SUMMARY
The response regulator CpdR couples phosphorylation events in Caulobacter crescentus with the AAA+ protease ClpXP to provide punctuated degradation of crucial substrates involved in cell cycle regulation. CpdR functions like an adaptor to alter substrate choice by ClpXP, however it remains unclear how CpdR influences its multiple targets. Here we show that, unlike canonical ClpXP adaptors, CpdR alone does not strongly bind its substrate. Instead, CpdR binds the N-terminal domain of ClpX and prepares (primes) the unfoldase for substrate engagement. This priming creates a recruitment interface that docks multiple substrates and additional adaptor components. We show that adaptor dependent priming of ClpX avoids concentration-dependent inhibition that limits traditional, scaffolding adaptors. Phosphosignaling disrupts the adaptor-protease interaction and mutations in CpdR that impact ClpX binding tune adaptor activity and biological function. Together, these results reveal how a single adaptor can command global changes in proteome composition through priming of a protease.
Graphical Abstract

INTRODUCTION
AAA+ (ATPases associated with diverse cellular activities)-protease regulation by adaptors plays central roles in a wide range of intracellular pathways (Kirstein et al., 2009; Battesti and Gottesman, 2013). AAA+ proteases are composed of unfoldase and peptidase components. Specific motifs in protein substrates are recognized by the unfoldase, and ATP hydrolysis drives substrate unfolding and translocation into the peptidase chamber where polypeptides are cleaved (Sauer et al., 2004). Adaptors modulate the responsibilities of their cognate AAA+ protease by modifying substrate selectivity of the unfoldase. Outcomes are diverse and important for cell physiology; ranging from discarding translationally-stalled proteins to regulation of protein factors involved in stress response, virulence or competence (Kirstein et al., 2009; Battesti and Gottesman, 2013).
The highly conserved AAA+ protease ClpXP regulates cell cycle-dependent proteolysis in the bacterial model system Caulobacter crescentus (Jenal, 2009). During the cell cycle, a non-replicative, motile swarmer cell differentiates into a replication-competent, stalked cell producing G1 and S stages analogous to the eukaryotic cell cycle stages (Degnen and Newton, 1972). The stalked cell then divides asymmetrically into swarmer and stalked cells that execute specific molecular programs for motility and replication respectively. Levels of many biomolecules such as mRNAs, proteins, and second messengers (e.g. cyclic di-GMP) are cell-cycle regulated (Kirkpatrick and Viollier, 2012). Interestingly, neither ClpX (the unfoldase component) nor ClpP (the peptidase component) levels change during cell cycle (Jenal and Fuchs, 1998), but numerous proteins are degraded by ClpXP in a cyclic manner dependent on the response regulator protein, CpdR (Biondi et al., 2006; Iniesta et al., 2006; Radhakrishnan et al., 2010; Abel et al., 2011; Bhat et al., 2013). CpdR activity is restrained through an intricate phosphorylation pathway during the swarmer cell stage (Biondi et al., 2006; Iniesta et al., 2006). Dephosphorylation of CpdR during the swarmer to stalk transition activates ClpXP protease activity (Iniesta et al., 2006; Chen et al., 2009).
CpdR is needed for the subcellular localization of ClpXP and this function was thought to promote degradation of similarly localized substrates such as the essential replication regulator CtrA (Iniesta et al., 2006; McGrath et al., 2006). In support of this, components needed for CtrA localization are also important for its degradation in vivo (McGrath et al., 2006; Duerig et al., 2009). However, subsequent biochemical work suggested that CpdR could work as an adaptor outside the internal organization of the bacterium to drive substrate degradation by ClpXP. In vitro reconstitution experiments with highly purified proteins showed that CpdR is necessary and sufficient to stimulate degradation of a cyclic-di-GMP phosphodiesterase PdeA by ClpXP (Abel et al., 2011; Rood et al., 2012). The cellular components needed for substrate localization can assemble as a multi-protein adaptor complex that enhances CtrA degradation in a CpdR-dependent manner in vitro (Smith et al., 2014). In both of these studies, it was shown that phosphorylation of CpdR inactivated delivery to ClpXP, mirroring what was observed in vivo. Taken together, these reconstituted systems clearly indicate that a localization mechanism alone does not drive cell cycle-dependent activity of ClpXP.
This present work focuses on the mechanism by which CpdR functions as an adaptor. ClpXP adaptors often work as scaffolds, such as in the case of SspB which binds to both the ssrA peptide and to the ClpX unfoldase, effectively tethering the substrate to the protease to enhance ClpXP degradation of translationally-stalled ssrA-tagged proteins (Levchenko et al., 2000; Flynn et al., 2001; Dougan et al., 2003). However, other adaptors do not appear to interact with their partners in a similar manner. For example, the RssB adaptor induces degradation of the RpoS stationary sigma factor by binding RpoS and promoting ClpX recognition, but RssB alone appears to bind poorly to ClpX (Zhou et al., 2001; Stüdemann et al., 2003; Hengge, 2009). Similarly, the FliT protein binds to the FlhC protein to accelerate FlhC degradation by ClpXP, but FliT alone does not bind ClpX (Sato et al., 2014). Therefore, adaptors are likely to operate as more than simple scaffolds, where binding by adaptors somehow prepares either the target or protease for successful degradation.
Here, we present a detailed characterization of a simple delivery system comprising the substrate PdeA, adaptor CpdR, and protease ClpXP (Figure 1A). In contrast with known adaptors of ClpXP that form stable complexes with cargo, CpdR does not interact with PdeA directly. Instead, CpdR interacts with ClpX to prepare the protease for engagement of PdeA, with phosphosignaling being critical for gating the interaction between CpdR and ClpX. Similar to the need for preparation of a surface to ensure binding of a second layer (e.g., priming a canvas before painting), we refer to this adaptor mechanism as priming. Using artificial tethers, we distinguish this priming mechanism from a simple scaffolding mechanism and show that the priming mechanism is insensitive to inhibitory effects due to excess adaptors as seen with scaffolding adaptors like SspB. Mutations in CpdR that modulate its binding to ClpX and its activity in vivo indicate that this mechanism extends beyond the substrate PdeA. In vitro experiments show that CpdR priming of ClpX applies to the degradation of McpA and the multi-adaptor system that regulates CtrA degradation by ClpXP. Together, these results reveal an unexpected mode of regulated protein degradation where an adaptor directly primes a protease to expand substrate specificity.
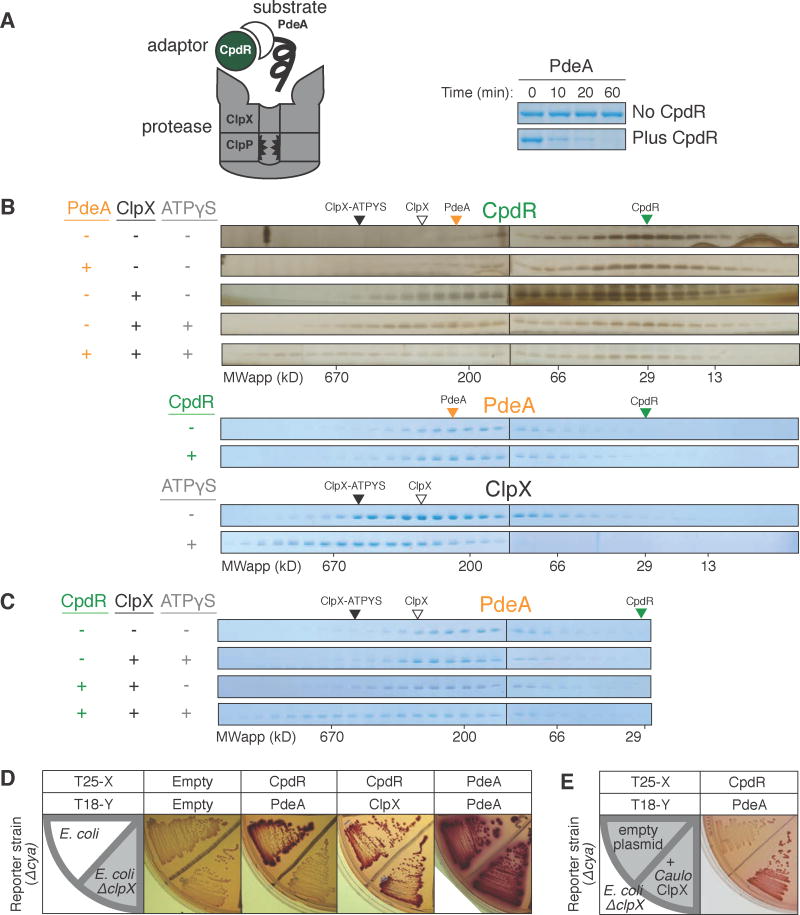
Figure 1. Interaction between adaptor CpdR and unfoldase ClpX enables recruitment of substrate PdeA.
