Abstract
Almost 30% of all acute myeloid leukemias (AML) are associated with an internal tandem duplication (ITD) in the juxtamembrane domain of FMS-like tyrosine kinase 3 receptor (FLT3). Patients with FLT3-ITD mutations tend to have a poor prognosis. MicroRNAs (miRNAs) have a pivotal role in myeloid differentiation and leukemia. MiRNA-155 (MiR-155) was found to be upregulated in FLT3-ITD-associated AMLs. In this study, we discovered that FLT3-ITD signaling induces the oncogenic miR-155. We show in vitro and in vivo that miR-155 expression is regulated by FLT3-ITD downstream targets nuclear factor-κB (p65) and signal transducer and activator of transcription 5 (STAT5). Further, we demonstrate that miR-155 targets the myeloid transcription factor PU.1. Knockdown of miR-155 or overexpression of PU.1 blocks proliferation and induces apoptosis of FLT3-ITD-associated leukemic cells. Our data demonstrate a novel network in which FLT3-ITD signaling induces oncogenic miR-155 by p65 and STAT5 in AML, thereby targeting transcription factor PU.1.
INTRODUCTION
Up to 30% of all acute myeloid leukemias (AMLs) are associated with an activating mutation in the FMS-like tyrosine kinase 3 receptor (FLT3).1 Two distinct groups of FLT3 mutations are found: (1) the most common are internal tandem duplications (ITDs) of the FLT3 juxtamembrane region, and (2) point mutations within the tyrosine kinase domains (TKDs).1,2 While FLT3-TKD mutations seem to have no prognostic relevance in AML, patients bearing an FLT3-ITD mutation have a significantly worse outcome compared with AML patients with wild-type FLT3 (FLT3-WT).3 FLT3-ITD constitutively activates several pathways such as MAPK/ERK, PI3K/AKT, NF-κB (nuclear factor-κB) and STAT5 (signal transducer and activator of transcription 5).4–9 It was shown that PU.1 (refs 4,10) and C/EBPα,4,10–11 two important transcription factors in myeloid differentiation, are repressed by FLT3-ITD. FLT3-ITD mutations induce proliferation, cell transformation and block myeloid differentiation.2,8,12
MicroRNAs (miRs) are small (~22 bp) noncoding RNAs, which regulate protein expression posttranscriptionally by recruitment of the RNA-induced silencing complex to the 3′-untranslated region (3′-UTR) of target mRNAs.13–14 It was shown that miRs are crucial regulators in myeloid differentiation15–17 and in leukemogenesis.18 Earlier publications reveal that AML patients bearing an FLT3-ITD mutation have an increased expression of miRNA-155 (miR-155).19–22 MiR-155 was found to be upregulated by NF-κB in inflammatory response.23,24 Further, it was shown that miR-155-knockout mice are immunodeficient.25 In addition, miR-155 functions as an oncomiR and is highly expressed in B-cell lymphoma,26,27 cervical cancer,28 pancreatic cancer,29 colon cancer30 and breast cancer.31 Furthermore, sustained expression of miR-155 in hematopoietic stem cells causes a myeloproliferative disorder.32 Marcucci et al.33 show that high miR-155 expression is associated with a worse clinical outcome in cytogenetically normal AML.
In this study, we report for the first time that miR-155 is induced by FLT3-ITD signaling. We demonstrate that NF-κB (p65) directly binds to the miR-155 promoter in FLT3-ITD-associated MV4;11 cells. In functional analyses, we show that miR-155 directly targets the transcription factor PU.1. In addition, we observed reduced miR-155 expression during myeloid differentiation, whereas overexpression of miR-155 blocks maturation of myeloid cells. Furthermore, we reveal that miR-155 is important for clonal growth of FLT3-ITD-associated leukemic cells. We demonstrate that overexpression of PU.1 or depletion of miR-155 in FLT3-ITD-associated MV4;11 cells induces apoptosis. In summary, our data identified miR-155 as a crucial player in maintenance and development of FLT3-ITD-associated AML. We propose that miR-155 could be a novel potential therapeutic target in FLT3-ITD-associated AML.
MATERIALS AND METHODS
Human cell samples from AML patients
AML patient samples (Table 1) were obtained from University Hospital Münster (Münster, Germany) and University Hospital Leipzig (Leipzig, Germany). The study protocols used for AML patient sample collection were approved by the ethics committees of the participating centers. All patients provided written informed consent in accordance with the Declaration of Helsinki. All samples were analyzed by cytogenetic and molecular genetic analyses.
Table 1.
Characteristics of AML patient samples used for miR-155 expression analysis
| Patient no. AML | Karyotype | FLT3 status | Age (years) | Gender | Material |
|---|---|---|---|---|---|
| 1 | Normal Karyotype | WT | 21 | M | Bone marrow |
| 2 | Normal karyotype | TKD | 39 | F | Bone marrow |
| 3 | Normal Karyotype | TKD | 58 | F | Bone marrow |
| 4 | Normal Karyotype | WT | 43 | F | Bone marrow |
| 5 | Normal karyotype | TKD | 58 | M | Bone marrow |
| 6 | Complex karyotype | ITD/TKD | 29 | M | Bone marrow |
| 7 | Complex karyotype | ITD | 30 | F | Bone marrow |
| 8 | t(8;21) | ITD | 42 | M | Bone marrow |
| 9 | t(8;21) | ITD/TKD | 67 | M | Bone marrow |
| 10 | Normal karyotype | ITD | 70 | F | Bone marrow |
| 11 | Complex karyotype | WT | 51 | F | Bone marrow |
| 12 | Trisomie 22, inv(16), CBF?-MYH11 pos | WT | 60 | F | Bone marrow |
| 13 | Unknown | WT | 48 | M | Bone marrow |
| 14 | del(5), t(15;17) | WT | 31 | F | Bone marrow |
| 15 | Trisomie 8; t(15;17) | TKD | 23 | F | Bone marrow |
| 16 | Trisomie 8; t(15;17) | WT | 50 | M | Bone marrow |
| 17 | t(15;17) | WT | 70 | M | Bone marrow |
| 18 | t(15;17) | WT | 71 | F | Bone marrow |
| 19 | inv(16) | TKD | 39 | M | Bone marrow |
| 20 | inv(16) | TKD | 42 | M | Bone marrow |
| 21 | inv(16) | TKD | 29 | M | Bone marrow |
| 22 | inv(16) | TKD | 41 | F | Bone marrow |
| 23 | inv(16) | WT | 45 | F | Bone marrow |
| 24 | t(8;21) | WT | 36 | F | Bone marrow |
| 25 | del(7), t(8;21) | WT | 26 | M | Bone marrow |
| 26 | del(9), t(8;21) | WT | 53 | F | Bone marrow |
| 27 | t(8;21) | WT | 42 | M | Bone marrow |
| 28 | del(7), inv(16) | TKD | 40 | M | Bone marrow |
| 29 | Normal karyotype | ITD | 61 | M | Bone marrow |
| 30 | Normal karyotype | ITD | 17 | F | Bone marrow |
| 31 | Normal karyotype | ITD | 26 | M | Bone marrow |
| 32 | Normal karyotype | ITD | 67 | F | Bone marrow |
| 33 | Normal karyotype | ITD | 72 | M | Bone marrow |
| 34 | Normal karyotype | ITD | 77 | F | Bone marrow |
| 35 | Normal karyotype | ITD | 48 | F | Bone marrow |
| 36 | Normal karyotype | ITD | 71 | M | Bone marrow |
| 37 | Normal karyotype | WT | 44 | M | Bone marrow |
| 38 | Normal karyotype | WT | 61 | M | Bone marrow |
| 39 | Normal karyotype | WT | 71 | F | Bone marrow |
| 40 | Normal karyotype | WT | 72 | M | Bone marrow |
| 41 | Normal karyotype | WT | 69 | M | Bone marrow |
| 42 | Normal karyotype | WT | 57 | F | Bone marrow |
| 43 | Normal karyotype | WT | 78 | M | Bone marrow |
| 44 | Complex karyotype | WT | 64 | F | Bone marrow |
| 45 | Normal karyotype | WT | 72 | M | Bone marrow |
| 46 | Normal karyotype | WT | 63 | M | Bone marrow |
| 47 | Normal karyotype | WT | 66 | F | Bone marrow |
| 48 | Normal karyotype | WT | 72 | F | Bone marrow |
| 49 | Normal karyotype | WT | 70 | F | Bone marrow |
| 50 | Normal karyotype | WT | 65 | M | Bone marrow |
| 51 | Normal karyotype | WT | 49 | F | Bone marrow |
| 52 | Normal karyotype | WT | 65 | F | Bone marrow |
| 53 | Normal karyotype | WT | 72 | F | Bone marrow |
| 54 | Normal karyotype | ITD/TKD | 60 | F | Bone marrow |
| 55 | Normal karyotype | ITD | 66 | F | Bone marrow |
| 56 | Normal karyotype | ITD | 67 | M | Bone marrow |
| 57 | Normal karyotype | ITD | 63 | M | Bone marrow |
| 58 | Normal karyotype | ITD | 59 | M | Bone marrow |
| 59 | Normal karyotype | WT | 52 | M | Bone marrow |
| 60 | Normal karyotype | ITD | 47 | F | Bone marrow |
| Healthy | |||||
| 61 | Normal karyotype | WT | 36 | F | Bone marrow |
| 62 | Normal karyotype | WT | 41 | M | Bone marrow |
| 63 | Normal karyotype | WT | 41 | M | Bone marrow |
Abbreviations: f, female; ITD, internal tandem duplication; m, male; TKD, tyrosine kinase domain; WT, wild type.
