Summary
Depression risk is exacerbated by genetic factors and stress exposure; however, the biological mechanisms through which these factors interact to confer depression risk are poorly understood. One putative biological mechanism implicates variability in the ability of cortisol, released in response to stress, to trigger a cascade of adaptive genomic and non-genomic processes through glucocorticoid receptor (GR) activation. Here, we demonstrate that common genetic variants in long-range enhancer elements modulate the immediate transcriptional response to GR activation in human blood cells. These functional genetic variants increase risk for depression and co-heritable psychiatric disorders. Moreover, these risk variants are associated with inappropriate amygdala reactivity, a transdiagnostic psychiatric endophenotype and an important stress hormone response trigger. Network modeling and animal experiments suggest that these genetic differences in GR-induced transcriptional activation may mediate the risk for depression and other psychiatric disorders by altering a network of functionally related stress-sensitive genes in blood and brain.
Video Abstract
Highlights
-
•
SNPs in long-range enhancers alter the transcriptional response of GR target genes
-
•
These functional SNPs predict risk for psychiatric but not other medical disorders
-
•
These variants are associated with differential neural circuit responses to threat
-
•
GR transcripts are strongly co-expressed and regulated in the brain of animal models
Using a stimulated eQTL approach, Arloth et al. show that common genetic variants that alter the initial transcriptome response to stress hormone receptor activation also cumulatively increase the risk for stress-related psychiatric disorders and predict a threat response from the amygdala.
Introduction
Major depressive disorder (MDD) has a lifetime prevalence of up to 17% (Kessler et al., 2005), resulting in one of the highest global burden of disease ratings by the World Health Organization (Ustün et al., 2004). Despite its prevalence and impact, the etiological and pathophysiological mechanisms underlying MDD are poorly understood, resulting in sub-optimal treatments with high rates of recurrence and treatment resistance (Warden et al., 2007). Family, twin, and population studies point to both genetic as well as environmental risk factors for depression. Genetic factors contribute up to 40% of the risk and are complemented largely by individual-specific environmental exposure to adverse life events (Kendler et al., 2006). Both sensitivity and resilience to the long-term effects of exposure to adverse life events may be modulated by genetic variation (Kendler, 2013).
Stress results in activation of the stress hormone system, which culminates in the activation of glucocorticoid receptors (GRs) by cortisol. The GR is a nuclear hormone receptor, and upon activation it translocates from the cytoplasm to the nucleus, where it binds to glucocorticoid response elements (GREs) and regulates gene expression (McKay and Cidlowski, 1999; Phuc Le et al., 2005). Activation of this receptor not only initiates adaptive physiological changes in the body to confront an imminent threat, but also facilitates the termination of these changes once the threat has been overcome. Thus, genetically driven variability in GR regulation of the stress hormone response may functionally interact with environmental risk factors, thereby producing individual differences in risk for MDD.
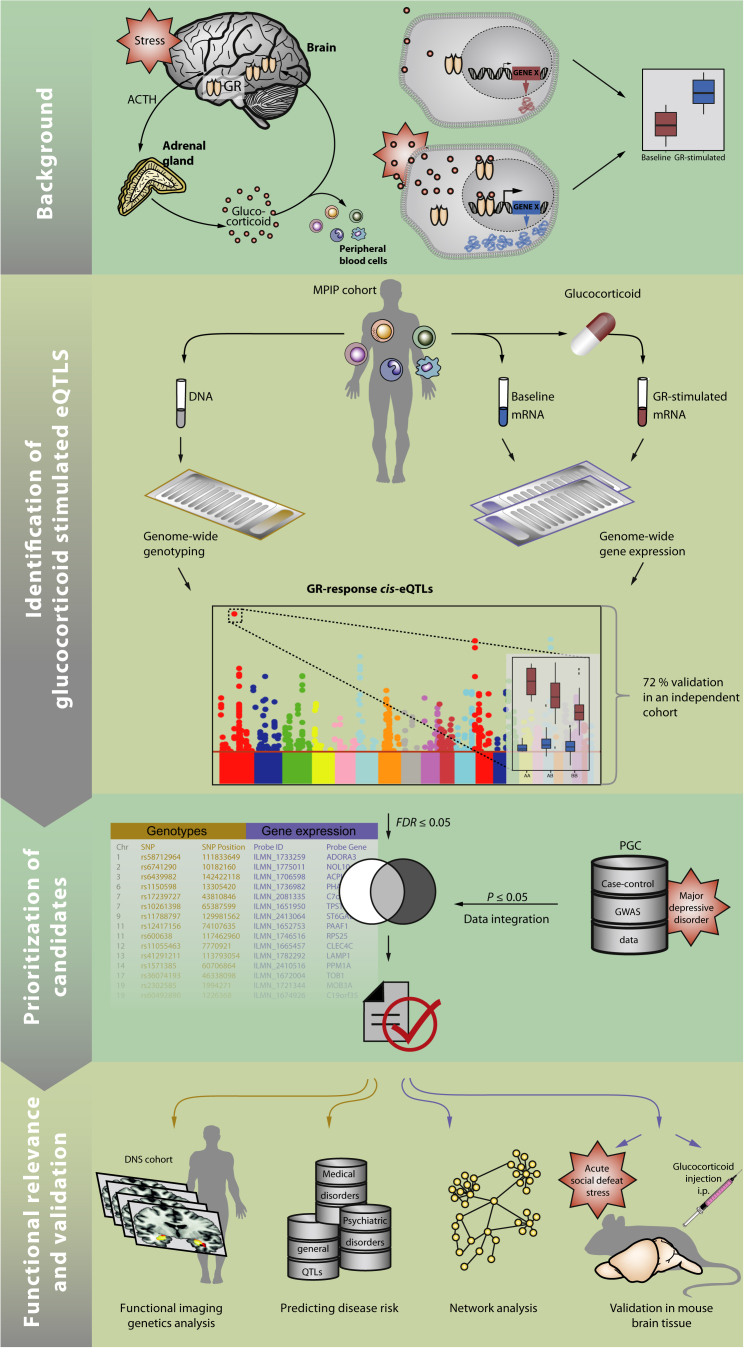
Consistent with this model, dysfunction of GR-mediated negative feedback has been reported in MDD (de Kloet et al., 2005) as well as in individuals exposed to early adversity (Heim and Binder, 2012; Wilkinson and Goodyer, 2011), one of the strongest risk factors for the development of MDD. Moreover, genetic variation in pathways regulating GR signaling has been linked with MDD risk (van Rossum et al., 2006). Here, we show that common genetic variants that modulate the initial transcriptional response to GR activation increase the risk for MDD as well as other psychiatric disorders. Gene network modeling and animal experiments suggest that these genetic differences in the transcriptional response to GR activation may mediate risk for depression and other psychiatric disorders by altering a network of co-expressed genes that are responsive to stress and glucocorticoids in the brain. In addition, these genetic variants shape the response of the amygdala, which is itself an important trigger of the stress hormone response and a functional neural phenotype implicated in the etiology and pathophysiology of depression and other forms of psychopathology (Jankord and Herman, 2008; Phillips et al., 2003). The main hypotheses and the experimental approach are summarized in Figure 1.
Figure 1.
Summary Figure Illustrating the Sequence of Experiments and Analyses Applied in This Study
The main hypothesis tested in this study is that common genetic variants that alter the short-term transcriptional response to GR activation also alter the risk for stress-related psychiatric disorders and related neural endophenotypes.
Results
Genetic Regulation of GR-Stimulated Gene Expression
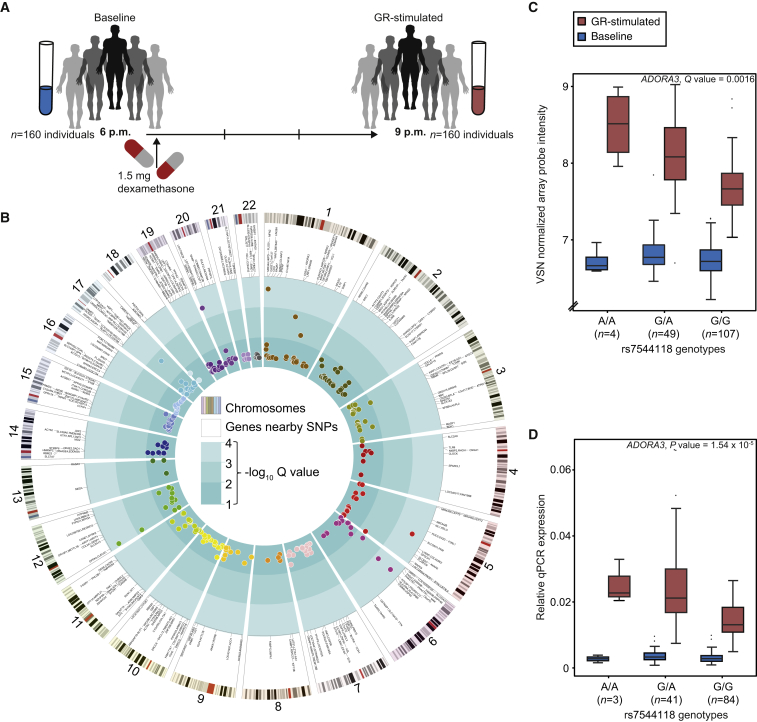
We first identified genetic variants that alter GR-stimulated gene expression changes by adopting a stimulated expression quantitative trait locus (eQTL) approach (Figure 2A). Gene expression profiles in peripheral blood cells from 160 male individuals of the Max-Planck Institute of Psychiatry (MPIP) cohort (91 cases and 69 controls, see Experimental Procedures) were obtained at baseline and 3 hr after stimulation with the selective GR agonist dexamethasone (Figure S1A) and combined with genome-wide SNP data. All individuals showed a strong endocrine response to dexamethasone (Cortisol: F1,159 = 43.93, p = 5.02 × 10−10 and ACTH: F1,158 = 37.96, p = 5.76 × 10−9; Figures S1B and S1C). After quality control, 4,447 gene expression probes that exhibited strong regulation following dexamethasone administration (absolute fold change in gene expression from baseline to 3 hr post-dexamethasone ≥ 1.3 in at least 20% of all samples) were combined with genotype data of ∼2 million imputed SNPs (see Experimental Procedures). Using the log fold change in gene expression standardized to baseline values as the outcome and restricting the analysis to a ± 1 Mb cis-region around each probe, we found that 3,820 GR-response-modulating cis-eQTLs (GR-response eQTLs) remained significant after accounting for disease status, age, and BMI and correction for multiple testing (see Experimental Procedures). These comprised 297 unique array probes and 3,662 unique SNPs. The 3,662 unique GR-response cis-expression SNPs (eSNPs) can be summarized in terms of independent tag SNPs into 296 uncorrelated GR-response cis-eSNP bins, i.e., sets of SNPs in linkage disequilibrium (LD; see Experimental Procedures). We defined the tag eSNP as the eSNP showing the highest association per bin (lowest Q value). These 296 GR-response cis-eSNP bins correspond to 320 GR-response cis-eQTL bins, i.e., cis-eSNP bin-probe combinations, as one cis-eSNP bin can be associated with the regulation of more than one transcript and vice versa. These GR-response cis-eQTL bins are listed in Table S1 and illustrated in Figures 2B–2D. Including dexamethasone serum levels or the blood cell count as covariate did not change the results, excluding any confounding effects of individual differences in dexamethasone concentration and cellular composition (see Supplemental Information).