(A) Degradation of PdeA by ClpXP when mediated by CpdR in the presence of an ATP regeneration system and 1mM GTP (which enhances PdeA degradation (Abel et al., 2011)). (B) Size exclusion chromatography (SEC) profiles of CpdR (±PdeA/ClpX/ATPγS). Profiles for PdeA (±CpdR) and ClpX (±ATPγS). Robust detection of CpdR required silver staining as it stained poorly by Coomassie (Figure S1). (C) SEC profiles of PdeA (±CpdR/ClpX/ATPγS). Colored triangles mark peak locations of individual proteins. (D) BACTH assay using McConkey agar results in red colonies when interacting proteins are fused to complementary fragments of adenylate cyclase (cya), T18 and T25 (Karimova et al., 1998). Fusions of CpdR, PdeA and ClpX (C. crescentus ortholog) examined in E. coli wild-type and ΔclpX (shaded) reporter strains. (E) Interaction between CpdR and PdeA examined in the ΔclpX reporter strain harboring a plasmid expressing C. crescentus ClpX (pCL1920-ClpX) or empty plasmid (pCL1920). For (B&C), 20μM CpdR/5μM PdeA/5μM ClpX6/5mM(sample mixture) or 1mM (in running buffer) ATPγS were used, with 1mM GTP throughout. See also Figure S1.
RESULTS
The adaptor CpdR interacts principally with ClpX to assemble a delivery complex
We originally set out to measure the adaptor/cargo affinities for CpdR and PdeA, given that other ClpX adaptors such as SspB directly bind their substrate prior to delivery (Levchenko et al., 2000; Kirstein et al., 2009) (Figure S1A). CpdR and PdeA did not form a stable complex as shown by size-exclusion chromatography (Figure 1B) in conditions where other adaptor-substrate complexes were readily detected (Figure S1A), suggesting that CpdR and PdeA do not strongly interact. This was surprising because prior work using a bacterial adenylate cyclase two-hybrid (BACTH) approach suggested that CpdR could directly bind PdeA and ClpX (Duerig et al., 2009; Abel et al., 2011). Consistent with the BACTH results, purified CpdR could bind ClpX in solution to form a stable complex in vitro (Figure 1B). Because two-hybrid approaches simply report on the proximity of the bait/prey proteins, we speculated that an endogenous factor in the Escherichia coli-based two-hybrid assay generated an apparent positive result between CpdR and PdeA. A natural candidate was ClpX because the E. coli ClpXP ortholog degrades PdeA in the presence of CpdR (Figure S1D). Consistent with this hypothesis, deletion of the E. coli clpX gene eliminated the apparent BACTH interaction between PdeA and CpdR without affecting either CpdR/ClpX or PdeA/PdeA interactions (Figure 1D), and introduction of a plasmid expressing the C. crescentus clpX in this ΔclpX background restores the interaction (Figure 1E & S1F).
Our BACTH results (Figure 1D) suggest that CpdR, ClpX and PdeA together form a higher order complex in vivo. This was verified in vitro as PdeA co-migrated with the CpdR-ClpX complex in the presence of ATPγS, a slowly hydrolyzed ATP analog, to generate a ternary complex of apparent molecular weight > 700 kDa (Figure 1C). ATPγS promotes formation of stable ClpX hexamers (Grimaud et al., 1998) without supporting PdeA unfolding for degradation (Figure S1C). In the absence of ATPγS, PdeA failed to incorporate into a ternary complex, while CpdR still bound ClpX under these conditions (Figures 1B,C). In the absence of CpdR, PdeA failed to interact with ClpX even when ATPγS was present. Taken together, these data suggest that CpdR, ClpX and PdeA are all required for the assembly of an adaptor-dependent substrate-engaged complex. It is also clear that CpdR alone does not bind strongly to its substrate, with interaction strengths substantially less than that of SspB/ssrA, which is easily detected in our assays (Figure S1A). This raises the possibility that rather than acting as a simple scaffold that stably binds both partners, CpdR binds and primes ClpX to prepare for subsequent substrate engagement.
Adaptor-dependent delivery requires the N-terminal domain of ClpX
The N-terminal domain of AAA+ proteases can act as an anchor for adaptors (Dougan et al., 2002, 2003; Kirstein et al., 2006). For example, the adaptor SspB binds the ssrA peptide and the N-terminal domain of ClpX (NTDClpX) to provide a tether that enhances ssrA substrate degradation (Wah et al., 2002, 2003; Dougan et al., 2003; Levchenko et al., 2003). To determine the importance of the NTDClpX with respect to CpdR function, we used a variant of ClpX lacking this domain (ΔN-ClpX). Unlike full-length ClpX, ΔN-ClpX failed to degrade PdeA even in the presence of CpdR (Figure 2B). ΔN-ClpX is functional since it degrades GFP-ssrA and as expected does not support SspB-enhanced degradation (Figure 2B) (Dougan et al., 2003). ΔN-ClpX also failed to bind CpdR in conditions where full-length ClpX forms a complex with CpdR (Figure 2C). Addition of excess NTDClpX to ClpXP competitively inhibited CpdR-mediated degradation of GFP-tagged PdeA, and as expected also inhibited SspB-mediated GFP-ssrA degradation (Figure 2D). Addition of NTDClpX did not significantly affect GFP-ssrA degradation in the absence of SspB, showing that there is not a general inhibition of ClpXP activity (Figure 2D). Consistent with a common need for NTDClpX, SspB and CpdR competitively inhibit each other (Figure S2E,F). Together, these data indicate that NTDClpX is necessary for the protease adaptor CpdR to bind the unfoldase ClpX and mediate degradation of PdeA by ClpXP.
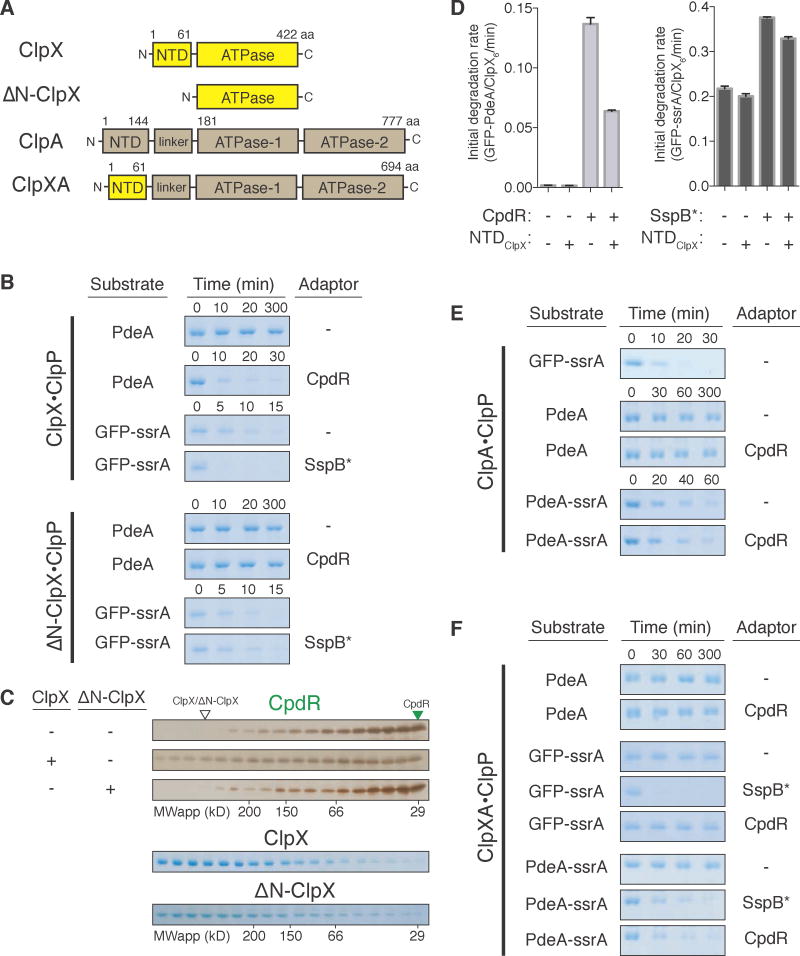
Figure 2. ClpX N-terminal domain is necessary and sufficient for CpdR adaptor function.
