the light guide structure to be implanted renders this alternative more invasive than fluorescent microprobes. For this reason, when imaging of cell population is not necessary, fiber-optic microprobes should be chosen.9,86,104 Although often sold as more flexible than all-electrical or opto-electrical solutions, all-optical solutions also have drawbacks. For example, the possible overlaps between the optical actuator and sensor spectra, the slow dynamics or weak fluorescence properties or sensors, intrinsic absorbance, fluorescence, and scattering properties of tissue for visible wavelengths, photodamage, photobleaching, and sparse or weak labeling.
5. Choosing the Right Probe for Each Application
Simultaneous optogenetics activation and electrophysiology can be achieved by different means. Optrodes should be used for in vivo applications, especially when access to the tissue of interest is limited. No single design offers a universal solution, but the combination of optical and electrical elements of different sizes and geometries can be permutated allowing for a large spectrum of optrodes designs addressing different needs (see Fig. 7). One can simply add to a standard extracellular electrode a commercial waveguide, available in different sizes with different NAs or fiber tip geometries. Large optical fibers (100 to ) are ideal to activate large population of cells [Fig 7(a)]. They can be placed at the brain surface or inserted deeper in the tissue via a cannula, but they are relatively invasive. As such, when the targeted region is deeper, a thinner or flexible structure should be envisaged to limit tissue damage. An important aspect to consider is that to activate a large area of the brain, multiple activation zones should be used instead of a using a single source to cover the same volume. This provides a more uniform illumination and helps prevent photodamage at the fiber output. Using a single point source to activate a large area of the brain produces a gradient of light across the brain structure and different regions/nuclei will be activated to different extents. This is mitigated on the excitation side because action potential generation is a nonlinear process but inhibition is a more “linear” process, so the gradient of illumination pattern for silencing is a major issue to keep in mind.
Fig. 7.
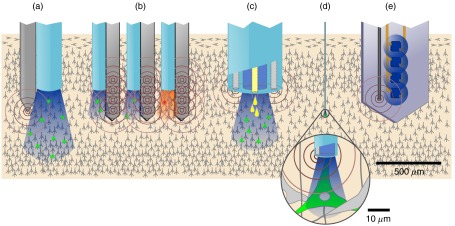
Schematic representation of different types of combined electrical and optical probe configuration, their illumination field (blue or red) as well as their electrical recording field (brown concentric circles). Drug delivery capability is represented by yellow drops. Drawings are to relative scales to illustrate the different bulkiness of each approach as well as the spatial relationships between the optical and electrical fields and cell populations being probed: (a) macro-optrode with silicon or metal probe, (b) multichannel penetrating optrode, (c) multifunctional optrode, (d) micro-optrode, and (e) implantable multimodal optoelectronic.
To gather more information, extracellular electrodes can be replaced by silicon probe electrode arrays, allowing simultaneous recordings of multiple sites upon optogenetic control [Fig. 7(b)]. They add a spatial component to optogenetics which can be instrumental for studying layer to layer interactions or the spatial propagation of brain waves. Custom made waveguides are ideal to incorporate electrophysiology, illumination, and drug delivery [Fig. 7(c)]. Yet the spatial resolution achieved, especially on the light control side, remains limited. Tapered or chemically etched optical fibers are valuable to achieve confined optogenetics activation and stimulate smaller population of cells, thin projection pathways or even single cells [Fig. 7(d)]. Optical waveguides also allow for fluorescence collection and offer flexibility for illumination. Importantly, several light sources can be coupled into the same implanted optical fiber. Further, microfabrication of waveguide arrays render possible the multiplexing of the activation zones.95
The choice of the electrical recording element is an important factor when choosing the electrode. The illumination of metal electrodes produces undesired photo-electric artifacts. The use of an all glass optrode9 is a good alternative to avoid such artifacts. All-glass micro-optrodes, because of their geometry, may emerge as an ideal tool for local, well-balanced light delivery, and especially for optical measurements, but these advantages apply to the control/monitoring of small brain areas. All-glass electrodes with hollow cores also offer access to all of the techniques associated with use of glass micropipettes, such as local drug or dye delivery by microiontophoresis or pressure ejection. However, they can hardly be used for chronic implantation.
For experimentation on freely behaving animals, wireless electrophysiology and micro-LEDs control should be envisaged. Although they remain bulky and thus rather invasive, implantable multimodal optoelectronic devices are emerging components [Fig. 7(e)]; the weight of their electronic board is becoming sufficiently small to be supported on a mouse head.110,117
6. Future Avenues
The booming field of optogenetics, with its enormous promises for cracking the neural code, understanding brain function, and eventually treating brain pathologies, is stimulating accelerated developments on the engineering side with the need for improved light delivery and readout strategies. The ultimate vision is that of miniaturized implants for seamless multimodal interrogation and control with unprecedented specificity in freely behaving subjects. Challenges on the hardware engineering side include: reducing the size of the optrodes and optoelectronic devices, minimizing their invasiveness and power consumptions, optimizing their data acquisition speed, and spatial resolution. New designs may include new material such as graphene (a material with promising optical and electrical properties),129 indium tin oxide optical fibers or coatings (transparent conductors),130 or organic transistors,131 and complex microstructuring strategies.132 Multicolor sensing and activation capabilities, within the limited spectral window offered by visible and near-infrared light, will be key to match the level of complexity that characterizes neural circuits. This will impose developments on the signal processing side as well as spectral control of light delivery.
