Significance
Based on several studies, including ours, we estimate that 40–70 million individuals carry a chromosomally integrated copy of the human herpesvirus 6 (HHV-6) genome in every cell of their body. This condition is referred to as inherited chromosomally integrated HHV-6 (iciHHV-6). The regions targeted for integration are telomeres, which play important roles in the self-renewal capacity of cells. Whether iciHHV-6 is associated with disease remains unknown. After conducting a population screen (n = 20,000), our results indicate that the prevalence of angina is three times greater in iciHHV-6+ subjects relative to iciHHV-6− ones. Furthermore, iciHHV-6+ subjects have shorter telomeres, a result that may explain, at least in part, how iciHHV-6 may contribute to the development of angina.
Keywords: HHV-6, telomere, angina, chromosomal integration, ciHHV-6
Abstract
Inherited chromosomally integrated human herpesvirus-6 (iciHHV-6) results in the germ-line transmission of the HHV-6 genome. Every somatic cell of iciHHV-6+ individuals contains the HHV-6 genome integrated in the telomere of chromosomes. Whether having iciHHV-6 predisposes humans to diseases remains undefined. DNA from 19,597 participants between 40 and 69 years of age were analyzed by quantitative PCR (qPCR) for the presence of iciHHV-6. Telomere lengths were determined by qPCR. Medical records, hematological, biochemical, and anthropometric measurements and telomere lengths were compared between iciHHV-6+ and iciHHV-6− subjects. The prevalence of iciHHV-6 was 0.58%. Two-way ANOVA with a Holm–Bonferroni correction was used to determine the effects of iciHHV6, sex, and their interaction on continuous outcomes. Two-way logistic regression with a Holm–Bonferroni correction was used to determine the effects of iciHHV6, sex, and their interaction on disease prevalence. Of 50 diseases monitored, a single one, angina pectoris, is significantly elevated (3.3×) in iciHHV-6+ individuals relative to iciHHV-6− subjects (P = 0.017; 95% CI, 1.73–6.35). When adjusted for potential confounding factors (age, body mass index, percent body fat, and systolic blood pressure), the prevalence of angina remained three times greater in iciHHV-6+ subjects (P = 0.015; 95%CI, 1.23–7.15). Analyses of telomere lengths between iciHHV-6− without angina, iciHHV-6− with angina, and iciHHV-6+ with angina indicate that iciHHV-6+ with angina have shorter telomeres than age-matched iciHHV-6− subjects (P = 0.006). Our study represents, to our knowledge, the first large-scale analysis of disease association with iciHHV-6. Our results are consistent with iciHHV-6 representing a risk factor for the development of angina.
Human herpesvirus (HHV)-6A (1) and HHV-6B (2) are worldwide ubiquitous human pathogens belonging to the β herpesvirus subfamily. Primary HHV-6B infection occurs early in life (6 mo to 3 y) and causes roseola or exanthem subitum (3). Fewer details exist on the epidemiology of HHV-6A, and at present, primary HHV-6A infections have not been linked to diseases. The fact that HHV-6B is more widely distributed, acquired earlier in life, and likely to provide cross-protective immunity (HHV-6A and HHV-6B proteins are 80% identical) could explain, at least in part, the apparent lack of disease association with primary HHV-6A infection.
All herpesviruses possess a bimodal replicative cycle that allow them to replicate and generate new infectious viral particles or allow them to enter a state of latency where viral gene expression is minimal and no viruses are produced. Latency allows these viruses to escape immune surveillance enabling them to persist for life within their host. Reactivation from latency and viral replication can be triggered by several stimuli such as UV exposure or hormonal changes. Depending on the immunological competency of the host, such secondary infections can either be contained (asymptomatic) or disseminated, often resulting in pathogenic outcomes (4). HHV-6A and HHV-6B latency is unique among human herpesviruses in that these viruses have developed the ability to integrate their genomes into human chromosomes (5), much like Marek’s disease virus (MDV), whose genomic integration is tightly associated with lymphoma development (6). HHV-6 integration can occur in several distinct chromosomes but invariably takes place within the telomeric regions (7). When integration occurs in germinal cells, integrated HHV-6A and HHV-6B can be passed on to descendants according to Mendelian genetics (8). Consequently, individuals with such inherited chromosomally integrated HHV-6 (iciHHV-6) carry a copy of the viral genome in every cell of their body. Diagnosis of iciHHV-6 is easily made by measuring and comparing the relative number of HHV-6 DNA copy per cell either using quantitative PCR (qPCR) and/or digital droplet PCR (9, 10). iciHHV-6+ subjects have approximately one million copies per million cells, whereas those that have acquired HHV-6 naturally typically have between 60 and 80 viral DNA copies per million cells (11). In vitro and in vivo studies have indicated that, once integrated, HHV-6 can express genes and reactivate from latency with pathogenic outcomes (12–15). Based on several studies, including the present one, we estimate that between 40 and 70 million individuals worldwide carry iciHHV-6 (7).