(A) Schematic of ClpX, ΔN-ClpX, ClpA, and chimeric ClpXA unfoldases. N-terminal domains (NTD) and ATPase domains are segmented; ClpX segments in yellow; ClpA segments in brown. (B) In vitro degradation of PdeA when mediated by CpdR, comparing ClpXP to ΔN-ClpXP. Degradation of GFP-ssrA by SspB* was similarly assayed. (C) SEC profiles of CpdR (±ClpX/ΔN-ClpX). Profiles of ClpX and ΔN-ClpX alone are also shown. Profiles were determined by silver staining (brown gels) or Coomassie staining (blue gels). (D) NTDClpX fragment addition to the in vitro degradation reactions of substrates GFP-PdeA and GFP-ssrA by ClpXP, ± respective adaptors CpdR/SspB*. Data are represented as mean ± SD, n = 3. (E) Degradation of GFP-ssrA, PdeA, and PdeA-ssrA (±CpdR) by ClpAP. (F) Degradation of PdeA, GFP-ssrA and PdeA-ssrA by the chimeric ClpXAP±CpdR/SspB*. nb. SspB* is the non-degradable variant of SspB (Figure S2A). See also Figure S2.
We further observed in BACTH studies that expressing the NTDClpX, similar to expressing full-length ClpX (Figure 1E & S1F), is sufficient to restore a positive bait/prey signal with CpdR and PdeA in a ΔclpX background (Figure S2G). Therefore, we asked if the NTDClpX could confer CpdR-dependent delivery of PdeA in the alternate AAA+ protease ClpAP, which does not normally recognize PdeA even in the presence of CpdR (Figure 2E). We generated a chimeric unfoldase (ClpXA) by transplanting NTDClpX onto ClpA (Figure 2A) and found that PdeA was not degraded by ClpXAP (ClpXA + ClpP) even in the presence of CpdR (Figure 2F). However, ClpXAP also failed to degrade GFP-ssrA (Figure 2F), a known substrate of both ClpXP (Figure 2B) and ClpAP (Figure 2E), reminiscent of prior work showing that modification of the N-terminal regions of ClpA can either enhance or repress delivery of GFP-ssrA for reasons not completely understood (Lo et al., 2001; Cranz-Mileva et al., 2008). Serendipitously, addition of SspB promotes GFP-ssrA degradation by ClpXAP (Figure 2F), suggesting that the NTDClpX in the ClpXA chimera can function with its cognate adaptors. We speculated that the PdeA C-terminus may be poorly recognized by the ClpA unfoldase of ClpXA chimera and that addition of a known ClpA degron could bypass this defect. In support of this reasoning, PdeA appended with ssrA (PdeA-ssrA) was degraded by ClpXAP in the presence of CpdR (Figure 2F). SspB also facilitated degradation of PdeA-ssrA (Figure 2F). However, CpdR did not improve degradation of GFP-ssrA by ClpXAP (Figure 2F), nor did CpdR stimulate PdeA-ssrA degradation by wildtype ClpAP (Figure 2E). These data suggest that CpdR enhances specific degradation, rather than increasing global protease activity upon binding NTDClpX in the ClpXA chimera. Overall, the results show that the NTDClpX is necessary and sufficient for CpdR-dependent degradation of PdeA given suitable substrate engagement by the associated unfoldase.
CpdR primes ClpX by binding the N-terminal domain to create a recruitment interface
The scaffolding adaptor SspB adaptor uses the NTDClpX as a simple tethering site (Dougan et al., 2003; Bolon et al., 2004; Park et al., 2007; Davis et al., 2009). As shown schematically in Figure 3 (box), if CpdR-dependent degradation also uses the NTDClpX in a similar manner, it should be possible to bypass the need for the NTDClpX by artificially tethering CpdR to ΔN-ClpX. However, if the NTDClpX serves as more than a passive anchor, simply tethering CpdR would not restore PdeA degradation. We adopted a previously reported tethering system where human FKBP12 protein is fused to E. coli ΔN-ClpX, and FKBP12-rapamycin-binding (FRB) domain of the rat mTOR protein is fused to E. coli SspB (Davis et al., 2009). Addition of rapamycin induces dimerization of FKBP12 and FRB, tethering SspB to the ΔN-ClpX fusion so adaptor function no longer requires NTDClpX (Davis et al., 2009).
Figure 3. CpdR binding to the ClpX N-terminal domain forms a recruitment interface.
Postulated models (box) for NTDClpX as a simple anchoring site for CpdR (left) or a unique partner in forming a composite recruitment interface for binding PdeA (right). (A) Cartoons illustrate the use of a rapamycin(rap)-induced FRB/FKBP dimer to tether CpdR or SspB (residues 10–125 that binds ssrA motif) directly to ΔN-ClpX. GFP-ssrA-SS (requires SspB for delivery) degradation by FKBP-ΔN-ClpXP when mediated by SspB-FRB. PdeA levels in the presence of FRB-CpdR/FKBP-ΔN-ClpXP/rap (detected by Western using α-PdeA due to overlapping bands). (B) PdeA levels when directly tethered to ΔN-ClpX via the FRB/FKBP dimer system (detected by Coomassie staining). FRB-PdeA C-terminus Arg-Gly is mutated to Asp-Asp to create FRB-PdeADD. (C) Impact of NTDClpX on PdeA levels in the presence of FRB-CpdR/FKBP-ΔN-ClpXP/rap (detected by α-PdeA Western blot). (D) Effect of NTDClpX fragment concentration on the delivery of GFP-PdeA to FRB-CpdR/FKBP-ΔN-ClpXP (rapamycin added). Apparent Kactivation is shown, see Methods. (E) Effect of phosphorylation on the NTDClpX-dependent delivery of GFP-PdeA to FRB-CpdR/FKBP-ΔN-ClpXP (rapamycin added). FRB-CpdR phosphorylated using its cognate phosphorelay CckA/ChpT (Biondi et al., 2006; Chen et al., 2009). (D&E) are represented as mean ± SD, n = 3 experiments. See also Figure S3.
We generated Caulobacter orthologs of the FRB-FKBP tethering system and validated that SspB-FRB could deliver GFP-ssrA-SS (a modified ssrA-tagged substrate that requires SspB for delivery) to FKBP-ΔN-ClpX upon addition of rapamycin (Figure 3A). We then examined the characteristics of the FRB-CpdR fusion. In contrast to SspB, tethering CpdR to ΔN-ClpX was insufficient to enhance PdeA degradation (Figure 3A) even though FRB-CpdR could deliver PdeA to full-length wildtype ClpXP and could bind FKBP-ΔN-ClpX (Figure S3A). Interestingly, FKBP-ΔN-ClpX could degrade PdeA when PdeA was directly tethered (using FRB-PdeA) to this protease by the FRB-FKBP dimer (Figure 3B). Recognition was not due to introduction of a new degron by the fusion proteins, as degradation required the same C-terminal degron needed for wildtype PdeA degradation (Rood, et al. 2012) (Figure 3B).
If our priming model is correct, then the NTDClpX acts as more than a passive docking site and binding of CpdR to NTDClpX is explicitly needed for PdeA recognition. Consistent with this hypothesis, addition of NTDClpX to reactions where FRB-CpdR is tethered to FKBP-ΔN-ClpX restored degradation of PdeA (Figure 3C). Increasing concentrations of NTDClpX increased substrate delivery of a GFP-PdeA reporter with an apparent Kactivation of 8.2 ± 0.6 μM (Figure 3D). The effect of NTDClpX addition is not due to a general increase in protease activity as addition of NTDClpX had no effect on GFP-ssrA-SS degradation (Figure S3B). Phosphorylation of CpdR is known to inactivate its adaptor function (Iniesta, et al. 2006; Biondi, et al., 2006; Abel, et al., 2011), and importantly, phosphorylation also inactivated FRB-CpdR, even in the presence of excess NTDClpX (Figure 3E). These results clearly show that CpdR does not use the NTDClpX as a simple tethering site. Rather, CpdR binding to NTDClpX is specifically necessary to prepare ClpX to engage PdeA.