Acknowledgments
This work was supported by the Canadian Institutes of Health Research and the Natural Science and Engineering Research Council of Canada. Y. D. K. was funded by a Chercheur National award from the Fonds de la Recherche en Santé du Québec. S. D. acknowledges the MITCAS Elevate Program, the SAVOY Foundation, and the Toronto Western Research Institute for postdoctoral fellowships. We thank S. Côté for expert assistance in generating some of the illustrations. We also thank P. Bazzigaluppi and Iliya Sigal for helpful comments on the manuscript.
Biographies
Suzie Dufour is a postdoctoral fellow at the Toronto Western Research Institute under the supervision of Dr. Peter L. Carlen and Dr. Ofer Levi. She received her BSc degree in physics from Laval University in 2004 and her PhD in neurobiology in 2012. Her PhD involved the design and fabrication of micro-optrodes for in vivo experimentation. Her current research interests include optical in vivo brain imaging, optogenetics, electrophysiology, brain ionic homeostasis, and epilepsy.
Yves De Koninck is professor of psychiatry and neurosciences at Laval University (Québec, Canada). He is scientific director of the Quebec Mental Health Institute and member of the Optic, Photonic and Laser Center (COPL). He obtained his PhD in physiology from McGill University. He founded the Neurophysics program (http://www.neurophysics.ca) and the Neurophotonics Centre (http://www.neurophotonics.ca) now core to the Canadian Neurophotonics Platform. His research interests are sensory processing, pain, and novel approaches to study brain function.
Appendix.
This appendix includes the MATLAB® script for the irradiance profile calculation at an optrode or optical fiber tip that was used for the preparation of Figs. 1 and 2. It is based on the model described in the Ref. 78.
function B = Intensity_profile(NA, r0)
% This function calculates the illumination profile at an optical fiber
% tip. It is based on the model described by T. J. Foutz (J Neurophysiol,
% 107(12), 2012).
% Function inputs: Optical fiber numerical aperture (NA), core radius in
% microns (r0).
% Function outputs: 2D calculated profile (B).
n0 = 1.36; % Tissue refractive index
rmax = 2*r0; % Transverse plot dimensions (micrometers)
zmax = 6*r0; % Longitudinal plot dimensions (micrometers)
dz = 10; % z increment (micrometers)
dr = 0.5; % r increment (micrometers)
K = 0.0001;% absorption coefficient (um^-1)
S = 0.005;% Scattering coefficient (um^-1)
l=0;
j=0; %indices
theta = asin(NA/n0); % Divergence half-angle
for z = 0:dz:zmax
z0(l+1)=l*dz;
l=l+1;
for r = -rmax:dr:rmax
j=j+1;
C = r0/(r0 + z*tan(theta)); % Geometric factor
G = (1/(2*pi))*exp(-2*(r/(r0 + z*tan(theta)))^2); % Gaussian factor
a = 1+(K/S);
b = ((a^2)-1)^0.5;
M = b/((a*sinh(b*S*((r^2+z^2)^0.5)))+(b*cosh(b*S*((r^2+z^2)^0.5)))); % Absorption and scattering
B(j,l)=C*G*M;
end
j=0;
end
imagesc(0:zmax,-rmax:rmax,B);
colormap(hot);
xlabel(‘Axial distance (microns)’);
ylabel(‘Transverse distance (micons)’);
title(‘Irradiance profile’);
figure();
[m,n]=size(B)
B0=B(round(m/2),:)/max(B(round(m/2),:));% Normalized value of the longitudinal profile for r=0;
plot(z0,B0);
xlabel(‘Axial distance (microns)’);
ylabel(‘Normalized intensity at fiber tip center (N.U.)’);
title(‘Longitudinal Irradiance profile’);
References
- 1.Deisseroth K., “Optogenetics,” Nat. Methods 8(1), 26–29 (2011). 10.1038/nmeth.f.324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stierl M., et al. , “Light modulation of cellular cAMP by a small bacterial photoactivated adenylyl cyclase, bPAC, of the soil bacterium Beggiatoa,” J. Biol. Chem. 286(2), 1181–1188 (2011). 10.1074/jbc.M110.185496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Masseck O. A., Mark M. D., Herlitze S., “Use of optogenetic approaches to control intracellular signaling of G protein-coupled receptors,” in G Protein-Coupled Receptor Genetics, pp. 149–160, Humana Press, New York: (2014). [Google Scholar]
- 4.Nelson S. B., Sugino K., Hempel C. M., “The problem of neuronal cell types: a physiological genomics approach,” Trends Neurosci. 29(6), 339–345 (2006). 10.1016/j.tins.2006.05.004 [DOI] [PubMed] [Google Scholar]
- 5.Smith I. T., Smith S. L., “Getting it through your thick skull,” Nat. Neurosci. 17(8), 1018–1019 (2014). 10.1038/nn.3766 [DOI] [PubMed] [Google Scholar]
