Abstract
The clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein (Cas) system is an adaptive immune system in bacteria and archaea that has recently been exploited for genome engineering. Mutant mice can be generated in one step through direct delivery of the CRISPR/Cas9 components into a mouse zygote. Although the technology is robust, delivery remains a bottleneck, as it involves manual injection of the components into the pronuclei or the cytoplasm of mouse zygotes, which is technically demanding and inherently low throughput. To overcome this limitation, we employed electroporation as a means to deliver the CRISPR/Cas9 components, including Cas9 messenger RNA, single-guide RNA, and donor oligonucleotide, into mouse zygotes and recovered live mice with targeted nonhomologous end joining and homology-directed repair mutations with high efficiency. Our results demonstrate that mice carrying CRISPR/Cas9-mediated targeted mutations can be obtained with high efficiency by zygote electroporation.
Keywords: CRISPR, Cas9, electroporation, mouse zygote
THE clustered regularly interspaced short palindromic repeat (CRISPR) locus and the CRISPR-associated protein (Cas) system, CRISPR/Cas9, is an acquired immune system in bacteria and archaea that has recently been exploited for genome engineering of many species and cell types (Jinek et al. 2012; Doudna and Charpentier 2014; Hsu et al. 2014). CRISPR/Cas9 is an RNA guided DNA endonuclease system in which Cas9 endonuclease forms a complex with two naturally occurring RNA species, CRISPR RNA (crRNA) and trans-activating CRISPR RNA (tracrRNA). This complex targets specific DNA sequences complementary to the 20-nt sequence residing at the 5′ end of the crRNA and generate DNA double stranded breaks (DSBs) at the target site (Jinek et al. 2012). Conveniently, crRNA and tracrRNA can be linked by an arbitrary stem loop sequence to generate a synthetic single-guide RNA (sgRNA). The other key element in determining target sequence specificity is the protospacer adjacent motif (PAM) that is adjacent to the target site at the genome locus, but is not a part of the sgRNA sequence (Supporting Information, Figure S1). DNA DSBs generated by CRISPR/Cas9 are repaired through either nonhomologous end joining (NHEJ) or the homology-directed repair (HDR) pathway, leading to varied DNA sequence modifications (Doudna and Charpentier 2014; Hsu et al. 2014).
We employed the CRISPR/Cas9 system and developed methods for one-step generation of mice carrying mutations in a single or multiple genes, as well as mice carrying reporter and conditional alleles (Wang et al. 2013; Yang et al. 2013). Using similar approaches, genetically modified animals of various species have been generated (Hwang et al. 2013; Li et al. 2013a,b; Hai et al. 2014; Niu et al. 2014). Compared to conventional gene targeting technology (Capecchi 2005), methods that employ nucleases, such as zinc finger nuclease (ZFN) (Urnov et al. 2010), transcription activator-like effector nuclease (TALEN) (Bogdanove and Voytas 2011), and CRISPR/Cas, have the advantage of generating genetically modified organisms by directly modifying the genome in the zygote, eliminating the need for a germline competent embryonic stem cell line, saving cost and shortening the time needed for creating mutant animals.
Microinjection has been the technology of choice for transgenic research for the last 30 years (Palmiter et al. 1982). To put CRISPR/Cas9 components into direct contact with target DNA, current methodology employs a microinjection needle to traverse the zona pelucida layer and the plasma membrane of the zygote and place the CRISPR/Cas9 components, including Cas9 messenger RNA (mRNA) or protein, sgRNA, single-stranded donor oligonucleotide, or double-stranded DNA template, into the pronuclei or cytoplasm of the mouse zygotes. Although effective, microinjection of the zygotes is technically demanding, labor intensive, and time consuming. It is also inherently low in throughput, requiring manipulation of the zygotes one at a time, limiting the size and scale of a transgenic experiment.
To improve the throughput of generating mice carrying targeted mutation, we explored electroporation as a means to deliver the CRISPR/Cas9 components into mouse zygotes. Here we report the development of zygote electroporation of nuclease (ZEN) technology to generate mice carrying targeted genetic modifications with high efficiency.
Materials and Methods
Production of Cas9 mRNA and sgRNA
The px330 plasmid carrying the wild-type Cas9 (Cong et al. 2013) was used as the DNA template for amplification of the Cas9 coding sequence in a polymerase chain reaction (PCR). The T7 promoter sequence was added to the forward primer and reverse primer from the coding sequence of the Cas9 gene. PCR product was amplified using the AccuPrime PCR system (Life Technologies) and purified using the QIAquick PCR purification kit (Qiagen) and in vitro transcription (IVT) performed using the mMESSAGE mMACHINE T7 ULTRA Transcription kit (Life Technologies). For sgRNA synthesis, the T7 promoter sequence was added to sgRNA template/forward primer and the IVT template generated by PCR amplification using primers listed in Table S5. The T7-sgRNA PCR product was purified and used as the template for IVT using MEGAshortscript T7 kit (Life Technologies). Both the Cas9 mRNA and the sgRNAs were purified using the MEGAclear kit (Life Technologies). Aliquots from an IVT reaction were separated on agarose gel to assess quality from a reaction. Single-stranded oligos were ordered as PAGE Ultramer from Integrated DNA Technologies.
