Abstract
Rotifers of Class Bdelloidea are common freshwater invertebrates of ancient origin whose apparent asexuality has posed a challenge to the view that sexual reproduction is essential for long-term evolutionary success in eukaryotes and to hypotheses for the advantage of sex. The possibility nevertheless exists that bdelloids reproduce sexually under unknown or inadequately investigated conditions. Although certain methods of population genetics offer definitive means for detecting infrequent or atypical sex, they have not previously been applied to bdelloid rotifers. We conducted such a test with bdelloids belonging to a mitochondrial clade of Macrotrachela quadricornifera. This revealed a striking pattern of allele sharing consistent with sexual reproduction and with meiosis of an atypical sort, in which segregation occurs without requiring homologous chromosome pairs.
Keywords: clonal erosion, cyclic parthenogenesis, meiosis, monogonont, Oenothera
BDELLOID rotifers are minute freshwater invertebrates commonly found in lakes, streams, and ponds and in ephemerally aquatic environments such as rock pools and the water films on moss and lichens where they are able to survive because of their unusual ability to withstand desiccation. Although typically only several tenths of a millimeter in size, bdelloids have ganglia; muscles; reproductive, digestive, excretory, and secretory systems; photosensitive and tactile sensory organs; and structures for crawling, feeding, and swimming. Characterized by their ciliated head and bilateral ovaries and classified in four families and hundreds of morphospecies, their ancient origin is indicated by the considerable synonymous sequence divergence between families and by the finding of bdelloid remains in 35- to 40-million year old amber (Mark Welch et al. 2009).
Despite much observation of field and laboratory populations and except for one heavily qualified account of having twice seen a male among many females in a sample from a lake (Wesenberg-Lund 1930), neither males, hermaphrodites, mating, nor meiosis have ever been reported within the class (Birky 2010). The only known mode of bdelloid reproduction is via eggs produced by two mitotic divisions from primary oocytes (Hsu 1956a,b). Most recently, further evidence suggestive of asexuality has come from genomic sequencing of the bdelloid Adineta vaga, showing it to be devoid of homologous chromosome pairs and therefore excluding the possibility of standard meiosis (Flot et al. 2013). Nevertheless, the possibility remains that bdelloids reproduce sexually under unknown or inadequately investigated conditions, employing a form of meiosis known in certain plants in which segregation occurs without homologous chromosome pairs—a possibility that can be tested by methods of population genetics.
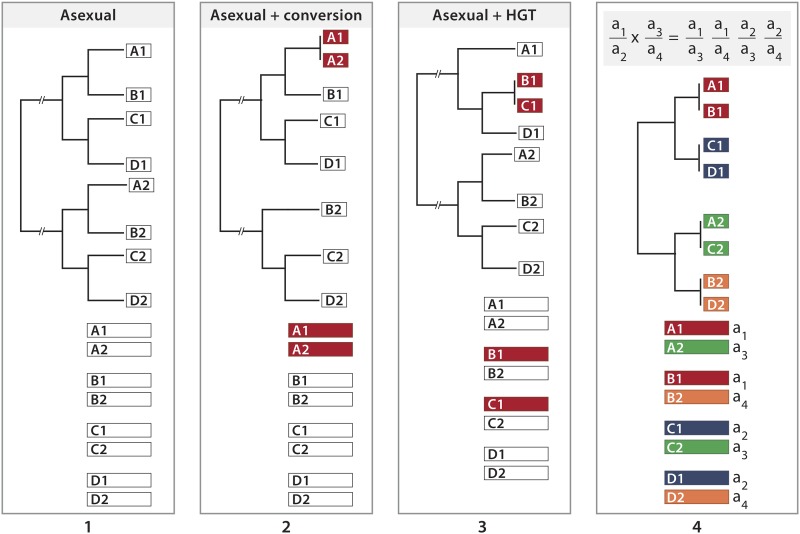
For any two individuals in a population descended from a common ancestor without sexual reproduction or any other sort of genetic transfer between individuals, the phylogenetic distance between a sequence in one individual and its homolog in the other is the same for all their sequences (Figure 1.1). Conversion within an asexual lineage replaces a sequence with its homolog but does not transfer genetic material between individuals (Figure 1.2). In contrast, a finding of two individuals closely related with respect to a given sequence but more distantly related with respect to its homolog, a form of phylogenetic noncongruence known as allele sharing, would imply that genetic material had been transferred between individuals or their progenitors (Judson and Normark 1996). This could occur by homologous transfer of DNA fragments (Figure 1.3), as in bacterial transformation, or as a consequence of sexual reproduction, which produces progeny that have parental sequences at every locus (Figure 1.4). A finding of two individuals with allele sharing at each of several genomic regions examined would therefore suggest sexual reproduction but would not be expected for transformation by DNA fragments. A further expectation for sexual reproduction but not for transfer of DNA fragments follows from the fact that among the four different F1 progeny of the cross a1/a2 × a3/a4 each individual shares one of its alleles with another individual and shares its homolog with a third individual at every diploid locus. Similarly, each of the F1 shares one of its alleles with one of the parents and shares its homolog with the other parent. A finding of three individuals with such a pattern of allele sharing at each of several genomic regions examined would strongly suggest sexual reproduction rather than transfer of DNA fragments. We have found such a pattern of relatively recent allele sharing and indications of earlier allele sharing in the lineages of individuals from a mitochondrial clade of bdelloids belonging to the morphospecies Macrotrachela quadricornifera.
Figure 1.
Allele phylogenies (top) for four diploid individuals designated A, B, C, and D and bars (bottom) depicting the constitution of each individual. 1, complete asexuality. Individuals descend from an ancient diploid ancestor in which sex was lost, giving rise to lineages 1 and 2. All alleles diverge independently so that for any two individuals the phylogenetic distance between a sequence in one individual and its homolog in the other will be the same for all their sequences. 2, asexuality with conversion (red) in individual A. 3, asexuality with homologous genetic transfer of an allele (red) from individual C to individual B. 4, a cross between heterozygotes a1/a2 and a3/a4. Each individual in the F1 shares one of its alleles at each locus with a second individual and shares its homolog with a third individual.