Phosphosignaling modulates ClpX binding through the CpdR output face
ClpXP-mediated protein degradation during the Caulobacter cell cycle depends on CpdR phosphorylation (Iniesta et al., 2006). Multiple upstream components control the hybrid histidine kinase CckA that transfers phosphate via the phosphotransferase ChpT onto Asp51 of CpdR (Biondi et al., 2006; Iniesta et al., 2006). Phosphorylation of CpdR inhibits degradation of PdeA (Abel et al., 2011) (Figure 4A), but how phosphorylation of CpdR modulates substrate recruitment was unclear. We find that phosphorylation of CpdR prevented its binding to ClpX (Figure 4B). This disruption required both ATP and Asp51 of CpdR, verifying that active phosphorylation of CpdR is needed to prevent ClpX binding (Figure 4B). Importantly, CpdR phosphorylation also prevents formation of the higher molecular weight PdeA/CpdR/ClpX complex (Figure 4C). This data suggests that loss of ClpX priming hinges upon the phosphorylation status of CpdR.
Figure 4. Phosphorylation modulates CpdR function through its response regulator output face.
(A) Effect of phosphorylation by the cognate CckA/ChpT phosphorelay on wildtype CpdR (WT) or the non-phosphorylatable variant CpdRD51A in mediating PdeA delivery to ClpXP. CpdR WT and CpdRD51A proteins were preincubated with the phosphorelay. PdeA detected by Western using α-PdeA due to overlapping bands. (B) SEC profiles of CpdR WT/CpdRD51A (±phosphorelay/ATP/ClpX). (C) SEC profiles of PdeA (detected by Western using α-PdeA) in the presence of CpdR WT/D51A (preincubated with phosphorelay/ATP) and ClpX/ATPγS. (D) Phyre homology model (RcsC receiver domain as template) for CpdR. Predicted α4-β5-α5 signaling output face (yellow), with conserved residues amongst α-proteobacteria CpdR orthologs (orange). (E) Alanine variants of CpdR H104 and R106 of CpdR were examined for their ability to mediate ClpXP degradation of GFP-PdeA. (F) CpdR variants characterized by fitting the initial rates of degradation of GFP-PdeA as a function of increasing adaptor concentration (Kactivation and Vmax reported; see Methods), data plotted as mean ± SD, n = 4. (G) SEC profiles of the CpdR variants (±ClpX). (H) Two-hybrid interactions between CpdR variants and ClpX/PdeA using E. coli Δcya reporter strains. Representative images on McConkey agar, and beta-galactosidase (β-gal) activity from 4 independent colonies (raw values, mean ± SD) are shown. See also Figure S4.
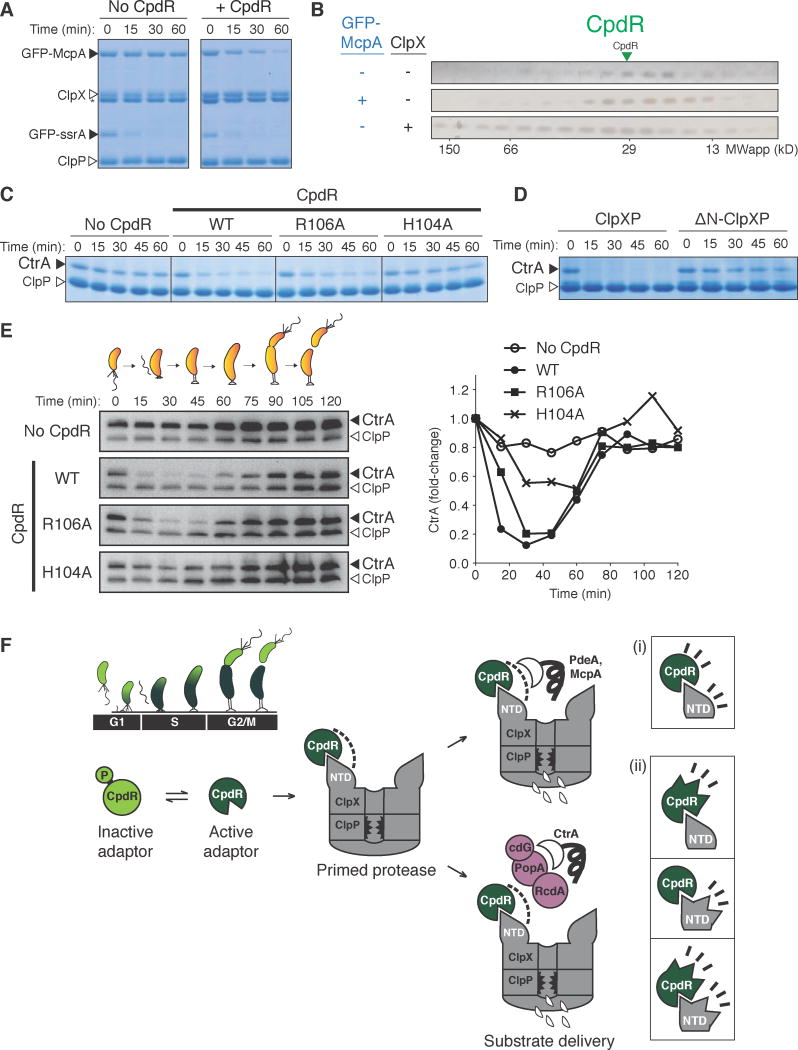
Phosphorylation often affects the function of response regulator receiver domains through conformational changes that are allosterically communicated to their conserved ‘signaling output face’ comprised of the α4-β5-α5 surface (Gao and Stock, 2010). CpdR homology modeling using Phyre (Kelley and Sternberg, 2009) revealed a topology similar to single-domain response regulators (Figure 4D) (Bourret, 2010). We mutated several conserved surface residues within the signaling output face (H104, R106, D107, E111, and K114) to alanine and identified two variants that were defective in enhancing PdeA degradation (Figures 4D,E). Both variants were properly folded based on proper recognition by the CckA/ChpT phosphorelay (Figure S4A). Although both variants were defective in mediating PdeA degradation, the R106A variant achieved wildtype activity at saturating adaptor concentrations while the H104A variant did not (Figure 4F). These mutations also inhibited the interaction between CpdR and ClpX in vitro (Figure 4G) reducing assembly of a CpdR/ClpX/PdeA complex as assessed by BACTH (Figure 4H) in the same ranked order as the loss in PdeA degradation (Figures 4E,F). Similar changes in activity were seen with the chimeric ClpXAP, supporting the important role of the NTDClpX in CpdR activity (Figure S4B). Whether CpdR residues H104 and R106 are involved in conformational changes that affect binding to ClpX, or if they directly constitute part of the interface with ClpX is unknown at this time.
Modifying the CpdR output face impacts its biological activity
The CpdR signaling output face variants (Figures 4D–H) provide the opportunity to tune ClpX binding in vivo and determine the biological consequences of altering the interaction between CpdR and ClpX. CpdR activity must be properly controlled in vivo. For example, both absence of CpdR and overproduction of a constitutively active, unphosphorylatable CpdRD51A variant both yield colonies with reduced sizes on soft agar plates (a combined outcome of changes in motility, chemotaxis, and growth) (Figures 5A,B) (Skerker et al., 2005; Iniesta et al., 2006). Strains expressing WT, R106A or H104A variants as their sole copies of CpdR produced colony sizes that followed the same rank order as their ability to deliver PdeA in vitro (Figure 5A). Similarly, overexpressing CpdRD51A variants carrying these point mutations reduced colony sizes in the same ranked order (Figure 5B). We also tested these CpdRD51A variants in ΔpdeA strains, which form smaller colonies than wildtype C. crescentus (Rood et al., 2012), and again observed effects in colony sizes with respect to CpdR variants (Figure 5C). Because CpdR influences cell-cycle dependent degradation of proteins beyond PdeA (Iniesta et al., 2006; Radhakrishnan et al., 2010; Bhat et al., 2013), these results suggest that additional ClpXP substrates rely on the same CpdR priming of ClpX as PdeA does.
Figure 5. CpdR signaling output face is important for its biological activity.