- 6.Lin J. Y., et al. , “ReaChR: a red-shifted variant of channelrhodopsin enables deep transcranial optogenetic excitation,” Nat. Neurosci. 16(10), 1499–1508 (2013). 10.1038/nn.3502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gradinaru V., et al. , “Targeting and readout strategies for fast optical neural control in vitro and in vivo,” J. Neurosci. 27(52), 14231–14238 (2007). 10.1523/JNEUROSCI.3578-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stark E., Koos T., Buzsáki G. , “Diode probes for spatiotemporal optical control of multiple neurons in freely moving animals,” J. Neurophysiol. 108(1), 349–363 (2012). 10.1152/jn.00153.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.LeChasseur Y., et al. , “A microprobe for parallel optical and electrical recordings from single neurons in vivo,” Nat. Methods 8(4), 319–325 (2011). 10.1038/nmeth.1572 [DOI] [PubMed] [Google Scholar]
- 10.Nabauer M., Morad M., “Ca2 (+)-induced Ca2+ release as examined by photolysis of caged Ca2+ in single ventricular myocytes,” Am. J. Physiol. 258(1), C189–C193 (1990). [DOI] [PubMed] [Google Scholar]
- 11.Kaplan J. H., “Sodium pump-mediated ATP: ADP exchange. The sided effects of sodium and potassium ions,” J. Gen. Physiol. 80(6), 915–937 (1982). 10.1085/jgp.80.6.915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zemelman B. V., et al. , “Selective photostimulation of genetically chARGed neurons,” Neuron 33(1), 15–22 (2002). 10.1016/S0896-6273(01)00574-8 [DOI] [PubMed] [Google Scholar]
- 13.Banghart M., et al. , “Light-activated ion channels for remote control of neuronal firing,” Nat. Neurosci. 7(12), 1381–1386 (2004). 10.1038/nn1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Volgraf M., et al. , “Allosteric control of an ionotropic glutamate receptor with an optical switch,” Nat. Chem. Biol. 2(1), 47–52 (2005). 10.1038/nchembio756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagel G., et al. , “Channelrhodopsin-1: a light-gated proton channel in green algae,” Science 296(5577), 2395–2398 (2002). 10.1126/science.1072068 [DOI] [PubMed] [Google Scholar]
- 16.Nagel G., et al. , “Channelrhodopsin-2, a directly light-gated cation-selective membrane channel,” Proc. Natl. Acad. Sci. 100(24), 13940–13945 (2003). 10.1073/pnas.1936192100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boyden E. S., et al. , “Millisecond-timescale, genetically targeted optical control of neural activity,” Nat. Neurosci. 8(9), 1263–1268 (2005). 10.1038/nn1525 [DOI] [PubMed] [Google Scholar]
- 18.Han X., Boyden E. S., “Multiple-color optical activation, silencing, and desynchronization of neural activity, with single-spike temporal resolution,” PloS One 2(3), e299 (2007). 10.1371/journal.pone.0000299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang F., et al. , “Multimodal fast optical interrogation of neural circuitry,” Nature 446(7136), 633–639 (2007). 10.1038/nature05744 [DOI] [PubMed] [Google Scholar]
- 20.Gradinaru V., Thompson K. R., Deisseroth K., “eNpHR: a natronomonashalorhodopsin enhanced for optogenetic applications,” Brain Cell Biol. 36(1–4), 129–139 (2008). 10.1007/s11068-008-9027-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kleinlogel S., et al. , “Ultra light-sensitive and fast neuronal activation with the Ca2+-permeable channelrhodopsin CatCh,” Nat. Neurosci. 14(4), 513–518 (2011). 10.1038/nn.2776 [DOI] [PubMed] [Google Scholar]
- 22.Zhang F., et al. , “Red-shifted optogenetic excitation: a tool for fast neural control derived from Volvoxcarteri,” Nat. Neurosci. 11(6), 631–633 (2008). 10.1038/nn.2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yizhar O., et al. , “Neocortical excitation/inhibition balance in information processing and social dysfunction,” Nature 477(7363), 171–178 (2011). 10.1038/nature10360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gunaydin L. A., et al. , “Ultrafast optogenetic control,” Nat. Neurosci. 13(3), 387–392 (2010). 10.1038/nn.2495 [DOI] [PubMed] [Google Scholar]
- 25.Lin J. Y., et al. , “Characterization of engineered channelrhodopsin variants with improved properties and kinetics,” Biophys. J. 96(5), 1803–1814 (2009). 10.1016/j.bpj.2008.11.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hochbaum D. R., et al. , “All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins,” Nat. Methods 11(8), 825–833 (2014). 10.1038/nmeth.3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klapoetke N. C., et al. , “Independent optical excitation of distinct neural populations,” Nat. Methods 11(3), 338–346 (2014). 10.1038/nmeth.2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berndt A., et al. , “Bi-stable neural state switches,” Nat. Neurosci. 12(2), 229–234 (2008). 10.1038/nn.2247 [DOI] [PubMed] [Google Scholar]
