Abstract
The degree of progressive chronic histological damage is associated with long-term renal allograft survival. In order to identify promising molecular targets for timely intervention, we examined renal allograft protocol and indication biopsies from 120 low-risk pediatric and adolescent recipients by whole-genome microarray expression profiling. In data-driven analysis, we found a highly regulated pattern of adaptive and innate immune gene expression that correlated with established or ongoing histological chronic injury, and also with development of future chronic histological damage, even in histologically pristine kidneys. Hence, histologically unrecognized immunological injury at a molecular level sets the stage for the development of chronic tissue injury, while the same molecular response is accentuated during established and worsening chronic allograft damage. Irrespective of the hypothesized immune or nonimmune trigger for chronic allograft injury, a highly orchestrated regulation of innate and adaptive immune responses was found in the graft at the molecular level. This occurred months before histologic lesions appear, and quantitatively below the diagnostic threshold of classic T-cell or antibody-mediated rejection. Thus, measurement of specific immune gene expression in protocol biopsies may be warranted to predict the development of subsequent chronic injury in histologically quiescent grafts and as a means to titrate immunosuppressive therapy.
Keywords: adaptive immunity, gene expression, histology, innate immunity, kidney transplantation, microarray, protocol biopsies
After renal transplantation, progressive chronic histological damage is observed in protocol biopsies performed at predefined time points after transplantation. Cumulative histological damage is associated with progressive renal graft dysfunction, reflected by a gradual rise in serum creatinine and/or development of proteinuria, and ultimately leads to complete loss of graft function.1–4 This histological damage has several phenotypes and its etiology is multifactorial. Both alloimmune (cellular rejection and antibody-mediated rejection) and non-alloimmune (for example, recurrent or de novo glomerular disease, calcineurin inhibitor nephrotoxicity) phenomena are recognized as major contributors of progressive scarring of the renal allografts, against the background of changing demographics of kidney transplant donors and recipients and the acceptance of lower-quality kidneys for transplantation.5,6
Early detection of chronic tubulointerstitial damage, especially with concomitant inflammation, has been associated with later allograft survival.1,7–9 Early development of subclinical interstitial fibrosis and tubular atrophy could therefore be used as predictive marker for long-term graft outcome.10 However, it has to be emphasized that the real cause of renal allograft loss is most often a specific disease process, and tubulointerstitial damage is often the consequence of these specific disease processes rather than the cause.4,11 Several disease processes can occur concomitantly, and interstitial fibrosis and tubular atrophy represent the cumulative burden of renal allograft injury, irrespective of its etiology.
In human kidney transplantation, using gene expression microarray technology on renal allograft biopsies, the molecular heterogeneity of renal allografts was demonstrated at the time of transplantation,12 at the time of acute rejection,13,14 and after chronic histological damage was established.15–18 In addition, gene expression analysis has been performed in protocol biopsies to investigate the molecular disturbances that accompany progressive chronic histological damage of renal allografts.19–23 In these studies, immune phenomena appear to have a significant role in the progression of chronic histological damage, and it is suggested that progressive histological damage is immune mediated. However, in these previous studies, there was either no information on the histological inflammatory burden in the early protocol biopsies that were used to predict progressive chronic damage,19,20 there were significant differences in chronic tubulointerstitial damage and in graft function at the early protocol biopsy time point,23 or there were differences in both total inflammatory burden and chronic histological damage at the early protocol biopsy time point.22 From these studies, it is unclear whether the immune gene signatures represent merely established injury, rather than future and ongoing injury. A study of gene expression at 1 month after transplantation showed increased expression of immune-related genes in the early biopsy of kidneys with progressive tubulointerstitial damage.21 However, this study did not exclude kidneys that experienced biopsy-proven acute rejection as overt cause of chronic histological damage progression. Interestingly, a recent study has demonstrated that the gene expression profile of early (6 weeks) protocol biopsies reflects mainly peritransplant injury such as delayed graft function, rather than predicting post-transplant histological evolution.24
Nevertheless, in many instances, inexorable chronic tubulo-interstitial injury is noted on the 1-year protocol biopsy,2,6,25,26 in the absence of any clinical or subclinical acute rejection, infection, or vascular detriment and with histologically quiescent early protocol biopsies. The question of what drives progressive chronic histological damage of renal allografts, specifically in the absence of overt post-transplant injury, thus remains largely unanswered. This carefully designed study of serial protocol biopsies performed at prescheduled time points, with simultaneously collated clinical, histological, and transcriptional data sets, was conducted to elucidate the gene expression dynamics associated with established, ongoing, and, most importantly, future histological damage in pediatric and adolescent patients who received good-quality kidneys, without the confounding interference of delayed graft function, graft rejection, or infection. Better insight in the early pathogenesis of chronic renal allograft damage, in the absence of clinically overt causes, could bring forward new targets for timely intervention and prevention of chronic scarring. In addition, prediction of future damage by early gene expression assessment could provide clinical guidance and may ultimately lead to individualized treatment of renal allograft recipients.