Cell cultures
U937 and 293 T cells were cultured in RPMI-1640 supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin. The 32D cells stably expressing FLT3-WT or FLT3-ITD were described before.12 FLT3-WT expressing 32D cells were cultured in RPMI-1640 supplemented with 10% fetal bovine serum and 10% WEHI supernatant, FLT3-ITD-expressing 32D cells were cultured in RPMI-1640 supplemented with 10% fetal bovine serum. MV4;11 cells were maintained in Iscove’s modified Dulbecco’s medium supplemented with 10% fetal bovine serum. Cells were incubated at 37 °C in 5% CO2. Recombinant interleukin-3 (IL-3) was purchased from Immunotools (Friesoythe, Germany). The tyrosine kinase inhibitors PKC412 and CEP701 were purchased from LC Laboratories (Woburn, MA, USA) and SU5614 from Calbiochem (Darmstadt, Germany).
MiRNA detection by quantitative real-time PCR
Total RNA was extracted from cells using TRIzol reagent (Invitrogen, Darmstadt, Germany). The miRNA quantification was performed as previously described15 by using RNUB6 or snoRNA-135 expression for normalization. Taqman reverse transcription (RT) and PCR primers for RNUB6, snoRNA-135, hsa-miR-155 and mmu-miR-155 were obtained from Applied Biosystems (Darmstadt, Germany).
DNA constructs and cloning
The FLT3-ITD, FLT3-WT and FLT3-TKD expression constructs were kindly provided by Professor Dr T Fischer.12 The miR-155 promoter luciferase constructs23 were provided by Professor Dr M Mallardo. The STAT5 and STAT5 1*6 expression constructs34 were a kind gift from Professor Dr T Kitamura.
For the PU.1–3′-UTR luciferase vector, the 3′-UTR was amplified from human genomic DNA and inserted into the unique XbaI restriction site 3′ to the luciferase gene in the pRL plasmid (Promega, Madison, WI, USA). For PU.1 3′-UTR amplification, the following primers were used: PU.1 3′-UTR XbaI forward, 5′-TCTAGATACGACTTCAGCGGCGAAGTGCTG-3′ and PU.1 3′-UTR XbaI reverse, 5′-TCTAGACGGATTGAGAATAACTTTACTTG-3′. For PU.1 3′-UTR mutagenesis, we used QuickChange II Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA, USA) following the manufacturer’s instructions. We used the following mutagenesis primers: PU.1 3′-UTR-mutant (MUT) forward, 5′-CTGCCATAATAATTAGCCCTCGCCCGGCC-3′ and PU.1 3′-UTR-MUT reverse, 5′-GGCCGGGCGAGGGCTAATTATTATGGCAG-3′. For the miR-155-expression vector (pcDNA6.2-miR-155), the miR-155 sequence was cloned into the pcDNA6.2-GW/EmGFP-miR plasmid using the BLOCK-iT Pol II miR RNAi Expression Vector Kit (Invitrogen). The following oligonucleotide sequences were used: miR-155-top, 5′-TGCTGTTAATGCTAATCGTGAT AGGGGTTTTTGCCTCCAACTGACTCCTACATATTAGCATTAA-3′ and miR-155-bottom, 5′-CCTGTTAATGCTAATATGTAGGAGTCAGTTGGAGGCAAAAACCCCTATCACGATTAGCAT TAA C-3′. The correct assembly of the vectors was verified by sequencing. pmiRZip constructs for miRNA knockdown were obtained from System Biosciences (Mountain View, CA, USA).
Transfections
Locked nucleic acids (LNAs) for miRNA knockdown were obtained from Exiqon (Vedbaek, Denmark). siRNAs for STAT5 and p65 knockdown were purchased from Qiagen (Hilden, Germany). Suspension cells were transfected with Nucleofector Kits (Lonza, Basel, Switzerland) according the manufacturer’s protocols for each cell line. Transfection efficiency, measured by FACS, was approximately 40–60% for MV4;11 and 50–60% for U937 cells. The 293 T cells were transfected using Lipofectamine LTX (Invitrogen) following the manufacturer’s instructions.
Lentiviral infections
Lentiviral infections were carried out using the pPACKH1 Lentiviral Packaging Kit (System Biosciences) following the manufacturer’s instructions.
Luciferase reporter assay
To test whether miR-155 directly targets PU.1, 293 T cells were transiently transfected with 0.7 μg of the PU.1 3′-UTR (pRL-PU.1–3′-UTR-WT or pRL-PU.1–3′-UTR-MUT) reporter construct and 1 μg pcDNA6.2-miR-155 or control plasmid.
For promoter luciferase assays, we co-transfected 293 T cells with 0.7 μg miR-155 promoter construct (pGL3-1783) or pGL3-control and 0.2 μg pcDNA3.1, p65 (pcDNA3.1-p65) or STAT5 (ref. 34) (pMX-STAT5A or pMX-STAT5A 1*6) expression constructs. Firefly luciferase and Renilla luciferase activities were determined 24 h after transfection using the Dual-Luciferase Reporter Assay System (Promega). Values were normalized by using firefly luciferase or Renilla luciferase, respectively.
Immunoblot analyses
For western blot analyses, the following antibodies were used: anti-phospho-STAT5 (Cell Signaling, Danvers, MA, USA), anti-STAT5, anti-PU.1, anti-p65 and anti-GAPDH (Santa Cruz, Dallas, TX, USA). Immunoblot analyses were performed as previously described.15 The immunoreactivity was determined using an enhanced chemiluminescence method (Amersham Biosciences, Glattbrugg, Switzerland) as per the manufacturer’s instructions. The band intensities were quantified using ImageJ software (National Institutes of Health, Bethesda, MD, USA).
Chromatin immunoprecipitation
For chromatin immunoprecipitation (ChIP) analyses, we used a protocol from the epigenome network of excellence: http://www.epigenome-noe.net/researchtools/protocol.php_protid=10.html.
For sonifcation a Branson Sonifier 450D (Branson Ultrasonics, Danbury, CT, USA) was used. For ChIP we used anti-p65 and anti-normal rabbit IgG antibodies (Santa Cruz). For amplification of enriched DNA, the following primers were used: p65 ChIP 1786 forward, 5′-TTCTGGGGATGAAAGGTCAC-3′ and p65 ChIP 1786 reverse, 5′-CCTGCTCAGATCCATGT-3′; p65 ChIP 1380 forward, 5′-TGCTCCCAAGTTCCTTAACC-3′ and p65 ChIP 1380 reverse, 5′-GTGACTGGGGCCTTTTTGTA-3′.
Flow cytometry
Cells were washed once with phosphate-buffered saline and stained for 20 min with the indicated antibodies. Subsequently, the cells were washed in phosphate-buffered saline and analyzed with BD LSR II cytometer using CellQuest software (BD Biosciences, Franklin Lakes, NJ, USA).
Mouse model and sorted mouse bone marrow
STAT5flox/flox and STAT5flox/flox Mx-Cre C57BL/6 mice35 were backcrossed into Balb/C mice for at least 10 generations. Bone marrow was isolated and retrovirally infected with FLT3-ITD construct. The transduced bone marrow was transplanted into lethally irradiated Balb/C mice. For the induction of the Mx1 promoter and subsequent deletion of STAT5, 250 μg high-molecular-weight polyinosinic:polycytidylic acid (InvivoGen) was intraperitoneally injected into mice at days 11, 14, 18 and 21 after bone marrow transplantation. Complete STAT5 knockout was observed at day 22 after bone marrow transplantation. STAT5flox/flox control mice were treated likewise. The animals were caged in a special caging system with autoclaved food and acidified water at the Technical University of Munich in accordance with National and Institutional Guidelines for Animal Care.
For expression analyses, mouse bone marrow sub-populations were isolated from WT C57BL/6 mice. LSK (Lin− Sca+ Kit+), CMP (common myeloid precursors), GMP (granulocytic-macrophage precursors), MEP (megakaryocyte–erythrocyte precursors) and granulocytes were sorted on FACS ARIA (BD Biosciences).
Clonal growth in methylcellulose
To analyze clonal growth after miR-155 knockdown, 32D cells stably expressing FLT3-ITD were transfected with LNA against miR-155 (LNA-155) or scramble control. Four hours after transfection, 5 × 103 cells were seeded per well of a 6-well culture dish. Cells were cultured in 3 ml of Iscove’s modified Dulbecco’s medium supplemented with 1% methylcellulose and 10% FCS. The assays were performed in triplicate, and colonies were photographed and counted on day 12. The results shown are representative of one of at least three independent experiments.