RFLP analysis and Sanger sequencing
Genomic DNA from mice or embryos was extracted and used in PCRs with gene-specific primers under the following conditions: 95° for 5 min; 35× (95° for 30 s, 58° for 30 s, 68° for 30 s); 68° for 2 min; hold at 4°. A total of 5 μl of Tet1 or Tet2 PCR products were digested with SacI (Tet1) or EcoRV (Tet2) and separated on a GelRed (Biotium)-stained agarose gel (1.5%), for detection of NHEJ-mediated mutation. In the samples electroporated with Tet2 donor oligonucleotide, the Tet2 PCR products were digested with EcoRI to detect the precise modification mediated by HDR. PCR products were processed by Sanger sequencing or cloned into the pCR2.1 vector from the Topo Cloning kit (Invitrogen), and sequences of individual clones were determined by Sanger sequencing.
Zygote isolation, microinjection, electroporation, culture, and transfer
All experiments adhered to the standards set forth by the National Institutes of Health in the Guide for the Care and Use of Laboratory Animals and were approved by the Jackson Laboratory Animal Care and Use Committee. B6D2F1/J donor female mice (3–4 weeks of age) were superovulated by administration of 5 IU (ip) of pregnant mare serum gonadotrophin (PMSG) (ProSpec HOR-272) followed 47 hr later by 5 IU (ip) human chorionic gonadotrophin (hCG) (ProSpec HOR-272). Immediately postadministration of hCG, the female was mated 1:1 with a B6D2F1/J stud male and 22 hr later checked for the presence of a copulation plug. Female mice displaying a copulation plug were sacrificed, the oviducts excised, and embryos collected.
Standard zygote microinjection procedure was performed on a Zeiss AxioObserver.D1 using Eppendorf NK2 micromanipulators in conjunction with Narashige IM-5A injectors. Injected zygotes were rinsed through three 30-μl drops of equilibrated K-RCVL (Cook, K-RVCL) before being placed into a separate 30-μl microdrop of equilibrated K-RCVL where they were either cultured to blastocyst (Cook, MINC benchtop incubator, 37°, 5% CO2, and 5% O2/nitrogen) or processed for embryo transfer via the oviduct on the day of injection.
For electroporation, zygotes were removed from K-RCVL media and placed in prewarmed M2 media (EmbryoMax, Millipore). In groups of 50, zygotes were placed in the acidic Tyrode’s solution (T1788, Sigma-Aldrich) for 10 sec and removed and washed through three 100-μl drops of prewarmed M2 media. Zygotes were then pipetted into 10 μl drops of Opti-MEM media (P/N 31985, Gibco) and the entire content mixed with 10 μl of the Cas9 mRNA/sgRNA/donor reconstituted in TE buffer (pH 7.5) and deposited into a 1-mm electroporation cuvette (P/N 45-0124, Harvard Apparatus) and electroporated in a ECM830 Square Wave Electroporation system (BTX, Harvard Apparatus) using 30 V, with 1-ms pulse duration and two pulses separated by 100-ms pulse interval. Following delivery of the pulse, a prewarmed 100-μl aliquot of KSOM/BSA media was deposited into the cuvette and zygotes were removed from the cuvette with a sterile plastic pipette, followed by three washes in prewarmed M2 media. Embryos were then cultured to blastocysts (MINC benchtop incubator, Cook; 37°C, 5% CO2, and 5% O2/nitrogen) or surgically transferred into CByB6F1/J pseudopregnant female mice.
Results
Zona pellucida is a glycoprotein layer surrounding the plasma membrane of an oocyte and may present a barrier preventing access of the CRISPR/Cas9 components to the zygote by electroporation. We confirmed that 10 sec of exposure of zygotes to acidic Tyrode’s solution (AT, pH 2.5) to weaken the zona does not retard development of the zygotes into blastocysts in vitro (Grabarek et al. 2002; Peng et al. 2012). To determine the optimal voltage and pulse interval compatible with embryo survival, we tested 30 V, 100 V, and 300 V with pulse duration of 1 ms, in combination with 100-ms, 500-ms, and 1000-ms intervals between the two pulses. We found that voltage has the most significant impact on embryo survival, and that zygotes can develop into blastocysts with high efficiency after being subjected to an electric shock of 30 V (Table S1). Therefore, we used 30 V with pulse duration of 1 ms, with a 100-ms interval between two pulses to deliver CRISPR/Cas9 components into the mouse zygotes.