Effects of CpdR output face variants (H104A and R106A) examined by soft agar colony assay. CpdR variants expressed using xylose-inducible plasmids in C. crescentus (A) ΔcpdR, (B) wildtype, and (C) ΔpdeA strains were examined by colony stabbings on soft agar (PYE 0.3% agar, respective antibiotics, and 0.2% xylose as inducer). Representative images of colonies are shown, and colony area (mean ± SD, n=8) normalized to the strain carrying the GFP-expressing control plasmid are shown. (D) Effects of CpdR variants on ClpX localization in C. crescentus. Xylose-inducible plasmids harboring CpdR variants were expressed in C. crescentus ΔcpdR PXyl-clpX-gfp (scale bar correspond to 2μm). ClpX foci intensities from 700 cells of each unsynchronized strain were analyzed using MicrobeTracker, and presented by histogram. See also Figure S5.
Previous studies suggested that CpdR-dependent degradation in vivo was due to the CpdR-dependent recruitment of ClpX to the incipient stalked pole of C. crescentus where substrates also transiently localize (Iniesta et al., 2006; McGrath et al., 2006). We found that loss in ClpX-GFP localization in ΔcpdR strains (Iniesta et al., 2006) could be overcome by expression of the R106A variant of CpdR, although with less intense foci (Figure 5D), while expression of the H104A variant resulted in diffuse localization of ClpX (Figure 5D) similar to that seen in the ΔcpdR strain (Iniesta et al., 2006). The results indicate that CpdR binding to ClpX is directly correlated with ClpX localization; however, it remains unclear if substrate degradation in vivo specifically requires ClpX localization.
Priming adaptors are resistant to inhibitory effects seen with scaffolding adaptors
Because general adaptor mechanisms may have different consequences, we considered how scaffolding and priming adaptors function at various adaptor concentrations. Scaffolds are often signal pathway components with optimal concentrations improving signal transduction, but excess scaffold reduces signal flux by titrating partner proteins away from one another (for examples, see Good et al., 2011). Similarly, as illustrated in Figure 6A, high concentrations of a scaffolding adaptor like SspB should reduce substrate delivery due to formation of complexes that do not contain both binding partners (i.e., complexes that contain adaptor/substrate only or adaptor/protease only). In contrast, excess concentrations of a priming adaptor like CpdR should show no inhibitory effect if substrates engage only a primed protease.
Figure 6. Adaptor effects at excess concentrations.
(A) Predicted changes in substrate delivery due to concentrations of a scaffolding adaptor or protease priming adaptor: Excess scaffolding adaptor inhibits substrate delivery due to formation of substrate-adaptor and protease-adaptor complexes. Excess protease-priming adaptor does not inhibit delivery. Degradation by ClpXP in vitro in the presence of varying adaptor concentrations were assayed as follows: (B) GFP-ssrA-SS (an SspB-obligate substrate) using adaptor SspB*; (C) GFP-PdeA (a CpdR-obligate substrate) using adaptor CpdRDD, (D) GFP-ssrA-SS using the substrate-binding domain (SBD) of SspB in the presence of active SspB*, and (E) GFP-PdeA using the ClpX-binding deficient adaptor variant CpdRH104ADD in the presence of active CpdR. For B and C, x-axis is adaptor concentration; For D and E, x-axis is ratio of mutant adaptor relative to wildtype. n.b. non-degradable adaptor variants (SspB* and CpdRDD) are used throughout to avoid trivial competition between adaptor and cargo substrates (Figures S2, S6A&B). Data represented as mean ± SD, n = 3 experiments. (F) Effect of SspB (SBD) on levels of GFP-ssrA-SS (detected by α-GFP) in WT and ΔsspB strains. (G) Effect of CpdRH104ADD on FLAG-PdeA (detected by α-FLAG) levels in WT and ΔcpdR strains. In (F) and (G), degradation reporters are expressed from the chromosomal vanA locus and adaptor variants from a xylose-inducible plasmid (JS14). See also Figure S2 and S6.
We tested our prediction by measuring substrate delivery of SspB and CpdR under conditions of excess adaptor, using substrates (GFP-ssrA-SS and GFP-PdeA) that are poorly recognized by ClpXP in the absence of their adaptors. To avoid concerns that the adaptors inhibit substrate delivery for the trivial reason of being degraded by ClpXP themselves (Chien et al., 2007; Iniesta, et al. 2008), we used the non-degradable, fully-active versions of these adaptors: SspB* (Chien et al., 2007) and CpdRDD (Figures S2A-D). At low concentrations, both adaptors effectively delivered their respective substrates, but high concentrations of SspB* inhibited delivery of GFP-ssrA-SS (Figures 6B; S6A). In contrast, excess CpdRDD did not suppress degradation of GFP-PdeA (Figures 6C; S6B).
Scaffolds are also sensitive to inhibition due to changes that eliminate one of the partner binding sites. Indeed, the substrate binding domain (SBD) of SspB is sufficient for ssrA binding but lacks the ClpX binding motif (Dougan et al., 2003; Bolon et al., 2004; Park et al., 2007; Davis et al., 2009). Addition of the SspB SBD strongly inhibited ssrA degradation even in the presence of full length adaptor (Figure 6D) and overexpression of the SspB SBD also stabilized an ssrA-tagged substrate even in the presence of wildtype SspB in vivo (Figure 6F and S6C). By contrast, addition of the ClpX-binding-deficient CpdRH104ADD did not inhibit CpdR-mediated degradation of GFP-PdeA (Figure 6E), and overexpression of this variant in vivo did not stabilize PdeA in the presence of endogenous CpdR (Figure 6G and S6E).
CpdR adaptor mechanism extends to other cell cycle-regulated substrates
Although many ClpXP substrates require CpdR for degradation in vivo, PdeA is the only substrate shown in vitro to be solely dependent on CpdR and ClpXP to date (Abel, et al. 2011; Rood, et al. 2012). Similar to PdeA, degradation of the chemoreceptor McpA in vivo is strictly dependent on CpdR (Iniesta, et al. 2006) and no other other regulators (McGrath et al., 2006; Duerig et al., 2009). Despite the discovery of cell cycle-dependent proteolytic control of McpA more than twenty years ago (Alley et al., 1993), degradation of McpA has yet to be shown in vitro. We purified the cytoplasmic portion of McpA as a GFP fusion and found that degradation of GFP-McpA in vitro by ClpXP was substantially enhanced by CpdR alone (Figure 7A). Similar to our results with PdeA (Figure 1B), McpA did not stably interact with CpdR alone in conditions where CpdR binds ClpX (Figure 7B). Degradation of McpA was affected by mutation of CpdR (Figure S7A) and depended on the NTDClpX as with PdeA (Figure S7B). Together, these results suggest that McpA degradation is governed by similar mechanisms that we have detailed for PdeA, and reveal a second member of ClpXP substrates that are directly delivered by CpdR.
Figure 7. Protease priming by CpdR affects degradation of other cell cycle-regulated substrates.
(A) In vitro degradation of GFP-tagged McpA by ClpXP when mediated by CpdR. GFP-ssrA degradation serves as a control for ClpXP activity. (B) SEC profiles of CpdR alone, and in the presence of GFP-McpA or ClpX. n.b. GFP-McpA elutes with an apparent molecular weight of ~400 kDa in this assay. (C) Impact of CpdR output face variants on in vitro degradation of CtrA by ClpXP in the presence of cognate DNA (fliF promoter), and its multi-adaptor complex RcdA, PopA and cyclic di-GMP (cdG). (D) CtrA degradation in vitro in the presence of components mentioned in (C) using CpdR WT to compare ClpXP to ΔN-ClpXP activity. (E) Cell cycle-dependent degradation of CtrA in C. crescentus ΔcpdR strain expressing CpdR variants. See Methods for synchronous swarmer cell preparation. CtrA quantification (normalized against ClpP and time zero) is shown on the right. (F) Model for the protease priming mechanism. During the Caulobacter cell cycle, the adaptor CpdR is kept phosphorylated in swarmer cells (G1 state) preventing interactions with ClpX. When the swarmer cell transitions into its stalked cell state (G1-S), dephosphorylation of CpdR promotes binding to the N-terminal domain of ClpX, priming it to recruit downstream components. This primed complex can directly recruit degradation substrates such as PdeA and McpA, and additional adaptors (RcdA, PopA-cdG) known to deliver CtrA for degradation. CpdR and NTDClpX could both be contributing individual weak interactions to promote cargo recruitment (i), or binding could be inducing conformational changes to promote cargo recruitment (ii). See also Figure S7.