It is well established that the self-renewal potential of cells is directly proportional to telomere lengths and telomerase activity (16, 17). It is also known that the shortest telomere, not average telomere length, is critical for cell viability and chromosome stability (18). When the number of telomeric repeated sequences (TRSs) falls below 13, chromosomal instability is observed (19). Integration of HHV-6 within the telomeric region is unlikely to be without consequences (7). In fact, Huang et al. recently reported that the chromosome harboring integrated HHV-6 is often the shortest, suggesting that integration affects telomeric integrity (20). Several diseases are linked with telomere dysfunctions and/or telomerase mutations such as cardiovascular diseases, hematopoietic dysfunction, pulmonary fibrosis, liver disease, degenerative diseases, and cancer (21–31). Alterations within telomeric regions are therefore a likely cause for cellular dysfunctions linked to diseases, but many of the factors affecting telomere integrity remain to be identified. Interestingly, by compiling several small independent studies, it has been reported that iciHHV-6 is 2.3 times more frequent (P < 0.001) in diseased (various diseases) individuals relative to healthy ones, suggesting that iciHHV-6 represents a risk factor in disease development (32). Knowing this, we undertook a large-scale population study to assess the prevalence of iciHHV-6 in the CARTaGENE (CaG) cohort. The CaG biobank (www.cartagene.qc.ca), consists of both biological samples and complete medical records on the health and lifestyles of randomly selected Quebecers (Canada) between 40 and 69 y of age. The bank contains in-depth information on nearly 20,000 individuals (17% born outside Canada), including sociodemographic, lifestyle, health data, and familial history of disease, 190 physiological parameters, biochemical analyses, and hematological analyses (33). Genomic DNA extracted from blood-spotted filters was used for HHV-6 qPCR and droplet digital PCR (ddPCR) analyses. Because iciHHV-6 individuals carry one viral genome copy per cell, identification of subjects with iciHHV-6 is easily made (9, 34). The overall prevalence of iciHHV-6 in the CaG cohort was determined, and disease prevalence in subjects with iciHHV-6 was compared with that of subjects without iciHHV-6.
Results
Our first objective was to determine the prevalence of iciHHV-6 in the province of Quebec by analyzing the CaG cohort. The results obtained are summarized in Tables 1 and 2. Of 19,597 subjects analyzed (51.3% females and 48.7% males), 113 were identified as iciHHV-6+, indicating an overall iciHHV-6 prevalence of 0.58% (95% CI, 0.34–0.67). Among iciHHV-6+ subjects, 56.6% were males and 43.4% were females (P = 0.094). Further analyses indicate that 41% of integrated cases were HHV-6A and 59% were HHV-6B, independent of sex. As could be expected from a nonlethal inherited condition, iciHHV-6 cases were distributed approximately evenly into three age groups (Table 1). The age (P = 0.08) and sex (P = 0.079) distributions of iciHHV-6− and iciHHV-6+ subjects were similar. Ninety-four of 113 (83%) iciHHV-6+ individuals were born in Canada, whereas the remaining 19 were born in 14 different countries (Table S1). More than 70% of iciHHV-6+ subjects parents and grandparents were also born in Canada. The ethnic background of the iciHHV-6+ subjects was white/European for the vast majority (88.3%), followed distantly by Arabic descendants (6.3%).
Table 1.
Demographics of the CARTaGENE cohort (>40 to <70 y old) along with iciHHV-6 prevalence
| Demographics | N screened (%) | N iciHHV-6− (%) | N iciHHV-6+ (%) [95% CI] |
| Sex | |||
| Female | 10,037 (51.22) | 9,988 (51.26) | 49 (43.36)[0.34–0.53] |
| Male | 9,560 (48.78) | 9,496 (48.74) | 64 (56.64)[0.47–0.67] |
| Total | 19,597 (100.00) | 19,484 (99.42) | 113 (0.58) |
| Age (y) | |||
| 40–49 | 6,786 (34.63) | 6,750 (34.64) | 36 (31.86)[0.23–0.41] |
| 50–59 | 7,465 (38.09) | 7,421 (38.09) | 44 (38.94)[0.30–0.49] |
| 60–69 | 5,346 (27.28) | 5,313 (27.27) | 33 (29.20)[0.21–0.39] |
| Total | 19,597 (100.00) | 19,484 (99.42) | 113 (0.58) |
Table 2.
Prevalence of iciHHV-6A and iciHHV-6B according to gender
| Sex | N iciHHV-6+ (%) | N iciHHV-6B+ (%) [95% CI] | N iciHHV-6A+ (%) [95% CI] |
| Female | 49 (43.36) | 30 (44.78)[0.33–0.57] | 19 (41.30)[0.27–0.57] |
| Male | 64 (56.64) | 37 (55.22)[0.43–0.67] | 27 (58.70)[0.43–0.73] |
| Total | 113 (100.00) | 67 (59.29) | 46 (40.71) |
We next proceeded to compare physical, hematological, and biochemical parameters between iciHHV-6− and iciHHV-6+ subjects. Values for several parameters differed between males and females (Table S2). However, all parameters examined were comparable (P = 0.068) between iciHHV-6− (males and females) and iciHHV-6+ subjects (males and females).
We next analyzed the prevalence of iciHHV-6 in various chronic diseases. As for biochemical parameters, the prevalence of certain diseases (such as thyroid disease, osteoarthritis, and allergy to medication) differed between males and females (Table S3). For analyses, the iciHHV-6A+ and iciHHV-6B+ subjects were fused into a single group. In addition to prevalence of disease, we analyzed prescribed medication intake. The prevalence of subjects (males or females) taking prescribed medication was similar between iciHHV-6− and iciHHV-6+ subjects (Table 3). We next analyzed the prevalence of chronic diseases using two-way logistic regression analyses followed by Holm–Bonferroni correction adjusted for multiple comparisons. Of the 50 diseases analyzed (Table S3), a single condition proved significantly more frequent in the iciHHV-6+ subjects (Table 3). Our results indicate that the prevalence of angina pectoris (stable and unstable forms) is significantly higher in the iciHHV-6+ subjects (9.7%) relative to iciHHV-6− subjects (3.2%) (OR, 3.31; 95% CI, 1.73–6.35; P = 0.017). There was no significant sex association (P = 0.839).