Materials and Methods
We collected 29 rotifers resembling bdelloids of the family Philodinidae from ground and tree moss at 10 sites in Connecticut; Massachusetts; North Carolina; and Long Island, New York in 2006. Initial cultures were started from single rotifers. A single egg from each initial culture was washed by 8–12 passages through 0.25 ml of sterile spring water in the wells of a microtiter dish and used to start a stock culture. Rotifers were cultured in sterile spring water in plastic petri dishes and fed Escherichia coli. For DNA preparation, rotifers and eggs from stock cultures were washed with spring water on a 10-μm nylon screen, resuspended in spring water, and kept overnight without food to allow digestion of ingested bacteria. After washing again on the nylon screen with 15 mM EDTA, 150 mM Tris (pH 8), rotifers and eggs were scraped off the screen and ground under liquid nitrogen. DNA was prepared by SDS–proteinase-K digestion, phenol-chloroform extraction, and ethanol precipitation and stored in 1 mM EDTA, 10 mM Tris (pH 8) at −20°. For mitochondrial sequencing, a 595-bp segment of the cox-1 gene was amplified with Taq DNA polymerase (Life Technologies), using primers described by Birky et al. (2005).
In selecting nuclear genomic regions to be examined for allele sharing we took advantage of previous studies of 25- to 70-kb segments of the genomes of the bdelloids A. vaga and Philodina roseola showing that in both species, along with numerous other genes, there are four such segments containing the histone gene cluster and four segments containing the hsp82 heat-shock gene (Mark Welch et al. 2004, 2008; Hur et al. 2009). The structure of the genome in these segments and, at this scale, in the A. vaga draft genome (Flot et al. 2013) is that of a degenerate tetraploid. Each group of four segments is composed of two fully collinear pairs with a few percent average synonymous divergence between genes belonging to a pair (homologs) and an order of magnitude greater synonymous divergence between genes in different pairs (homeologs). Many genes are present in only one or the other pair of homologs, rendering the genome partially diploid and partially tetraploid. Within homologous pairs there are occasional tracts of identity generally up to several hundred nucleotide pairs in length attributable to conversion and longer tracts that may be due to break-induced replication. There are no such tracts between homeologs. Aside from genes that are present in only one species or the other, the sequenced segments of A. vaga are identical in gene order and orientation to the corresponding segments of P. roseola. Further, there is no case in which a gene present in only one collinear pair is in the homeologous pair in the other species. Thus, the sequence structure in these segments is largely conserved across the two bdelloid families, Adinetidae and Philodinidae, indicating that degenerate tetraploidy was established before the families diverged (Hur et al. 2009).
Fluorescent in situ hybridization shows that in A. vaga and P. roseola the four segments containing hsp82 are all on separate chromosomes and that homologous segments containing the histone gene cluster are on two additional chromosomes in P. roseola (Mark Welch et al. 2004; Hur et al. 2009). The chromosomal locations of the other pair of histone gene clusters in P. roseola and of the four histone gene clusters in A. vaga have not been investigated although all eight regions reside on separate scaffolds in the A. vaga draft genome.
Each of the four regions we sequenced to test for allele sharing lies within and is fully syntenic to the corresponding segment in A. vaga (Figure 2). The four regions, each comprising a homologous pair of sequences, are designated hspA, hspB, hisA, and hisB, where A and B designate homeologs and the homologs in each region are numbered 1 and 2. For amplification of each region we first designed primers based on sequences conserved between A. vaga and P. roseola that lie just outside each region and are present in only one or the other homeologous pair, allowing us to amplify the two homologs in each region without interference from their homeologs. The four pairs of primers, each pair specific for a different region, were then used to amplify ∼300-bp intervals at both ends of each region. Alignments of the resulting sequences from each region from all isolates to be sequenced were used to design four pairs of consensus primers for amplification of each of the four regions (see Supporting Information, Table S1, Table S2).
Figure 2.
The sequenced hisA, hisB, hspA, and hspB regions (red) showing complete synteny in gene order and orientation with the corresponding segments of Adineta vaga (blue). The designations A and B distinguish homeologs. Annotation and gene numbers are from Hur et al. (2009). Genes 14–17 are his4, his3, his2B, and hisH2Av. Gene 33 is hsp82. Altogether, 13 annotated genes are represented in the sequenced regions. For all gene identifications, see Table S2.
The hisA, hisB, and hspA regions were amplified with the Expand Long Template PCR System (Roche). The hspB region was amplified with PfuUltra II fusion HS DNA Polymerase (Stratagene, La Jolla, CA). Amplicons were cloned with the StrataClone Ultra Blunt PCR Cloning Kit (Agilent Technologies). Sequences of the hisA, hisB, and hspA regions were determined from cloned amplicons from two separate amplifications while sequences of the hspB region are from cloned amplicons from one amplification. At least nine cloned amplicons from each amplification of each region were sequenced to a depth of ∼600 nt at each end of the region to identify the two homologs for full-length sequencing. At least three cloned amplicons of each homolog from each amplification were then fully sequenced by primer walking with BigDye Terminator v3.1 Cycle Sequencing Kits (Applied Biosystems, Foster City, CA).
Particular care was taken to ensure that our results were not influenced by contamination or sequencing error. End sequencing and full-length sequencing of cloned amplicons, altogether hundreds of sequence determinations, always revealed the same two and only two sequences specific for each region in each isolate, as expected for allelic pairs. Sequences from a different isolate were never found, ruling out cross-isolate contamination. The sequences of both homologs of the hisB region in each of the six isolates of clade 1 were determined by one author and were independently verified starting with new single-egg cultures by a second author, who also sequenced the hspB regions in isolates MA, MM, and MQ. Sequences of the hisA and hspA regions were determined by a third author. Sequences were assembled with Phred/Phrap, manually inspected, and verified in Consed and, for each region, homologous sequences were aligned with ClustalX. Inspection of the aligned sequences identified a few sites with a nucleotide different from the consensus, indicating an average sequencing error rate of about one nucleotide per kilobase.