Statistical analyses
We used the Student’s t-test to determine the statistical significance of experimental results. A P-value of 0.05 or less was considered significant. The results were represented as the average ± s.d. from at least three independent experiments.
RESULTS
FLT3-ITD induces miR-155 expression
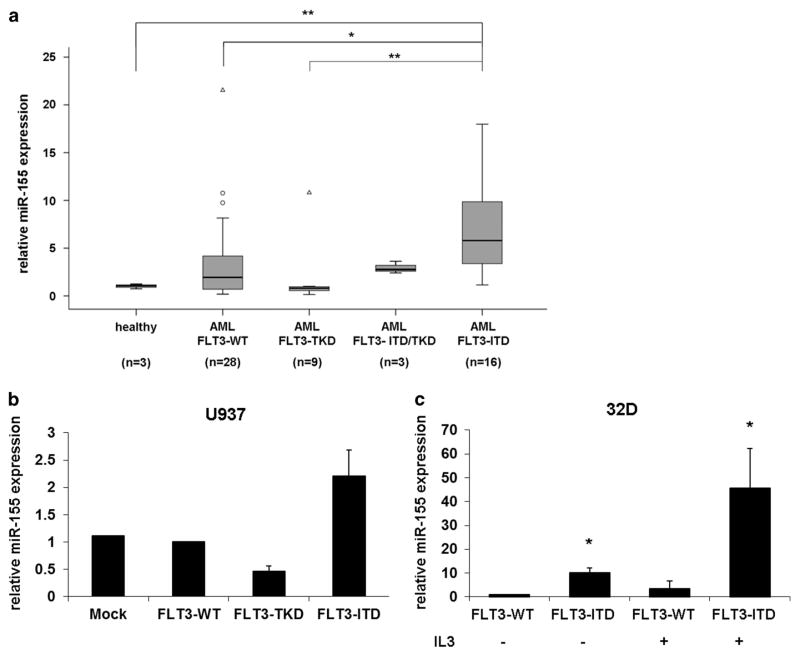
Previous studies have shown a correlation between miR-155 expression and FLT3-ITD mutations in AML patients.19–22 To further investigate the relationship between miR-155 expression and FLT3 mutations, we compared the miR-155 expression in 56 AML patient samples with different FLT3 status (FLT3-WT, FLT3-TKD, FLT3-TKD/ITD and FLT3-ITD) and 3 healthy donors by quantitative RT-PCR. We found that miR-155 was expressed significantly higher in FLT3-ITD-associated patients compared with healthy donors, FLT3-WT and FLT3-TKD patients (Figure 1a). To investigate the regulation of miR-155 by FLT3-ITD in AML, we transiently overexpressed FLT3-WT, FLT3-TKD or FLT3-ITD in U937 cells and measured the miR-155 expression after 24 h. We observed that FLT3-ITD induced the miR-155 expression as compared with FLT3-WT (Figure 1b). On the other hand, we found that the FLT3-TKD-MUT failed to upregulate miR-155 expression (Figure 1b). In addition, we measured miR-155 expression in murine 32D cells, stably expressing either FLT3-WT or FLT3-ITD.12 Our data show that miR-155 expression was approximately 10-fold higher in FLT3-ITD-expressing 32D cells, independent of IL-3 (Figure 1c). These results illustrates that FLT3-ITD signaling induces miR-155 expression.
Figure 1.
FLT3-ITD induces miR-155 expression. (a) MiR-155 expression is significantly higher in FLT3-ITD-associated AML patients. RT-qPCR analyses of miR-155 expression in bone marrow cells of 3 healthy donors and 56 AML patients categorized as FLT3-WT (n = 28), FLT3-TKD (n = 9), FLT3-ITD/TKD (n = 3) and FLT3-ITD (n = 16). (b) Overexpression of FLT3-ITD induces miR-155 expression in U937 cells. U937 cells were transfected with mock, FLT3-WT, FLT3-TKD or FLT3-ITD. After 24 h, the expression of miR-155 was measured by qPCR. (c) qPCR for miR-155 in 32D cells stably expressing FLT3-WT or FLT3-ITD cultured for 24 h with or without IL-3. All values were normalized to U6. (° mild outliers; Δ extreme outliers; *P ≤ 0.05; **P ≤ 0.01).
Block of FLT3-ITD signaling reduces miR-155 expression
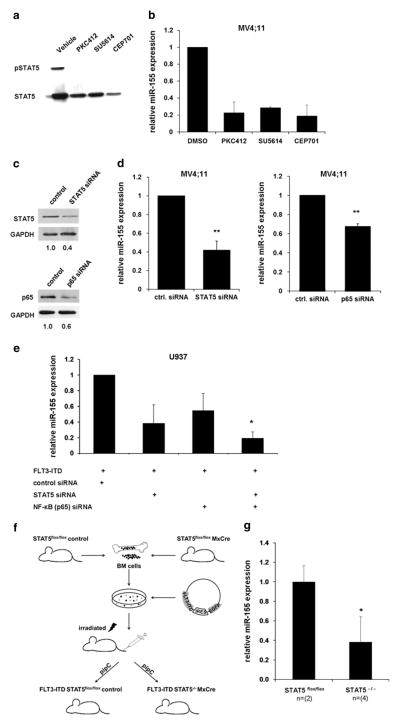
To further investigate that miR-155 expression is dependent upon FLT3-ITD signaling, we treated MV4;11 cells with different protein kinase inhibitors (PKIs). PKC412 (ref. 36), SU5614 (refs 37,38) and CEP701 (ref. 39) have been shown to be effective inhibitors of FLT3-ITD tyrosine kinase activity. MV4;11 cells were cultured for 24 h in medium containing PKC412, CEP701 or SU5614. Control cells were treated with dimethyl sulfoxide. STAT5 is known to be constitutively activated by FLT3-ITD signaling,5,6,12 so we analyzed the phosphorylation of STAT5 as a measure of inhibition of FLT3-ITD tyrosine kinase activity. Our data show that STAT5 phosphorylation disappeared after treatment with PKIs (Figure 2a). To determine miR-155 expression after PKI treatment of MV4;11 cells, we isolated total RNA and performed quantitative RT-PCR. We detected a significant decrease in miR-155 expression (~80% reduction compared with control treated) after treatment of MV4;11 cells with PKIs (Figure 2b). These data confirm that miR-155 is a downstream target of FLT3-ITD. It is well known that FLT3-ITD constitutively activates the STAT5 pathway, whereas FLT3-TKD and FLT3-WT only marginally induce it.4,12,40 Because of this and our previous finding that FLT3-ITD induces miR-155 expression in comparison with FLT3-TKD and FLT3-WT, we hypothesized that STAT5 is involved in miR-155 regulation. To answer this question, MV4;11 cells were transfected with STAT5-specific small interfering RNA (siRNA) and cultured for 24 h. The STAT5 knockdown was analyzed by western blot and showed ~ 60% reduction of the STAT5 protein level (Figure 2c). STAT5 knockdown resulted in a highly significant decrease in miR-155 expression in comparison with control siRNA-transfected cells (Figure 2d).
Figure 2.
Block of FLT3-ITD signaling reduces miR-155 expression. MV4;11 cells were treated with PKIs PKC412 (100 nM), SU5614 (1 μM) and CEP701 (100 nM) for 24 h. (a) Western blot shows decreased STAT5 phosphorylation after FLT3-ITD kinase activity inhibition. (b) Expression of miR-155 is decreased after PKI treatment of MV4;11 cells for 24 h. (c) Knockdown of STAT5 or p65 reduces miR-155 expression. Western blot on lysates from MV4;11 cells transfected for 24 h with STAT5 or p65 siRNA. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as loading control. (d) Expression of miR-155 24 h after transfection of siRNA targeting STAT5 or p65 in MV4;11 cells. (e) Knockdown of STAT5 or p65 inhibits FLT3-ITD-mediated miR-155 induction. U937 cells were co-transfected with FLT3-ITD and siRNAs (control, STAT5, p65 and combination of STAT5 and p65 siRNA). After 24 h, miR-155 expression was analyzed by qPCR. (f) Schematic of workflow to generate an Mx-Cre-inducible STAT5flox/flox knockout mouse model constitutively expressing FLT3-ITD. (g) Analyses of miR-155 expression in bone marrow of FLT3-ITD-inducible Mx-Cre STAT5flox/flox knockout mouse model without and with STAT5 knockout. All values were normalized to U6 (**P ≤ 0.01; *P ≤ 0.05).
Previous publications describe that p65 induces miR-155 expression during the inflammatory response.23,41 It is known that NF-κB pathways are activated by FLT3 (refs 9,42) and in AML blasts.9,43,44 To answer the question if p65 is also involved in miR-155 induction in FLT3-ITD-associated AML, we analyzed miR-155 expression in MV4;11 cells after siRNA-mediated p65 knockdown. We transfected MV4;11 cells with p65 siRNA or control siRNA. Total RNA was isolated after 24 h. We verified reduction in p65 protein level using western blot (Figure 2c). The miR-155 expression correlated with the p65 knockdown and was significantly decreased (Figure 2d). Our findings suggest that miR-155 expression is mediated through p65 and STAT5.