Table 3.
Prevalence of disease in iciHHV-6− and iciHHV-6+ subjects
| Variable | iciHHV-6− (%) | iciHHV6+ (%) | Adjusted P value* | OR (95% CI) |
| Currently taking prescribed medication | 12,829/19,542 (65.6) | 70/113 (61.9) | 1.00 | 0.86 (0.58–1.27) |
| Disease of the circulatory system | ||||
| High blood pressure occcurence | 4,820/19,436 (24.8) | 29/113 (25.7) | 1.00 | 1.00 (0.65–1.56) |
| Myocardial infarct | 542/19,520 (2.8) | 3/113 (2.7) | 1.00 | 1.11 (0.33–3.74) |
| Stroke occurrence | 311/19,520 (1.6) | 5/112 (4.5) | 1.00 | 2.87 (1.14–7.23) |
| Angina occurrence | 624/19,473 (3.2) | 11/113 (9.7) | 0.017 | 3.31 (1.73–6.35) |
Statistically significant results (P < 0.05) are in italic.
P values for iciHHV-6 from two-way logistic regression were adjusted for multiple comparisons (n = 56) using Holm–Bonferroni.
Knowing that cardiovascular diseases are influenced by several factors such as body mass index (BMI), age, systemic blood pressure, and percent body fat (35), additional analyses were conducted to take into account these factors. As shown in Table 4, BMI, age, and systolic blood pressure represented independent risk factors associated with the development of angina. When adjusted for these three factors and percent body fat, the prevalence of angina in iciHHV-6+ individuals remained significantly elevated (P = 0.015), with an OR of 2.97 (95% CI, 1.23–7.15). These results suggest that iciHHV-6 is a predisposing risk factor for the development of angina.
Table 4.
Multivariate analyses on the impact of iciHHV-6 on angina adjusted for age, BMI, systemic blood pressure, percent body fat, and sex
| Factor/characteristic | OR (95% CI) | P value |
| Age | 1.11 (1.09–1.13) | <0.001 |
| BMI | 1.024 (1.02–1.09) | <0.001 |
| Systolic blood pressure | 0.99 (0.99–0.99) | 0.004 |
| Percent body fat | 1.01 (0.99–1.04) | 0.321 |
| Sex | 0.81 (0.32–2.04) | 0.656 |
| iciHHV-6+ | 2.97 (1.23–7.15) | 0.015 |
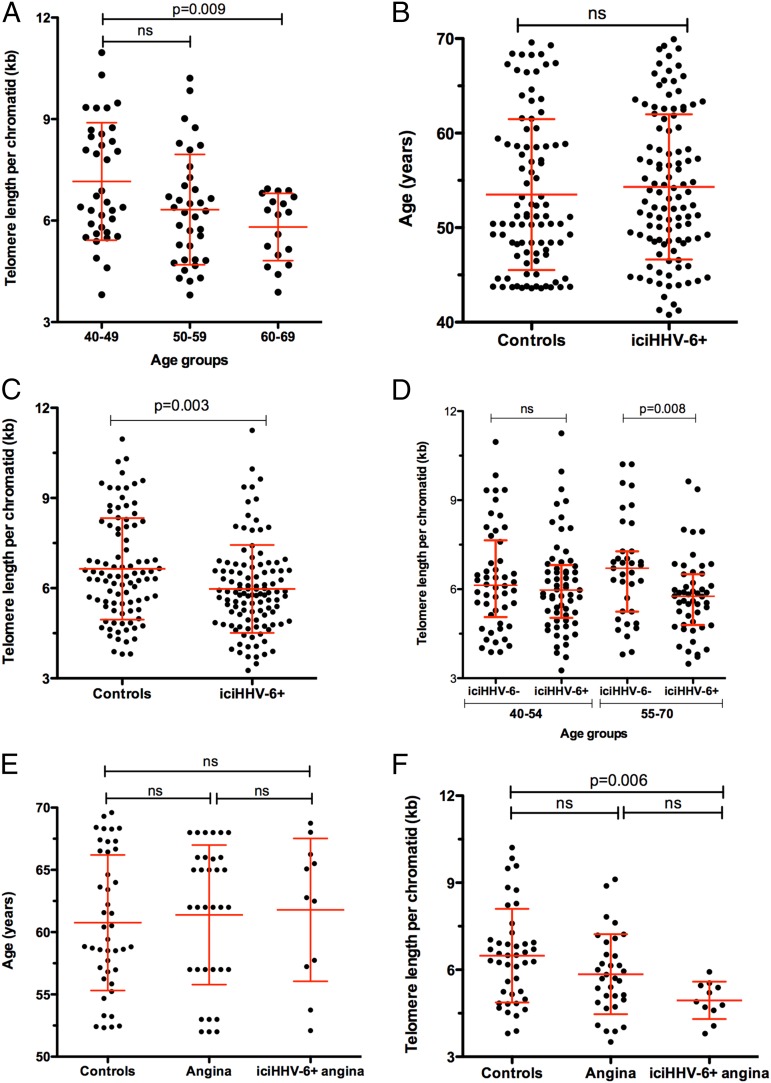
A recent meta-analysis concluded that telomere length is inversely associated with the risk of coronary heart disease, independently of conventional vascular risk factors (36). Considering this, we sought to determine the mean telomere length of leukocyte DNA from iciHHV-6+ subjects relative to iciHHV-6− ones. To validate the assay (37), we first determined telomere lengths as a function of age in iciHHV-6− subjects. As shown in Fig. 1A, an inverse correlation between age and telomere length was observed, as expected and previously reported (38). DNA samples from randomly selected and individually aged-matched iciHHV-6+ (n = 111) and iciHHV-6− (n = 93) individuals (Fig. 1B) were analyzed for telomere length determination. As shown in Fig. 1C, the overall length of telomeres of iciHHV-6+ subjects is significantly shorter than that of iciHHV-6− subjects (P = 0.003). When analyzed as a function of age, the average telomere length of iciHHV-6+ subjects aged between 55 and 70 y was shorter than individually age-matched iciHHV-6− subjects (P = 0.008; Fig. 1D). Analyses of telomere lengths between age-matched iciHHV-6− without angina (n = 43), iciHHV-6− with angina (n = 33), and iciHHV-6+ with angina (n = 11) indicate that iciHHV-6+ with angina have shorter telomeres than age-matched controls (P = 0.006; Fig. 1 E and F). Relative to controls, the mean telomere length of iciHHV-6− subjects with angina is 10% shorter. In comparison, the overall telomere length of iciHHV-6+ subjects with angina was 24% and 15% shorter than iciHHV-6− subjects and iciHHV-6− subjects with angina, respectively. These results indicate that iciHHV-6+ subjects have shorter telomeres than age-matched iciHHV-6- subjects.