Figure 2.
GR-Response-Modulating cis-eQTLs
(A) Study design for GR-stimulated gene expression in whole blood of 160 male individuals from the Max Planck Institute of Psychiatry cohort.
(B) Circularized Manhattan plot displaying cis-associations for GR-response eQTL bins (n = 320) and their respective significance (−log10 Q values). Displayed from the outer to the inner circle are the number of chromosomes, the ideograms for the human karyotype (hg18), genes nearby eSNPs, and Manhattan plots for the eQTL bins that survived correction for multiple testing.
(C and D) Boxplots of human gene expression values for ADORA3, which is an example of a significant GR-response eQTL. Expression levels are stratified based on the eSNP genotypes for ADORA3. Baseline (6 p.m.) measures are displayed in blue and GR-stimulated measures (9 p.m.) in red. Microarrays data are displayed in (C) and their qPCR validation in (D). Q value in (C) is derived from GR-response cis-eQTL analysis and the p value in (D) from the qPCR linear regression model.
To assess the robustness of these GR-response eQTLs, we validated them in an independent sample of n = 58 (see Experimental Procedures) by performing a sample size-weighted Z score meta-analysis across both samples. In this analysis, 72% of the GR-response eQTLs could be validated, i.e., showed a meta-analysis p value equal to or more significant than in the discovery sample alone (see Experimental Procedures). This method accounts for the small size of the validation sample and suggests the robustness of most of the GR-response eQTLs.
Characterization of GR-Response eSNPs
To better understand the properties of these GR-response eQTLs, we first mapped the GR-eSNPs (n = 3,662 SNPs) to GR binding regions as defined by ChIP-seq peaks in lymphoblastoid cell line (LCL) GM12878 (see Experimental Procedures). We observed a significant enrichment of GR-response eSNPs in GR binding sites as compared to random SNPs (fold enrichment = 2.4, permutation-based FDR ≤ 0.001).
Next, we mapped the distance of the 320 GR-response eQTL bins to the genomic location of the probe sequence of the respective regulated transcript (utilizing the closest SNP within a bin) and compared this to the probe distance for baseline cis-eQTL bins, i.e., the eSNP-probe combinations that showed a significant association of the genotype with transcription levels at baseline (see Supplemental Information). The GR-response eSNP bin-to-probe distance (mean = 406 kb, standard deviation [SD] = 303 kb, n = 320 bins) was significantly longer (Wilcoxon p value = 1.03 × 10−50) than baseline eSNP bin-to-probe distance (mean = 149 kb, SD = 232 kb, n = 1,148 bins; Supplemental Information). This suggests that GR stimulation is associated with significantly more long-range transcriptional regulation than baseline gene expression and that distinct regulatory elements may be involved in baseline versus GR-stimulated gene transcription.
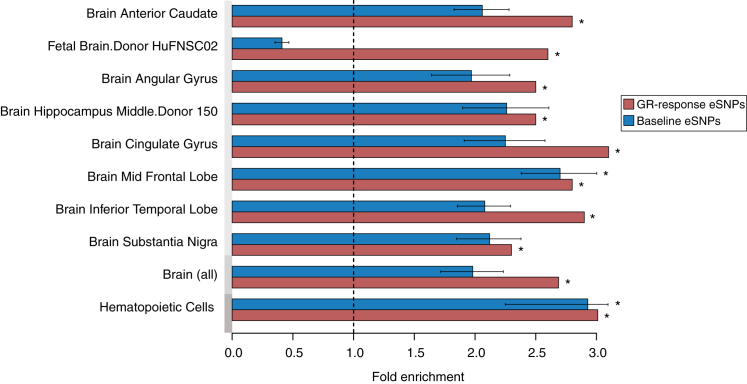
To determine the regulatory potential of GR-response eSNPs, we investigated whether they are enriched within enhancer regions as defined by the Roadmap Epigenome Project (Kundaje et al., 2015) (see Experimental Procedures). GR-response tag eSNPs were significantly enriched within enhancers in 62 different tissues, including blood cells, but also non-hematopoietic tissue such as brain (see Figure S2). When testing baseline tag eSNPs, we only observed an enrichment in enhancers in 54% of these 62 tissues. Whether combined enrichment of both GR-response tag eSNPs and baseline tag eSNPs was observed seemed to be tissue specific (see Figure S2). In fact, GR-response eSNPs were more enriched in brain enhancers than baseline eSNPs, i.e., only one of the eight brain enhancers significantly enriched with GR-response eSNPs also displayed a significant enrichment for baseline eSNPs (see Figure 3). In contrast, we observed equal enrichment for GR-response as well as baseline eSNPs in primary hematopoietic tissues (see Figure S2). These results further support the viewpoint that GR-response eSNPs affect different transcriptional regulators than baseline eSNPs and suggest possible cross-tissue effects of these SNPs.
Figure 3.
GR-Response eSNPs Are Enriched in Enhancer Regions in Multiple Tissues
Bar graph illustrating the enrichment of GR-response eSNPs for enhancers in multiple tissues from the Roadmap Epigenome Project, including brain tissue. The x axis shows the fold enrichment and the y axis all brain enhancers all well as the mean fold enrichment among all hematopoietic cells (see Figure S2) and brain enhancers. The fold enrichment for GR-response eSNPs is illustrated in red and for the permuted baseline eSNPs in blue. Only the GR-response eSNP enrichment, which passes a Bonferroni corrected significance threshold (corrected for the number of all tested tissues or cells, n = 62) is illustrated. ∗ p ≤ 0.05, obtained by binomial enrichment test and Bonferroni correction, error bars ± SD.
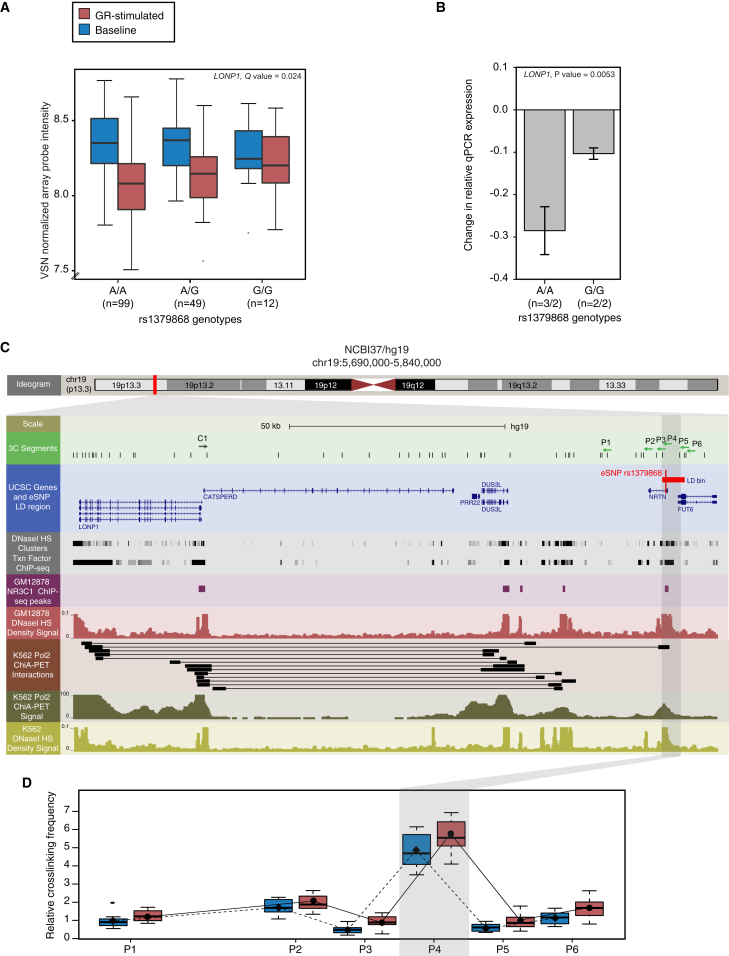
To evaluate whether the long-range regulation of GR-response eQTLs may be associated with long-range physical chromatin interaction, we compared our data with that from a chromatin interaction analysis with paired-end tag sequencing (ChIA-PET) generated by ENCODE (ENCODE Project Consortium, 2011) in the leukemia cell line K562. For this, we examined whether regions containing the GR-response eSNP bin and the corresponding probe gene overlap with physically interacting ChIA-PET tags (see Experimental Procedures). Twenty-five percent of the GR-response eSNP bin-probe gene combinations overlapped with chromatin interaction signals. This was significantly greater than 1,000 equally sized sets of randomly distributed GR-response eSNP bin, especially when restricting the analysis to more long-range eSNP bin-probe gene pairs with distances > 100 kb (fold enrichment>100kb = 1.57, permutation-based FDR>100kb = 0.007; see Experimental Procedures). To validate these long-range chromatin interactions, we used a chromatin conformation capture (3C) assay to confirm a physical interaction between the eSNP bin regions of the GR-response eSNP tag rs1379868 in the NRTN locus and the corresponding GR-stimulated transcript LONP1 (see Figures 4A and 4B), which is over 130 kb upstream. This eSNP bin includes a GR binding site and ChIA-PET tags (see Figure 4C), which interact with the transcription start site of the LONP1 gene. The 3C assay confirmed an increased chromatin interaction (p = 3.35 × 10−23, χ2 = 115.15 at baseline) of the eSNP bin with the TSS of the LONP1 gene (P4 in Figures 4C and 4D) in five LCLs. The average interaction frequency of these two sites was higher following stimulation with the GR-agonist dexamethasone (4.83 versus 5.65). These results suggest that long-range regulation of GR-response eQTLs could be mediated by direct chromatin interaction of enhancer regions with the respective transcription start sites.