RESULTS
Immune cell activation gene expression in chronic allograft injury (Project 1)
In Project 1, a case–control analysis was conducted on 48 protocol biopsies with varying degrees of chronic allograft injury (graded by the Chronic Allograft Damage Index (CADI) score,1 which was used as surrogate end point for graft survival), obtained from 48 different patients, and split equally into a discovery set (protocol biopsies at various time points) and a validation set (protocol biopsies at 24 months after transplantation). Mean recipient age and donor age were 10.9±6.1 years and 33.6±11.0 years, respectively, and 35/48 (73.9%) of the kidneys were obtained from living donors (Table 1 and Figure 1). The distribution of the individual components of the CADI scores in the different biopsy groups is represented in Supplementary Figure S1 online.
Table 1.
Donor–recipient demographics and histology of the 120 biopsies included in the different analyses
| Project 1 |
Projects 2 and 3 |
Project 4 |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Discovery set |
Validation set |
|||||||||||||
| All biopsies (N=120) |
Low CADI | High CADI | Low CADI | High CADI | Stable CADI score over time (nonprogressors) |
Increasing CADI score over time (progressors) |
Low CADIa | High CADIa | Rejectionb | |||||
| Donor-recipient demographics | ||||||||||||||
| Number of patients | 67 | 12 | 12 | 12 | 12 | 12 | 12 | 24 | 24 | 24 | ||||
| Steroid-free/steroid-based | 74/46 | 12/0 | 12/0 | 4/8 | 7/5 | 4/8 | 7/5 | 16/8 | 19/5 | 17/7 | ||||
| Donor gender (M/F) | 65/55 | 5/7 | 8/4 | 9/3 | 6/6 | 9/3 | 6/6 | 14/10 | 14/10 | 17/7 | ||||
| Donor age (years) | 32±11 | 36±10 | 41±9.4 | 25±9.4 | 34±9.7* | 24.7±9.41 | 34.0±9.69* | 30±11 | 37±10* | 33±11 | ||||
| Recipient gender (M/F) | 69/51 | 9/3 | 7/5 | 6/6 | 7/5 | 6/6 | 7/5 | 15/9 | 14/10 | 14/10 | ||||
| Recipient age (years) | 11±5.6 | 13±6.8 | 7.7±5.6* | 14±4.0 | 9.1±6.1* | 13.9±3.97 | 9.10±6.10* | 13.4±5.4 | 8.4±5.8* | 11.3±4.6 | ||||
| Cause of ESRD (1/2/3/4/5/6) | 4/0/2/1/3/2 | 4/1/3/2/2/0 | 3/2/0/2/1/4 | 2/0/3/2/2/3 | 3/2/0/2/1/4 | 2/0/3/2/2/3 | 7/2/2/3/4/6 | 6/1/6/4/4/3 | 5/0/3/7/3/6 | |||||
| Living/deceased | 75/45 | 11/1 | 12/0 | 4/5 | 8/4 | 4/8 | 8/4 | 15/9 | 20/4 | 16/8 | ||||
| Histology of protocol biopsies | ||||||||||||||
| Number of biopsies | 120 | 12 | 12 | 12 | 12 | 12 | 12 | 12 | 12 | 12 | 12 | 24 | 24 | 24 |
| Time after transplantation (months) | 13±16 | 15±7.4 | 14±7.8 | 24 | 24 | 0 | 6 | 24c | 0 | 6 | 24d | 19±7.0 | 19±7.3 | 22±27 |
| Tubulitis score >0 | 27/120 (22%) |
0/12 (0%) |
1/12 (8.3%) |
0/12 (0%) |
2/12 (17%) |
0/12 (0%) |
0/12 (0%) |
0/12 (0%) |
0/12 (0%) |
0/12 (0%) |
2/12 (17%) |
0/24 (0%) |
3/24 (13%) |
24/24 (100%)* |
| Interstitial inflammation score >0 | 28/120 (23%) |
0/12 (0%) |
1/12 (8.3%) |
0/12 (0%) |
2/12 (17%) |
0/12 (0%) |
0/12 (0%) |
0/12 (0%) |
0/12 (0%) |
1/12 (8.3%) |
2/12 (17%) |
0/24 (0%) |
3/24 (13%) |
24/24 (100%)* |
| Vasculitis score >0 | 4/120 (33%) |
0/12 (0%) |
0/12 (0%) |
0/12 (0%) |
0/12 (0%) |
0/12 (0%) |
0/12 (0%) |
0/12 (0%) |
0/12 (0%) |
0/12 (0%) |
0/12 (0%) |
0/24 (0%) |
0/24 (0%) |
4/24 (17%)* |
| IF/TA grade >1 | 24/120 (20%) |
0/12 (0%) |
8/12 (67%)* |
1/12 (8.3%) |
9/12 (75%)* |
0/12 (0%) |
0/12 (0%) |
1/12 (8.3%) |
0/12 (0%) |
2/12 (17%) |
9/12 (75%)* |
1/24 (4%) |
17/24 (71%)* |
4/24 (17%) |
| Arteriolar hyalinosis (present) | 7/120 (5.8%) |
2/12 (17%) |
2/12 (17%) |
0/12 (0%) |
1/12 (8.3%) |
1/12 (8.3%) |
0/12 (0%) |
0/12 (0%) |
1/12 (8.3%) |
0/12 (0%) |
1/12 (8.3%) |
2/24 (8%) |
3/24 (13%) |
0/24 (0%) |
| Intimal vascular thickening (present) | 33/120 (27%) |
4/12 (33%) |
9/12 (75%)* |
3/12 (25%) |
6/12 (50%) |
1/12 (8.3%) |
1/12 (8.3%) |
3/12 (25%) |
0/12 (0%) |
4/12 (33%) |
6/12 (50%) |
7/24 (29%) |
13/24 (63%)* |
5/24 (21%) |
| Glomerulosclerosis (present) | 38/120 (32%) |
3/12 (25%) |
8/12 (67%)* |
1/12 (8.3%) |
8/12 (67%)* |
1/12 (8.3%) |
0/12 (0%) |
1/12 (8.3%) |
3/12 (25%) |
2/12 (17%) |
8/12 (67%)* |
4/24 (18%) |
16/24 (67%)* |
12/24 (50%)* |
| Remuzzi score | NA | NA | NA | NA | NA | 0.25±0.45 | NA | NA | 0.75±0.87 | NA | NA | NA | NA | NA |
| CADI score | 3.5±2.9 | 2.4±1.0 | 6.7±1.0* | 1.8±1.4 | 7.3±1.6* | 0.25±0.45 | 0.75±0.75 | 1.8±1.4 | 1.00±1.21 | 3.17±1.80* | 7.3±1.6* | 2.1±1.2 | 7.0±1.3* | 5.6±2.4* |
| Graft functione | ||||||||||||||
| Schwartz GFR at day 0 | 13.0±7.51 | 11.2±3.59 | 13.7±6.2 | 12.1±9.65 | 14.1±8.14 | 12.1±9.65 | 14.1±8.14 | 11.6±7.13 | 13.9±7.06 | 13.4±5.52 | ||||
| Schwartz GFR at month 6 | 103±30.7 | 94.4±23.8 | 101±21.9 | 91.4±27.0 | 117±30.5* | 91.4±27.0 | 117±30.5* | 92.9±24.9 | 109±27.2 | 104±39.2 | ||||
| Schwartz GFR at month 12 | 105±30.1 | 95.4±19.4 | 98.6±17.3 | 109±31.9 | 117±28.1 | 109±31.9 | 117±28.1 | 103±27.0 | 108±24.7 | 81.9±31.8* | ||||
| Schwartz GFR at month 24 | 95.7±30.8 | 92.9±24.4 | 86.4±15.8 | 105±33.5 | 102±20.6 | 105±33.5 | 102±20.6 | 100±30.3 | 95.5±19.9 | 65.9±37.4* | ||||
Abbreviations: CADI, Chronic Allograft Damage Index; ESRD, end-stage renal disease; GFR, glomerular filtration rate; IF/TA, interstitial fibrosis/tubular atrophy; NA, not applicable.
ESRD (1/2/3/4/5/6): 1, glomerulonephritis, 2, polycystic kidney disease, 3, renal dysplasia, 4, reflux nephropathy, 5, obstructive uropathy, 6=other or unknown.
P<0.05 versus the comparison group.
Compilation of discovery sets and validation sets from Project 1.
Obtained at time of graft dysfunction in a different patient cohort, independent of the patients included in the other projects.
The same data set as the low CADI group of the validation set in Project 1.
The same data set as the high CADI group of the validation set in Project 1.
Schwartz GFR (ml/min per 1.73 m2).49
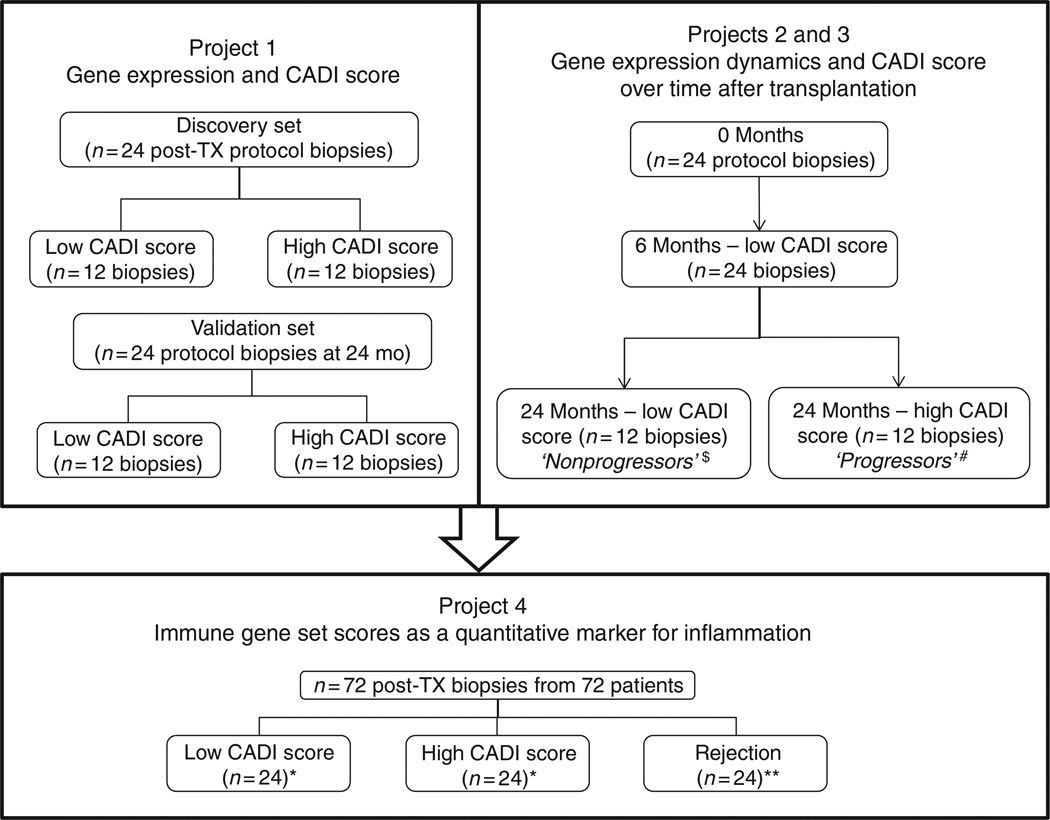
Figure 1. Classification of the 120 renal allograft biopsy samples included in the current study.
*Compilation of discovery sets and validation sets from Project 1. **Patients different from those in Projects 1–3. CADI, Chronic Allograft Damage Index; mo, months; TX, transplantation. $The same data set as the low CADI group of the validation set of Project 1. #The same data set as the high CADI group of the validation set of Project 1.