We previously showed that overexpression of FLT3-ITD in U937 cells induces miR-155 expression (Figure 1b). For a better understanding of the regulatory network, we analyzed if a knockdown of p65 and STAT5 is able to overcome FLT3-ITD-dependent miR-155 induction. Therefore, we co-transfected FLT3-ITD and different siRNAs (control siRNA, p65 siRNA, STAT5 siRNA and p65/STAT5 siRNA) into U937 cells. After 24 h, we isolated total RNA and analyzed the miR-155 expression. The results show a decreased induction of miR-155 in the cells co-transfected with siRNA targeting STAT5 or p65. Furthermore, a combination of p65 and STAT5 siRNAs exerted the strongest inhibition of miR-155 expression (Figure 2e). These data indicate that both STAT5 and p65 are crucial for FLT3-ITD to induce miR-155 expression.
To investigate the impact of STAT5 on miR-155 expression in vivo, we analyzed STAT5-deficient mouse bone marrow (Figure 2f). We isolated bone marrow of STAT5flox/flox Mx-Cre and STAT5flox/flox control mice, retrovirally transduced with FLT3-ITD and transplanted into lethally irradiated Balb/C mice. To induce the STAT5 knockout, the mice were treated with poly (I:C). The expression of miR-155 was significantly decreased in the STAT5−/− bone marrow as compared with STAT5flox/flox cells (Figure 2g). In conclusion, our results prove the important role of STAT5 and p65 in FLT3-ITD-induced miR-155 expression in vitro and in vivo.
NF-κB (p65) binds to the miR-155 promoter and constitutively active STAT5 enhances transcriptional induction
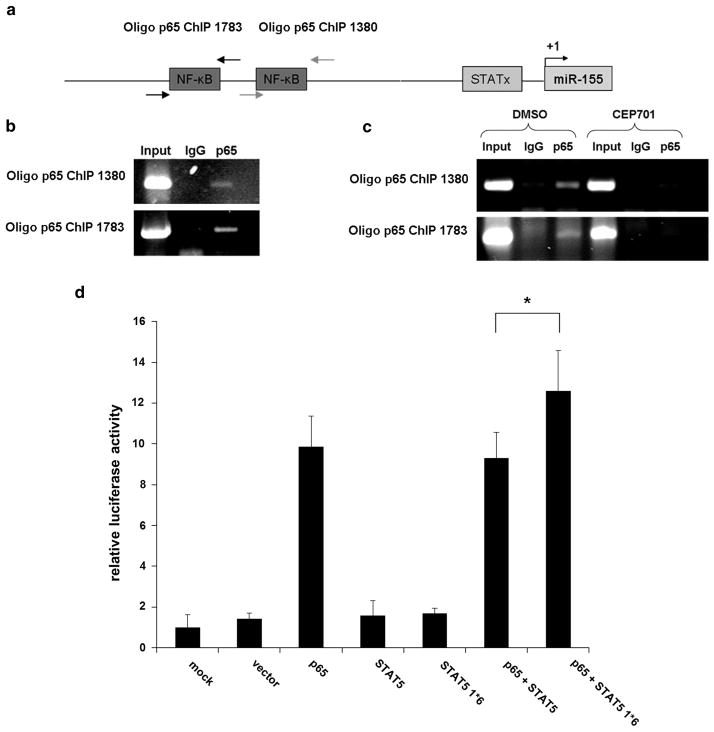
Promoter analyses identified two potential p65 binding sites and one STAT binding site upstream of the miR-155 locus (Figure 3a). Gatto et al.23 have described that p65 regulates the miR-155 in Epstein–Barr virus infection by directly binding to the promoter of the miR-155 host gene BIC. It is known that NF-κB is activated by the PI3K/AKT pathway45,46 which is induced by FLT3-ITD.46,47 Thus, we analyzed p65 binding to the miR-155 promoter in the FLT3-ITD-associated AML cell line MV4;11 by ChIP. Our findings reveal that p65 binds to the miR-155 promoter in FLT3-ITD-associated MV4;11 cells (Figure 3b). We also investigated p65 promoter binding after PKI treatment. We incubated MV4;11 cells for 24 h with CEP701 (100 nM) or dimethyl sulfoxide (control) and analyzed the p65 binding to the miR-155 promoter by ChIP. We could not identify any p65 binding to the miR-155 promoter after treatment with CEP701 (Figure 3c). These data support the idea that miR-155 expression is regulated by p65 in an FLT3-ITD-dependent manner. However, in our ChIP analyses, we could not provide evidence for STAT5 binding to the putative STAT binding site in the miR-155 promoter (data not shown). To further analyze the induction of miR-155 expression by p65 and STAT5, we performed a promoter luciferase activity assay. The 293 T cells were co-transfected with an miR-155 promoter construct (pGL3- 1783) or control (pGL3) along with empty vector, p65, STAT5, constitutively active STAT5 (STAT5 1*6) or mock. After 24 h, luciferase activity was measured. We could show that p65 activated the miR-155 promoter, whereas neither STAT5 nor STAT5 1*6 showed any influence on the miR-155 promoter activity. However, co-transfection of p65 together with constitutively active STAT5 showed an enhanced activation of the miR-155 promoter compared with p65 and STAT5 WT co-transfection (Figure 3d). Taken together, our data indicate that p65 induces the miR-155 expression by direct promoter binding, whereas constitutively active STAT5 increases transcription without direct binding to the core promoter.
Figure 3.
NF-κB (p65) binds to the miR-155 promoter and constitutively active STAT5 enhances transcriptional induction. (a) Scheme showing putative binding sites for NF-κB (p65) and STAT proteins in the miR-155 promoter. (b) Chromatin derived from MV4;11 cells was immunoprecipitated with anti-p65 or immunoglobulin G antibodies. Recovered DNA was PCR amplified with specific primers for p65 binding sites. (c) MV4;11 cells were treated for 24 h with the tyrosin kinase inhibitor CEP701 or control (dimethyl sulfoxide (DMSO)). Chromatin was immunoprecipitated with anti-p65 or immunglobulin G antibodies and DNA was amplified by PCR as described (b). (d) P65 and STAT5 show synergistic effects in miR-155 promoter activation. Luciferase activity assays were performed in 293 T cells. Cells were co-transfected with miR-155 promoter construct (pGL3-1783) or control (pGL3) and empty vector, p65, STAT5 and constitutively active STAT5. After 24 h Renilla luciferase activity was measured. Bars represent the average of three independent experiments ±s.d. All values were normalized to firefly luciferase and calculated to pGL3 (*P ≤ 0.05).
MiR-155 downregulation is necessary for myeloid differentiation
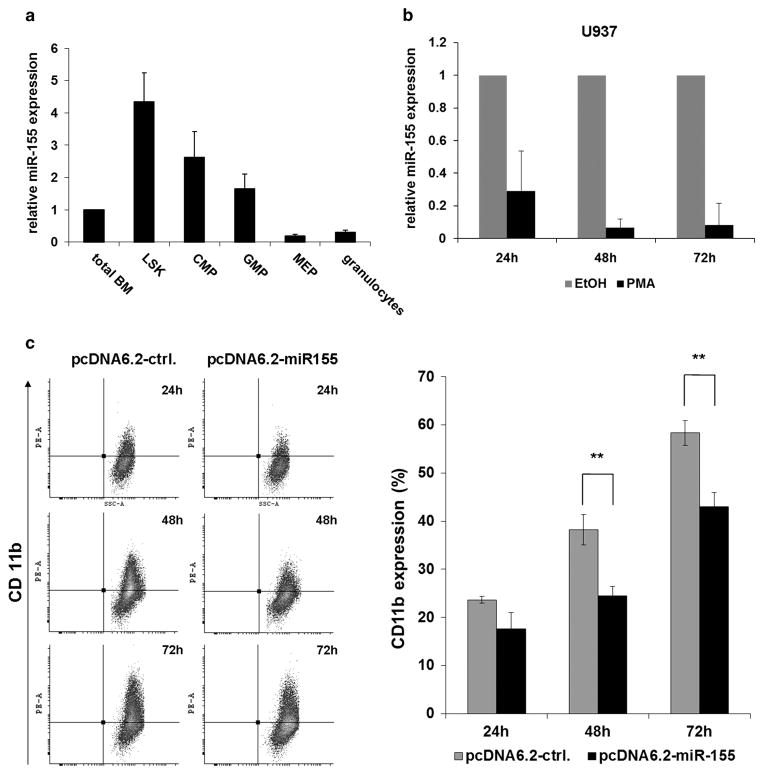
Former publications show that overexpression of miR-155 in hematopoietic stem cells leads to a myeloproliferative disorder32 and blocks the differentiation to myeloid and erythroid cells.48 Thus, we analyzed miR-155 expression during myeloid differentiation. We compared miR-155 expression in sorted sub-populations from mouse bone marrow (LSKs, CMPs, GMPs, MEPs and granulocytes) (Figure 4a). We found that miR-155 was expressed at the highest levels in the LSK population and the expression was decreased upon myeloid differentiation. Further, we analyzed miR-155 expression in U937 cells stimulated with phorbol 12-myristate 13-acetate (PMA) for macrophage differentiation. Cells were collected 24, 48 and 72 h after PMA stimulation. We observed miR-155 expression declined significantly during differentiation (Figure 4b). To further analyze the role of miR-155 in differentiation, we transfected U937 cells with miR-155 (pcDNA6.2-miR-155) or control (pcDNA6.2-ctrl.) construct. Twenty-four hours after transfection, the cells were stimulated with PMA. Transfected cells were analyzed by flow cytometry for CD11b expression 24, 48 and 72 h after PMA stimulation. We observed that overexpression of miR-155 in U937 cells inhibited myeloid differentiation upon PMA treatment (Figure 4c). Cells overexpressing miR-155 showed approximately 25% less CD11b expression. These data indicate that miR-155 downregulation is necessary for myeloid differentiation.