For electroporation, we used 20 μl total volume to accommodate the CRISPR/Cas reagents (Cas9 mRNA, sgRNA, donor oligonucleotide) reconstituted in TE buffer (10 mM Tris-HCl, 0.1 mM EDTA, pH 7.5) and embryos suspended in Opti-MEM, a medium commonly used in transfection experiments. As TE buffer is hypotonic and Opti-MEM isotonic, we tested different combinations of the TE and Opti-MEM volumes to identify a combination that would support embryo survival. As shown in Table S2, a 1:1 volume ratio of TE buffer and Opti-MEM supports embryo survival and development into blastocysts in vitro. Based on these results, for all subsequent experiments we placed embryos into 10 μl of Opti-MEM media and added 10 μl of CRISPR/Cas9 reagents reconstituted in TE buffer in preparation for electroporation.
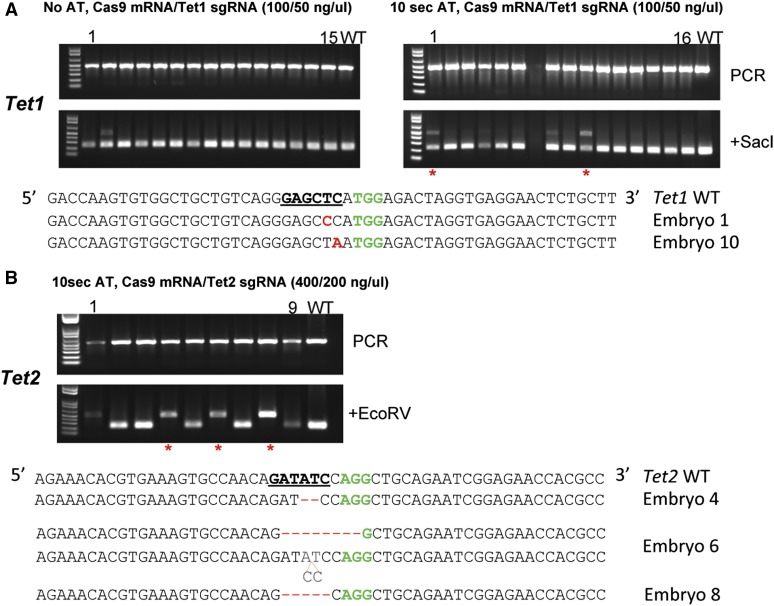
To test for the efficiency of electroporation to deliver the CRISPR/Cas9 components, we chose the Tet1 and Tet2 genes as our targets, targeting these two loci with sgRNAs as previously described (Wang et al. 2013) (Figure S1). Mouse zygotes isolated from the B6D2F2/J strain were treated with AT for 10 sec, deposited into 10 μl of Opti-MEM, and then mixed with 10 μl of Cas9 mRNA/Tet1 sgRNA reagent at 40/20 and 100/50 ng/μl final concentration. Zygotes bathed in the CRISPR/Cas9 reagents were electroporated in a cuvette at the settings as described above. The embryos were washed three times with M2 solution and cultured in vitro for 3.5 days until blastocysts developed. Blastocysts were harvested and PCR products encompassing the target site were amplified and examined by RFLP analysis using the SacI site, which is part of the target sequence in Tet1. When a mixture of Cas9 mRNA at 40 ng/μl and Tet1 sgRNA at 20 ng/μl was used, no mutation was detected among 10 embryos treated with AT or 8 embryos not treated with AT (data not shown). A mixture of Cas9 mRNA at 100 ng/μl and Tet1 sgRNA at 50 ng/μl yielded 3 of 15 AT-treated and 1 of 15 AT-untreated embryos with mutations destroying the SacI site in the Tet1 locus (Figure 1A and Figure S1). Electroporating Cas9 mRNA at 400 ng/μl and Tet2 sgRNA at 200 ng/μl into mouse zygotes under the same conditions yielded 4 of 9 AT-treated embryos that carry mutations disrupting the EcoRV site in the Tet2 locus, as indicated by the presence of PCR products resistant to EcoRV digestion (Figure 1B and Figure S1). All PCR products were sequenced and samples with overlapping sequencing traces were subcloned and individual alleles determined (Figure 1B). We conclude that both Tet1 and Tet2 loci can be efficiently targeted in mouse embryos using electroporation to deliver the CRISPR/Cas9 reagents.
Figure 1.