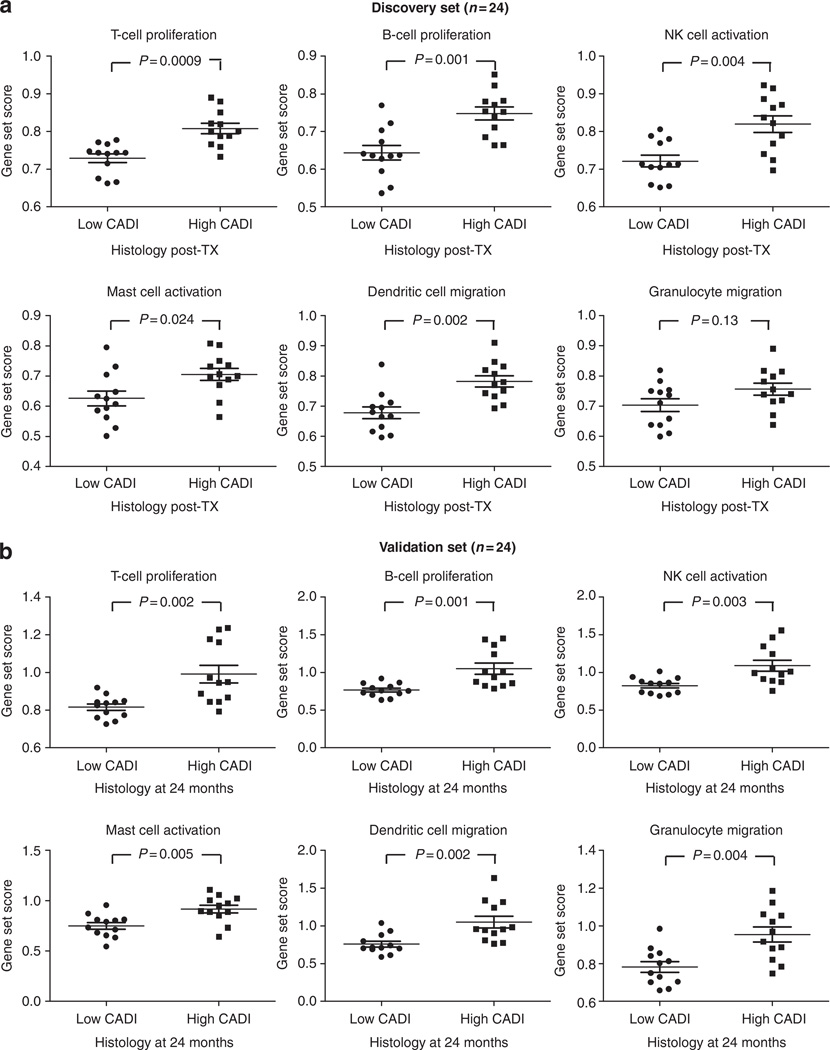
In the discovery set, 864 probe sets were significantly differently expressed between biopsies with high CADI score and biopsies with low CADI score (Table 2); 397 of these were significantly upregulated in the high CADI score group (340 unique genes; q<0.05). In biological function assessment of these 397 genes using Ingenuity Pathway Analysis (IPA), a highly significant enrichment of cell-specific immune response genes was observed (P=7.99 × 10−43), with significant enrichment of T-lymphocyte (P=1.06 × 10−23) and B-lymphocyte (P=1.01 × 10−11) proliferation transcripts, transcripts involved in natural killer (NK) cell activation (P=4.75 × 10−7), mast cell activation (P=4.95 × 10−8), dendritic cell migration (P=6.65 × 10−6), and granulocyte migration (P=5.64 × 10−11; Table 2 and Figure 2a).
Table 2.
Number of probe sets differently expressed in SAM (q<0.05)a, together with the pathway enrichment P-values of the upregulated transcripts, obtained from IPAb
| Enrichment P-value (IPA) of upregulated genes (and the number of differently expressed upregulated individual genes that participate in the IPKB function/total number of differently expressed upregulated individual genes in each analysis) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Probe sets differentially expressed (N)a |
Probe sets upregulated (N)a |
Mapped individual genes of upregulated probe sets (N)a |
Immune response (overall) |
T-cell proliferation |
B-cell proliferation |
NK cell activation |
Mast cell activation |
Dendritic cell migration |
Granulocyte migration |
|||
| Project 1 | CADI score discovery set |
12 High vs. 12 low CADI (N=24 biopsies) |
864 | 397 | 340 | 7.99 × 10−43 (85/340) |
1.06 × 10−23 (46/340) |
1.01 × 10−11 (21/340) |
4.75 × 10−7 (11/340) |
4.95 × 10−8 (9/340) |
6.65 × 10−6 (9/340) |
5.64 × 10−11 (23/340) |
| CADI score validation set |
12 High vs. 12 low CADI (N=24 biopsies) |
804 | 703 | 576 | 2.77 × 10−74 (140/576) |
1.20 × 10−29 (64/576) |
6.03 × 10−18 (33/576) |
3.15 × 10−10 (17/576) |
1.71 × 10−8 (11/576) |
1.26 × 10−7 (13/576) |
5.63 × 10−15 (34/576) |
|
| Project 2 | CADI score dynamics over time |
12 Progressors vs. 12 nonprogressors (N=72 biopsies) |
601 | 601 | 539 | 1.35 × 10−91 (153/539) |
1.03 × 10−31 (65/539) |
1.89 × 10−21 (36/539) |
1.63 × 10−12 (19/539) |
4.94 × 10−11 (13/539) |
3.57 × 10−21 (25/539) |
2.19 × 10−28 (48/539) |
| Project 3 | CADI score prediction at six months |
12 Progressors vs. 12 non progressors (N=24 biopsies) |
92 | 92 | 92 | 1.03 × 10−4 (11/92) |
3.21 × 10−3 (6/92) |
3.63 × 10−3 (4/92) |
3.82 × 10−2 (2/92) |
NS | 1.06 × 10−3 (3/92) |
NS |
Abbreviations: CADI, Chronic Allograft Damage Index; IPA, Ingenuity Pathway Analysis; IPKB, Ingenuity Pathway Knowledge Base; NK, natural killer; NS, nonsignificant; SAM, significance analysis of microarray.
The probe set lists are obtained using SAM. In Projects 1 and 3, two-class unpaired SAM was used. In Project 2, we used two-class unpaired time course analysis.
The enrichment analysis showed a highly significant overrepresentation of immune genes, involved in both innate and adaptive immunity. Also scattered nonimmune biological functions were significantly enriched; however, without clear relevance for organ transplantation (Supplementary Table S1–S4 online).
Figure 2. Gene expression and CADI score (Project 1).
After significance analysis of microarrays, comparing gene expression of biopsies with high versus low CADI score, the probe set lists were analyzed using the Ingenuity Pathway Analysis program (Ingenuity Systems) and demonstrated highly significant enrichment of immune genes. The immune gene set scores represented in the graphs were calculated as the geometric mean of fold changes (unlogged values) across all probe sets (not only the significant probe sets) within the Ingenuity Pathways Knowledge Base (IPKB) lists for the different immune cell functions (Supplementary Table S5 online). (a) Comparison of the immune gene set scores between biopsies with high versus low CADI scores in a discovery set (n=24) and (b) in a validation set (n=24). P-values represented in the graphs were calculated using Mann–Whitney tests. The error bars represent mean±standard error of the mean. CADI, Chronic Allograft Damage Index; NK, natural killer; TX, transplantation.
In the validation set, again comparing biopsies with high versus low CADI score, we found differential expression of 804 probe sets (703 probe sets upregulated; q<0.05). Pathway analysis confirmed a highly significant enrichment of immune response genes and enrichment of the different immune cell types (Table 2 and Figure 2b).
Although enrichment analysis of the 397 and 703 probe set lists showed remarkable overrepresentation of immunity-related functions, scattered nonimmune biological functions were also significantly enriched, without clear immediate relevance for organ transplantation (Supplementary Tables S1 and S2 online).
Schwartz glomerular filtration rate (GFR; ml/min per 1.73 m2) was not associated with these gene set scores, neither in the discovery set nor in the validation set (data not shown).
Gene expression dynamics and CADI score progression over time (Project 2)
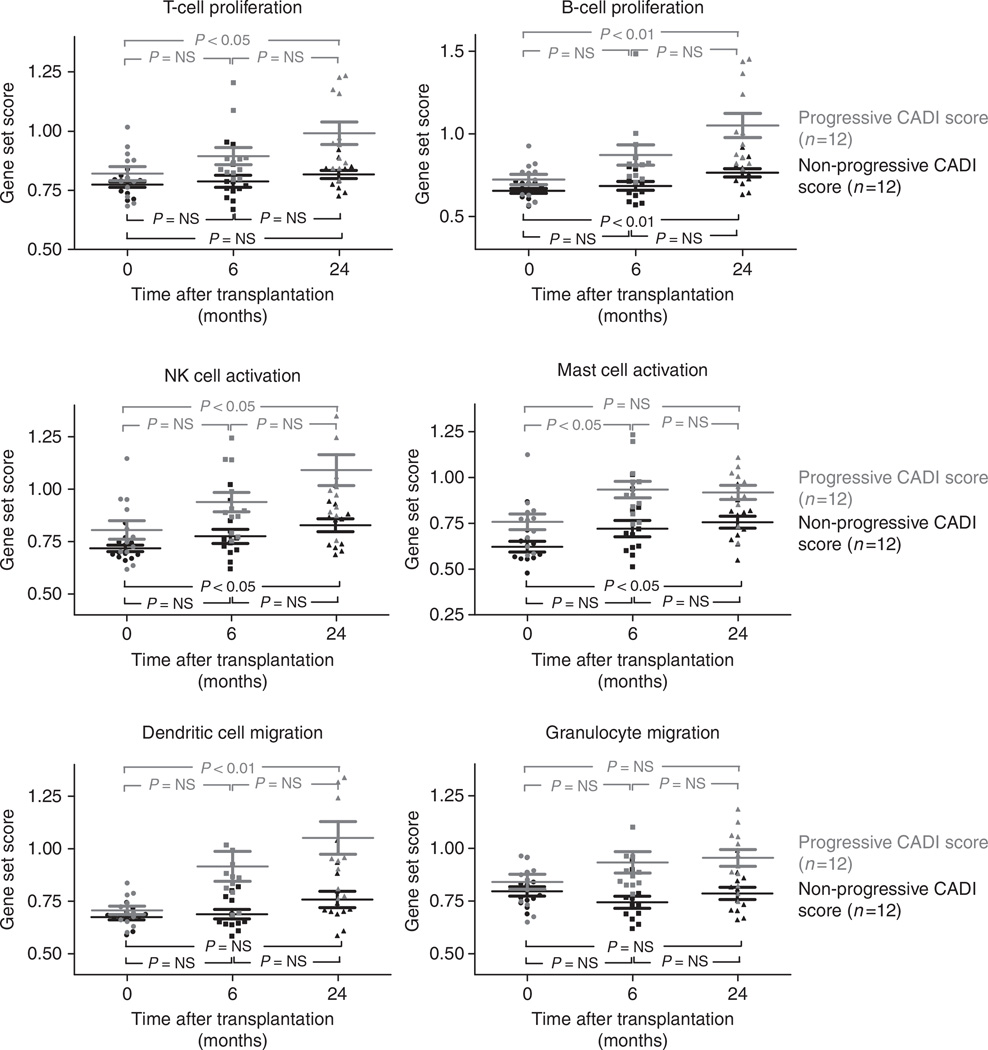
Project 2 consisted of a longitudinal analysis of 72 serial protocol biopsies from 24 patients, each of whom underwent biopsies at implantation, at 6 and 24 months. None of these patients experienced Banff-grade acute T-cell-mediated rejection within the first 2 years after transplantation. To mitigate the confounding influence of donor quality and extended ischemia time on gene expression changes, previously shown by our group,12 the sample set was carefully selected such that kidney graft quality was excellent at implantation, with minimal chronic damage and a mean Remuzzi score27 of only 0.63±0.81 (median 0, range 0–2). Despite the pristine tissue quality at engraftment, as also previously shown by our group,28 the effect of increasing post-transplant time, even in the absence of interval acute rejection, results in a significant increase in chronic histological damage in all histological compartments. At 24 months after transplantation, 50% of the group (12 patients; 36 biopsies) had significantly greater histological progression, scored by the incremental CADI score, and for analysis purposes, have been labeled the progressor group, and gene expression data in this group have been compared with the remainder of the patients, who, for purposes of analysis, have been called the nonprogressor group. At 6 months, the progressors had higher mean CADI scores than the nonprogressors (3.2±1.8 vs. 0.75±0.75; P=0.0003), and similar trends were seen for mean CADI scores for the 24-month biopsies (7.3±1.6 in progressors, in contrast to 1.8±1.4 in nonprogressors; P<0.0001). No other Banff qualifiers were significantly different at any time point (Table 1 and Supplementary Figure S2 online).