Figure 4.
Long-Range Chromatin Interaction of GR-Response eQTLs
(A) Long-range chromatin interaction as exemplified by the eSNP region containing the NRTN locus (chr10: 5,690,000–5,840,000; hg19) was confirmed by 3C in five lymphoblastoid cell lines (LCLs) each, homozygous for the two opposite SNP alleles, both in the presence and absence of dexamethasone. A SNP in the NRTN locus (rs1379868) affects the differentially regulated gene expression of LONP1 in human whole blood cells (based on GR-response eQTL analysis). Baseline (6 p.m.) measures are displayed in blue and GR-stimulated measures (9 p.m.) in red.
(B) SNP effect on GR-dependent gene transcription was validated by qPCR in the LCLs used for the 3C assay.
(C) Characterization of the eSNP locus. Top panel, ideogram for chromosome 19 (p13.3). A red box isolates the region shown (enlarged) in the bottom panel. Bottom panel: 3C-primers (green track) were designed at the LONP1 TSS (C1, anchor) and multiple regions (P1–P6) in and around the eSNP bin. The eSNP bin includes a GR binding site in blood cells (pink track). ChIA-PET tags from the leukemia cell line (brown and green tracks) validate a direct chromatin interaction between the NRTN eSNP locus and the regulated gene LONP1. The paired ChIA-PET tags coincide with DNaseI hypersensitivity sites in the leukemia cell line (red track) and blood cells (yellow track).
(D) Chromatin conformation capture interaction data. A 3C physical interaction between the LONP1 TSS and eSNP bin (P4), emphasized by a gray box, was found in the 3C libraries made from LCLs (p = 3.35 × 10−23, χ2 = 115.15) with a stronger interaction following stimulation with the GR-agonist (p = 0.06, χ2 = 3.35). Q values in (A) are derived from GR-response cis-eQTL analysis, and p values in (B) and (D) are derived from linear mixed model; error bars ± SD.
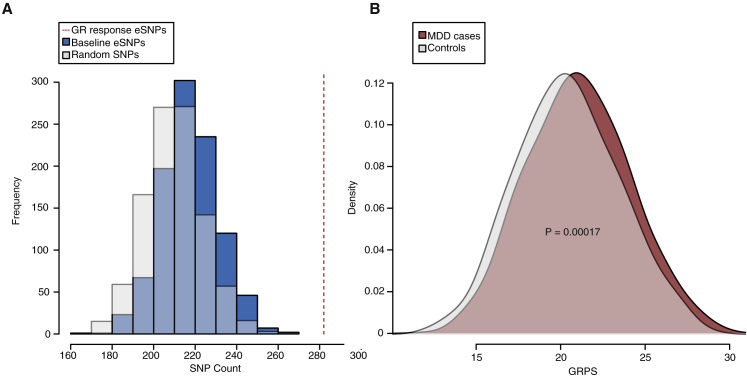
GR-Response eSNPs Are Enriched in Loci Nominally Associated with MDD and Other Psychiatric Disorders as well as in Genome-wide Significant Schizophrenia Loci
Besides their functional characterization, an important question was to assess whether the genetic variants that alter the immediate transcriptional response to GR activation (GR-response eSNPs) would also be associated with risk for stress-related psychiatric disorder. To assess this, we first tested whether our GR-response eSNPs were overrepresented among SNPs associated with MDD in the genome-wide association study (GWAS) results of the Psychiatric Genomics Consortium (PGC), which includes approximately 9,000 cases and the same number of controls (Ripke et al., 2013). Among nominally associated loci with MDD (at meta-analysis p value ≤ 0.05), 282 SNPs also represent a GR-response eSNP. Permutation analysis (see Experimental Procedures) predicted an expected mean overlap of 210 SNPs from 1,000 randomly selected SNP sets (fold enrichment = 1.34, permutation-based FDR < 0.001; Figure 5A). We next investigated whether GR-response eSNPs were also enriched over baseline eSNPs, as SNPs associated with transcriptional changes have been shown to be more enriched in GWASs in general (Roussos et al., 2014). Again the mean overlap for 1,000 permuted baseline cis-eSNP sets (218 SNPs) was significantly lower than the actual overlap of GR-response eSNPs (fold enrichment = 1.29, permutation-based FDR < 0.001; Figure 5A). These enrichments remain significant when using only the tag eSNPs (n = 285) to control for possible confounding due to linkage disequilibrium (LD) structure (fold enrichment = 1.31, permutation-based FDR = 0.082).
Figure 5.
GR-Response eSNPs Are Enriched among Variants Associated with MDD
(A) The dotted red line shows the enriched number of GR-response eSNPs that overlap with SNPs in our meta-analysis for MDD (= MDD-related GR eSNPs; 8,864 cases and 8,982 controls). The distribution of the observed overlap for sets of 1,000 random SNPs (gray) and 1,000 random baseline eSNPs (blue) are represented as histograms (null distributions). Both permuted data sets never reached the same overlap with MDD-associated SNPs as the GR-response eSNPs.
(B) The distribution of the MDD-related GR eSNP genetic risk profile scores (GRPSs) for an independent sample of MDD cases (n = 1,005 cases; red) and controls (n = 478; gray) are represented as density plots. Individuals with MDD display higher GRPSs (p = 0.00017). p value by logistic regression model.
The 282 GR-response eSNPs that overlap with MDD-associated SNPs correspond to 23 unique eSNP bins (reflecting 26 eQTL bins) that regulate 25 unique transcripts (Table S2). We call these 23 eSNP bins “MDD-related GR eSNP bins” in the remainder of the manuscript to refer to GR-response eSNPs that also show a nominal association with MDD.
Validation of Enrichment and Extension to Other Psychiatric Disorders
We next examined whether these MDD-related GR eSNPs would also be associated with MDD in an independent sample. For this we constructed a genetic risk profile score (GRPS) using the tagging SNPs of the 23 MDD-related GR eSNP bins for each individual in an independent validation sample of 1,005 MDD cases and 478 controls (Table S3; see also Experimental Procedures). We found these GRPSs to be significantly associated with MDD and that individuals with higher GRPSs were overrepresented in the case group (Z = 3.76, p = 0.00017; Figure 5B). This GRPS explains about 2.6% of the total variance for MDD in this sample, and the association of these MDD-related GR eSNP GRPSs was more significant than GRPSs constructed from 1,000 randomly generated SNP profiles (permutation-based FDR = 0.008; see Experimental Procedures).
As exposure to stressful life events is a strong risk factor not only for MDD but also for other psychiatric disorders, including bipolar disorder (BPD) and schizophrenia (SCZ) (Dohrenwend and Egri, 1981; Kendler and Karkowski-Shuman, 1997), we tested whether the GR-response eSNPs were also overrepresented among SNPs associated with other psychiatric disorders utilizing meta-analysis data from the PGC. Using this approach, we tested for for significant GR-response eSNP enrichment compared to 1,000 randomly generated baseline eSNP sets in the PGC for four additional psychiatric disorders and the cross-disorders analysis including also MDD (see Table 1). In the latest multi-stage SCZ GWAS, which includes up to 36,989 cases and 113,075 controls (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014), we detected a significant enrichment of GR-response eSNPs compared to baseline eSNPs with SNPs associated with SCZ at p ≤ 0.05 (fold enrichment 1.29, permutation-based FDR ≤ 0.001). When we limited the enrichment analysis to genome-wide significant SCZ loci, we detected 134 GR-response eSNPs that overlapped SNPs associated with SCZ at p ≤ 5 × 10−8. This corresponds to a 10-fold enrichment over baseline eSNPs and is 7.75-fold higher than for nominally associated SCZ SNPs (see Table 1). A significant negative enrichment was identified for loci associated with attention deficit-hyperactivity disorder (permutation-based FDR ≤ 0.012; ADHD; 840 cases and 1,947 trio cases) and autism spectrum disorder (ASD; permutation-based FDR ≤ 0.001; 161 cases and 4,788 trio cases) but not BPD (see Table 1).
Table 1.
