Abstract
Saccharomyces cerevisiae telomeres have been a paradigm for studying telomere position effects on gene expression. Telomere position effect was first described in yeast by its effect on the expression of reporter genes inserted adjacent to truncated telomeres. The reporter genes showed variable silencing that depended on the Sir2/3/4 complex. Later studies examining subtelomeric reporter genes inserted at natural telomeres hinted that telomere position effects were less pervasive than previously thought. Additionally, more recent data using the sensitive technology of chromatin immunoprecipitation and massively parallel sequencing (ChIP-Seq) revealed a discrete and noncontinuous pattern of coenrichment for all three Sir proteins at a few telomeres, calling the generality of these conclusions into question. Here we combined the ChIP-Seq of the Sir proteins with RNA sequencing (RNA-Seq) of messenger RNAs (mRNAs) in wild-type and in SIR2, SIR3, and SIR4 deletion mutants to characterize the chromatin and transcriptional landscape of all native S. cerevisiae telomeres at the highest achievable resolution. Most S. cerevisiae chromosomes had subtelomeric genes that were expressed, with only ∼6% of subtelomeric genes silenced in a SIR-dependent manner. In addition, we uncovered 29 genes with previously unknown cell-type-specific patterns of expression. These detailed data provided a comprehensive assessment of the chromatin and transcriptional landscape of the subtelomeric domains of a eukaryotic genome.
Keywords: Sir complex, telomeres, ChIP-Seq, RNA-Seq, mating-type regulation
TELOMERES are specialized structures at the ends of eukaryotic chromosomes that are critical for various biological functions. Telomeres bypass the problem of replicating the ends of linear DNA, protect chromosome ends from exonucleases and nonhomologous end joining, prevent the linear DNA ends from activating a DNA-damage checkpoint, and exhibit suppressed recombination [reviewed in Wellinger and Zakian (2012)]. In Saccharomyces cerevisiae, telomeres are composed of three sequence features: telomeric repeats, which consist of 300 ± 75 bp of (TG1–3)n repeated units produced by telomerase; X elements; and Y′ elements, which contain an ORF for a putative helicase gene. The X elements are subdivided into a core X [consisting of an autonomously replicating sequence (ARS) consensus sequence and an Abf1-binding site] and subtelomeric repeats that have variable numbers of repeated units containing a binding site for Tbf1 (Louis 1995). All telomeres contain telomeric repeats plus an X element, and about half of S. cerevisiae’s 32 telomeres also contain a Y′ element (X-Y′ telomeres). X-only telomeres contain an X element but not a Y′ element. Unlike the Y′ elements, the telomeric repeats and X elements are bound by proteins that are critical for maintenance of telomeres. Rap1 binds the TG1–3 telomeric repeats and recruits the Sir2/3/4 protein complex, the trio of heterochromatin structural proteins critical for repression of the silent mating loci HMLα and HMRa. Sir proteins are also recruited to the core X sequence through interactions with Abf1 and the origin recognition complex (ORC), which binds the ARS consensus sequence within the core X. Thus telomeres have a heterogeneous sequence composition, recruit proteins that can form heterochromatin-like structures, and are critical for maintaining the genomic integrity of the cell.
As first described in Drosophila (Schultz 1947; Hazelrigg et al. 1984), the heterochromatic structure of telomeric chromatin results in the transcriptional silencing of adjacent genes, an effect known as telomere position effect. Since its description, telomere position effect has been observed in other organisms, where it can be an important means of regulating gene expression. For example, the malarial parasite Plasmodium falciparum genome contains subtelomeric var genes that encode cell surface antigens that use Sir2-dependent telomeric heterochromatin for their repression (Guizetti and Scherf 2013). var genes are selectively expressed, one at a time, and switch expression states, allowing Plasmodium to stay ahead of the host’s immune response. This selective expression of one antigen over all the other antigen genes is maintained by the epigenetic silencing of all var copies except the expressed one (Tonkin et al. 2009; Guizetti and Scherf 2013). Similarly, in Candida glabrata, the EPA adhesion genes essential for colonization of the host urinary tract are located in subtelomeric regions, and their expression is regulated by a Sir-protein-based silencing mechanism that is responsive to the differences in niacin concentration in the bloodstream vs. the urinary track (De Las Peñas et al. 2003; Domergue et al. 2005). In S. cerevisiae, genes encoding cell wall components and genes required for the metabolism of certain nutrients tend to be located in subtelomeric regions and are expressed specifically under certain stressful conditions (Ai et al. 2002).
Telomere position effect was first described in S. cerevisiae by the attenuated expression of reporter genes placed adjacent to a synthetic telomere on either the left arm of chromosome VII or the right arm of chromosome V (Gottschling et al. 1990; Renauld et al. 1993; Fourel et al. 1999). Reminiscent of general epigenetic silencing, the effect was concluded to be independent of gene identity and promoter sequence. Furthermore, much like silencing at the mating-type cassettes HMLα and HMRa, the silenced state of telomere-adjacent URA3 and ADE2 was heritable and depended on the silent information regulator proteins Sir2, Sir3, and Sir4. Unlike HMLα and HMRa, deletion of SIR1 had no effect on telomeric silencing (Aparicio et al. 1991). These and other early studies led to the view that Sir proteins were in a continuous gradient, highest at the telomere and extending inward for a few kilobase pairs, depending in particular on the level of Sir3 protein (Renauld et al. 1993; Hecht et al. 1996; Strahl-Bolsinger et al. 1997).
More recent findings have questioned the earlier view of telomere position effect in S. cerevisiae. For example, when inserted adjacent to the native telomeres TEL10R, TEL04L, and TEL03R, the same URA3 reporter detects little transcriptional repression (Pryde and Louis 1999). For the few natural telomeres at which URA3 appears repressed (TEL13R, TEL11L, and TEL02R), silencing is discontinuous across the length of the telomere and largely restricted to positions close to the X element. Similarly, Sir proteins also associate discretely at select natural telomeres, with the highest levels of enrichment proximal to the X element (Zill et al. 2010; Radman-Livaja et al. 2011; Thurtle and Rine 2014). The natural telomeres that repress the URA3 transgene exhibit a characteristic array of phased nucleosomes specific to those telomeres (Loney et al. 2009). Additionally, some Y′ elements are transcribed, a fact that is inconsistent with Sir protein–mediated repression of all Y′ elements (Fourel et al. 1999; Pryde and Louis 1999). In addition to these discrepancies, metabolic reporters are not biologically neutral, and some complexity regarding these reporters has emerged (Rossmann et al. 2011; Takahashi et al. 2011). For example, DOT1, SWI4, and ARD1, all of which abrogate H3K79 methylation, had been implicated in telomeric silencing, as assayed by the URA3 reporter at artificial telomeres. However, transcription of native genes at telomeres, as measured by microarray analysis, revealed little change in expression level in a dot1 mutant and other mutants proposed to disrupt H3K79 methylation (Takahashi et al. 2011). Subsequent interrogation of the URA3 reporter found that dot1 and other mutants are actually differentially sensitized to the drug 5-FOA used to monitor URA3 expression (Rossmann et al. 2011). Therefore, the phenotypes of these mutants, as measured by 5-FOA sensitivity, do not reliably reflect the transcriptional status of URA3 at telomeres.
In summary, establishing the prevalence of telomere position effect and identifying the genes and proteins that mediate it have been complicated by three issues: (1) nonsystematic studies of different telomeres in S. cerevisiae, (2) the influence of metabolism on telomeric reporters, and (3) limitations on the resolution of chromatin immunoprecipitation (ChIP) and microarray analysis. To resolve these confounding issues, we undertook a high-resolution analysis of chromatin architecture and expression state at all natural S. cerevisiae telomeres, free of reporter genes, by using chromatin immunoprecipitation and massively parallel sequencing (ChIP-Seq) analysis of Sir proteins combined with RNA sequencing (RNA-Seq) analysis of wild-type (WT) cells and sir2Δ, sir3Δ, and sir4Δ mutants. ChIP-Seq of acetylated H4K16, a histone mark anticorrelated with silencing, was also analyzed to further evaluate specific histone modifications with respect to expression data from RNA-Seq. This study provided a definitive analysis of the chromatin landscape and degree of silencing at telomeres in S. cerevisiae and highlighted the functional variation among telomeres, befitting the accelerated sequence changes seen in these cauldrons of genetic innovation.
Materials and Methods
Yeast strains
Yeast strains and plasmid-containing strains are listed in Supporting Information, Table S5. All yeast strains were generated in the W303 background. Deletion alleles were constructed via one-step integration of knockout cassettes (Longtine et al. 1998).
RNA isolation
Cells were grown at 30° in rich medium (YPD) to an A600 of 0.8. RNA was extracted from 15 A600 units of cells using the hot acid–phenol and chloroform method (Collart and Oliviero 2001). Briefly, cells were incubated in TES buffer (10 mM Tris HCl, pH 7.5, 10 mM EDTA, and 0.5% SDS) and citrate-saturated phenol (pH 4.3) for 1 hr at 65° and vortexed every 10 min. RNA was isolated from lysed cells with two rounds of phenol-chloroform extraction, pelleted, and then resuspended in RNase-free water and treated with DNase I (Roche) to digest genomic DNA. A final round of phenol-chloroform extraction was performed prior to library preparation and/or complementary DNA (cDNA) synthesis.
RNA library preparation and sequencing
Paired-end sequencing was performed to accurately assign reads. 100-bp paired-end RNA-Seq libraries were prepared using the Illumina TruSeq Stranded mRNA Sample Prep Kit with 4 μg of total RNA as starting material, as described in the TruSeq Stranded mRNA Sample Prep Kit protocol. Libraries were quantified using a Bioanalyzer (Agilent) and sequenced on an Illumina HiSeq 2000 machine. Reads have been deposited in the NCBI Sequence Read Archive (SRA) at http://www.ncbi.nlm.nih.gov/sra under accession no. SRP055208.
Quantitative reverse-transcriptase-PCR (qRT-PCR) analysis
cDNA was prepared from 2 μg of total RNA using the SuperScript III Reverse Transcriptase Kit (Invitrogen). qRT-PCR was performed using SYBR Green Real-Time PCR Master Mix (Thermofisher) and was quantified using a Stratagene Mx3000 qPCR System. Standard curves were generated from a WT strain and a sir2Δ strain, and all expression values were normalized to ACT1. Values shown are the average of three biological replicates. Error bars reflect the standard error. Two-tailed Student’s t-test was performed to evaluate the significance of the observed differences in expression. Oligos used are listed in Table S6.