We and others recently reconstituted the regulated degradation of the master regulator CtrA in vitro using a multi-protein adaptor complex that includes CpdR, RcdA and PopA (Smith et al., 2014). We find that ClpX-binding-deficient variants of CpdR were also deficient in adaptor-dependent degradation of CtrA (Figure 7C) and the degradation required the NTDClpX (Figures 7D; S7C&D). These CpdR variants support similarly timed but less dramatic oscillations in CtrA levels during cell cycle when present as the sole copy of CpdR in vivo (Figure 7E). Thus, we propose that the interaction between CpdR and ClpX generates a composite recruitment interface for binding multiple substrates directly (e.g. PdeA and McpA) in addition to recruiting additional adaptors (e.g. RcdA/PopA) that further expand substrate specificity (Figure 7F).
DISCUSSION
Cell cycle-regulated proteolysis in C. crescentus depends on an intricate network of phosphosignaling proteins such as CpdR (Kirkpatrick and Viollier, 2012). Although CpdR functions as a ClpXP adaptor, it was unclear how this molecule couples its phosphorylation status to a mechanism for delivering multiple substrates. In this study, we show that phosphorylating CpdR prevents its binding to ClpX. Dephosphorylated CpdR binds ClpX N-terminal domain to prime, or prepare, a recruitment interface that affords engagement of multiple substrates as well as additional adaptors that aid substrate delivery (Figure 7F). We propose that this mechanism underlies how changes in CpdR phosphorylation during the cell cycle causes dynamic assembly of complexes needed for regulated degradation of many substrates with biological consequences. For example, the phosphodiesterase PdeA normally maintains low levels of cyclic-di-GMP levels in swarmer cells for improved motility (Abel et al., 2011). The chemoreceptor McpA helps swarmer cells search for nutrient replete regions in the environment (Alley et al., 1991). CtrA plays a critical role as an inhibitor of replication (Quon et al., 1998), and must be maintained to prevent inappropriate replication initiation while swarmer cells are searching for a suitable habitat. Degradation controlled by modulating CpdR activity could facilitate cell reprogramming by removing these specific proteins when cells are ready to enter its replicative sessile state.
Our working model is that phosphorylation affects the equilibrium among CpdR conformations, similar to other response regulators (Bourret, 2010). In the case of CpdR, phosphorylation favors an output face configuration that blocks ClpX binding. We do not know specifically how CpdR primes ClpX; however, as illustrated in Figure 7F, binding of CpdR to the NTDClpX could induce conformational changes in CpdR, the NTDClpX, or both to promote recruitment of substrates and additional adaptors. Alternatively, CpdR and ClpX may each provide weak interactions that collectively result in strong binding of additional factors. Finally, the oligomeric nature of ClpX could promote a polyvalent display of CpdR that increases binding to substrates or downstream adaptors. How CpdR-NTDClpX recruits multiple downstream components remains an outstanding question.
CpdR was originally shown to be a key localization factor for the protease ClpXP that promotes protease and substrate colocalization, and this mechanism was thought to be needed for regulated degradation (Iniesta, et. al 2006). However, later work suggested that localization of ClpXP substrates could be decoupled from their degradation in vivo (Taylor et al., 2009). Here, we show that CpdR variants that weaken ClpX binding affect both localization of ClpX and CpdR-dependent ClpXP activity (Figure 6D). This data could support the model where localization promotes degradation because effects on degradation mirror changes in localization. Alternatively, CpdR activation at the incipient pole (Tsokos and Laub, 2012) could be temporarily producing protease-substrate-adaptor foci, a transient process that is relieved when substrates are degraded or adaptors are inactivated. This implies that loss of adaptors should result in loss of protease foci. In support of this alternative model, PopA, a component of the CtrA adaptor complex (Smith, et al. 2014), also contributes to ClpX foci formation (Duerig et al., 2009). Therefore, we favor a model where foci attributed to localization of the protein factors are a manifestation of the adaptor system, rather than driving degradation itself.
In conclusion, through this work we show how a protease-priming adaptor regulates cell cycle-dependent proteolysis in Caulobacter. This unexpected mechanism has an advantage over scaffolding adaptors in that an excess concentration of adaptor does not inhibit target degradation. Activation of CpdR function relies on its ability to translate cues from an intricate network of phosphosignaling proteins into a decision on whether to bind ClpX. This decision is an important one because it creates a recruitment interface that regulates engagement of substrates and downstream adaptors that further expands substrate selectivity. Finally, we note that CpdR is critical in the α-proteobacterium Sinorhizobium meliloti for nitrogen fixation in legumes (Kobayashi et al., 2009) and can function even with ClpX from E. coli, a γ-proteobacterium. Given the fact that almost all bacteria encode ClpX, we speculate that priming adaptors for ClpX will exist in other species.
Experimental Procedures
In vitro degradation assay and phosphotransfer reaction
Degradation assays and phosphotransfer reactions using recombinant protein components were performed in H-Buffer (20mM HEPES, pH 7.5, 100mM KCl, 10mM MgCl2, 10% glycerol, 5mM β-mercaptoethanol) at 30°C. Buffer used for ClpA and ClpXA degradation contained 40mM HEPES, pH 7.5, 220mM NaCl, 27mM KCl, 17mM MgCl2, 10% glycerol, and 5mM β-mercaptoethanol. In general, concentrations were 0.4μM unfoldases (ClpX, ClpA, ClpXA) (hexamer concentration), 0.8μM ClpP14, (GFP-ssrA and PdeA-ssrA by ClpAP used 0.1μM ClpA6 and 0.2μM ClpP14) in the presence of 4mM ATP (+ regeneration system); 1–2μM adaptors; 1–3μM substrates. When noted 25μM NTDClpX, 10 μM rapamycin, 10 μM fliF dsDNA 1mM GTP (for PdeA degradation) were used. Additional details are in Supplemental Information. Gel-based reactions were sampled at indicated times, quenched with SDS loading dye, and frozen immediately. Samples were heated at 95°C for 5 min prior to SDS-PAGE and Coomassie blue staining, Silver staining, or Western blotting. GFP-tagged substrates were detected using a 384-well black plate using a Spectramax M5 (Molecular Devices) microplate reader at excitation wavelength 460nm, and collection of emission wavelength at 540nm with a cutoff of 515 nm. Initial rates of degradation (Figures 3D & 4F) were fit to an activation model (GraphPad Prism) where binding of CpdR to ClpX activates degradation: v = Vmax*[CpdR]/(Kactivation+[CpdR]) and in Figure 3D using (v = Vmax*[NTD]/(Kactivation+[NTD])).
Analytical size exclusion chromatography (SEC) assay
Recombinant proteins were purified to more than 95% homogeneity and concentrations used for each experiment are found in Supplementary Experimental Procedures. To assay for interactions in vitro, 50μL mixtures containing recombinant proteins were incubated for 20 minutes at 25°C in H-Buffer prior to injection onto a Superdex 200 5/150 GL size exclusion chromatography column (GE Healthcare Bio-Sciences, 3mL bed volume) equilibrated with H-Buffer. CpdR phosphorylation reaction mixtures for SEC studies were validated by checking for inhibition of adaptor activity when degrading GFP-PdeA via ClpXP. The rest of the mixture components were added to the phosphorelay-CpdR mix prior to the SEC injection. SEC fractionation profiles (50μL per fraction) were examined by SDS-PAGE, followed by Coomassie blue staining, silver staining, or Western blotting. SEC experiments were performed independently at least twice.
Bacterial adenylate cyclase two-hybrid (BACTH) assay
Protein-protein interactions assayed by BACTH (Karimova et al., 1998) were performed using pKT25 and pUT18C (Euromedex) in the E. coli cya reporter strain BTH101 (Euromedex). An E. coli cya clpX strain was generated from BTH101 (see Supplementary Experimental Procedures for additional details).
C. crescentus soft agar plate assay
Colonies of cells that were transformed with plasmids expressing epitope tagged CpdR (WT or variants) from xylose-driven promoters were inoculate into PYE media containing 0.3% agar with 0.2% xylose with the appropriate antibiotic (5μg/mL kanamycin or 50μg/mL spectinomycin). After incubation at 30°C, colony sizes were determined using the threshold feature in ImageJ (NIH).