Figure 4.
MiR-155 downregulation is necessary for myeloid differentiation. MiR-155 expression is downregulated during myeloid differentiation. (a) Expression of miR-155 in sorted sub-populations of mouse bone marrow (LSK, Lin− Sca+ Kit+; CMP, common myeloid progenitor; GMP, granulocyte–macrophage progenitor; MEP, megakaryocyte erythroid progenitor and granulocytes). Bone marrow of three C57BL/6 mice were pooled in each set. The bars represent the average ± s.d. of miR-155 expression of three independent sets. Values were normalized to snoRNA-135. (b) miR-155 expression is decreased during PMA-induced myeloid differentiation of U937 cells. U937 cells were treated with 10 nM PMA or control (dimethyl sulfoxide (DMSO)) for 24, 48 and 72 h. (c) Overexpression of miR-155 inhibits PMA-induced myeloid differentiation of U937 cells. Flow cytometric analyses of U937 cells stimulated with 1 nM PMA 24 h after pcDNA6.2-ctrl. or pcDNA6.2-miR-155 transfection. Representative dot blots show the expression of the myeloid differentiation marker CD11b at different duration after PMA induction. Bars represent the percentages of cells expressing CD11b (average ± s.d.) of three independent experiments (**P ≤ 0.01).
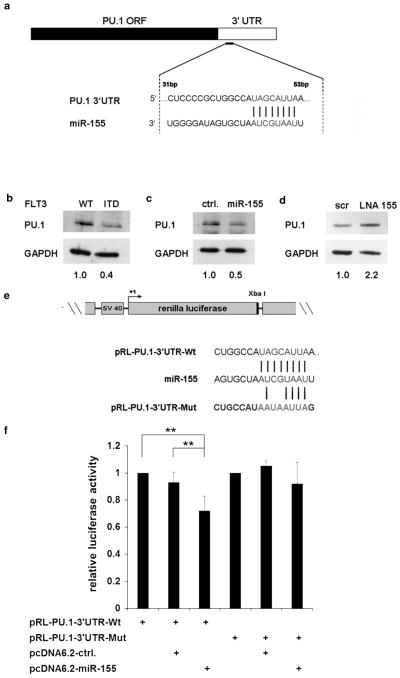
PU.1 is a direct target of miR-155 in FLT3-ITD-associated AML
The transcription factor PU.1 is a key regulator of myeloid differentiation.49 In in silico analyses, we found PU.1 as a putative target of miR-155 (Figure 5a). As it was shown that PU.1 is repressed by FLT3-ITD,5,10 we wanted to examine if FLT3-ITD overexpression leads to decreased PU.1 protein. Therefore, we transiently overexpressed FLT3-WT or FLT3-ITD in U937 cells and analyzed PU.1 by western blot (Figure 5b). We observed that overexpression of FLT3-ITD decreased PU.1 protein level. As we have shown that miR-155 is induced by FLT3-ITD overexpression in U937 cells, we hypothesized that FLT3-ITD targets PU.1 via miR-155. To determine whether PU.1 is posttranscriptionally regulated by miR-155, we overexpressed miR-155 in U937 cells and analyzed PU.1 protein by western blot (Figure 5c). Here, we found that miR-155 overexpression downregulates PU.1 protein level. To investigate the significance of miR-155 in FLT3-ITD-mediated downregulation of PU.1, we blocked miR-155 by specific LNAs in MV4;11 cells and analyzed the PU.1 protein level 24 h after transfection (Figure 5d). The LNA-mediated block of miR-155 increased PU.1 protein. To find out whether PU.1 is a direct target of miR-155, we introduced the WT as well as a mutated 3′-UTR of PU.1 into a luciferase construct (pRL-PU.1–3′-UTR-WT and pRL-PU.1–3′-UTR-MUT) and performed luciferase assays in 293 T cells (Figure 5e). We observed that miR-155 overexpression resulted in a significant reduction in luciferase activity of the pRL-PU.1–3′-UTR-WT in comparison with control, whereas the pRL-PU.1–3′-UTR-MUT showed no change in the luciferase activity (Figure 5f). Our data demonstrate that miR-155 directly targets PU.1 in an FLT3-ITD-dependent manner.
Figure 5.
PU.1 is a direct target of miR-155 in FLT3-ITD associated AML. (a) Schematic overview of miR-155 seed sequence in the PU.1 3′-UTR. (b) Western blot for PU.1 was performed in U937 cells 24 h after transient overexpression of FLT3-WT or FLT3-ITD. (c) Western blot analysis for PU.1, 24 h after overexpression of miR-155 in U937 cells. (d) Knockdown of miR-155 by LNAs in MV4;11 cells increases PU.1 protein level 24 h after transfection. (e) MiR-155 directly binds to the PU.1 3′-UTR. Schematic representation of WT and mutated PU.1 3′-UTR cloned into pRL vector. (f) Luciferase activity assays were performed in 293 T cells. Cells were co-transfected with WT (pRL-PU.1–3′-UTR-WT) or MUT (pRL-PU.1–3′UTR-MUT) PU.1 3′-UTR construct and miR-155 (pcDNA6.2-miR-155) or control (cDNA6.2-ctrl.). After 24 h Renilla luciferase activity was measured and values were normalized to firefly luciferase. Bars represent the average ±s.d. of five independent experiments (**P ≤ 0.01).
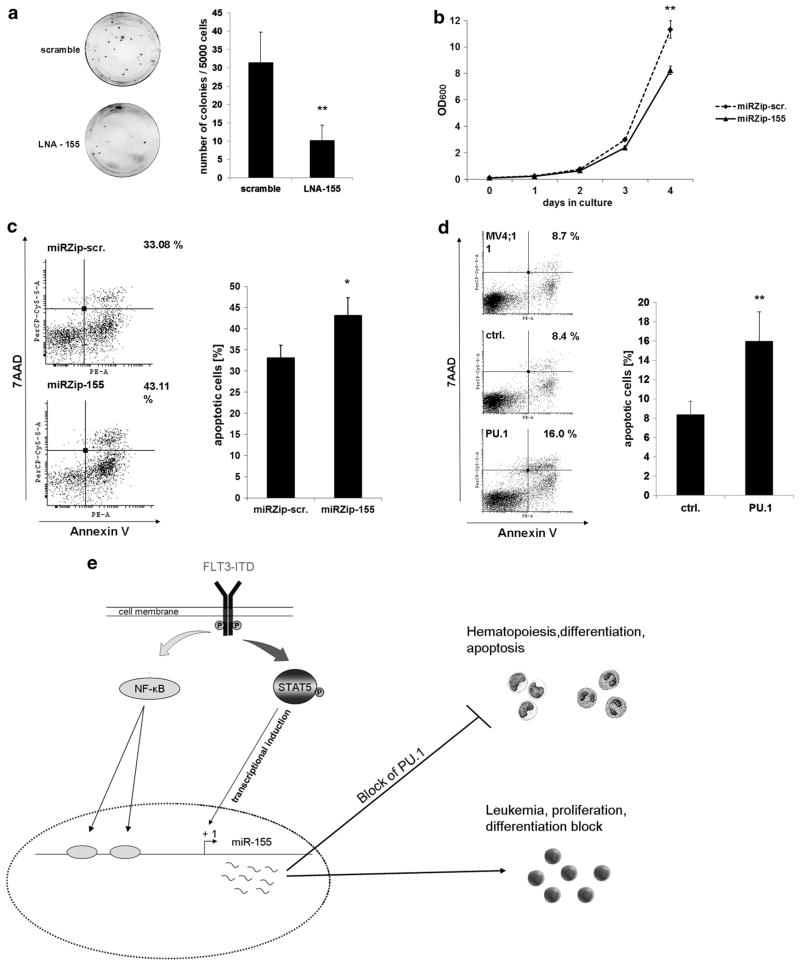
MiR-155 expression is important for FLT3-ITD induced proliferation and survival in AML
To further investigate the effects of miR-155 on proliferation and survival, we examined the colony-forming ability of 32DFLT3-ITD cells transfected with scramble or LNA against miR-155. Four hours after transfection, 5000 cells were seeded per dish in methylcellulose medium to perform colony-forming unit assays. Colonies were enumerated after 12 days. We observed a highly significant reduction in the number of colonies formed by the 32DFLT3-ITD cells transfected with LNA against miR-155 (Figure 6a). Further, we generated a stable miR-155 knockdown in 32DFLT3-ITD cells by lentiviral infection using miRZip-155 or miRZip-scr as control. By measurement of the optical density at 600 nm we could observe, that after 4 days in culture the stable miR-155 knockdown reduced the cell growth of the 32DFLT3-ITD cells to ~ 72% of the control (Figure 6b), which indicates that miR-155 is important for FLT3-ITD induced proliferation. Further, we analyzed if a knockdown of miR-155 can induce apoptosis in MV4;11 cells. Therefore, we transfected MV4;11 cells with a miR-155 knockdown construct (pmiRZip-155) or control (pmiRZip-scr). Forty-eight hours after transfection, we measured Annexin V expression on the transfected cells by flow cytometry. We detected 30% more apoptotic cells after miR-155 knockdown compared with control (Figure 6c). As we showed that a knockdown of miR-155 increases PU.1 protein and apoptosis in FLT3-ITD bearing leukemic cells, we hypothesized that enhanced PU.1 expression might trigger apoptosis of these cells. Thus, we overexpressed PU.1 in MV4;11 cells and analyzed Annexin V expression after 24 h. We found that overexpression of PU.1 resulted in 50% more apoptotic cells compared with control (Figure 6d). Taken together, our data show that miR-155 expression is important for proliferation and survival of FLT3-ITD leukemic cells, whereas overexpression of the miR-155 target PU.1 induces apoptosis.