Data analysis
ChIP-Seq read mapping:
ChIP-Seq reads analyzed were from previous Sir protein ChIP studies (Teytelman et al. 2013; Thurtle and Rine 2014), deposited in the NCBI Sequence Read Archive under accession nos. SRP030670 and SRP034921, respectively. Reads were mapped using BWA (Li and Durbin 2009) to a modified sacCer2 genome in which the MAT locus was replaced with the Hyg-MX cassette. Duplicate reads were removed using Picard (http://picard.sourceforge.net). Because of the repeated sequences shared among telomeres, some reads could not be mapped to specific telomeres. Making the simplifying assumption that all copies of a repeat sequence contributed to the production of sequence reads of that repeat, reads that mapped to repeated sequences were randomly assigned to copies of that repeat, allowing for an estimation of Sir protein association even at the repetitive elements of the telomeres. However, to indicate which reads were uniquely mapped and which mapped more than once, we graphed the percentage of reads within each telomere that did not map uniquely (Figure S3). This analysis clearly showed that Y′ elements at all telomeres are difficult to distinguish from each other except at positions of polymorphisms unique to individual Y′ elements. Additionally, almost the entire 20-kbp regions of TEL01R, TEL04L, TEL09L, TEL10L, TEL10R, TEL14L, TEL15R, and TEL16L are not unique. The laboratory strain (derived from W303) on which the ChIP-Seq experiments were performed had deletions in subtelomeric regions compared to the S288C reference genome (TEL07L, TEL14R, and small gaps on TEL01R and TEL13R). These missing regions in the sequenced strain were indicated in the figures. Reads were mapped to the S288C genome to allow direct reference to the annotated features on the Saccharomyces Genome Database (SGD). For each sample, per-base-read counts were determined using SAMtools (Li et al. 2009). Enrichment was determined as the number of IP reads divided by the number of input reads for that base-pair position.
MACS peak calling:
MACS peak calling was performed on the default settings, except that no model was used to optimize for the broader peaks typical of chromatin-interacting proteins. For each Sir protein chromatin sample, MACS was run on two biological replicates of ChIP-Seq data from chromatin sheared by sonication and on a third sample for each Sir protein in which the chromatin sample was prepared by enzymatic digestion with MNase (Thurtle and Rine 2014). For each chromatin sample analyzed with MACS, the IP sample was the “treatment,” and the input sample was the “control.” We defined peaks as reproducible if they were called in at least two of the three data sets, as noted in Table S1.
RNA-Seq:
Reads were mapped using Tophat2, and per-gene transcript quantification was performed using Cufflinks and reported as fragments per kilobase per million reads (FPKM) (Trapnell et al. 2009, 2012). Locations of multi-mapped RNA reads are indicated in Figure S8. Genome-wide RNA read pileups per base pair were calculated using SAMtools (Li et al. 2009). The DESeq pipeline was used to perform differential gene expression analysis, as outlined in the following steps: (1) raw read counts per gene were determined using htseq-count, which discards multimapped paired-end read fragments (Anders and Huber 2010); therefore, only uniquely mapped reads were included in tests for differential expression of genes; and (2) read counts were normalized and subjected to differential expression analysis using the DESeq package in R (Anders and Huber 2013). Genes that showed statistically significant differences in expression of twofold or greater relative to WT with a P-value of less than 0.05 and a false-discovery rate of less than 10% were included in the final list of candidate genes under SIR2/3/4 repression or as possible haploid-specific genes.
Comparison of transcription at telomeres vs. nontelomeric loci:
Genes were classified as either falling within (telomeric) or not falling within (nontelomeric) 20 kbp of a chromosome end, resulting in two distributions of FPKM values. A Wilcoxon rank-sum test was performed to compare the telomeric vs. nontelomeric distributions.
MEME analysis:
The MAST program within the MEME package was used to scan the coding sequence, plus and minus 1000 base pairs, for a1/α2 and α2/Mcm1 binding sites in candidate haploid-specific genes (Bailey et al. 2009). Results were filtered for E-values < 10.
Scanning motif binding sites on the yeast transcription factor specificity compendium:
The Binding Site Genome Browser (http://nbrowse.ccbr.utoronto.ca/mgb2/gbrowse/yetfasco/) was used to search for a1/α2 and α2/Mcm1 binding sites within 1 kbp of each candidate gene. All a1/α2 and α2 binding sites with a score greater than 80% of the motif’s maximum position-weighted matrix-score threshold were noted.
Results
Sir proteins associated at discrete positions at natural telomeres
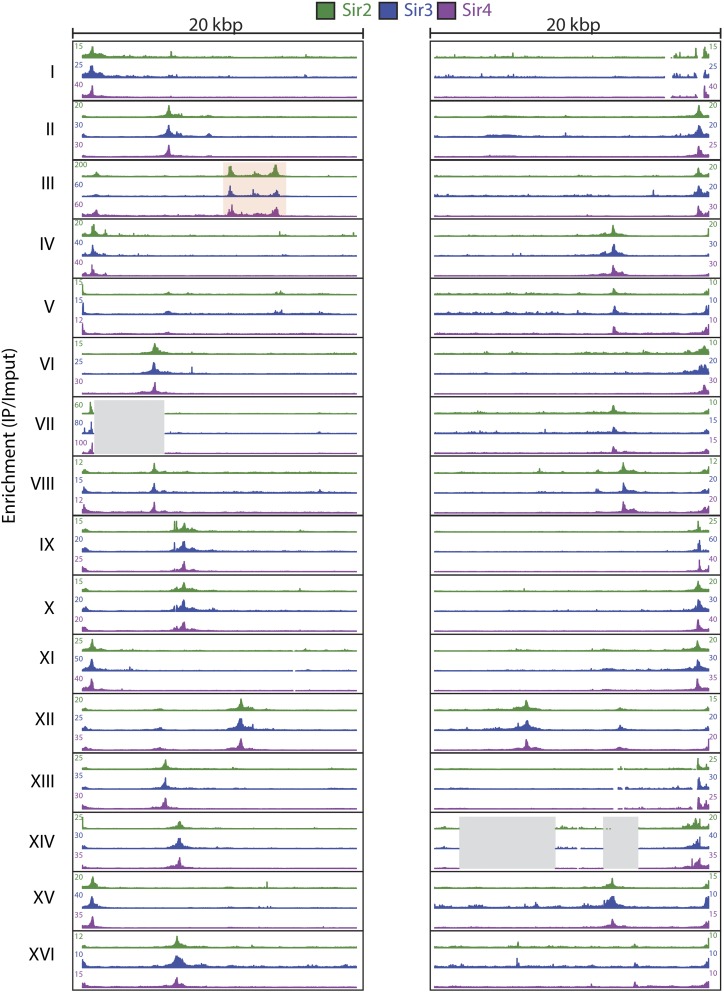
To investigate Sir protein association at the 32 natural telomeres of S. cerevisiae, we analyzed ChIP-Seq data sets in the 20-kbp subtelomeric region of Myc-tagged Sir2, Sir3, and Sir4 from our previous Sir ChIP-Seq studies (Thurtle and Rine 2014) (Figure 1). Additionally, we analyzed ChIP-Seq data sets for green fluorescent protein endowed with a nuclear localization signal (GFP-NLS) and a no-tag sample immunoprecipitated with the Myc antibody as controls for artifacts of ChIP-Seq analyses and nonspecific enrichment, respectively (Teytelman et al. 2013) (Figure S1 and Figure S2). The telomeric regions are difficult to analyze because of their repetitive nature and incomplete sequencing at some of the telomere ends. Thus we made simplifying assumptions about ambiguously mapped reads, as outlined in Materials and Methods and Figure S3. The peaks at the TEL05L and TEL14L chromosomes, for example, for which no telomeric repeats are annotated, presumably arose from ChIP-Seq reads that extended from telomeric repeats into sufficiently unique flanking sequences to allow mapping. Where the telomerase-generated repeats are present, the Rap1 protein-binding sites embedded in those repeats were presumably responsible for the Sir protein enrichment at those positions (e.g., TEL08R and TEL08L). Most strikingly, at the 32 natural telomeres, the enrichment patterns of the three Sir protein complex members were highly similar, illustrating both the remarkable degree of reproducibility of the enrichment patterns and the discontinuous nature of the Sir protein enrichments at each and every telomere (Figure 1). There was no evidence of a gradient of Sir proteins, as envisioned by early models of telomere position effect (Hecht et al. 1996). The discontinuous distribution of Sir proteins has been reported previously for specific telomeres (Zill et al. 2010; Thurtle and Rine 2014). Overall, this analysis clearly established the generality of the discrete nature of Sir protein association at all 32 telomeres.
Figure 1.
Sir2, Sir3, and Sir4 enrichment at all 32 yeast telomeres. ChIP-Seq of Myc-tagged Sir2, Sir3, and Sir4 was analyzed at all yeast telomeres. (Left) The first 20 kbp of each chromosome. (Right) The last 20 kbp of each chromosome. IP/input enrichment values for Sir2 (green), Sir3 (blue), and Sir4 (green) are shown for each telomere. On chromosome III, HML is boxed in red, and regions absent in the sequenced W303 strain relative to the S288C sacCer2 genome are represented by a gray-shaded box.
To provide a statistical evaluation of the Sir2, Sir3, and Sir4 peaks detected by eye, we called peaks of significant enrichment with MACS using the default P-value cutoff of 0.00001 (Zhang et al. 2008). To control for nonspecific enrichment, we also called peaks of enrichment with MACS on a ChIP-Seq data set from a heterologous protein control, GFP-NLS. For the GFP-NLS, only one small region on the TEL02L (base-pair positions 8824–10,250) showed overlapping enrichment with Sir protein peaks. Thus the Sir protein peak was adjusted to account for this nonspecific enrichment. Otherwise, nonspecific enrichment from highly expressed transcripts did not confound the ChIP enrichment at telomeres, in contrast to other places in the genome (Teytelman et al. 2013). As determined by the MACS peak calling, all but 5 of the 32 yeast telomere X elements exhibited significant enrichment of Sir proteins (Table S1). For those five telomeres in which MACS did not identify a peak (TEL1R, TEL2R, TEL10R, TEL13R, and TEL14R), there appeared to be ample enrichment by eye (Figure 1). All five of these telomeres were X-only telomeres in which the enrichment abutted the end of the chromosome, possibly resulting in MACS not calling the peak because of its abrupt end and the presence of a repetitive sequence. Hence Sir protein enrichment appeared to be a property of all, or nearly all, X elements. For 15 of the 19 X-Y′ telomeres, MACS positioned the peak of Sir protein enrichment as extending all the way from the chromosome end to the internal X element, spanning the entire Y′ element (Table S1). To determine whether there actually was detectable Sir protein enrichment within the Y′ element or whether these large peaks called were due to the proximity of two distinct peaks, we calculated the average enrichment (IP/input) for all the X elements and all the Y′ elements for Sir2, Sir3, and Sir4 (Figure S4). For the three Sir proteins, the average X element enrichment was fourfold for Sir2 and eightfold for Sir3 and Sir4. In contrast, the Y′ elements all showed IP/input values of less than 1 for all three Sir proteins, indicating that the IP values for this region all were below background. Thus, as reported previously for specific telomeres (Zhu and Gustafsson 2009; Zill et al. 2010; Takahashi et al. 2011; Thurtle and Rine 2014), the Y′ elements did not exhibit any Sir protein enrichment. In summary, Sir proteins showed the highest level of association at the core X element, with average enrichment values between 4.5 and 8.2 for the three Sir proteins, where ORC and Abf1 bind, whether at an X-element-only telomere or at an X-Y′ telomere (Figure 1 and Figure S4).