CRISPR/Cas9-mediated indel mutations in mouse embryos delivered by electroporation. (A) Genotyping of mouse embryos targeted at the Tet1 locus. Mouse embryos were electroporated with Cas9 mRNA (100 ng/μl) and sgRNA targeting the Tet1 locus (50 ng/μl), cultured to blastocyst stage of development, and RFLP analysis performed as shown in the top panel. PCR products are sequenced and those showing overlapping sequencing traces were cloned and individual clones sequenced. In the bottom half, mutant alleles are shown for embryos 1 and 10 (indicated by red star) from the group treated with AT for 10 sec. The SacI restriction site at the target region, used for RFLP analysis, is bold and underlined. The protospacer adjacent motif (PAM) sequence is colored in green. Mutated bases are labeled in red. (B) Genotyping of mouse embryos targeted at theTet2 locus. Mutant alleles identified from embryos 4, 6, and 8 (indicated by red star) are shown. Only one mutant allele was recovered from embryos 4 and 8. The EcoRV site located within the target sequence is bold and underlined and PAM sequence colored in green.
To determine if live mice could be obtained from electroporated embryos, we performed an embryo transfer experiment. Zygotes were treated with AT for 10 sec, electroporated with the CRISPR/Cas9 mixture at different concentrations (Cas9 mRNA/Tet2 sgRNA at 200/100, 400/200, and 600/300 ng/μl), and immediately transferred into pseudopregnant female mice, and live mice were derived. PCR products were amplified from genomic DNA isolated from tail biopsies, analyzed for RFLPs, and mutations were subsequently confirmed by Sanger sequencing. As summarized in Table 1, we derived live mice from the ZEN protocol and observed a Cas9 mRNA/sgRNA concentration-dependent improvement in founder efficiency. When Cas9 mRNA/sgRNA was used at 200/100 ng/μl, one founder was identified among the 32 mice screened (3%). Increasing the concentration of Cas9 mRNA/sgRNA to 400/200 ng/μl improved founder efficiency to 16% (5/31); further increase of the Cas9 mRNA/sgRNA concentration to 600/300 ng/μl yielded 57% efficiency (8/14) (Table 1).
Table 1. CRISPR/Cas9-mediated Tet2 targeting in B6D2F2/J mice delivered by electroporation.
| Cas9/sgRNA conc. (ng/μl) | Pseudo ID | Embryos transferred | Mice born | Mutant mice | Total mice analyzed | Percentage (%) |
|---|---|---|---|---|---|---|
| 200/100 | 1 | 17 | 11 | 0 | 11 | 3 |
| 2 | 17 | 12 | 0 | 11 | ||
| 3 | 17 | 10 | 1 | 10 | ||
| 400/200 | 4 | 18 | 11 | 1 | 10 | 16 |
| 5 | 16 | 13 | 2 | 13 | ||
| 6 | 16 | 11 | 2 | 8 | ||
| 600/300 | 7 | 18 | 7 | 4 | 6 | 57 |
| 8 | 21 | 11 | 4 | 8 | ||
| 0/0 | 9 | 17 | 11 | 0 | 8 | 0 |
| 10 | 12 | 7 | 0 | 3 |
Zygotes were derived from B6D2F2/J and treated with acidic Tyrode’s solution for 10 sec before electroporation. Embryos were divided into groups of 20 or 40 and electroporated in a cuvette of 1-mm gap size in 20 μl of TE/Opti-MEM at 1:1 volume ratio. Following electroporation, the embryos were transferred to 100 μl M2 media and transferred into pseudopregnent female mice and live mice were derived. For each live born mouse, PCR product at the target site was amplified from genomic DNA isolated from tail biopsies and analyzed by RFLP and Sanger sequencing.
To test whether our ZEN protocol is applicable to other genes, we used electroporation to target 10 genes that have been previously targeted using microinjection. For each experiment, 30 zygotes were mixed with CRISPR/Cas9 reagents to achieve a final concentration of 600 ng/μl Cas9 mRNA and 300 ng/μl gene-specific sgRNA, electroporated, and immediately transferred into pseudopregnant female mice, and the pups were genotyped. We were able to derive mutant mice in 5 of 10 experiments, indicating the ZEN technology is robust (Table 2). Although the current ZEN protocol was not as efficient as a skilled microinjectionist who produced mutant mice for 8 of this same set of 10 genes (Table 2), we believe performance of the ZEN protocol could be improved with further understanding and optimization of the parameters critical in an electroporation experiment. We should note that B6D2F2 embryos were used in the ZEN experiments, while NOD/ShiLtJ were used in microinjection. This is because we have been using the B6D2F2 strain to optimize the ZEN protocol and still have to work out the conditions for the NOD/ShiLtJ strain.