Results
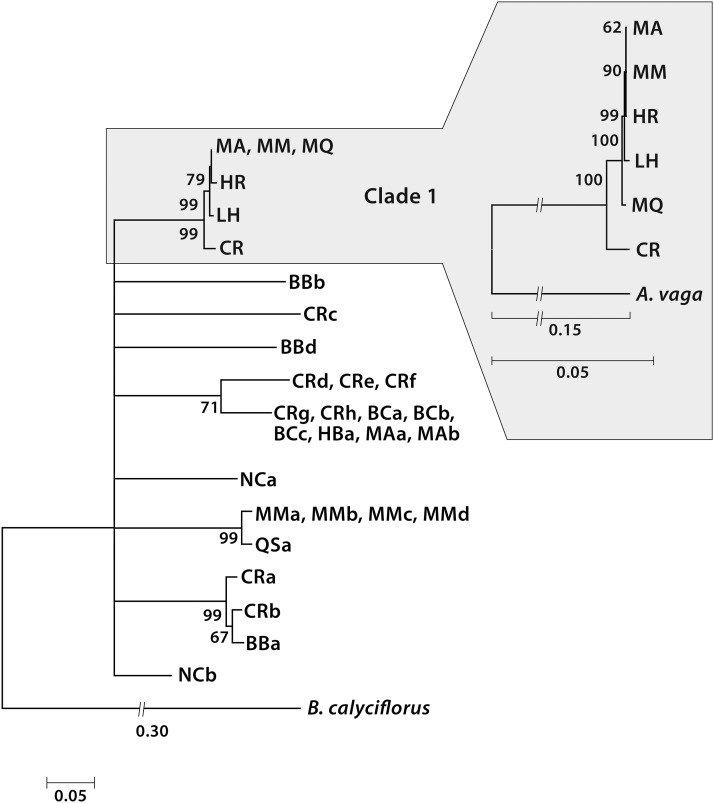
Rotifers of the cyclically parthenogenetic Class Monogononta belonging to the same mitochondrial clade, even when from locations thousands of kilometers apart, yield fertile progeny when crossed, while monogonont rotifers from different clades often do not (Suatoni et al. 2006). Therefore considering that a mitochondrial clade of bdelloids might correspond to a population within which sexual reproduction could occur, we sequenced a 595-bp segment of the mitochondrial cox-1 gene of each of the 29 isolates collected in the field. As has consistently been found in other collections of bdelloid rotifers (Birky et al. 2005; Fontaneto et al. 2009, 2011) the phylogeny of the mitochondrial sequences revealed distinct clades, monophyletic groups of individuals with closely related sequences well separated from other groups (Figure 3). As found in other such bdelloid clades, there is wide geographic dispersion within clades, consistent with the minute size of bdelloids and their ability to survive desiccation and be transported by wind (Wilson and Sherman 2013).
Figure 3.
Bottom left, phylogeny of a 595-bp segment of the mitochondrial cox-1 gene of 29 bdelloid isolates and M. quadricornifera with the monogonont rotifer Brachionus calyciflorus as outgroup. Collection sites: MA, Huntington, Long Island, New York; BC, CR, LH, and QS, four different sites in Cambridge, Massachusetts; HR, Centerville, Massachusetts; MM, Woods Hole, Massachusetts; MQ, near Milan, Italy; BB and HB, two different sites in Easton, Connecticut; NC, Durham, North Carolina. Top right, phylogeny of the full 14-kb mitochondrial genome of the 6 isolates belonging to clade 1 with the bdelloid A. vaga as outgroup. Full mitochondrial sequences are from Lasek-Nesselquist (2012). Phylogenetic analysis was performed by maximum likelihood with 1000 replications implemented in MEGA 5, using the general time-reversible substitution model.
For sequencing nuclear genomic regions, we chose the group designated clade 1. It includes five isolates we collected in eastern Massachusetts and Long Island, New York and our laboratory strain of M. quadricornifera, isolated by C. Ricci from moss in Italy (Ricci 1991). All six members of the clade were found to have 10 chromosomes of similar size, as previously shown for this species (Pagani et al. 1993; Mark Welch and Meselson 1998). The oocyte DNA content of M. quadricornifera is ∼1500 Mb (Pagani et al. 1993; Mark Welch and Meselson 2003). The species is traditionally assigned on the basis of morphology to the family Philodinidae although phylogenetic analysis of its mitochondrial DNA sequence is ambiguous as to whether it belongs to Philodinidae or Adinetidae (Lasek-Nesselquist 2012) and its structure in the histone cluster and hsp82 regions is more closely similar to that of A. vaga than to that of P. roseola (Hur et al. 2009).
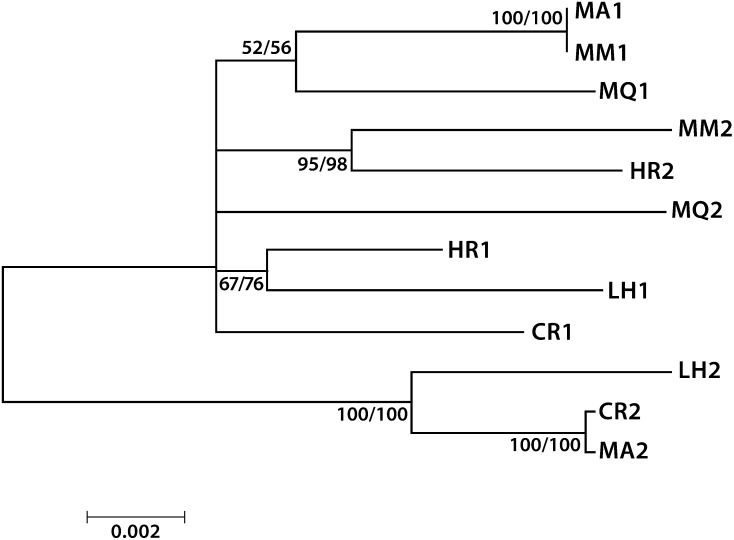
We tested for allele sharing by first sequencing both homologs of the 9.7-kb hisB region in the six members of clade 1. The phylogeny of the resulting 12 sequences shows striking examples of allele sharing between isolates MA and MM and between MA and CR (Figure 4). One sequence of MA is identical to a sequence from MM and the other sequence of MA differs from CR by two single-nucleotide substitutions 815 bp apart, as verified by amplifying and sequencing a 1-kb region including them. All other pairs of sequences are much more distantly related, with substitutions and indels distributed throughout the region (see Figure S1 and Figure S2). Earlier sharing events are suggested by the clustering of LH2 with CR2/MA2, MM2 with HR2, MQ1 with MA1/MM1, and HR1 with LH1, with the two more recent of these having high bootstrap support. Sharing between ancestors of CR, MM, HR, LH, and MQ and lineages not represented in our sample may have occurred more recently than the sharing suggested by the phylogeny of Figure 4 but would have had to involve the nonshared homologs of these lines and would not be evident in a phylogeny based only on the six isolates of clade 1.