Catalytic activity of Sir2 at telomeres
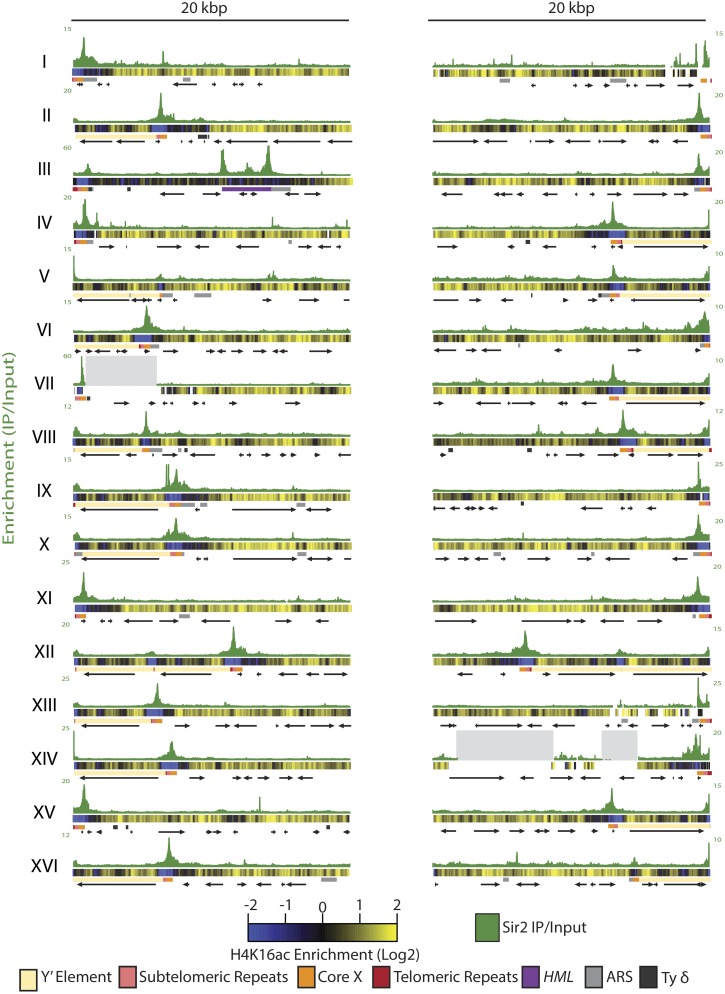
To determine whether positions of H4K16 hypoacetylation overlapped with Sir2 distribution at telomeres, we analyzed ChIP-Seq of H4K16-acetyl and compared Sir2 ChIP-Seq profiles at all 32 telomeres to the H4K16-acetyl ChIP-Seq profiles (Figure 2). H4K16 was hypoacetylated in regions slightly larger than the X element, with the lowest levels of H4K16-acetyl at the core X sequence. Additionally, X-Y′ telomeres showed a variable amount of H4K16 hypoacetylation within the Y′ region. We also observed regions of H4K16 hypoacetylation without detectable Sir2 association, which presumably reflected the action of a different histone deacetylase such as Rpd3 or Hst1. Both have been shown to associate with subtelomeric chromatin (Kurdistani et al. 2002; Ehrentraut et al. 2010; Li et al. 2013). Alternatively, the hyopacetylation of H4K16 in these regions could be due to transient Sir2 association not captured by ChIP-Seq. Previous studies have shown that Sir2, but not Sir3 or Sir4, controls some origins of replication (Pappas et al. 2004; Crampton et al. 2008; Yoshida et al. 2014). However, MACS did not detect any significant enrichment for Sir2 at subtelomeric ARSs outside the core X element.
Figure 2.
H4K16 exhibited hypoacetylation in regions greater than Sir2 protein association. Sir2 enrichment is shown for each telomere as the IP/input for that base-pair position. Below the Sir2 enrichment track for each telomere is a heat map representing the log2 of H4K16 IP/input. Blue represents regions of hypoacetylation where the IP value is below the input value, and yellow represents IP/input values greater than 1, which indicate acetylated regions. Salient features for each telomere are shown: telomeric repeats as red boxes, subtelomeric repeats as pink boxes, the core X as orange boxes, and HML as a dark-purple rectangle. Origins of replication and Ty δ elements are marked in light gray and dark gray, respectively. ORFs are represented by black arrows. All features were mapped as annotated in the SGD.
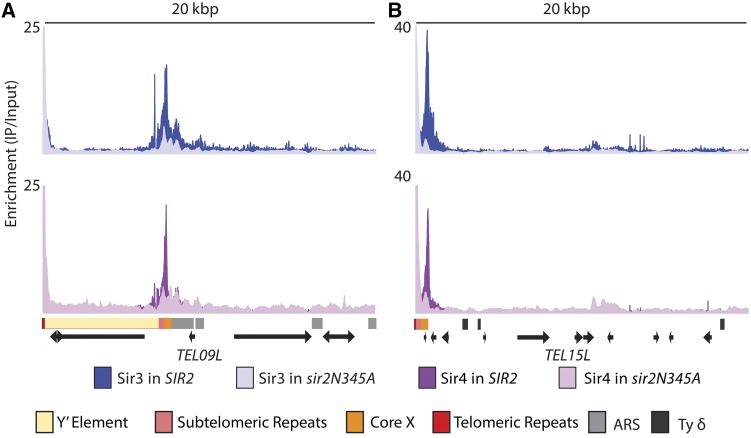
The deacetylation of H4K16-acetyl by Sir2 is thought to be key for the spreading of Sir proteins (Hecht et al. 1996; Rusche et al. 2002; Hoppe et al. 2002). In the standard model for spreading [reviewed in Rusche et al. (2002)], Sir proteins are recruited to nucleation sites via protein interactions among ORC, Abf1, and Rap1, which are bound to DNA, Sir3, and a Sir2-Sir4 dimer. According to the model, Sir2 deacetylates nearby nucleosomes, which creates high-affinity binding sites for Sir3 and Sir4, resulting in the spread of additional copies of the Sir protein complex. Thus this model predicts that Sir protein enrichment should be continuously distributed along the length of a telomere. However, the distribution of Sir proteins at the telomeres was discrete (Figure 1 and Figure 2) and therefore not in support of the spreading model. To determine the role of Sir2’s catalytic activity in Sir protein association at the telomeres, Sir3 and Sir4 enrichment was examined at the telomeres in a strain lacking Sir2 catalytic activity (Thurtle and Rine 2014). As shown for a representative X-only telomere (TEL15L), there seemed to be some indications of spreading for Sir3 because the association of Sir3 in the WT background extended about 800 bp beyond where Sir3 associated in a strain lacking Sir2 catalytic activity (Figure 3). This extended distribution was less prominent for Sir4 at the X-only telomere and both Sir3 and Sir4 at the internal X element of the X-Y′ telomere (TEL09L) (Figure 3). These results indicate that if Sir complex spreading occurred at telomeres, it did so only to a slight extent. The prominent feature of all telomeres was the overall reduced Sir3 and Sir4 association at the core X element in a strain lacking Sir2 catalytic activity, indicating that Sir2’s catalytic activity is necessary for the association and/or stability of the Sir protein complex with ORC and Abf1. Both Sir3 and Sir4 showed enrichment in the telomeric repeats in a strain lacking Sir2 catalytic activity. However, as reported previously (Zill et al. 2010; Teytelman et al. 2013), the telomeric repeats showed enrichment in the no-tag ChIP-Seq control sample as well, indicating that the telomeric repeats, whether at the chromosome ends of X-only telomeres or at internal locations of X-Y′ telomeres, interact nonspecifically with the anti-Myc antibody (Figure S2). This interaction seemed to be specific for the Myc antibody because the GFP-NLS immunoprecipitated with an anti-GFP antibody did not show enrichment at the telomeric repeats (Figure S1). It was surprising that the no-tag ChIP-Seq control sample and the Sir3 and Sir4 samples in strains lacking Sir2 catalytic activity indicated greater enrichment at the telomeric repeats than the level of Sir protein enrichment at the telomeric repeats in WT strains. However this apparent greater enrichment may be a consequence of increasing the signal-to-noise ratio: there are fewer sites with lower amounts of Sir3 and Sir4 enrichment in a strain lacking Sir2 catalytic activity and very little association in the no-tag sample; thus more Myc antibody is available to associate nonspecifically with the telomeric repeats. Overall, Sir2’s catalytic activity at telomeres was important for association of the Sir protein complex at the core X nucleation sites and less implicated in the spreading of the Sir complex into subtelomeric regions.
Figure 3.
Sir3 and Sir4 association in strains lacking Sir2 catalytic activity. ChIP-Seq reads of Myc-tagged Sir3 and Sir4 in a strain expressing a catalytically inactive point mutant SIR2 allele, SIR2N345A, were analyzed at the telomeres. A representative X-Y′ telomere is shown in A, and a representative X-only telomere is shown in B. The upper panel shows Sir3 association in the WT SIR2 strain (dark blue) and the mutant sir2N345A background (light blue). The lower panel shows Sir4 association in the WT SIR2 strain (dark purple) and the mutant sir2N345A background (light purple). Salient features for each telomere are as in Figure 2.