In an unpaired timeline analysis (signed area test) of the serial biopsies (0, 6, and 24 months after transplantation), we found significantly different evolution in the expression of 601 probe sets that mirrored the histological evolution of chronic injury over time (q<0.05). Using IPA, the histological evolution of chronic histological injury in the graft, even in the absence of any histological evidence of acute rejection, had a strong signal for an activation state of different immune cells (P=1.35 × 10−91) involved in T- and B-cell proliferation (P=1.03 × 10−31 and 1.89 × 10−21, respectively), NK cell (P=1.63 × 10−12 ), mast cell (P=4.49 × 10−11), dendritic cell activation (P=3.57 × 10−21), and granulocyte migration (P=2.19 × 10−28; Table 2 and Figure 3). Although enrichment analysis of the 601 probe set list showed remarkable overrepresentation of immunity-related functions, scattered nonimmune biological functions were significantly enriched, without clear immediate relevance for organ transplantation (Supplementary Table S3 online).
Figure 3. Gene expression dynamics and CADI score over time after transplantation (Project 2).
Enrichment analysis of the probe set list that was generated from unpaired time course analysis (signed area test on 72 serial protocol biopsies at 0, 6, and 24 months after transplantation) comparing progressors (N=12 patients) with nonprogressors (N=12 patients) showed highly significant enrichment of immune genes involved in both innate and adaptive immunity. The immune gene set scores represented in the graphs were calculated as the geometric mean of fold changes (unlogged values) across all probe sets (not only the significant probe sets) within the Ingenuity Pathways Knowledge Base lists for the different immune cell functions (Supplementary Table S5 online). P-values represented in the graphs were calculated using repeated measures analysis of variance. The error bars represent mean±standard error of the mean. The significant P-values of the enrichment analysis from the comparison between progressors and non-progressors are given in Table 2 (Project 2). CADI, Chronic Allograft Damage Index; NK, natural killer; NS, nonsignificant.
There was no correlation between gene set scores at implantation and post-transplantation graft function. Gene set scores obtained at 6 months correlated significantly but positively with Schwartz GFR (ml/min per 1.73 m2) at 6 months (P < 0.05 for T-cell proliferation, B-cell proliferation, NK cell activation, granulocyte migration, and dendritic cell migration). Gene set scores at 6 months were, however, not associated with later graft function. There was no correlation between gene set scores obtained at 24 months and graft function at this time point (data not shown).
Association of 6 months gene expression with CADI score at 24 months (Project 3)
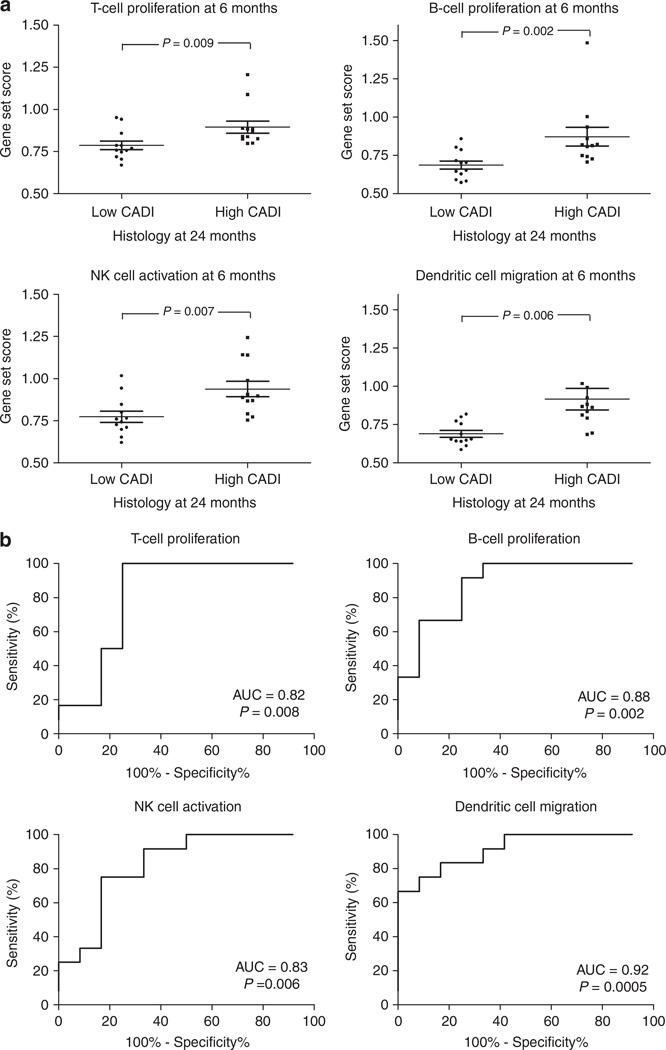
We next interrogated the gene expression changes in the 6-month biopsies as a means to predict the evolution of histological injury in the same graft, when sampled by biopsy 18 months later. We compared gene expression of 6-month biopsies of the progressor group with gene expression of 6-month biopsies of the nonprogressor group and performed an associative significance analysis of microarray (SAM) analysis. A total of 92 probe sets were significantly over-expressed in the 6-month biopsies of the progressor group (q<0.05) and again, by IPA, there was a significant enrichment of similar immune genes (P=0.0001), having a role in T- and B-cell proliferation (P=0.003 and 0.003, respectively), NK cell activation (P=0.04), and migration of dendritic cells (P=0.001; Table 2 and Figure 4a). Although enrichment analysis of the 92 probe set list again showed overrepresentation of immunity-related functions, scattered nonimmune biological functions were significantly enriched, without clear immediate relevance for organ transplantation (Supplementary Table S4 online).
Figure 4. Association of 6 months gene expression with CADI score at 24 months (Project 3).
(a) After significance analysis of microarrays, comparing gene expression of 6-month biopsies of progressors versus nonprogressors, the probe set list was analyzed using the Ingenuity Pathway Analysis program (Ingenuity Systems) and demonstrated significant enrichment of immune genes. The immune gene set scores represented in the graphs were calculated as the geometric mean of fold changes (unlogged values) across all probe sets (not only the significant probe sets) within the Ingenuity Pathways Knowledge Base lists for the different immune cell functions (Supplementary Table S5 online). P-values represented in the graphs were calculated using Mann–Whitney tests. The error bars represent mean±standard error of the mean. (b) Receiver operating characteristic-curve analysis demonstrates good accuracy of the 6-month immune gene set scores for prediction of subsequent damage. AUC, area under the curve; CADI, Chronic Allograft Damage Index; NK, natural killer.
There was a significant association between these immune gene set scores in these 6-month biopsies and future histological damage progression (Figure 4a). Classification models can be built with area under the receiver operating characteristic curves greater than 0.82, for predicting the downstream evolution of chronic histological allograft injury by these gene set scores (Figure 4b).