Fig. 1.
Telomere length analyses of DNA from iciHHV-6− and iciHHV-6+ subjects. (A) Blood leukocyte DNA samples from iciHHV-6− and iciHHV-6+ subjects were analyzed by qPCR for telomere length. A scatter plot with mean ± SD telomere length according to age is presented. (B) Age distribution of iciHHV-6− and iciHHV-6+ subjects used for telomere length analyses. (C) DNA samples from iciHHV-6− subjects and iciHHV-6+ individuals were analyzed for telomere length by qPCR. Results are presented as a scatter plot with mean ± SD outlined. (D) A scatter plot with mean ± SD telomere length of iciHHV-6− and iciHHV-6+ subjects according to age. (E) Age distribution of iciHHV-6− subjects, iciHHV-6− subjects with angina and iciHHV-6+ subjects with angina used for telomere length analyses. Results are presented as a scatter plot with mean ± SD outlined. (F) DNA samples from iciHHV-6− subjects, iciHHV-6− subjects with angina and iciHHV-6+ subjects with angina were used for telomere length analyses by qPCR. Results are presented as a scatter plot with mean ± SD outlined. Statistical analyses were conducted using one-way ANOVA with a Bonferroni multiple comparison test (A, E, and F) or t test (B–D). Results with P < 0.05 were considered significant.
All other diseases including overall cancer prevalence occurred at similar frequencies between iciHHV-6− and iciHHV-6+ individuals. Analyses of individual cancer types (e.g., leukemia and prostate) were not conducted based on the observation that only a total of 12 cancer cases representing diverse cancer types were present in the iciHHV-6+ population.
Discussion
The first cases of chromosomally integrated HHV-6 were reported in 1993 (5), and not until 1998 was the inheritance of integrated HHV-6 demonstrated (8). Although initially considered an oddity, integration of HHV-6 into host chromosomes is now considered a part of the HHV-6 life cycle. In fact, integration is one way HHV-6 viruses establish latency. Several reports suggested that HHV-6 might reactivate once it has integrated the human genome (12, 14, 15, 39). Furthermore, recent compelling in vivo evidence indicates that iciHHV-6 does reactivate in severely immunocompromised subject with pathogenic outcomes (13).
Whether iciHHV-6 predisposes humans to certain diseases is presently unknown, but considering that across the world, several million individuals are iciHHV-6+, this is an important subject. In a recent review, Pellett et al. reported that iciHHV-6 is more prevalent in diseased (various diseases) individuals (32), suggesting that iciHHV-6 represents a risk factor for the development of certain medical conditions. One potential caveat of the previous study lies in the fact that such assumption was made by pooling data from several small independent studies from various geographical regions where iciHHV-6 incidence and environmental factors may differ, which is likely to affect analysis outcome. The worldwide prevalence of iciHHV6+ individuals is estimated to be ∼1% with variations observed (0.2–2.9%) depending on the geographical regions and population sampled (healthy vs. diseased) (40–44). A relatively large sample size is therefore needed to have a sufficient number of subjects with iciHHV-6 to determine potential disease associations. Much of what is known about the causes of chronic disorders comes from large epidemiological studies (45, 46). To determine whether iciHHV-6 represents a predisposing factor for particular diseases, we analyzed a total of 19,597 individuals from the CaG cohort (33). A special feature of this cohort is the common ancestry of the bulk of the 20,000 volunteers. The cohort includes Quebecers from various ethnic and racial backgrounds (especially in the Montreal subgroup), but the great majority are French Canadians. The common genetic heritage of participants makes it easier to rule out potential environmental contributors to either good health or the development of chronic disorders. In the CaG cohort, participants were not recruited for a particular disease, but instead represent a random selection among the population, minimizing the need to correct for bias in measured phenotypes.
The results obtained here indicate that the prevalence of iciHHV-6 in Quebec falls within the 0.2–2.9% prevalence previously reported for other region of the globe (40–44). The main finding of the current study is that prevalence of angina pectoris is found elevated 3.3 times in iciHHV-6+ subjects relative to iciHHV-6− subjects (P = 0.017). When adjusted for age, BMI, percent body fat, and systolic blood pressure, the prevalence of angina in iciHHV-6+ individuals remains elevated (2.97×), suggesting that this inherited trait may predispose to the development of angina (P = 0.015).