Figure 6.
MiR-155 expression is important for FLT3-ITD induced proliferation and survival in AML. (a) LNA-mediated block of miR-155 in 32D cells, stably expressing FLT3-ITD, reduces clonal growth. Colony-forming assays were performed in methylcellulose medium to analyze influence of miR-155 knockdown on the clonal growth capacity. The 32D FLT3-ITD cells were transfected with LNAs (200 nM) against miR-155 or scramble control. Four hours after transfection, cells were plated in triplicate at a density of 5000 cells per dish. Colonies were analyzed on day 12. Each bar represents the average ±s.d. of three independent experiments. (b) Knockdown of miR-155 reduces proliferation of FLT3-ITD associated 32D cells. Growth curve of 32DFLT3-ITD cells stably infected with miRZip-155 for miR-155 knockdown or miRZip-scr as control. A total of 105 cells were seeded and cultured for 4 days. Optical density was measured every day by 600 nm to detect proliferation. The data represents the average ±s.d. of three independent experiments (**P ≤ 0.01). (c) MiR-155 knockdown in FLT3-ITD-associated MV4;11 cells enhances apoptosis. Flow cytometric analyses for Annexin V expression in MV4;11 cells 48 h after transfection with miRZip-scr or miRZip-155. The bars represent the average ±s.d. of three independent experiments (*P ≤ 0.05). (d) PU.1 overexpression in FLT3-ITD-associated MV4;11 cells induces apoptosis. Flow cytometric analyses for Annexin V expression in MV4;11 cells 24 h after transfection of control or PU.1. The bars represents the average ±s.d. of three independent experiments (**P ≤ 0.01). (e) A model depicting transcriptional regulation and function of miR-155 in FLT3-ITD-associated AML. FLT3-ITD induces the oncogenic miR-155 by STAT5 and NF-κB. High expression of miR-155 decreases PU.1 protein level, which leads to differentiation block, proliferation and transformation.
DISCUSSION
Our investigations provide new insights into a novel FLT3-ITD/miR-155/PU.1 network in AML. This is the first study to identify that oncogenic miR-155 is an FLT3-ITD downstream target. It has been previously shown that miR-155 expression correlates with FLT3-ITD in AML patients.19–22 Garzon et al.19 could not prove an FLT3-ITD impact on miR-155 expression. However, we demonstrate that expression of FLT3-ITD induces miR-155, whereas inhibition of FLT3-ITD tyrosine kinase activity represses the miR-155 expression. We found an elevated miR-155 expression in FLT3-ITD-associated patient samples, whereas AML patients with FLT3-TKD or FLT3-WT showed no increased miR-155 expression. These data illustrate that miR-155 expression is related to FLT3-ITD signaling.
To our knowledge, no functional studies exist for the miR-155 in FLT3-ITD AML. It was shown that p65 is constitutively active in AML.43,44,50 Further, it has been shown that STAT5 (refs 5,6,12) and NF-κB (p65)42,47 are crucial mediators in FLT3-ITD signaling. Hence, we proposed that the transcription factors p65 and STAT5 are potential regulators of miR-155. Using ChIP assay, we found that p65 directly binds to the miR-155 promoter. Other groups reported that p65 regulates miR-155 via direct promoter binding during Epstein–Barr virus infections.23,24 In luciferase assay analyses, we showed a strong induction of miR-155 promoter activity by p65. Further, we could show that STAT5 is involved in miR-155 induction. STAT5 knockdown in FLT3-ITD model systems reduced miR-155 expression in vitro and in vivo. In silico analyses predicted an STAT binding site in the miR-155 promoter. Kopp et al.51 showed that STAT5 induces the miR-155 expression by direct promoter binding in cutaneous T-cell lymphoma. However, we were not able to detect binding of STAT5 to the predicted binding site in the miR-155 promoter (data not shown). Instead, luciferase assay analyses showed that constitutively active STAT5 (STAT5 1*6) enhanced the miR-155 promoter activity induction by p65, whereas WT STAT5 did not. We also observed that double knockdown of p65 and STAT5 shows the strongest repression of FLT3-ITD-induced miR-155. From our data, we conclude that miR-155 is regulated by p65 via direct promoter binding, which can be enhanced by constitutively activated STAT5. Similar results from other groups claim that activated STAT5 enhances the DNA-binding affinity of p65 on target promoter sequences.34,52,53 Further, we could show that treatment of MV4;11 cells with CEP701, which blocks FLT3-ITD kinase activity and STAT5 phosphorylation, inhibits p65 binding to the miR-155 promoter. In summary, we propose a regulatory network in which FLT3-ITD activates p65 and STAT5. The constitutively activated STAT5 enhances the DNA binding of p65 to the promoter of miR-155 inducing the expression of miR-155.
In sorted bone marrow populations, we could show that miR-155 expression was decreased gradually during myeloid differentiation in mice. Additionally, we found reduced miR-155 expression during PMA-induced myeloid differentiation of U937 cells. In contrast, enforced overexpression of miR-155 blocked the myeloid differentiation of PMA-induced U937 cells. O’Connell et al.32 reported that sustained expression of miRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. Further, it was shown that miR-155 inhibits the generation of myeloid and erythroid colonies by primary human CD34+ cells.48 Therefore, we conclude that miR-155 downregulation is important for normal myeloid differentiation.
In in silico analyses, we found PU.1 as a potential target of miR-155. PU.1 was shown to be targeted by miR-155 in dendritic cells54 and B cells.55 Further, it was already reported that a knockdown of PU.1 leads to leukemic disease in mice56 and blocks myeloid differentiation.57,58 Furthermore, it was shown that PU.1 is downregulated in FLT3-ITD-associated leukemia,4,10 but no mechanism has been previously described. Here we demonstrate that PU.1 is repressed by miR-155 in FLT3-ITD-associated AML. Overexpression of FLT3-ITD or miR-155 reduced PU.1 protein level, while LNA-mediated knockdown of miR-155 increased PU.1 protein. In luciferase assays, we could confirm that miR-155 directly targets the PU.1 3′-UTR.
Additionally, we studied the biologic relevance of miR-155 in FLT3-ITD-associated AML. Knockdown of miR-155 or overexpression of its target PU.1 in FLT3-ITD-bearing leukemic cells induces apoptosis. Babar et al.59 demonstrated that an miR-155 knockdown in B-cell lymphoma increases cell mortality. Further, it has been shown that PU.1 induces apoptosis in myeloma cells by direct transactivation of TRAIL.60 From this, we conclude that miR-155-mediated PU.1 repression is important for survival of AML blasts. In proliferation assays, we showed that LNA-mediated knockdown of miR-155 in stably FLT3-ITD-expressing 32D cells blocks clonal growth. Because it was demonstrated that FLT3-ITD expression induces IL3-independent clonal growth of 32D cells,4,12 we conclude that miR-155 is important for FLT3-ITD to induce proliferation.
Taken together, our data show that miR-155 is expressed in an FLT3-ITD-dependent manner. The main downstream regulatory factors are p65 and STAT5. High expression of miR-155 blocks PU.1 and thereby reduces myeloid differentiation and apoptosis (Figure 6e). Furthermore, miR-155 is important for FLT3-ITD-induced proliferation. Both FLT3-ITD3 and miR-155 (ref. 33) are described as prognostic factors in AML. Targeting miR-155 in mice with lymphoma reduces proliferation, tumor mass and induces apoptosis.59,61 Hence, we propose miR-155 as a novel therapeutic target in FLT3-ITD-associated AMLs.