Proportion of GR-Response eSNPs Overlapping with GWAS SNPs with Nominal Significance
| GR-Response eSNPs |
Random SNPs |
Fold enrichment | Baseline eSNPs |
Fold enrichment | |||||
|---|---|---|---|---|---|---|---|---|---|
| Count | Mean counta | Range | FDR | Mean counta | Range | FDR | |||
| CDA | 115 | 86.5 ± 8.99 SD | 61–119 | 0.001 | 1.33 | 102.03 ± 8.57 SD | 71–130 | 0.066 | 1.13 |
| BPD | 91 | 70.36 ± 8.34 SD | 44–100 | 0.009 | 1.29 | 86.18 ± 8.2 SD | 59–115 | 0.295 | 1.05 |
| SCZ | 157 | 84.08 ± 8.79 SD | 61–111 | <0.001 | 1.87 | 129.07 ± 9.61 SD | 99–158 | 0.027 | 1.22 |
| SCZ2 | 948 | 533.29 ± 21.59 SD | 469–615 | <0.001 | 1.78 | 736.55 ± 22.32 SD | 676–813 | < 0.001 | 1.29 |
| SCZ2 (5 × 10−8)b | 134 | 6.43 ± 2.52 SD | 0–18 | <0.001 | 20.94 | 13.37 ± 3.32 SD | 4–24 | < 0.001 | 10.02 |
| ADHD | 29 | 55.69 ± 7.14 SD | 36-79 | <0.001c | −1.89 | 42.23 ± 5.78 SD | 25–63 | 0.012c | −1.44 |
| ASD | 34 | 63.73 ± 7.62 SD | 44–91 | <0.001c | −1.85 | 114.94 9.09 SD | 80–147 | < 0.001c | −3.35 |
| MDD | 282 | 210 ± 13.9 SD | 168–255 | <0.001 | 1.34 | 218.11 ± 13.49 SD | 174–268 | < 0.001 | 1.29 |
| CD | 149 | 83.16 ± 8.89 SD | 61–112 | <0.001 | 1.8 | 150.5 ± 10.27 SD | 121–182 | 0.591 | −1.006 |
| RA | 396 | 71.9 ± 8.06 SD | 46–100 | <0.001 | 5.56 | 372.37 ± 16.08 SD | 323–430 | 0.078 | 1.06 |
| Height | 350 | 146.01 ± 11.9 SD | 108–188 | <0.001 | 2.4 | 340.84 ± 14.91 SD | 294–390 | 0.268 | 1.03 |
Schizophrenia, SCZ; bipolar disorder, BPD; attention deficit-hyperactivity disorder, ADHD; Crohn’s disease, CD; autism spectrum disorder, ASD; major depressive disorder, MDD; cross-disorder associations, CDA; rheumatoid arthritis, RA.
Proportion of the number of GR-response eSNPs observed for 1,000 permuted random SNPs and baseline eSNPs.
Overlap at genome-wide significance level.
Negative enrichment and inverse fold enrichment.
To test whether these enrichments of GR-response eSNPs as compared to baseline eSNPs are specific to psychiatric disorders, we mapped these variants to GWAS for rheumatoid arthritis, Crohn’s disease, and height but found no enrichment more than 1.06-fold (see Table 1). These analyses suggest that GR-response eSNPs are unrelated to these medical disorders or general quantitative traits but specifically contribute to the risk for MDD and SCZ.
Functional Relevance of Transcripts Regulated by MDD-Related GR eSNPs
Gene Network Analysis of MDD-Related GR Genes
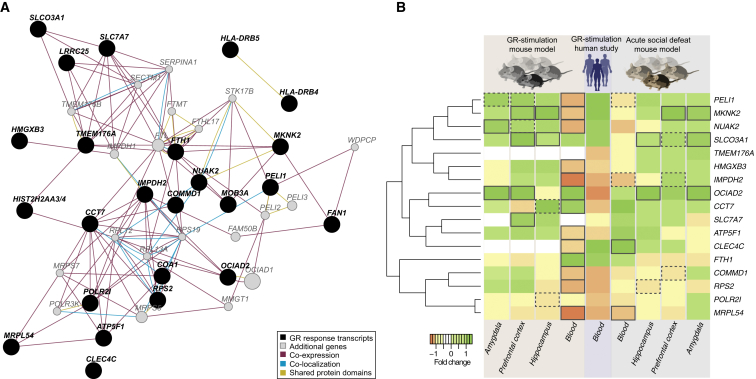
Next, we investigated whether the probe genes (n = 24), regulated by the MDD-related GR eSNPs, are part of specific pathways that may be relevant for the pathophysiology of psychiatric disorders. Using the GeneMANIA tool (Montojo et al., 2014), we were able to generate a gene network containing 23 of the 24 MDD-related GR genes (see Figure 6A and Experimental Procedures). Within this network, the type of interactions between the MDD-related GR genes that were most enriched were: co-expression (1.21 times the number expected when using other GR-stimulated transcript sets), co-localization (genes are expressed in the same tissue or proteins are found in the same location; fold enrichment = 1.21), and shared protein domains (fold enrichment = 3.77). Several genes, e.g., FTH1, CCT7, RPS2, IMPDH2, and PELI1, presented more than ten interactions. Additional co-expression analysis identified that the MDD-related GR genes are more tightly co-regulated in blood than in 1,000 sets of randomly chosen transcripts selected from all GR-responsive transcripts (fold enrichment = 1.04, permutation-based FDR = 0.078). These data provide support that the MDD-related GR genes functionally interact to perform an orchestrated function, i.e., they are coordinated in their transcriptional response to GR activation or stress. A limited network analysis through manually curated interactions from the scientific literature (Lechner et al., 2012) revealed that these genes show associations with MDD, SCZ, BPD, neurodevelopmental disorders, posttraumatic stress disorder, and response to antidepressant treatment in independent datasets (see Figure S3). In addition, they seem predominantly involved in pathways associated with ubiquitination and proteasome degradation and the inflammatory response, systems that have been implicated in the pathophysiology of MDD and SCZ, as well as in stress-related changes in synaptic plasticity (Miller et al., 2009; Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014; Tai and Schuman, 2008).
Figure 6.
Functional Annotation of Transcripts Regulated by MDD-Related GR Risk Variants
(A) Gene network produced using GeneMANIA. The network consists of 43 genes (circles) connected by 164 interactions (edges). Genes that are within a black filled circle indicate our MDD-related GR transcripts (n = 24), while those within a gray filled circle indicate additional genes (n = 20). The interactions found between these genes, which were more enriched than expected, are shown (co-expression: purple lines, shared protein domains: yellow lines, and co-localization: blue lines).
(B) Heatmap of gene expression changes (log2) between stress versus vehicle groups of mouse in brain and blood (n = 17 mice, left panel) as well as between baseline and GR-stimulation in human blood cells (blue, middle panel) and in mouse brain and blood (n = 22 mice, right panel). Investigated tissues are labeled within the bottom row of the heatmap (prefrontal cortex [PFC], hippocampus [HC], and amygdala [AM]). p values were computed by using linear regression model, and significance is indicated by a black box (FDR ≤ 0.1, dotted box p ≤ 0.05).
Convergent Functional Genomics: Integrating Human MDD-Related GR Genes with Relevant Mouse Models
To establish whether the transcripts regulated by acute GR activation in blood are also regulated in the brain within a similar time frame, we investigated whether the orthologs of the 24 MDD-related GR transcripts were differentially regulated in mouse blood and brain (prefrontal cortex [PFC], hippocampus [HC], and amygdala [AM]) 4 hr following dexamethasone administration (10 mg/kg dexamethasone i.p.). In this experiment, 17 of the 24 genes had a mouse orthologous gene, and 16 were expressed above microarray detection threshold. One-third of the genes showed significant changes at FDR ≤ 0.1 and 53.3% at p ≤ 0.05 in one or more of the investigated brain regions. Over 86% of the genes were significantly regulated (FDR ≤ 0.1) in mouse blood (see Figure 6B left panel).
In order to extend these results from pharmacologic GR agonism, we further evaluated whether acute social defeat stress, which is commonly used to induce depressive-like behavior, differentially regulates these same 24 MDD-related genes in mice. In this experiment, 17 orthologous genes were analyzed in blood, PFC, AM, and HC samples 4 hr after exposure to an aggressive resident mouse with short attack latency (Wagner et al., 2013). Here, three (MKNK2, SLCO3A1, and OCIAD2) of the five genes that were significantly differently regulated after dexamethasone stimulation were also significantly regulated following social defeat (FDR ≤ 0.1) in in one or more of the analyzed brain regions (see Figure 6B right panel). This suggests that a subset of MDD-related GR genes is also regulated by acute social defeat, providing an important extension to stress-related risk for depression.
Cumulative Risk Scores for the MDD-Related GR eSNPs Correlate with Dysfunctional Amygdala Reactivity
To investigate the relationship between MDD-related GR eSNPs and variability in stress-related brain function in humans, we applied an imaging genetics strategy to data from 647 participants (171 individuals with current or past DSM-IV Axis I disorders and 476 controls; 306 of participants were self-reported European-Americans [EUR-AM]; Table S5 and see also Experimental Procedures) of the Duke Neurogenetics Study (DNS) (see Experimental Procedures). Our analyses focused on centromedial amygdala reactivity to canonical threat-related angry and fearful facial expressions (Figure 7A), because this phenotype is clearly implicated in the etiology and pathophysiology of stress-related disorders, including depression (Phillips et al., 2003). Moreover, amygdala reactivity can trigger rapid physiological and behavioral responses to threat, including activation of the stress hormone response via projections from the medial division of the central nucleus of the amygdala, (captured in our analysis by our centromedial amygdala region of interest) to the paraventricular nucleus of the hypothalamus (Ulrich-Lai and Herman, 2009). Lastly, amygdala function is influenced by the slow-acting, presumably genomic effects of hydrocortisone administration (Henckens et al., 2010), further highlighting its importance as a systems-level phenotype sensitive to our observed GR-induced transcriptional responses.