Infections, whether they are of bacterial or viral origin, including herpesviruses such as cytomegalovirus, are thought to play important roles in atherosclerosis development (47). One hypothesis is that endothelial cells from iciHHV-6 subjects express, at some point in time, viral proteins and/or produce virions that can lead to immune activation (of monocytes, lymphocytes, and/or platelets) and cell damage, initiating and fueling the development of atherosclerotic lesions and, eventually leading to angina (48). In support, a recent study indicates reactivation of iciHHV-6 in subjects with myocardial dysfunctions. Virus particles were identified in degenerating myocytes and interstitial cells and antiviral treatment abolished viral mRNA and ameliorated cardiac symptoms (49). The authors hypothesize that damage of endothelial cells caused by HHV-6 reactivation in iciHHV-6 patients might explain complaints of angina as a sequela of myocyte and vascular endothelial dysfunction, as previously reported for parvovirus B19 (50). Experiments addressing these issues are currently ongoing.
Alternatively, a vast body of literature also exists linking short telomere lengths and cardiovascular disease development (51, 52). A recent meta-analysis of 24 studies involving more than 40,000 subjects concluded that telomere length was inversely correlated with the incidence of cardiovascular diseases (36). Considering that the shortest chromosome is responsible for triggering cell damage (senescence or chromosomal instability) (18, 19) and that the chromosome carrying the integrated HHV-6 is often the shortest (20), it can be speculated that angina pectoris can result from an accelerated shortening of the telomere of the chromosome carrying HHV-6 integration. When analyzed for overall length, the telomeres of DNA from iciHHV-6+ subjects were 10% shorter than those of age-matched iciHHV-6− individuals (P = 0.003). Our results therefore support the observation that reduced telomere length is associated with a higher prevalence of cardiovascular disease. Disease may appear more or less rapidly based on the chromosome targeted for integration and whether large or small telomeric deletions were introduced.
Although there is no convincing evidence linking HHV-6 infection or iciHHV-6 to any cancer type, weak associations have been made (53). In the CaG cohort, a total of 1,553 subjects experienced at least one cancer type (8.0%). When all cancer types were considered, no association with iciHHV-6 was noted. A larger cohort could have helped identify iciHHV-6 association with low incidence cancers.
Integration of HHV-6 into host DNA is likely to affect the genomic architecture with somewhat unpredictable outcomes. By altering the telomeric regions, HHV-6 may affect the long-term ability of a cell to divide and cause premature senescence, leading to organ dysfunctions or alternatively provoke chromosomal instability favoring cancer development. The fact that HHV-6 targets several distinct chromosomes for integration complicates analysis as disease outcomes may vary depending on the integrated chromosomes. In addition to its effects on cell homeostasis, the fact that the integrated virus can reactivate and/or express viral mRNA and proteins can lead to inflammation and immune mediated reactions that can lead to tissue destruction and diseases.
This study has the following limitations. First, the association between iciHHV6+ and the risk of developing angina will need to be confirmed in a large longitudinal cohort design; second, although nearly 20,000 subjects were analyzed, the number of iciHHV-6+ subjects prevented us from conducting in depth analyses on diseases of low prevalence. Larger population studies will possibly identify other medical conditions of lower incidence in which iciHHV-6 would be the first event predisposing to disease development according to the second hit theory (54); and third, not having access to cells from iciHHV-6+ subjects prevented us from identifying the chromosomes carrying the integrated virus. Depending on chromosome carrying the integration, diseases may or may not become apparent. Additional studies are therefore needed, including the mapping of chromosomes integrated by HHV-6 and experiments designed to explain the underlying mechanisms linking iciHHV-6 to angina.
In summary, our results are consistent with iciHHV-6 representing a predisposing risk factor for the development of angina pectoris. Cardiovascular diseases are widespread diseases and represent a major cause of mortality in North America. Fortunately, for such diseases, effective medicine exists [nitrates, statins, β-blockers, calcium channel blockers, angiotensin-converting-enzyme inhibitors, oral antiplatelet medicines, or anticoagulants (blood thinners)]. Considering that iciHHV-6+ subjects show increased risk of developing angina and considering the ease by which an iciHHV-6 diagnosis can be made, it can be argued that systematic iciHHV-6 screening should be considered (possibly at birth along with other genetic testing) to identify subjects at greater risks of developing cardiovascular diseases.
Materials and Methods
Subjects.
The institutional ethics review boards of the Centre Hospitalier Universitaire de Quebec Research Center and Sainte-Justine hospital reviewed and approved this study. All participants signed a consent form.
Cohort.
We analyzed a total of 19,597 DNA samples from the CaG cohort consisting of men and women between 40 and 69 y of age, residing in metropolitan areas of the province of Quebec (Montreal, Quebec, Sherbrooke, and Saguenay). A description of the cohort is provided in Description of the CARTaGENE Cohort, and complete details can be found by consulting Awadalla et al. (33).
DNA Isolation and iciHHV-6 Determination.
Blood from participants were stored onto Flinders Technology Associates (FTA) filters at the Genome Quebec facility in Chicoutimi, Quebec. Once obtained, DNA was retrieved from the FTA filter using the prepGem storage card blood reagents according to the manufacturer’s guidelines (Zygem). The eluted DNA was diluted 10-fold with water and subjected to duplex TaqMan qPCR amplification using validated HHV-6– (55) and GAPDH-specific primers and probes. Amplification of GAPDH, a single copy human gene, was used as internal control. Positive and negative controls were included in each run. The DNA, primers, and probes were mixed with an equal volume of 2× Rotor-Gene Multiplex PCR kit (Qiagen) and subjected to 40 cycles of amplification. DNA isolated from known iciHHV-6A+ and iciHHV-6B+ individuals was used as positive controls in every run. A standard curve generated from DNA isolated from an iciHHV-6+ individual was used to estimate the number of HHV-6 copies. Samples with estimated HHV-6 copy number ≥0.5 per cell were initially considered positive. A total of 115 samples were identified as iciHHV-6+. These samples were reanalyzed by TaqMan qPCR for HHV-6A or HHV-6B determination (56). All these samples were reanalyzed by ddPCR, as previously described (10). Of the 115 samples initially identified, 113 were confirmed as iciHHV-6+ by ddPCR and considered for analyses.