Immune gene set scores as quantitative parameters for inflammation (Project 4)
Given the strong immune cell activation signal associated with the detection and evolution of chronic histological injury alone, we next included gene expression analysis for indication biopsies with histological evidence of Banff-graded acute rejection to compare and contrast the extent of immune activation signals between acute rejection and chronic graft injury without acute rejection. For this purpose, we conducted Project 4, which consisted of 72 unique biopsies; 48 biopsies from patients with chronic graft injury (from Project 1) and 24 for-cause biopsies with Banff-grade acute T-cell-mediated rejection. As expected, histological parameters were significantly different between the 48 rejection-free protocol biopsies and the 24 biopsies with acute rejection (Table 1 and Supplementary Figure S3 online).
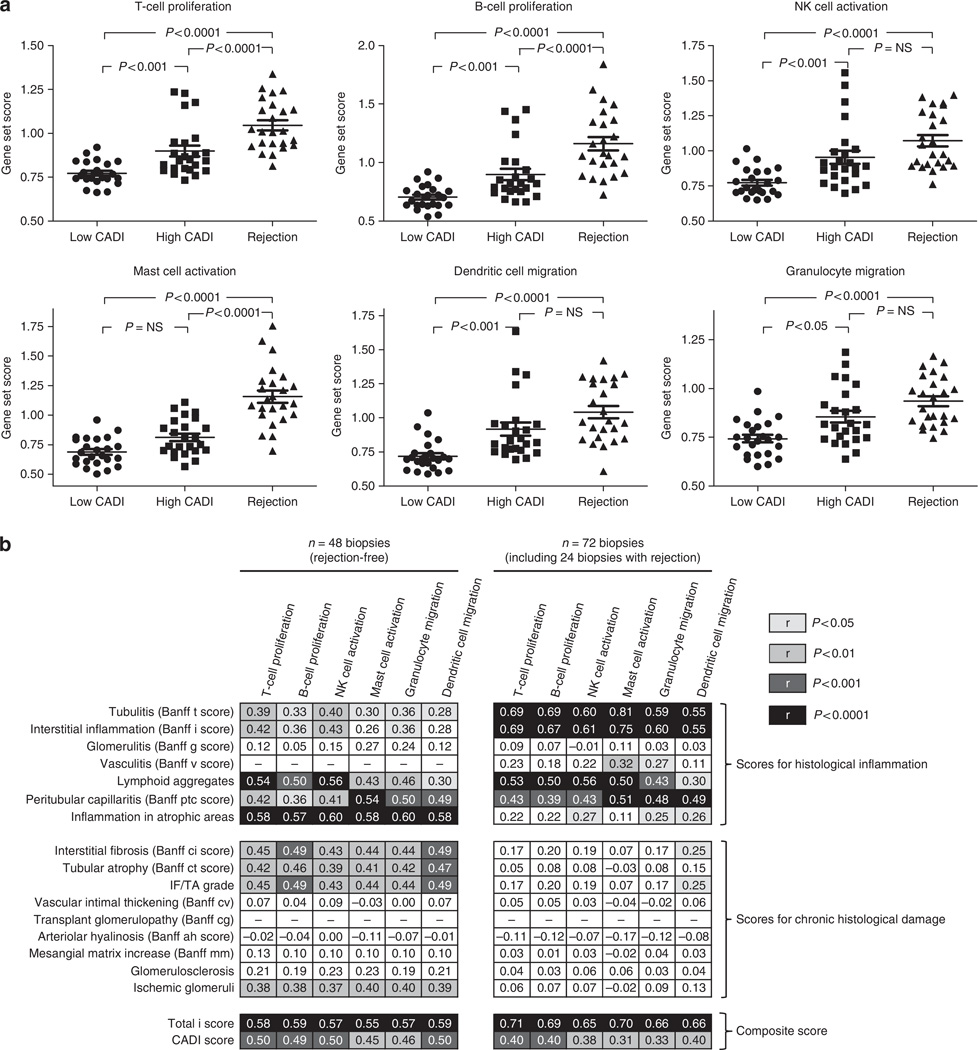
The immune gene set scores for chronic histological injury signals, developed in the earlier Projects, demonstrate that there is a highly regulated environment of subclinical inflammation in the graft that associates with the progression of this injury. We wanted to elucidate whether immune gene set scores are also increased in clinically relevant inflammation (Banff-graded T-cell-mediated acute rejection). We noted significantly higher immune gene set scores in acute rejection biopsies compared to biopsies with low CADI score. The immune gene set scores in acute rejection biopsies were comparable with or even higher than the gene set scores observed in biopsies with a high CADI score (Figure 5a). The correlation between the different immune gene set scores and the different histological lesions is shown in Figure 5b. The correlation was best for total i-score, which represents the total inflammatory burden in the graft. These data suggest that the extent of graft inflammation, either clinical or subclinical, follows the same paradigm of inflammatory cell-specific signals, and the persistence of this injury, undetected by histological evaluations, programs the host tissue to inexorable cellular injury and fibrosis. It is thus clear that the gene expression signature of clinical acute cellular rejection with tubulitis is very similar to the signature of progressive chronic tissue damage in the absence of tubulitis, and the extent of inflammatory burden may be a threshold effect that could be a factor that determines the development of tubulitis and clinical acute rejection. Throughout the study (N=120 biopsies), there was no association between steroid use and intragraft immune gene expression.
Figure 5. Immune gene set scores as quantitative marker for inflammation (Project 4).
(a) There was a gradual association between the CADI score/T-cell-mediated rejection and the different immune gene set scores, as calculated from the Ingenuity Pathways Knowledge Base. Therefore, these immune gene set scores not only represent a good marker for ongoing and future chronic histological damage, but also offer a quantifiable measure for other types of inflammation in the graft. P-values represented in the graphs were calculated using Kruskall–Wallis tests. The error bars represent mean±standard error of the mean. (b) Spearman correlation analysis between histological features and gene set scores in the 48 rejection-free biopsies (left panel) and the 72 biopsies (48 rejection-free biopsies + 24 biopsies with T-cell-mediated rejection) included in Project 4. The numbers given in the boxes represent correlation coefficients; the filled colors correspond to the statistical significance of the correlation. Even in the absence of rejection (left panel), the correlation between the scores of histological inflammation and inflammatory gene set scores was high (especially for inflammation in atrophic areas and the total i-score), which demonstrates that the gene sets capture subtle inflammation. When rejection biopsies were included in the correlation analysis (right panel), interstitial inflammation and tubulitis correlated highly significantly with the gene set scores. The gene set scores therefore represent a global assessment of the inflammatory burden (without information of the exact anatomical localization of the cells that express the inflammatory genes) and a quantitative marker for ongoing inflammation, from moderate subclinical inflammation associated with chronic histological damage to marked inflammation of Banff-grade acute T-cell-mediated rejection. CADI, Chronic Allograft Damage Index; IF/TA, interstitial fibrosis/tubular atrophy; NK, natural killer; NS, nonsignificant.
DISCUSSION
Slowly progressive chronic histological damage associates with long-term renal allograft survival. This injury is accelerated in grafts that undergo acute rejection; however, in the absence of the injury of interval allograft rejection, the driving molecular forces behind progressive histological damage are not known.
This paper reports on an in-depth study on the correlation between the subclinical histological evolution of renal allografts and whole-genome gene expression in renal allograft recipients with low immunological risk. By access to serially collected protocol biopsy samples, with centralized, blinded semiquantitative histological scores, a highly selected set of biopsies was used for interrogation of the molecular processes that associate with chronic histological injury, in the absence of any graft dysfunction. We used microarray technology applied on kidney biopsy tissue samples obtained at preset time points in the first 2 years after transplantation. The very strict selection of only pristine kidneys at time of transplantation without any donor pathology, the absence of delayed graft function, the exclusion of patients with allograft rejection in the main analysis, the exclusion of patients with de novo or recurrent glomerular disease and donor-specific antibodies, and in-depth statistical analysis integrating pathology scores, expression measurements, and gene set scores allowed us to uncover the molecular mechanisms associated with established histological injury and to predict histological damage progression in the early post-transplant period.
Using a cross-validated genomewide transcript analytical approach, we confirm the highly significant association of established chronic histological damage with the coordinated regulation of adaptive and innate immune response genes,16,22,29,30 in the absence of Banff-grade acute T-cell-mediated or antibody-mediated rejection. Despite the absence of rejection in these protocol biopsies, the expression profile of the biopsies with established chronic damage is remarkably similar to biopsies with acute rejection. The highly significant correlation between the immune cell-associated transcript scores and the Banff interstitial inflammation and tubulitis scores, inflammation in atrophic areas and especially the recently proposed31 total i-score in our study, suggest that it is this cellular infiltration and inflammation that largely explains the gene expression profile variability. However, the finding that the correlation between immune cell infiltrates (inflammation in atrophic areas and total i-score) and immune cell transcript scores remains highly significant in the absence of inflammation in viable tissue (Banff i- and t-scores equal to zero; data not shown) demonstrates that microarray analysis performed on whole tissue cores is able to detect even subtle inflammation, below the diagnostic threshold of histological diagnosis as defined by the current Banff classification. This is consistent with previous studies also showing the good correlation between the total i-score and T-cell-associated transcripts, γ-interferon-associated transcripts, B-cell transcripts, and mast cell increase,11,17,32 and highlights the fact that the gene expression of tissue biopsies represents a global assessment of the inflammatory burden within a biopsy.