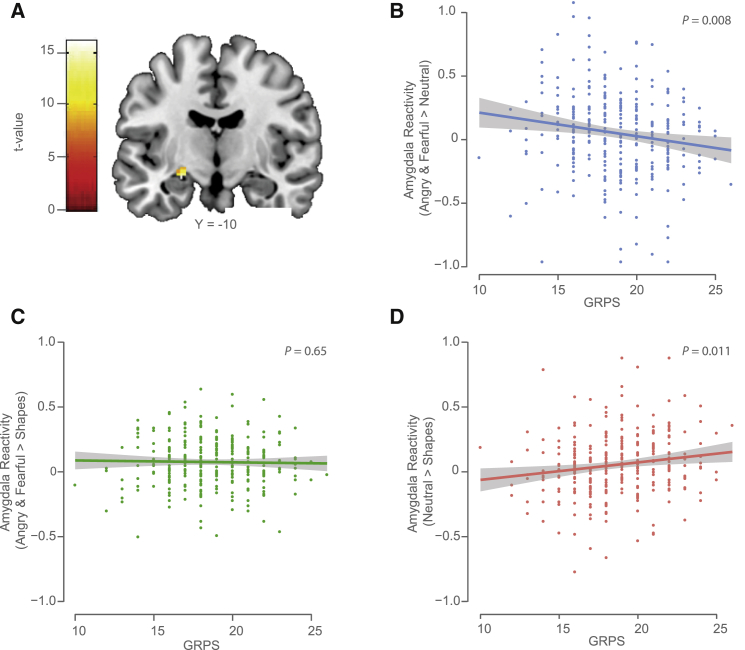
Figure 7.
GR-Response MDD-Related eSNP GRPS Correlate with Overgeneralized Amygdala Reactivity
(A) Statistical parametric map illustrating left centromedial amygdala reactivity to facial expressions with an “Angry & Fearful > Neutral” contrast in the entire sample (15 contiguous voxels; max voxel MNI coordinate, x = −24, y = −10, z = −14, t = 4.35, p = 7.76 × 10−6).
(B) Higher MDD-related GR eSNP genetic risk profile scores (GRPSs) in the European-American subsample of the DNS cohort (n = 306) predicted amygdala reactivity to threat-related facial expressions in comparison to neutral facial expressions.
(C and D) Post hoc analyses revealed that GRPSs did not predict amygdala reactivity to threat-related expressions (C), but that higher GRPSs predicted elevated amygdala reactivity to neutral facial expressions (D) in comparison to non-face control stimuli. The 95% confidence interval is displayed as gray shaded band in (B)–(D).
Higher MDD-related GR tag eSNP GRPSs (Table S4; see also Experimental Procedures) were associated with blunted centromedial amygdala response to angry and fearful facial expressions relative to neutral expressions in the EUR-AM subsample, even after accounting for age, sex, and the presence of an Axis I disorder (F1,301 = 7.06, p = 0.008; Figure 7B). This effect was also observed in the entire sample after accounting for population stratification (F1,637 = 6.05, p = 0.014; Figure S4A). Permutation analyses that formed random SNP profiles (n = 1,000; matched for MAF and not exceeding the maximum correlation among profile SNPs; see Experimental Procedures) indicated that the actual GRPS were more likely to be associated with these differences in amygdala reactivity than 1,000 sets of random SNP profiles (EUR-AM subsample: permutation-based FDR = 0.003; entire sample: permutation-based FDR = 0.012). Post hoc analyses revealed that this differential effect was driven by higher centromedial amygdala reactivity to neutral facial expressions relative to our control condition in participants with higher GRPS (EUR-AM subsample: F1,301 = 6.47, p = 0.011; Figure 7D; entire sample: F1,637 = 8.52, p = 0.004; Figures S4A and S4C). There were no effects of GRPS on amygdala reactivity to angry and fearful facial expressions relative to our control condition (EUR-AM subsample: F1,301 = 0.2, p = 0.65; Figure 7C and entire sample: F1,637 = 0.09, p = 0.76; Figures S4A and S4B).
This pattern of altered amygdala reactivity in individuals with higher GRPS is suggestive of impaired threat-related cue learning with inappropriately increased reactivity to neutral expressions, which do not convey threat (Britton et al., 2011; Oliveira et al., 2013). Thus, higher GRPS may be associated with non-specific or overgeneralized threat and stress responses, which are consistently observed in depression as well as other mood and anxiety disorders (Britton et al., 2011; Oliveira et al., 2013).
Discussion
We have shown that common variants in long-range enhancer elements alter the transcriptional responsiveness of GR target genes to the GR and that these variants cumulatively increase the risk for psychiatric disorders, including MDD and SCZ. These findings suggest that the risk of developing MDD after adverse life events may be influenced by an individual’s sensitivity to the downstream, transcriptional effects of cortisol released during the stressful adverse events. In addition, the findings suggest that the changes seen in the initial transcriptional response to stress may influence how an individual processes stressful exposures. Indeed, the risk variants were also associated with overgeneralized centromedial amygdala reactivity to non-threat stimuli. This is consistent with dysfunctional behavioral and physiological hyper-responsiveness to threat in MDD and other psychiatric disorders.
One of our notable genetic findings is that the distance between the GR-response eSNPs and the regulated gene expression probe was significantly longer than the distances previously reported for baseline eQTLs (149 kb baseline eQTLs versus 406 kb for GR-response eQTLs in our dataset). Our data support and extend previous observations that indicated a long-range transcriptional regulation by the GR (Hakim et al., 2011; John et al., 2011; So et al., 2007). In fact, a combined analysis of our GR-response eQTLs and ChIA-PET data from the ENCODE project (ENCODE Project Consortium, 2011) as well as a validation using 3C analysis suggests that there could be a physical long-range interaction between the eSNP locus and the promoter of the GR-regulated transcript for at least 25% of the GR-response eQTLs. Additional experiments are necessary in order to investigate the direct effects of the different alleles on the enhancer function and chromatin conformation in other tissues, including the brain, to further validate this.
More broadly, our results indicate that stimulated eQTL approaches using disease-risk-relevant transcriptional stimuli (in our case GR activation and stress) can identify novel risk genes for common disorders that may otherwise go undetected. Previous studies have used eQTLs or DNA methylation QTLs (mQTLs) for the annotation of GWAS results and indicated the importance of using eQTLs and mQTLs from disease-relevant tissues (Gamazon et al., 2013; Nicolae et al., 2010). While we do not observe a significant enrichment of baseline blood eQTLs, GR-response eQTLs from this tissue were significantly enriched, even over baseline SNPs, among the variants associated with MDD and SCZ (see Table 1). Interestingly, GR-response eSNPs identified in whole blood were enriched in enhancers specific to brain tissue, while this was not the case for baseline eSNPs identified in blood (see Figures 3 and S2). This suggests that GR-response eSNPs may have more relevance for cross-tissue effects, especially in the brain. This pattern may underlie the observation that GR-response eSNPs were associated with psychiatric disorders and amygdala function, but not with other medical disorders or height. Our findings support the notion that not only the tissue but also the type of stimulation, e.g., mimicking aspects of stress in our experiments, can be relevant for using such QTL studies in annotating GWAS results.
While these common genetic variants were discovered in peripheral blood cells, we provide evidence for their importance in neural circuits that are critical for generating and regulating the stress axis response to adversity. First, GR-response eSNP regions are enriched in enhancers relevant in brain tissue. Second, a number of the transcripts affected by these MDD-related GR eSNPs in their GR-regulated gene expression in human blood were also regulated by short-term GR activation or following exposure to acute social defeat stress in the mouse hippocampus, prefrontal cortex, or amygdala. Third, using imaging genetics, we demonstrate that the cumulative MDD-related GR tag eSNP genetic risk profile predicts overgeneralized reactivity of the human amygdala. It has to be noted, however, that while the GR-response eQTLs were identified using the selective GR agonist dexamethasone, the GR shares response elements with other steroid receptors, especially the mineralocorticoid receptor, so that we cannot exclude an important contribution of these other receptors.
Furthermore, the MDD-related GR genes formed a strongly interconnected gene network (over 85% of the genes are co-expressed; Figure 6A). Within this network, inflammation was the pathway with the highest connectivity (see Figure S3), and a number of studies indicate the pathophysiological relevance of this system in the development of MDD and SCZ (Haroon et al., 2012; Keller et al., 2013; Miller et al., 2009). The role of the immune system was also supported by results of the latest GWAS meta-analysis for SCZ (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014). The connectivity of this system was followed in strength by the connectivity of proteasome degradation. It has been shown, for example, that activation of GRs enhances ubiquitin/proteasome-mediated degradation of glutamate receptor subunits and thereby mediates cognitive impairment induced by repeated stress exposure (Yuen et al., 2012). Genetic modulation of GR effects on the immune system in addition to ubiquitin/proteasome-mediated degradation thus provide a mechanistic link between risk for psychiatric disorders and the genetic differences in GR-induced gene expression.
Most importantly, our GR-response eQTL analysis revealed an enrichment of these GR-response eSNPs among MDD-associated SNPs over baseline eSNP sets as well as random SNP sets. This suggests that SNPs altering the initial transcriptional response to stress also influence the risk for MDD. The association was verified in an independent cohort. Furthermore, the increased risk conferred by these functional variants may extend to SCZ. This is consistent with evidence from recent studies of psychiatric disorders, which suggest shared genetic risk loci, with MDD having the highest co-heritability with BPD followed by SCZ (Lee et al., 2013). The fact that we do not detect a significant enrichment of GR-response eSNPs with BPD, despite the large SNP co-heritability of this disorder with both SCZ and MDD, may in part be due to the smaller sample size in this meta-analysis and thus insufficient power to detect true associations (n = 6,704 cases for BPD versus over 9,000 cases for MDD and SCZ1). The fact that the fold enrichment of GR-response eSNPs with SCZ increases with sample size (SCZ1: n = 9,087 cases versus SCZ2: n = 36,989 cases) and p value cut-off (p < 0.05 and p < 5 × 10−8) suggests that the strategy of using stimulated eQTL approaches may help in identifying true associations for disease. Interestingly, we find that four MDD-related GR eQTL bins, which are not only associated with MDD but also with SCZ, and the cross-disorder associations from the PGC analyses (Table 1) reach genome-wide significance for SCZ in the most recent meta-analysis (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014). Besides the extended MHC region (chr6: 26–34 Mb, hg19), these overlapping risk loci include a region on chr1: 149,998,890–150,242,490 (hg19) that now ranks 48th for association with SCZ and overlaps with the MDD-related GR eQTL bin, ANP32/PLEKHO1, that regulates the probe gene HIST2H2AA3/4. This GR-response eQTL drives the overlap with genome-wide SCZ2 associations, and it has been validated using qPCR (Supplemental Information). These findings suggest that GR-response eSNPs may contribute to the shared risk between psychiatric disorders, especially MDD and SCZ, and that this approach may delineate between shared and specific risk factors for these disorders.