Telomere Length Analysis.
Overall telomere length analysis was determined by qPCR as previously described (37). Subjects were individually aged matched at a 1:1 ratio for telomere length analyses between iciHHV-6− and iciHHV-6+ individuals. For analyses involving subjects with angina, iciHHV-6− and iciHHV-6+ subjects were aged matched individually at a 3:1 ratio, respectively.
Statistical Analyses.
A χ2 test was used to compare the distribution of sex and age between iciHHV-6− and iciHHV6+ subjects. A one-way χ2 goodness-of-fit test was used to compare the distribution of sex and age among iciHHV6+ subjects to those of the general population.
Two-way ANOVA was used to determine the effects of iciHHV6, sex, and their interaction on continuous outcomes. Different variances for each group were estimated when variances were heterogeneous. P values were adjusted using Holm–Bonferroni correction for multiple comparisons (36 comparisons for each; Table S2). Means and SEs are presented for each combination of iciHHV6 and sex, as well as by iciHHV6.
Frequency and proportions are presented for each binary outcome according to sex and iciHHV6. Two-way logistic regression was used to determine the effects of iciHHV6, sex, and their interaction. The prevalence of disease was determined at the time of blood donation. In presence of quasi-separation, Firth bias correction was used (57). P values were adjusted using Holm–Bonferroni correction for multiple comparisons (54 comparisons for sex and interaction and 56 comparisons for iciHHV-6; Table 3 and Table S3). ORs and their 95% CI are presented for iciHHV6+ effect. Outcomes with significant results were also analyzed adjusting for age, BMI, systolic blood pressure, and percent body fat (Table 4).
Comparison of telomere lengths and ages between iciHHV-6− and iciHHV-6+ subjects was determined using a t test. Analysis of telomere lengths among age-matched iciHHV-6− subjects, iciHHV-6− subjects with angina, and iciHHV-6+ subjects with angina was determined using one-way ANOVA followed by the Bonferroni multiple comparison test (three comparisons).
P < 0.05 was considered significant. All statistical analyses were performed with SAS, version 9.3 (SAS Institute Inc., Cary, NC).
Supplementary Material
Acknowledgments
We thank the volunteers that participated in the CARTaGENE project and made this study possible. We thank Anne Sophie Julien from the Centre Hospitalier Universitaire de Quebec clinical and evaluative research service for statistical analyses. This study was supported by Canadian Institutes of Health Research Grant MOP_123214 (to L.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1502741112/-/DCSupplemental.
References
- 1.Salahuddin SZ, et al. Isolation of a new virus, HBLV, in patients with lymphoproliferative disorders. Science. 1986;234(4776):596–601. doi: 10.1126/science.2876520. [DOI] [PubMed] [Google Scholar]
- 2.Lopez C, et al. Characteristics of human herpesvirus-6. J Infect Dis. 1988;157(6):1271–1273. doi: 10.1093/infdis/157.6.1271. [DOI] [PubMed] [Google Scholar]
- 3.Yamanishi K, et al. Identification of human herpesvirus-6 as a causal agent for exanthem subitum. Lancet. 1988;1(8594):1065–1067. doi: 10.1016/s0140-6736(88)91893-4. [DOI] [PubMed] [Google Scholar]
- 4.Zerr DM, et al. Clinical outcomes of human herpesvirus 6 reactivation after hematopoietic stem cell transplantation. Clin Infect Dis. 2005;40(7):932–940. doi: 10.1086/428060. [DOI] [PubMed] [Google Scholar]
- 5.Luppi M, et al. Three cases of human herpesvirus-6 latent infection: Integration of viral genome in peripheral blood mononuclear cell DNA. J Med Virol. 1993;40(1):44–52. doi: 10.1002/jmv.1890400110. [DOI] [PubMed] [Google Scholar]
- 6.Osterrieder N, Kamil JP, Schumacher D, Tischer BK, Trapp S. Marek’s disease virus: From miasma to model. Nat Rev Microbiol. 2006;4(4):283–294. doi: 10.1038/nrmicro1382. [DOI] [PubMed] [Google Scholar]
- 7.Morissette G, Flamand L. Herpesviruses and chromosomal integration. J Virol. 2010;84(23):12100–12109. doi: 10.1128/JVI.01169-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daibata M, Taguchi T, Sawada T, Taguchi H, Miyoshi I. Chromosomal transmission of human herpesvirus 6 DNA in acute lymphoblastic leukaemia. Lancet. 1998;352(9127):543–544. doi: 10.1016/S0140-6736(05)79251-5. [DOI] [PubMed] [Google Scholar]
- 9.Gravel A, Sinnett D, Flamand L. Frequency of chromosomally-integrated human herpesvirus 6 in children with acute lymphoblastic leukemia. PLoS ONE. 2013;8(12):e84322. doi: 10.1371/journal.pone.0084322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sedlak RH, et al. Identification of chromosomally integrated human herpesvirus 6 by droplet digital PCR. Clin Chem. 2014;60(5):765–772. doi: 10.1373/clinchem.2013.217240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geraudie B, et al. 2012. Quantitation of human herpesvirus-6A, -6B and -7 DNAs in whole blood, mononuclear and polymorphonuclear cell fractions from healthy blood donors. J Clin Virol 53(2):151–155.