Figure 4.
Allele phylogeny of the 9.7-kb hisB region. Isolate MA shares one of its homologs with MM and its other homolog with CR. Earlier sharing events are suggested by the clustering of LH2 with CR2/MA2, MM2 with HR2, MQ1 with MA1/MM1, and HR1 with LH1, with high support for the two most recent of these earlier clusterings. Bootstrap support is shown for maximum likelihood (above) and minimum evolution (below). One thousand replications were implemented in MEGA 5, using the general time-reversible substitution model for ML and the maximum composite-likelihood model for ME.
To determine whether allele sharing occurs at multiple genomic regions, as expected for sexual reproduction but not for transfer by DNA fragments, we sequenced both homologs in three additional regions, hisA, hspA, and hspB, in isolates MA, MM, and CR, the isolates exhibiting the most recent sharing of the hisB region. The occurrence of allele sharing in each of the four regions is immediately evident upon inspection of the 15 pairwise differences between the six sequences at each of the four regions (Table 1). In each region, a sequence of MA is identical to a sequence of MM and its homolog is identical or nearly identical to a sequence of CR.
Table 1. The number of differences (above the diagonals) and percentage of differences (below the diagonals, in italics) between pairs of homologous sequences within and between isolates MA, MM, and CR in each of the four sequenced regions.
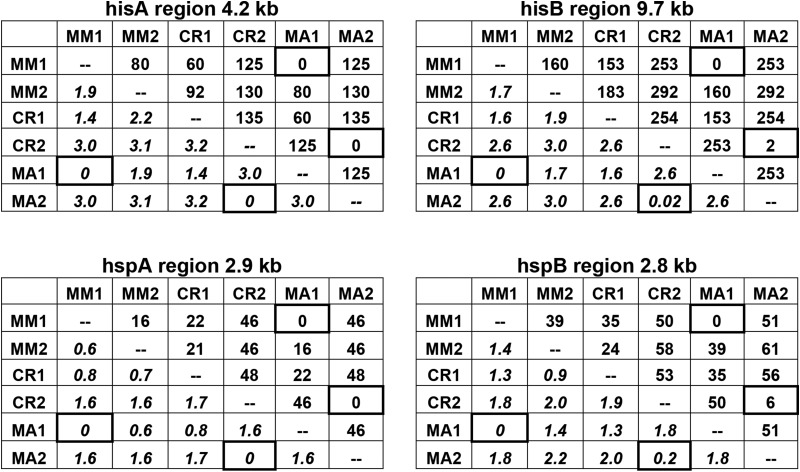 |
Pairs of sequences were aligned in ClustalW with default parameters. Substitutions and indels in the paired sequences were identified with a custom Perl script (see File S1), counting each indel as a single difference. The two differences between MA2 and CR2 at the hisB region, SNPs 400 bp apart, were confirmed by amplifying and sequencing a 1-kb segment containing them. The six differences between MA2 in the hisA region, five SNPs and one indel, are all within an interval of 400 bp. Differences between MA1 and MM1 and between MA2 and CR2 are in cells with heavy outlines. The distribution of differences within each region and the locations of individual genes are shown in Figure S1 and Figure S2.
The shared sequences differ on average by 0.04% (range 0–0.20) while all other differences between homologous sequences are much larger, averaging 1.96% (range 0.55–3.2), and are mainly SNPs spread throughout each region and a smaller number of indels (see Figure S2). The six differences between MA2 and CR2 in the 2.8-kb hspB region, all in gene 32, are five SNPs in exons and a single-base-pair indel in an intron. Some of these differences distinguish MA from the region consensus while others distinguish CR, indicating that mutations occurred in both lineages after allele sharing.
Discussion
Bdelloid asexuality
Before considering what process could have produced the observed allele sharing, it is useful to review the observations that have been viewed as evidence that bdelloids, unlike at least the great majority of animals and plants, have evolved without sexual reproduction. Aside from the single account of Wesenberg-Lund (1930), the failure to find males despite extensive observation (Birky 2010) has long been seen as evidence that bdelloids are entirely asexual, although this does not exclude the possibility that bdelloids produce males under conditions that have not been adequately investigated. Further support for asexuality was once seen in the presence in bdelloid genomes of highly diverged gene copies, initially thought to be homologs, although an alternative explanation based on polyploidy was cited (Mark Welch and Meselson 2000). Subsequent investigation, however, showed that bdelloids are indeed polyploid, being degenerate tetraploids, and that the highly diverged gene copies are homeologs rather than homologs and are therefore not evidence of asexuality (Mark Welch et al. 2008; Hur et al. 2009). Also, the paucity of vertically transmitted transposons in bdelloid genomes (Arkhipova and Meselson 2000), predicted by theory for asexuals, may alternatively be explained as the result of selection against aneuploidies produced by ectopic repair of DNA double-strand breaks (Gladyshev and Meselson 2008) or by enhanced silencing mechanisms (Flot et al. 2013). Another finding that has been seen as suggestive of asexuality is the presence of odd chromosome numbers in the bdelloids P. roseola (Hsu 1956a; Mark Welch and Meselson 1998) and Habrotrocha tridens (Hsu 1956b). Probes that hybridize to the single long chromosome of P. roseola do so at two sites and to no other chromosomes, suggesting that it may be an isochromosome (Mark Welch 2001; Mark Welch et al. 2009). If it contains essential genes not present elsewhere and exists as a balanced polymorphism throughout the population with no mechanism for avoiding homozygosis, there could be 50% sterility in crosses, a reduction in fertility unlikely to have a significant effect on overall fitness if sexual reproduction is infrequent.
Of particular importance to the question of whether bdelloids are capable of sexual reproduction is the recent finding by sequencing and partial assembly of the A. vaga genome that it has the structure of a degenerate tetraploid that has undergone extensive structural change incompatible with the existence of homologous chromosome pairs (Flot et al. 2013). There are many scaffolds with no continuous homolog and some scaffolds that include homologous pairs of numerous genes in palindromes or direct repeats without homologous genes elsewhere in the genome. Although these findings show that the genome is devoid of homologous chromosome pairs and therefore preclude standard meiosis, they do not exclude the possibility that bdelloids reproduce sexually using an atypical form of meiosis in which segregation occurs without requiring homologous chromosome pairs, as discussed below.