Most S. cerevisiae telomeres have expressed genes
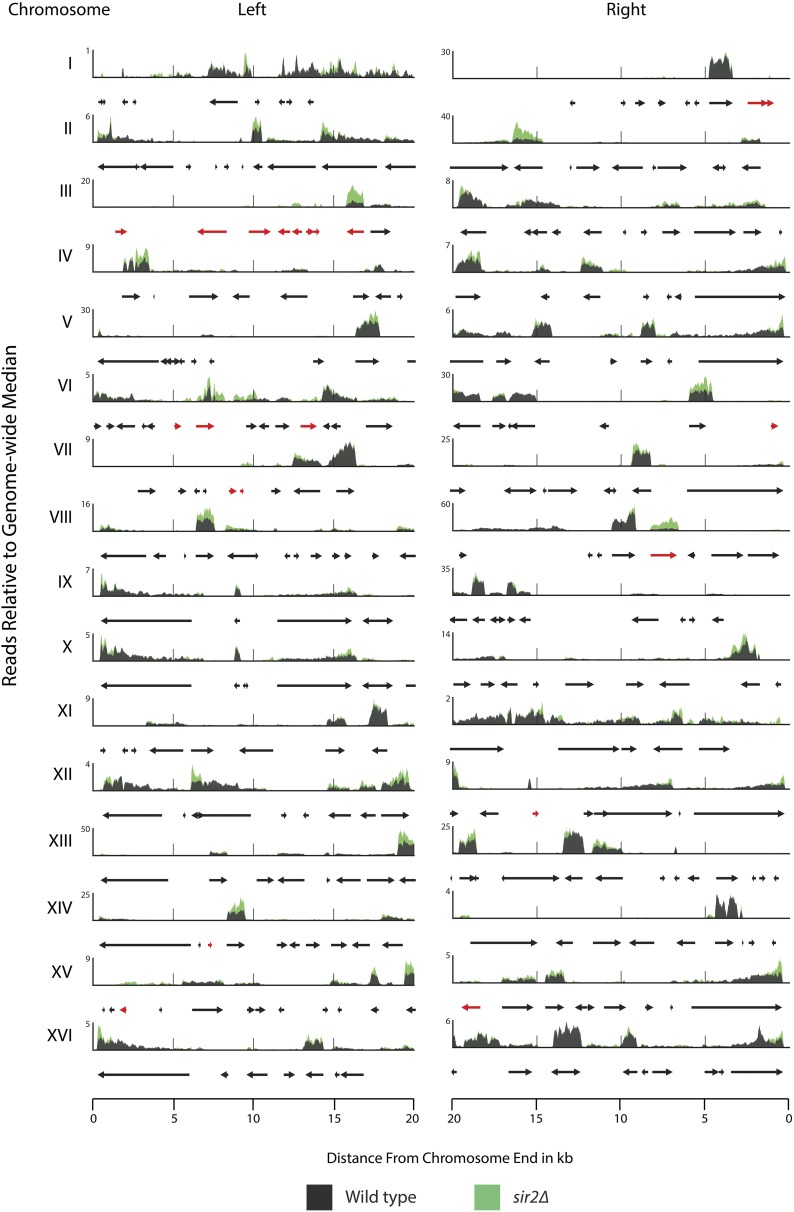
To determine the expression state of all genes at all 32 S. cerevisiae telomeres, we performed mRNA-Seq on RNA samples from WT and sir2Δ, sir3Δ, and sir4Δ strains. The MAT locus, which specifies mating type, was deleted in these strains to allow nearly complete, unambiguous read mapping between the two silent-mating-type cassettes HMLα and HMRa. Analysis of mRNAs in WT and sir2Δ strains across all subtelomeric regions revealed several important generalizations (Figure 4 and Figure S5; the highly similar results for sir3Δ and sir4Δ are shown in Figure S6 and Figure S7). All chromosomes had numerous genes within 20 kbp of the ends that were expressed. Transcription occurred within 5 kbp of most ends. Thus there was no evidence supporting widespread Sir-based repression of most genes near telomeres. For most of the transcripts detected in subtelomeric regions, there was no detectable increase in transcript number in sir2∆ relative to WT strains. For some loci, transcription increased modestly in sir2∆ strains (ORFs shown in red; genes listed in Table 1). An important and expected exception was HMLα1 and HMLα2; these genes showed a substantial increase in expression in sir2∆ strains (see TEL03L 15 kbp from end). Interestingly, repression at TEL03L extended approximately 12 kbp beyond HMLα to the end of chromosome III because all annotated ORFs in this region increased in expression in sir2∆ strains (Table 1). Sir2 was found to be enriched across this entire domain as well, along with hypoacetylated H4K16. Thus the expression status in WT strains correlated with these two marks of heterochromatin. This was the only telomere for which there was evidence of a Sir protein–mediated domain of repression.
Figure 4.
Transcription at all 32 telomeres in WT and sir2Δ strains. RNA-Seq was performed on WT and sir2Δ strains. Shown are read pileups from WT (black) and sir2Δ (green). Read pileups are normalized to the median genome-wide coverage and are the average of three biological replicates. Genes that showed a twofold or greater increase in expression in all three sir mutants (sir2Δ, sir3Δ, and sir4Δ) are colored as red arrows. Genes that showed no significant change in expression between WT and all three sir mutants are in black.
Table 1. Subtelomeric genes under Sir2/3/4 repression.
| Gene | Systematic name | Wild type | sir2Δ | sir3Δ | sir4Δ | Distance to nearest sir peak (bp) |
|---|---|---|---|---|---|---|
| IMD1 | YAR073W | 0.1 | 1.1 | 1.1 | 1.1 | 1,575 |
| YAR075W | YAR075W | 1.6 | 26 | 21.9 | 25 | 846 |
| YCL076W | YCL076W | 0 | 3.3 | 2.8 | 3.5 | 0 |
| YCL075W | YCL075W | 0 | 1.9 | 2.6 | 2.8 | 0 |
| YCL074W | YCL074W | 0 | 4.5 | 6.5 | 4.9 | 0 |
| GEX1 | YCL073C | 0.1 | 0.4 | 0.5 | 0.5 | 0 |
| VBA3 | YCL069W | 0.4 | 3.5 | 3.9 | 4.5 | 0 |
| YCL068C | YCL068C | 0.1 | 4.8 | 0.5 | 7.4 | 0 |
| YCL065W | YCL065W | 0 | 14.9 | 9.1 | 9.2 | 0 |
| CHA1 | YCL064C | 51.2 | 148 | 229.4 | 242.2 | 0 |
| YFL063W | YFL063W | 0 | 1.7 | 1.2 | 0.4 | 175 |
| COS4 | YFL062W | 5 | 12.5 | 15.3 | 18.1 | 1,527 |
| THI5 | YFL058W | 1.3 | 4.4 | 3.8 | 3.1 | 7,972 |
| YFR057W | YFR057W | 0.2 | 12 | 9.7 | 10.8 | 529 |
| YPS5 | YGL259W | 0.2 | 2.9 | 3.3 | 2.7 | 2,836 |
| YGL258W-A | YGL258W-A | 3.4 | 13.1 | 27.8 | 29.7 | 3,396 |
| IMD2 | YHR216W | 61.5 | 234.2 | 331.9 | 352.5 | 989 |
| PAU4 | YLR461W | 0.5 | 1.1 | 1.6 | 1.9 | 1,239 |
| YNL337W | YNL337W | 0 | 2.2 | 0.4 | 0.6 | 77 |
| AAD15 | YOL165C | 2.1 | 7.2 | 10.1 | 10.4 | 0 |
| FDH1 | YOR388C | 1.4 | 2.7 | 2.5 | 2.7 | 11,622 |
Shown in this table are the expression values in FPKM for the 21 subtelomeric genes that increased in expression in sir2Δ, sir3Δ, and sir4Δ. Genes are ordered by chromosome number and map position. FPKM values represent the average of three biological replicates. Distances to nearest Sir peaks were calculated by taking the difference of the midpoint of the gene and the genomic coordinate of the highest nearby Sir protein IP/input enrichment value.
Telomeres produced significantly fewer transcripts than nontelomeric loci
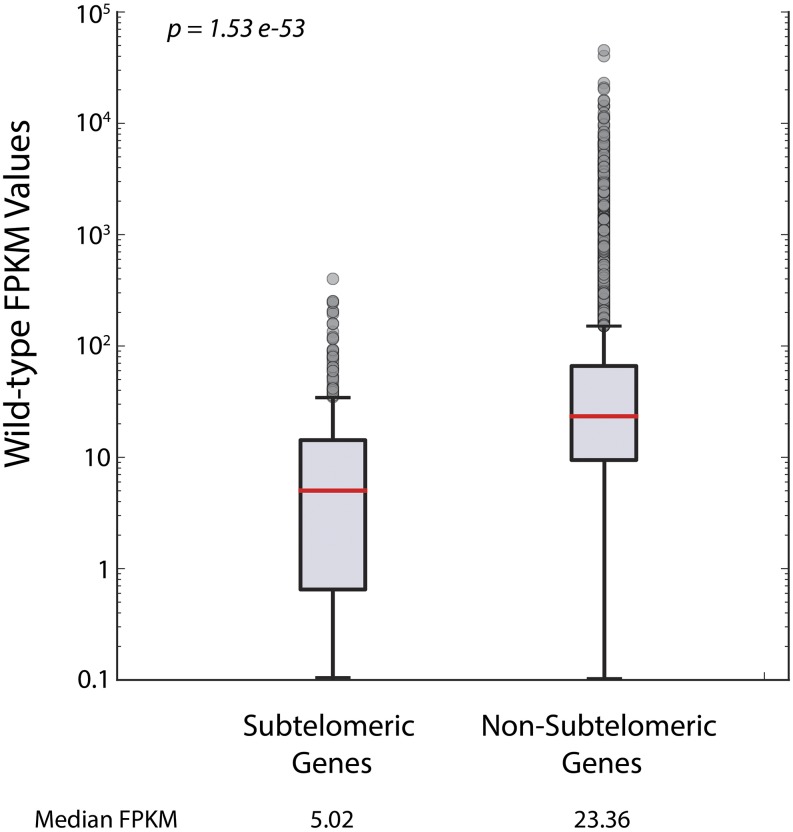
Once we observed transcription at subtelomeric domains, we wanted to determine how transcription at telomeres and subtelomeric domains compared to transcription at nontelomeric loci. Though transcripts were detected from many of the genes at subtelomeric regions, these genes had lower expression levels (FPKM values) on average than nontelomeric genes. We compared the distribution of FPKM values of subtelomeric protein-coding genes to non-subtelomeric protein-coding genes and found a statistically significant lower level of FPKM values among subtelomeric genes (Figure 5). These data corroborate previous subtelomeric transcript quantification in S. cerevisiae (Wyrick et al. 1999; Teytelman et al. 2008). This decreased transcription at telomeres could be attributed, in part, to decreased ORF density at telomeres (Louis 1995).
Figure 5.
FPKM values for subtelomeric genes were significantly lower than FPKM values for non-subtelomeric genes. The distribution in FPKM values of subtelomeric genes was compared to the distribution of FPKM values of non-subtelomeric genes in the WT genetic background using the Wilcoxon rank-sum test. The median FPKM value for subtelomeric genes was 5.02, whereas the median FPKM value for non-subtelomeric genes was 23.4 (P-value = 1.53−53).