Acknowledgments
We thank Professor Dr M Mallardo for providing miR-155 promoter luciferase constructs, Professor Dr T Kitamura for STAT5 and STAT5 1*6 expression constructs and Professor Dr T Fischer for providing FLT3-WT, FLT3-ITD and FLT3-TKD expression constructs. This study was supported by grants from DFG (German Research Foundation, BE 2042/7-1), Deutsche José Carreras Leukämie-Stiftung e.V. (DJCLS R 11/17), Deutsche Krebshilfe and Translational Centre for Regenerative Medicine Leipzig (to GB), Deutsche José Carreras Leukämie-Stiftung e.V. (DJCLS F 08/05) (to DG), Deutsche José Carreras Leukämie-Stiftung e.V. (DJCLS F 12/03) (to AAW) and the National Institute of Health (CA118316) (to DGT).
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
References
- 1.Gilliland DG, Griffin JD. The roles of FLT3 in hematopoiesis and leukemia. Blood. 2002;100:1532–1542. doi: 10.1182/blood-2002-02-0492. [DOI] [PubMed] [Google Scholar]
- 2.Zheng R, Small D. Mutant FLT3 signaling contributes to a block in myeloid differentiation. Leuk Lymphoma. 2005;46:1679–1687. doi: 10.1080/10428190500261740. [DOI] [PubMed] [Google Scholar]
- 3.Bienz M, Ludwig M, Leibundgut EO, Mueller BU, Ratschiller D, Solenthaler M, et al. Risk assessment in patients with acute myeloid leukemia and a normal karyotype. Clin Cancer Res. 2005;11:1416–1424. doi: 10.1158/1078-0432.CCR-04-1552. [DOI] [PubMed] [Google Scholar]
- 4.Choudhary C, Schwable J, Brandts C, Tickenbrock L, Sargin B, Kindler T, et al. AML-associated Flt3 kinase domain mutations show signal transduction differences compared with Flt3 ITD mutations. Blood. 2005;106:265–273. doi: 10.1182/blood-2004-07-2942. [DOI] [PubMed] [Google Scholar]
- 5.Choudhary C, Brandts C, Schwable J, Tickenbrock L, Sargin B, Ueker A, et al. Activation mechanisms of STAT5 by oncogenic Flt3-ITD. Blood. 2007;110:370–374. doi: 10.1182/blood-2006-05-024018. [DOI] [PubMed] [Google Scholar]
- 6.Spiekermann K, Bagrintseva K, Schwab R, Schmieja K, Hiddemann W. Overexpression and constitutive activation of FLT3 induces STAT5 activation in primary acute myeloid leukemia blast cells. Clin Cancer Res. 2003;9:2140–2150. [PubMed] [Google Scholar]
- 7.Hayakawa F, Towatari M, Kiyoi H, Tanimoto M, Kitamura T, Saito H, et al. Tandem-duplicated Flt3 constitutively activates STAT5 and MAP kinase and introduces autonomous cell growth in IL-3-dependent cell lines. Oncogene. 2000;19:624–631. doi: 10.1038/sj.onc.1203354. [DOI] [PubMed] [Google Scholar]
- 8.Tse KF, Mukherjee G, Small D. Constitutive activation of FLT3 stimulates multiple intracellular signal transducers and results in transformation. Leukemia. 2000;14:1766–1776. doi: 10.1038/sj.leu.2401905. [DOI] [PubMed] [Google Scholar]
- 9.Chalandon Y, Schwaller J. Targeting mutated protein tyrosine kinases and their signaling pathways in hematologic malignancies. Haematologica. 2005;90:949–968. [PubMed] [Google Scholar]
- 10.Mizuki M, Schwable J, Steur C, Choudhary C, Agrawal S, Sargin B, et al. Suppression of myeloid transcription factors and induction of STAT response genes by AML-specific Flt3 mutations. Blood. 2003;101:3164–3173. doi: 10.1182/blood-2002-06-1677. [DOI] [PubMed] [Google Scholar]
- 11.Radomska HS, Basseres DS, Zheng R, Zhang P, Dayaram T, Yamamoto Y, et al. Block of C/EBP alpha function by phosphorylation in acute myeloid leukemia with FLT3 activating mutations. J Exp Med. 2006;203:371–381. doi: 10.1084/jem.20052242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mizuki M, Fenski R, Halfter H, Matsumura I, Schmidt R, Muller C, et al. Flt3 mutations from patients with acute myeloid leukemia induce transformation of 32D cells mediated by the Ras and STAT5 pathways. Blood. 2000;96:3907–3914. [PubMed] [Google Scholar]
- 13.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 14.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 15.Pulikkan JA, Dengler V, Peramangalam PS, Peer Zada AA, Muller-Tidow C, Bohlander SK, et al. Cell-cycle regulator E2F1 and microRNA-223 comprise an autoregulatory negative feedback loop in acute myeloid leukemia. Blood. 2010;115:1768–1778. doi: 10.1182/blood-2009-08-240101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katzerke C, Madan V, Gerloff D, Brauer-Hartmann D, Hartmann JU, Wurm AA, et al. Transcription factor C/EBPalpha-induced microRNA-30c inactivates Notch1 during granulopoiesis and is downregulated in acute myeloid leukemia. Blood. 2013;122:2433–2442. doi: 10.1182/blood-2012-12-472183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pulikkan JA, Peramangalam PS, Dengler V, Ho PA, Preudhomme C, Meshinchi S, et al. C/EBPalpha regulated microRNA-34a targets E2F3 during granulopoiesis and is down-regulated in AML with CEBPA mutations. Blood. 2010;116:5638–5649. doi: 10.1182/blood-2010-04-281600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schotte D, Pieters R, Den Boer ML. MicroRNAs in acute leukemia: from biological players to clinical contributors. Leukemia. 2012;26:1–12. doi: 10.1038/leu.2011.151. [DOI] [PubMed] [Google Scholar]
- 19.Garzon R, Garofalo M, Martelli MP, Briesewitz R, Wang L, Fernandez-Cymering C, et al. Distinctive microRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin. Proc Natl Acad Sci USA. 2008;105:3945–3950. doi: 10.1073/pnas.0800135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garzon R, Volinia S, Liu CG, Fernandez-Cymering C, Palumbo T, Pichiorri F, et al. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood. 2008;111:3183–3189. doi: 10.1182/blood-2007-07-098749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jongen-Lavrencic M, Sun SM, Dijkstra MK, Valk PJ, Lowenberg B. MicroRNA expression profiling in relation to the genetic heterogeneity of acute myeloid leukemia. Blood. 2008;111:5078–5085. doi: 10.1182/blood-2008-01-133355. [DOI] [PubMed] [Google Scholar]
- 22.Cammarata G, Augugliaro L, Salemi D, Agueli C, La Rosa M, Dagnino L, et al. Differential expression of specific microRNA and their targets in acute myeloid leukemia. Am J Hematol. 2010;85:331–339. doi: 10.1002/ajh.21667. [DOI] [PubMed] [Google Scholar]
- 23.Gatto G, Rossi A, Rossi D, Kroening S, Bonatti S, Mallardo M. Epstein–Barr virus latent membrane protein 1 trans-activates miR-155 transcription through the NF-kappaB pathway. Nucleic Acids Res. 2008;36:6608–6619. doi: 10.1093/nar/gkn666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu F, Weidmer A, Liu CG, Volinia S, Croce CM, Lieberman PM. Epstein–Barr virus-induced miR-155 attenuates NF-kappaB signaling and stabilizes latent virus persistence. J Virol. 2008;82:10436–10443. doi: 10.1128/JVI.00752-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, et al. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci USA. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kluiver J, Poppema S, de Jong D, Blokzijl T, Harms G, Jacobs S, et al. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J Pathol. 2005;207:243–249. doi: 10.1002/path.1825. [DOI] [PubMed] [Google Scholar]
- 28.Wang X, Tang S, Le SY, Lu R, Rader JS, Meyers C, et al. Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. PLoS One. 2008;3:e2557. doi: 10.1371/journal.pone.0002557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee EJ, Gusev Y, Jiang J, Nuovo GJ, Lerner MR, Frankel WL, et al. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer. 2007;120:1046–1054. doi: 10.1002/ijc.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 32.O’Connell RM, Rao DS, Chaudhuri AA, Boldin MP, Taganov KD, Nicoll J, et al. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J Exp Med. 2008;205:585–594. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marcucci G, Maharry KS, Metzeler KH, Volinia S, Wu YZ, Mrozek K, et al. Clinical role of microRNAs in cytogenetically normal acute myeloid leukemia: miR-155 upregulation independently identifies high-risk patients. J Clin Oncol. 2013;31:2086–2093. doi: 10.1200/JCO.2012.45.6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kawashima T, Murata K, Akira S, Tonozuka Y, Minoshima Y, Feng S, et al. STAT5 induces macrophage differentiation of M1 leukemia cells through activation of IL-6 production mediated by NF-kappaB p65. J Immunol. 2001;167:3652–3660. doi: 10.4049/jimmunol.167.7.3652. [DOI] [PubMed] [Google Scholar]