Table 2. CRISPR/Cas9-mediated gene editing for 10 genes delivered by microinjection and electroporation.
| Microinjection (NOD/ShiLtJ) | Electroporation (B6D2F2/J) | |||||||
|---|---|---|---|---|---|---|---|---|
| Target gene | Embryos transferred | Mice born | Mutant mice | Mutant percentage (%) | Embryos transferred | Mice born | Mutant mice | Mutant percentage (%) |
| Cd69 | 59 | 23 | 0 | 0 | 29 | 16 | 0 | 0 |
| Cd226 | 61 | 19 | 4 | 21 | 20 | 13 | 0 | 0 |
| Clec16a | 57 | 0 | NA | NA | 25 | 16 | 2 | 13 |
| Cyp27b1 | 64 | 23 | 12 | 52 | 28 | 20 | 0 | 0 |
| Fut2 | 64 | 25 | 7 | 28 | 29 | 25 | 9 | 36 |
| Ormdl3 | 62 | 19 | 17 | 89 | 26 | 15 | 2 | 13 |
| Rgs1 | 62 | 18 | 8 | 44 | 28 | 19 | 5 | 26 |
| Tlr7 | 66 | 22 | 6 | 27 | 30 | 15 | 0 | 0 |
| Tlr8 | 60 | 15 | 1 | 7 | 29 | 22 | 3 | 14 |
| Tnfsf9 | 61 | 21 | 15 | 71 | 25 | 14 | 0 | 0 |
| Total | 616 | 185 | Live birth rate 30% | 296 | 175 | Live birth rate 59% | ||
Zygotes were derived from B6D2F2/J and treated with acidic Tyrode’s solution for 10 sec before electroporation. A total of 30 embryos for each target gene were electroporated in a cuvette of 1-mm gap size and in 20 μl of TE/Opti-MEM at 1:1 volume ratio. Following electroporation, the embryos were transferred to 100 μl of M2 media and transferred into pseudopregnent female mice. For each live born mouse, PCR product at the target site was amplified from genomic DNA isolated from tail biopsies and analyzed by Sanger sequencing. Guide sequences for the 10 genes were chosen from the CRISPR Design software (crispr.mit.edu), targeting in-frame “ATG” downstream from the starting “ATG” if possible, with varied scores ranging from “41” for Cd69 to “90” for Fut2. NA, not applicable.
Inbred strains of mice have great value in biomedical research, owing to their controlled genetic background. To test the ZEN protocol on inbred strains of mice, zygotes were isolated from the C57BL/6NJ strain and electroporated with a mixture of Cas9 mRNA (600 ng/μl) and sgRNA (300 ng/μl) targeting the Tet2 locus and live mice were derived. RFLP analysis and Sanger sequencing of PCR products encompassing the target site showed that two of seven mice from replicate 1 and all six mice from replicate 2 carried mutations in the Tet2 locus (Table 3, Figure S2). These results showed that the C57BL/6NJ inbred strain could be efficiently targeted by CRISPR/Cas9 delivered by electroporation.
Table 3. CRISPR/Cas9-mediated gene editing at the Tet2 Locus in C57BL/6NJ embryos delivered by electroporation.
| Group | Cas9/sgRNA conc. (ng/μl) | Embryos electroporated | Embryos transferred | Mice born | Mutant mice | Efficiency (%) |
|---|---|---|---|---|---|---|
| 1 | 600/300 | 20 | 18 | 7 | 2 | 29 |
| 2 | 600/300 | 20 | 19 | 6 | 6 | 100 |
| 3 | 0/0 | 20 | 20 | 11 | NA | NA |
Zygotes were derived from C57BL/6NJ and treated with acidic Tyrode’s solution for 10 sec before electroporation. A total of 20 embryos for each concentration were electroporated in a cuvette of 1-mm gap size and in 20 μl of TE/Opti-MEM at 1:1 volume ratio. Following electroporation, the embryos were transferred to 100 μl of M2 media and transferred into pseudopregnent female mice. For each live born mouse, PCR product at the target site was amplified from genomic DNA isolated from tail biopsies and analyzed by RFLP and Sanger sequencing. conc., concentration.