Fluorescence microscopy
Exponential-phased cells were immobilized on 1% (wt/vol) agarose pads made with PYE + 0.2% xylose. Microscopy was carried out using an epifluorescent Nikon E600 microscope. An ORCA-ER-cooled charge-coupled device (CCD) camera (Hamamatsu) and Openlabs software (Improvision) were used for all image acquisition. A phase-contrast image and z-stack of GFP fluorescence images (10 steps) was acquired with z-step of 0.3 μm around the focal plane of the cell. The z-stack images were deconvolved using Volocity 4.0 (Improvision, Inc.) software. Phase contrast images of C. crescentus cells were outlined using MicrobeTracker. Stalked pole fluorescence foci were first detected using the SpotFinderZ tool in MicrobeTracker, and then verified visually. Measurements of the foci intensity from SpotFinderZ were extracted and plotted in histograms.
Adaptor overexpression studies in vivo
For experiments shown in Figures 6F,G and S6C,E, strains from PYE agar plates (2μg/mL chloramphenicol, 0.2% glucose (to suppress JS14 expression)) were inoculated into PYE media (plus 1μg/mL chloramphenicol and 0.2% glucose) for overnight growth at 30°C in a shaking incubator. Cells sufficient for a final resuspension of cell density OD600=0.2 were pelleted. The supernatant media was removed, and the cells were resuspended into PYE containing 1μg/mL chloramphenicol, 1mM vanillate pH7.5, and 0.2% xylose or glucose, and grown for 4 hours at 30°C in a shaking incubator. Cells were at exponential phase (OD600<1) at harvest. Equivalent cell density amounts were sampled for Western analysis.
Synchrony for in vivo stability assay
C. crescentus cells expressing CpdR variants were grown overnight in PYE media (100μg/mL spectinomycin, 0.2% xylose) and back-diluted into fresh PYE media (100μg/mL spectinomycin, 0.2% xylose) at OD600 0.05 for growth to an OD600 of 0.3–0.5. Swarmer cells were isolated by Percoll density centrifugation, and cells were released into the same media. Samples were taken at specified time points for Western blot analysis using antibodies against CtrA and ClpP.
Supplementary Material
HIGHLIGHTS.
The CpdR adaptor alone does not form stable complexes with cargo
CpdR primes ClpX upon binding to create a recruitment interface for cargo
Phosphosignaling alters CpdR binding to ClpX via its signaling output face
Priming adaptors are insensitive to inhibition due to excess unfulfilled adaptors
Acknowledgments
We thank members of the Chien lab, Laura Francis, Indu Santhanagopalan, Eric Strieter and Steve Glynn for helpful feedback. We thank Steven Sandler and Ben Waldman for help with fluorescence microscopy, image analysis, and gifts of bacteriophage and plasmids. We thank Kevin Griffith for aid in the phosphorylation reactions and general advice. We thank Kamal Joshi (Chien Lab) for contributing protein reagents and affinity purified PdeA antibody. This work was sponsored in part by funding from a Chemistry Biology Interface Program Training Grant (NIH T32GM08515) to JL and RHV, the Initiative for Maximizing Student Diversity fellowship to LH-A (NIH R25GM099649), and NIH grants R00GM084157 and R01GM111706 to PC.
Footnotes
Supplementary Experimental Procedures include details on bacteria strains and plasmid constructions, bacterial two-hybrid details, protein purification methods, Western blotting, radioactive phosphotransfer and additional experimental details.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abel S, Chien P, Wassmann P, Schirmer T, Kaever V, Laub MT, Baker TA, Jenal U. Regulatory Cohesion of Cell Cycle and Cell Differentiation through Interlinked Phosphorylation and Second Messenger Networks. Mol Cell. 2011;43:550–560. doi: 10.1016/j.molcel.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alley MR, Maddock JR, Shapiro L. Requirement of the Carboxyl Terminus of a Bacterial Chemoreceptor for Its Targeted Proteolysis. Science. 1993;259:1754–1757. doi: 10.1126/science.8456303. [DOI] [PubMed] [Google Scholar]
- Alley MRK, Gomes SL, Alexander W, Shapiro L. Genetic Analysis of a Temporally Transcribed Chemotaxis Gene Cluster in Caulobacter crescentus. Genetics. 1991;129:333–342. doi: 10.1093/genetics/129.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battesti A, Gottesman S. Roles of adaptor proteins in regulation of bacterial proteolysis. Curr Opin Microbiol. 2013;16:140–147. doi: 10.1016/j.mib.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat NH, Vass RH, Stoddard PR, Shin DK, Chien P. Identification of ClpP substrates in Caulobacter crescentus reveals a role for regulated proteolysis in bacterial development. Mol Microbiol. 2013;88:1083–1092. doi: 10.1111/mmi.12241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biondi EG, Reisinger SJ, Skerker JM, Arif M, Perchuk BS, Ryan KR, Laub MT. Regulation of the bacterial cell cycle by an integrated genetic circuit. Nature. 2006;444:899–904. doi: 10.1038/nature05321. [DOI] [PubMed] [Google Scholar]
- Bolon DN, Wah DA, Hersch GL, Baker TA, Sauer RT. Bivalent Tethering of SspB to ClpXP is Required for Efficient Substrate Delivery: A Protein-Design Study. Mol Cell. 2004;13:443–449. doi: 10.1016/s1097-2765(04)00027-9. [DOI] [PubMed] [Google Scholar]
- Bourret RB. Receiver domain structure and function in response regulator proteins. Curr Opin Microbiol. 2010;13:142–149. doi: 10.1016/j.mib.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YE, Tsokos CG, Biondi EG, Perchuk BS, Laub MT. Dynamics of Two Phosphorelays Controlling Cell Cycle Progression in Caulobacter cresccentus. J Bacteriol. 2009;191:7417–7429. doi: 10.1128/JB.00992-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien P, Perchuk BS, Laub MT, Sauer RT, Baker TA. Direct and adaptor-mediated substrate recognition by an essential AAA+ protease. Proc Natl Acad Sci U S A. 2007;104:6590–6595. doi: 10.1073/pnas.0701776104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cranz-Mileva S, Imkamp F, Kolygo K, Maglica Ž, Kress W, Weber-Ban E. The Flexible Attachment of the N-Domains to the ClpA Ring Body Allows their Use On Demand. J Mol Biol. 2008;378:412–424. doi: 10.1016/j.jmb.2008.02.047. [DOI] [PubMed] [Google Scholar]
- Davis JH, Baker TA, Sauer RT. Engineering Synthetic Adaptors and Substrates for Controlled ClpXP Degradation. J Biol Chem. 2009;284:21848–21855. doi: 10.1074/jbc.M109.017624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnen ST, Newton A. Chromosome replication during development in Caulobacter crescentus. J Mol Biol. 1972;64:671–680. doi: 10.1016/0022-2836(72)90090-3. [DOI] [PubMed] [Google Scholar]
- Dougan DA, Reid BG, Horwich AL, Bukau B. ClpS, a substrate modulator of the ClpAP machine. Mol Cell. 2002;9:673–683. doi: 10.1016/s1097-2765(02)00485-9. [DOI] [PubMed] [Google Scholar]
- Dougan DA, Weber-Ban E, Bukau B. Targeted delivery of an ssrA-tagged substrate by the adaptor protein SspB to its cognate AAA+ protein ClpX. Mol Cell. 2003;12:373–380. doi: 10.1016/j.molcel.2003.08.012. [DOI] [PubMed] [Google Scholar]
- Duerig A, Abel S, Folcher M, Nicollier M, Schwede T, Amiot N, Giese B, Jenal U. Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression. Genes Dev. 2009;23:93–104. doi: 10.1101/gad.502409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JM, Levchenko I, Seidel M, Wickner SH, Sauer RT, Baker TA. Overlapping recognition determinants within the ssrA degradation tag allow modulation of proteolysis. Proc Natl Acad Sci U S A. 2001;98:10584–10589. doi: 10.1073/pnas.191375298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R, Stock AM. Molecular strategies for phosphorylation-mediated regulation of response regulator activity. Curr Opin Microbiol. 2010;13:160–167. doi: 10.1016/j.mib.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good MC, Zalatan JG, Lim WA. Scaffold proteins: hubs for controlling the flow of cellular information. Science. 2011;332:680–686. doi: 10.1126/science.1198701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith KL, Wolf RE. Measuring Beta-Galactosidase Activity in Bacteria: Cell Growth, Permeabilization, and Enzyme Assays in 96-Well Arrays. Biochem Biophys Res Commun. 2002;290:397–402. doi: 10.1006/bbrc.2001.6152. [DOI] [PubMed] [Google Scholar]