Transfer of DNA fragments
It might be thought that the observed allele sharing is the result of homologous transfer of DNA fragments. Horizontal genetic transfer of limited regions is known to occur in bdelloids as the accumulation of foreign genes in telomeric regions, possibly by a mechanism restricted to telomeres. Homologous transfer was not excluded, as the work was done with DNA representing a single individual (Gladyshev et al. 2008). To account for the allele sharing we have found as a consequence of transfer of DNA fragments, transfer would have had to occur between ancestors of MA, MM, and CR and fully cover all four regions. It also must not have been followed by further transfer, causing replacement of any of the shared sequences by more divergent sequences. Moreover, transfer would have had to be truly massive to have even a small probability of producing the observed allele sharing in all four regions of each of the three isolates. Consider, for example, that one-quarter of the DNA in MM and CR had entered independently as fragments from MA. As MA must not contribute the same homolog from any of its four sequenced regions to both MM and CR and as each of the eight regions examined (four in each of the two presumed recipients) must have one sequence transferred from MA and one sequence not from MA, the chance of horizontal transfer of DNA fragments from MA giving the observed result is [1/2]4 × [2(1/4)(3/4)]8 = 2.4 × 10−5. If any less DNA had been transferred horizontally or if one-quarter of the DNA in MA had come from MM and CR, the calculated probability would be lower. We conclude that the transfer of DNA fragments is not a plausible explanation of the observed allele sharing.
Parasexuality
It has been suggested (Flot et al. 2013) that the absence of homologous chromosome pairs in the sequenced strain of A. vaga, although ruling out standard meiosis, may nevertheless be compatible with parasexuality. In the parasexual cycle, known only in certain fungi and protozoans, nuclei from different individuals fuse, forming nuclei of doubled ploidy that during subsequent cell divisions undergo occasional mitotic recombination and, over many divisions, lose chromosomes through misdivision, occasionally yielding viable euploids (Heitman 2010). For bdelloids, parasexuality would require that nuclei of compatible individuals fuse and give rise through a series of aneuploids, all of which must be viable, to euploid individuals. The extreme inefficiency of parasexual reproduction; the difficulty in imagining how germline nuclei from different individuals, normally sequestered in well-differentiated ovaries, might come together; and the fact that parasexual chromosome loss is more or less random and therefore unlikely to result in the observed pattern of allele sharing at all four regions mean that parasexuality provides no explanation for our findings.
Relative abundance of lineages from recent outcrossing
The sharing of identical sequences by MA and MM and the sharing of nearly identical sequences by MA and CR in the four investigated genomic regions, altogether covering 39.2 kb, must mean not only that sharing occurred relatively recently but also, considering the smallness and wide geographic dispersion of our sample, that descendants of such sharing constitute a major proportion of the clade we sampled. The association of such predominance with relatively recent allele sharing suggests the possibility that sexual reproduction imparts enhanced fitness and that, without it, fitness decays at a rate sufficiently high that only lineages from relatively recent crosses are abundant in the mitochondrial clade that we sampled. Until recently, however, there was no evidence for such rapid demise of asexuals. Instead, it had been thought that clonal erosion is driven by more gradual processes, such as the continuing occurrence of deleterious mutation and the advance of Muller’s ratchet (Muller 1964). Contrary to this expectation, asexual lines of the cyclic parthenogenetic water flea Daphnia pulex are found to be surprisingly young (Tucker et al. 2013) as may also be the case for asexual lines of the brine shrimp Artemia urmania (Maccari et al. 2013).
Even the oldest of 11 asexual lines of D. pulex collected at diverse sites in Canada and the United States appears to be separated by only ∼300 generations from the time of crossing with males that carry the meiosis-suppressing elements that rendered them asexual, as estimated by Tucker et al. (2013) from the proportion of male-specific markers that have become homozygous and the rate of conversion per generation measured in mutation accumulation lines of asexual D. pulex (Omilian et al. 2006; Xu et al. 2011). At ∼7 generations per year in the field, the age of the oldest asexual line would be ∼40 years. To account for the unexpectedly recent origin of the asexual lines, Tucker et al. (2013) propose that conversion, occurring at a rate of ∼3.3 × 10−5 per nucleotide site per generation, progressively uncovers accumulated deleterious recessive mutations, driving asexual lines to early extinction, while the restoration of heterozygosity by outcrossing provides cyclically parthenogenetic D. pulex with an escape from such genetic decay.
The implication of the findings in D. pulex for bdelloids is that they too may be cyclic parthenogens and that clones arising from relatively recent outcrossing, having enhanced fitness owing to the restoration of heterozygosity, become predominant within the mitochondrial clade to which they belong while the parental types and other lines that have had no recent restoration of heterozygosity continue to suffer clonal erosion. A bdelloid mitochondrial clade might then correspond to a genetically compatible group of cyclic parthenogens, with lines from recent outcrossing predominating. In this picture, the abundance of lineages MA, MM, and CR in clade 1 would mean that they are descendants of the F1 progeny of relatively recent crossing.
Apparent lack of assortment
In each of the four sequenced regions, MA shares one sequence with MM and shares its homolog with CR. Such uniformity across the four regions would be unlikely in the progeny of a cross if the regions had assorted independently, as would result from independent assortment of chromosomes or from an approximation to linkage equilibrium. An indication that the regions have not assorted independently may be obtained by considering the likelihood that three isolates drawn from a population descended from the F1 progeny of a cross will have the particular pattern of sharing exhibited by isolates MA, MM, and CR. If even two regions had assorted independently, the number of F1 genotypes from the corresponding cross would be 16. The number of combinations of n things taken three at a time regardless of order and allowing repetition is (n + 2)!/3!(n − 1)!, which, for n = 16, is 816. Of these 816 sets of three individuals, only 4 or 4.9 × 10−3 have the property that at each region each of its members shares one of its alleles with another member and its other allele with the third, as may be seen by inspecting the corresponding Punnett square. If three or all four of the regions had assorted independently, the resulting number of different genotypes would be 64 or 256, respectively, and the corresponding chance of drawing three individuals with the observed pattern of allele sharing would be 8.7 × 10−5 or 1.4 × 10−6. For comparison, if there were no assortment, there would be only 4 F1 genotypes and the chance of drawing three individuals with the observed pattern of sharing would be 0.2. It therefore appears that the four regions have not assorted independently.