Most importantly, our study shows that the upregulation and overrepresentation of immunity genes is not solely a feature of biopsies with established chronic damage. Instead, upregulation of adaptive (T- and B-cell signatures) and innate immune cell transcripts (dendritic cell and NK cell transcripts) is already present in biopsies of kidneys several months before chronic histological damage occurs. This illustrates that the immune gene set scores used in the current study represent a quantifiable parameter of total inflammatory burden in a biopsy with a very wide dynamic range: from subtle inflammatory activity before development of histological lesions, over moderate subclinical inflammatory gene expression associated with ongoing and established chronic histological damage, to marked overexpression of inflammatory genes in biopsies with Banff-grade acute T-cell-mediated rejection. The association of later histological damage with molecular disturbances of immune genes in early protocol biopsies was also seen in adult renal transplantation programs.20–23 Although these previous studies adequately illustrate the importance of immune-related gene expression tracking together with progression of chronic histological damage, our study is the first to illustrate the association of total inflammatory burden in pristine early protocol biopsies with progression of chronic tubulointerstitial damage, below the current histological diagnostic thresholds and in the absence of acute rejection episodes. The studies in adult renal transplant recipients either included patients with interval acute rejection21 or predictor biopsies with significant inflammation or chronic damage,22,23 which could have influenced the results considerably as the gene expression signatures in these studies are essentially derived from comparisons between biopsies with and without histologically evident damage.
The coregulation of both innate and adaptive immune genes illustrates the complex interplay between innate and adaptive immune responses in a transplantation setting.33 These subtle inflammatory changes in renal allografts are likely very important, as an association between T-lymphocyte-mediated inflammation in scarred areas and graft outcome was demonstrated previously,9 which suggests continuing effects of these inflammatory infiltrates on the scarring process. Importantly, however, especially in the light of possible therapeutic interventions, the current study supports the growing theory that other immune cell types such as B cells,13,34,35 NK cells,36–38 dendritic cells,39–41 mast cells,17,42,43 and granulocytes44,45 are involved in chronic renal allograft damage and may be driving the immunological processes after transplantation, potentially by their implication in antigen presentation, in the secretion of inflammatory cytokines, and in the regulation of T cells.
The preemptive finding of both cell-specific enrichment for a panel of immune cells and immune activation signatures in the graft suggests that both infiltration and activation of the infiltrating cells are critical triggers for the development and progression of alloreactive chronic injury. Nevertheless, in this human study, it is not possible to distinguish the relative impact of alloreactivity from nonspecific injury mechanisms in the graft. We emphasize that the CADI score used in our study does not help to differentiate between the different pathogenic processes that have a role in the progression of the histological damage, as the lesions included in the CADI score are all nonspecific. After the description of these gene signatures in nonspecific chronic histological damage progression and in acute cellular rejection, as was done in the current study, it will be necessary to study the potential usefulness of these immune gene signatures in other inflammatory disease processes such as de novo or recurrent glomerular disease, polyomavirus nephropathy, antibody-mediated rejection and pyelonephritis, and in noninflammatory renal pathology such as acute tubular necrosis or postrenal obstruction. However, from the current study, it could be hypothesized that much of the progression of chronic damage is inflammation mediated, irrespective of the specific disease process. The immune gene signatures could potentially be used as nonspecific but very sensitive markers for future and ongoing subclinical injury. Whether this is sufficient to yield a useful clinical biomarker to guide therapeutic decisions is another question that can only be answered through further refinement and simplification of the gene expression biomarker, and extensive validation studies.
Although it is tantalizing to hypothesize that increased immunosuppression in the patients with high immune gene set scores could prevent or even reverse progressive chronic histological damage, further studies in animals and controlled immunosuppression trials in humans are suggested and necessary to elucidate whether the spectrum of histological damage responds to increased or, more likely, targeted immunosuppression. This study was performed in pediatric and young adult renal allograft recipients with low immunological risk. Moreover, these patients were treated with the combination of tacrolimus and mycophenolate mofetil, the most common immunosuppressive drug combination in current clinical practice. Therefore, additional validation of the intragraft injury responses should be done in patients with higher immunological risk, in patients treated with other immunosuppressive drug regimens and in adult recipients, as the mechanisms contributing to chronic histological damage could differ between populations, both in terms of risk for subclinical alloimmune problems as in terms of nonimmune injury such as relative renal hypoperfusion in adult-sized kidneys transplanted into infant recipients.28
Clinical follow-up time of the current study is too short to make any conclusion about the impact of early molecular disturbance or histological damage progression on long-term outcome and hard clinical end points. Much larger studies with longer clinical follow-up time and a cross-sectional trial design will be needed to address this issue. The Schwartz GFR (ml/min per 1.73 m2) did not correlate with the molecular disturbances, apart from a positive correlation of higher Schwartz GFR with increased expression of immune genes at 6 months after transplantation. Although this finding was not consistent throughout the study projects, this significant correlation could be explained by the relatively high Schwartz GFR in the youngest patients, who not only have the best Schwartz GFR but also most histological damage, likely through chronic ischemia secondary to size-discrepant kidney transplantation.28
The careful selection of patients for inclusion in the current study was necessary to avoid interference of clinical factors contributing to chronic allograft damage, as it was confirmed recently that gene expression in the earliest phase post-transplantation (at 6 weeks) reflects mainly peritransplant injury, without information of the fate of the kidney transplant in the long term.24 Our study supports the idea that a protocol biopsy performed at a later stage (6 months), when peritransplant injury has largely faded away, provides a better view on the future of a kidney allograft. However, even in this study of completely pristine kidneys without delayed graft function and with exclusion of overt post-transplantation complications, all kidney allografts inevitably went through some injury during harvest and reperfusion. It can therefore not be completely ruled out that the observed changes are still a consequence of this early injury, and the molecular disturbances observed could still reflect the healing of tissue instead of new injury processes. In this light, it should be taken into account that in clinical practice different injury processes can occur simultaneously and patients do not always have a clear and straightforward histological evolution. Apart from peritransplant injury, transplanted kidneys also experience rejection episodes, recurrence of the original disease, viral or bacterial infections, episodes of dehydration or ischemia, cardiovascular events, and other factors that affect renal allograft histology, renal graft function, and graft survival. It is therefore clear that a biopsy performed at 6 months after transplantation cannot foresee the future of every graft. Nevertheless, this study provides a road map of the molecular basis of the tissue injury that may be programmed to occur.
CONCLUSION
In conclusion, the current study demonstrates that progressive chronic histological damage after kidney transplantation is associated with regulation of both innate and adaptive immune responses that cannot be currently evaluated by histology. There is involvement of a broad spectrum of immune mediators such as T cells, B cells, NK cells, mast cells, granulocytes, and dendritic cells. This study therefore underscores the complexity of the immunological processes in human kidney transplantation, and corroborates the idea that inflammation that is quantitatively below the diagnostic threshold of acute T-cell-mediated rejection is involved in early subclinical stages of progressive renal allograft damage. Timely intervention aimed at influencing these early immune responses or turn these inflammatory signals into a more adaptive state could well be the clue to slow or abort the progression of chronic renal graft scarring and improve long-term graft survival.
MATERIALS AND METHODS
Patients and biopsies
The study comprised 120 renal allograft biopsies from 67 pediatric and adolescent kidney allograft recipients (1–21 years of age). All biopsies included in this study were selected on the basis of tissue availability, conduct of blinded and centralized histological evaluation, and adequate RNA quality (see below). All patients received an immunosuppressive regimen consisting of a combination of tacrolimus (Prograf, Astellas Pharma US, Deerfield, IL), mycophenolate mofetil (Cellcept, Hoffman-La Roche, Nutley, NJ), and daclizumab (Zenapax, Hoffman-La Roche). Some patients received a steroid-avoidance regimen, whereas others received a steroid-based immunosuppressive regimen, as described previously.46 Excellent quality deceased donor kidneys were used following rigid donor selection criteria and living donor kidneys were obtained from young adults (mostly parental donors). There was no delayed graft function in patients included in this study; delayed graft function in our center was defined as the absence of a >50% decline in the recipient serum creatinine in the first 72 h and/or requirement for dialysis in the first week post-transplant. All patients or their parents gave written informed consent, and the study was approved by the institutional review board of Stanford University. For inclusion in the study, patients needed to have a clear histological evolution, that is, either absence of progression of chronic histological damage or clear progression of histological damage. Patients who had unexplained regression of chronic damage (likely due to sampling error problems) and patients with de novo or recurrent glomerular disease or donor-specific antibodies were excluded from the study.