- 29.Airan R. D., et al. , “Temporally precise in vivo control of intracellular signalling,” Nature 458(7241), 1025–1029 (2009). 10.1038/nature07926 [DOI] [PubMed] [Google Scholar]
- 30.Chow B. Y., et al. , “High-performance genetically targetable optical neural silencing by light-driven proton pumps,” Nature 463(7277), 98–102 (2010). 10.1038/nature08652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han X., et al. , “A high-light sensitivity optical neural silencer: development and application to optogenetic control of non-human primate cortex,” Front. Syst. Neurosci. 5, 18 (2011). 10.3389/fnsys.2011.00018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sudo Y., et al. , “A blue-shifted light-driven proton pump for neural silencing,” J. Biol. Chem. 288(28), 20624–20632 (2013). 10.1074/jbc.M113.475533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Flytzanis N. C., et al. , “Archaerhodopsin variants with enhanced voltage-sensitive fluorescence in mammalian and Caenorhabditiselegans neurons,” Nat. Commun. 5, 4894 (2014). 10.1038/ncomms5894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wietek J., et al. , “Conversion of channelrhodopsin into a light-gated chloride channel,” Science 344(6182), 409–412 (2014). 10.1126/science.1249375 [DOI] [PubMed] [Google Scholar]
- 35.Berndt A., et al. , “Structure-guided transformation of channelrhodopsin into a light-activated chloride channel,” Science 344(6182), 420–424 (2014). 10.1126/science.1252367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chuong A. S., et al. , “Noninvasive optical inhibition with a red-shifted microbial rhodopsin,” Nat. Neurosci. 17(8), 1123–1129 (2014). 10.1038/nn.3752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yizhar O., et al. , “Optogenetics in neural systems,” Neuron 71(1), 9–34 (2011). 10.1016/j.neuron.2011.06.004 [DOI] [PubMed] [Google Scholar]
- 38.Miesenböck G., “The optogenetic catechism,” Science 326(5951), 395–399 (2009). 10.1126/science.1174520 [DOI] [PubMed] [Google Scholar]
- 39.Mutoh H., et al. , “Optogenetic monitoring of membrane potentials,” Exp. Physiol. 96(1), 13–18 (2011). 10.1113/expphysiol.2010.053942 [DOI] [PubMed] [Google Scholar]
- 40.Looger L. L., Griesbeck O., “Genetically encoded neural activity indicators,” Curr. Opin. Neurobiol. 22(1), 18–23 (2012). 10.1016/j.conb.2011.10.024 [DOI] [PubMed] [Google Scholar]
- 41.Kralj J. M., et al. , “Optical recording of action potentials in mammalian neurons using a microbial rhodopsin,” Nat. Methods 9(1), 90–95 (2012). 10.1038/nmeth.1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao Y., et al. , “An expanded palette of genetically encoded Ca2+ indicators,” Science 333(6051), 1888–1891 (2011). 10.1126/science.1208592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakai J., Ohkura M., Imoto K., “A high signal-to-noise Ca2+ probe composed of a single green fluorescent protein,” Nat. Biotechnol. 19(2), 137–141 (2001). 10.1038/84397 [DOI] [PubMed] [Google Scholar]
- 44.Siegel M. S., Isacoff E. Y., “A genetically encoded optical probe of membrane voltage,” Neuron 19(4), 735–741 (1997). 10.1016/S0896-6273(00)80955-1 [DOI] [PubMed] [Google Scholar]
- 45.Sakai R., et al. , “Design and characterization of a DNA–encoded, voltage–sensitive fluorescent protein,” Eur. J. Neurosci. 13(12), 2314–2318 (2001). 10.1046/j.0953-816x.2001.01617.x [DOI] [PubMed] [Google Scholar]
- 46.Akemann W., et al. , “Imaging brain electric signals with genetically targeted voltage-sensitive fluorescent proteins,” Nat. Methods 7(8), 643–649 (2010). 10.1038/nmeth.1479 [DOI] [PubMed] [Google Scholar]
- 47.Ataka K., Pieribone V. A. , “A genetically targetable fluorescent probe of channel gating with rapid kinetics,” Biophys. J. 82(1), 509–516 (2002). 10.1016/S0006-3495(02)75415-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chanda B., et al. , “A hybrid approach to measuring electrical activity in genetically specified neurons,” Nat. Neurosci. 8(11), 1619–1626 (2005). 10.1038/nn1558 [DOI] [PubMed] [Google Scholar]