Only ∼6% of subtelomeric genes were silenced by Sir proteins
To determine the extent to which Sir proteins affect the expression of subtelomeric genes, we performed a differential gene expression analysis using the DESeq package in R (Anders and Huber 2013). Genes showing a statistically significant difference in expression from WT (as indicated by a P-value < 0.05), a greater than twofold change in expression, and a false discovery rate of less than 10% (to control for the multiple-testing problem) were included in the final list of differentially expressed genes. Using these criteria, 42 genes appeared to be up-regulated in all three sir mutants (for a complete list of all statistically significant observed expression changes, see Table S7). In principle, these 42 genes were expected to fall into either of two categories: (1) genes directly subject to Sir-based repression (e.g., genes at HMLα, HMRa, and subtelomeric regions) and (2) genes normally expressed more highly in a/α diploids as a result of simultaneous HML and HMR de-repression in sir mutants. Of these 42 genes, 21 (50%) were in subtelomeric regions (Table 1 and Figure 4, red arrows). Of these, 13 were completely repressed or averaged less that 1 FPKM among replicate experiments in WT strains. However, even in sir mutant conditions, many of these genes had low expression levels, averaging at ∼3.8 FPKM (Table 1). The remaining genes were expressed from two- to sixfold higher in sir mutants than in WT strains, with some highly expressed even in WT strains (e.g., CHA1 and HXK1). A previous study found BNA1 to increase in sir2Δ strains (Bernstein et al. 2000); our data did not reproduce this finding.
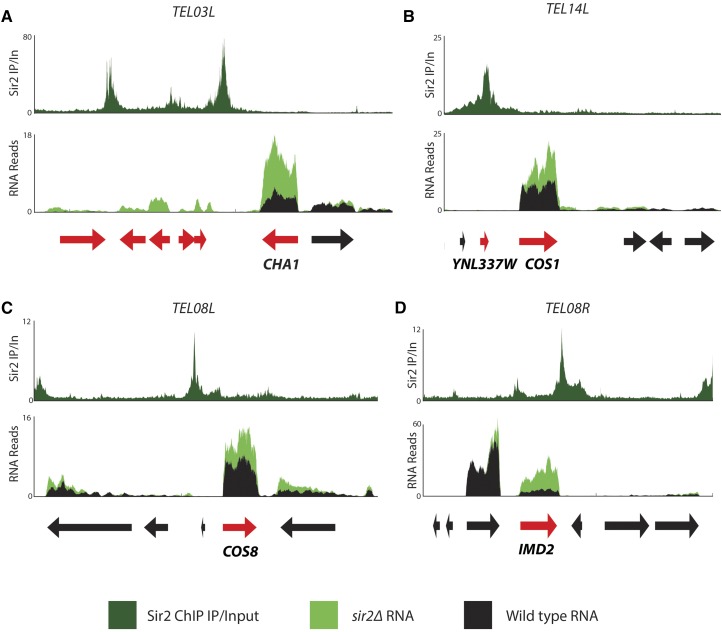
For the 21 subtelomeric genes that were up-regulated in all three sir mutants, we evaluated whether proximity to Sir proteins influenced repression. First, we determined whether the genes that increased expression in all three mutants were within peaks as defined by MACS. Most (15 of 21) of the genes whose expression changes in all three sir mutants (Table 1) were within MACS peaks (Table S1). For 17 of these up-regulated genes, the distance between the midpoint of the gene and the midpoint of the nearest prominent Sir protein peak was less than 2 kbp (Table 1, last column). Four such examples of Sir-repressed coding genes adjacent to Sir peaks are shown in Figure 6. Another gene, COS6, displayed a significantly enriched peak for only Sir4, and the expression of this gene increased ∼1.4-fold relative to WT in the sir4∆ strain (because it did not increase in sir2∆ and sir3∆ strains, this gene is not included in Table 1). Proximity to a Sir protein peak was not, however, predictive of whether or not a gene would be de-repressed in a sir mutant. There were many genes that either fell under a Sir protein peak or fell within 2 kbp of a Sir protein peak but did not change in expression in a sir mutant. Of the 101 coding genes that fell within 2 kbp of Sir2 peaks, 84 (∼83%) were not de-repressed in a sir2∆ strain. Additionally, there were three genes that MACS called as significantly enriched for at least one of the three Sir proteins but whose expression did not change in the sir mutants: IRC7, VBA5, and PAU20. PAU20 was previously implicated as a secondary recruitment site for Sir3 (Radman-Livaja et al. 2011). Thus Sir proteins can be recruited to a loci without repressing the adjacent gene.
Figure 6.
Genes that were de-repressed in sir mutants tended to be located near peaks of Sir binding. For each panel, the top horizontal axis shows Sir2 ChIP IP/input. The lower panel shows expression in the form of RNA read pileups in WT (black) and sir2Δ (green) strains. Genes that showed a statistically significant increase in expression in sir2Δ relative to WT are colored in red. (A) Left arm of chromosome III, TEL03L. CHA1 is adjacent to a peak of Sir2 present at the HML E silencer. (B) Left arm of chromosome XIV, TEL14L. Both YNL337W and COS1 are adjacent to a peak of Sir2 and were de-repressed in the sir2Δ mutant. (C and D) Left and right arms of chromosome VIII, TEL08L and TEL08R, respectively. Both COS8 and IMD2 are adjacent to a peak of Sir2 and showed increased expression in the sir2Δ mutant.
At least 13 Y′ elements were expressed
There are 19 annotated Y′ elements, all near the telomeres in the S288C genome. A small percentage (0.010–0.058%) of the total reads in each RNA-Seq library mapped to Y′ elements (Table S3), corroborating previous work on the expression of Y′ elements (Pryde and Louis 1999). The high degree of sequence similarity among Y′ elements precluded microarray experiments from being able to determine which of the Y′ elements were expressed. Likewise, most of our reads from Y′ elements (∼81%) did not map uniquely to specific Y′ elements. Using the ∼19% that mapped uniquely due to SNPs that distinguish Y′ elements (read counts shown in Table S4), we found that 13 Y′ elements were expressed. Absolute differences in read counts were difficult to interpret because the number of uniquely mapped reads per Y′ element varies as a function of the number of unique SNPs within its sequence. Nevertheless, in no case was the level of expression significantly higher or lower in a sir mutant relative to WT strains (Table S4). Six Y′ elements (TEL04R-YP, TEL16L-YP, TEL07R-YP, TEL12R-YP1, TEL14L-YP, and TEL15R-YP) contributed no uniquely mapped reads.
Others have detected telomere-repeat-containing RNAs (TERRAs) originating from the repeated sequences within X elements (Iglesias et al. 2011). We detected a small percentage of sequence reads that mapped to sufficiently polymorphic X elements and found that X elements present at TEL02L, TEL06L, TEL06R, TEL07R, and TEL11R increased in expression in all three sir mutants. However, the transcripts we detected originated from the core X, which contains the Abf1 and ORC binding sites, not the repeats within X elements.
Newly identified haploid- or diploid-regulated genes
S. cerevisiae cell type is specified by the activity of transcription factors encoded by alleles of the MAT locus [reviewed in Haber (2012)]. These transcription factors activate or repress transcriptional programs in each of the three cell types. Haploid yeast mutants for SIR2, SIR3, or SIR4 simultaneously express the α2 and a1 proteins as a result of de-repression of HMLα and HMRa, respectively. Dimerization of a1 and α2 leads to the a1/α2 repressor complex, which represses haploid-specific genes by directly binding to their promoters. α2 also dimerizes with Mcm1 and represses a-specific genes. Our data provided an opportunity to use the enhanced resolving power and sensitivity of RNA-Seq to obtain a potentially full catalog of haploid-specific genes and a/α-specific genes. Therefore, any previously undiscovered a-specific genes also may be included among the haploid-specific genes because of their decreased expression in sir mutants relative to WT strains.
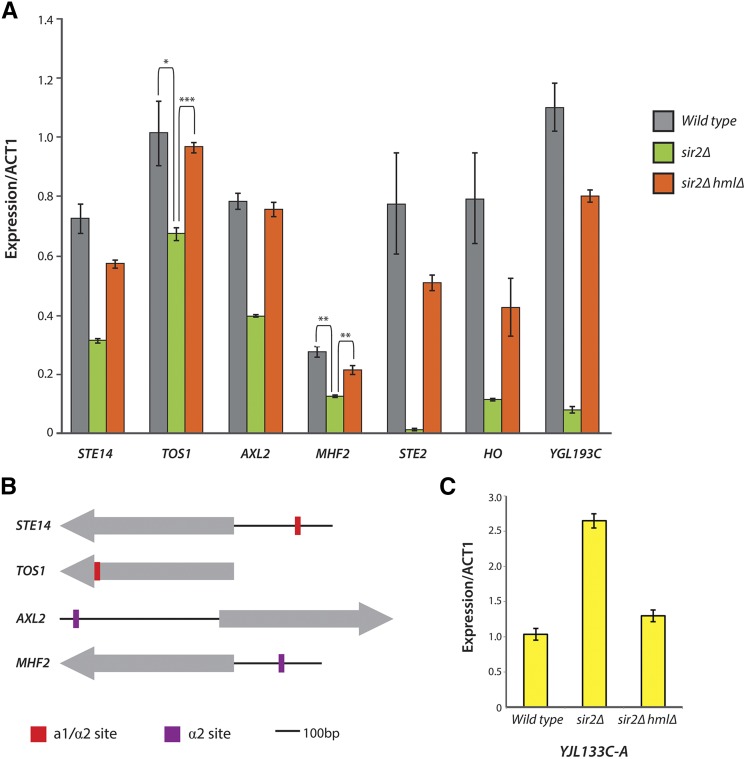
We applied the following criteria to obtain a list of candidate cell-type-specific genes: (1) the gene increased or decreased in all three sir mutants compared to WT strains, (2) the gene’s expression level had a twofold or greater statistically significant change, and (3) the gene was not directly bound by Sir2, Sir3, or Sir4. Using these criteria, we identified 16 genes with elevated expression in sir mutants (Table 2). Six of these genes have mitochondrial functions (FMP43, SFC1, CYC7, CYC1, NCA3, and YJL133C-A) and are clearly expressed in haploids as well. Hence these genes were more accurately interpreted as having a/α-enhanced expression. No common functions were found for the remaining 11 genes, nor have any diploid functions been attributed to these. To evaluate the dependence of these expression changes on the presence of the a1/α2 dimer, HMLα was deleted in the sir2Δ background, and expression changes were measured using qRT-PCR. The expression increase for YJL133C-A depended on the presence of α2 (Figure 7C), making it a candidate for indirect regulation by a1/α2 (perhaps through RME1, for example).