Oenothera-like meiosis
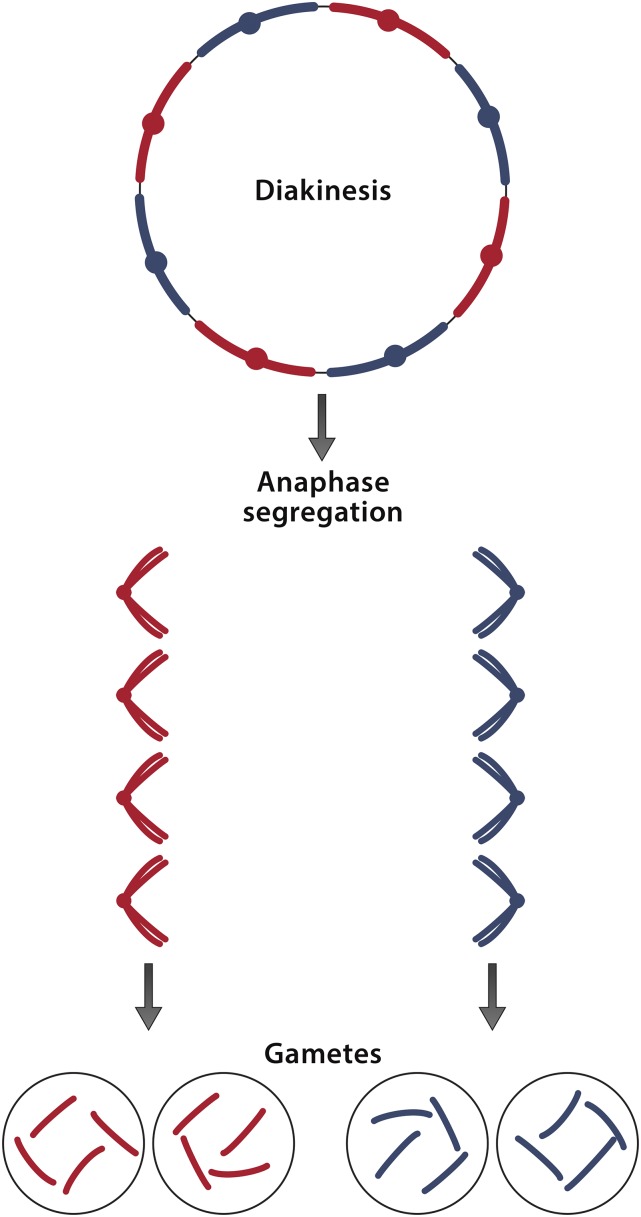
Although the sequenced A. vaga genome lacks homologous chromosome pairs and consequently cannot undergo standard meiosis (Flot et al. 2013), a possible explanation for how meiotic segregation and sexual reproduction might nevertheless occur is suggested by an atypical form of meiosis known in some species of the genus Oenothera and in certain other plants (Cleland 1972; Holsinger and Ellstrand 1984; Golczyk 2011; Rauwolf et al. 2011; Golczyk et al. 2014). In these plants and perhaps in some animals (Chinnappa and Victor 1979; Gross et al. 2009; Schneider et al. 2009), instead of synapsing side by side, chromosomes join end to end to form rings in which paternal and maternal chromosomes alternate. At first meiotic anaphase, maternal and paternal chromosomes segregate to opposite poles without genetic exchange except in telomeric regions, thereby keeping chromosomes of the same parentage together generation after generation as independently inherited units, known as Renner complexes (Figure 5). The ring-forming plants are almost exclusively self-fertilizing, with the production of homozygotes prevented by a system of balanced zygotic and gamete-specific lethals, while infrequent outcrossing gives diverse combinations of existing complexes.
Figure 5.
Oenothera-like meiosis depicted in a diploid genome of eight chromosomes. At diakinesis, all eight chromosomes are found joined end to end, forming a ring in which chromosomes derived from one parent (red) alternate with chromosomes derived from the other parent (blue). The two groups of chromosomes are referred to as complexes. There is no crossing over between complexes except possibly in telomeric regions where genetic exchange occasionally occurs. At first anaphase, chromosomes belonging to different complexes segregate to opposite poles. Entire haploid genomes therefore pass through meiosis intact, generation after generation. Rearrangements within a complex erode chromosome structural homology but do not change the genetic content of the complex in which they occur. Segregation is therefore accomplished without requiring homologous chromosome pairs. Oenothera-like meiosis in the present case would suggest that the genomes of isolates MA, MM, and CR are each composed of two complexes and that each complex includes an allele of each of the four sequenced regions. Owing to the identity in each region of MA1 with MM1 and the near identity of MA2 with CR2, there would be four distinct complexes represented in the three isolates: MA1/MM1, MA2/CR2, MM2, and CR1. Designating these complexes as 1, 2, 3, and 4, the genomes of MA, MM, and CR would be 1/2, 1/3, and 4/2, respectively. The multigeneration stability of each complex would then account for the apparent lack of assortment among the sequenced regions of isolates MA, MM, and CR.
While structural rearrangements entirely within individual complexes would erode chromosome structural homology, they would leave each complex with a full complement of intact genes, except for genes fragmented by breakpoints. In bdelloids, Oenothera-like meiosis might therefore provide a mechanism for meiotic segregation compatible with the lack of homologous chromosome pairs seen in the A. vaga draft genome and could also account for the apparent lack of assortment of homologs at the four regions of isolates MA, MM, and CR. In addition to structural changes that merely rearrange genes within individual complexes, therefore not preventing their regular segregation, the A. vaga draft genome also includes some hundreds of genes in palindromes or direct repeats without homologs elsewhere in the genome. Half or all the progeny from selfing would be the same as their parents, depending on whether there is a system for preventing the production of homozygotes. A complication arises, however, if a genome with essential genes in a palindrome or repeat in one complex and lacking them in its other complex is crossed with a genome with a normal complement of these genes in both of its complexes. In that case, half the progeny would have one copy of such genes and half would have three copies. Viability would then require that the imbalance in gene expression be either tolerated or prevented by autosomal gene dosage compensation (Birchler and Veitia 2007), as must have occurred in the evolution of bdelloid degenerate tetraploidy.