The classification of the biopsies is given in Figure 1 and in Table 1. The study comprised four projects: (1) case–control analysis of the differences in gene expression between biopsies with high versus low CADI score; (2) repeated measures analysis of the dynamics of gene expression over time after transplantation, in kidneys with versus without progression of the CADI score; (3) prediction of future histological damage in six months biopsy samples from a longitudinal cohort of patients with serial biopsies; (4) case–control analysis of the gene expression signature of acute T-cell-mediated rejection in indication biopsies versus rejection-free protocol biopsies with low or high CADI scores. In Project 1, 48 protocol biopsies with low (N=24) and high (N=24) CADI scores were included, divided into a discovery and validation set. In Project 2, 72 protocol biopsies were examined from 24 unique patients at each of the following time points: at implantation, 6 months, and 24 months. Patients were categorized into a progressor group when the histological CADI score reached at least 6 within the first 2 years after transplantation. All other patients were categorized as nonprogressors. Project 3 comprised the 24 protocol biopsies in Project 2 obtained at 6 months. In Project 4, gene expression profiles of 24 for-cause biopsies showing acute T-cell-mediated rejection (indication biopsies performed because of graft dysfunction), obtained in a separate group of patients, were compared with the profiles of the 48 protocol biopsies from Project 1 (Figure 1 and in Table 1). The biopsies were classified into the respective groups by manual selection based on clinical demographics and histological parameters to match cases and controls. This grouping was performed independent of the gene expression results. No biopsies with antibody-mediated rejection or patients with donor-specific antibodies were included in this study, and patients with BK nephropathy and specific glomerular disease (recurrent or de novo) were excluded.
Histological evaluation
All biopsy slides (n=120) were rescored semiquantitatively according to the revised Banff criteria31 for the severity of acute histological lesions (interstitial inflammation (i), tubulitis (t), intimal arteritis (v), glomerulitis (g), inflammation in atrophic areas) and of chronic histological lesions (tubular atrophy, TA (ci), interstitial fibrosis, IF (ct), vascular intimal thickening (cv), arteriolar hyalinosis (ah), increase in mesangial matrix (mm), transplant glomerulopathy (cg), with the addition of the number of globally sclerosed glomeruli (gs), the ‘total i-score’,31 and the CADI score1) (Supplementary Methods online). For implantation biopsies, we calculated the ‘Remuzzi score’.27
RNA extraction, quality control, amplification, and hybridization
RNA was extracted from each kidney allograft biopsy and hybridized onto Affymetrix GeneChip Human Genome U133 Plus 2.0 Arrays after RNA integrity was ensured and after RNA amplification (Supplementary Methods online). For processing and normalization of the scanned images, dChip 2006 software was used, with perfect match (PM)/mismatch (MM) difference modeling and invariant set normalization.47
Study design, data processing, and analysis
Significance analysis of microarrays (SAM) for two-class unpaired data (Projects 1 and 3) or two-class unpaired time course data (signed area test; Project 2) was performed to detect expression differences based on q-values (false discovery rates).48 SAM is an unbiased statistical technique for finding significant genes in a set of microarray experiments. The input to SAM is gene expression measurements from a set of microarray experiments, as well as a response variable from each experiment. SAM uses repeated permutations of the data to determine whether the expression of each gene is significantly related to the response. Significance levels were set at a q-value of 5%.
After identifying and validating the differentially expressed genes, these probe sets were analyzed using IPA (Ingenuity Systems, Redwood City, CA) in order to assess their biological functions and examine canonical pathways based on the Ingenuity Pathways Knowledge Base. Gene set scores were calculated as the geometric mean of fold changes (unlogged values) across all probe sets (not only the significant probe sets) within the Ingenuity Pathways Knowledge Base lists for the different immune cell functions (Supplementary Table S5 online). Gene set scores were compared between groups with Mann–Whitney test, Kruskall–Wallis test, and repeated measures analysis of variance, as appropriate. The raw data sets for the 120 biopsies included are deposited at the Gene Expression Omnibus under GSE25902.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge the invaluable and persisting efforts of all people involved in the pediatric kidney transplant program at Stanford University, the clinicians, renal nurses, transplant coordinators, the children, and their families. We also acknowledge Dr Neeraja Kambham from the Department of Pathology at Stanford University for her great effort in scoring the biopsies according to the Banff classification. This project was funded by Beta Sigma Phi (OS, MS) and the Packard Foundation (MMS, LL).
Footnotes
DISCLOSURE
All the authors declared no competing interests.
AUTHOR CONTRIBUTIONS
MN, MMS, and OS designed the study; MN, TKS, and MJV performed the experiments; MN, PK, LL, and RC performed the analysis; MN, PK, AJB, and MMS wrote the manuscript.
SUPPLEMENTARY MATERIAL
Figure S1. Distribution of the different components of the CADI score in the different biopsy groups included in the discovery set and the validation set of Project 1.
Figure S2. Distribution of the different components of the CADI score in the different biopsy groups included in Project 2–3.
Figure S3. Distribution of the different components of the CADI score in the different biopsy groups included in Project 4.
Table S1. Details of the IPA enrichment analysis (biological functions) of the 397 upregulated probe sets in the discovery set of Project 1.
Table S2. Details of the IPA enrichment analysis (biological functions) of the 703 upregulated probe sets in the validation set of Project 1.
Table S3. Details of the IPA enrichment analysis (biological functions) of the 601 upregulated probe sets in Project 2.
Table S4. Details of the IPA enrichment analysis (biological functions) of the 92 upregulated probe sets in Project 3.
Table S5. Probe sets (and their corresponding genes on Affymetrix GeneChip Human Genome U133 Plus 2.0 Arrays) included in the Ingenuity Pathways Knowledge Base lists, which were used to calculate the different immune gene set scores.
Supplementary material is linked to the online version of the paper at http://www.nature.com/ki
REFERENCES
- 1.Yilmaz S, Tomlanovich S, Mathew T, et al. Protocol core needle biopsy and histologic Chronic Allograft Damage Index (CADI) as surrogate end point for long-term graft survival in multicenter studies. J Am Soc Nephrol. 2003;14:773–779. doi: 10.1097/01.asn.0000054496.68498.13. [DOI] [PubMed] [Google Scholar]
- 2.Nankivell B, Richard J, Borrows R, et al. The natural history of chronic allograft nephropathy. New Engl J Med. 2003;349:2326–2333. doi: 10.1056/NEJMoa020009. [DOI] [PubMed] [Google Scholar]
- 3.Nankivell BJ, Chapman JR. Chronic allograft nephropathy: current concepts and future directions. Transplantation. 2006;81:643–654. doi: 10.1097/01.tp.0000190423.82154.01. [DOI] [PubMed] [Google Scholar]
- 4.El-Zoghby ZM, Stegall MD, Lager DJ, et al. Identifying specific causes of kidney allograft loss. Am J Transplant. 2009;9:527–535. doi: 10.1111/j.1600-6143.2008.02519.x. [DOI] [PubMed] [Google Scholar]
- 5.Ojo AO, Hanson JA, Meier-Kriesche H, et al. Survival in recipients of marginal cadaveric donor kidneys compared with other recipients and wait-listed transplant candidates. J Am Soc Nephrol. 2001;12:589–597. doi: 10.1681/ASN.V123589. [DOI] [PubMed] [Google Scholar]
- 6.Naesens M, Lerut E, de Jonge H, et al. Donor age and renal P-glycoprotein expression associate with chronic histological damage in renal allografts. J Am Soc Nephrol. 2009;20:2468–2480. doi: 10.1681/ASN.2009020192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nankivell BJ, Fenton-Lee CA, Kuypers DR, et al. Effects of histological damage on long-term kidney transplant outcome. Transplantation. 2001;71:515–523. doi: 10.1097/00007890-200102270-00006. [DOI] [PubMed] [Google Scholar]
- 8.Cosio FG, Grande JP, Wadei H, et al. Predicting subsequent decline in kidney allograft function from early surveillance biopsies. Am J Transplant. 2005;5:2464–2472. doi: 10.1111/j.1600-6143.2005.01050.x. [DOI] [PubMed] [Google Scholar]
- 9.Moreso F, Ibernon M, Goma M, et al. Subclinical rejection associated with chronic allograft nephropathy in protocol biopsies as a risk factor for late graft loss. Am J Transplant. 2006;6:747–752. doi: 10.1111/j.1600-6143.2005.01230.x. [DOI] [PubMed] [Google Scholar]