Table 2. Mating-type regulated genes.
| Gene | Systematic name | Wild type | sir2Δ | sir3Δ | sir4Δ |
|---|---|---|---|---|---|
| Genes increasing in expression | |||||
| YJL047C-Aa | YJL047C-A | 0 | 39.2 | 9.5 | 11.8 |
| YER053C-Aa | YER053C-A | 0 | 777.5 | 1640.7 | 371.2 |
| SFC1a | YJR095W | 0.8 | 1.6 | 1.4 | 1.8 |
| FMP43a | YGR243W | 1.3 | 10.4 | 8.5 | 8.3 |
| JID1a | YPR061C | 3.2 | 9.1 | 8.3 | 8.5 |
| GTO3a | YMR251W | 3.7 | 7.6 | 8.5 | 10.6 |
| YDR042Ca | YDR042C | 4.6 | 19.4 | 14.6 | 10.7 |
| HMX1 | YLR205C | 6.7 | 29.3 | 44 | 24.5 |
| MTH1a | YDR277C | 6.8 | 14.3 | 18.8 | 16.6 |
| YKR075Ca | YKR075C | 8.1 | 24.6 | 36.1 | 38.5 |
| NCA3a | YJL116C | 10.1 | 24.4 | 28.4 | 25.8 |
| YJR115Wa | YJR115W | 11.2 | 20.9 | 21.3 | 24.5 |
| CYC7a | YEL039C | 11.2 | 26.8 | 99.7 | 62.9 |
| YDR119W-Aa | YDR119W-A | 27 | 70.8 | 145.3 | 136.2 |
| YJL133C-Aa | YJL133C-A | 67.2 | 183.8 | 152 | 303.5 |
| CYC1a | YJR048W | 130.8 | 444.2 | 513.5 | 267.8 |
| AHP1a | YLR109W | 218.2 | 480.6 | 438.6 | 526.6 |
| Genes decreasing in expression | |||||
| SNO3a | YFL060C | 7.8 | 2 | 2.4 | 3.1 |
| HUA2a | YOR284W | 10 | 3.8 | 4.3 | 4.6 |
| HO | YDL227C | 10.7 | 1.7 | 0.8 | 1.1 |
| AXL1 | YPR122W | 15 | 4.3 | 3.6 | 2.9 |
| STE5 | YDR103W | 15.1 | 1.7 | 2.7 | 2.3 |
| YPR027Ca | YPR027C | 16.1 | 2.7 | 3.5 | 4.1 |
| YDR170W-A | YDR170W-A | 16.1 | 3.9 | 4.6 | 3.5 |
| SST2a | YLR452C | 16.8 | 7 | 7.5 | 6.5 |
| RDH54 | YBR073W | 16.9 | 3.3 | 3.7 | 2.7 |
| NEJ1 | YLR265C | 19 | 2.4 | 2.1 | 1.6 |
| YDR034C-Da | YDR034C-D | 25.8 | 6.1 | 15.4 | 12 |
| STE6 | YKL209C | 25.9 | 2.9 | 4.1 | 3.6 |
| GPA1 | YHR005C | 26.1 | 3.5 | 2.8 | 2.8 |
| ICS2 | YBR157C | 31.4 | 5.8 | 4.6 | 5.1 |
| VBA2a | YBR293W | 35.1 | 8.2 | 10 | 8 |
| BAR1 | YIL015W | 44.7 | 4.3 | 3.2 | 3.2 |
| FUS3 | YBL016W | 49.1 | 1.1 | 0.8 | 0.9 |
| MHF2a | YDL160C-A | 49.7 | 19.9 | 13.5 | 18.6 |
| AXL2a | YIL140W | 49.7 | 14.8 | 21.8 | 14.9 |
| CLN2a | YPL256C | 50.3 | 21.9 | 20.6 | 19.6 |
| IME4 | YGL192W | 53.8 | 6 | 8 | 7.4 |
| STE14a | YDR410C | 75.6 | 23.5 | 21.5 | 17 |
| STE4 | YOR212W | 75.8 | 8 | 7.3 | 5.8 |
| YGL193C | YGL193C | 79.2 | 2.6 | 3.3 | 4.2 |
| STE18 | YJR086W | 82.8 | 10.8 | 10.7 | 5.3 |
| AGA2 | YGL032C | 87.8 | 0.5 | 2 | 2.3 |
| DDR2 | YOL052C-A | 97.3 | 39.2 | 41.2 | 29.8 |
| AMN1 | YBR158W | 102.5 | 39.4 | 39.5 | 33.6 |
| RME1 | YGR044C | 108.2 | 5.1 | 6.7 | 4.8 |
| MFA1 | YDR461W | 227.3 | 0 | 0 | 0 |
| SUN4a | YNL066W | 311.4 | 125.2 | 122.1 | 136.1 |
| STE2 | YFL026W | 327.7 | 5.5 | 5.5 | 5.8 |
| ZRT1a | YGL255W | 389.9 | 110.8 | 117.2 | 160.9 |
| TOS1a | YBR162C | 1143.3 | 437.3 | 557.7 | 478.5 |
| MFA2b | YNL145W | 3465.9 | 0 | 71.6 | 0 |
All genes in this table (1) changed significantly in expression in all three sir mutants relative to WT and (2) are not located at HML, HMR, or subtelomeric regions. Seventeen genes increased in expression, and 35 decreased in expression.
Genes not found in previous lists of haploid-specific or haploid-enhanced genes Expression levels are in units of FPKM, and genes are ordered by increasing FPKM levels in WT.
The FPKM value for MFA2 in the sir3Δ strain, though greater than 0, is not statistically different from the value of 0 FPKM seen in sir2Δ and sir4Δ strains. Similar numbers of raw reads mapped to the MFA2 locus in all three mutants (18, 19, and 11 average reads for sir2Δ, sir3Δ, and sir4Δ, respectively). The inflated FPKM value seen in the sir3Δ strain is likely a consequence of the FPKM normalization method used by Cufflinks, which, because of the substantially larger library size of the sir3Δ strains (Table S2), may have overestimated the FPKM value for the lowly expressed MFA2 gene.
Figure 7.
Expression confirmation via qRT-PCR and promoter analysis of candidate haploid-specific genes. (A) STE14, TOS1, AXL2, and MHF2 were weakly repressed in an α2-dependent manner. The strongly a1/α2-repressed genes STE2, HO, and YGL193C are shown for comparison. (B) Annotated binding sites for the a1/α2 heterodimer and α2 itself are shown in relation to the protein-coding sequences (gray arrows) for STE14, TOS1, AXL2, and MHF2 (coding regions are not drawn to scale). STE14 contains a weak a1/α2 binding site 232 bp upstream from its coding sequence. TOS1 contains a weak a1/α2 binding site within its gene body. Both AXL2 and MHF2 contain weak α2 binding sites 578 and 174 bp, respectively, upstream from their coding regions. (C) YJL133C-A, a gene of unknown function, increases in expression in a α2-dependent manner.
Thirty-five genes decreased in expression in sir mutants relative to WT strains. We compared this list to known haploid-specific genes as found by chromatin immunoprecipitation of α2 in a/α diploids followed by hybridization of immunoprecipitated DNA to a genome-wide array (Galgoczy et al. 2004). That study found 20 haploid-specific genes, all of which were reproduced in our data set (Table 2, genes without footnote markers). YGL193C and the anti-sense transcript of IME4, which are positioned in tandem, are also known a1/α2 targets that were reproduced in our data set (Hongay et al. 2006; Valencia-Burton et al. 2006). An additional known indirect a1/α2 target reproduced in our data set was the G1 cyclin gene CLN2. CLN2 is weakly activated by RME1 and therefore, as expected, decreased in expression in sir mutants presumably because of the repression of RME1 itself (Table 2) (Toone et al. 1995).
The remaining 13 of 35 genes in the decreasing-genes list represented genes with previously unrecognized haploid-specific or a-specific expression (Table 2, genes with footnote markers). To further evaluate whether these genes were direct targets of a1/α2 or α2/Mcm1 repression, we performed two additional tests: (1) a scan of each gene’s promoter sequences for the presence of annotated a1/α2 or α2/Mcm1 binding motifs using the motif discovery program MEME and the Yeast Transcription Factor Specificity Compendium (YeTFaSpCo) (Bailey et al. 2009; De Boer and Hughes 2012) and (2) measurement of the expression of each gene via qRT-PCR in a sir2Δ hmlΔ strain. If the observed expression change were in fact due to the presence of a1/α2, deleting α2 should abolish the effect. For both tests, known a1/α2 and α2/Mcm1 targets served as positive controls. Four genes with previously unrecognized haploid-specific expression were confirmed with these two tests: STE14, TOS1, AXL2, and MHF2. Interestingly, none of the four were under strong a1/α2 repression. Instead, they appeared to be weakly repressed by α2 (Figure 7A). Consistent with this observation, none possessed clear a1/α2 binding motifs of the kind found in the strongly repressed haploid-specific genes STE2 and HO. However, weak a1/α2 or α2 binding sites, as annotated in the YeTFaSpCo, were found for all four (Figure 7B).
Discussion
This study provided a comprehensive evaluation of both the molecular topology of Sir protein distribution at telomeres and subtelomeric regions and of the extent of telomere position effects on gene expression mediated by Sir-based gene silencing. The URA3 reporter gene and other reporter genes near truncated telomeres have served as an assay for telomere position effects for many years. Their use has enabled multiple discoveries, including the gene for the RNA component of telomerase (Singer and Gottschling 1994), and has implicated many chromatin factors and histone modifications as key players in silencing genes near telomeres. However, because repression of the URA3 reporter at the truncated telomere of TEL07L is robust, there exists a commonly held view that all natural telomeres of S. cerevisiae are transcriptionally silent and that most, if not all, subtelomeric genes are strongly repressed by the Sir protein complex. By measuring expression at native telomeres using the highly sensitive RNA-Seq method, we found that many genes near telomeres are transcribed, albeit at lower levels than the rest of the genome, supporting and extending earlier data that expression of genes in subtelomeric regions of S. cerevisiae is largely uninfluenced by Sir proteins (Takahashi et al. 2011). Moreover, we found that Sir-based silencing was not a widespread phenomenon at telomeres despite strong enrichment of Sir proteins at telomeric repeats and core X elements. Twenty-one genes in the vicinity of Sir proteins are de-repressed, but most genes are not, resulting in only 6% of subtelomeric genes repressed by Sir proteins. Qualitatively, these data are in agreement with a high-density microarray-based genome-wide expression study of WT strains and sir2∆, sir3∆, and sir4∆ mutants (Wyrick et al. 1999).