Hypothesis
To have a definite hypothesis to suggest possible population genetic and life-history investigations, we speculate that isolates MA, MM, and CR descend from the F1 of a mass cross between two sexually compatible heterozygous lines that colonized the same niche, both reproducing clonally to give a mixed population at high density. That bdelloid population density can sometimes be very great is seen in the report of Wesenberg-Lund (1930, p. 186) of finding bdelloids in a lake “present in almost incredible numbers” and the finding of bdelloids in rock pools at densities as great as 300,000/liter (Elizabeth J. Walsh, personal communication). We suppose that as population density increased, in response to each other’s presence, mediated by strain-specific chemical signals as in monogonont rotifers (Snell 2011), and perhaps also by environmental factors (Pourriot and Clément 1975), descendants of both founders underwent Oenothera-like meiosis, both producing, as in monogonont rotifers (Gilbert 2004), short-lived haploid males and females carrying haploid eggs. Reciprocal fertilization (Suatoni et al. 2006) and consequent restoration of heterozygosity might then impart enhanced fitness to the progeny while, as in monogononts, parental types that had entered the sexual cycle would be incapable of further reproduction. Any parental types not entering the sexual cycle, having had no restoration of heterozygosity, would suffer continued clonal erosion.
It may be noted that the hypothesized reciprocal fertilization implies the production of two sets of F1 progeny with respect to mitochondrial origin, depending on the mother, as occurs in reciprocal crosses of monogonont rotifers (Suga et al. 2007). This may account for the fact that while MA and MM have nearly identical mitochondrial genomes, differing by only 5 substitutions and one indel, they differ from CR by 191 substitutions with two indels and 192 substitutions with three indels, respectively (see Table S3). A different possibility is that there was subsequent outcrossing of an ancestor of CR with a line carrying the divergent mitochondria and that the sequences shared with MA were not displaced.
Summary
We have obtained clear evidence for recent allele sharing among three lineages represented in a mitochondrial clade of the bdelloid rotifer M. quadricornifera. The finding of allele sharing at each of the four genomic regions examined is not consistent with transfer of DNA fragments but is in agreement with expectation for the result of a cross and sexual reproduction. A possible explanation for how bdelloid sexual reproduction may be reconciled with the finding by Flot et al. (2013) that the bdelloid A. vaga is devoid of homologous chromosome pairs is offered by Oenothera-like meiosis, in which segregation is achieved without side-by-side chromosome pairing. Such meiosis, in which entire maternal and paternal haploid genomes segregate without assortment or crossing over, is consistent with the apparent lack of assortment among the four sequenced regions of the allele-sharing isolates MA, MM, and CR. Our finding of descendants of recent allele sharing within a small sample of what must be a very large population may have its explanation in the evidence of Tucker et al. (2013) that asexual lineages of D. pulex are surprisingly short lived owing to the progressive loss of heterozygosity and the implication that its restoration by outcrossing in cyclic parthenogens imparts enhanced fitness to progeny lines while parental lines continue to suffer clonal erosion, leaving the descendants of recent outcrossing as the predominant members of a sexually compatible group.
Supplementary Material
Acknowledgments
Karyotype analysis was done by Mei Hsu. We are grateful to Deborah Charlesworth and anonymous reviewers for critical reading of earlier versions of the manuscript, to Marc Johnson and Stephan Greiner for discussion of meiosis in Oenothera and other ring-forming plants, and to the U.S. National Science Foundation for initial support.
Footnotes
Communicating editor: J. Heitman
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.176719/-/DC1.
Literature Cited
- Arkhipova I., Meselson M., 2000. Transposable elements in sexual and ancient asexual taxa. Proc. Natl. Acad. Sci. USA 97: 14473–14477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler J. A., Veitia R. A., 2007. The gene balance hypothesis: from classical genetics to modern genomics. Plant Cell 19: 395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birky C. W., Jr, 2010. Positively negative evidence for asexuality. J. Hered. 101(Supp1.): S42–S45. [DOI] [PubMed] [Google Scholar]
- Birky C. W., Jr, Wolf C., Maughan H., Herbertson L., Henry E., 2005. Speciation and selection without sex. Hydrobiologia 546: 29–45. [Google Scholar]
- Chinnappa C. C., Victor R., 1979. Achiasmatic meiosis and complex heterozygosity in female cyclopoid copepods. Chromosoma 71: 227–236. [Google Scholar]
- Cleland R. E., 1972. Oenothera Cytogenetics and Evolution. Academic Press, New York. [Google Scholar]
- Flot J. F., Hespeels B., Li X., Noe B., Arkhipova I., et al. , 2013. Genomic evidence for ameiotic evolution in the bdelloid rotifer Adineta vaga. Nature 500: 453–457. [DOI] [PubMed] [Google Scholar]
- Fontaneto D., Kaya M., Herniou E. A., Barraclough T. G., 2009. Extreme levels of hidden diversity in microscopic animals (Rotifera) revealed by DNA taxonomy. Mol. Phylogenet. Evol. 53: 182–189. [DOI] [PubMed] [Google Scholar]
- Fontaneto D., Iakovenko N., Eyres E., Kaya M., Wyman M., et al. , 2011. Cryptic diversity in the genus Adineta Hudson & Gosse, 1886 (Rotifera: Bdelloidea: Adinetidae): a DNA taxonomy approach. Hydrobiologia 662: 27–33. [Google Scholar]
- Gilbert J. J., 2004. Population density, sexual reproduction and diapause in monogonont rotifers: new data for Brachionus and a review. J. Limnol. 63: 32–36. [Google Scholar]
- Gladyshev E., Meselson M., 2008. Extreme resistance of bdelloid rotifers to ionizing radiation. Proc. Natl. Acad. Sci. USA 105: 5139–5144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladyshev E. A., Meselson M., Arkhipova I. R., 2008. Massive horizontal gene transfer in bdelloid rotifers. Science 320: 1210–1213. [DOI] [PubMed] [Google Scholar]