- 49.Barnett L., et al. , “A fluorescent, genetically-encoded voltage probe capable of resolving action potentials,” PLoS One 7(9), e43454 (2012). 10.1371/journal.pone.0043454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jin L., et al. , “Single action potentials and subthreshold electrical events imaged in neurons with a fluorescent protein voltage probe,” Neuron 75(5), 779–785 (2012). 10.1016/j.neuron.2012.06.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kralj J. M., et al. , “Electrical spiking in Escherichia coli probed with a fluorescent voltage-indicating protein,” Science 333(6040), 345–348 (2011). 10.1126/science.1204763 [DOI] [PubMed] [Google Scholar]
- 52.Gong Y., et al. , “Imaging neural spiking in brain tissue using FRET-opsin protein voltage sensors,” Nat. Commun. 5, 3674 (2014). 10.1038/ncomms4674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zou P., et al. , “Bright and fast multicoloured voltage reporters via electrochromic FRET,” Nat. Commun. 5, 4625 (2014). 10.1038/ncomms5625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ding J., et al. , “Structural basis of the ultrasensitive calcium indicator GCaMP6,” Sci. China Life Sci. 57(3), 269–274 (2014). 10.1007/s11427-013-4599-5 [DOI] [PubMed] [Google Scholar]
- 55.Akerboom J., et al. , “Genetically encoded calcium indicators for multi-color neural activity imaging and combination with optogenetics,” Front. Mol. Neurosci. 6, 2 (2013). 10.3389/fnmol.2013.00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Inoue M., et al. , “Rational design of a high-affinity, fast, red calcium indicator R-CaMP2,” Nat. Methods 12, 64–70 (2015). 10.1038/nmeth.3185 [DOI] [PubMed] [Google Scholar]
- 57.Thestrup T., et al. , “Optimized ratiometric calcium sensors for functional in vivo imaging of neurons and T lymphocytes,” Nat. Methods 11(2), 175–182 (2014). 10.1038/nmeth.2773 [DOI] [PubMed] [Google Scholar]
- 58.Wu J., et al. , “A long Stokes shift red fluorescent Ca2+ indicator protein for two-photon and ratiometric imaging,” Nat. Commun. 5, 5262 (2014). 10.1038/ncomms6262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nagai T., et al. , “Expanded dynamic range of fluorescent indicators for Ca2+ by circularly permuted yellow fluorescent proteins,” Proc. Natl. Acad. Sci. U. S. A. 101(29), 10554–10559 (2004). 10.1073/pnas.0400417101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mank M., et al. , “A genetically encoded calcium indicator for chronic in vivo two-photon imaging,” Nat. Methods 5(9), 805–811 (2008). 10.1038/nmeth.1243 [DOI] [PubMed] [Google Scholar]
- 61.Okumoto S., et al. , “Detection of glutamate release from neurons by genetically encoded surface-displayed FRET nanosensors,” Proc. Natl. Acad. Sci. U. S. A. 102(24), 8740–8745 (2005). 10.1073/pnas.0503274102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hires S. A., Zhu Y., Tsien R. Y., “Optical measurement of synaptic glutamate spillover and reuptake by linker optimized glutamate-sensitive fluorescent reporters,” Proc. Natl. Acad. Sci. 105(11), 4411–4416 (2008). 10.1073/pnas.0712008105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kuner T., Augustine G. J., “A genetically encoded ratiometric indicator for chloride,” Neuron 27(3), 447–459. 10.1016/S0896-6273(00)00056-8 [DOI] [PubMed] [Google Scholar]
- 64.Grimley J. S., et al. , “Visualization of synaptic inhibition with an optogenetic sensor developed by cell-free protein engineering automation,” J. Neurosci. 33(41), 16297–16309 (2013). 10.1523/JNEUROSCI.4616-11.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nguyen Q.-T., et al. , “An in vivo biosensor for neurotransmitter release and in situ receptor activity,” Nat. Neurosci. 13(1), 127–132 (2010). 10.1038/nn.2469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tantama M., Hung Y. P., Yellen G., “Imaging intracellular pH in live cells with a genetically encoded red fluorescent protein sensor,” J. Am. Chem. Soc. 133(26), 10034–10037 (2011). 10.1021/ja202902d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shen Y., et al. , “pHuji, a pH-sensitive red fluorescent protein for imaging of exo-and endocytosis,” J. Cell Biol. 207(3), 419–432 (2014). 10.1083/jcb.201404107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Berg J., Hung Y. P., Yellen G. , “A genetically encoded fluorescent reporter of ATP: ADP ratio,” Nat. Methods 6(2), 161–166 (2009). 10.1038/nmeth.1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mehmeti I., Lortz S., Lenzen S., “The -sensitive HyPer protein targeted to the endoplasmic reticulum as a mirror of the oxidizing thiol-disulfide milieu,” Free Radicals Biol. Med. 53(7), 1451–1458 (2012). 10.1016/j.freeradbiomed.2012.08.010 [DOI] [PubMed] [Google Scholar]