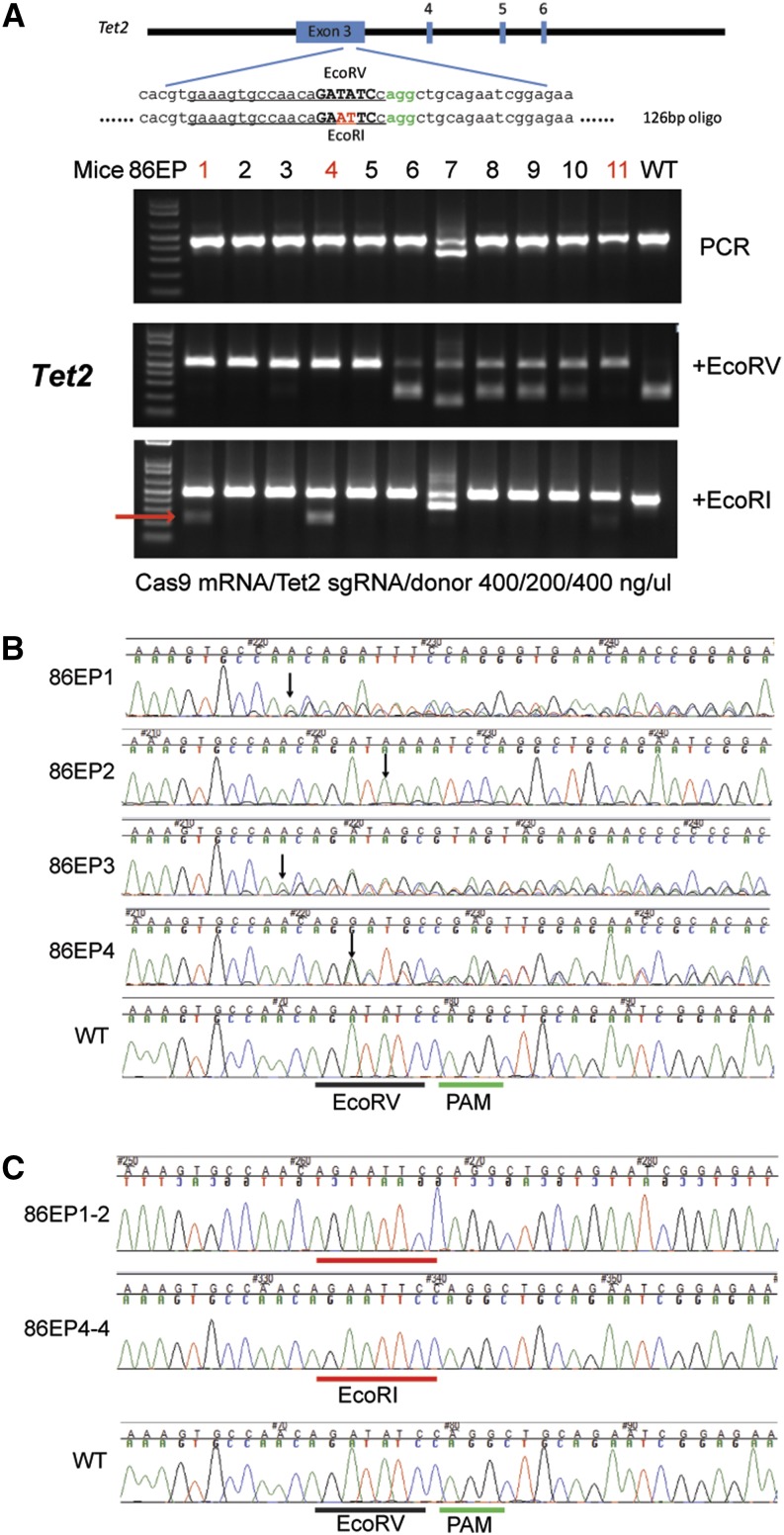
To address whether the ZEN method can generate mice with precise genome modifications, we coelectroporated Cas9 mRNA, sgRNA targeting Tet2, and a 126-nt donor oligonucleotide with DNA sequence flanking the target site and a 2-nt mutation converting the naturally occurring EcoRV site (GATATC) in the target sequence to an EcoRI site (GAATTC) and recovered live mice. To identify CRISPR/Cas9-mediated NHEJ and HDR mutations, we digested the 466-bp PCR product encompassing the target site with EcoRV and with EcoRI independently (Figure 2). Using Cas9 mRNA at 400 ng/μl, Tet2 sgRNA at 200 ng/μl, and single-stranded DNA (ssDNA) donor at 400 ng/μl, we achieved 100% efficiency for introduction of the NHEJ mutation (loss of the EcoRV site; 11 positive mice of 11 mice analyzed). In addition, we identified three founder mice carrying HDR alleles as evident by acquisition of the EcoRI site (Figure 2, samples 86EP1, 86EP4, and 86EP11). We cloned the PCR products from 86EP1 and 86EP4, determined their DNA sequences, and confirmed that both mice acquired the EcoRI site (Figure 2C). In addition to this HDR allele, two other indel alleles were present in mouse 86EP1 and one indel allele in 86EP4, while no wild-type allele was found. Similar to mutant mice generated by microinjection, mice derived using the ZEN method often are mosaic. Using the TIDE software (Brinkman et al. 2014) to decompose sequencing traces, we found four mice carrying more than two Tet2 alleles among the 11 shown in Figure 2. Using a higher concentration of the donor oligonucleotide (1000 ng/μl), we obtained similar results with >70% NHEJ efficiency and >30% HDR efficiency at the Tet2 locus (Table S3).
Figure 2.