Results from our imaging genetics study provide one potential neural pathway by which MDD-related GR eSNPs may increase the risk for the development of stress-related psychopathology, including depression. Interestingly, MDD-related GR eSNPs predict heightened amygdala reactivity to stimuli that do not inherently signal threat (i.e., neutral facial expressions). This suggests that MDD-related GR eSNPs associated with the immediate transcriptome response to stress may impair the neural circuitry that supports the learning of threat-related cues and, possibly, thereby contribute to the overgeneralization of threat-related stress responses. Indeed, in healthy individuals, the genomic effects of hydrocortisone result in more specific reactivity to threatening stimuli (Henckens et al., 2010). As such, MDD-related GR eSNPs may underpin a less adaptive and overgeneralized amygdala response that leaves individuals more likely to perceive threat in the absence of unambiguous cues; this in turn may lead to the development of cognitive biases associated with depression, or perhaps even paranoia, in the context of schizophrenia.
The data presented in this study show that common genetic variants that change the GR-mediated immediate transcriptome response to stress are linked, in the long-term, to both changes in neural processing of threat and increased risk for MDD and SCZ. Our data lend further support to the notion of a possible shared genetic liability of some psychiatric disorders and specifically point to stress-responsive genes as common risk factors. Studies dissecting how these genetic variants alter the molecular, cellular, and neural response to glucocorticoids in the short and long term could inform the development of novel strategies for the prevention and treatment of stress-related psychiatric disorders.
Experimental Procedures
Samples and Study Designs
MPIP Cohort
The subject pool for the eQTL analysis consisted of 164 male Caucasian individuals: 93 healthy probands and 71 in-patients with depressive disorders treated at the Max Planck Institute of Psychiatry’s hospital in Munich, Germany (MPIP cohort; see Supplemental Experimental Procedures; Hennings et al., 2009; Menke et al., 2012 for details). Baseline whole-blood samples (for plasma and RNA) were obtained at 6 p.m. after 2 hr of fasting and abstention from coffee and physical activity. Immediately afterward the participants were given 1.5 mg dexamethasone orally. A second blood draw was performed 3 hr later at 9 p.m. (see Figure 2A). Cortisol and ACTH serum levels were determined using previously described radioimmunoassays (Hennings et al., 2009; Menke et al., 2012). Plasma dexamethasone concentrations were assessed in serum samples drawn at 9 p.m. using liquid chromatography-tandem mass spectrometry on API4000 (AB Sciex).
MARS Cohort
This sample included 1,483 participants with European ancestry (1,005 with MDD) recruited for the MARS project at the MPIP in Munich. All individuals used within the eQTL study (MPIP cohort) were not part of this sample (see Supplemental Experimental Procedures and Hennings et al., 2009 for details).
DNS Cohort
The imaging genetics analysis was conducted on data from (1) a European-American subsample of 306 participants (63 with DSM-IV Axis I disorder) and (2) a full sample of 647 participants (117 with DSM-IV Axis I disorder) of the ongoing Duke Neurogenetics Study (see Supplemental Experimental Procedures). All participants completed a widely utilized functional magnetic resonance imaging (fMRI) paradigm assessing threat-related amygdala reactivity (see Supplemental Experimental Procedures).
Mouse Models
Twenty-two male C57BL/6N mice were used for the dexamethasone-stimulation test (DEX-mouse). The experiment was performed twice with two separate batches of mice (n = 22 per batch). Animals were injected i.p. with either vehicle (VEH, n = 11) or 10 mg/kg dexamethasone (DEX, n = 11) between 9 a.m. and 11 a.m. Animals were sacrificed 4 hr post-injection.
The acute social defeat sample included 17 male C57BL/6N mice (n = 8 control and n = 9 acute stress mice) taken from a larger study that were used for this experiment. Mice underwent the acute social defeat stress once exactly 4 hr preceding sacrifice and tissue collection. The acute social defeat paradigm was performed as described previously (Wagner et al., 2013) on a single day between 9 a.m. and 12 p.m. (see Supplemental Experimental Procedures).
From both animal models described above, blood was collected and the brain was carefully extracted and dissected (see Supplemental Experimental Procedures). The following brain regions were collected: hippocampus (HC), prefrontal cortex (PFC), and the amygdala (AM).
All human studies have been approved by the respective local ethics committees and all individuals gave written informed consent. Details about the individual studies are listed below or in the Supplemental Experimental Procedures. The mouse model protocols were approved by the Committee for the Care and Use of Laboratory Animals of the Government of Upper Bavaria, Germany.
Gene Expression Data
The human whole-blood RNA of the MPIP cohort samples was hybridized to Illumina HumanHT-12 v3.0 array. All array probes have been subjected to an extensive quality control (QC; see Supplemental Experimental Procedures). For the GR-response eQTL analysis, only transcripts that showed a difference in gene expression between the samplings at 6 p.m. and 9 p.m. with an absolute fold change ≥ 1.3 in at least 20% of all samples were categorized as robustly effected by dexamethasone stimulation (n = 4,630 transcripts) and further used in the analysis. The position of the array probes and possible SNPs within these sequences were annotated using ReMOAT version August 2009 (Barbosa-Morais et al., 2010), leaving 4,447 autosomal array probes for the GR-response eQTL analysis (see Supplemental Experimental Procedures).
DEX-mouse RNA samples were hybridized on Illumina MouseRef-8 v2.0 chips, and the mouse RNA from the acute social defeat mouse model was hybridized on Illumina MouseWG-8 v2.0 chips. QC was applied separately for each tissue and experiment as described in Supplemental Experimental Procedures.
Genotype Data
Human DNA from MPIP and MARS cohort subjects was extracted from EDTA blood samples and genotyped on Illumina Human610-Quad/Human660W-Quad arrays (MPIP cohort) and Illumina Sentix Human-1/HumanHap300/Human610-Quad/HumanOmniExpress arrays (MARS cohort). From the SNP data surviving QC, imputation of additional variants was performed using IMPUTE v2 (Howie et al., 2009; see Supplemental Experimental Procedures for more detail on genotyping QC and imputation).
The MARS GRPSs included alleles from 20 of the 23 tag eSNPs (three SNPs diverged from HWE in the MARS sample, see Table S3). See also Supplemental Experimental Procedures.
Human DNA from participants of the DNS cohort was isolated from saliva and genotyped on the Illumina HumanOmniExpress array as well as a custom array containing an additional ∼300,000 SNPs. The DNS GRPSs included alleles from 19 of the 23 tag eSNPs (four SNPs not present on genotyping array; see Table S4 and Supplemental Experimental Procedures).
Statistical Analysis
The eQTL analysis (MPIP cohort) was restricted to those SNP-probe pairs that map within a region of 1 Mb upstream or downstream of the gene expression probe, in order to detect cis-eQTLs. To measure the transcriptional response, we used the log fold change in gene expression changes between 6 p.m. (baseline) and 9 p.m. (GR-stimulation) standardized to baseline.
PLINK v1.07 (Purcell et al., 2007) was used to test for cis-association between all imputed SNPs and transcriptional response. As eQTL data were composed of two kinds of data, genotyping and expression data, we used two stages of multiple testing correction: (1) SNP level correction: for each cis-region (array probe), we performed a permutation test. The sample identifiers in the gene expression data were shuffled in order to preserve the structure in the genotype data (LD). A total of 500,000 permutations were carried out per probe, and the empirical p values were adjusted using the Westfall-Young correction for the number of SNPs per probe, i.e., maxT procedure of Westfall-Young (Westfall and Young, 1993). (2) Probe level correction: cis-regions with an extensive LD structure will increase the number of false positive eQTLs (Westra et al., 2013). Therefore, we applied the Benjamini-Hochberg method to correct the maxT-adjusted p value significance by using only the most significant and independent SNPs per probe (tag SNPs). The number of tag eSNPs per cis-region was identified by LD pruning and “clumping” the SNPs using the “clump” command in PLINK (using distance < 1 Mb and r2 ≤ 0.2 as setting). Each tag SNP forms a SNP bin by aggregating SNPs at r2 ≤ 0.2 and distance < 1 Mb. SNPs within a given bin were correlated to the tag SNP, but not to any other tag SNP of an other SNP bin. We limited the false-positive SNP-probe pairs to less than 5% and therefore considered the FDR analog of the p value (Q value) < 5% as statistically significant.
Validation of GR-response cis-eQTL results was carried out with a sample size-weighted Z score meta-analysis (Evangelou and Ioannidis, 2013) in an additional independent dataset using peripheral blood samples of 58 individuals (see Supplemental Experimental Procedures). A GR-response cis-eQTL was validated if the meta-analysis p value was less than the actual maxT-adjusted p value in the discovery sample alone.
The genomic control inflation factor (λgc; Devlin and Roeder, 1999) was calculated for every GR-response eQTL gene expression probe (n = 297) based on the genome-wide genotype data (λgc). The inflation factor was computed in PLINK as median χ2 statistic. The median λgc over all probes is 1, which implies no large inflation was present.