- 70.Vo-Dinh T., Biomedical Photonics Handbook, CRC Press, United States: (2010). [Google Scholar]
- 71.Pisanello F., et al. , “Multipoint-emitting optical fibers for spatially addressable in vivo optogenetics,” Neuron 82(6), 1245–1254 (2014). 10.1016/j.neuron.2014.04.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gradinaru V., et al. , “Optical deconstruction of parkinsonian neural circuitry,” Science 324(5925), 354–359 (2009). 10.1126/science.1167093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Han X., “In vivo application of optogenetics for neural circuit analysis,” ACS Chem. Neurosci. 3(8), 577–584 (2012). 10.1021/cn300065j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cardin J. A., et al. , “Targeted optogenetic stimulation and recording of neurons in vivo using cell-type-specific expression of channelrhodopsin-2,” Nat. Protoc. 5(2), 247–254 (2010). 10.1038/nprot.2009.228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ayling O. G., et al. , “Automated light-based mapping of motor cortex by photoactivation of channelrhodopsin-2 transgenic mice,” Nat. Methods 6(3), 219–224 (2009). 10.1038/nmeth.1303 [DOI] [PubMed] [Google Scholar]
- 76.Zhu Z. Y., Yappert M. C., “Determination of effective depth and equivalent pathlength for a single-fiber fluorometric sensor,” Appl. Spectrosc. 46(6), 912–918 (1992). 10.1366/0003702924124411 [DOI] [Google Scholar]
- 77.Tai D. C., et al. , “Illumination and fluorescence collection volumes for fiber optic probes in tissue,” J. Biomed. Opt. 12(3), 034033 (2007). 10.1117/1.2750288 [DOI] [PubMed] [Google Scholar]
- 78.Foutz T. J., Arlow R. L., McIntyre C. C., “Theoretical principles underlying optical stimulation of a channelrhodopsin-2 positive pyramidal neuron,” J. Neurophysiol. 107(12), 3235–3245 (2012). 10.1152/jn.00501.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Deisseroth Lab, “Optogenetics online ressources,” 2014, http://web.stanford.edu/group/dlab/cgi-bin/graph/chart.php (14 June 2015).
- 80.Utzinger U., Richards-Kortum R. R., “Fiber optic probes for biomedical optical spectroscopy,” J. Biomed. Opt. 8(1), 121–147 (2003). 10.1117/1.1528207 [DOI] [PubMed] [Google Scholar]
- 81.Castonguay A., et al. , “Repetitive and retinotopically restricted activation of the dorsal lateral geniculate nucleus with optogenetics,” PloS One 9(4), e94633 (2014). 10.1371/journal.pone.0094633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ward M. P., et al. , “Toward a comparison of microelectrodes for acute and chronic recordings,” Brain Res. 1282, 183–200 (2009). 10.1016/j.brainres.2009.05.052 [DOI] [PubMed] [Google Scholar]
- 83.Humphrey D. R., Schmidt E. M., “Extracellular single-unit recording methods,” Neuromethods 15, 1–64 (1990). [Google Scholar]
- 84.Duff Davis M., Schmidt J. J., “In vivo spectrometric calcium flux recordings of intrinsic Caudate-Putamen cells and transplanted IMR-32 neuroblastoma cells using miniature fiber optrodes in anesthetized and awake rats and monkeys,” J. Neurosci. Methods 99(1), 9–23 (2000). 10.1016/S0165-0270(00)00209-0 [DOI] [PubMed] [Google Scholar]
- 85.Kühl M., “Optical microsensors for analysis of microbial communities,” Methods Enzymol. 397, 166–199 (2005). 10.1016/S0076-6879(05)97010-9 [DOI] [PubMed] [Google Scholar]
- 86.Dufour S., et al. , “In vivo simultaneous intra-and extracellular potassium recordings using a micro-optrode,” J. Neurosci. Methods 194(2), 206–217 (2011). 10.1016/j.jneumeth.2010.10.004 [DOI] [PubMed] [Google Scholar]
- 87.Katz Y., et al. , “Optopatcher—an electrode holder for simultaneous intracellular patch-clamp recording and optical manipulation,” J. Neurosci. Methods 214(1), 113–117 (2013). 10.1016/j.jneumeth.2013.01.017 [DOI] [PubMed] [Google Scholar]