CRISPR/Cas9-mediated HDR mutation in live mice delivered by electroporation. (A) Top panel, schematic of the target sequence and donor oligonucleotide from mouse Tet2 locus. The protospacer sequence is underlined and PAM sequence colored in green. Oligonucleotide-directed 2-bp changes are colored in red. Lower panel, RFLP analysis of 11 mice from the group electroporated with Cas9 mRNA, sgRNA targeting the Tet2 locus, and donor oligonucleotide at 400/200/400 ng/μl as indicated at the bottom of the panel. The cleaved band from the introduced EcoRI site is indicated by a red arrow. (B) Sequencing traces of PCR products encompassing the Tet2 target region from four mutant mice presented in A (86EP1–4). The PAM sequence is underlined by a green bar and the wild-type EcoRV site, a black bar. Overlapping sequencing traces among the mutant mice indicate existence of more than one allele among these mice, as compared to the WT mouse. The positions of the start of mutations are indicated by black arrows. (C) PCR products from two mice, 86EP1 and 86EP4, were cloned and individual clones sequenced. Sequences from clones 86EP1-2 and 86EP4-4 are shown. Precise modification converting EcoRV (underlined by a black bar) to EcoRI (underlined by a red bar) site was confirmed in these two mice.
To assess the off-target profile of CRISPR/Cas9 delivered by ZEN, we predicted the top 14 off-target sites for the Tet2 sgRNA (with up to three base pair mismatches) based on published principles (Yang et al. 2013) and analyzed all 14 loci in four founder mice carrying Tet2 targeted mutations (86EP1–4). Sanger sequencing of PCR products encompassing these loci showed none of these regions contain mutations at the predicted off-target sites (Table S4), suggesting CRISPR/Cas9 delivered by ZEN did not produce significant off-target effect, similar to our previous results using microinjection (Wang et al. 2013; Yang et al. 2013).
Discussion
Microinjection, invented in the early 1980s, has been the technology of choice to deliver exogenous genetic material into mouse zygotes for the generation of genetically modified mouse models. Although effective, it is technically demanding and low in throughput. In addition, it requires significant investment in equipment and operators with years of training and experience. As such, the capacity for generating genetically modified mice is often limiting, even for the most well-funded laboratories.
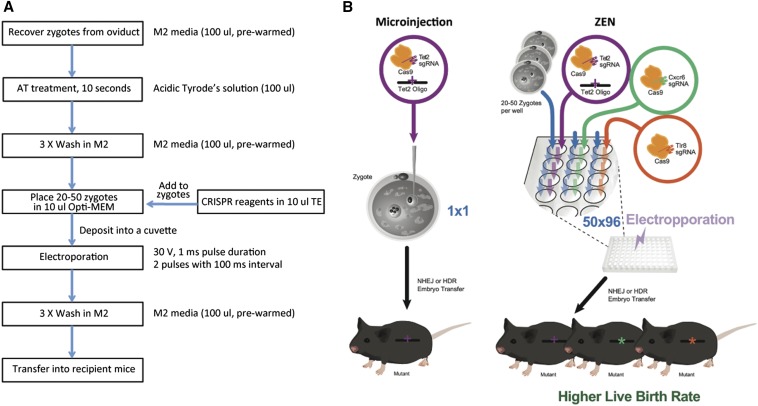
We demonstrated electroporation is an effective method for delivering CRISPR/Cas9 material into the mouse zygotes. Using our zygote electroporation of nuclease (ZEN) protocol (Figure 3A), in one experimental setting, we were able to generate mice carrying NHEJ mutations at 100% efficiency (11/11) and HDR-mediated modifications at 27% efficiency (3/11) at the Tet2 locus. To our surprise, for hybrid embryos, electroporation appears to be less invasive, as compared to the microinjection method. The live birth rate of ZEN-treated embryos is 59% (Table 2), a level approaching that of untreated embryos, and significantly higher than the 30% live birth rate from microinjected embryos in our hands. In addition, an obvious advantage is that a large number of embryos can be processed with the ZEN protocol (20–50 embryos were loaded into a single cuvette in our study) in one single run and multiple projects in parallel (Figure 3B).
Figure 3.
ZEN enables high throughput genome editing in mice. (A) Flow chart of CRISPR/Cas9-mediated genome editing delivered by ZEN. (B) ZEN compared to microinjection.
Targeting efficiency using ZEN can be excellent, although there are significant variations among experiments we performed: 29% NHEJ mutation rate in one experiment (2/7), 100% in another (6/6), processed at the same time and under the seemingly identical conditions. This variation could be due to the subtleties of electroporation that still need to be understood, including the developmental stages of the embryos, variations in the AT treatment, and electroporation settings. Understanding the parameters and further optimization should lead to more consistent performance of the technology.
While we were preparing this manuscript, another group reported successful use of electroporation to deliver endonuclease mRNA to generate genetically modified rats (Kaneko et al. 2014). We achieved a high NHEJ mutation rate in mice with our ZEN protocol with a much lower concentration of Cas9 mRNA (600 ng/μl) and sgRNA (300 ng/μl), and precise genetic modification by coelectroporation of a donor oligonucleotide. Zona weakening by AT treatment in our protocol, which does not seem to compromise survival of the embryos, might improve access to the embryos by CRISPR/Cas9 reagents, thereby achieving a higher targeting efficiency.