- Golczyk H., 2011. Cytogenetics of the permanent translocation heterozygote Rhoeo spathacea var. variegata. Implications for complex chromosome rearrangements in Rhoeo. Caryologia 64: 325–334. [Google Scholar]
- Golczyk H., Massouh A., Greiner S., 2014. Translocations of chromosome end-segments and facultative heterochromatin promote ring formation in evening primroses. Plant Cell 26: 1280–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross M. C., Feldberg E., Cella D. M. E., Schneider M. C., Schneider C. H., et al. , 2009. Intriguing evidence of translocations in Discus fish (Symphysodon, Cichlidae) and a report of the largest meiotic chromosomal chain observed in vertebrates. Heredity 102: 435–441. [DOI] [PubMed] [Google Scholar]
- Heitman J., 2010. Evolution of eukaryotic microbial pathogens via covert sexual reproduction. Cell Host Microbe 8: 86–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsinger K. E., Ellstrand N. C., 1984. The evolution and ecology of permanent translocation heterozygotes. Am. Nat. 124: 48–71. [Google Scholar]
- Hsu W. S., 1956a Oogenesis in the Bdelloidea rotifer Philodina roseola. Ehrenberg. Cellule 57: 283–296. [Google Scholar]
- Hsu W. S., 1956b Oogenesis in Habrotrocha tridens (Milne). Biol. Bull. 111: 364–374. [Google Scholar]
- Hur J., Van Doninck K., Mandigo M. L., Meselson M., 2009. Degenerate tetraploidy was established before bdelloid rotifer families diverged. Mol. Biol. Evol. 26: 375–383. [DOI] [PubMed] [Google Scholar]
- Judson O. P., Normark B. B., 1996. Ancient asexual scandals. Trends Ecol. Evol. 11: A41–A46. [DOI] [PubMed] [Google Scholar]
- Lasek-Nesselquist E., 2012. A mitogenomic re-evaluation of the bdelloid phylogeny and relationships among the syndermata. PLoS ONE 7(8): e43554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccari M. F., Amat F., Gomez A., 2013. Origin and genetic diversity of diploid parthenogenetic Artemia in Eurasia. PLoS ONE 8: e43348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark Welch D. B., Meselson M., 2000. Evidence for the evolution of bdelloid rotifers without sexual reproduction or genetic exchange. Science 288: 1211–1215. [DOI] [PubMed] [Google Scholar]
- Mark Welch D. B., Meselson M., 2003. Oocyte nuclear DNA content and GC proportion in rotifers of the anciently asexual class Bdelloidea. Biol. J. Linn. Soc. Lond. 79: 85–91. [Google Scholar]
- Mark Welch D. B., Mark Welch J. L., Meselson M., 2008. Evidence for degenerate tetraploidy in bdelloid rotifers. Proc. Natl. Acad. Sci. USA 105: 5145–5149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark Welch, D. B., C. Ricci, and M. Meselson, 2009 Bdelloid rotifers: understanding the success of an evolutionary scandal, pp. 259–280 in Lost Sex, edited by I. Schön, K. Martens, and P. van Dijk. Springer-Verlag, Heidelberg, Germany. [Google Scholar]
- Mark Welch, J. L., 2001 Cytological Evidence for the Absence of Meiosis in Bdelloid Rotifers. Doctoral dissertation, Harvard University, Cambridge, MA. [Google Scholar]
- Mark Welch J. L., Meselson M., 1998. Karyotypes of bdelloid rotifers from three families. Hydrobiologia 387/388: 403–407. [Google Scholar]
- Mark Welch J. L., Mark Welch D. B., Meselson M., 2004. Cytogenetic evidence for asexual evolution of bdelloid rotifers. Proc. Natl. Acad. Sci. USA 101: 1618–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H. J., 1964. The relation of recombination to mutational advance. Mutat. Res. 106: 2–9. [DOI] [PubMed] [Google Scholar]
- Omilian A. R., Cristescu M. E. A., Dudycha J. L., Lynch M., 2006. Ameiotic recombination in asexual lineages of Daphnia. Proc. Natl. Acad. Sci. USA 103: 18638–18643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagani M., Ricci C., Redi C. A., 1993. Oogenesis in Macrotrachela quadricornifera (Rotifera, Bdelloidea) I. Germarium, eutely, karyotype and DNA content. Hydrobiologia 255/256: 225–230. [Google Scholar]
- Pourriot R., Clément P., 1975. Influence de la durée de l’éclairement quotidien sur le taux de femelles mictiques chez Notommata copeus Ehr. (Rotifere). Oecologia 22: 67–77. [DOI] [PubMed] [Google Scholar]
- Rauwolf U., Greiner S., Mracek J., Rauwolf M., Golcyk H., et al. , 2011. Uncoupling of sexual reproduction from homologous recombination in homozygous Oenothera species. Heredity 107: 87–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci C., 1991. Comparison of five strains of a parthenogenetic species, Macrotrachela quadricornifera (Rotifera, Bdelloidea) I. Life history traits. Hydrobiologia 211: 147–155. [Google Scholar]
- Schneider M. C., Zacaro A. A., Pinto-da-Rocha R., Candido D. M., Cella D. M., 2009. Complex meiotic configuration of the holocentric chromosomes: the intriguing case of the scorpion Tityus bahiensis. Chromosome Res. 17: 883–898. [DOI] [PubMed] [Google Scholar]
- Snell T. W., 2011. A review of the molecular mechanisms of monogonont rotifer reproduction. Hydrobiologia 662: 89–97. [Google Scholar]
- Suatoni E., Vicario V., Rice S., Snell T., Caccone A., 2006. An analysis of species boundaries and biogeographic patterns in a cryptic species complex: the rotifer–Brachionus plicatilis. Mol. Phylogenet. Evol. 41: 86–98. [DOI] [PubMed] [Google Scholar]
- Suga K., Tanaka Y., Sakahura Y., Hagiwara A., 2007. Inheritance of mitochondrial DNA in the rotifer Brachionus plicatilis. Hydrobiologia 593: 167–173. [Google Scholar]
- Tucker A. E., Ackerman M. S., Eads B. D., Xu S., Lynch M., 2013. Population-genomic insights into the evolutionary origin and fate of obligately asexual Daphnia pulex. Proc. Natl. Acad. Sci. USA 110: 15740–15745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesenberg-Lund C., 1930. Contributions to the Biology of the Rotifera, Part II: The Periodicity and Sexual Periods. A. F. Host and Son, Copenhagen. [Google Scholar]
- Wilson C. G., Sherman P. W., 2013. Spatial and temporal escape from fungal parasitism in natural communities of anciently asexual bdelloid rotifers. Proc. Biol. Sci. 280: 20131255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S., Omelian A. R., Cristescu M. E., 2011. High rate of large-scale hemizygous deletions in asexually propagating Daphnia: implications for the evolution of sex. Mol. Biol. Evol. 28: 335–342. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.