We used NR3C1 ChIP-seq data obtained from the ENCODE Project (ENCODE Project Consortium, 2011) to determine actual GR binding at GR-response eSNPs (see Supplemental Experimental Procedures).
To determine whether GR-response eSNPs were enriched for functional regions, we annotated them using HaploReg (Ward and Kellis, 2012) and compared the results to a realistic null distribution based on permuted baseline eSNP sets (see Supplemental Experimental Procedures).
ChIA-PET data were obtained from the UCSC Genome Browser (http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeGisChiaPet; see Supplemental Experimental Procedures).
To identify whether GR-response eSNPs were enriched for association with MDD, SCZ, BPD, ADHD, ASD, Crohn’s disease, rheumatoid arthritis, and the GWAS loci for height, we integrated our data with results from the previously published GWAS analysis (see Supplemental Experimental Procedures). To prove the significance of the MDD-related GR SNPs for MDD, we used a logistic regression model to test the association of the MDD-related GR tag eSNP GRPSs for disease status in the independent MARS cohort. Gender, BMI, and age were used as covariates. To establish the null distribution, we generated 1,000 random SNP profiles by swapping individual labels to provide new SNP profiles under the null hypothesis. To further account for the genomic LD structure, we limited the analyses to tag SNPs (tag SNP = SNP showing the highest association per cis-eQTL bin) and generated 1,000 randomized SNP sets; conditional on MAF and each of the same size as the GR-response tag SNPs overlap with MDD associations (n = 285).
The gene network analysis was performed using the online tool GeneMANIA (Montojo et al., 2014). To establish the null-distribution, we calculated the gene network for ten sets of randomly chosen GR-response transcripts (n = 4,422). Finally, we determined the average gene network results in order to establish the relationship between MDD-relevant GR-response transcripts and non-MDD-relevant but GR-response transcripts. Network categories showing a fold enrichment > 1 are reported in Figure 6A.
For the co-expression analysis, we used the GR-response residuals from all array probes (n = 4,422) to determine if the 25 MDD-related GR array probes are more co-regulated than 1,000 sets of randomly chosen GR-stimulated transcripts (see Supplemental Experimental Procedures).
A disease-related network was built by manual curation and literature mining using the CIDeR database (Lechner et al., 2012) and the yED software (yWorks GmbH, Tübingen).
To test the relationship of the GR-response eSNPs and threat-related amygdala reactivity, we used an imaging genetics strategy as described in the Supplemental Experimental Procedures.
Chromatin Conformation Capture Analysis
3C was carried out in five LCLs as described in Hagège et al., 2007 and detailed in the Supplemental Experimental Procedures.
qPCR Validation
Quantitative real-time PCR (qPCR) was used to validate the association between eSNPs and GR-stimulated gene expression of ADORA3 (the probe with the most significant GR-response eQTL) and HIST2H2AA3/HIST2H2AA4 (the probe with the most eSNPs overlapping with data from our meta-analysis for MDD) in whole blood cells and for a long-range GR-response eQTL-NRTN in five LCLs, which were also used with the 3C assay. More details are provided in Supplemental Experimental Procedures.
Acknowledgments
This study has received its main financial support from the European Union under European Research Council GA no. 281338 (to E.B.B.) and was also supported by the BMBF within the Program for Medical Genome Research with the Research Grant FKZ 01GS08151 (to W.W.) and financed by the DFG within the Cooperation Clinical Group Molecular Neurogenetics (WU 164/3-2) and the Helmholtz Alliances of Systems Biology and of Mental Health in an Ageing Society Grant HA-215 (W.W.). J.A. was supported in part by the NeuroNova gGmbH, A.R.H. by NIH (NIDA R01-DA031579), R.B. by the Klingenstein Third Generation Foundation and McDonnell Center for Systems Neuroscience, and C.E.C. by an NSF pre-doctoral grant (DGE-1143954). The Duke Neurogenetics Study (DNS) is supported by Duke University and the NIH (NIDA R01-DA033369). We thank D. Spengler, A. Hoffmann, M. Rex-Haffner, S. Darchinger, A. Löschner, and M. Ködel for excellent technical support. We are grateful to S. Röh for processing the NR3C1 ChIP-seq data. We also thank L. Preis and A. Eichelkraut for recruitment and sample ascertainment of the Max-Planck Institute of Psychiatry (MPIP) cohort and Y.S. Nikolova, A.X. Gorka, B. Brigidi, K. Faig, S. Jacobson, and A. Knodt for assistance with DNS cohort data collection.
See Supplemental Information for the full list of collaborators of the Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium (PGC).
Published: June 3, 2015
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Contributor Information
Elisabeth B. Binder, Email: binder@psych.mpg.de.
Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium (PGC):
Stephan Ripke, Naomi R. Wray, Cathryn M. Lewis, Steven P. Hamilton, Myrna M. Weissman, Gerome Breen, Enda M. Byrne, Douglas H.R. Blackwood, Dorret I. Boomsma, Sven Cichon, Andrew C. Heath, Florian Holsboer, Susanne Lucae, Pamela A.F. Madden, Nicholas G. Martin, Peter McGuffin, Pierandrea Muglia, Markus M. Noethen, Brenda P. Penninx, Michele L. Pergadia, James B. Potash, Marcella Rietschel, Danyu Lin, Bertram Müller-Myhsok, Jianxin Shi, Stacy Steinberg, Hans J. Grabe, Paul Lichtenstein, Patrik Magnusson, Roy H. Perlis, Martin Preisig, Jordan W. Smoller, Kari Stefansson, Rudolf Uher, Zoltan Kutalik, Katherine E. Tansey, Alexander Teumer, Alexander Viktorin, Michael R. Barnes, Thomas Bettecken, Elisabeth B. Binder, René Breuer, Victor M. Castro, Susanne E. Churchill, William H. Coryell, Nick Craddock, Ian W. Craig, Darina Czamara, Eco J. De Geus, Franziska Degenhardt, Anne E. Farmer, Maurizio Fava, Josef Frank, Vivian S. Gainer, Patience J. Gallagher, Scott D. Gordon, Sergey Goryachev, Magdalena Gross, Michel Guipponi, Anjali K. Henders, Stefan Herms, Ian B. Hickie, Susanne Hoefels, Witte Hoogendijk, Jouke Jan Hottenga, Dan V. Iosifescu, Marcus Ising, Ian Jones, Lisa Jones, Tzeng Jung-Ying, James A. Knowles, Isaac S. Kohane, Martin A. Kohli, Ania Korszun, Mikael Landen, William B. Lawson, Glyn Lewis, Donald MacIntyre, Wolfgang Maier, Manuel Mattheisen, Patrick J. McGrath, Andrew McIntosh, Alan McLean, Christel M. Middeldorp, Lefkos Middleton, Grant M. Montgomery, Shawn N. Murphy, Matthias Nauck, Willem A. Nolen, Dale R. Nyholt, Michael O’Donovan, Högni Oskarsson, Nancy Pedersen, William A. Scheftner, Andrea Schulz, Thomas G. Schulze, Stanley I. Shyn, Engilbert Sigurdsson, Susan L. Slager, Johannes H. Smit, Hreinn Stefansson, Michael Steffens, Thorgeir Thorgeirsson, Federica Tozzi, Jens Treutlein, Manfred Uhr, Edwin J.C.G. van den Oord, Gerard Van Grootheest, Henry Völzke, Jeffrey B. Weilburg, Gonneke Willemsen, Frans G. Zitman, Benjamin Neale, Mark Daly, Douglas F. Levinson, and Patrick F. Sullivan
Accession Numbers
Data from the human gene expression microarray experiment were deposited at the GEO repository under GEO: GSE46743.