Transcription occurs near telomeres but at lower levels than at nontelomeric regions
Although transcription does occur in subtelomeric regions, it produces fewer transcripts per gene than nontelomeric regions of the genome. This global observation is consistent with previous studies that found telomeres to be both gene-poor and, for the genes present, to have lower levels of transcription than is typical for the rest of the genome, as measured by hybridization studies with high-density microarrays (Louis 1995; Wyrick et al. 1999). A limitation of all RNA-based studies to date is their reliance on mRNA samples from a large population of cells. Hence high-level expression in a small fraction of cells but no expression in most would have been missed. Indeed, the epigenetic inheritance of expression states observed for reporter genes at telomeres underscores the existence of such cell-to-cell variation.
Importantly, however, transcript levels at subtelomeric regions in sir mutants did not match transcript levels from non-subtelomeric regions. Therefore, Sir protein binding at telomeres was not solely responsible for the low transcript levels from most genes in subtelomeric regions. Other factors potentially responsible for the lower expression of subtelomeric genes include (1) other non–Sir protein chromatin factors that might confer an additional tier of repression on subtelomeric genes and (2) sequence-specific reasons for low subtelomeric expression, such as the use of intrinsically weak promoters. In support of the first possibility, histone H4 depletion increases expression of 15% of subtelomeric genes, whereas sir mutations increase expression of only 7–9% of genes within subtelomeric regions (Wyrick et al. 1999; Martin et al. 2004). Our data show that a similar percentage (∼6%) of subtelomeric genes are repressed by Sir proteins. Perhaps other chromatin factors targeting histone H4 confer an additional repressive effect on subtelomeric regions. Silencing at different telomeres also might be more or less sensitive to distinct histone-modifying enzymes. For example, the subtelomeric gene FLO10, which encodes a cell wall glycoprotein, is repressed by the action of deacetylases Hst1 and Hst2, two paralogs of Sir2 (Halme et al. 2004). Additionally, there is almost no agreement as to the identity of the genes repressed by Dot1 (Takahashi et al. 2011), the enzyme that catalyzes H3K79 methylation, and those repressed by SIR2 (this study), which deacetylates H4K16-acetyl.
The second possible reason that subtelomeric domains exhibit lower levels of transcription could be that subtelomeric genes, on average, have weaker promoters than centromere-proximal genes. If subtelomeric genes tend to have weaker promoters and lack transcriptional activator binding sites, it would be expected that most are weakly expressed regardless of chromatin state. Interestingly, subtelomeric genes are among the most highly divergent genes in the yeast genome and are often up-regulated under stressful conditions (Harrison et al. 2002; Teytelman et al. 2008). Previous studies have shown that part of the reason for this elevated rate of divergence is the ability of Sir proteins to interfere with certain types of DNA repair, highlighting a functional consequence of Sir protein association (Terleth et al. 1989). Our data implied that this mechanism could not account for all the enhanced divergence in these regions because the distribution of Sir proteins was focal rather than throughout the region. However, given that some mechanisms of DNA repair are transcription coupled (Svejstrup 2002), perhaps the low expression level of genes (or cell-to-cell variation in expression) in the subtelomeric regions leads to the absence of transcription-coupled repair and thereby contributes to their rapid divergence. If so, the higher mutation rate could, in turn, result in reduced functioning of promoter elements. Furthermore, a higher proportion of ORFs at telomeres are categorized as “dubious” or “uncharacterized,” with ∼56% of subtelomeric genes falling into these two categories as opposed to ∼24% of non-subtelomeric genes. Thus many subtelomeric ORFs may not be functional protein-coding genes whose expression is needed for general cellular function.
Only a small fraction of subtelomeric genes were repressed by Sir proteins
Overall, we found that Sir proteins repressed only 6% of all subtelomeric genes. Why are some subtelomeric genes repressed by Sir proteins and others not? Certain strong transcription activators can efficiently escape Sir-based repression (D. Steakley and J. Rine, unpublished results). Perhaps genes with increased expression in the absence of Sir proteins possess promoters with binding sites for weak transcriptional activators or weak binding sites for strong activators. In the absence of Sir proteins, these weakly binding activators would gain access and promote transcription. If so, the promoters of these Sir protein–sensitive genes might contain transcription factor binding sites that are distinct from binding sites present at genes that are not repressed by Sir proteins. To explore this possibility, we cataloged the transcription factor binding profiles for the promoters of the 21 SIR-sensitive subtelomeric genes and compared them to each other as well as to the transcription factor binding profiles from all other subtelomeric genes. Overall, we found no differences in transcription factor binding profiles between SIR-sensitive and SIR-resistant subtelomeric genes, though the small number of genes involved limited any statistical power of the analysis (data not shown). Motifs for the Mot2 and Ash1 transcription factors were the most commonly found sequences in the data set for all subtelomeric genes analyzed regardless of whether they were Sir repressed or not. Furthermore, 13 of the 21 SIR-sensitive genes are annotated as “dubious,” and the remaining 8 shared no common functional annotations, consistent with an absence of common transcription factor binding sites. In sum, we were unable to find differences in promoter sequence or transcription factor binding sites between the genes that were repressed by Sir proteins and those that were not.
The functional significance of Sir proteins at telomeres
At present, one clear function of Sir proteins at telomeres is to repress, or at least lower, the expression of a small subset of genes in this part of the genome. But why would a cell want to simply lower the expression of genes in this way, as opposed to simply having a weaker promoter for such genes? Perhaps subtelomeric genes regulated by Sir proteins in S. cereivisiae, like those in C. glabrata (De Las Peñas et al. 2003; Domergue et al. 2005; Ma et al. 2009), are involved in regulating the transcription of genes that are necessary only under certain conditions. In support of this model, seven genes encoding metabolic enzymes increased in expression in all three sir mutants: CHA1, AAD15, IMD2, FDH1, THI5, VBA3, and PAU4. It is possible that S. cerevisiae encounters some condition in nature that would inhibit Sir-based silencing like nicotinamide does in the laboratory. If so, perhaps these enzymes are part of an as yet undiscovered response mechanism to such agents or conditions.
A second hypothesis is that Sir proteins at telomeres contribute to the suppression of recombination at telomeric repeats, much like Sir2 suppresses recombination at the recombinant DNA repeats (Gottlieb and Esposito 1989; Smith and Boeke 1997). While yeast Ku proteins, which associate with Sir proteins at the subtelomeric core X sequences, do suppress recombination between telomeric repeats (Marvin et al. 2009), so far there is no direct evidence that Sir proteins are involved in this suppression. Additionally, a previous observation that Sir proteins associate with Ku70/Ku80 (suggesting a role for Sir proteins in preventing nonhomologous end joining), as reported by Tsukamoto et al. (1997), has since been shown to be an artifact of the a/α state of sir mutants (Åström et al. 1999).
Discovery of novel haploid-specific genes
Historically, elucidation of transcriptional regulatory circuits of S. cerevisiae has relied on microarray-based technologies, which are limited in sensitivity and dynamic range (Galgoczy et al. 2004). The sensitivity of RNA-Seq and the “pseudodiploid” state of sir mutants allowed us to evaluate the completeness of the identification of cell-type-regulated genes, particularly genes that are potential targets of a1/α2 and α2/Mcm1 regulation. We confirmed all previously identified genes of these classes. In addition, we found 29 new candidate haploid-specific or a/α-specific genes. Of these 29 genes, the expression of YJL133C-A, STE14, TOS1, AXL2, and MHF2 was verified by qRT-PCR and found to be moderately repressed in an α2-dependent manner, thus revealing a new class of genes that are partially but not fully repressed in the a/α cell type. The remaining 24 genes were too low in expression to be verified by qRT-PCR. The cell-type regulation of these genes was likely missed in previous studies precisely because they are not strongly repressed and thus exhibit a less dramatic fold change in expression than other a/α-regulated genes. At least three of the five genes were verified by qRT-PCR function in processes unrelated to cell-type determination. For example, STE14 encodes a methyltransferase that methylates a-factor in MATa cells and Ras proteins in all cell types (Marr et al. 1990; Hrycyna et al. 1991). On a per-cell basis, it is likely that more a-factor is produced in MATa cells than Ras proteins in all cell types, consistent with the partial reduction in STE14 expression in cells that do not make a-factor because of the expression of α2. We speculate that the Tos1, Mhf2, and Axl1 proteins have functions needed in all cell types, leading to their modest repression in a/α diploids.
Supplementary Material
Acknowledgments
We thank Minyong Chung and the Vincent J. Coates Genomics Sequencing Laboratory at the University of California, Berkeley, supported by National Institutes of Health S10 Instrumentation grants S10-RR029668 and S10-RR027303. This work was supported by a grant from the National Institutes of Health (GM-31105 to J.R.), National Science Foundation Predoctoral Fellowships (to A.E. and D.T.), a Berkeley Fellowship to A.E., and the University of California, Berkeley’s Cellular, Biochemical and Molecular Training Grant T32 GM 007127 from the National Institutes of Health.
Footnotes
Communicating editor: A. Gasch
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.175711/-/DC1
Literature Cited
- Ai W., Bertram P. G., Tsang C. K., Chan T., Zheng X. F. S., 2002. Regulation of subtelomeric silencing during stress response. Mol. Cell 10: 1295–1305. [DOI] [PubMed] [Google Scholar]
- Anders S., Huber W., 2010. Differential expression analysis for sequence count data. Genome Biol. 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., and W. Huber, 2013 Differential expression of RNA-Seq data at the gene level: the DESeq package. Available at: http://www.bioconductor.org/packages/devel/bioc/vignettes/DESeq/inst/doc/DESeq.pdf Accessed: March 1, 2014.