Compared to hybrid strains, genome editing has been proven to be more challenging in many inbred strains of mice, likely due to the fact that zygotes from inbred strains of mice are more prone to damage from microinjection procedure. Here we show that, using electroporation to deliver CRISPR/Cas9 reagents, mice with targeted mutations can be efficiently derived from the inbred C57BL/6NJ, a background strain used for the Knockout Mouse projects (KOMP). We are in the process of testing and optimizing our ZEN protocol on other inbred strains and hope to establish ZEN as a delivery method for CRISPR/Cas9-mediated genome editing for various inbred strains of mice.
Development of the electroporation protocol brings high efficiency, high throughput genome engineering in animal models within closer reach. By combining the simplicity and robustness of the CRISPR/Cas9 system with the throughput of the ZEN technology, we may now be in a position to generate mouse models of human disease alleles with unprecedented efficiency and throughput. This convergence of the power of reading and writing the genome promises to accelerate biological research at an unprecedented pace in the coming years.
Supplementary Material
Acknowledgments
We thank the Microinjection Service group at The Jackson Laboratory (JAX) for their excellent assistance with embryo generation, transfer, and sample collection and the Cell Biology Service group at JAX for in vitro embryo culture. We thank Jesse Hammer for the art work. This work is supported in part by the National Cancer Institute under award number P30CA034196 and an institutional grant from The Jackson Laboratory (H.W.). A.C. is supported by a JAX postdoctoral scholar fellowship. A.M.G. and Y.-G.C. are supported by National Institute of Diabetes and Digestive and Kidney Diseases grant DK097605. H.W. is supported by “National Natural Science Foundation of China” (31471215), “Strategic Priority Research Program” of the Chinese Academy of Sciences (XDA01010409), and the State 863 Project 2015AA020307.
Footnotes
Communicating editor: S. K. Sharan
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.176594/-/DC1.
Literature Cited
- Bogdanove A. J., Voytas D. F., 2011. TAL effectors: customizable proteins for DNA targeting. Science 333: 1843–1846. [DOI] [PubMed] [Google Scholar]
- Brinkman E. K., Chen T., Amendola M., van Steensel B., 2014. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 42: e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capecchi M. R., 2005. Gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century. Nat. Rev. Genet. 6: 507–512. [DOI] [PubMed] [Google Scholar]
- Cong L., Ran F. A., Cox D., Lin S., Barretto R., et al. , 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna J. A., Charpentier E., 2014. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346: 1258096. [DOI] [PubMed] [Google Scholar]
- Grabarek J. B., Plusa B., Glover D. M., Zernicka-Goetz M., 2002. Efficient delivery of dsRNA into zona-enclosed mouse oocytes and preimplantation embryos by electroporation. Genesis 32: 269–276. [DOI] [PubMed] [Google Scholar]
- Hai T., Teng F., Guo R., Li W., Zhou Q., 2014. One-step generation of knockout pigs by zygote injection of CRISPR/Cas system. Cell Res. 24: 372–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P. D., Lander E. S., Zhang F., 2014. Development and applications of CRISPR-Cas9 for genome engineering. Cell 157: 1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang W. Y., Fu Y., Reyon D., Maeder M. L., Tsai S. Q., et al. , 2013. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 31: 227–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A., et al. , 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko T., Sakuma T., Yamamoto T., Mashimo T., 2014. Simple knockout by electroporation of engineered endonucleases into intact rat embryos. Sci Rep. 4: 6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Qiu Z., Shao Y., Chen Y., Guan Y., et al. , 2013a Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat. Biotechnol. 31: 681–683. [DOI] [PubMed] [Google Scholar]
- Li W., Teng F., Li T., Zhou Q., 2013b Simultaneous generation and germline transmission of multiple gene mutations in rat using CRISPR-Cas systems. Nat. Biotechnol. 31: 684–686. [DOI] [PubMed] [Google Scholar]
- Niu Y., Shen B., Cui Y., Chen Y., Wang J., et al. , 2014. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell 156: 836–843. [DOI] [PubMed] [Google Scholar]
- Palmiter R. D., Brinster R. L., Hammer R. E., Trumbauer M. E., Rosenfeld M. G., et al. , 1982. Dramatic growth of mice that develop from eggs microinjected with metallothionein-growth hormone fusion genes. Nature 300: 611–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H., Wu Y., Zhang Y., 2012. Efficient delivery of DNA and morpholinos into mouse preimplantation embryos by electroporation. PLoS ONE 7: e43748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov F. D., Rebar E. J., Holmes M. C., Zhang H. S., Gregory P. D., 2010. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 11: 636–646. [DOI] [PubMed] [Google Scholar]
- Wang H., Yang H., Shivalila C. S., Dawlaty M. M., Cheng A. W., et al. , 2013. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153: 910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Wang H., Shivalila C. S., Cheng A. W., Shi L., et al. , 2013. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154: 1370–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.