- 88.Zhang J., et al. , “Integrated device for optical stimulation and spatiotemporal electrical recording of neural activity in light-sensitized brain tissue,” J. Neural Eng. 6(5), 055007 (2009). 10.1088/1741-2560/6/5/055007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Royer S., et al. , “Multi-array silicon probes with integrated optical fibers: light-assisted perturbation and recording of local neural circuits in the behaving animal,” Eur. J. Neurosci. 31(12), 2279–2291 (2010). 10.1111/(ISSN)1460-9568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wu F., et al. , “An implantable neural probe with monolithically integrated dielectric waveguide and recording electrodes for optogenetics applications,” J. Neural Eng. 10(5), 056012 (2013). 10.1088/1741-2560/10/5/056012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zalocusky K., Deisseroth K., “Optogenetics in the behaving rat: integration of diverse new technologies in a vital animal model,” Optogenetics 1, 1–17 (2013). 10.2478/optog-2013-0001 [DOI] [Google Scholar]
- 92.Stark E., et al. , “Inhibition-induced theta resonance in cortical circuits,” Neuron 80(5), 1263–1276 (2013). 10.1016/j.neuron.2013.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang J., et al. , “Integrated device for combined optical neuromodulation and electrical recording for chronic in vivo applications,” J. Neural Eng. 9(1), 016001 (2012). 10.1088/1741-2560/9/1/016001 [DOI] [PubMed] [Google Scholar]
- 94.Tamura K., et al. , “A glass-coated tungsten microelectrode enclosing optical fibers for optogenetic exploration in primate deep brain structures,” J. Neurosci. Methods 211(1), 49–57 (2012). 10.1016/j.jneumeth.2012.08.004 [DOI] [PubMed] [Google Scholar]
- 95.Abaya T., et al. , “A 3D glass optrode array for optical neural stimulation,” Biomed. Opt. Express 3(12), 3087–3104 (2012). 10.1364/BOE.3.003087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Im M., et al. , “Neural probes integrated with optical mixer/splitter waveguides and multiple stimulation sites,” in Proc. 2011 IEEE 24th Int. Conf. on Micro Electro Mechanical Systems (MEMS), pp. 1051–1054 (2011). [Google Scholar]
- 97.Zorzos A. N., Boyden E. S., Fonstad C. G., “Multiwaveguide implantable probe for light delivery to sets of distributed brain targets,” Opt. Lett. 35(24), 4133–4135 (2010). 10.1364/OL.35.004133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zorzos A. N., et al. , “Three-dimensional multiwaveguide probe array for light delivery to distributed brain circuits,” Opt. Lett. 37(23), 4841–4843 (2012). 10.1364/OL.37.004841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chen S., et al. , “A fiber-based implantable multi-optrode array with contiguous optical and electrical sites,” J. Neural Eng. 10(4), 046020 (2013). 10.1088/1741-2560/10/4/046020 [DOI] [PubMed] [Google Scholar]
- 100.Stark E., et al. , “Pyramidal cell-interneuron interactions underlie hippocampal ripple oscillations,” Neuron 83(2), 467–480 (2014). 10.1016/j.neuron.2014.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Beltramo R., et al. , “Layer-specific excitatory circuits differentially control recurrent network dynamics in the neocortex,” Nat. Neurosci. 16(2), 227–234 (2013). 10.1038/nn.3306 [DOI] [PubMed] [Google Scholar]
- 102.Roux L., et al. , “In vivo optogenetic identification and manipulation of GABAergic interneuron subtypes,” Curr. Opin. Neurobiol. 26, 88–95 (2014). 10.1016/j.conb.2013.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zorzos A., et al. , “Light-proof neural recording electrodes,” in Program No. 338.12, 2009 Neuroscience Meeting Planner, Society for Neuroscience, Chicago, Illinois: (2009). [Google Scholar]
- 104.Dufour S., et al. , “A multimodal micro-optrode combining field and single unit recording, multispectral detection and photolabeling capabilities,” PloS One 8(2), e57703 (2013). 10.1371/journal.pone.0057703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lin S.-T., et al. , “Flexible optitrode for localized light delivery and electrical recording,” Opt. Lett. 37(11), 1781–1783 (2012). 10.1364/OL.37.001781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Canales A., et al. , “Multifunctional fibers for simultaneous optical, electrical and chemical interrogation of neural circuits in vivo,” Nat. Biotechol. 33(3), 277–284 (2015). 10.1038/nbt.3093 [DOI] [PubMed] [Google Scholar]
- 107.Klorig D. C., Godwin D. W., “A magnetic rotary optical fiber connector for optogenetic experiments in freely moving animals,” J. Neurosci. Methods 227, 132–139 (2014). 10.1016/j.jneumeth.2014.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ung K., Arenkiel B., “Fiber-optic implantation for chronic optogenetic stimulation of brain tissue,” JoVE (68), e50004 (2012). 10.3791/50004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Doric S., “Independent dual path optical rotary joint,” US Patent No. 20140226932 A1 (2014).