- Aparicio O. M., Billington B. L., Gottschling D. E., 1991. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell 66: 1279–1287. [DOI] [PubMed] [Google Scholar]
- Åström S. U., Okamura S. M., Rine J., 1999. Yeast cell-type regulation of DNA repair. Nature 397: 310. [DOI] [PubMed] [Google Scholar]
- Bailey T. L., Boden M., Buske F. A., Frith M., Grant C. E., et al. , 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37: W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein B. E., Tong J. K., Schreiber S. L., 2000. Genomewide studies of histone deacetylase function in yeast. Proc. Natl. Acad. Sci. USA 97: 13708–13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer C. G., Hughes T. R., 2012. YeTFaSCo: a database of evaluated yeast transcription factor sequence specificities. Nucleic Acids Res. 40: D169–D179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart M. A., Oliviero S., 2001. Preparation of yeast RNA, pp. 13.12.1–13.12.5 in Current Protocols in Molecular Biology, Wiley, Hoboken, NJ. [DOI] [PubMed] [Google Scholar]
- Crampton A., Chang F., Pappas D. L. J., Frisch R. L., Weinreich M., 2008. An ARS element inhibits DNA replication through a SIR2-dependent mechanism. Mol. Cell 30: 156–166. [DOI] [PubMed] [Google Scholar]
- De Las Peñas A., Pan S., Castaño I., Alder J., Cregg R, et al. , 2003. Virulence-related surface glycoproteins in the yeast pathogen Candida glabrata are encoded in subtelomeric clusters and subject to RAP1- and SIR-dependent transcriptional silencing. Genes Dev. 17: 2245–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domergue R., Castan I., De Las Peñas A., Zupancic M., Lockatell V., et al. , 2005 Nicotinic acid limitation regulates silencing of Candida adhesins during UTI. Science 308: 866–870. [DOI] [PubMed] [Google Scholar]
- Ehrentraut S., Weber J. M., Dybowski J. N., Hoffmann D., Ehrenhofer-Murray A. E., 2010. Rpd3-dependent boundary formation at telomeres by removal of Sir2 substrate. Proc. Natl. Acad. Sci. USA 107: 5522–5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourel G., Revardel E., Koering C. E., Gilson É., 1999. Cohabitation of insulators and silencing elements in yeast subtelomeric regions. EMBO J. 18: 2522–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galgoczy D. J., Cassidy-Stone A., Llinas M., O’Rourke S. M., Herskowitz I., et al. , 2004. Genomic dissection of the cell-type-specification circuit in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 101: 18069–18074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb S., Esposito R. E., 1989. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell 56: 771–776. [DOI] [PubMed] [Google Scholar]
- Gottschling D. E., Aparicio O. M., Billington B. L., Zakian V. A., 1990. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell 63: 751–762. [DOI] [PubMed] [Google Scholar]
- Guizetti J., Scherf A., 2013. Silence, activate, poise and switch! Mechanisms of antigenic variation in Plasmodium falciparum. Cell. Microbiol. 15: 718–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber J. E., 2012. Mating-type genes and MAT switching in Saccharomyces cerevisiae. Genetics 191: 33–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halme A., Bumgarner S., Styles C., Fink G. R., 2004. Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell 116: 405–415. [DOI] [PubMed] [Google Scholar]
- Harrison P., Kumar A., Lan N., Echols N., Snyder M., et al. , 2002. A small reservoir of disabled ORFs in the yeast genome and its implications for the dynamics of proteome evolution. J. Mol. Biol. 316: 409–419. [DOI] [PubMed] [Google Scholar]
- Hazelrigg T., Levis R., Rubin G. M., 1984. Transformation of white locus DNA in Drosophila: dosage compensation, zeste interaction, and position effects. Cell 36: 469–481. [DOI] [PubMed] [Google Scholar]
- Hecht A., Strahl-Bolsinger S., Grunstein M., 1996. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature 383: 92–96. [DOI] [PubMed] [Google Scholar]
- Hongay C. F., Grisafi P. L., Galitski T., Fink G. R., 2006. Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell 127: 735–745. [DOI] [PubMed] [Google Scholar]
- Hoppe G. J., Tanny J. C., Rudner A. D., Gerber S. A., Danaie S., et al. , 2002. Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol. Cell. Biol. 22: 4167–4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrycyna C. A., Sapperstein S. K., Clarke S., Michaelis S., 1991. The Saccharomyces cerevisiae STE14 gene encodes a methyltransferase that mediates C-terminal methylation of a-factor and RAS proteins. The EMBO Journal 10(5): 1699–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias N., Redon S., Pfeiffer V., Dees M., Lingner J., et al. , 2011. Subtelomeric repetitive elements determine TERRA regulation by Rap1/Rif and Rap1/Sir complexes in yeast. EMBO Rep. 12: 587–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurdistani S. K., Robyr D., Tavazoie S., Grunstein M., 2002. Genome-wide binding map of the histone deacetylase Rpd3 in yeast. Nat. Genet. 31: 248–254. [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Valsakumar V., Poorey K., Bekiranov S., Smith J. S., 2013. Genome-wide analysis of functional sirtuin chromatin targets in yeast. Genome Biol. 14: R48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loney E. R., Inglis P. W., Sharp S., Pryde F. E., Kent N. A., et al. , 2009. Repressive and non-repressive chromatin at native telomeres in Saccharomyces cerevisiae. Epigenet. Chromatin 2: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., III, Demarini D. J., Shah N. G., Wach A., et al. , 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- Louis E. J., 1995. The chromosome ends of Saccharomyces cerevisiae. Yeast 11: 1553–1573. [DOI] [PubMed] [Google Scholar]
- Ma B., Pan S.-J., Domergue R., Rigby T., Whiteway M., et al. , 2009. High-affinity transporters for NAD+ precursors in Candida glabrata are regulated by Hst1 and induced in response to niacin limitation. Mol. Cell. Biol. 29: 4067–4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marr R. S., Blair L. C., Thorner J., 1990. Saccharomyces cerevisiae STE14 gene is required for COOH-terminal methylation of a-factor mating pheromone. J. Biol. Chem. 265(33): 20057–20060. [PubMed] [Google Scholar]
- Martin A. M., Pouchnik D. J., Walker J. L., Wyrick J. J., 2004. Redundant roles for histone H3 N-terminal lysine residues in subtelomeric gene repression in Saccharomyces cerevisiae. Genetics 167: 1123–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvin M. E., Becker M. M., Noel P., Hardy S., Bertuch A. A., et al. , 2009. The association of yKu with subtelomeric core X sequences prevents recombination involving telomeric sequences. Genetics 183: 453–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappas D. L. J., Frisch R., Weinreich M., 2004. The NAD(+)-dependent Sir2p histone deacetylase is a negative regulator of chromosomal DNA replication. Genes Dev. 18: 769–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryde F. E., Louis E. J., 1999. Limitations of silencing at native yeast telomeres. EMBO J. 18: 2538–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radman-Livaja M., Ruben G., Weiner A., Friedman N., Kamakaka R., et al. , 2011. Dynamics of Sir3 spreading in budding yeast: secondary recruitment sites and euchromatic localization. EMBO J. 30: 1012–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renauld H., Aparicio O. M., Zierath P. D., Billington B. L., Chhablani S. K., et al. , 1993. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes Dev. 7: 1133–1145. [DOI] [PubMed] [Google Scholar]
- Rossmann M. P., Luo W., Tsaponina O., Chabes A., Stillman B., 2011. A common telomeric gene silencing assay is affected by nucleotide metabolism. Mol. Cell 42: 127–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche L. N., Kirchmaier A. L., Rine J., 2002. Ordered nucleation and spreading of silenced chromatin in Saccharomyces cerevisiae. Mol. Biol. Cell 13: 2207–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J., 1947. The nature of heterochromatin. Cold Spring Harb. Symp. Quant. Biol. 12: 179–191. [Google Scholar]
- Singer M. S., Gottschling D. E., 1994. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science 266: 404–409. [DOI] [PubMed] [Google Scholar]
- Smith J. S., Boeke J. D., 1997. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 11: 241–254. [DOI] [PubMed] [Google Scholar]
- Strahl-Bolsinger S., Hecht A., Luo K., Grunstein M., 1997. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 11: 83–93. [DOI] [PubMed] [Google Scholar]
- Svejstrup J. Q., 2002. Mechanisms of transcription-coupled DNA repair. Nat. Rev. Mol. Cell Biol. 3: 21–29. [DOI] [PubMed] [Google Scholar]
- Takahashi Y.-H., Schulze J. M., Jackson J., Hentrich T., Seidel C., et al. , 2011. Dot1 and histone H3K79 methylation in natural telomeric and HM silencing. Mol. Cell 42: 118–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terleth C., van Sluis C. A., van de Putte P., 1989. Differential repair of UV damage in Saccharomyces cerevisiae. Nucleic Acids Res. 17: 4433–4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teytelman L., Eisen M. B., Rine J., 2008. Silent but not static: accelerated base-pair substitution in silenced chromatin of budding yeasts. PLoS Genet. 4: e1000247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teytelman L., Thurtle D. M., Rine J., Oudenaarden A., van, 2013. Highly expressed loci are vulnerable to misleading ChIP localization of multiple unrelated proteins. Proc. Nat. Acad. Sci. USA 110: 18602–18607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurtle D. M., Rine J., 2014. The molecular topography of silenced chromatin in Saccharomyces cerevisiae. Genes Dev. 28: 245–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonkin C. J., Carret C. K., Duraisingh M. T., Voss T. S., Ralph S. A., et al. , 2009. Sir2 paralogues cooperate to regulate virulence genes and antigenic variation in Plasmodium falciparum. PLoS Biol. 7: e1000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toone W. M., Johnson A. L., Banks G. R., Toyn J. H., Stuart D., et al. , 1995. Rmel, a negative regulator of meiosis, is also a positive activator of G1 cyclin gene expression. EMBO J. 14: 5824–5832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S. L., 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., et al. , 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto Y., Kato J., Ikeda H., 1997. Silencing factors participate in DNA repair and recombination in Saccharomyces cerevisiae. Nature 388: 900–903. [DOI] [PubMed] [Google Scholar]
- Valencia-Burton M., Oki M., Johnson J., Seier T. A., Kamakaka R., et al. , 2006. Different mating-type-regulated genes affect the DNA repair defects of Saccharomyces RAD51, RAD52 and RAD55 mutants. Genetics 174: 41–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellinger R. J., Zakian V. A., 2012. Everything you ever wanted to know about Saccharomyces cerevisiae telomeres: beginning to end. Genetics 191: 1073–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyrick J. J., Holstege F. C. P., Jennings E. G., Causton H. C., Shore D., et al. , 1999. Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature 402: 418–421. [DOI] [PubMed] [Google Scholar]
- Yoshida K., Bacal J., Desmarais D., Padioleau I., Tsaponina O., et al. , 2014. The histone deacetylases Sir2 and Rpd3 act on ribosomal DNA to control the replication program in budding yeast. Mol. Cell 54: 691–697. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., et al. , 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Gustafsson C. M., 2009. Distinct differences in chromatin structure at subtelomeric X and Y′ elements in budding yeast. PLoS ONE 4: e6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zill O. A., Scannell D., Teytelman L., Rine J., 2010. Co-evolution of transcriptional silencing proteins and the DNA elements specifying their assembly. PLoS Biol. 8: e1000550. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.