Supplemental Information
References
- Barbosa-Morais N.L., Dunning M.J., Samarajiwa S.A., Darot J.F.J., Ritchie M.E., Lynch A.G., Tavaré S. A re-annotation pipeline for Illumina BeadArrays: improving the interpretation of gene expression data. Nucleic Acids Res. 2010;38:e17. doi: 10.1093/nar/gkp942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton J.C., Lissek S., Grillon C., Norcross M.A., Pine D.S. Development of anxiety: the role of threat appraisal and fear learning. Depress. Anxiety. 2011;28:5–17. doi: 10.1002/da.20733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kloet E.R., Joëls M., Holsboer F. Stress and the brain: from adaptation to disease. Nat. Rev. Neurosci. 2005;6:463–475. doi: 10.1038/nrn1683. [DOI] [PubMed] [Google Scholar]
- Devlin B., Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- Dohrenwend B.P., Egri G. Recent stressful life events and episodes of schizophrenia. Schizophr. Bull. 1981;7:12–23. doi: 10.1093/schbul/7.1.12. [DOI] [PubMed] [Google Scholar]
- ENCODE Project Consortium A user’s guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelou E., Ioannidis J.P.A. Meta-analysis methods for genome-wide association studies and beyond. Nat. Rev. Genet. 2013;14:379–389. doi: 10.1038/nrg3472. [DOI] [PubMed] [Google Scholar]
- Gamazon E.R., Badner J.A., Cheng L., Zhang C., Zhang D., Cox N.J., Gershon E.S., Kelsoe J.R., Greenwood T.A., Nievergelt C.M. Enrichment of cis-regulatory gene expression SNPs and methylation quantitative trait loci among bipolar disorder susceptibility variants. Mol. Psychiatry. 2013;18:340–346. doi: 10.1038/mp.2011.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagège H., Klous P., Braem C., Splinter E., Dekker J., Cathala G., de Laat W., Forné T. Quantitative analysis of chromosome conformation capture assays (3C-qPCR) Nat. Protoc. 2007;2:1722–1733. doi: 10.1038/nprot.2007.243. [DOI] [PubMed] [Google Scholar]
- Hakim O., Sung M.-H., Voss T.C., Splinter E., John S., Sabo P.J., Thurman R.E., Stamatoyannopoulos J.A., de Laat W., Hager G.L. Diverse gene reprogramming events occur in the same spatial clusters of distal regulatory elements. Genome Res. 2011;21:697–706. doi: 10.1101/gr.111153.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haroon E., Raison C.L., Miller A.H. Psychoneuroimmunology meets neuropsychopharmacology: translational implications of the impact of inflammation on behavior. Neuropsychopharmacology. 2012;37:137–162. doi: 10.1038/npp.2011.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim C., Binder E.B. Current research trends in early life stress and depression: review of human studies on sensitive periods, gene-environment interactions, and epigenetics. Exp. Neurol. 2012;233:102–111. doi: 10.1016/j.expneurol.2011.10.032. [DOI] [PubMed] [Google Scholar]
- Henckens M.J.A.G., van Wingen G.A., Joëls M., Fernández G. Time-dependent effects of corticosteroids on human amygdala processing. J. Neurosci. 2010;30:12725–12732. doi: 10.1523/JNEUROSCI.3112-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennings J.M., Owashi T., Binder E.B., Horstmann S., Menke A., Kloiber S., Dose T., Wollweber B., Spieler D., Messer T. Clinical characteristics and treatment outcome in a representative sample of depressed inpatients - findings from the Munich Antidepressant Response Signature (MARS) project. J. Psychiatr. Res. 2009;43:215–229. doi: 10.1016/j.jpsychires.2008.05.002. [DOI] [PubMed] [Google Scholar]
- Howie B.N., Donnelly P., Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankord R., Herman J.P. Limbic regulation of hypothalamo-pituitary-adrenocortical function during acute and chronic stress. Ann. N Y Acad. Sci. 2008;1148:64–73. doi: 10.1196/annals.1410.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John S., Sabo P.J., Thurman R.E., Sung M.-H., Biddie S.C., Johnson T.A., Hager G.L., Stamatoyannopoulos J.A. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat. Genet. 2011;43:264–268. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller W.R., Kum L.M., Wehring H.J., Koola M.M., Buchanan R.W., Kelly D.L. A review of anti-inflammatory agents for symptoms of schizophrenia. J. Psychopharmacol. (Oxford) 2013;27:337–342. doi: 10.1177/0269881112467089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler K.S. What psychiatric genetics has taught us about the nature of psychiatric illness and what is left to learn. Mol. Psychiatry. 2013;18:1058–1066. doi: 10.1038/mp.2013.50. [DOI] [PubMed] [Google Scholar]
- Kendler K.S., Karkowski-Shuman L. Stressful life events and genetic liability to major depression: genetic control of exposure to the environment? Psychol. Med. 1997;27:539–547. doi: 10.1017/s0033291797004716. [DOI] [PubMed] [Google Scholar]
- Kendler K.S., Gatz M., Gardner C.O., Pedersen N.L. A Swedish national twin study of lifetime major depression. Am. J. Psychiatry. 2006;163:109–114. doi: 10.1176/appi.ajp.163.1.109. [DOI] [PubMed] [Google Scholar]
- Kessler R.C., Berglund P., Demler O., Jin R., Merikangas K.R., Walters E.E. Lifetime prevalence and age-of-onset distributions of DSM-IV disorders in the National Comorbidity Survey Replication. Arch. Gen. Psychiatry. 2005;62:593–602. doi: 10.1001/archpsyc.62.6.593. [DOI] [PubMed] [Google Scholar]
- Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., Ziller M.J., Roadmap Epigenomics Consortium Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner M., Höhn V., Brauner B., Dunger I., Fobo G., Frishman G., Montrone C., Kastenmüller G., Waegele B., Ruepp A. CIDeR: multifactorial interaction networks in human diseases. Genome Biol. 2012;13:R62. doi: 10.1186/gb-2012-13-7-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.H., Ripke S., Neale B.M., Faraone S.V., Purcell S.M., Perlis R.H., Mowry B.J., Thapar A., Goddard M.E., Witte J.S., Cross-Disorder Group of the Psychiatric Genomics Consortium. International Inflammatory Bowel Disease Genetics Consortium (IIBDGC) Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat. Genet. 2013;45:984–994. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay L.I., Cidlowski J.A. Molecular control of immune/inflammatory responses: interactions between nuclear factor-kappa B and steroid receptor-signaling pathways. Endocr. Rev. 1999;20:435–459. doi: 10.1210/edrv.20.4.0375. [DOI] [PubMed] [Google Scholar]
- Menke A., Arloth J., Pütz B., Weber P., Klengel T., Mehta D., Gonik M., Rex-Haffner M., Rubel J., Uhr M. Dexamethasone stimulated gene expression in peripheral blood is a sensitive marker for glucocorticoid receptor resistance in depressed patients. Neuropsychopharmacology. 2012;37:1455–1464. doi: 10.1038/npp.2011.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller A.H., Maletic V., Raison C.L. Inflammation and its discontents: the role of cytokines in the pathophysiology of major depression. Biol. Psychiatry. 2009;65:732–741. doi: 10.1016/j.biopsych.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montojo J., Zuberi K., Rodriguez H., Bader G.D., Morris Q. GeneMANIA: Fast gene network construction and function prediction for Cytoscape. F1000Res. 2014;3:153. doi: 10.12688/f1000research.4572.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolae D.L., Gamazon E., Zhang W., Duan S., Dolan M.E., Cox N.J. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira L., Ladouceur C.D., Phillips M.L., Brammer M., Mourao-Miranda J. What does brain response to neutral faces tell us about major depression? evidence from machine learning and fMRI. PLoS ONE. 2013;8:e60121. doi: 10.1371/journal.pone.0060121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips M.L., Drevets W.C., Rauch S.L., Lane R. Neurobiology of emotion perception II: Implications for major psychiatric disorders. Biol. Psychiatry. 2003;54:515–528. doi: 10.1016/s0006-3223(03)00171-9. [DOI] [PubMed] [Google Scholar]
- Phuc Le P., Friedman J.R., Schug J., Brestelli J.E., Parker J.B., Bochkis I.M., Kaestner K.H. Glucocorticoid receptor-dependent gene regulatory networks. PLoS Genet. 2005;1:e16. doi: 10.1371/journal.pgen.0010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S., Wray N.R., Lewis C.M., Hamilton S.P., Weissman M.M., Breen G., Byrne E.M., Blackwood D.H., Boomsma D.I., Cichon S., Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium A mega-analysis of genome-wide association studies for major depressive disorder. Mol. Psychiatry. 2013;18:497–511. doi: 10.1038/mp.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussos P., Mitchell A.C., Voloudakis G., Fullard J.F., Pothula V.M., Tsang J., Stahl E.A., Georgakopoulos A., Ruderfer D.M., Charney A. A role for noncoding variation in schizophrenia. Cell Rep. 2014;9:1417–1429. doi: 10.1016/j.celrep.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So A.Y., Chaivorapol C., Bolton E.C., Li H., Yamamoto K.R. Determinants of cell- and gene-specific transcriptional regulation by the glucocorticoid receptor. PLoS Genet. 2007;3:e94. doi: 10.1371/journal.pgen.0030094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai H.-C., Schuman E.M. Ubiquitin, the proteasome and protein degradation in neuronal function and dysfunction. Nat. Rev. Neurosci. 2008;9:826–838. doi: 10.1038/nrn2499. [DOI] [PubMed] [Google Scholar]
- Ulrich-Lai Y.M., Herman J.P. Neural regulation of endocrine and autonomic stress responses. Nat. Rev. Neurosci. 2009;10:397–409. doi: 10.1038/nrn2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ustün T.B., Ayuso-Mateos J.L., Chatterji S., Mathers C., Murray C.J.L. Global burden of depressive disorders in the year 2000. Br. J. Psychiatry. 2004;184:386–392. doi: 10.1192/bjp.184.5.386. [DOI] [PubMed] [Google Scholar]
- van Rossum E.F., Binder E.B., Majer M., Koper J.W., Ising M., Modell S., Salyakina D., Lamberts S.W., Holsboer F. Polymorphisms of the glucocorticoid receptor gene and major depression. Biol. Psychiatry. 2006;59:681–688. doi: 10.1016/j.biopsych.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Wagner K.V., Hartmann J., Mangold K., Wang X.-D., Labermaier C., Liebl C., Wolf M., Gassen N.C., Holsboer F., Rein T. Homer1 mediates acute stress-induced cognitive deficits in the dorsal hippocampus. J. Neurosci. 2013;33:3857–3864. doi: 10.1523/JNEUROSCI.4333-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward L.D., Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warden D., Rush A.J., Trivedi M.H., Fava M., Wisniewski S.R. The STAR∗D Project results: a comprehensive review of findings. Curr. Psychiatry Rep. 2007;9:449–459. doi: 10.1007/s11920-007-0061-3. [DOI] [PubMed] [Google Scholar]
- Westfall P.H., Young S.S. On Adjusting P-Values for Multiplicity. Biometrics. 1993;49:941–944. [Google Scholar]
- Westra H.-J., Peters M.J., Esko T., Yaghootkar H., Schurmann C., Kettunen J., Christiansen M.W., Fairfax B.P., Schramm K., Powell J.E. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet. 2013;45:1238–1243. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson P.O., Goodyer I.M. Childhood adversity and allostatic overload of the hypothalamic-pituitary-adrenal axis: a vulnerability model for depressive disorders. Dev. Psychopathol. 2011;23:1017–1037. doi: 10.1017/S0954579411000472. [DOI] [PubMed] [Google Scholar]
- Yuen E.Y., Wei J., Liu W., Zhong P., Li X., Yan Z. Repeated stress causes cognitive impairment by suppressing glutamate receptor expression and function in prefrontal cortex. Neuron. 2012;73:962–977. doi: 10.1016/j.neuron.2011.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.