- 35.Cui Y, Riedlinger G, Miyoshi K, Tang W, Li C, Deng CX, et al. Inactivation of Stat5 in mouse mammary epithelium during pregnancy reveals distinct functions in cell proliferation, survival, and differentiation. Mol Cell Biol. 2004;24:8037–8047. doi: 10.1128/MCB.24.18.8037-8047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weisberg E, Boulton C, Kelly LM, Manley P, Fabbro D, Meyer T, et al. Inhibition of mutant FLT3 receptors in leukemia cells by the small molecule tyrosine kinase inhibitor PKC412. Cancer Cell. 2002;1:433–443. doi: 10.1016/s1535-6108(02)00069-7. [DOI] [PubMed] [Google Scholar]
- 37.Yee KW, O’Farrell AM, Smolich BD, Cherrington JM, McMahon G, Wait CL, et al. SU5416 and SU5614 inhibit kinase activity of wild-type and mutant FLT3 receptor tyrosine kinase. Blood. 2002;100:2941–2949. doi: 10.1182/blood-2002-02-0531. [DOI] [PubMed] [Google Scholar]
- 38.Spiekermann K, Dirschinger RJ, Schwab R, Bagrintseva K, Faber F, Buske C, et al. The protein tyrosine kinase inhibitor SU5614 inhibits FLT3 and induces growth arrest and apoptosis in AML-derived cell lines expressing a constitutively activated FLT3. Blood. 2003;101:1494–1504. doi: 10.1182/blood-2002-04-1045. [DOI] [PubMed] [Google Scholar]
- 39.Levis M, Allebach J, Tse KF, Zheng R, Baldwin BR, Smith BD, et al. A FLT3-targeted tyrosine kinase inhibitor is cytotoxic to leukemia cells in vitro and in vivo. Blood. 2002;99:3885–3891. doi: 10.1182/blood.v99.11.3885. [DOI] [PubMed] [Google Scholar]
- 40.Grundler R, Miething C, Thiede C, Peschel C, Duyster J. FLT3-ITD and tyrosine kinase domain mutants induce 2 distinct phenotypes in a murine bone marrow transplantation model. Blood. 2005;105:4792–4799. doi: 10.1182/blood-2004-11-4430. [DOI] [PubMed] [Google Scholar]
- 41.Thompson RC, Herscovitch M, Zhao I, Ford TJ, Gilmore TD. NF-kappaB downregulates expression of the B-lymphoma marker CD10 through a miR-155/PU.1 pathway. J Biol Chem. 2011;286:1675–1682. doi: 10.1074/jbc.M110.177063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takahashi S, Harigae H, Ishii KK, Inomata M, Fujiwara T, Yokoyama H, et al. Overexpression of Flt3 induces NF-kappaB pathway and increases the expression of IL-6. Leuk Res. 2005;29:893–899. doi: 10.1016/j.leukres.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 43.Guzman ML, Neering SJ, Upchurch D, Grimes B, Howard DS, Rizzieri DA, et al. Nuclear factor-kappaB is constitutively activated in primitive human acute myelogenous leukemia cells. Blood. 2001;98:2301–2307. doi: 10.1182/blood.v98.8.2301. [DOI] [PubMed] [Google Scholar]
- 44.Bueso-Ramos CE, Rocha FC, Shishodia S, Medeiros LJ, Kantarjian HM, Vadhan-Raj S, et al. Expression of constitutively active nuclear-kappa B RelA transcription factor in blasts of acute myeloid leukemia. Hum Pathol. 2004;35:246–253. doi: 10.1016/j.humpath.2003.08.020. [DOI] [PubMed] [Google Scholar]
- 45.Grandage VL, Gale RE, Linch DC, Khwaja A. PI3-kinase/Akt is constitutively active in primary acute myeloid leukaemia cells and regulates survival and chemoresistance via NF-kappaB, Mapkinase and p53 pathways. Leukemia. 2005;19:586–594. doi: 10.1038/sj.leu.2403653. [DOI] [PubMed] [Google Scholar]
- 46.Brandts CH, Sargin B, Rode M, Biermann C, Lindtner B, Schwable J, et al. Constitutive activation of Akt by Flt3 internal tandem duplications is necessary for increased survival, proliferation, and myeloid transformation. Cancer Res. 2005;65:9643–9650. doi: 10.1158/0008-5472.CAN-05-0422. [DOI] [PubMed] [Google Scholar]
- 47.Grosjean-Raillard J, Ades L, Boehrer S, Tailler M, Fabre C, Braun T, et al. Flt3 receptor inhibition reduces constitutive NFkappaB activation in high-risk myelodysplastic syndrome and acute myeloid leukemia. Apoptosis. 2008;13:1148–1161. doi: 10.1007/s10495-008-0243-4. [DOI] [PubMed] [Google Scholar]
- 48.Georgantas RW, III, Hildreth R, Morisot S, Alder J, Liu CG, Heimfeld S, et al. CD34+ hematopoietic stem-progenitor cell microRNA expression and function: a circuit diagram of differentiation control. Proc Natl Acad Sci USA. 2007;104:2750–2755. doi: 10.1073/pnas.0610983104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fisher RC, Scott EW. Role of PU.1 in hematopoiesis. Stem Cells. 1998;16:25–37. doi: 10.1002/stem.160025. [DOI] [PubMed] [Google Scholar]
- 50.Baumgartner B, Weber M, Quirling M, Fischer C, Page S, Adam M, et al. Increased IkappaB kinase activity is associated with activated NF-kappaB in acute myeloid blasts. Leukemia. 2002;16:2062–2071. doi: 10.1038/sj.leu.2402641. [DOI] [PubMed] [Google Scholar]
- 51.Kopp KL, Ralfkiaer U, Gjerdrum LM, Helvad R, Pedersen IH, Litman T, et al. STAT5-mediated expression of oncogenic miR-155 in cutaneous T-cell lymphoma. Cell Cycle. 2013;12:1939–1947. doi: 10.4161/cc.24987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nakamura T, Ouchida R, Kodama T, Kawashima T, Makino Y, Yoshikawa N, et al. Cytokine receptor common beta subunit-mediated STAT5 activation confers NF-kappa B activation in murine proB cell line Ba/F3 cells. J Biol Chem. 2002;277:6254–6265. doi: 10.1074/jbc.M109878200. [DOI] [PubMed] [Google Scholar]
- 53.Nagy ZS, LeBaron MJ, Ross JA, Mitra A, Rui H, Kirken RA. STAT5 regulation of BCL10 parallels constitutive NFkappaB activation in lymphoid tumor cells. Mol Cancer. 2009;8:67. doi: 10.1186/1476-4598-8-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martinez-Nunez RT, Louafi F, Friedmann PS, Sanchez-Elsner T. MicroRNA-155 modulates the pathogen binding ability of dendritic cells (DCs) by downregulation of DC-specific intercellular adhesion molecule-3 grabbing non-integrin (DC-SIGN) J Biol Chem. 2009;284:16334–16342. doi: 10.1074/jbc.M109.011601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vigorito E, Perks KL, Abreu-Goodger C, Bunting S, Xiang Z, Kohlhaas S, et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27:847–859. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Metcalf D, Dakic A, Mifsud S, Di Rago L, Wu L, Nutt S. Inactivation of PU.1 in adult mice leads to the development of myeloid leukemia. Proc Natl Acad Sci USA. 2006;103:1486–1491. doi: 10.1073/pnas.0510616103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rosenbauer F, Wagner K, Kutok JL, Iwasaki H, Le Beau MM, Okuno Y, et al. Acute myeloid leukemia induced by graded reduction of a lineage-specific transcription factor, PU.1. Nat Genet. 2004;36:624–630. doi: 10.1038/ng1361. [DOI] [PubMed] [Google Scholar]
- 58.Voso MT, Burn TC, Wulf G, Lim B, Leone G, Tenen DG. Inhibition of hematopoiesis by competitive binding of transcription factor PU.1. Proc Natl Acad Sci USA. 1994;91:7932–7936. doi: 10.1073/pnas.91.17.7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Babar IA, Cheng CJ, Booth CJ, Liang X, Weidhaas JB, Saltzman WM, et al. Nanoparticle-based therapy in an in vivo microRNA-155 (miR-155)-dependent mouse model of lymphoma. Proc Natl Acad Sci USA. 2012;109:E1695–E1704. doi: 10.1073/pnas.1201516109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ueno S, Tatetsu H, Hata H, Iino T, Niiro H, Akashi K, et al. PU.1 induces apoptosis in myeloma cells through direct transactivation of TRAIL. Oncogene. 2009;28:4116–4125. doi: 10.1038/onc.2009.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Y, Roccaro AM, Rombaoa C, Flores L, Obad S, Fernandes SM, et al. LNA-mediated anti-miR-155 silencing in low-grade B-cell lymphomas. Blood. 2012;120:1678–1686. doi: 10.1182/blood-2012-02-410647. [DOI] [PubMed] [Google Scholar]