- Grimaud R, Kessel M, Beuron F, Steven AC, Maurizi MR. Enzymatic and Structural Similarities between the Escherichia coli ATP-dependent Proteases, ClpXP and ClpAP. J Biol Chem. 1998;273:12476–12481. doi: 10.1074/jbc.273.20.12476. [DOI] [PubMed] [Google Scholar]
- Hengge R. Proteolysis of RpoS and the general stress response in Escherichia coli. Res Microbiol. 2009;160:667–676. doi: 10.1016/j.resmic.2009.08.014. [DOI] [PubMed] [Google Scholar]
- Iniesta AA, McGrath PT, Reisenauer A, McAdams HH, Shapiro L. A phospho-signaling pathway controls the localization and activity of a protease complex critical for bacterial cell cycle progression. Proc Natl Acad Sci U S A. 2006;103:10935–10940. doi: 10.1073/pnas.0604554103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iniesta AA, Shapiro L. A bacterial control circuit integrates polar localization and proteolysis of key regulatory proteins with a phosphosignaling cascade. Proc Natl Acad Sci USA. 2008;105:16602–16607. doi: 10.1073/pnas.0808807105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal U. The role of proteolysis in the Caulobacter crescentus cell cycle and development. Res Microbiol. 2009;160:687–695. doi: 10.1016/j.resmic.2009.09.006. [DOI] [PubMed] [Google Scholar]
- Jenal U, Fuchs T. An essential protease involved in bacterial cell-cycle control. EMBO J. 1998;17:5658–5669. doi: 10.1093/emboj/17.19.5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimova G, Pidoux J, Ullmann A, Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci U S A. 1998;95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJE. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick CL, Viollier PH. Decoding Caulobacter development. FEMS Microbiol Rev. 2012;36:193–205. doi: 10.1111/j.1574-6976.2011.00309.x. [DOI] [PubMed] [Google Scholar]
- Kirstein J, Schlothauer T, Dougan DA, Lilie H, Tischendorf G, Mogk A, Bukau B, Turgay K. Adaptor protein controlled oligomerization activates the AAA+ protein ClpC. EMBO J. 2006;25:1481–1491. doi: 10.1038/sj.emboj.7601042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirstein J, Molière N, Dougan DA, Turgay K. Adapting the machine: adaptor proteins for Hsp100/Clp and AAA+ proteases. Nat Rev Microbiol. 2009;7:589–599. doi: 10.1038/nrmicro2185. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, De Nisco NJ, Chien P, Simmons LA, Walker GC. Sinorhizobium meliloti CpdR1 is critical for co-ordinating cell cycle progression and the symbiotic chronic infection. Mol Microbiol. 2009;73:586–600. doi: 10.1111/j.1365-2958.2009.06794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levchenko I, Seidel M, Sauer RT, Baker TA. A Specificity-Enhancing Factor for the ClpXP Degradation Machine. Science. 2000;289:2354–2356. doi: 10.1126/science.289.5488.2354. [DOI] [PubMed] [Google Scholar]
- Levchenko I, Grant RA, Wah DA, Sauer RT, Baker TA. Structure of a delivery protein for an AAA+ protease in complex with a peptide degradation tag. Mol Cell. 2003;12:365–372. doi: 10.1016/j.molcel.2003.08.014. [DOI] [PubMed] [Google Scholar]
- Lo JH, Baker TA, Sauer RT. Characterization of the N-terminal repeat domain of Escherichia coli ClpA—A class I Clp/HSP100 ATPase. Protein Sci. 2001;10:551–559. doi: 10.1110/ps.41401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath PT, Iniesta AA, Ryan KR, Shapiro L, McAdams HH. A Dynamically Localized Protease Complex and a Polar Specificity Factor Control a Cell Cycle Master Regulator. Cell. 2006;124:535–547. doi: 10.1016/j.cell.2005.12.033. [DOI] [PubMed] [Google Scholar]
- Park EY, Lee BG, Hong SB, Kim HW, Jeon H, Song HK. Structural Basis of SspB-tail Recognition by the Zinc Binding Domain of ClpX. J Mol Biol. 2007;367:514–526. doi: 10.1016/j.jmb.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Quon KC, Yang B, Domian IJ, Shapiro L, Marczynski GT. Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin. Proc Natl Acad Sci U S A. 1998;95:120–125. doi: 10.1073/pnas.95.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan SK, Pritchard S, Viollier PH. Coupling Prokaryotic Cell Fate and Division Control with a Bifunctional and Oscillating Oxidoreductase Homolog. Cell. 2010;18:90–101. doi: 10.1016/j.devcel.2009.10.024. [DOI] [PubMed] [Google Scholar]
- Rood KL, Clark NE, Stoddard PR, Garman SC, Chien P. Adaptor-Dependent Degradation of a Cell-Cycle Regulator Uses a Unique Substrate Architecture. Structure. 2012;20:1–10. doi: 10.1016/j.str.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Takaya A, Mouslim C, Hughes KT, Yamamoto T. FliT Selectively Enhances Proteolysis of FlhC Subunit in FlhD4C2 Complex by an ATP-dependent Protease ClpXP. J Biol Chem. 2014 doi: 10.1074/jbc.M114.593749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer RT, Bolon DN, Burton BM, Burton RE, Flynn JM, Grant RA, Hersch GL, Joshi SA, Kenniston JA, Levchenko I, et al. Sculpting the Proteome with AAA+ Proteases and Disassembly Machines. Cell. 2004;119:9–18. doi: 10.1016/j.cell.2004.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerker JM, Prasol MS, Perchuk BS, Biondi EG, Laub MT. Two-Component Signal Transduction Pathways Regulating Growth and Cell Cycle Progression in a Bacterium: A System-Level Analysis. PLoS Biol. 2005;3:1770–1788. doi: 10.1371/journal.pbio.0030334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SC, Joshi KK, Zik JJ, Trinh K, Kamajaya A, Chien P, Ryan KR. Cell cycle-dependent adaptor complex for ClpXP-mediated proteolysis directly integrates phosphorylation and second messenger signals. Proc Natl Acad Sci. 2014;111:14229–14234. doi: 10.1073/pnas.1407862111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stüdemann A, Noirclerc-Savoye M, Klauck E, Becker G, Schneider D, Hengge R. Sequential recognition of two distinct sites in sigma(S) by the proteolytic targeting factor RssB and ClpX. EMBO J. 2003;22:4111–4120. doi: 10.1093/emboj/cdg411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JA, Wilbur JD, Smith SC, Ryan KR. Mutations that Alter RcdA Surface Residues Decouple Protein Localization and CtrA Proteolysis in Caulobacter cresccentus. J Mol Biol. 2009;394:46–60. doi: 10.1016/j.jmb.2009.08.076. [DOI] [PubMed] [Google Scholar]
- Tsokos CG, Laub MT. Polarity and cell fate asymmetry in Caulobacter crescentus. Curr Opin Microbiol. 2012;15:744–750. doi: 10.1016/j.mib.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wah DA, Levchenko I, Baker TA, Sauer RT. Characterization of a Specificity Factor for an AAA+ ATPase: Assembly of SspB Dimers with ssrA-Tagged Proteins and the ClpX Hexamer. Cell. 2002;9:1237–1245. doi: 10.1016/s1074-5521(02)00268-5. [DOI] [PubMed] [Google Scholar]
- Wah DA, Levchenko I, Rieckhof GE, Bolon DN, Baker TA, Sauer RT. Flexible Linkers Leash the Substrate Binding Domain of SspB to a Peptide Module that Stabilizes Delivery Complexes with the AAA+ ClpXP Protease. Mol Cell. 2003;12:355–363. doi: 10.1016/s1097-2765(03)00272-7. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Gottesman S, Hoskins JR, Maurizi MR, Wickner S. The RssB response regulator directly targets sigma(S) for degradation by ClpXP. Genes Dev. 2001;15:627–637. doi: 10.1101/gad.864401. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.