- 12.Arbuckle JH, et al. The latent human herpesvirus-6A genome specifically integrates in telomeres of human chromosomes in vivo and in vitro. Proc Natl Acad Sci USA. 2010;107(12):5563–5568. doi: 10.1073/pnas.0913586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Endo A, et al. Molecular and virological evidence of viral activation from chromosomally integrated human herpesvirus 6A in a patient with X-linked severe combined immunodeficiency. Clin Infect Dis. 2014;59(4):545–548. doi: 10.1093/cid/ciu323. [DOI] [PubMed] [Google Scholar]
- 14.Gravel A, Hall CB, Flamand L. Sequence analysis of transplacentally acquired human herpesvirus 6 DNA is consistent with transmission of a chromosomally integrated reactivated virus. J Infect Dis. 2013;207(10):1585–1589. doi: 10.1093/infdis/jit060. [DOI] [PubMed] [Google Scholar]
- 15.Hill JA, et al. 2015. Prevalence of chromosomally integrated human herpesvirus 6 in patients with human herpesvirus 6-central nervous system dysfunction. Biol Blood Marrow Transplant 21(2):371–373.
- 16.Bodnar AG, et al. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279(5349):349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 17.Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345(6274):458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 18.Hemann MT, Strong MA, Hao LY, Greider CW. The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell. 2001;107(1):67–77. doi: 10.1016/s0092-8674(01)00504-9. [DOI] [PubMed] [Google Scholar]
- 19.Capper R, et al. The nature of telomere fusion and a definition of the critical telomere length in human cells. Genes Dev. 2007;21(19):2495–2508. doi: 10.1101/gad.439107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang Y, et al. Human telomeres that carry an integrated copy of human herpesvirus 6 are often short and unstable, facilitating release of the viral genome from the chromosome. Nucleic Acids Res. 2014;42(1):315–327. doi: 10.1093/nar/gkt840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sahin E, et al. Telomere dysfunction induces metabolic and mitochondrial compromise. Nature. 2011;470(7334):359–365. doi: 10.1038/nature09787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Artandi SE, et al. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature. 2000;406(6796):641–645. doi: 10.1038/35020592. [DOI] [PubMed] [Google Scholar]
- 23.Vulliamy T, Marrone A, Dokal I, Mason PJ. Association between aplastic anaemia and mutations in telomerase RNA. Lancet. 2002;359(9324):2168–2170. doi: 10.1016/S0140-6736(02)09087-6. [DOI] [PubMed] [Google Scholar]
- 24.Yamaguchi H, et al. Mutations in TERT, the gene for telomerase reverse transcriptase, in aplastic anemia. N Engl J Med. 2005;352(14):1413–1424. doi: 10.1056/NEJMoa042980. [DOI] [PubMed] [Google Scholar]
- 25.Wilkie AO, Lamb J, Harris PC, Finney RD, Higgs DR. A truncated human chromosome 16 associated with alpha thalassaemia is stabilized by addition of telomeric repeat (TTAGGG)n. Nature. 1990;346(6287):868–871. doi: 10.1038/346868a0. [DOI] [PubMed] [Google Scholar]
- 26.Mitchell JR, Wood E, Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999;402(6761):551–555. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]
- 27.Qazilbash MH, et al. A new syndrome of familial aplastic anemia and chronic liver disease. Acta Haematol. 1997;97(3):164–167. doi: 10.1159/000203674. [DOI] [PubMed] [Google Scholar]
- 28.Armanios MY, et al. Telomerase mutations in families with idiopathic pulmonary fibrosis. N Engl J Med. 2007;356(13):1317–1326. doi: 10.1056/NEJMoa066157. [DOI] [PubMed] [Google Scholar]
- 29.O’Sullivan JN, et al. Chromosomal instability in ulcerative colitis is related to telomere shortening. Nat Genet. 2002;32(2):280–284. doi: 10.1038/ng989. [DOI] [PubMed] [Google Scholar]
- 30.Calado RT, et al. Constitutional hypomorphic telomerase mutations in patients with acute myeloid leukemia. Proc Natl Acad Sci USA. 2009;106(4):1187–1192. doi: 10.1073/pnas.0807057106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohyashiki JH, et al. Telomere shortening associated with disease evolution patterns in myelodysplastic syndromes. Cancer Res. 1994;54(13):3557–3560. [PubMed] [Google Scholar]
- 32.Pellett PE, et al. Chromosomally integrated human herpesvirus 6: Questions and answers. Rev Med Virol. 2012;22(3):144–155. doi: 10.1002/rmv.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Awadalla P, et al. CARTaGENE Project Cohort profile of the CARTaGENE study: Quebec’s population-based biobank for public health and personalized genomics. Int J Epidemiol. 2013;42(5):1285–1299. doi: 10.1093/ije/dys160. [DOI] [PubMed] [Google Scholar]
- 34.Ward KN, et al. Human herpesvirus 6 chromosomal integration in immunocompetent patients results in high levels of viral DNA in blood, sera, and hair follicles. J Clin Microbiol. 2006;44(4):1571–1574. doi: 10.1128/JCM.44.4.1571-1574.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yusuf S, et al. INTERHEART Study Investigators Effect of potentially modifiable risk factors associated with myocardial infarction in 52 countries (the INTERHEART study): Case-control study. Lancet. 2004;364(9438):937–952. doi: 10.1016/S0140-6736(04)17018-9. [DOI] [PubMed] [Google Scholar]