- 10.Seron D. Interstitial fibrosis and tubular atrophy in renal allograft protocol biopsies as a surrogate of graft survival. Transplant Proc. 2009;41:769–770. doi: 10.1016/j.transproceed.2008.12.027. [DOI] [PubMed] [Google Scholar]
- 11.Halloran PF, De Freitas DG, Einecke G, et al. An integrated view of molecular changes, histopathology and outcomes in kidney transplants. Am J Transplant. 2010;10:2223–2230. doi: 10.1111/j.1600-6143.2010.03268.x. [DOI] [PubMed] [Google Scholar]
- 12.Naesens M, Li L, Ying L, et al. Expression of complement components differs between kidney allografts from living and deceased donors. J Am Soc Nephrol. 2009;20:1839–1851. doi: 10.1681/ASN.2008111145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sarwal M, Chua MS, Kambham N, et al. Molecular heterogeneity in acute renal allograft rejection identified by DNA microarray profiling. N Engl J Med. 2003;349:125–138. doi: 10.1056/NEJMoa035588. [DOI] [PubMed] [Google Scholar]
- 14.Mueller TF, Einecke G, Reeve J, et al. Microarray analysis of rejection in human kidney transplants using pathogenesis-based transcript sets. Am J Transplant. 2007;7:2712–2722. doi: 10.1111/j.1600-6143.2007.02005.x. [DOI] [PubMed] [Google Scholar]
- 15.Mas V, Maluf D, Archer K, et al. Establishing the molecular pathways involved in chronic allograft nephropathy for testing new noninvasive diagnostic markers. Transplantation. 2007;83:448–457. doi: 10.1097/01.tp.0000251373.17997.9a. [DOI] [PubMed] [Google Scholar]
- 16.Park W, Griffin M, Grande JP, et al. Molecular evidence of injury and inflammation in normal and fibrotic renal allografts one year posttransplant. Transplantation. 2007;83:1466–1476. doi: 10.1097/01.tp.0000265501.33362.d3. [DOI] [PubMed] [Google Scholar]
- 17.Mengel M, Reeve J, Bunnag S, et al. Molecular correlates of scarring in kidney transplants: the emergence of mast cell transcripts. Am J Transplant. 2009;9:169–178. doi: 10.1111/j.1600-6143.2008.02462.x. [DOI] [PubMed] [Google Scholar]
- 18.Nakorchevsky A, Hewel JA, Kurian SM, et al. Molecular mechanisms of chronic kidney transplant rejection via large-scale proteogenomic analysis of tissue biopsies. J Am Soc Nephrol. 2010;21:362–373. doi: 10.1681/ASN.2009060628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scherer A, Krause A, Walker JR, et al. Early prognosis of the development of renal chronic allograft rejection by gene expression profiling of human protocol biopsies. Transplantation. 2003;75:1323–1330. doi: 10.1097/01.TP.0000068481.98801.10. [DOI] [PubMed] [Google Scholar]
- 20.Scherer A, Gwinner W, Mengel M, et al. Transcriptome changes in renal allograft protocol biopsies at 3 months precede the onset of interstitial fibrosis/tubular atrophy (IF/TA) at 6 months. Nephrol Dial Transplant. 2009;24:2567–2575. doi: 10.1093/ndt/gfp183. [DOI] [PubMed] [Google Scholar]
- 21.Vitalone MJ, O’Connell PJ, Wavamunno M, et al. Transcriptome changes of chronic tubulointerstitial damage in early kidney transplantation. Transplantation. 2010;89:537–547. doi: 10.1097/TP.0b013e3181ca7389. [DOI] [PubMed] [Google Scholar]
- 22.Park WD, Griffin MD, Cornell LD, et al. Fibrosis with inflammation at one year predicts transplant functional decline. J Am Soc Nephrol. 2010;21:1987–1997. doi: 10.1681/ASN.2010010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scian MJ, Maluf DG, Archer KJ, et al. Gene expression changes are associated with loss of kidney graft function and interstitial fibrosis and tubular atrophy: diagnosis versus prediction. Transplantation. 2011;91:657–665. doi: 10.1097/TP.0b013e3182094a5a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mengel M, Chang J, Kayser D, et al. The molecular phenotype of 6-week protocol biopsies from human renal allografts: reflections of prior injury but not future course. Am J Transplant. 2010;11:708–718. doi: 10.1111/j.1600-6143.2010.03339.x. [DOI] [PubMed] [Google Scholar]
- 25.Cosio FG, Grande JP, Larson TS, et al. Kidney Allograft Fibrosis and Atrophy Early After Living Donor Transplantation. Am J Transplant. 2005;5:1130–1136. doi: 10.1111/j.1600-6143.2005.00811.x. [DOI] [PubMed] [Google Scholar]
- 26.Ortiz F, Paavonen T, Tornroth T, et al. Predictors of renal allograft histologic damage progression. J Am Soc Nephrol. 2005;16:817–824. doi: 10.1681/ASN.2004060475. [DOI] [PubMed] [Google Scholar]
- 27.Remuzzi G, Cravedi P, Perna A, et al. Long-term outcome of renal transplantation from older donors. N Engl J Med. 2006;354:343–352. doi: 10.1056/NEJMoa052891. [DOI] [PubMed] [Google Scholar]
- 28.Naesens M, Kambham N, Concepcion W, et al. The evolution of non-immune histological injury and its clinical relevance in adult-sized kidney grafts in pediatric recipients. Am J Transplant. 2007;7:2504–2514. doi: 10.1111/j.1600-6143.2007.01949.x. [DOI] [PubMed] [Google Scholar]
- 29.Flechner SM, Kurian SM, Head SR, et al. Kidney transplant rejection and tissue injury by gene profiling of biopsies and peripheral blood lymphocytes. Am J Transplant. 2004;4:1475–1489. doi: 10.1111/j.1600-6143.2004.00526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hotchkiss H, Chu TT, Hancock WW, et al. Differential expression of profibrotic and growth factors in chronic allograft nephropathy. Transplantation. 2006;81:342–349. doi: 10.1097/01.tp.0000195773.24217.95. [DOI] [PubMed] [Google Scholar]
- 31.Solez K, Colvin RB, Racusen LC, et al. Banff 07 classification of renal allograft pathology: updates and future directions. Am J Transplant. 2008;8:753–760. doi: 10.1111/j.1600-6143.2008.02159.x. [DOI] [PubMed] [Google Scholar]
- 32.Mengel M, Reeve J, Bunnag S, et al. Scoring total inflammation is superior to the current Banff inflammation score in predicting outcome and the degree of molecular disturbance in renal allografts. Am J Transplant. 2009;9:1859–1867. doi: 10.1111/j.1600-6143.2009.02727.x. [DOI] [PubMed] [Google Scholar]
- 33.Kim IK, Bedi DS, Denecke C, et al. Impact of innate and adaptive immunity on rejection and tolerance. Transplantation. 2008;86:889–894. doi: 10.1097/TP.0b013e318186ac4a. [DOI] [PubMed] [Google Scholar]
- 34.Zarkhin V, Kambham N, Li L, et al. Characterization of intra-graft B cells during renal allograft rejection. Kidney Int. 2008;74:664–673. doi: 10.1038/ki.2008.249. [DOI] [PubMed] [Google Scholar]
- 35.Porcheray F, Wong W, Saidman SL, et al. B-cell immunity in the context of T-cell tolerance after combined kidney and bone marrow transplantation in humans. Am J Transplant. 2009;9:2126–2135. doi: 10.1111/j.1600-6143.2009.02738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blank U, Essig M, Scandiuzzi L, et al. Mast cells and inflammatory kidney disease. Immunol Rev. 2007;217:79–95. doi: 10.1111/j.1600-065X.2007.00503.x. [DOI] [PubMed] [Google Scholar]
- 37.Beilke JN, Gill RG. Frontiers in nephrology: the varied faces of natural killer cells in transplantation-contributions to both allograft immunity and tolerance. J Am Soc Nephrol. 2007;18:2262–2267. doi: 10.1681/ASN.2007040423. [DOI] [PubMed] [Google Scholar]
- 38.van der Touw W, Bromberg JS. Natural killer cells and the immune response in solid organ transplantation. Am J Transplant. 2010;10:1354–1358. doi: 10.1111/j.1600-6143.2010.03086.x. [DOI] [PubMed] [Google Scholar]
- 39.Steinman RM, Banchereau J. Taking dendritic cells into medicine. Nature. 2007;449:419–426. doi: 10.1038/nature06175. [DOI] [PubMed] [Google Scholar]
- 40.Colvin BL, Matta BM, Thomson AW. Dendritic cells and chemokine-directed migration in transplantation: where are we headed? Clin Lab Med. 2008;28:375–384. doi: 10.1016/j.cll.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomson AW. Tolerogenic dendritic cells: all present and correct? Am J Transplant. 2010;10:214–219. doi: 10.1111/j.1600-6143.2009.02955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roberts IS, Brenchley PE. Mast cells: the forgotten cells of renal fibrosis. J Clin Pathol. 2000;53:858–862. doi: 10.1136/jcp.53.11.858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jahanyar J, Koerner MM, Loebe M, et al. The role of mast cells after solid organ transplantation. Transplantation. 2008;85:1365–1371. doi: 10.1097/TP.0b013e31816fc0a3. [DOI] [PubMed] [Google Scholar]
- 44.Nickeleit V, Mihatsch M. Kidney transplants, antibodies and rejection: is C4d a magic marker? Nephrol Dial Transplant. 2003;18:2232–2239. doi: 10.1093/ndt/gfg304. [DOI] [PubMed] [Google Scholar]
- 45.Nickeleit V, Andreoni K. Inflammatory cells in renal allografts. Front Biosci. 2008;13:6202–6213. doi: 10.2741/3148. [DOI] [PubMed] [Google Scholar]
- 46.Li L, Chang A, Naesens M, et al. Steroid-free immunosuppression since 1999: 129 pediatric renal transplants with sustained Graft and patient benefits. Am J Transplant. 2009;9:1362–1372. doi: 10.1111/j.1600-6143.2009.02640.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schwartz GJ, Haycock GB, Edelmann CM, Jr, et al. A simple estimate of glomerular filtration rate in children derived from body length and plasma creatinine. Pediatrics. 1976;58:259–263. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.