- 110.Wentz C. T., et al. , “A wirelessly powered and controlled device for optical neural control of freely-behaving animals,” J. Neural Eng. 8(4), 046021 (2011). 10.1088/1741-2560/8/4/046021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Grossman N., et al. , “Multi-site optical excitation using ChR2 and micro-LED array,” J. Neural Eng. 7(1), 016004 (2010). 10.1088/1741-2560/7/1/016004 [DOI] [PubMed] [Google Scholar]
- 112.McGovern B., et al. , “A new individually addressable micro-LED array for photogenetic neural stimulation,” IEEE Trans. Biomed. Circuits Syst. 4(6), 469–476 (2010). 10.1109/TBCAS.2010.2081988 [DOI] [PubMed] [Google Scholar]
- 113.McAlinden N., et al. , “Thermal and optical characterization of micro-LED probes for in vivo optogenetic neural stimulation,” Opt. Lett. 38(6), 992–994 (2013). 10.1364/OL.38.000992 [DOI] [PubMed] [Google Scholar]
- 114.Nurmikko A. V., et al. , “Listening to brain microcircuits for interfacing with external world—progress in wireless implantable microelectronic neuroengineering devices,” Proc. IEEE 98(3), 375–388 (2010). 10.1109/JPROC.2009.2038949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.McCall J. G., et al. , “Fabrication and application of flexible, multimodal light-emitting devices for wireless optogenetics,” Nat. Protoc. 8(12), 2413–2428 (2013). 10.1038/nprot.2013.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ameli R., et al. , “A wireless and batteryless neural headstage with optical stimulation and electrophysiological recording,” in Proc. 2013 35th Annual Int. Conf. of the IEEE Engineering in Medicine and Biology Society (EMBC), pp. 5662–5665 (2013). [DOI] [PubMed] [Google Scholar]
- 117.Kim T.-i., et al. , “Injectable, cellular-scale optoelectronics with applications for wireless optogenetics,” Science 340(6129), 211–216 (2013). 10.1126/science.1232437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pashaie R., Falk R., “Single optical fiber probe for fluorescence detection and optogenetic stimulation,” IEEE Trans. Biomed. Eng. 60(2), 268–280 (2013). 10.1109/TBME.2012.2221713 [DOI] [PubMed] [Google Scholar]
- 119.Hochbaum D. R., et al. , “All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins,” Nat. Methods 11, 825–833 (2014). 10.1038/nmeth.3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tokuda T., et al. , “CMOS on-chip bio-imaging sensor with integrated micro light source array for optogenetics,” Electron. Lett. 48(6), 312–314 (2012). 10.1049/el.2011.4087 [DOI] [Google Scholar]
- 121.Klimas A., Entcheva E., “Toward microendoscopy-inspired cardiac optogenetics in vivo: technical overview and perspective,” J. Biomed. Opt. 19(8), 080701 (2014). 10.1117/1.JBO.19.8.080701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bélanger E., et al. , “Live animal myelin histomorphometry of the spinal cord with video-rate multimodal nonlinear microendoscopy,” J. Biomed. Opt. 17(2), 021107 (2012). 10.1117/1.JBO.17.2.021107 [DOI] [PubMed] [Google Scholar]
- 123.Hayashi Y., et al. , “Spatio‐temporal control of neural activity in vivo using fluorescence microendoscopy,” Euro. J. Neurosci. 36(6), 2722–2732 (2012). 10.1111/ejn.2012.36.issue-6 [DOI] [PubMed] [Google Scholar]
- 124.Rickgauer J. P., Deisseroth K. , Tank D. W. , “Simultaneous cellular-resolution optical perturbation and imaging of place cell firing fields,” Nat. Neurosci. 17(12), 1816–1824 (2014). 10.1038/nn.3866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Vogt N., “Neuroscience: all-optical electrophysiology in behaving animals,” Nat. Methods 12(2), 101–101 (2015). 10.1038/nmeth.3272 [DOI] [PubMed] [Google Scholar]
- 126.Packer A. M., et al. , “Simultaneous all-optical manipulation and recording of neural circuit activity with cellular resolution in vivo,” Nat. Methods 12, 140–144 (2015). 10.1038/nmeth.3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Jung J. C., et al. , “In vivo mammalian brain imaging using one-and two-photon fluorescence microendoscopy,” J. Neurophysiol. 92(5), 3121–3133 (2004). 10.1152/jn.00234.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Levene M. J., et al. , “In vivo multiphoton microscopy of deep brain tissue,” J. Neurophysiol. 91(4), 1908–1912 (2004). 10.1152/jn.01007.2003 [DOI] [PubMed] [Google Scholar]
- 129.Bonaccorso F., et al. , “Graphene photonics and optoelectronics,” Nat. Photonics 4(9), 611–622 (2010). 10.1038/nphoton.2010.186 [DOI] [Google Scholar]
- 130.Lee J., et al. , “Transparent micro-optrode arrays for simultaneous multichannel optical stimulation and electrical recording,” CLEO Sci. Innovations CTh3I, 7 (2013). 10.1364/CLEO_SI.2013.CTh3I.7 [DOI] [Google Scholar]
- 131.Khodagholy D., et al. , “In vivo recordings of brain activity using organic transistors,” Nat. Commun. 4, 1575 (2013). 10.1038/ncomms2573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Dochow S., et al. , “Multicore fiber with integrated fiber Bragg gratings for background-free Raman sensing,” Opt. Express 20(18), 20156–20169 (2012). 10.1364/OE.20.020156 [DOI] [PubMed] [Google Scholar]