- 36.Haycock PC, et al. Leucocyte telomere length and risk of cardiovascular disease: Systematic review and meta-analysis. BMJ. 2014;349:g4227. doi: 10.1136/bmj.g4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O’Callaghan NJ, Fenech M. A quantitative PCR method for measuring absolute telomere length. Biol Proced Online. 2011;13:3. doi: 10.1186/1480-9222-13-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Slagboom PE, Droog S, Boomsma DI. Genetic determination of telomere size in humans: A twin study of three age groups. Am J Hum Genet. 1994;55(5):876–882. [PMC free article] [PubMed] [Google Scholar]
- 39.Prusty BK, Krohne G, Rudel T. Reactivation of chromosomally integrated human herpesvirus-6 by telomeric circle formation. PLoS Genet. 2013;9(12):e1004033. doi: 10.1371/journal.pgen.1004033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hubacek P, et al. Prevalence of HHV-6 integrated chromosomally among children treated for acute lymphoblastic or myeloid leukemia in the Czech Republic. J Med Virol. 2009;81(2):258–263. doi: 10.1002/jmv.21371. [DOI] [PubMed] [Google Scholar]
- 41.Leong HN, et al. The prevalence of chromosomally integrated human herpesvirus 6 genomes in the blood of UK blood donors. J Med Virol. 2007;79(1):45–51. doi: 10.1002/jmv.20760. [DOI] [PubMed] [Google Scholar]
- 42.Potenza L, et al. Prevalence of human herpesvirus-6 chromosomal integration (CIHHV-6) in Italian solid organ and allogeneic stem cell transplant patients. Am J Transplant. 2009;9(7):1690–1697. doi: 10.1111/j.1600-6143.2009.02685.x. [DOI] [PubMed] [Google Scholar]
- 43.Tanaka-Taya K, et al. Human herpesvirus 6 (HHV-6) is transmitted from parent to child in an integrated form and characterization of cases with chromosomally integrated HHV-6 DNA. J Med Virol. 2004;73(3):465–473. doi: 10.1002/jmv.20113. [DOI] [PubMed] [Google Scholar]
- 44.Ward KN, Leong HN, Thiruchelvam AD, Atkinson CE, Clark DA. Human herpesvirus 6 DNA levels in cerebrospinal fluid due to primary infection differ from those due to chromosomal viral integration and have implications for diagnosis of encephalitis. J Clin Microbiol. 2007;45(4):1298–1304. doi: 10.1128/JCM.02115-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Doll R. Cohort studies: History of the method. I. Prospective cohort studies. Soz Praventivmed. 2001;46(2):75–86. doi: 10.1007/BF01299724. [DOI] [PubMed] [Google Scholar]
- 46.Willett WC, Colditz GA. Approaches for conducting large cohort studies. Epidemiol Rev. 1998;20(1):91–99. doi: 10.1093/oxfordjournals.epirev.a017975. [DOI] [PubMed] [Google Scholar]
- 47.Chatzidimitriou D, Kirmizis D, Gavriilaki E, Chatzidimitriou M, Malisiovas N. Atherosclerosis and infection: Is the jury still not in? Future Microbiol. 2012;7(10):1217–1230. doi: 10.2217/fmb.12.87. [DOI] [PubMed] [Google Scholar]
- 48.Libby P, Lichtman AH, Hansson GK. Immune effector mechanisms implicated in atherosclerosis: From mice to humans. Immunity. 2013;38(6):1092–1104. doi: 10.1016/j.immuni.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kühl U, et al. Chromosomally integrated human herpesvirus 6 in heart failure: Prevalence and treatment. Eur J Heart Fail. 2015;17(1):9–19. doi: 10.1002/ejhf.194. [DOI] [PubMed] [Google Scholar]
- 50.Schmidt-Lucke C, et al. Interferon beta modulates endothelial damage in patients with cardiac persistence of human parvovirus b19 infection. J Infect Dis. 2010;201(6):936–945. doi: 10.1086/650700. [DOI] [PubMed] [Google Scholar]
- 51.Butt HZ, Atturu G, London NJ, Sayers RD, Bown MJ. 2010. Telomere length dynamics in vascular disease: A review. Eur J Vasc Endovasc Surg 40(1):17–26.
- 52.Saliques S, et al. 2010. Telomere length and cardiovascular disease. Arch Cardiovasc Dis 103(8-9):454–459.
- 53.Crawford JR. HHV-6A and HHV-6B in cancer. In: Flamand L, Lautenschlager I, Krueger G, Ablashi D, editors. Human Herpesviruses HHV-6A, HHV-6B and HHV-7: Diagnostic and Clinical Mangement. 3rd Ed. Elsevier, New York; 2014. pp. 297–310. [Google Scholar]
- 54.Knudson AG., Jr Mutation and cancer: Statistical study of retinoblastoma. Proc Natl Acad Sci USA. 1971;68(4):820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Flamand L, et al. Multicenter comparison of PCR assays for detection of human herpesvirus 6 DNA in serum. J Clin Microbiol. 2008;46(8):2700–2706. doi: 10.1128/JCM.00370-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boutolleau D, et al. Identification of human herpesvirus 6 variants A and B by primer-specific real-time PCR may help to revisit their respective role in pathology. J Clin Virol. 2006;35(3):257–263. doi: 10.1016/j.jcv.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 57.Heinze G, Schemper M. A solution to the problem of separation in logistic regression. Stat Med. 2002;21(16):2409–2419. doi: 10.1002/sim.1047. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.